Legal et al. use cryo-electron tomography to study the tips of motile cilia. They reveal novel structural features of the central pair, expand on the functional role of CEP104/FAP256, and identify potential new proteins that are important for the stability of the ciliary tip.
Abstract
Cilia are essential organelles that protrude from the cell body. Cilia are made of a microtubule-based structure called the axoneme. In most types of cilia, the ciliary tip is distinct from the rest of the cilium. Here, we used cryo-electron tomography and subtomogram averaging to obtain the structure of the ciliary tip of the ciliate Tetrahymena thermophila. We show that the microtubules at the tip are highly crosslinked with each other and stabilized by luminal proteins, plugs, and cap proteins at the plus ends. In the tip region, the central pair lacks typical projections and twists significantly. By analyzing cells lacking a ciliary tip–enriched protein CEP104/FAP256 by cryo-electron tomography and proteomics, we discovered candidates for the central pair cap complex and explained the potential functions of CEP104/FAP256. These data provide new insights into the function of the ciliary tip and the mechanisms of ciliary assembly and length regulation.
Introduction
Cilia are microtubule-based cellular protrusions that extend from the surface of most cells in the human body. Cilia are conventionally classified into two categories: motile cilia and primary cilia. Motile cilia propel cells such as sperm cells or move fluids such as mucus in the respiratory tract, while primary cilia function as sensory organelles (Satir and Christensen, 2007).
Due to their important roles, mutations in ciliary proteins can cause multiple disorders called ciliopathies. These disorders can lead to infertility, brain malformations, or respiratory problems and affect approximately 1 in 2,000 individuals (Reiter and Leroux, 2017).
The microtubules inside the cilia assemble into a core structure known as the axoneme. They are templated from the basal body, a circular structure of nine triplet microtubules. The basal body is followed by a transition zone in which the triplet microtubules become doublets made of a complete A-tubule and a partial B-tubule (Ichikawa et al., 2017). The doublet microtubules become singlet microtubules at the ciliary tip as the B-tubules terminate (Fisch and Dupuis-Williams, 2011). Motile cilia also contain two microtubule singlets in the center, known as the central pair (CP) or central apparatus (Gibbons and Grimstone, 1960). The CP microtubules are called C1 and C2, as they differ significantly in structure. C1 is chemically more stable than C2, and the C1 projections are longer than those of C2 (Hopkins, 1970). Ciliary beating is driven by outer and inner dynein arms (Lin and Nicastro, 2018). They are found on the outside of doublets and slide them apart when activated. The sliding motion is then converted into the bending motion of the cilia because of the nexin–dynein regulatory complex linking adjacent doublet microtubules (Summers and Gibbons, 1971). Doublets are linked to the CP by radial spokes that coordinate dynein activity (Smith and Sale, 1992).
The ciliary tip region has special importance because it is the zone of ciliary growth (Rosenbaum et al., 1969). Tubulins and other ciliary subcomplexes are transported by intraflagellar transport (IFT) trains to the tips of cilia, where they assemble to build the cilia (Qin et al., 2004). Anterograde trains (walking towards the tip) are reorganized and converted to retrograde trains (walking towards the base) at the ciliary tip (Chien et al., 2017). In Chlamydomonas reinhardtii, anterograde trains move on the B-tubule, while retrograde trains move on the A-tubule of doublet microtubules within the doublet zone of the axoneme (Stepanek and Pigino, 2016).
Most of our current knowledge about the ciliary tip comes from early negative-staining experiments. The CP is capped by a protein complex shaped like a ball attached to the membrane preceded by two plates (Dentler and Rosenbaum, 1977). The spherical plate directly capping the two microtubules is attached to two structures that go inside the microtubules (Suprenant and Dentler, 1988). The ends of A-tubules are plugged by a Y-shaped filament bundle that extends 70 nm inside the lumens, namely, A-tubule plugs (Suprenant and Dentler, 1988). Interestingly, these cap and plug structures are conserved among the unicellular organisms Chlamydomonas and Tetrahymena and mammalian tracheal cilia. However, they are not found at the tips of human sperm cells or human primary cilia (Soares et al., 2019). Therefore, the organization of the ciliary tip may differ depending on the functional specialization of cilia. The functions of ciliary caps remain to be explored. Additionally, it is unknown how tubulin subunits are added to the plus ends of microtubules while the capping structures are present. Interestingly, the Chlamydomonas cap complex shown by cryo-electron tomography (cryo-ET) appears as a much smaller complex, unlike described previously (Höög et al., 2014). This study also shows that most microtubules end in a flared conformation near the membrane. The identities of the cap proteins remain to be discovered.
In addition, studies have shown that the CP structure changes in the tip region. In Tetrahymena, CP projections terminate at the region where the B-tubules and radial spokes end. In this species, the tip CP region is ∼500 nm long (Sale and Satir, 1977). In Chlamydomonas, the tip CP region is much shorter, ∼200 nm (Pratelli et al., 2022). The structures of both doublet-zone CP and tip region CP in Tetrahymena remain to be determined.
Several proteins have been identified to localize to the ciliary tip. Kinesins from the kinesin-4, -8, and -13 subfamilies localize to the ciliary tips, where they regulate microtubule length (He et al., 2014; Niwa et al., 2012; Piao et al., 2009; Schwarz et al., 2017; Vasudevan et al., 2015; Wang et al., 2013). EB1, a microtubule plus-end binding protein, localizes to the ciliary tips of Chlamydomonas (Pedersen et al., 2003). The TOG domain proteins CHE-12/Crescerin and CEP104/FAP256 were also shown to localize to the ciliary tips of motile cilia in Tetrahymena (Louka et al., 2018). CEP104/FAP256 also localizes to the tips of primary cilia in humans (Satish Tammana et al., 2013). TOG domains bind to tubulin and promote microtubule elongation (Al-Bassam et al., 2007). Interestingly, CEP104/FAP256 is conserved in mammalian primary cilia, where it promotes ciliary elongation (Yamazoe et al., 2020). Mutations in CEP104/FAP256 cause Joubert syndrome, a neurological disorder characterized by brain malformation (Srour et al., 2015). In Tetrahymena, knockout of FAP256 reduces cell swimming velocity and ciliary beat frequency (Louka et al., 2018). How all these proteins come together to regulate ciliary length and the ends of A- and B-tubules is still unclear.
In recent years, multiple structures of ciliary complexes have been published. Using single-particle cryo-electron microscopy (cryo-EM), the structures of the doublet microtubules, the radial spokes, the outer dynein arms, and the CP were solved to high resolution. At slightly lower resolution, cryo-ET has been used to study the structures of doublet and singlet microtubules in mammalian sperm and identify proteins de novo. Importantly, the ciliary tip has not been studied at high resolution apart from the singlet-A-tubule of sperm cells (reviewed in Grossman-Haham [2023]).
Here, we used cryo-ET and subtomogram averaging to obtain the structure of the ciliary tip of the wild-type and FAP256-knockout (KO) Tetrahymena thermophila mutant. We show the three-dimensional (3D) organization of the intact ciliary tip, elaborating on the cap, plugs, and tip microtubule interactions. We also show that the CP is highly stabilized by multiple proteins binding to the inside and outside of its microtubules. In addition, using mass spectrometry analysis of FAP256-KO mutants, we generated a list of potential candidates for the CP cap complex and described the potential roles of FAP256.
Results
Cryo-ET of the ciliary tip confirms previous observations
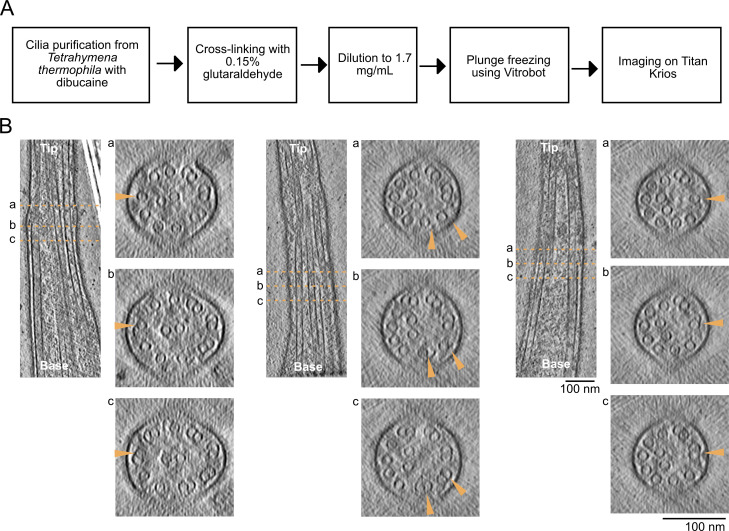
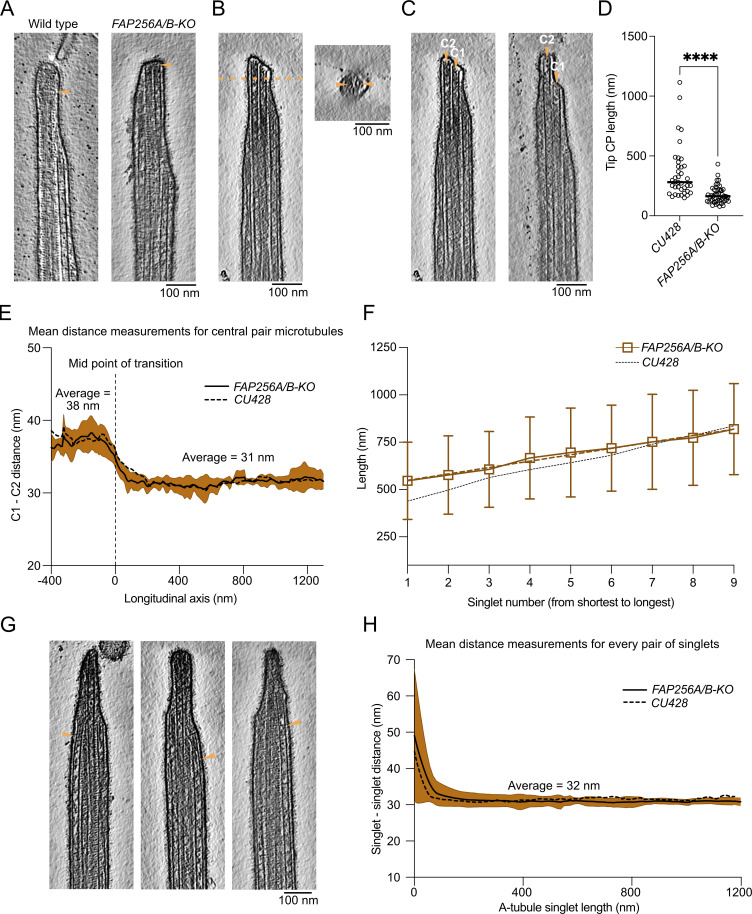
To obtain a high-resolution 3D view of the intact ciliary tip, we deciliated Tetrahymena thermophila cells and lightly crosslinked the cilia before plunge-freezing and collecting cryo-electron tomograms (Table S1 and Fig. S1 A). We collected ∼150 tomograms of ciliary tips and ciliary narrowing regions in three datasets: two with the membrane intact for visualization of membrane connections and one without the membrane for better visualization of microtubules. Segmentation of these tomograms after subtomogram averaging (see Materials and methods) allowed us to observe the organization of the CP, the singlet A-tubules, and the CP cap complex within the ciliary tip (Fig. 1 A).
Figure S1.
Cilia preparation and incomplete doublets. (A) Diagram of cilia preparation workflow. (B) Longitudinal slices through tomograms and corresponding cross-sections as in Fig. 1 E. Arrowheads point to the same doublet that becomes incomplete in the tip region.
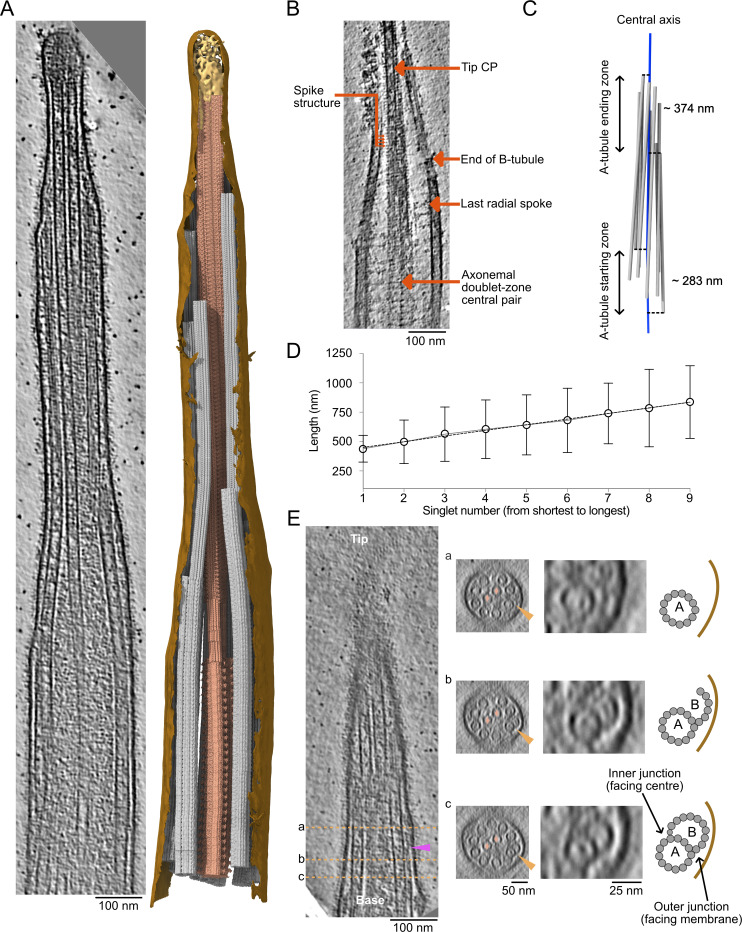
Figure 1.
