Abstract
Although systolic function characteristically shows gradual impairment in pressure overload (PO)-evoked left ventricular (LV) hypertrophy (LVH), rapid progression to congestive heart failure (HF) occurs in distinct cases. The molecular mechanisms for the differences in maladaptation are unknown. Here, we examined microRNA (miRNA) expression and miRNA-driven posttranscriptional gene regulation in the two forms of PO-induced LVH (with/without systolic HF). PO was induced by aortic banding (AB) in male Sprague–Dawley rats. Sham-operated animals were controls. The majority of AB animals demonstrated concentric LVH and slightly decreased systolic function (termed as ABLVH). In contrast, in some AB rats severely reduced ejection fraction, LV dilatation and increased lung weight-to-tibial length ratio was noted (referred to as ABHF). Global LV miRNA sequencing revealed fifty differentially regulated miRNAs in ABHF compared to ABLVH. Network theoretical miRNA-target analysis predicted more than three thousand genes with miRNA-driven dysregulation between the two groups. Seventeen genes with high node strength value were selected for target validation, of which five (Fmr1, Zfpm2, Wasl, Ets1, Atg16l1) showed decreased mRNA expression in ABHF by PCR. PO-evoked systolic HF is associated with unique miRNA alterations, which negatively regulate the mRNA expression of Fmr1, Zfmp2, Wasl, Ets1 and Atg16l1.
Subject terms: Molecular medicine, Molecular biology, Non-coding RNAs, Transcriptomics
Introduction
Sustained pressure overload (PO) of the left ventricle (LV) (e.g., aortic stenosis [AS], aortic coarctation or arterial hypertension) induces the development of pathological LV myocardial hypertrophy (LVH). PO-evoked LVH is typically characterized by LV wall thickening, impaired diastolic function and preserved, or mildly reduced, systolic function1. These characteristic features develop relatively soon after PO is established, and in some patients and experimental animals none or only a slight and slow further progression could be observed2. On the other hand, rapid transition from pathological LVH into congestive systolic heart failure (HF) might take place3.
Importantly, the severity of PO has been identified as a strong predictor for the development of systolic HF by prior experimental studies. Accordingly, in small animal models of aortic banding (AB), the more severe constriction of the aorta (leading to a greater degree of PO) was found to be associated with a higher risk for LV dilatation and contractile dysfunction4. Nevertheless, it is also worth mentioning that even under laboratory conditions (where the severity of PO is standardized and the potential confounding effects of different comorbidities do not prevail) only a subgroup of experimental animals manifest dilated cardiomyopathy with decompensated systolic function3,5.
Data are scarce regarding the molecular mechanisms that drive the differences in maladaptation of the myocardium in response to chronic PO. Over the last decades, the parallel improvement of omics technologies and bioinformatics has made it possible to approach this scientific question in an unbiased manner6. Among the different omics methods, myocardial microRNA (miRNA) transcriptomics in particular has gained lot of attention7. miRNA are short, non-coding RNA sequences that regulate gene expression on the posttranscriptional level. Of great interest, screening of myocardial miRNA expression followed by miRNA-based target gene prediction has been found to be a useful tool for the identification of novel molecular targets in different cardiovascular pathologies8–12.
It also worth mentioning that the dysregulation of numerous miRNAs has been already reported in PO-induced pathological LVH13,14. Nonetheless, whether the two forms of PO-evoked LVH (the more progressive form with LV dilatation and decompensated systolic function versus the classical form with concentric LVH and preserved systolic function, respectively) show characteristic differences on the miRNA level leading to distinct gene expressional changes has not yet been investigated. Importantly, this type of comparison might help to filter out those maladaptive alterations of PO-evoked molecular changes which might be responsible for the deterioration of systolic function and the dilation of the LV.
Upon this, in the current study we aimed to investigate LV miRNA expressional changes and miRNA-driven posttranscriptional gene regulations in AB rats with and without systolic HF in order to identify novel molecular targets of systolic HF.
Results
Model characterization
Invasive hemodynamic measurement indicated markedly increased arterial blood pressure in the ABLVH (systolic blood pressure [SBP]: 218 ± 8 vs. 146 ± 6 mmHg, diastolic blood pressure [DBP]: 153 ± 6 vs. 116 ± 4 mmHg and mean arterial pressure [MAP]: 174 ± 6 vs. 126 ± 5 mmHg; P < 0.001 for each comparisons) and ABHF (SBP: 214 ± 6 vs. 146 ± 6 mmHg, DBP: 155 ± 3 vs. 116 ± 4 mmHg and MAP: 174 ± 4 vs. 126 ± 5 mmHg; P < 0.001 for each comparison) groups compared to the sham group. Nevertheless, SBP, DBP and MAP did not differ between the ABLVH and ABHF groups (P = 0.713 for SBP, P = 0.791 for DBP and P = 0.988 for MAP).
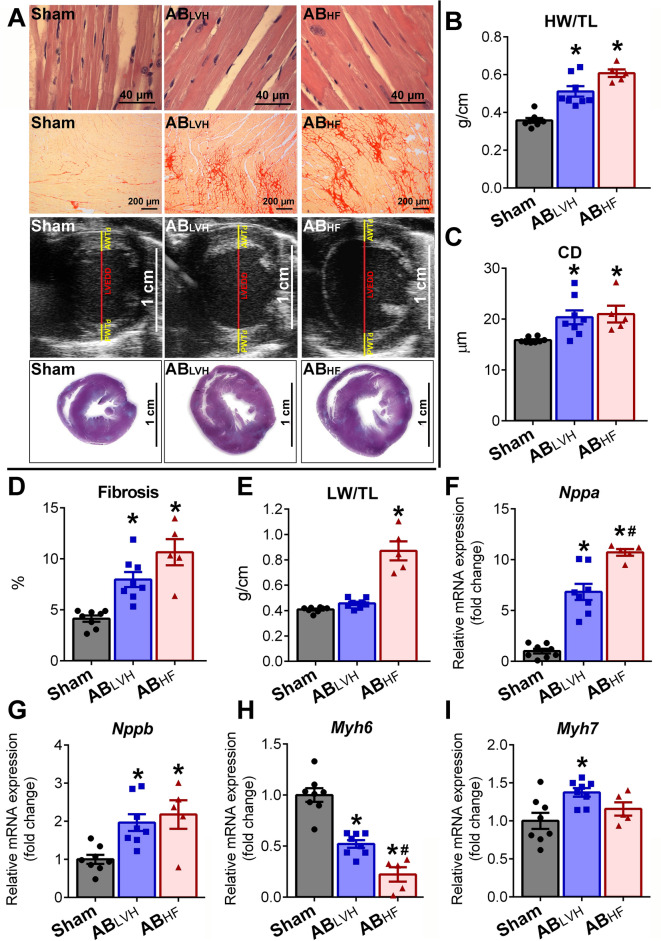
Furthermore, echocardiography revealed increased anterior wall thickness in diastole (AWTd), posterior wall thickness in diastole (PWTd) and LV mass in the ABLVH and the ABHF groups at week 6 and 12, when compared to sham group (Table 1). Heart weight-to-tibial length (HW/TL) (Fig. 1A), cardiomyocyte diameter (CD) (Fig. 1B), interstitial myocardial fibrosis (Fig. 1C) as well as the mRNA levels of natriuretic peptides (natriuretic peptide type A [Nppa] and natriuretic peptide type B [Nppb]) (Fig. 1E,F) also showed significantly increased values in ABLVH and ABHF, confirming the development of pathological LVH in both groups. In addition, the robust decrease in α-myosin heavy chain (Myh6) mRNA expression (Fig. 1G) coupled with slight changes in β-myosin heavy chain (Myh7) transcription (Fig. 1H) led to an increased Myh7/Myh6 ratio on the mRNA level in both the ABLVH (Sham vs. ABLVH, P = 0.027) and in the ABHF (Sham vs. ABHF P = 0.002) groups.
