Abstract
Purpose:
Real-world evidence for radiation therapy (RT) is limited because it is often documented only in the clinical narrative. We developed a natural language processing system for automated extraction of detailed RT events from text to support clinical phenotyping.
Methods and Materials:
A multi-institutional data set of 96 clinician notes, 129 North American Association of Central Cancer Registries cancer abstracts, and 270 RT prescriptions from HemOnc.org was used and divided into train, development, and test sets. Documents were annotated for RT events and associated properties: dose, fraction frequency, fraction number, date, treatment site, and boost. Named entity recognition models for properties were developed by fine-tuning BioClinicalBERT and RoBERTa transformer models. A multiclass RoBERTa-based relation extraction model was developed to link each dose mention with each property in the same event. Models were combined with symbolic rules to create a hybrid end-to-end pipeline for comprehensive RT event extraction.
Results:
Named entity recognition models were evaluated on the held-out test set with F1 results of 0.96, 0.88, 0.94, 0.88, 0.67, and 0.94 for dose, fraction frequency, fraction number, date, treatment site, and boost, respectively. The relation model achieved an average F1 of 0.86 when the input was gold-labeled entities. The end-to-end system F1 result was 0.81. The end-to-end system performed best on North American Association of Central Cancer Registries abstracts (average F1 0.90), which are mostly copy–paste content from clinician notes.
Conclusions:
We developed methods and a hybrid end-to-end system for RT event extraction, which is the first natural language processing system for this task. This system provides proof-of-concept for real-world RT data collection for research and is promising for the potential of natural language processing methods to support clinical care.
Introduction
The widespread investment in and uptake of electronic medical records (EMR) holds potential to accelerate cancer research and improve patient care by providing massive real-world data. However, this promise has been curtailed for more complex studies with finer-level details because much of patients’ clinical data remain documented primarily as free text in clinic notes, radiology and pathology reports, and other clinical narratives.1–3 This critical information is not amenable to direct extraction and analysis, and therefore cannot easily be used to generate real world evidence.
This is especially salient in radiation therapy (RT), a cornerstone of cancer treatment that is a part of the management of at least 50% of patients with cancer.4,5 Although structured RT data exist, they are often in siloed treatment planning systems and record and verify systems available only to the internal radiation oncology department.6 These systems often do not interface with an institution’s primary EMR, and may not include older treatments or those delivered at another institution. Instead, much of the RT information that is widely accessible to clinicians and researchers is documented only as unstructured text in clinic notes, presenting challenges for research and clinical care.1,2,7,8 For example, national registries, including the Surveillance, Epidemiology, and End Results Program (SEER) cancer registry and the National Cancer Database, do not include robust RT information due to data abstraction challenges, limiting health outcomes and patterns-of-care research from real-world data.9–11 On the clinical side, manual review of the ever-growing EMR for prior RT is time-intensive and error-prone, potentially affecting RT safety, quality, and clinician burnout.12
Natural language processing (NLP), a rapidly advancing field of artificial intelligence that converts human language to representations that computers can process for downstream tasks, presents an opportunity to address these challenges by automating treatment extraction, including RT extraction, from text.1–3 However, most clinical NLP systems to date have focused on extracting specific clinical entities from notes, but not relating them together into comprehensive treatment events. Event extraction, which is the NLP task of identifying and extracting incidents of interest and their relevant associated details from text,13,14 is needed for robust treatment phenotyping to support research and care delivery. Specifically, substantive details of RT treatment, including date, dose, fraction frequency, fraction number, treatment site, and whether it is a boost course, need to be extracted and linked together as part of the same RT event to meaningfully capture a patient’s treatment.15 As an example, understanding RT dose alone is insufficient for clinical care and research. One must know the dose along with the number of fractions it was delivered over and the fraction frequency to understand the effective radiation dose delivered to a patient—essential information for both treatment decision-making and outcomes research. The site of RT and timing are equally essential to understanding how a patient’s cancer was treated. These details are often found separated across multiple sentences and paragraphs within a clinical document. Furthermore, multiple RT courses are often described in sequence using RT-specific jargon and abbreviations, making it difficult for nonexperts to accurately extract course-level information.
To address the need for improved real-world RT data extraction from clinical documents, we developed a fully automated, end-to-end NLP pipeline that can (1) identify key RT details and (2) link details that are part of the same RT course for comprehensive RT event extraction. This is the first effort to extract detailed RT events from text and serves as a proof-of-concept of the ability of modern NLP to automatically capture complex information about patients’ cancer treatments to support research and, in the future, clinical care.
Methods and Materials
Data
We created a corpus of 495 documents describing RT from several complementary data sets. Data sets included free-text RT regimen descriptions from HemOnc.org, a publicly available oncology ontology16,17; Kentucky Cancer Registry (Institution 1) North American Association of Central Cancer Registries (NAACCR) abstracts from patients with lung, breast, prostate, and colorectal cancer; and lung, breast, prostate, and colorectal cancer clinical notes from the University of Kentucky (Institution 2), brain and colorectal cancer clinical notes from the THYME corpus from Clinical TempEval,18 and breast cancer clinical notes from University of Pittsburgh Medical Center (Institution 3). All clinician notes included at least 1 mention of RT. Five of the clinician notes were RT treatment summary notes; the remainder were consult and progress notes. NAACCR abstracts are abstractions of patients’ medical records within a case-level XML and are often copy–pasted from clinician notes by tumor registrars. Although RT descriptions from HemOnc.org do not contain text written for clinical use, they are written in a similar style to how clinicians describe RT events and allowed us to augment our clinical data for system development and make them publicly available. All data sets included instances of grammatical errors and abbreviations (eg, fx instead of fraction, pt instead of patient, PTV instead of planning tumor volume), and brief summarizations commonly seen in clinical documentation (eg, 1/1–1/15/22, WBRT, 10FXs, 6X, 2500CGY [anonymized NAACCR example]); the clinician notes included instances with RT information documented in tabular format. Table 1 shows the data distribution per data set, which we refer to as HemOnc, NAACCR abstracts, and EMR. Additionally, we divided the data sets into training, development and test splits used for model training, parameter optimization and final testing respectively.