Overview of the ciliary tip. (A) Tomogram slice of a representative ciliary tip. 3D model of the same cilium generated with the subtomogram averages described in this study. The membrane is in brown, the CP is in dark salmon, doublets and singlet A-tubules are in gray, and the cap complex is in gold. The doublet and singlet at the front of the image were removed to allow the visualization of the CP. (B) Representative slice of a demembranated cilium showing the tip CP, the end of one B-tubule, the last radial spoke, the spike structure, and the axonemal doublet-zone CP. Scale bar: 100 nm. (C) 3D representation of the A-tubules (gray) from a representative tomogram. The central axis of the cilium (center of the CP) is in blue. (D) Length of A-tubules numbered from shortest to longest. The dotted line shows a linear trend line going through all points. Error bars show the standard deviation. n = 6 cilia. (E) Longitudinal slice through a representative tomogram at the narrowing zone. Orange dotted lines show cross-sections in panels a, b, and c. Orange arrowheads point toward the disassembling/incomplete B-tubule. Insets show a doublet with a disassembling/incomplete B-tubule. The CP microtubules are marked by a salmon diamond. Cartoons indicate what is shown in the insets. The purple arrowhead shows a filament capping the end of the B-tubule.
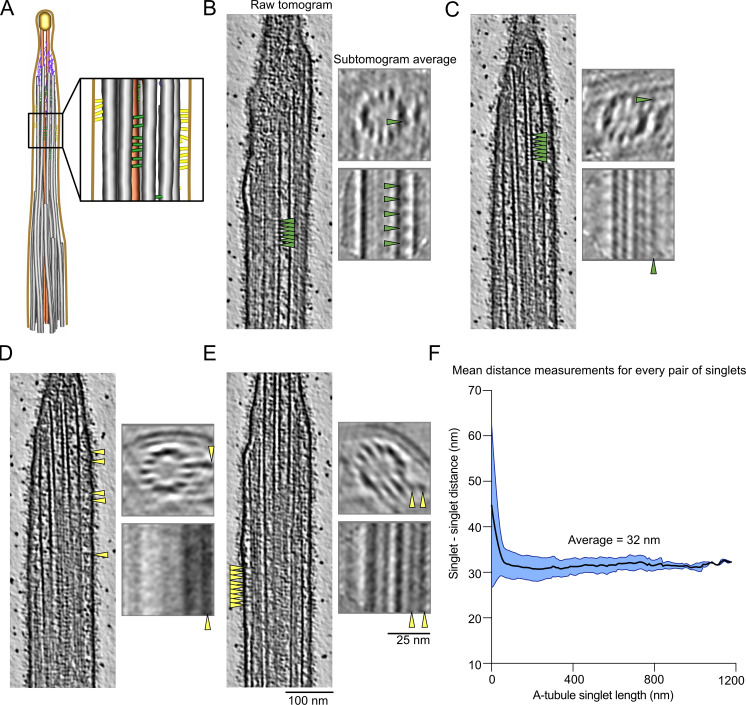
Denoising these tomograms allowed us to visualize the narrowing zones and ciliary tips in detail (Fig. 1 B). We revealed structural differences in CP organization between the tip region and the rest of the axoneme. The characteristic projections on the CP stop just before the distance between C1 and C2 microtubules decreases. Within the tip region, the CP projections are replaced by a characteristic 8-nm repeat short-spike protein. Doublet microtubules become singlets when they reach the ciliary tip. The B-tubules, radial spokes, and dynein arms all disappear within a short distance from each other (Fig. 1 B). The ends of the B-tubules coincide with a narrowing of the cilium diameter, which we refer to as the narrowing zone.
Doublets do not become singlets at the same level. By projecting the start position of each singlet A-tubule (equivalent to the end of each B-tubule) to the cilium’s central axis (Fig. 1 C, dotted lines), we measured the A-tubule starting zone to span over 282.6 ± 61.8 nm. Similarly, the A-tubule ending zone showed substantial variation between cilia, with an average of 374.1 ± 277.4 nm (n = 6 cilia). The length of singlet A-tubules also varied significantly from 0 to 1,428 nm (Fig. 1 D). Interestingly, when the A-tubules are ordered from shortest to longest, their lengths increase linearly. We did not find any correlation between the A-tubule length and their starting and ending points. Finally, some B-tubules separated from the inner junction (Fig. 1 E and Fig. S1 B). Furthermore, some B-tubules end with a filamentous plug (Fig. 1 E, purple arrow).
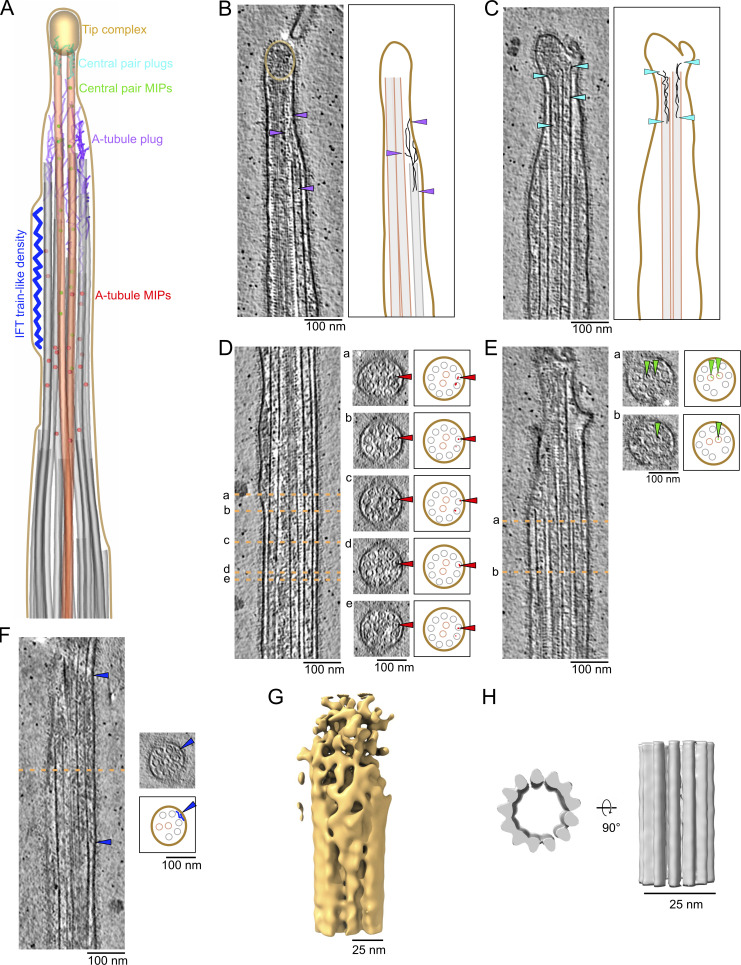
We next focused on protein densities specific to the ciliary tip region, which we summarized in a model figure and a movie (Fig. 2 A and Video 1). We first examined the ends of A-tubules and CP microtubules. A-tubules end with slightly curved protofilaments (Fig. 2 B and Fig. S2 A), similar to the plus-ends of microtubules observed in situ in axons or fibroblasts (Foster et al., 2022; Koning et al., 2008). Interestingly, the plug seen at the end of A-tubules appears to be composed of multiple filamentous proteins that extend 110 ± 30 nm (n = 29) inside the A-tubule. The filaments make connections with the membrane and the CP microtubules (Fig. 2 B and Fig. S2 A, purple arrows). We did not observe the “ball” seen in previous negative staining experiments (Suprenant and Dentler, 1988). The ends of CP microtubules were difficult to visualize because of the ciliary cap. In most cases, we believe both microtubules are roughly the same length. The CP microtubule cap appears as a mesh of entangled proteins (Fig. 2 B, golden circle). We did not observe the plug structures or the characteristic plates described in previous negative staining studies (Suprenant and Dentler, 1988). Filamentous proteins extend from the cap to the inside of CP microtubules with an average length of 127 ± 34.7 nm (n = 10; Fig. 2 C and Fig. S2 B).
Figure 2.
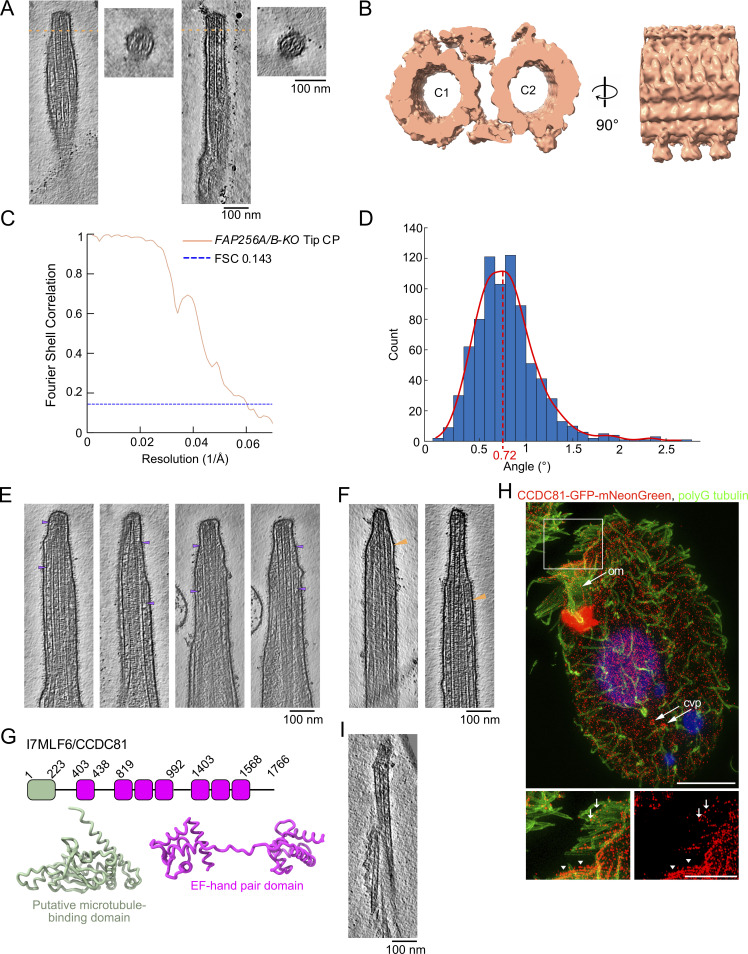
The ciliary tip contains many MAPs. (A) 3D model of the various tip proteins is shown in this figure. (B and C) Representative tomogram slices along with corresponding diagrams highlighting what is shown in the tomograms. Arrowheads point to the A-tubule plug (B) and CP plug (C). (D–F) Representative tomogram slices showing A-tubule MIPs (D), CP MIPs (E), and IFT train-like particles (F) along with cross-sections indicated by the dotted orange lines. The diagram highlights what is shown in the cross-sections. (B–F) Scale bars: 100 nm. (G and H) Subtomogram averages of the cap complex (G) and singlet A-tubule (H).
Video 1.
Segmentation of a representative tomogram summarizing the main findings of this study.
Figure S2.
Examples of plugs, MIPs, and IFT-train-like particles. (A and B) Examples of filamentous plugs in A-tubules (A) and CP microtubules (B). Arrowheads point to the extremities of the filamentous plugs. (C–E) Examples of MIPs found in A-tubules (C) and CP microtubules (D). (E) Examples of IFT train-like particles. Longitudinal slices along tomograms. Dotted lines indicate cross-sections. (F) Fourier shell correlation (FSC) curves of the cap complex and the singlet A-tubules.
We also observed many microtubule inner proteins (MIPs) in the A-tubules. Unlike the A-tubules of the doublet zone, these MIPs do not seem to have a regular repeating pattern. Some were associated with the microtubule wall, while others were inside the microtubule lumen (Fig. 2 D and Fig. S2 C). Many of these MIPs are similar in size and shape to the MIPs observed in mammalian primary cilia (Kiesel et al., 2020) and Drosophila melanogaster neurons (Foster et al., 2022). In addition, we observed that MIPs in both CP microtubules were also either associated with the microtubule wall or floating inside the lumen (Fig. 2 E and Fig. S2 D).
IFT trains convert from the anterograde to retrograde direction at the ciliary tip (Chien et al., 2017). IFT train-like particles were bound to the A-tubule in five tomograms (Fig. 2 F and Fig. S2 E). While IFT trains on the A-tubules were found in primary cilia (Kiesel et al., 2020), this is the first observation of IFT trains on the A-tubules of the singlet zone in motile cilia. Due to the insufficient number of particles in our tomograms, we could not carry out subtomogram averaging on IFT trains.
To gain insights into the CP cap structure, we carried out subtomogram averaging. The resulting averaged map has a resolution of ∼130 Å. This unsatisfying result reflects the structural character of the cap complex, which is not rigidly attached to the tip of the two CP microtubules (Fig. 2 G; Fig. S2 F; and Table S2). It has been reported that A-tubules in primary cilia are decorated by a protein that repeats every 8 nm, interpreted by the authors as the microtubule tip-binding protein EB1 (Kiesel et al., 2020). We did not observe any similar decorations on the outside of A-tubules in our tomograms (Fig. 2 D and Fig. S2 C). To confirm this observation, we used subtomogram averaging to obtain the structure of the A-tubule. Our average structure did not reveal MIPs or microtubule-associated proteins (MAPs) fully decorating the outside of the A-tubules at the tip at this resolution (Fig. 2 H; Fig. S2 F; and Table S2).
The ciliary tip singlets are maintained parallel to each other
While carrying out subtomogram averaging of the A-tubules, we observed densities on the outside of some A-tubules (Fig. 3, A–E). As these densities are not found on all A-tubules and are positioned differently on each A-tubule, they were not seen in the final subtomogram average of all A-tubules. We observed two types of densities: proteins that seem to link two A-tubules (Fig. 3, B and C; and Fig. S3, A–C) and proteins that link the A-tubule to the membrane (Fig. 3, D and E; and Fig. S3 D). Due to the limited number of particles, the periodicity of these proteins is difficult to determine, but some are repeated every 8 nm (Fig. 3, B, C, and E, arrows). Interestingly, we only observed these densities along short portions of the A-tubules. These observations led us to ask whether the A-tubules at the tip were maintained parallel to each other by these links. To answer this question, we measured the distance between all adjacent A-tubules. We found that the average distance between two A-tubules was 31 nm and was constant along the length of the A-tubules (Fig. 3 F). Therefore, we believe that these linker densities act as spacers that maintain the distance between the A-tubules at the tip.
Figure 3.