Table 1.
Echocardiographic follow-up.
| Week 6 | Week 12 | |||||
|---|---|---|---|---|---|---|
| Sham (n = 8) | ABLVH (n = 8) | ABHF (n = 5) | Sham (n = 8) | ABLVH (n = 8) | ABHF (n = 5) | |
| Body weight, g | 460 ± 13 | 480 ± 9 | 450 ± 15 | 549 ± 14 | 576 ± 12 | 543 ± 23 |
| Heart rate, beats/min | 360 ± 9 | 349 ± 19 | 365 ± 16 | 356 ± 14 | 353 ± 8 | 340 ± 13 |
| LV AWTd, mm | 1.92 ± 0.07 | 2.55 ± 0.10* | 2.69 ± 0.11* | 1.99 ± 0.06 | 2.65 ± 0.05* | 2.78 ± 0.06* |
| LV AWTs, mm | 3.44 ± 0.08 | 4.50 ± 0.21* | 3.84 ± 0.12# | 3.57 ± 0.11 | 4.25 ± 0.12* | 3.92 ± 0.09 |
| LVEDD, mm | 8.05 ± 0.16 | 8.56 ± 0.14 | 8.71 ± 0.27 | 8.32 ± 0.13 | 8.87 ± 0.15 | 9.93 ± 0.46*,# |
| LVESD, mm | 4.68 ± 0.19 | 4.97 ± 0.20 | 6.14 ± 0.45*,# | 4.83 ± 0.12 | 5.53 ± 0.26 | 7.59 ± 0.60*,# |
| LV PWTd, mm | 2.00 ± 0.08 | 2.73 ± 0.09* | 2.60 ± 0.12* | 2.00 ± 0.07 | 2.50 ± 0.05* | 2.46 ± 0.11* |
| LV PWTs, mm | 3.24 ± 0.08 | 4.24 ± 0.10* | 3.53 ± 0.21# | 3.19 ± 0.10 | 4.13 ± 0.06* | 3.26 ± 0.17# |
| LV mass, mg | 1.25 ± 0.03 | 2.11 ± 0.07* | 2.15 ± 0.10* | 1.34 ± 0.04 | 2.15 ± 0.07* | 2.65 ± 0.15*,# |
| LVEDV, µl | 460 ± 14 | 518 ± 27 | 551 ± 48 | 511 ± 25 | 572 ± 25 | 734 ± 80*,# |
| LVESV, µl | 202 ± 14 | 243 ± 21 | 369 ± 52*,# | 211 ± 10 | 307 ± 21 | 605 ± 69*,# |
| SV, µl | 258 ± 12 | 275 ± 26 | 182 ± 16# | 300 ± 19 | 265 ± 15 | 130 ± 20*,# |
| CO, ml/min | 97.9 ± 13.0 | 93.2 ± 5.7 | 65.9 ± 4.9 | 106.8 ± 7.9 | 93.8 ± 6.0 | 43.1 ± 5.8*,# |
| Ejection fraction, % | 56 ± 2 | 53 ± 3 | 34 ± 5*,# | 59 ± 2 | 47 ± 2* | 18 ± 3*,# |
LV left ventricular, AWTd anterior wall thickness in diastole, AWTs anterior wall thickness in systole, LVEDD LV end-diastolic diameter, LVESD LV end-systolic diameter, PWTd LV posterior wall thickness in diastole, PWTs LV posterior wall thickness in systole, LVEDV LV end-diastolic volume, LVESV LV end-systolic volume, SV stroke volume, CO cardiac output. The sample numbers are indicated in the table. Depending on the distribution of the datasets, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test or Kruskal–Wallis test followed by Dunn’s post hoc test was carried out. *P < 0.05 vs. Sham. #P < 0.05 vs. ABLVH.
Figure 1.
Model characterization. (A) Representative photomicrographs of hematoxylin and eosin (magnification 200 ×, scale bar: 40 µm) and picrosirius red staining (magnification 50 ×, scale bar: 200 µm), parasternal short axis echocardiographic recordings at the midpapillary muscle level and cross-sectional whole heart images are shown demonstrating myocardial hypertrophy and intensified fibrosis in the aortic-banded (AB) groups. (B) Heart weight-to-tibial length (HW/TL), (C) cardiomyocyte diameter (CD), (D) interstitial fibrosis, (F) atrial (Nppa) and (G) B-type natriuretic peptide (Nppb) mRNA expression were increased, while (H) myosin heavy chain alpha (Myh6) mRNA expression was decreased in the aortic-banded (AB) groups with systolic heart failure (ABHF) and also without systolic heart failure (ABLVH) compared to the sham group. The decrease in Myh6 and the increase in Nppa was more severe in the ABHF compared to the ABLVH group. In contrast, (I) Myh7 showed elevated levels only in the ABLVH group when compared to its control group. Lung weight-to-tibial length (LW/TL) (E) increased only in the ABHF group, indicating severe pulmonary congestion. Sample numbers were the following: Sham: n = 8, ABLVH: n = 8, ABHF: n = 5. Depending on the distribution of the datasets, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test or Kruskal–Wallis test followed by Dunn’s post hoc test was carried out. *P < 0.05 vs. Sham. #P < 0.05 vs. ABLVH.
Despite the fact that pathological LVH developed in both the ABLVH and ABHF groups, chamber dilatation along with progressive decompensation of LV systolic function only occurred in the ABHF and not in the ABLVH group. Accordingly, while in the ABLVH group systolic function was found to be only slightly deteriorated at week 12, ejection fraction (EF), stroke volume (SV) and cardiac output (CO) were massively reduced in the ABHF group (Table 1). Similarly, LV end-diastolic volume (LVEDV) and LV end-diastolic diameter (LVEDD) were elevated in the ABHF, but not in the ABLVH group (Table 1). The manifestation of congestive HF in the ABHF group was also reinforced by an elevated lung weight-to-tibial length (LW/TL) ratio (Fig. 1D).
miRNA screening
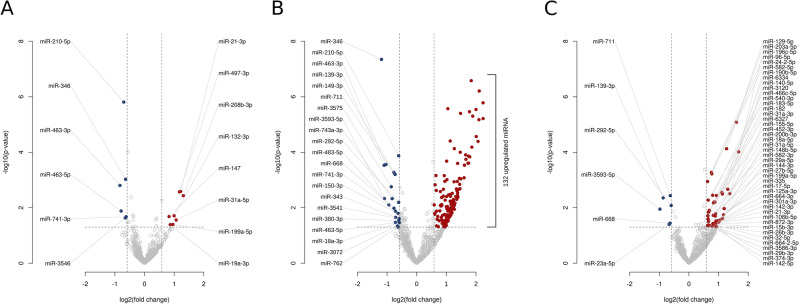
miRNA sequencing identified 14 differentially expressed miRNAs in the ABLVH group (Fig. 2A) and 153 in the ABHF group (Fig. 2B) when compared to the Sham group (for the detailed lists please see Supplementary Tables 1 and 2). Importantly, 50 miRNAs showed different expression (upregulated miRNAs: rno-miR-106b-5p, 125a-3p, 129-5p, 140-5p, 142-3p, 142-5p, 144-3p, 148b-5p, 155-5p, 15b-3p, 17-5p, 182, 183-5p, 18a-5p, 190b-5p, 196c-5p, 199a-5p, 200b-3p, 203a-5p, 24-2-5p, 26b-3p, 27b-5p, 29a-5p, 29b-3p, 301a-3p, 3120, 31a-3p, 31a-5p, 32-5p, 335, 3586-3p, 374-3p, 452-3p, 466c-5p, 540-3p, 582-3p, 582-5p, 6327, 6334, 664-2-5p, 664-3p, 872-3p, 96-5p and downregulated miRNAs: rno-miR-139-3p, 23a-5p, 292-5p, 3593-5p, 668, 711) in the ABHF compared to the ABLVH group (Supplementary Table 3 and Fig. 2C). Considering that in the present study our aim was to uncover novel molecular targets which might contribute to the development of systolic HF in case of PO-evoked LVH, we focused on the comparison of ABHF vs. ABLVH.
Figure 2.
miRNA screening. Volcano plots presenting altered miRNA expression profiles of the different conditions. In the comparison of (A) aortic-banded (AB) groups without systolic heart failure (ABLVH) versus sham group, (B) AB groups with systolic heart failure (ABHF) versus sham group and (C) ABHF group versus ABLVH group 14, 153 and 50 miRNAs showed significant differential expression, respectively. Up- (red) and downregulated (blue) miRNAs that meet the specified significance threshold conditions (P-value < 0.05 and fold change > 1.5 or < 1/1.5) are listed (for the complete list of upregulated miRNAs in ABHF group versus sham group see the Supplementary files).
Target prediction
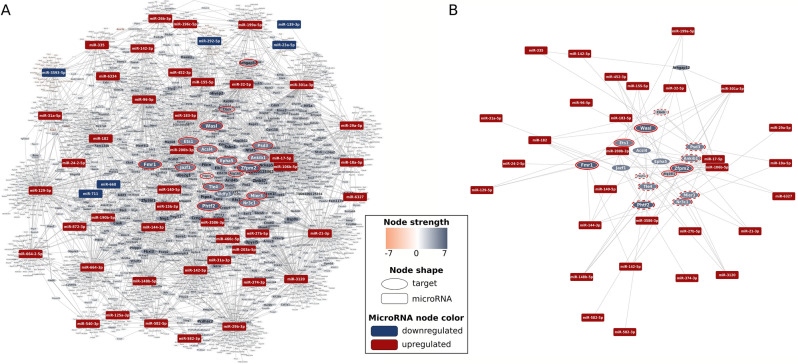
Based on the results of miRNA transcriptomics, network theoretical miRNA-target analysis predicted 3007 genes (Fig. 3), the expression of which might have been differentially regulated by miRNAs in the ABHF group compared to the ABLVH group (for the complete list of genes please see Supplementary Table 4).
Figure 3.
Target prediction. MiRNA-target interaction network presenting genes predicted to be involved in the maladaptive transition from left ventricular (LV) myocardial hypertrophy (LVH) into heart failure (HF). Up- and downregulated miRNA nodes are indicated with uniformly sized red and blue rectangles, respectively. Orange and blue ovals represent up- and downregulated target gene nodes, respectively. Node size and color intensity of target gene nodes increases according to the absolute node strength values. (A) Whole interaction network with red circles indicating genes selected to be validated on mRNA level. (B) Regulatory subnetwork of targets selected for validation. Targets with significant and non-significant expression change in the predicted direction are highlighted by continuous and dotted red circles, respectively. Expression change of target gene nodes without marking could not be observed.
Target validation
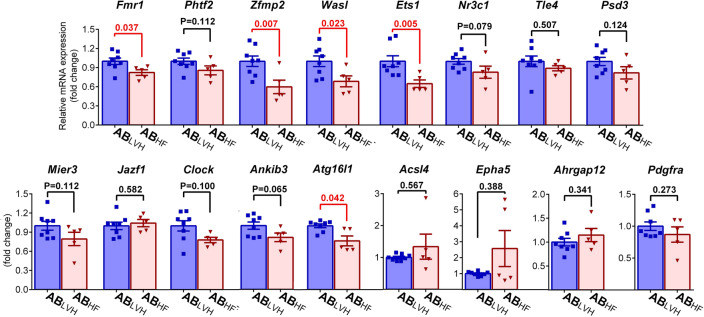
For validation purposes, target genes with the highest absolute node strength values were selected. 4 genes (FMRP translational regulator 1/fragile X messenger ribonucleoprotein 1 [Fmr1]; putative homeodomain transcription factor 2 [Phtf2]; zinc finger protein, multitype 2 [fZfpm2] and WASP like actin nucleation promoting factor [Wasl]) showed a node strength value of 7. Furthermore, expression of another ten genes (acyl-CoA synthetase long-chain family member 4 [Acsl4], ETS proto-oncogene 1[Ets1], nuclear receptor subfamily 3, group C, member 1 [Nr3c1], TLE family member 4, transcriptional corepressor [Tle4], WD repeat and FYVE domain containing 3 [Wdfy3], pleckstrin And Sec7 Domain Containing 3 [Psd3], MIER family member 3 [Mier3], ankyrin repeat and IBR domain-containing protein 1 [Ankib1], JAZF zinc finger 1 [Jazf1], EPH receptor A5 [Epha5]) were found to be unidirectionally modulated by 6 miRNAs. The mRNA expression level of all these genes (except for Wdfy3, the expression of which could not be measured due to methodological reasons) coupled with four randomly selected genes, with predicted strong regulation (autophagy related 16-like 1 [Atg16l1], Rho GTPase Activating Protein 12 [Arhgrap12], clock circadian regulator [Clock], Platelet Derived Growth Factor Receptor Alpha [Pdgfra]) were measured. Out of the tested 17 genes, Fmr1, Zfmp2, Wasl, Ets1 and Atg16l1 showed reduced mRNA expression (Fig. 4).
Figure 4.