The A-tubules are linked to the membrane and to each other in the ciliary tip. (A) 3D model of the ciliary tip with inset showing A-tubule links in green and membrane links in yellow. (B–E) Representative raw tomogram slices along with cross (top) and longitudinal (bottom) slices of subtomogram averages of individual A-tubules. Green arrowheads point to A-tubule links (B and C) and yellow arrowheads point to membrane links (D and E). Scale bars tomograms: 100 nm, subtomogram averages: 25 nm. (F) Graph showing the distance between pairs of A-tubule singlets in the ciliary tip (n = 6 cilia). Standard deviation is shown in light blue.
Figure S3.
A-tubule and membrane links. (A–C) Examples of A-tubule links (A–C) and membrane links (D) as in Fig. 3. (B–E) Representative raw tomogram longitudinal slices along with cross- and longitudinal sections of subtomogram averages of individual A-tubules. Green and yellow arrowheads point to A-tubule and membrane links, respectively.
The tip CP has a pseudo twofold symmetric structure
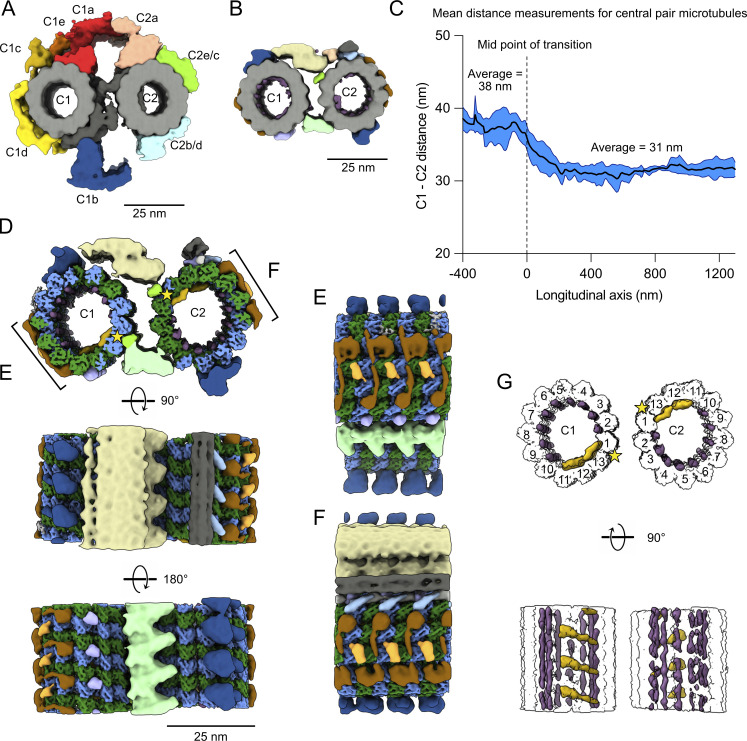
The difference in CP morphology observed in our tomograms (Fig. 1, A and B) before and after the narrowing zone motivated us to obtain subtomogram averages of both the doublet-zone CP (hereafter referred to as “main CP”) and the tip CP (Fig. 4, A and B; and Fig. S4 A). We obtained a subtomogram average of the 32-nm repeat of the main CP at a resolution of ∼23 Å (Fig. 4 A, Fig. S4 B, and Table S2). It shows the canonical architecture of known CP structures (Carbajal-González et al., 2013; Fu et al., 2019; Leung et al., 2021). The CP of Tetrahymena is similar to the mammalian CP in that the C1f projection is missing. When comparing our structure with other species, we found that the Tetrahymena CP is the most similar to sea urchin CP (Carbajal-González et al., 2013), especially when comparing C1d and C1c projections (Fig. S4 C). C2 has similar projections to known CP structures. C1 and C2 are made up of 13 protofilaments each and linked by a bridge. Tetrahymena CP does not contain large globular MIPs inside C1 or C2 as opposed to pig and horse CPs (Leung et al., 2021).
Figure 4.
The tip CP has a different structure from the main CP. (A) Subtomogram average of the main CP. (B) Subtomogram average of the tip CP. In A and B, structures are lowpass-filtered to 20 Å to be comparable. Microtubules are in gray and projections are colored. (C) Graph of the distance between C1 and C2 microtubules. The reference on the x-axis is the middle of the narrowing zone. The average is shown as a thick blue line. The standard deviation is shown in light blue (n = 5 cilia). (D) Subtomogram average of the tip CP. Microtubules are colored according to tubulin subunits. α-tubulin is colored in green and β-tubulin is colored in blue. C1 is on the left and C2 is on the right. The yellow stars indicate the seams. (E and F) Side view of the tip CP as shown in D. (G) Tip CP rendering showing the MIPs in purple and yellow. The microtubules are white and protofilaments are numbered from the seam. The yellow stars indicate the seams.
Figure S4.
Data processing and analyses of the CP. (A) Diagram of the subtomogram averaging workflow used in this study. (B) FSC curves of the tip CP and main CP structures. (C) Front and side views of Tetrahymena main CP (this study, dark salmon), sea urchin CP (EMD 9385, navy blue), and both overlaid (Tetrahymena CP is transparent). (D) Measured twist angles for WT main (blue bars) and tip (gray bars, transparent) CP. The dotted line is drawn at the highest point of the curve fitted to the histogram (green line for main CP, dark salmon line for tip CP). (E) Example of measurement of the distance between both CP microtubules along the length of a tomogram. The blue dots represent the coordinates of the center of a microtubule subtomogram. The red lines represent the shortest distance between two blue points. (F) Masks were used to refine the structure of the tip CP in this study. The resolution obtained after masking is indicated on the figure for each mask. (G) Fitting of a tubulin dimer in the electron density map of the subtomogram average of the tip CP. α-Tubulin is colored green and the β-tubulin is colored blue. For comparison, the same tubulin dimer was fitted in a map at 4 Å resolution, which was then lowpass filtered to 9 Å. (H) Local resolution map for C1 and C2 colored according to the scale bar on the left-hand side.
We then compared the structure of the main CP to the subtomogram average of the tip CP filtered to a similar resolution (Fig. 4 B). Strikingly, the characteristic projections on the main CP are absent. The tip CP has smaller MAPs bound to the outside of C1 and C2. The spike densities observed in our tomograms (Fig. 1 B) are shown in blue and localized to the top of the C1- and bottom of the C2-microtubule (Fig. 4 B, blue). The bridge between C1 and C2 is also different. The tip CP microtubules are linked by two densities: one at the top and one at the bottom (yellow and green densities, Fig. 4 B).
When we visualized the subtomogram averages of the CP in the original tomograms (Fig. 1 A), we clearly observed that the tip CP twists in an anti-clockwise direction, confirming previous observations (Omoto and Kung, 1979; Reynolds et al., 2018). We measured the twist of the CP in the main part and compared it to that of the tip. We found that the tip CP twists almost twice as much in the tip region: 0.72° every 8 nm versus 0.43° for the main CP (Fig. S4 D).
The distance between C1 and C2 microtubules is significantly reduced at the tip CP. On average, the centers of C1 and C2 are 38.2 nm apart in the main region and 31.2 nm apart in the tip region (Fig. 4 C and Fig. S4 E). The distance between C1 and C2 at the tip is similar to the distance between A-tubules (Fig. 3 F). Interestingly, the narrowing zone seems consistent in all the tomograms with a length of ∼100 nm. The shorter distance between C1 and C2 suggests that there exist linkers that tightly bundle C1 and C2 and singlet A-tubules in the tip region.
To increase the resolution of the tip CP, we averaged the tip CP subtomograms picked every 8 nm and obtained a subtomogram average at 9 Å resolution. This resolution allowed us to distinguish between α- and β-tubulin subunits (Fig. 4 D, Fig. S4, F–H; and Table S2). This allowed us to identify the seams on C1 and C2 microtubules (yellow stars). Both seams face the inside of the CP. The seam on C2 is at the same position as the main CP in Chlamydomonas; however, the seam on C1 is one protofilament lower in the tip CP (Gui et al., 2022b; Han et al., 2022). C2 has more MAPs than C1 and is topped by a filament bundle, which is perhaps similar to FAP7 found on top of C1 in the Chlamydomonas CP (Gui et al., 2022b; Han et al., 2022).
A closer inspection of the tip CP structure shows that some MAPs bind laterally to C1 and C2, contacting four protofilaments. These MAPs have a very similar shape, suggesting that they might be the same proteins (Fig. 4, E and F, brown). We identified filamentous MIPs inside both C1 and C2 between adjacent protofilaments (Fig. 4 G, purple). There are also arc-MIPs making lateral contact with four protofilaments in both C1 and C2 (Fig. 4 G, yellow). Although we cannot be certain about the repeating unit of the filamentous MIPs due to our imposed 8-nm periodicity averaging, our structure shows that all protofilaments are stabilized by either longitudinal or arc-MIPs.
Surprisingly, we noticed that C1 and C2 are symmetric apart from the linker between the two microtubules in the tip CP, as opposed to the main CP. The MIPs and MAPs on C1 overlay with the MIPs and MAPs on C2 when it is rotated 180° around the longitudinal axis (Fig. 4, D–G), suggesting that the same proteins assemble in and around both C1 and C2.
FAP256A/B-KO mutants have multiple ciliary tip defects
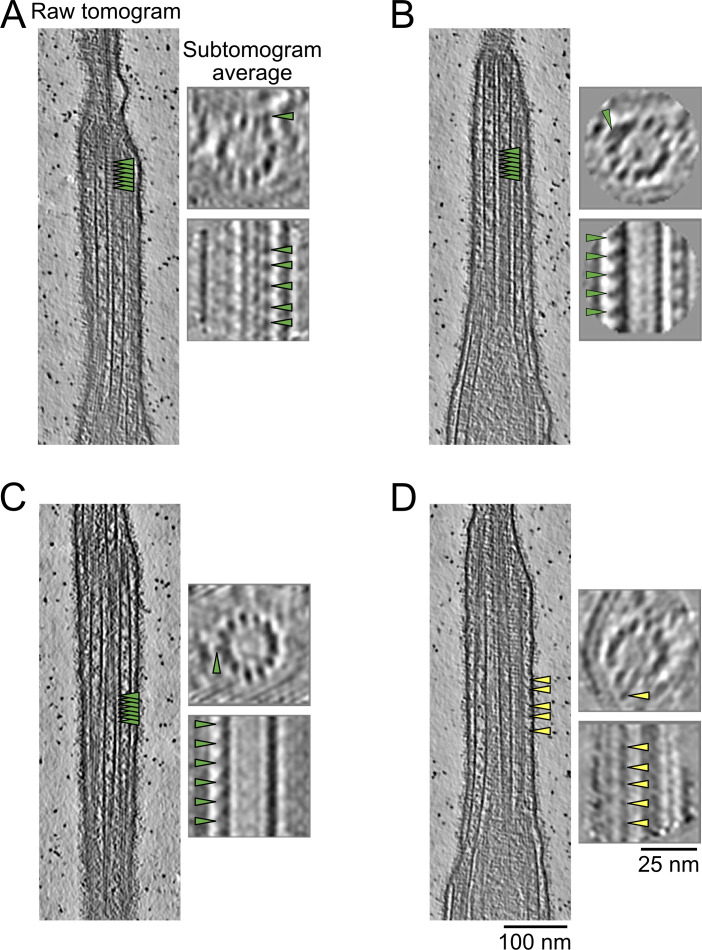
The current resolution of our subtomogram averages does not allow us to identify proteins de novo. Only a few proteins are known to localize to the ciliary tip in Tetrahymena, including FAP256 (A and B), CHE-12 (A and B), and ARMC9 (A and B). We decided to use cryo-ET to study the cilia of cells lacking FAP256 (FAP256A/B-KO, same strain as described in Louka et al., 2018) because of the lack of ciliary cap complex reported. Analysis of the tomograms revealed various defects at the tips. First, we observed that the CP cap was absent in all our tomograms, while the cilia had a normal overall shape (Fig. 5 A and Fig. S5 A). The filaments inside the CP microtubules were still present (Fig. 5 B, Fig. S5 A, and Fig. 2 C). Although their molecular identities remain unknown, the similarity to the plug observed at the end of the A-tubules suggests that they are composed of the same proteins. Looking closely at the tip in FAP256A/B-KO mutants, we observed that the CP microtubules end with slightly curved protofilaments that contact the membrane directly (Fig. 5, B and C). The curvature of the protofilaments seems higher than that of sperm singlets when they reach the membrane (Zabeo et al., 2018, 2019). We did not measure the curvature of individual protofilaments, as we cannot resolve them in the WT tomograms due to the ciliary cap and therefore cannot compare the curvature to a control. Although microtubules appeared flared at their ends, we did not observe any tapered ends. The CP MIPs were also present in a similar amount as WT cells, although due to their lack of a repeating pattern, we did not quantify them.
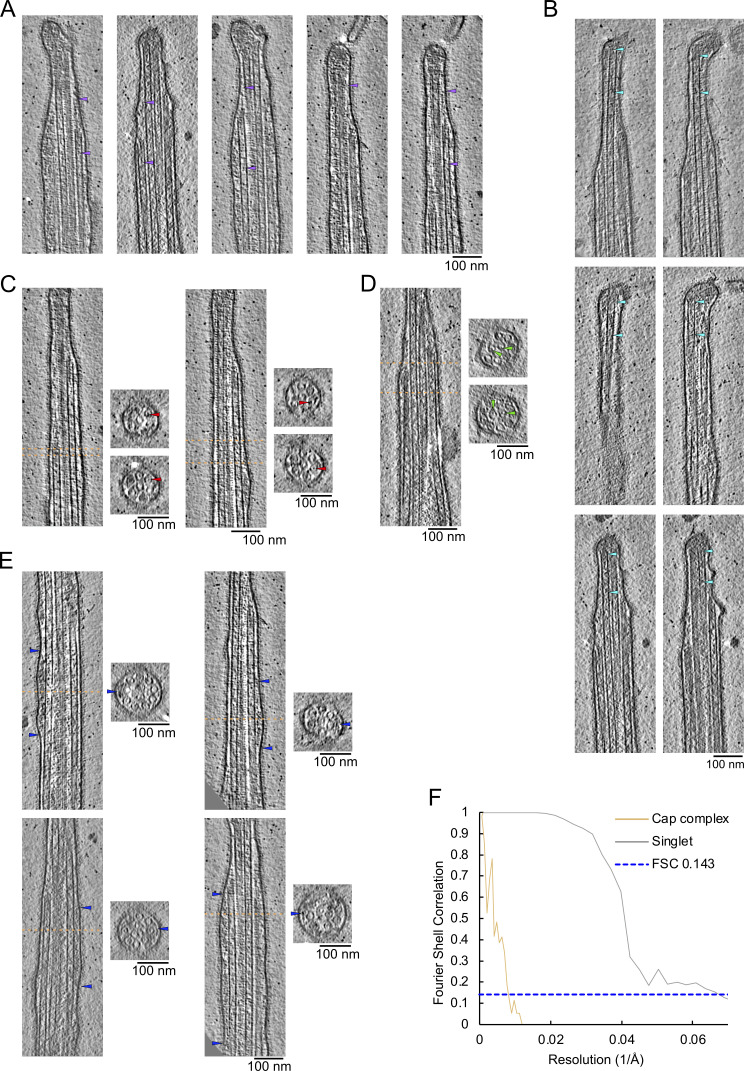
Figure 5.