Target validation. Out of the tested 17 targets, five genes (Fmr1: FMRP translational regulator 1/fragile X messenger ribonucleoprotein 1, Zfmp2: zinc finger protein, multitype 2, Wasl: WASP like actin nucleation promoting factor, Ets1: ETS proto-oncogene 1 and Atg16l1: autophagy related 16-like 1) showed reduced mRNA expression in the ABHF (aortic banded group with systolic heart failure) compared to the ABLVH (aortic banded group without systolic heart failure) group. Furthermore, a tendency towards decreased values without reaching the level of significance was observed in case of Phtf2 (putative homeodomain transcription factor 2), Nr3c1 (nuclear receptor subfamily 3, group C, member 1). Mier3 (MIER family member 3), Clock (clock circadian regulator) and Ankib3 (ankyrin repeat and IBR domain-containing protein 1). No changes could be detected in mRNA expression between the two study groups in case of Acsl4 (acyl-CoA synthetase long-chain family member 4), Tle4 (TLE family member 4, transcriptional corepressor), Wdfy3 (WD repeat and FYVE domain containing 3), Psd3 (pleckstrin And Sec7 Domain Containing 3), Jazf1 (JAZF zinc finger 1) and Epha5 (EPH receptor A5). Sample numbers were n = 8 in the ABLVH and n = 5 in the ABHF groups. Depending on the distribution of the datasets, two sample Student’s t test or Mann–Whitney tests were carried out. Data normalized to the ABLVH are depicted. *P < 0.05.
Discussion
The present paper confirmed that LV miRNA expression is characteristically different in PO-induced LVH with systolic HF compared to PO-evoked LVH without systolic HF. Furthermore, bioinformatic analysis revealed that the altered miRNA profile in PO-evoked systolic HF might have led to the dysregulation of several genes. Importantly, out of the most promising predicted targets, Fmr1, Zfpm2, Wasl, Ets1 and Atg16l1 showed reduced expression on the mRNA level in PO-induced LVH with systolic HF.
The maladaptation of the LV in response to pure chronic PO (without an ischemic insult or severe valvular regurgitation leading to concomitant LV volume overload) characteristically involves the development of pathological LVH along with cardiomyocyte hypertrophy, interstitial fibrosis, impaired diastolic function but preserved or only mildly reduced systolic function2,15,16. On the other hand, advanced systolic HF with marked chamber dilatation manifests in a smaller portion of patients/experimental animals3,17. Data is scarce regarding the reasons and pathological mechanism for the different remodeling processes in case of sustained LV PO. Experimental studies indicate that the severity of PO is a major determinant of systolic functional impairment. Indeed, a higher degree of PO was found to predispose for contractility deterioration and LV dilatation in small animal models18. Furthermore, Richards et al. demonstrated that systolic dysfunction and elevated HF markers (plasma BNP level as well as LV ANP and BNP mRNA expression) were only present in mice with transverse aortic constriction (TAC) when the severity of PO exceeded a distinct level4. Based on these results, it could be hypothesized that if the extent of the pathological stimulus of PO reaches a critical “threshold” level, a more fulminant disease progression takes place. However, this notion is further complicated by the fact that only a portion of the animals developed decompensated systolic HF in well-controlled preclinical studies (where the degree of PO was highly standardized)3,5. In addition, in patients with AS, the degree of pathological LVH was only weakly related to the severity of valve obstruction16, but its extent was strongly associated with systolic dysfunction and HF development19. Therefore, it could be more precisely stated that a more severe PO increases the risk for systolic HF on the basis of individual susceptibility.
In our study the abdominal AB rat model was utilized, in which the LV was subjected to a moderately severe PO. In recent years, our research group has provided a detailed characterization of the natural progression of PO-evoked LVH in this particular model20,21. During these consecutive studies we have observed that myocardial hypertrophy along with typical pathological gene expressional changes (reactivation of the fetal gene program), interstitial fibrosis and impaired diastolic function (predominantly with prolonged active relaxation) develops relatively early after AB induction (at week 6)21. At this early stage, global systolic function is characteristically preserved. However, a moderate but progressive decline in LV contractility could be noted at later stages (after week 12)21. In parallel to our prior findings, the majority of the AB animals (43/49, 87.8%) demonstrated preserved systolic LV function at week 6 and slightly decreased EF at week 12 (Table 1). Furthermore, as also found earlier, development of LVH (as indicated by increased HW/TL, LV mass, AWTd, PWTd and CD) in the ABLVH was associated with reactivated fetal genes (elevated nppa, nppb and myh7 expression and decreased myh6 expression) and interstitial collagen accumulation (Fig. 1). In contrast to this classical course of PO-induced LVH, in a small subpopulation of the AB animals (6/49, 12.2%), rapid progression into decompensated systolic HF with massive pulmonary congestion (as reflected by the increased wet LW/TL ratio [Fig. 1D]) was noted. Analysis of the echocardiographic data revealed that systolic function was severely impaired already at week 6 in these rats, indicating that the remodeling processes were fundamentally separated from each other at a relatively early time point. Importantly, wall thickening did not differ between the ABHF and ABLVH groups. Similarly, no differences in pathological mechanism could be observed either in interstitial collagen accumulation, natriuretic peptide expression (indicating a comparable level of ventricular wall stress) or in MHC isoform switch (although myh6 showed more reduction in ABHF and myh7 demonstrated more increase in ABLVH, the overall shift in the ratio did not differ [P = 0.242] between the groups [not depicted]). It is also worth highlighting that the extent of afterload increment (as reflected by SBP, DBP and MAP) was similar in the AB groups, indicating that the observed phenotypic differences could not simply originate from differences in hemodynamic loading conditions.
To better understand the underlying molecular mechanisms for the observed phenotypic differences in the ABLVH and ABHF groups, LV miRNA transcriptomic profiling followed by network theoretical miRNA-target analysis was carried out. Our decision to use a miRNA-based, unbiased target prediction approach to identify genes, the altered expression of which could have contributed to the decompensation of systolic function in the ABHF group was rationalized by two reasons. First, a great body of scientific evidence supports the fact that LV miRNA expression is substantially dysregulated in PO-evoked pathological LVH13,14. Hence, marked changes in the LV miRNAome could have been anticipated to occur in our model as well. Secondly, the effectiveness of miRNA interactome analysis in the identification of important, post-transcriptionally regulated genes in different cardiovascular pathologies have been well established by earlier studies8–12.
The NGS-derived miRNA expressional data was compared between the study groups in all the possible scenarios (ABHF vs. Sham, ABLVH vs. Sham and ABHF vs. ABLVH; see also Fig. 2). Considering the fact that the direct comparison of PO-induced LVH with and without systolic HF carries the potential to filter out those pathological molecular alterations that might have specifically contributed to the deterioration of systolic function and chamber dilatation (and not to the development of LVH), the comparison of ABHF to ABLVH was chosen for further evaluation. This comparison revealed 44 upregulated and 6 downregulated miRNAs in the ABHF compared to the ABLVH group (Fig. 2C). Based on these results, our network analysis method predicted more than 3000 differentially regulated genes (Fig. 3). Given the big number of targets, only 17 genes with high node strength values were analyzed on the mRNA level. This validation process confirmed reduced expression of the following five genes: Fmr1, Zfpm2, Wasl, Ets1 and Atg16l1 in the ABHF group compared to the ABLVH group (Fig. 4), suggesting a potential role of these genes in the development of systolic HF.
Among these target genes, Fmr1 encodes a protein called fragile X messenger ribonucleoprotein 122, which is mainly required for physiological cognitive development. Mutation of the Fmr1 gene (classically due to trinucleotide repeat expansion) leads to fragile X syndrome (FXS)23. Although cardiovascular abnormalities are not uncommon in FXS24, the exact role of Fmr1 in the heart is not completely understood. A recent study suggested that Fmr1 might have some cardioprotective properties, since its overexpression alleviated oxidative stress and apoptosis in an in vitro model of lipopolysaccharide-induced myocardial injury25. Furthermore, previous experiments confirmed that genetic silencing of Fmr1 (by knocking the gene out [KO]) was associated with increased iron content in the adult myocardium26, and premature closure of the mitochondrial permeability transition pore in the developing heart27. Nevertheless, the significance of these observations on the cardiovascular system warrants further investigations, as newborn Fmr1 KO mice do not exhibit LV dysfunction27.
Another interesting target, the reduced expression of which was successfully validated in our study, was the multizinc finger protein Zfpm2 gene (also known as the friend of GATA [FOG]-2). Importantly, the essential role of ZFPM2/FOG-2 in the normal morphogenesis of the heart and in the effective vascularization of the myocardium has been well documented28. Correspondingly, genetic deletion of Zfpm2 during embryonic development results in severe defects in cardiac structure and coronary artery formation29. Of particular significance, cardiomyocyte-specific loss of the Zfpm2 gene product in adult mice did not lead to anatomical disturbances, but resulted in severely depressed ventricular function and premature mortality29. Taking these data together, it is tempting to hypothesize that reduction in Zfpm2 expression might have inhibited the normal vascularization of the hypertrophied myocardium, leading to tissue hypoxia and consequently to reduced LV contractility in the ABHF group.
The protein coded by Wasl associates with a variety of signaling molecules that regulate the actin cytoskeleton system. Genetic mutation of Wasl results in Wiskott-Aldrich syndrome (WAS), which is characterized by immunodeficiency, thrombocytopenia and atopic dermatitis30. The cardiovascular system is characteristically not affected in WAS. Unfortunately, previous publications did not provide a detailed characterization of cardiac function and structure in Wasl KO mice models. Hence, further experimentation is required to uncover the potential role of Wasl in HF.
Ets1 encodes a transcriptional factor that plays a pivotal role in cardiac lineage commitment from a pluripotent stage during early embryonic development31. Its defect has been also associated with LV non-compaction32. Furthermore, ETS1 is required for the proper development of the coronary artery system by regulating epithelial-mesenchymal transformation33. This later finding suggests that (besides Zfpm2) the reduced expression of Ets1 could have also resulted in inappropriate vascularization of the pathologically remodeled LV in the ABHF group.
Finally, the protein product of Atg16l1 connects to the ATG5–ATG12 conjugate to form the ATG5–ATG12–ATG16L1 complex, which is essential for proper autophagosome formation34. Of great significance, the deleterious effect of autophagy inhibition in PO-evoked systolic HF was elegantly demonstrated in a prior publication, in which Atg5 deficient mice exhibited LV dilatation and severe systolic dysfunction early after PO induction35. Although, the same experiment was not carried out with Atg16l1 KO mice, one might speculate that the reduced expression of Atg16l1 would also result in impaired function of the ATG5–ATG12–ATG16L1 complex and hence it would also negatively affect the remodeling process in response to sustained PO.
Study limitations
Due to the low appearance rate of the ABHF phenotype (~ 10%), relatively small sample sizes could be utilized in the present study. To avoid imbalances in sample numbers among the study groups, 8 out of 43 AB animals demonstrating conventional disease progression were randomly selected to form the ABLVH. Therefore, molecular analysis was only performed in the minority of ABLVH rats. Furthermore, it is also worth mentioning that the presence of air-filled intestines placed around the operated abdominal aorta did not allow us to directly assess the severity of aortic constriction by non-invasive Doppler ultrasound. Hence, the extent of PO could be only determined via invasive hemodynamic measurement at the end of the study protocol. In addition, the interpretation of results is limited to young male rats. Therefore, further experimentation is required to investigate whether our observations could also be extended to female and older animals as well. Finally, it also has to be considered that the presence of numerous comorbidities (most importantly myocardial ischemia) and pharmacological HF therapies as well as the fundamental differences between the human pathophysiological states of sustained LV PO (long standing AS or arterial hypertension) and the AB rat model significantly limit the direct translation of our molecular findings into the clinical arena. Indeed, further validation of our results on human myocardial samples is necessary.
Besides the above-mentioned model-associated concerns, our molecular measurements also contain limitations. Of particular significance, the mRNA levels of the predicted target genes were measured in order to directly detect the miRNA-driven post-transcriptional regulations. Nevertheless, further investigation is needed to assess whether the observed gene expressional alterations indeed manifest in altered protein levels. Furthermore, since the global myocardial miRNA transcriptomic profiles were only assessed to provide input for the network theoretical prediction of the most relevant targets, our work did not focus either on individual miRNA level measurements by qPCR or on functional interaction analysis between distinct miRNAs and target genes.
Conclusion
LV miRNA expression is characteristically different in PO-evoked LVH with and without systolic HF in a rat model of abdominal AB. Furthermore, the changes of myocardial miRNA expression are associated with the dysregulation of a great number of genes in the ABHF compared to the ABLVH groups. Out of the most prominent targets, reduced expression was confirmed in the following 5 genes: Fmr1, Zfpm2, Wasl, Ets1 and Atg16l1.
Methods
Animals
The investigation conformed to the EU Directive 2010/63/EU and the Guide for the Care and Use of Laboratory Animals used by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and all of the methods were performed according to these proper guidelines and regulations. The experiments were approved by the ethics committee of the Regional Council of Karlsruhe for Animal Experimentation (G-94/15). Furthermore, the study is also interpreted in accordance with the ARRIVE (Animals in Research: Reporting in Vivo Experiments) guidelines36. Male Sprague–Dawley rats (n = 63; 5–6 weeks-old; 160–180 g; Janvier Labs, Saint Berthevin, France) were kept under standard conditions (22 ± 2 °C with 12 h light/dark cycles) and were allowed access to laboratory rat diet and water ad libitum.
Abdominal aortic banding
After a one-week-long acclimatization period, abdominal AB (n = 55) or sham operation (n = 8) was performed. In brief, under isoflurane anesthesia a midline laparotomy was carried out. Then, the intestinal tract was gently placed aside and the peritoneal layer was dissected in order to gain access to the retroperitoneal space. The abdominal aorta between the right renal artery and the superior mesenteric artery was carefully cleaned from the surrounding connective tissue. After isolation, a blunted 22-gauge needle (with the external diameter of 0.72 mm) was placed on the anterior surface of the abdominal aorta and a surgical thread was looped below the vessel and the needle. In rats undergoing AB surgery, the surgical thread was then subsequently tightened and the needle was removed, inducing a constriction of the size of the applied needle. After AB was completed, the intestines were placed back to the abdominal cavity and the abdominal muscle layer was sutured in single interrupted fashion. Finally, the skin wound was closed by applying surgical clips. Following surgery, analgesia was provided by subcutaneously administered buprenorphine in the dose of 0.05 mg/kg BW. Sham-operated animals were subjected to the same surgical procedure, except the aortic constriction.