FAP256A/B-KO ciliary tips show multiple structural defects. (A) Slices of WT and FAP256A/B-KO tomograms. The arrowhead points to the end of the CP microtubules to highlight the lack of cap in mutant tomograms. (B) Tomogram slice of FAP256A/B-KO cilium along with cross-section (dotted line). The arrowheads indicate the densities inside the CP microtubules that correspond to the plugs. (C) Tomogram slices from the same FAP256A/B-KO cilia in B. The arrowheads indicate the end of the CP microtubules. (D) Quantification of tip CP length in WT (n = 38) and FAP256A/B-KO (n = 50) cilia. Each dot represents one CP. ****P value < 0.0001, Mann‒Whitney test. Black line indicates the median. (E) Graph of the distance between C1 and C2 microtubules. The reference on the x-axis is the middle of the narrowing zone. The average is shown with a solid black line. Standard deviation is shown in brown (n = 6 cilia). For comparison, the corresponding CU428 data are indicated by a dotted line. (F) Length of A-tubules numbered from shortest to longest. The brown dotted line shows a linear trend line going through all points. Error bars show the standard deviation. n = 8 cilia. For comparison, the corresponding CU428 data are indicated by a dotted line. (G) Tomogram slice of a FAP256A/B-KO mutant cilium. The arrowhead points to a bent singlet A-tubule. (H) Graph showing the distance between pairs of A-tubule singlets in the ciliary tip (n = 6 cilia). Standard deviation is shown in brown. For comparison, the corresponding CU428 data are indicated by a dotted line.
Figure S5.
FAP256A/B-KO tip defects and CCDC81 localisation. (A) Slices of FAP256A/B-KO tomograms along with cross-sections (dotted line) showing examples of the filamentous plugs in both CP microtubules. (B) Subtomogram average of FAP256A/B-KO tip CP (dark salmon). (C) FSC curve of the FAP256A/B-KO tip CP structure. (D) Measured twist angles for FAP256A/B-KO CP. The dotted line is drawn at the highest point of the curve fitted to the histogram. (E) Gallery of A-tubule plugs. The arrowheads point to the extremities of the plugs. (F) Gallery of bent A-tubules indicated by arrowheads. (G) Predicted structures of CCDC81 putative microtubule-binding domain (green) and EF-hand pair domains (purple). (H) Representative SR-SIM images of cells expressing CCDC81-GFP-mNeonGreen under the native promoter fixed and labeled with AXO49 anti-polyglycylated (polyG) tubulin monoclonal antibody that labels ciliary axonemes (green), anti-GFP polyclonal antibodies (red), and DAPI (Blue). Inset highlights CCDC81 enriched along the length of oral membranelles (arrowheads) and at the tips of anterior cilia (arrows). Scale bars, 10 μm; inset, 5 μm. (I) Slice through an example tomogram of demembranated CU428 showing that the cap complex is still present.
While analyzing our tomograms, we also observed that C1 and C2 often had different lengths (Fig. 5 C), while they would typically have the same length in WT cilia. In the two examples shown, C2 is longer than C1. These defects in CP microtubules prompted us to measure the length of the tip CP, that is, the CP portion covered by the spike protein (Fig. 5 D). Indeed, the tip CP was significantly shorter in FAP256A/B-KO than in WT, agreeing with a previous study (Louka et al., 2018). To verify whether the CP in FAP256A/B-KO had defects, we used subtomogram averaging to obtain a structure of the tip CP. At the same resolution, both WT and FAP256A/B-KO tip CP were identical (Fig. S5, B and C; and Table S2). The distances between C1 and C2 in the doublet zone and at the tip were comparable with those of WT (Fig. 5 E). We also measured the twist of the CP and obtained the same value as in WT cells (0.72° per 8 nm, Fig. S5 D and Fig. S4 D).
We then focused our attention on the A-tubules in FAP256A/B-KO cells. They mostly appeared identical to WT. The plugs at their ends were also present (Fig. S5 E). Their lengths were almost the same as those of WT cells (Fig. 5 F), and the starting and ending zones measured were 268 ± 88 nm and 194 ± 74 nm, respectively (n = 8 cilia). Interestingly, we observed that multiple A-tubule singlets were curved toward the CP when they were normally straight in WT cells (Fig. 5 G and Fig. S5 F). However, when we measured the average distance between A-tubules in the mutant tips, we did not observe any difference compared with WT (Fig. 5 H). This is because A-tubules only seem to bend at their ends, and as they do not end at the same position, the smaller inter-A-tubule distances are not sufficient to influence the average of multiple A-tubules significantly.
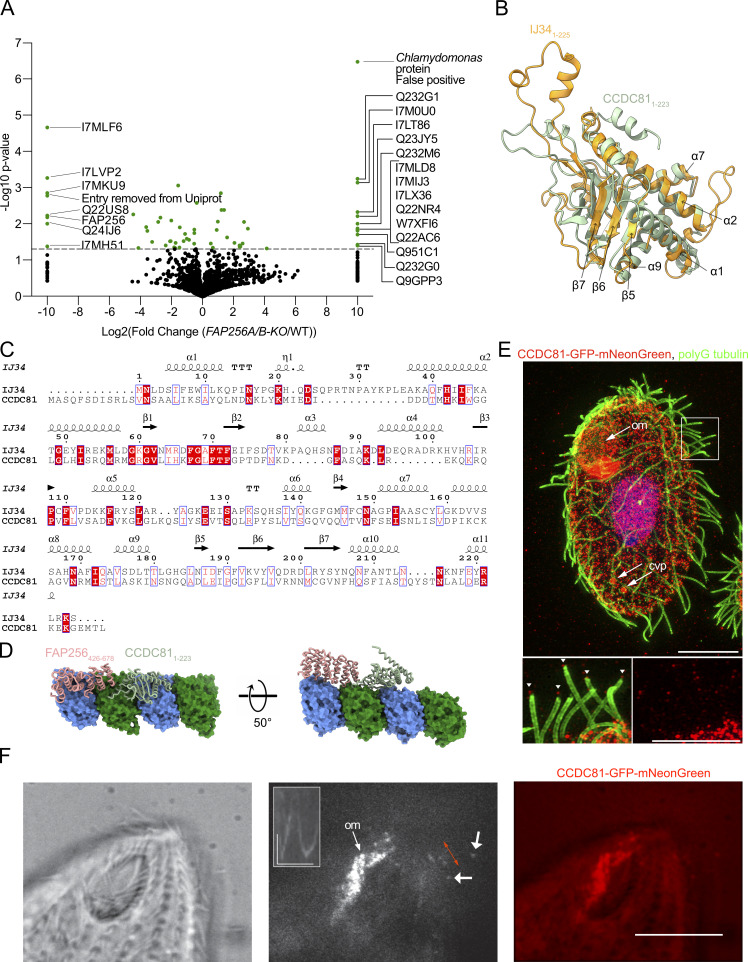
As the ciliary cap complex was absent from FAP256A/B-KO cilia, we next sought to identify the proteins found in this complex. To this end, we carried out mass spectrometry to identify the missing proteins in mutant cilia (Fig. 6 A and Table S3). Both FAP256A and FAP256B were indeed absent, confirming that our assay worked. We identified six other proteins that were absent from FAP256A/B-KO cilia (Table 1). However, none of the proteins that we identified as absent from FAP256A/B-KO were found in the previously published screen in Chlamydomonas (Satish Tammana et al., 2013). This observation is likely because we did not find homologs for the proteins we identified. We also listed in Table 1 the proteins that had at least a fourfold reduction in FAP256A/B-KO cilia. These proteins include a predicted kinesin (UniProtID: I7LVY1) that is likely to bind the CP microtubules in the cap complex. This kinesin also contains a CCDC78 domain. Interestingly, CCDC78 localizes to the deuterosome in the cytoplasm of Xenopus multiciliated cells and mouse tracheal epithelial cells. Knockdown of the gene causes the number of centrioles to decrease (Klos Dehring et al., 2013). There were also multiple proteins that were present only in FAP256A/B-KO or were enriched fourfold or more in the mutant (Table 2). These proteins were perhaps expressed to compensate for the loss of FAP256, although based on their human homologs and known localization, it is difficult to draw conclusions from this list.
Figure 6.
The ciliary cap complex contains CCDC81. (A) Volcano plot of the peptides identified in FAP256A/B-KO versus WT cilia. The proteins absent in FAP256A/B-KO are listed on the left-hand side of the graph. The proteins only found in FAP256A/B-KO are listed on the right-hand side of the graph. (B) Atomic models of IJ341–225 (red) and the N-terminal domain of CCDC811–223 (green) are shown in Fig. S5 H as well as the two models overlaid. (C) Alignment of Tetrahymena IJ341–225 and CCDC811–223. Residues in white on a red background are conserved, and residues in red have similar properties. The secondary structure of IJ34 is shown above the alignment. (D) Predicted AlphaFold atomic model of the putative microtubule-binding domain CCDC811–223 (light green) along with α-tubulin (green) and β-tubulin (blue). The TOG domain of FAP256426–678 (pink) is shown bound to an adjacent curved tubulin dimer. (E) Representative SR-SIM images of cells expressing CCDC81-GFP-mNeonGreen under the native promoter fixed and labeled with AXO49 anti-polyglycylated (polyG) tubulin monoclonal antibody that labels ciliary axonemes (green), anti-GFP polyclonal antibodies (red), and DAPI (blue). Inset highlights CCDC81 at the tips of locomotory lateral cilia. Scale bars 10 μm, inset 5 μm. Abbreviations: om, oral membranelle; cvp, contractile vacuole pore. Arrowheads point to ciliary tips. (F) Live imaging of CCDC81. Brightfield, TIRF, and merged images of the anterior regions of Tetrahymena cells expressing CCDC81-GFP-mNeonGreen. Short arrows point to signals present in the distal region of moving locomotory cilia. Inset displays a kymograph showing movements of the CCDC81 signal at the distal portion of a locomotory cilium. Red bilateral arrow marks the areas used for the kymograph. The images shown were extracted from Video 2. Scale bar: 10 μm. Horizontal scale bar for kymograph: 2 s, vertical scale bar 2 μm.
Table 1.
List of proteins significantly absent and reduced at least fourfold from FAP256A/B-KO compared with CU428 along with their human homologs, known localization, percent coverage, emPAI score, and number of peptides
| Absent or decreased at least fourfold in FAP256A/B-KO cells | Uniprot ID | Name | Amino acids | Human homolog? | % identity | Known localization? | Percent coverage (highest of repeats) | emPAI score | Number of peptides (average of WT) |
|---|---|---|---|---|---|---|---|---|---|
| Absent | I7MLF6 | EF hand protein | 1766 | CCDC81 | 23.74 | Centrosome | 7.53 | 0.169 | 8.7 |
| Absent | Q22US8 | Uncharacterized protein | 798 | 2.88 | 0.054 | 1.3 | |||
| Absent | I7MKU9 | Uncharacterized protein | 1730 | 2.08 | 0.046 | 2.3 | |||
| Absent | Q24IJ6 | Tetratricopeptide repeat protein | 519 | TPR repeat protein 28 | 24.07 | Microtubule organizing center | 8.09 | 0.225 | 3 |
| Absent | Q23FN3 | FAP256 | 872 | CEP104 | 29.05 | Cilia and centriole | 20.3 | 0.392 | 8.3 |
| Absent | I7LVP2 | Divergent AAA domain protein | 273 | SLFNL1 | 30.77 | Sperm cells | 16.5 | 0.438 | 3.3 |
| Absent | I7MH51 | Uncharacterized protein | 474 | 10.1 | 0.123 | 1.7 | |||
| Absent | I7LTB1 | FAP256B | 1017 | CEP104 | 25.98 | Cilia and centriole | 2.75 | 0.015 | 0.7 |
| Decreased at least fourfold | Q22Y55 | Phosphatase 2A | 579 | PP2A | 34.15 | Ubiquitous | 50.4 | 1.419 | 16.3 |
| Decreased at least fourfold | Q229R5 | Ser/Thr phosphatase | 313 | PP2A | 70.1 | 17.3 | 0.497 | 3.7 | |
| Decreased at least fourfold | I7LX00 | Group 1 truncated haemoglobin | 121 | 56.2 | 2.929 | 4 | |||
| Decreased at least fourfold | I7LVY1 | Kinesin motor catalytic domain protein | 1066 | Kif16B | 29.75 | Early endosomes | 14.9 | 0.284 | 7.7 |
| Decreased at least fourfold | Q22D06 | WD domain, G-beta repeat protein | 2585 | Dynein assembly factor with WDR repeat domains | 34.2 | Cilia | 8.55 | 0.171 | 13.7 |
| Decreased at least fourfold | Q24CF9 | WD domain, G-beta repeat protein | 2207 | Dynein assembly factor with WDR repeat domains | 25.82 | Cilia | 27.5 | 0.79 | 38 |
| Decreased at least fourfold | I7M9E4 | ATP:guanido phosphotransferase, carboxy-terminal domain protein | 370 | Creatine kinase | 41.87 | Ubiquitous | 41.4 | 3.474 | 12.3 |
| Decreased at least fourfold | Q23DC5 | Thioredoxin glutathione reductase | 491 | Thioredoxin reductase | 55.7 | Ubiquitous | 7.74 | 0.177 | 2.3 |
| Decreased at least fourfold | I7LU65 | C2 domain protein | 841 | Coiled-coil domain-containing protein 33 | 26.24 | Peroxisome | 9.99 | 0.278 | 5.7 |
| Decreased at least fourfold | I7MMW4 | STOP Protein | 592 | 19.8 | 0.441 | 6.7 | |||
| Decreased at least fourfold | Q22F01 | Phospholipid scramblase | 277 | 14.8 | 0.718 | 3.3 |
Table 2.
List of proteins significantly only found and increased at least fourfold from FAP256A/B-KO compared with CU428 along with their human homologs, known localization, percent coverage, emPAI score, and number of peptides
| Only found or increased at least fourfold in FAP256A/B-KO cells | Uniprot ID | Name | Amino acids | Human homolog? | % identity | Known localization? | Percent coverage (highest of repeats) | emPAI score | Number of peptides (average of FAP256A/B-KO) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Only in mutant | Q66T67 | Chlamydomonas - Heat shock protein 90C - False positive - one peptide from ABC transporter, one peptide from HSP82 | 180 | |||||||
| Only in mutant | I7MLD8 | 14-3-3 family protein 14-3-3 beta/zeta | 254 | 14-3-3 protein | 46.94 | Ubiquitous | 46.1 | 1.43 | 7.3 | |
| Only in mutant | I7MIJ3 | Phospholipid scramblase | 270 | 5.19 | 0.099 | 1 | ||||
| Only in mutant | W7XFI6 | Cation channel family transporter | 2914 | Voltage-dependent L-type calcium channel | 43 | Membrane | 4.7 | 0.078 | 8.3 | |
| Only in mutant | I7M0U0 | Mg2+-specific channel-like exchanger, putative | 625 | Sodium/potassium/calcium exchanger 3 | 30 | Membrane | 8.16 | 0.209 | 4.7 | |
| Only in mutant | Q9GPP3 | Immobilization antigen LB | 316 | 55.1 | 2.028 | 8 | ||||
| Only in mutant | Q951C1 | Ymf66 | 446 | 6.28 | 0.057 | 1 | ||||
| Only in mutant | I7LX36 | C2 NT-type domain-containing protein | 832 | 1.2 | 0.03 | 1 | ||||
| Only in mutant | Q22AC6 | PAS domain S-box protein | 1177 | 0.77 | 0.021 | 1 | ||||
| Only in mutant | Q22NR4 | Arrestin | 331 | 11.2 | 0.105 | 1.3 | ||||
| Only in mutant | Q232M6 | 14-3-3 family epsilon domain protein | 246 | 14-3-3 protein | 47.03 | Ubiquitous | 40.7 | 0.714 | 5.3 | |
| Only in mutant | Q232G0 | Immobilization antigen | 316 | EPH receptor B1 | 33.33 | Membrane | 55.1 | 1.488 | 5 | |
| Only in mutant | Q232G1 | Immobilization antigen | 148 | EPH receptor B1 | 28.24 | Membrane | 50 | 2.302 | 2 | |
| Only in mutant | Q23JY5 | Arrestin | 391 | 5.63 | 0.114 | 1.7 | ||||
| Only in mutant | I7LT86 | Transmembrane protein, putative | 871 | 9.41 | 0.155 | 4.7 | ||||
| Increased at least fourfold | I7M849 | ABC transporter family protein | 1831 | ABC3 | 31.97 | Membrane | 17 | 0.295 | 17 | |
| Increased at least fourfold | I7MJL8 | Uncharacterized protein | 851 | 3.06 | 0.05 | 1.7 | ||||
| Increased at least fourfold | Q234Y0 | Uncharacterized protein | 115 | 31.3 | 0.471 | 1.7 | ||||
| Increased at least fourfold | Q22KL2 | Sel1 domain protein | 766 | Protein sel-1 homolog 2 | 28.74 | Endoplasmic reticulum | 4.31 | 0.056 | 1.7 | |
| Increased at least fourfold | I7MMI9 | Multidrug resistance protein-like transporter family ABC domain protein | 1289 | ATP-dependent translocase ABCB1 | 36.38 | Membrane | 12.6 | 0.221 | 9.3 | |
| Increased at least fourfold | Q23Q30 | Regulator of Vps4 activity in the MVB pathway protein | 402 | 3.48 | 0.136 | 1 | ||||
| Increased at least fourfold | W7X3N9 | LITAF-like zinc ribbon domain protein | 170 | LITAF | 35.21 | Membrane | 15.3 | 0.361 | 2 | |
| Increased at least fourfold | Q239Z2 | Calpain family cysteine protease | 1171 | Calpain-15 | 31.67 | Ubiquitous | 16.3 | 0.196 | 8 |
The proteins that are absent from FAP256A/B-KO include CCDC81, which is found in humans but not Chlamydomonas. It is found in centrosomes in human cells but is not found in human primary cilia (Firat-Karalar et al., 2014). The predicted structure of the CCDC81 protein (TTHERM_00500930, UniProtID: I7MLF6) contains an N-terminal CCDC81 domain and several EF-hand domain pairs in the C-terminus (Fig. S5 G). IJ34 (UniProtID: I7M9T0) is another CCDC81 homolog that was recently identified as a B-tubule MIP binding to the surface of the A-tubule at the inner junction (Kubo et al., 2023). The N-terminal CCDC81 domain overlays well with the structure of IJ34, notably with helices α1, α2, α7, α8, and α10 and strands β1, β2, β3, β5, β6, and β7 of IJ34, suggesting that it is also a microtubule-binding protein (Fig. 6 B). Although the predicted structure of CCDC81 is similar to IJ34, sequence alignment reveals that their sequences are not highly conserved with the exception of β1 (Fig. 6 C).
The loss of CCDC81 when FAP256 is knocked out suggests that these proteins might interact. We, therefore, tested this possibility in silico using different constructs and AlphaFold multimer. Some of the results pointed toward a direct interaction between the two proteins, although with low confidence. We assessed the likelihood of their interaction on the same protofilament by positioning the TOG domain of FAP256 on an adjacent tubulin dimer, similar to the crystal structure of Stu2 bound to tubulins (PDB: 4FFB; Ayaz et al., 2012). When CCDC81 and IJ34 are placed on the same protofilament, their structures do not clash and are close to each other, supporting a potential interaction (Fig. 6 D).
We also constructed a Tetrahymena strain expressing CCDC81-GFP-mNeonGreen under the native promoter. Using super-resolution structured illumination microscopy (SR-SIM), we observed CCDC81 fluorescence along the entire length of cilia in oral rows or membranelles (Fig. 6 E and Fig. S5 H, marked “om”; inset in Fig. S5 H, arrowheads) and around the microtubule-based contractile vacuole pores (Fig. 6 E and Fig. S5 H, marked “cvp”). In addition, we detected dots of CCDC81 at the tips of some cilia, including a subset of lateral locomotory cilia (inset in Fig. 6 E) and short anterior cilia (inset in Fig. S5 I, arrows). We also performed total internal reflection fluorescence (TIRF) microscopy imaging of live Tetrahymena cells expressing CCDC81-GFP-mNeonGreen. Our results confirmed the presence of CCDC81 in the beating oral membranelles and in the distal portions of some anterior locomotory cilia (Fig. 6 F; and Videos 2 and 3).
Video 2.
Brightfield (left) and TIRF (right) imaging of a live partially immobilized Tetrahymena cell expressing CCDC81-GFP-mNeonGreen. Prominent fluorescence is visible in oral cilia. Arrows point to dots of fluorescence in the distal portions of motile anterior locomotory cilia. Stills are shown in Fig. 6 E. The video was recorded at a speed of 15 fps and is shown 1.4× faster.
Video 3.
Brightfield (left) and TIRF (right) imaging of a live partially immobilized Tetrahymena cell expressing CCDC81-GFP-mNeonGreen. The strongest fluorescence is present in the beating oral membranelle. The video was recorded at a speed of 15 fps and is shown 1.4× faster.
Sperm-tail PG-rich repeat protein (UniProtID: Q239A2) and tubulin-tyrosine ligase (UniProt ID: Q22BT7) were absent, although not significant. Although the human homologs of these two proteins are unclear, sperm-tail PG-rich proteins were identified as MAPs binding both inside and outside microtubules (I7M2G0 and Q24GM1; Kubo et al., 2023). The other proteins that are missing from FAP256A/B-KO cells are predicted to be long and disordered and have not been characterized previously.
All the missing proteins in the FAP256A/B-KO mutant are present in the proteome of demembranated WT cilia, in which the ciliary cap is still present (Fig. S5 I; Kubo et al., 2021). I7LVP2 is also absent from RIB72A/B-KO mutant cells, which still have the CP cap, so it is unlikely to be a tip protein but rather a chaperone (Kubo et al., 2023). Based on these results, we propose that the proteins absent from FAP256A/B-KO, except I7LVP2, make up part of the ciliary cap complex.
Discussion
Length regulation of the ciliary tip
The ciliary tips of Tetrahymena were first studied using negative staining electron microscopy. The cap complex and A-tubule plugs have been described extensively (Dentler, 1980b). Ciliary tips of motile cilia from Tetrahymena, Chlamydomonas, Trypanosoma, and mammalian sperm cells were then studied by cryo-ET (Höög et al., 2014; Reynolds et al., 2018; Zabeo et al., 2018, 2019). Using cryo-ET, we confirmed these data and provided more accurate information on the overall architecture of the motile ciliary tips in Tetrahymena (summarized in Table 3).
Table 3.
Summary of the differences in cilia structures between the medial region and the tip
| Region Structure |
Medial region | Tip region |
|---|---|---|
| Doublets | Intact Linked to each other by the N-DRC complex |
B-tubules are absent Inner junction is incompletely assembled |
| A-tubules | Present as part of the doublet | Singlets Parallel to each other Linked to each other Linked to the membrane Have different lengths |
| Central pair | Big projections on both microtubules | The two microtubules are closer to each other The projections disappear and are replaced by small projections |
| Capping structures | None | A-tubules and CP microtubules are capped by filamentous structures |
There exists a variety in the length and ending points of the singlet A-tubule in the tip region. Recently, three proteins were proposed as regulators of the length of A- and B-tubules. FAP256 promotes the elongation of A-tubules, while CHE-12 and ARMC9 promote the elongation and depolymerization of B-tubules (Louka et al., 2018). The outside of the doublet microtubule has a periodicity of 96 nm that is regulated by the coiled-coil proteins CCDC39 and CCDC40 (Oda et al., 2014a). It is likely that the availability of these proteins regulates the assembly of the last radial spokes and dynein arms. Interestingly, the doublet microtubules near the narrowing zone show the lack of the inner junction but the existence of CFAP52, an inner junction MIP that is linked to ciliopathies in humans (Ta-Shma et al., 2015). This suggests that the inner junction protofilament proteins PACRG and CFAP20 assemble last to close and stabilize the B-tubule. It is likely that ARMC9, as a negative regulator of B-tubules, prevents the assembly of the inner junction between PACRG and CFAP20. Glutamylation and other microtubule posttranslational modifications might also play a role in the termination of B-tubules, as they are known to be highly glutamylated (Lechtreck and Geimer, 2000).
Interestingly, both A-tubules and CP microtubules contain non-periodic MIPs. MIPs of similar size have been identified in neurons and primary cilia (Bouchet-Marquis et al., 2007; Foster et al., 2022; Garvalov et al., 2006; Kiesel et al., 2020). The identities of these MIPs remain a mystery, but some could be microtubule-modifying enzymes, such as acetyltransferases. They might also be important for the stability of the microtubules, but we think this is an unlikely scenario for two reasons: one because the CP microtubules are already stabilized by filamentous and arc-MIPs, and two because the other microtubule-stabilizing proteins found in cilia have a constant periodicity. Intriguingly, our subtomogram average of the A-tubule does not show any stabilizing protein, raising the question of how these are stabilized. In human, bull, pig, and horse sperm cells, SPACA9 provides stability to the A-tubule by forming a helix inside the lumen (Gui et al., 2022a; Leung et al., 2021, 2023; Zabeo et al., 2018). This difference likely reflects the role of the ciliary tip. The tips of sperm cells might need to be more resistant to forces as opposed to the tips of Tetrahymena.
Microtubules are dynamic and unstable at their plus ends. They must therefore be stabilized in the cilia. The plugs seen at the ciliary tip are likely to play this role. Proteins stabilizing the plus ends of centriolar microtubules have been studied previously. CEP110 and CEP97 act as a plug that can be visualized in vitro using tomography (Ogunmolu et al., 2022 Preprint).
IFT trains convert from anterograde to retrograde at the tips. The observation that anterograde trains walk on the B-tubules raises the question of how these trains reach the ciliary tip when the B-tubules end. The IFT train-like densities we observed seem more similar to those of anterograde than retrograde trains. Anterograde trains have already been observed on the A-tubules in primary cilia (Kiesel et al., 2020). In our tomograms, we did not observe retrograde-train-looking particles (Jordan et al., 2018). Whether the densities we observed are active anterograde trains on A-tubules remains to be determined.
Assembly of the CP
When reaching the ciliary tip, the CP is entirely reshaped. At our resolution, we were unable to identify densities shared between the main and tip portions of the CP. The mechanism that regulates the lengths of the two CP segments remains to be discovered. The different structures suggest a dual role for the CP. In the main portion of the axoneme, the CP acts as a signal transducer between the radial spokes and its large projections (Oda et al., 2014b). In the tip region, the CP may act as a strengthening rod for the ciliary tip. Interestingly, when cilia grow, the CP reaches its final length before the doublets and singlets, which continue to grow until they reach theirs (Reynolds et al., 2018).
The striking symmetry observed between C1 and C2 suggests that the same proteins assemble around the two CP microtubules. The seams facing each other are consistent with the seam-centric model for the assembly of the CP (Gui et al., 2022b). Interestingly, the seam on C1 in the tip CP is shifted by one protofilament compared with the Chlamydomonas main CP (Gui et al., 2022b; Han et al., 2022). It is possible that the seams in the main CP of Tetrahymena are consistent with the tip CP and different from those of Chlamydomonas. Alternatively, the C1 microtubule might rotate slightly during the main-to-tip-CP transition. A high-resolution structure of the main CP in Tetrahymena will be needed to address this uncertainty.