Experimental groups
From the 55 rats undergoing AB, 5 died in the subacute phase (within the first 72 h) and one during the follow-up period. Hence, the final sample size in the AB group was 49 at the end of the experimental period. From these 49 AB rats, progressive transition from pathological LVH (ABLVH) into systolic HF occurred in 6 rats (12.2%) (termed as ABHF group) at week 12. An ejection fraction of less than 30% measured by echocardiography at week 12 was determined as an inclusion criterion for the ABHF (based on the current definition of advanced systolic HF)37. Accordingly, the experimental groups were the following:
Sham group (n = 8): these rats underwent sham operation and a 12-week long follow-up period.
ABLVH group (n = 8): these rats underwent AB operation and a 12-week long follow-up period and an EF of more than 30% was measured by echocardiography at the end of the experimental period. 43 rats met this criterion, however to match the sample size of this group to the other two experimental groups, only 8 rats were randomly selected for further analysis.
ABHF group (n = 5): these rats underwent AB operation and a 12-week long follow-up period and an EF of less than 30% was measured by echocardiography at the end of the experimental period. In one of the ABHF rats a sudden death occurred during the final echocardiographic measurement and therefore invasive blood pressure monitoring as well as tissue collection for histological and molecular analysis could not be carried out. Therefore, this animal was excluded from further analysis.
Echocardiography
Echocardiography was carried out using the Vevo® 2100 imaging system (FujiFilm VisualSonics Inc., Toronto, Ontario, Canada) equipped with a 21-MHz linear probe according the previously published protocol38. Repetitive measurements were performed at 6 and 12 weeks after AB/Sham operation. In brief, rats were anesthetized with 5% isoflurane in a chamber. After the induction of anesthesia, rats were placed on an automatic heating pad in a supine position with core temperature maintained at 37 °C. The thorax of the animals was shaved to obtain an optimal acoustic window. Anesthesia was maintained by inhalation of 1–1.5% isoflurane gas in 100% O2. Images in two-dimensional parasternal long-axis (PLAX) and short-axis views (PSAX) as well as M-mode recordings at the midpapillary level were taken. From PLAX acoustic window, LV longitudinal diameters (from LV apex to the aortic root) and transverse diameters (at the mid-papillary level) were measured at LVESD and LVEDD. On M-mode recordings (also at the mid-papillary level), AWT and PWT in diastole (d) and systole (s) were determined. LVESV and LVEDV were determined by the biplane ellipsoid model. SV and CO were calculated based on the following equations: SV = LVEDV-LVESV, CO = SV*heart rate (HR). LV mass was assessed according to the Deveroux formula.
Invasive hemodynamics
After completion of the echocardiographic protocol, in vivo hemodynamic measurements were performed. Briefly, rats were tracheotomized and intubated to facilitate breathing. Anesthesia was supported by mechanical ventilation with 1–1.5% isoflurane gas in 100% O2. A 2-Fr pressure-conductance microcatheter (SPR-838; Millar Instruments, Houston, TX) was then inserted into the right carotid artery and advanced into the ascending aorta. After stabilization, aortic pressure curves were recorded to measure and calculate SBP, DBP and MAP.
Post-mortem organ measurements
Following the invasive hemodynamic measurement, median laparotomy was performed and the abdominal aorta was isolated and cannulated. The blood of the animals was collected via the inserted cannula. To remove the residual blood cells from myocardial tissue, retrograde perfusion with cold physiological saline was applied. After perfusion, the hearts and the lungs of the animals were removed from the thorax and their weights were quickly measured on a scale. This was followed by conservation of LV myocardial tissue. Accordingly, transverse segments (the middle third part) of the right and the LV were fixed in buffered paraformaldehyde solution (4%) and embedded in paraffin for histological analysis. Furthermore, the apex of the LV was cut into small pieces (40–50 mg) and subsequently snap frozen in liquid nitrogen for molecular measurements. After tissue conservation was completed, tibial length (TL) was measured and the ratios of HW/TL and LW/TL were calculated.
Histology
Transverse, transmural, 5-µm thick slices of the ventricles were cut and placed on adhesive slides. These sections were stained with hematoxylin and eosin staining to determine CD as a cellular marker of myocardial hypertrophy38. In each sample, 100 longitudinally oriented cardiomyocytes from the LV were examined, and the diameters at transnuclear positions were defined with ImageJ software (National Institutes of Health, Bethesda, MD). The mean value of 100 measurements represented one sample.
The extent of interstitial myocardial fibrosis was assessed on picrosirius-stained sections, as also described earlier39. ImageJ software (National Institutes of Health, Bethesda, MD) was used to identify the picrosirius-red positive area. Three images (magnification 50 ×) were randomly taken from the free LV wall on each sections. After background subtraction, eye-controlled auto-threshold has been determined to detect positive areas. The collagen area (picrosirius red positive area-to-total area ratio) was determined on each image, and the mean value of three images represents each animal. The evaluation of the histological sections was performed by an independent observer who was blinded to the experimental design.
LV miRNA and mRNA expression analysis
Sample preparation
RNA isolation was performed from 50 mg of LV myocardial tissue using the RNeasy Plus Universal kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions paying special attention to appendix D in order to ensure purification of total RNA including small RNAs. Isolated RNA was subsequently eluted in 30 µl RNase-free water.
Quality control of RNA
RNA quality and concentration were determined and reviewed using a NanoDrop® spectrophotometer (Thermo Scientific™, Waltham, MA, USA) to measure the concentration and optical density (OD) 260/280 values of the samples and an Agilent RNA TapeStation® (Agilent Technologies, Santa Clara, USA) to check RNA quality via RINe number. RINe > 5 were accepted as input for library preparation. Quantification was performed via absorbance measurements using Qubit RNA broad range (BR) Assay Kit (Thermo Scientific™, Waltham, MA, USA).
Sequencing library
Library preparation was done using the QIAseq® miRNA Library Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. A total of 500 ng of RNA per sample was converted into miRNA next generation sequencing (NGS) libraries. Primers containing UMIs were used during reverse transcription. The resulting cDNA was amplified using PCR (13 cycles) with universal forward and sample index assigning reverse primers. The PCR product was then purified. Quality control of the library preparation was attained by capillary electrophoresis using the High Sensitivity Tape D1000 (Agilent Technologies, Santa Clara, USA). Based on the quality of the inserts and the concentration measurements, the libraries were pooled in equimolar ratios. The library pool(s) were quantified using qPCR and were then sequenced on a NextSeq® Sequencing System (Illumina, San Diego, USA) according to the manufacturer instructions with 75 bp read length for read 1 and 8 bp for the index read (1 × 75, 1 × 8). Raw data was demultiplexed and FASTQ files for each pool were generated using the bcl2fastq Conversion Software (Illumina, San Diego, USA).
cDNA synthesis and qPCR
For cDNA synthesis the Qiagen RT2 First Strand Kit (cat. no. 330401) was used. This kit provides a rapid and convenient procedure for efficient first strand cDNA synthesis. The cDNA was mixed with an appropriate RT2 SYBR® Green ROX Mastermix (cat. no. 330529). This mixture was aliquoted into the wells of the Custom RT2 Rat PCR Array (CLAR40805). PCR was performed on the ViiA 7 Real-Time PCR System (Thermo Scientific™, Waltham, MA, USA), and finally relative expression (for the following genes: Acsl4, Arhgap12, Ankib1, Atg16l1, Clock, Ets, Epha5, Fmr1, Jazf1, Mier3, Myh6, Myh7, Nppa, Nppb, Nr3c1, Pdgfra, Phtf2, Psd3, Tle4, Wasl, Zfpm2) normalized to housekeeping genes was determined using data from the real-time cycler and the 2−ΔCT method. Regarding the qPCR measurements, one sample from the ABHF group was excluded from data analysis due to technical issues.
Network theoretical miRNA-target prediction
After selecting significantly differentially expressed miRNAs identified in different comparisons the network theoretical miRNAtarget™ (http://mirnatarget.com; Pharmahungary, Szeged, Hungary) software was used for miRNA-target prediction as in previous works8–12,40. miRNAtarget™ software integrates both experimentally validated (miRTarBase v7.041) and predicted (miRDB v5.0 with score > 80.042 and microRNA.org release of August 2010 with mirSVR score < − 1.243) miRNA-target interaction databases to enable reliable predictions. In the predicted miRNA-target interaction networks nodes and edges represent miRNA and target entities and interactions between them, respectively. Weight of the edges originating from up- and downregulated miRNA nodes were defined to be + 1 and − 1, respectively. Node strength values are provided as the measure of the predicted expression change of the given target node, calculated as the sum of edge weights of the associated edges. Therefore, negative and positive node strength values indicate predicted up- and downregulation of the targets, respectively. miRNA-target interaction networks were visualized using Cytoscape software platform44 with the EntOptLayout plugin45.
Statistics
The distribution of the datasets was tested by Kolgomorov–Smirnov normality test. All values are expressed as mean ± standard error of the mean.
An unpaired two-tailed Student’s t-test in case of normal distribution or Mann–Whitney U test in case of non-normal distribution was used to compare two independent groups.
To compare three independent groups, one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test (in case of normal distribution) or Kruskal–Wallis test followed by Dunn’s post hoc test (in case of not normal distribution) were carried out.
A P-value of < 0.05 was used as the criterion of statistical significance. In addition, for declaring significant differential expressions a fold change threshold of 1.5 (fold change > 1.5 or < 1/1.5) was also considered.
Supplementary Information
Acknowledgements
The excellent technical assistance of Patricia Kraft, Tobias Mayer, Karin Sonnenberg, Lutz Hoffmann, Henriett Biró and Benjamin Prokaj is greatly acknowledged.
Author contributions
Conception and design: M.R., S.K.-I., B.M., P.F., G.S.Z. and T.R. Performed the experiments: M.R., B.A., B.B. D.N., T.B., A.O., K.B., A.A.S., B.A.B. Analysis and interpretation of data: M.R., B.B., B.A. Drafting of the manuscript: M.R. Revising the manuscript for important intellectual content: M.K., S.K.-I., B.M., P.F., G.Sz. and T.R. All authors read and approved the final manuscript.
Funding
Open access funding provided by Semmelweis University. Project no. RRF-2.3.1-21-2022-00003 has been implemented with the support provided by the European Union. TKP2021-EGA-23 has been implemented with the support provided by the Ministry of Innovation and Technology of Hungary from the National Research, Development and Innovation Fund, financed under the TKP2021-EGA funding scheme. The study was supported by the National Research, Development and Innovation Office of Hungary (National Heart Program; NVKP-16-1-2016-0017), by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (to M.R.), by a grant from the National Research, Development and Innovation Office (NKFIH) of Hungary (K134939 to T.R.) and by the Medical Faculty of the University of Heidelberg Germany (to S. Korkmaz-Icöz). BÁ and MR were further supported by the New National Excellence Program of the Ministry for Innovation and Technology from the National Research, Development and Innovation Fund source (ÚNKP-20-4-I-SE-7, ÚNKP-21-4-II-SE-18, ÚNKP-22-5-SE-20).
Data availability
The datasets from the RNA sequencing experiments are deposited and available at ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress) under the accession number of E-MTAB-12671.
Competing interests
P.F. is the founder and CEO of, and B.Á. is employed by Pharmahungary Group, a group of R&D companies and the provider of miRNAtarget.com. All the other authors have nothing to declare.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tamás Radovits and Gábor Szabó.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-43171-1.
References
- 1.Nemeth BT, et al. Cinaciguat prevents the development of pathologic hypertrophy in a rat model of left ventricular pressure overload. Sci. Rep. 2016;6:37166. doi: 10.1038/srep37166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litwin SE, et al. Serial echocardiographic-Doppler assessment of left ventricular geometry and function in rats with pressure-overload hypertrophy. Chronic angiotensin-converting enzyme inhibition attenuates the transition to heart failure. Circulation. 1995;91:2642–2654. doi: 10.1161/01.cir.91.10.2642. [DOI] [PubMed] [Google Scholar]
- 3.Norton GR, et al. Heart failure in pressure overload hypertrophy. The relative roles of ventricular remodeling and myocardial dysfunction. J. Am. Coll. Cardiol. 2002;39:664–671. doi: 10.1016/s0735-1097(01)01792-2. [DOI] [PubMed] [Google Scholar]
- 4.Richards DA, et al. Distinct phenotypes induced by three degrees of transverse aortic constriction in mice. Sci. Rep. 2019;9:5844. doi: 10.1038/s41598-019-42209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemaly ER, Chaanine AH, Sakata S, Hajjar RJ. Stroke volume-to-wall stress ratio as a load-adjusted and stiffness-adjusted indicator of ventricular systolic performance in chronic loading. J. Appl. Physiol. 2012;1985(113):1267–1284. doi: 10.1152/japplphysiol.00785.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrino C, et al. Epigenomic and transcriptomic approaches in the post-genomic era: Path to novel targets for diagnosis and therapy of the ischaemic heart? Position paper of the European Society of Cardiology Working Group on cellular biology of the heart. Cardiovasc. Res. 2017;113:725–736. doi: 10.1093/cvr/cvx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz R, Agg B, Ferdinandy P. Survival pathways in cardiac conditioning: Individual data vs. meta-analyses. What do we learn? Basic Res. Cardiol. 2018;113:4. doi: 10.1007/s00395-017-0661-y. [DOI] [PubMed] [Google Scholar]