This study raises questions on the mode of assembly of the CP. There is evidence suggesting that the CP assembles in a unique manner, distinct from the peripheral axonemal microtubules. In Chlamydomonas dikaryons that repair their CP under conditions of genetic complementation, the CP initially assembles as short segments that are positioned away from either the basal body or the ciliary distal end. It is possible that these fragments later fuse to form the CP (Lechtreck et al., 2013). Whether the CP assembles as multiple fragments during the normal course of ciliary elongation is still unknown. In Tetrahymena, a recent study showed that the CP was present at the tips of short-growing cilia; however, its precise structure and elongation mechanism remain undetermined (Reynolds et al., 2018). Regardless of the mode of CP assembly, it remains to be determined how the boundary between the main and tip segments of the CP is positioned during ciliary assembly.
The tip is a highly stable entity
This distal region must be very stable to resist the various forces at the tips of Tetrahymena cilia. Various lines of evidence point in that direction. First, we see that the CP microtubules are closer to each other, indicating that the bridge has shortened to restrict movement between the two microtubules. Second, the spike proteins binding to the outside of the CP microtubules repeat every 8 nm as opposed to 16 or 32 nm for the main CP. Third, the conformation of MAPs binding on the outside of C1 and C2 is such that multiple protofilaments are engaged and therefore strengthen the lateral interaction of tubulins. Microtubule interactions are stronger in the longitudinal direction (VanBuren et al., 2002). Lateral MIPs and MAPs are likely to be important to stabilize microtubules while cilia are moving in different directions. These different MAPs with a shorter periodicity might explain the higher twist seen in the ciliary tip. Fourth, we observed links between A-tubules themselves and between A-tubules and the membrane in the tip that contribute to their stability. The identity of these links remains unknown, but a previous study identified a tubulin-like protein in solubilized Tetrahymena membranes (Dentler, 1980a). This protein might be the one that links A-tubules to the membrane.
FAP256 contributes to ciliary tip stability
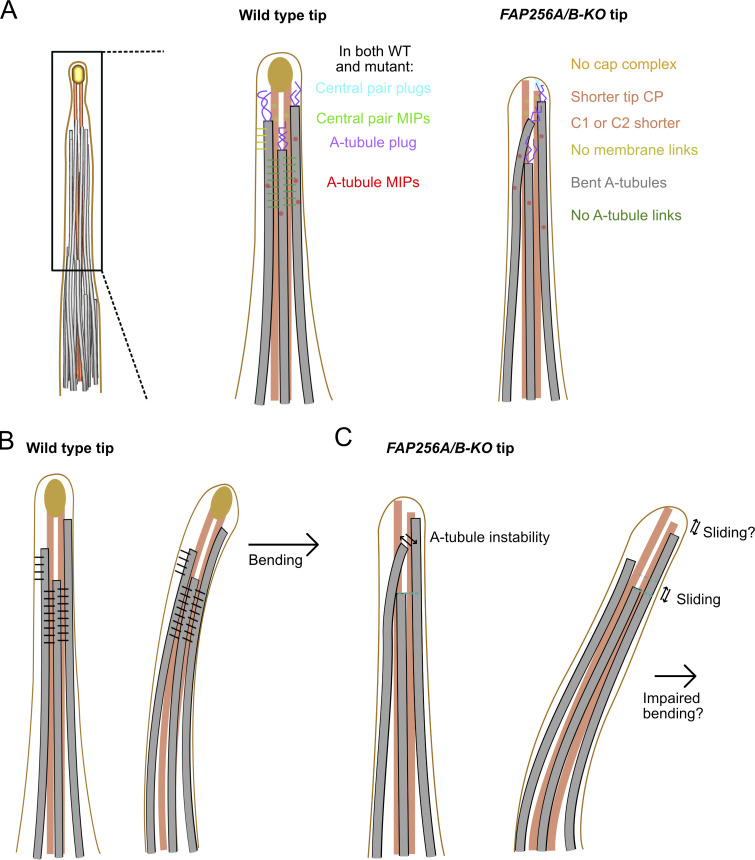
The main two defects we observed in FAP256A/B-KO mutant cells are the bending of singlets and one of the CP microtubules longer than the other (Fig. 7 A). The difference in length observed between the two CP microtubules might be due to the sliding of one microtubule next to the other. Alternatively, either C1 or C2 microtubules might disassemble abnormally or not form entirely at the tip. The CP microtubules are capped by the cap complex in WT cells, which is absent in mutant cells. FAP256, along with the proteins of the cap complex, might therefore prevent the sliding or depolymerization of CP microtubules and likely serve as an anchor for the CP microtubules to the membrane.
Figure 7.
The A-tubule links stabilize the A-tubules and restrict sliding. (A) Diagram summarizing the differences between CU428 and FAP256A/B-KO cilia. (B) Diagram of WT cilia with the A-tubule and membrane links drawn in black in straight and bending cilia. (C) Diagram of FAP256A/B-KO mutant cilia without the A-tubule links showing bent A-tubules and sliding A-tubules and CP microtubules during bending.
The observation that CCDC81 and FAP256 are predicted to be microtubule-binding proteins and the fact that CCDC81 is absent when FAP256 is knocked out suggests that these two proteins might interact to regulate microtubule polymerization. As our imaging localizes CCDC81 in the ciliary tip, we propose that CCDC81 is a novel cap complex protein.
We did not observe links between A-tubules or between A-tubules and the membrane in the subtomogram averages of FAP256A/B-KO singlets. Furthermore, the bending of the A-tubules observed in our FAP256A/B-KO tomograms suggests that some links are missing. We, therefore, propose that FAP256, along with its interacting proteins, contributes to stabilizing the ciliary tip microtubules by preventing sliding and/or bending of these microtubules. Our model explains the observed reduced functionality of motile cilia caused by the loss of FAP256 in Tetrahymena (Louka et al., 2018; Fig. 7, B and C). C2 in Chlamydomonas can slide regarding C1 in the medial region of the cilium. The sliding distance measured was up to 24 nm (Han et al., 2022). It is currently unclear whether the same sliding occurs in other species and in the tip of Tetrahymena thermophila. The ciliary cap is possibly essential to maintain the association of C1 and C2 during sliding.
To conclude, this study represents the most detailed description of the ciliary tip of Tetrahymena thermophila to date. Future studies will be needed to identify the proteins that bind to the CP microtubules and shed light on their precise functions.
Materials and methods
Cell culture and gene editing
All Tetrahymena strains (CU428 wild-type, FAP256A/B-KO mutant) used in this study were grown in SPP media (Williams et al., 1980) in a shaker incubator at 30°C and 150 rpm. Cultures were harvested at an OD600 of ∼0.6. Refer to (Black et al., 2021) for further details. The FAP256A/B-KO mutant was created in a previous study (Louka et al., 2018).
To tag CCDC81 with GFP-mNeonGreen, the TTHERM_0500930 gene was edited using homologous DNA recombination. Primers 930-5F (5′-AGAGCTTGACGGGGAAAGCCAGCAAAACTCAAACAGTGTGC-3′) and 930-5R (5′-TCATCAAGCTTGCCATCCGCGGATTCTTTATTAATTAATTTGCAGTTTA-3′) were used to amplify a terminal portion of the coding region and primers 930-3F (5′-TATCAAGCTTATCGATACCGGGTCTGTCTTTCACTGGATCAAG-3′) and 930-3R (5′-AGATTTAAATAAGCTCCTCCTGCTTCAGCTAAGTGTCCCA-3′) were used for amplification a portion of the 3′ UTR. The two fragments were cloned on the sides of the GFP-mNeonGreen-BTU2-3′UTR-neo5 module on the plasmid pCDAH_GFP_mNeon. The resulting edited fragment TTHERM_0500930 was targeted to the native locus using biolistic bombardment of starved Tetrahymena cells and paromomycin selection.
Cilia purification
Tetrahymena cells were harvested and deciliated as previously described (Black et al., 2021). Briefly, 2 liter of cell culture was harvested by centrifugation at 2,000 × g for 10 min at 23°C. Whole cells were resuspended in fresh SPP and adjusted to a total volume of 24 ml. Dibucaine (25 mg in 1 ml SPP) was added and the culture was gently swirled for 45 s. To stop the dibucaine treatment, 75 ml of ice-cold SPP supplemented with 1 mM EGTA was added and the dibucaine-treated cultures were centrifuged for 15 min at 2,500 g and 4°C. The supernatant (cilia) was collected and centrifuged at 25,000 × g for 45 min at 4°C. The pellet (cilia) was gently washed and resuspended in cilia wash buffer (50 mM HEPES at pH 7.4, 3 mM MgSO4, 0.1 mM EGTA, 1 mM DTT, and 250 mM sucrose) before snap-freezing in liquid nitrogen for long-term storage. Cilia were resuspended in cilia final buffer (50 mM HEPES at pH 7.4, 3 mM MgSO4, 0.1 mM EGTA, 1 mM DTT, 0.5% trehalose) before grid freezing. NP-40 Alternative (#492016; Millipore Sigma) was added to a final concentration of 1.5% for 45 min on ice to remove the membrane. Axonemes were then centrifuged and resuspended in cilia final buffer.
Cryo-ET preparation
The axonemes for cryo-ET were crosslinked with glutaraldehyde (final concentration 0.15%) for 30 min on ice and quenched with 35 mM Tris pH 7.5. The axoneme concentration was ∼3.6 mg/ml when mixed with 5 (Cytodiagnostics) or 10 (Aurion) nm gold beads in a 1:1 ratio for a final axoneme concentration of 1.8 mg/ml. Crosslinked axoneme samples (4 μl) were applied to negatively glow-discharged (10 mA, 10 s) C-Flat Holey thick carbon grids (#CFT312-100; Electron Microscopy Services) inside the Vitrobot Mk IV (Thermo Fisher Scientific) chamber. The sample was incubated on the grid for 45 s at 23°C and 100% humidity before being blotted with force 0 for 8 s and plunged-frozen in liquid ethane.
Tilt series acquisition
Tilt series were acquired on a Titan Krios electron microscope (Thermo Fisher Scientific) operated at 300 kV equipped with a BioQuantum energy filter (Gatan, Inc.) and a K3 Summit (Gatan, Inc.) electron detector. Tilt series were collected using SerialEM (Mastronarde, 2005) at 42,000× and 26,000× magnifications using the grouped dose-symmetric scheme from −60° to 60° with an increment of 3° (136, 64, and 33 tilt series of demembranated WT, membranated WT, and FAP256A/B-KO, Table S1). The defocus for each tilt series ranged from −2 to −6 μm. The total dose for each tilt series was 164 e− per Å2. For each view, a movie of 10–13 frames was collected. The pixel sizes at 42,000× and 26,000× were 2.12 and 3.37 Å, respectively. Frame alignment was performed with Alignframes (Mastronarde and Held, 2017). Tomograms were reconstructed using IMOD (Kremer et al., 1996). Aligned tilt series were manually inspected for quality and sufficient fiducial presence. Batch reconstruction was performed in IMOD.
Subtomogram averaging
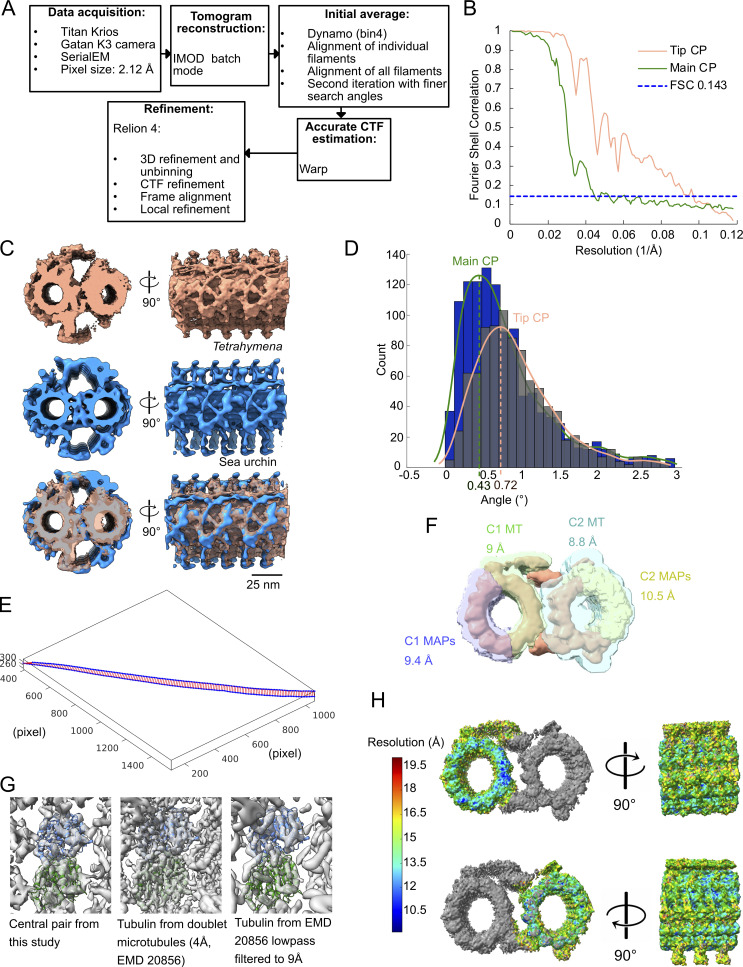
CTF estimation of the tomogram was performed with WARP (Tegunov and Cramer, 2019). The picking of the CP and A-tubules was performed using IMOD by tracing lines along the microtubules (Kremer et al., 1996). The subtomograms were then picked at 8-nm (tip CP and A-tubules) or 16-nm (main CP) intervals along the traced lines. The prealignment of the four-times binned 8-nm repeating unit of the tip CP of WT and FAP256A/B-KO mutants (11,534 and 4,407 subtomograms, respectively) was performed using a set of Dynamo (Castaño-Díez et al., 2012) scripts for filament alignment (https://github.com/builab/dynamoDMT). Then, the alignment parameters were converted into Relion 4.0 format. Refinement of the subtomogram averages was performed with the Relion 4.0 pipeline (Kimanius et al., 2021).