- 8.Saghy E, et al. Cardiac miRNA expression and their mRNA targets in a rat model of prediabetes. Int. J. Mol. Sci. 2020;21:2128. doi: 10.3390/ijms21062128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bencsik P, et al. Sensory neuropathy affects cardiac miRNA expression network targeting IGF-1, SLC2a-12, EIF-4e, and ULK-2 mRNAs. Int. J. Mol. Sci. 2019;20:991. doi: 10.3390/ijms20040991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreckenberg R, et al. Ischaemic post-conditioning in rats: Responder and non-responder differ in transcriptome of mitochondrial proteins. J. Cell. Mol. Med. 2020;24:5528–5541. doi: 10.1111/jcmm.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makkos A, et al. Molecular network approach reveals rictor as a central target of cardiac protectomiRs. Int. J. Mol. Sci. 2021;22:9539. doi: 10.3390/ijms22179539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agg B, et al. MicroRNA interactome analysis predicts post-transcriptional regulation of ADRB2 and PPP3R1 in the hypercholesterolemic myocardium. Sci. Rep. 2018;8:10134. doi: 10.1038/s41598-018-27740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatsuguchi M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell. Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker AL, et al. Pressure-volume loops in patients with aortic stenosis. J. Heart Valve Dis. 2003;12:325–332. [PubMed] [Google Scholar]
- 16.Dweck MR, Boon NA, Newby DE. Calcific aortic stenosis: A disease of the valve and the myocardium. J. Am. Coll. Cardiol. 2012;60:1854–1863. doi: 10.1016/j.jacc.2012.02.093. [DOI] [PubMed] [Google Scholar]
- 17.Gunther S, Grossman W. Determinants of ventricular function in pressure-overload hypertrophy in man. Circulation. 1979;59:679–688. doi: 10.1161/01.cir.59.4.679. [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Ma LL, Kong FJ, Qiao Z. Distinct phenotypes induced by different degrees of transverse aortic constriction in C57BL/6N mice. Front. Cardiovasc. Med. 2021;8:641272. doi: 10.3389/fcvm.2021.641272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupari M, Turto H, Lommi J. Left ventricular hypertrophy in aortic valve stenosis: Preventive or promotive of systolic dysfunction and heart failure? Eur. Heart J. 2005;26:1790–1796. doi: 10.1093/eurheartj/ehi290. [DOI] [PubMed] [Google Scholar]
- 20.Ruppert M, et al. Incomplete structural reverse remodeling from late-stage left ventricular hypertrophy impedes the recovery of diastolic but not systolic dysfunction in rats. J. Hypertens. 2019;37:1200–1212. doi: 10.1097/HJH.0000000000002042. [DOI] [PubMed] [Google Scholar]
- 21.Ruppert M, et al. Myofilament Ca(2+) sensitivity correlates with left ventricular contractility during the progression of pressure overload-induced left ventricular myocardial hypertrophy in rats. J. Mol. Cell. Cardiol. 2019;129:208–218. doi: 10.1016/j.yjmcc.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Schneider A, Hagerman RJ, Hessl D. Fragile X syndrome—From genes to cognition. Dev. Disabil. Res. Rev. 2009;15:333–342. doi: 10.1002/ddrr.80. [DOI] [PubMed] [Google Scholar]
- 23.Huddleston LB, Visootsak J, Sherman SL. Cognitive aspects of Fragile X syndrome. Wiley Interdiscip. Rev. Cogn. Sci. 2014;5:501–508. doi: 10.1002/wcs.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tassanakijpanich N, Cohen J, Cohen R, Srivatsa UN, Hagerman RJ. Cardiovascular problems in the Fragile X premutation. Front. Genet. 2020;11:586910. doi: 10.3389/fgene.2020.586910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao J, Ye C, Zheng Z, Zhou Z. Fmr1 protects cardiomyocytes against lipopolysaccharide-induced myocardial injury. Exp. Ther. Med. 2018;16:1825–1833. doi: 10.3892/etm.2018.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talvio K, Kanninen KM, White AR, Koistinaho J, Castren ML. Increased iron content in the heart of the Fmr1 knockout mouse. Biometals. 2021;34:947–954. doi: 10.1007/s10534-021-00320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barajas M, et al. The newborn fmr1 knockout mouse: A novel model of excess ubiquinone and closed mitochondrial permeability transition pore in the developing heart. Pediatr. Res. 2021;89:456–463. doi: 10.1038/s41390-020-1064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tevosian SG, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhou B, et al. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Investig. 2009;119:1462–1476. doi: 10.1172/JCI38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Notarangelo LD, Miao CH, Ochs HD. Wiskott–Aldrich syndrome. Curr. Opin. Hematol. 2008;15:30–36. doi: 10.1097/MOH.0b013e3282f30448. [DOI] [PubMed] [Google Scholar]
- 31.Ruan H, et al. Single-cell reconstruction of differentiation trajectory reveals a critical role of ETS1 in human cardiac lineage commitment. BMC Biol. 2019;17:89. doi: 10.1186/s12915-019-0709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Lin L, Qi H, Chen J, Grossfeld P. Endothelial loss of ets1 impairs coronary vascular development and leads to ventricular non-compaction. Circ. Res. 2022;131:371–387. doi: 10.1161/CIRCRESAHA.121.319955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lie-Venema H, et al. ETS-1 and ETS-2 transcription factors are essential for normal coronary and myocardial development in chicken embryos. Circ. Res. 2003;92:749–756. doi: 10.1161/01.RES.0000066662.70010.DB. [DOI] [PubMed] [Google Scholar]
- 34.Walczak M, Martens S. Dissecting the role of the Atg12–Atg5–Atg16 complex during autophagosome formation. Autophagy. 2013;9:424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 36.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The arrive guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metra M, et al. Advanced chronic heart failure: A position statement from the study group on advanced heart failure of the heart failure association of the European Society of Cardiology. Eur. J. Heart Fail. 2007;9:684–694. doi: 10.1016/j.ejheart.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Ruppert M, et al. Pressure-volume analysis reveals characteristic sex-related differences in cardiac function in a rat model of aortic banding-induced myocardial hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2018;315:H502–H511. doi: 10.1152/ajpheart.00202.2018. [DOI] [PubMed] [Google Scholar]
- 39.Olah A, et al. Physiological and pathological left ventricular hypertrophy of comparable degree is associated with characteristic differences of in vivo hemodynamics. Am. J. Physiol. Heart Circ. Physiol. 2016;310:H587–597. doi: 10.1152/ajpheart.00588.2015. [DOI] [PubMed] [Google Scholar]
- 40.Makkos A, et al. Systematic review and network analysis of microRNAs involved in cardioprotection against myocardial ischemia/reperfusion injury and infarction: Involvement of redox signalling. Free Radic. Biol. Med. 2021;172:237–251. doi: 10.1016/j.freeradbiomed.2021.04.034. [DOI] [PubMed] [Google Scholar]
- 41.Huang HY, et al. MiRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2022;50:D222–D230. doi: 10.1093/nar/gkab1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Wang X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microrna.org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agg B, et al. The entoptlayout cytoscape plug-in for the efficient visualization of major protein complexes in protein-protein interaction and signalling networks. Bioinformatics. 2019;35:4490–4492. doi: 10.1093/bioinformatics/btz257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets from the RNA sequencing experiments are deposited and available at ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress) under the accession number of E-MTAB-12671.