After importing the prealigned subtomogram into Relion 4.0, the following workflow was followed: (i) generation of pseudosubtomograms; (ii) refinement of the subtomograms; (iii) CTF refinement; (iv) frame alignment; (v) pseudotomogram generation; and (vi) refinement of the polished subtomograms (Fig. S4 A).
The resolutions for the 8-nm repeating unit of the tip CP of WT and FAP256A/B-KO are 9 and 19 Å, respectively. Local resolution maps were obtained using ResMap (Kucukelbir et al., 2014).
For the main CP, after obtaining the 16-nm averages, we performed a 3D classification into 32-nm repeats of the main CP. The resolution of the main CP is 23 Å.
FAP256A/B-KO tip CP was first processed in Dynamo and refined in Relion 3.1 and 4.0. C1 and C2 were masked and refined independently.
Visualization/segmentation
For denoising, tomograms were CTF-deconvolved and missing-wedge corrected using IsoNet (Liu et al., 2022). The IMOD model of the ciliary tip was generated from both by automatically converting the aligned subtomogram coordinates (microtubules and cap complex) and manually tracing the tomograms (MIPs, plugs, and links). Membrane segmentation was performed on the denoised tomograms using TomoSegMemTV (Martinez-Sanchez et al., 2014). For visualization of the subtomogram averages inside the tomogram coordinates (Fig. 1 A), Subtomo2Chimera (Bui, 2022) was used.
For subtomogram averages, surface rendering, segmentation, and fitting were performed using ChimeraX (Pettersen et al., 2021).
Measurement of distance and twist between C1 and C2 microtubules and distance between A-tubules
The distance and twist between C1 and C2 microtubules were measured using a custom MATLAB script. These microtubules were first picked independently in the tomograms. Subtomogram averaging of each microtubule was then performed using particles picked every 16 nm to properly center the C1 and C2 microtubules. Using the center of each particle, a spline line was extrapolated along the C1 microtubule and the shortest distance from equally spaced 8-nm points of the C2 microtubule to the C1 microtubule line was measured (Fig. S4 E). The distance measured between C1 and C2 from different tomograms was plotted on a graph (Fig. 4 C). In the tomograms containing the narrowing zone, the inflection point (midpoint) was identified automatically by determining the points with the most negative slope. Each CP graph was shifted such that the midpoint was situated at length 0. A mean graph was finally generated from multiple tomograms.
A similar analysis was carried out to measure the distance between A-tubule pairs
For the twist analysis, the angle between vectors representing the shortest distance between consecutive points of the C2 microtubule and the C1 microtubule was calculated. A histogram of all the twist angles measured from multiple tomograms containing the tip CP and main CP was then created and a curve was fitted using the smoothing function.
Measurements of CP and A-tubule lengths
CP and A-tubule lengths were measured manually in IMOD by clicking two points at the start and end of the tip CP or the A-tubules and measuring the distance in 3D between those two points. The endpoint was always the end of the longest CP microtubule that could be seen in the tomogram.
Sequence alignment
Sequences of IJ341–225 (UniProtID: I7M9T0) and CCDC811–223 (UniProtID: I7MLF6) were aligned using Clustal Omega (Madeira et al., 2022), modified manually so that the sequences whose structures overlay would be aligned with each other and colored using ESPript (Robert and Gouet, 2014).
Generation of CCDC81 and FAP256 models
AlphaFold2 was used to predict the structure of CCDC81 and FAP256. The TOG domain of FAP256 was overlaid with the crystal structure of Stu2 with α- and β-tubulins (PDB: 4FFB). The putative microtubule domain of CCDC81 was overlaid with the structure of IJ34 from Kubo et al. (2023).
Mass spectrometry
Mass spectrometry was performed as described in a previous publication (Khalifa et al., 2020). Approximately 25–30 μg of membranated cilia was loaded on the SDS-PAGE gel from three biological replicates. The electrophoresis run was terminated before the proteins entered the resolving gel. The band containing all proteins was cut out and subjected to in-gel digestion. After that, the peptides were separated on a Dionex Ultimate 3000 UHPLC and loaded onto a Thermo Acclaim Pepmap (75 μm ID × 2 cm with 3 μm C18 beads; Thermo Fisher Scientific) precolumn and then onto an Acclaim Pepmap Easyspray (75 μm × 25 cm with 2 μm C18 beads; Thermo Fisher Scientific) analytical column. The peptides were separated with a flow rate of 200 nl/min with a gradient of 2–35% solvent (acetonitrile containing 0.1% formic acid) over 2 h. Peptides of charge 2+ or higher were recorded using a Thermo Orbitrap Fusion mass spectrometer. The data were searched against the Tetrahymena thermophila protein dataset from UniProt (https://www.uniprot.org/).
MS data were analyzed by Scaffold_4.8.4 (Proteome Software Inc.). A t-test on the total spectral count was carried out to identify statistically significant proteins missing in the FAP256A/B-KO mutant compared with the WT.
Immunofluorescence and TIRF microscopy
For immunofluorescence, T. thermophila cells (wild-type CU428 and expressing CCDC81-GFP-mNeonGreen) were fixed and stained using the quick method that includes simultaneous fixation/permeabilization and drying of cells on the cover glass (Gaertig et al., 2013). The primary antibodies used were the mouse monoclonal anti-polyglycylated tubulin AXO49 (diluted 1:200; Bré et al., 1996) and polyclonal anti-GFP antibodies (600-401-215, 1:800; Rockland). The secondary antibodies used were goat-anti-mouse IgG-FITC (115-095-146; Jackson Immunoresearch) and goat-anti-rabbit-Cy3 antibodies (111-165-003; Jackson Immunoresearch). SR-SIM imaging was conducted on an ELYRA S1 microscope equipped with a 63 × NA 1.4 Oil Plan-Apochromat DIC objective. The optical slices were analyzed by Fiji/ImageJ (Z project tool).
TIRF imaging of live cells was done as described (Jiang et al., 2015) except that cells were immobilized by entrapment in a small volume and not with NiCl2. In brief, the TIRF system was house-built around a Nikon Eclipse Ti-U microscope equipped with a GFP/mCherry TIRF filter cube, a 60 × NA 1.49 TIRF objective, and an Andor iXON × 3 DU897 EMCCD camera (with a C mount and a 2.5 × lens). Images and videos were digitally recorded with the Nikon elements emission software module. ImageJ was used to generate kymographs. Cells were imaged in 10 mM Tris HCl, pH 7.5 at room temperature.
Statistical analyses
Statistical tests were calculated using Prism 9.5.1 (GraphPad Software, LLC). Normal distribution was assessed using a D’Agostino & Pearson test. Tip CP length difference was calculated using a Mann–Whitney test.
Online supplemental material
Fig. S1 shows our preparation workflow and examples of incomplete doublets. Fig. S2 shows examples of plugs, MIPs, IFT-train-like particles, and FSC curves for the cap complex and singlet A-tubules. Fig. S3 shows examples of A-tubule and membrane links. Fig. S4 shows our subtomogram averaging workflow, FSC curves of the tip and main CPs, a comparison of Tetrahymena and sea urchin CPs, the twist angle of the CP, a diagram explaining the measurement of the distance between both CP microtubules, masks used for the tip CP, the fitting of a tubulin dimer in the tip CP map, and the local resolution of the tip CP structure. Fig. S5 shows examples of plugs in FAP256A/B-KO, the subtomogram average of FAP256A/B-KO tip CP and the FSC curve related to this structure, the twist angle of the CP and examples of bent A-tubules as well as the predicted structure of CCDC81, and a microscopy image showing it at the ciliary tip. Table S1 shows the data collection parameters. Table S2 shows the characteristics of the subtomogram averages shown in this study, and Table S3 contains all the peptides identified in the mass spectrometry experiment. Video 1 shows a summary of the findings presented in this study. Videos 2 and 3 show CCDC81 in moving cilia.
Supplementary Material
shows data collection parameters for the various datasets used in this study.
shows characteristics of the subtomogram averages shown in this study.
shows peptide count, percentage coverage, and emPAI score of all the proteins identified in CU428 and FAP256A/B-KO cells.
Acknowledgments
We thank all the members of the Bui lab for the critical reading of the manuscript. We thank Drs. Mike Strauss, Kaustuv Basu, and Kelly Sears (Facility for Electron Microscopy Research at McGill University) for helping with data collection. We thank Amy Wong, Lorne Taylor, and Jean-François Trempe (RI-MUHC Proteomics Platform) for their help with mass spectrometry.
K.H. Bui is supported by grants from the Canadian Institutes of Health Research (PJT-156354) and Natural Sciences and Engineering Research Council of Canada (RGPIN-2022-04774). J. Gaerting and K. Lechtrek are supported by National Institutes of Health grants R01GM135444 and R01GM139856, respectively.
Author contributions: Conceptualization: K.H. Bui. Methodology: T. Legal. Formal analysis: T. Legal and M. Tong. Investigation: T. Legal, M. Tong, C. Black, M. Parra, E. Joachimiak, K. Lechtrek, M. Valente-Paterno, and K.H. Bui. Resources: K. Lechtrek, J. Gaertig, and K.H. Bui. Writing–original draft: T. Legal and K.H. Bui. Writing–review & editing: T. Legal, K.H. Bui, C. Black, and J. Gaertig.
Data availability
The tomography data produced in this study have been deposited at the Electron Microscopy Data Bank. The cryo-EM maps of CU428 main CP, singlet A-tubules, cap complex have accession codes EMD-40891, EMD-40894, and EMD-40892, respectively. The CU428 tip CP maps have accession codes EMD-40896, 40897, 40898, 40899, and 40900. The FAP256A/B-KO mutant tip CP maps have accession codes: EMD-40909, 40910, 40911, and 40912. The reconstructed tomogram used in Fig. 1 A (bin 4, before and after IsoNet denoising) has also been deposited and has accession code EMD-40913.
The mass spectrometry data of FAP256A/B-KO and CU428 (WT) have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD042582 and https://doi.org/10.6019/PXD042582.
The MATLAB code for subtomogram averaging in Dynamo and distance analysis is available at https://github.com/builab/dynamoDMT.
References
- Al-Bassam, J., Larsen N.A., Hyman A.A., and Harrison S.C.. 2007. Crystal structure of a TOG domain: Conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 15:355–362. 10.1016/j.str.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Ayaz, P., Ye X., Huddleston P., Brautigam C.A., and Rice L.M.. 2012. A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 337:857–860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, C., Dai D.C., Peri K., Ichikawa M., and Bui K.H.. 2021. Preparation of doublet microtubule fraction for single particle cryo-electron microscopy. Bio Protoc. 11. e4041. 10.21769/BioProtoc.4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet-Marquis, C., Zuber B., Glynn A.M., Eltsov M., Grabenbauer M., Goldie K.N., Thomas D., Frangakis A.S., Dubochet J., and Chrétien D.. 2007. Visualization of cell microtubules in their native state. Biol. Cell. 99:45–53. 10.1042/BC20060081 [DOI] [PubMed] [Google Scholar]
- Bré, M.-H., Redeker V., Quibell M., Darmanaden-Delorme J., Bressac C., Cosson J., Huitorel P., Schmitter J.M., Rossler J., Johnson T., et al. 1996. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: Widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J. Cell Sci. 109:727–738. 10.1242/jcs.109.4.727 [DOI] [PubMed] [Google Scholar]
- Bui, K H., 2022. Builab/subtomo2Chimera v0.12. zenodo. 10.5281/zenodo.6820119 [DOI] [Google Scholar]
- Carbajal-González, B.I., Heuser T., Fu X., Lin J., Smith B.W., Mitchell D.R., and Nicastro D.. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton. 70:101–120. 10.1002/cm.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Díez, D., Kudryashev M., Arheit M., and Stahlberg H.. 2012. Dynamo: A flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178:139–151. 10.1016/j.jsb.2011.12.017 [DOI] [PubMed] [Google Scholar]
- Chien, A., Shih S.M., Bower R., Tritschler D., Porter M.E., and Yildiz A.. 2017. Dynamics of the IFT machinery at the ciliary tip. Elife. 6:e28606. 10.7554/eLife.28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler, W.L. 1980a. Microtubule-membrane interactions in cilia. I. Isolation and characterization of ciliary membranes from Tetrahymena pyriformis. J. Cell Biol. 84:364–380. 10.1083/jcb.84.2.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler, W.L. 1980b. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J. Cell Sci. 42:207–220. 10.1242/jcs.42.1.207 [DOI] [PubMed] [Google Scholar]
- Dentler, W.L., and Rosenbaum J.L.. 1977. Flagellar elongation and shortening in Chlamydomonas. III. structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 74:747–759. 10.1083/jcb.74.3.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar, E.N., Sante J., Elliott S., and Stearns T.. 2014. Proteomic analysis of mammalian sperm cells identifies new components of the centrosome. J. Cell Sci. 127:4128–4133. 10.1242/jcs.157008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch, C., and Dupuis-Williams P.. 2011. Ultrastructure of cilia and flagella - back to the future!. Biol. Cell. 103:249–270. 10.1042/BC20100139 [DOI] [PubMed] [Google Scholar]
- Foster, H.E., Ventura Santos C., and Carter A.P.. 2022. A cryo-ET survey of microtubules and intracellular compartments in mammalian axons. J. Cell Biol. 221:e202103154. 10.1083/jcb.202103154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G., Zhao L., Dymek E., Hou Y., Song K., Phan N., Shang Z., Smith E.F., Witman G.B., and Nicastro D.. 2019. Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. J. Cell Biol. 218:4236–4251. 10.1083/jcb.201906006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig, J., Wloga D., Vasudevan K.K., Guha M., and Dentler W.. 2013. Discovery and functional evaluation of ciliary proteins in Tetrahymena thermophila. Methods Enzymol. 525:265–284. 10.1016/B978-0-12-397944-5.00013-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov, B.K., Zuber B., Bouchet-Marquis C., Kudryashev M., Gruska M., Beck M., Leis A., Frischknecht F., Bradke F., Baumeister W., et al. 2006. Luminal particles within cellular microtubules. J. Cell Biol. 174:759–765. 10.1083/jcb.200606074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, I.R., and Grimstone A.V.. 1960. On flagellar structure in certain flagellates. J. Biophys. Biochem. Cytol. 7:697–716. 10.1083/jcb.7.4.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman-Haham, I. 2023. Towards an atomic model of a beating ciliary axoneme. Curr. Opin. Struct. Biol. 78:102516. 10.1016/j.sbi.2022.102516 [DOI] [PubMed] [Google Scholar]
- Gui, M., Croft J.T., Zabeo D., Acharya V., Kollman J.M., Burgoyne T., Höög J.L., and Brown A.. 2022a. SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules. Proc. Natl. Acad. Sci. USA. 119:e2207605119. 10.1073/pnas.2207605119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, M., Wang X., Dutcher S.K., Brown A., and Zhang R.. 2022b. Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Nat. Struct. Mol. Biol. 29:483–492. 10.1038/s41594-022-00770-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L., Rao Q., Yang R., Wang Y., Chai P., Xiong Y., and Zhang K.. 2022. Cryo-EM structure of an active central apparatus. Nat. Struct. Mol. Biol. 29:472–482. 10.1038/s41594-022-00769-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M., Subramanian R., Bangs F., Omelchenko T., Liem K.F. Jr, Kapoor T.M., and Anderson K.V.. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16:663–672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höög, J.L., Lacomble S., O’Toole E.T., Hoenger A., McIntosh J.R., and Gull K.. 2014. Modes of flagellar assembly in Chlamydomonas reinhardtii and Trypanosoma brucei. ELife. 3:e01479. 10.7554/eLife.01479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, J.M.. 1970. Subsidiary components of the flagella of Chlamydomonas reinhardii. J. Cell Sci. 7:823–839. 10.1242/jcs.7.3.823 [DOI] [PubMed] [Google Scholar]
- Ichikawa, M., Liu D., Kastritis P.L., Basu K., Hsu T.C., Yang S., and Bui K.H.. 2017. Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat. Commun. 8:15035. 10.1038/ncomms15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y.Y., Lechtreck K., and Gaertig J.. 2015. Total internal reflection fluorescence microscopy of intraflagellar transport in Tetrahymena thermophila. Methods Cell Biol. 127:445–456. 10.1016/bs.mcb.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.A., Diener D.R., Stepanek L., and Pigino G.. 2018. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20:1250–1255. 10.1038/s41556-018-0213-1 [DOI] [PubMed] [Google Scholar]
- Khalifa, A.A.Z., Ichikawa M., Dai D., Kubo S., Black C.S., Peri K., McAlear T.S., Veyron S., Yang S.K., Vargas J., et al. 2020. The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications. Elife. 9:e52760. 10.7554/eLife.52760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesel, P., Alvarez Viar G., Tsoy N., Maraspini R., Gorilak P., Varga V., Honigmann A., and Pigino G.. 2020. The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat. Struct. Mol. Biol. 27:1115–1124. 10.1038/s41594-020-0507-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius, D., Dong L., Sharov G., Nakane T., and Scheres S.H.W.. 2021. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478:4169–4185. 10.1042/BCJ20210708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring, D.A., Vladar E.K., Werner M.E., Mitchell J.W., Hwang P., and Mitchell B.J.. 2013. Deuterosome-mediated centriole biogenesis. Dev. Cell. 27:103–112. 10.1016/j.devcel.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning, R.I., Zovko S., Bárcena M., Oostergetel G.T., Koerten H.K., Galjart N., Koster A.J., and Mieke Mommaas A.. 2008. Cryo electron tomography of vitrified fibroblasts: Microtubule plus ends in situ. J. Struct. Biol. 161:459–468. 10.1016/j.jsb.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kubo, S., Black C.S., Joachimiak E., Yang S.K., Legal T., Peri K., Khalifa A.A.Z., Ghanaeian A., McCafferty C.L., Valente-Paterno M., et al. 2023. Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins. Nat. Commun. 14:2168. 10.1038/s41467-023-37868-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, S., Yang S.K., Black C.S., Dai D., Valente-Paterno M., Gaertig J., Ichikawa M., and Bui K.H.. 2021. Remodeling and activation mechanisms of outer arm dyneins revealed by cryo-EM. EMBO Rep. 22:e52911. 10.15252/embr.202152911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir, A., Sigworth F.J., and Tagare H.D.. 2014. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 11:63–65. 10.1038/nmeth.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.F., and Geimer S.. 2000. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil. Cytoskeleton. 47:219–235. [DOI] [PubMed] [Google Scholar]
- Lechtreck, K.F., Gould T.J., and Witman G.B.. 2013. Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia. 2:15. 10.1186/2046-2530-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, M.R., Roelofs M.C., Ravi R.T., Maitan P., Henning H., Zhang M., Bromfield E.G., Howes S.C., Gadella B.M., Bloomfield-Gadêlha H., and Zeev-Ben-Mordehai T.. 2021. The multi-scale architecture of mammalian sperm flagella and implications for ciliary motility. EMBO J. 40:e107410. 10.15252/embj.2020107410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, M.R., Zeng J., Wang X., Roelofs M.C., Huang W., Zenezini Chiozzi R., Hevler J.F., Heck A.J.R., Dutcher S.K., Brown A., et al. 2023. Structural specializations of the sperm tail. Cell. 186:2880–2896.e17. 10.1016/j.cell.2023.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., and Nicastro D.. 2018. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science. 360:eaar1968. 10.1126/science.aar1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.T., Zhang H., Wang H., Tao C.L., Bi G.Q., and Zhou Z.H.. 2022. Isotropic reconstruction for electron tomography with deep learning. Nat. Commun. 13:6482. 10.1038/s41467-022-33957-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka, P., Vasudevan K.K., Guha M., Joachimiak E., Wloga D., Tomasi R.F., Baroud C.N., Dupuis-Williams P., Galati D.F., Pearson C.G., et al. 2018. Proteins that control the geometry of microtubules at the ends of cilia. J. Cell Biol. 217:4298–4313. 10.1083/jcb.201804141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira, F., Pearce M., Tivey A.R.N., Basutkar P., Lee J., Edbali O., Madhusoodanan N., Kolesnikov A., and Lopez R.. 2022. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50:W276–W279. 10.1093/nar/gkac240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez, A., Garcia I., Asano S., Lucic V., and Fernandez J.J.. 2014. Robust membrane detection based on tensor voting for electron tomography. J. Struct. Biol. 186:49–61. 10.1016/j.jsb.2014.02.015 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., and Held S.R.. 2017. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197:102–113. 10.1016/j.jsb.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S., Nakajima K., Miki H., Minato Y., Wang D., and Hirokawa N.. 2012. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev. Cell. 23:1167–1175. 10.1016/j.devcel.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Oda, T., Yanagisawa H., Kamiya R., and Kikkawa M.. 2014a. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science. 346:857–860. 10.1126/science.1260214 [DOI] [PubMed] [Google Scholar]
- Oda, T., Yanagisawa H., Yagi T., and Kikkawa M.. 2014b. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J. Cell Biol. 204:807–819. 10.1083/jcb.201312014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunmolu, F.E., Moradi F., Volkov V.A., van Hoorn C., Wu J., Andrea N., Hua S., Jiang K., Vakonakis I., Potočnjak M., et al. 2022. Microtubule plus-end regulation by centriolar cap proteins. bioRxiv. 10.1101/2021.1112.1129.474442 (Preprint posted January 20, 2022) [DOI]
- Omoto, C.K., and Kung C.. 1979. The pair of central tubules rotates during ciliary beat in Paramecium. Nature. 279:532–534. 10.1038/279532a0 [DOI] [PubMed] [Google Scholar]
- Pedersen, L.B., Geimer S., Sloboda R.D., and Rosenbaum J.L.. 2003. The Microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 13:1969–1974. 10.1016/j.cub.2003.10.058 [DOI] [PubMed] [Google Scholar]
- Perez-Riverol, Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M., et al. 2022. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50:D543–D552. 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., and Ferrin T.E.. 2021. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30:70–82. 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao, T., Luo M., Wang L., Guo Y., Li D., Li P., Snell W.J., and Pan J.. 2009. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 106:4713–4718. 10.1073/pnas.0808671106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli, A., Corbo D., Lupetti P., and Mencarelli C.. 2022. The distal central pair segment is structurally specialised and contributes to IFT turnaround and assembly of the tip capping structures in Chlamydomonas flagella. Biol. Cell. 114:349–364. 10.1111/boc.202200038 [DOI] [PubMed] [Google Scholar]
- Qin, H., Diener D.R., Geimer S., Cole D.G., and Rosenbaum J.L.. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, M.J., Phetruen T., Fisher R.L., Chen K., Pentecost B.T., Gomez G., Ounjai P., and Sui H.. 2018. The Developmental process of the growing motile ciliary tip region. Sci. Rep. 8:7977. 10.1038/s41598-018-26111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, X., and Gouet P.. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J.L., Moulder J.E., and Ringo D.L.. 1969. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41:600–619. 10.1083/jcb.41.2.600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale, W.S., and Satir P.. 1977. The termination of the central microtubules from the cilia of Tetrahymena pyriformis. Cell Biol. Int. Rep. 1:45–49. 10.1016/0309-1651(77)90008-X [DOI] [PubMed] [Google Scholar]
- Satir, P., and Christensen S.T.. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400. 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- Satish Tammana, T.V., Tammana D., Diener D.R., and Rosenbaum J.. 2013. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J. Cell Sci. 126:5018–5029. 10.1242/jcs.133439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, N., Lane A., Jovanovic K., Parfitt D.A., Aguila M., Thompson C.L., da Cruz L., Coffey P.J., Chapple J.P., Hardcastle A.J., and Cheetham M.E.. 2017. Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum. Mol. Genet. 26:3451. 10.1093/hmg/ddx245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E.F., and Sale W.S.. 1992. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 257:1557–1559. 10.1126/science.1387971 [DOI] [PubMed] [Google Scholar]
- Soares, H., Carmona B., Nolasco S., Viseu Melo L., and Gonçalves J.. 2019. Cilia distal domain: Diversity in evolutionarily conserved structures. Cells. 8:160. 10.3390/cells8020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srour, M., Hamdan F.F., McKnight D., Davis E., Mandel H., Schwartzentruber J., Martin B., Patry L., Nassif C., Dionne-Laporte A., et al. 2015. Joubert syndrome in French Canadians and identification of mutations in CEP104. Am. J. Hum. Genet. 97:744–753. 10.1016/j.ajhg.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek, L., and Pigino G.. 2016. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 352:721–724. 10.1126/science.aaf4594 [DOI] [PubMed] [Google Scholar]
- Summers, K.E., and Gibbons I.R.. 1971. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 68:3092–3096. 10.1073/pnas.68.12.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suprenant, K.A., and Dentler W.L.. 1988. Release of intact microtubule-capping structures from Tetrahymena cilia. J. Cell Biol. 107:2259–2269. 10.1083/jcb.107.6.2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta-Shma, A., Perles Z., Yaacov B., Werner M., Frumkin A., Rein A.J., and Elpeleg O.. 2015. A human laterality disorder associated with a homozygous WDR16 deletion. Eur. J. Hum. Genet. 23:1262–1265. 10.1038/ejhg.2014.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov, D., and Cramer P.. 2019. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 16:1146–1152. 10.1038/s41592-019-0580-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren, V., Odde D.J., and Cassimeris L.. 2002. Estimates of lateral and longitudinal bond energies within the microtubule lattice. Proc. Natl. Acad. Sci. USA. 99:6035–6040. 10.1073/pnas.092504999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, K.K., Jiang Y.Y., Lechtreck K.F., Kushida Y., Alford L.M., Sale W.S., Hennessey T., and Gaertig J.. 2015. Kinesin-13 regulates the quantity and quality of tubulin inside cilia. Mol. Biol. Cell. 26:478–494. 10.1091/mbc.E14-09-1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Piao T., Cao M., Qin T., Huang L., Deng H., Mao T., and Pan J.. 2013. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J. Cell Sci. 126:1531–1540. 10.1242/jcs.124255 [DOI] [PubMed] [Google Scholar]
- Williams, N.E., Wolfe J., and Bleyman L.K.. 1980. Long-term maintenance of Tetrahymena spp. J. Protozool. 27:327. 10.1111/j.1550-7408.1980.tb04270.x [DOI] [PubMed] [Google Scholar]
- Yamazoe, T., Nagai T., Umeda S., Sugaya Y., and Mizuno K.. 2020. Roles of TOG and jelly-roll domains of centrosomal protein CEP104 in its functions in cilium elongation and Hedgehog signaling. J. Biol. Chem. 295:14723–14736. 10.1074/jbc.RA120.013334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeo, D., Croft J.T., and Höög J.L.. 2019. Axonemal doublet microtubules can split into two complete singlets in human sperm flagellum tips. FEBS Lett. 593:892–902. 10.1002/1873-3468.13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeo, D., Heumann J.M., Schwartz C.L., Suzuki-Shinjo A., Morgan G., Widlund P.O., and Höög J.L.. 2018. A lumenal interrupted helix in human sperm tail microtubules. Sci. Rep. 8:2727. 10.1038/s41598-018-21165-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows data collection parameters for the various datasets used in this study.
shows characteristics of the subtomogram averages shown in this study.
shows peptide count, percentage coverage, and emPAI score of all the proteins identified in CU428 and FAP256A/B-KO cells.
Data Availability Statement
The tomography data produced in this study have been deposited at the Electron Microscopy Data Bank. The cryo-EM maps of CU428 main CP, singlet A-tubules, cap complex have accession codes EMD-40891, EMD-40894, and EMD-40892, respectively. The CU428 tip CP maps have accession codes EMD-40896, 40897, 40898, 40899, and 40900. The FAP256A/B-KO mutant tip CP maps have accession codes: EMD-40909, 40910, 40911, and 40912. The reconstructed tomogram used in Fig. 1 A (bin 4, before and after IsoNet denoising) has also been deposited and has accession code EMD-40913.
The mass spectrometry data of FAP256A/B-KO and CU428 (WT) have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD042582 and https://doi.org/10.6019/PXD042582.
The MATLAB code for subtomogram averaging in Dynamo and distance analysis is available at https://github.com/builab/dynamoDMT.