Table 1.
Number of documents, entity instances, and relation instances from each source and data provenance in the total corpus
| Number of documents | ||||
|---|---|---|---|---|
|
| ||||
| Document source | Total (N = 495) | Train set (n = 282) | Development set (n = 102) | Test set (n = 111) |
|
| ||||
| EMR | ||||
| University of Kentucky (Institution 2) | 16 (3.2%) | 9 (3.2%) | 4 (3.8%) | 3 (2.7%) |
| University of Pittsburgh (Institution 3) | 9 (1.8%) | 5 (1.8%) | 2 (2.0%) | 2 (1.8%) |
| THYME Corpus | 71 (14.3%) | 28 (9.9%) | 16 (15.7%) | 27 (24.3%) |
| NAACCR abstracts | 129 (26.1%) | 78 (27.7%) | 26 (25.5%) | 25 (22.5%) |
| HemOnc | 270 (54.5%) | 162 (57.4%) | 54 (52.9%) | 54 (48.6%) |
| Number of entity instances | ||||
| Entity | Total (N = 7981) | Train set (n = 4855) | Development Set (n = 1510) | Test set (n = 1616) |
| Boost | 513 (6.4%) | 332 (6.8%) | 110 (7.3%) | 71 (4.4%) |
| Date | 649 (8.1%) | 406 (8.4%) | 128 (8.5%) | 115 (7.1%) |
| Dose | 917 (11.5%) | 540 (11.1%) | 166 (11.0%) | 211 (13.1%) |
| Fraction frequency | 905 (11.3%) | 589 (12.1%) | 167 (11.1%) | 149 (9.2%) |
| Fraction number | 2585 (32.4%) | 1600 (33.0%) | 473 (31.3%) | 512 (31.7%) |
| Site | 2412 (30.2%) | 1388 (28.6%) | 466 (30.9%) | 558 (34.5%) |
| Number of relation instances | ||||
| Relation | Total (N = 12,727) | Train set (n = 7839) | Development Set (n = 2401) | Test set (n = 2487) |
| None | 10,466 (82.2%) | 6480 (82.7%) | 1989 (82.8%) | 1997 (80.3%) |
| Dose-Boost | 89 (0.7%) | 54 (0.7%) | 22 (0.9%) | 13 (0.5%) |
| Dose-Date | 297 (2.3%) | 177 (2.3%) | 62 (2.6%) | 58 (2.3%) |
| Dose-Dose | 484 (3.8%) | 310 (4.0%) | 78 (3.2%) | 96 (3.9%) |
| Dose-Fraction frequency | 152 (1.2%) | 105 (1.3%) | 23 (1.0%) | 24 (1.0%) |
| Dose-Fraction number | 655 (5.1%) | 391 (5.0%) | 115 (4.8%) | 149 (6.0%) |
| Dose-Site | 584 (4.6%) | 322 (4.1%) | 112 (4.7%) | 150 (6.0%) |
Entity and relation instance counts are reported at the window level, not the document level.
Abbreviations: EMR = electronic medical record; NAACCR = National American Association of Central Cancer Registries.
This study was approved by the NAACCR institutional review board, and consent was waived as this was deemed exempt human subjects research.
Task definition and data labeling
We defined our task as the automated extraction of RT events with their associated treatment-related properties from text, with the ultimate intended use of assisting19 EMR review and populating cancer registry records in a human-in-the-loop setting. Specifically, we aimed to extract substantive event descriptions containing the RT details needed for clinical decision-making and research. An RT event (also referred to as RT instance) was defined as a treatment phase to align with NAACCR RT data elements, which defines a phase as “one or more consecutive treatments delivered to the same anatomic volume with no change in the treatment technique.”20 Properties associated with events were Dose, Fraction Frequency, Fraction Number, Date, Treatment Site, and Boost, referred to hereafter as Dose, FxFreq, FxNo, Date, Site, and Boost, respectively. These properties were chosen based on NAACCR data elements20 and American Society for Radiation Oncology Minimum Data Elements.15 Dose is any description of radiation dosage in the text, either the total dose or fractional doses, generally described using the unit gray (Gy) or centigray (cGy). FxNo is any mention of the number of fractions delivered, and FxFreq is the frequency of fraction delivery. Site is the anatomic site, relative region, or RT volume or field; an example of an anatomic site would be “prostate,” a relative region would be “tumor bed,” and a volume would be “clinical tumor volume.” Boost is a mention that conveys the treatment instance is a second phase of RT that brings a smaller treatment site to a higher dose. Date is a date of RT administration.
Annotation guidelines to guide gold standard labeling of RT events used for training and evaluation were developed and have been made publicly available.21 In brief, the annotation guidelines described the annotation task in these steps:
Identify and label the RT Instance text, which is the text span that describes an RT event.
Identify and label RT properties resulting in named entity mentions of type Dose, FxFreq, FxNo, Date, Site, and Boost.
Link each RT property with the RT instance that it describes.
As most RT instances included Dose, we chose to classify the relations between each Dose mention and every other property mentioned in the RT instance. Thus, Dose mentions served as the anchor for the relations within an RT instance. Properties that were linked to the same RT instance in step 3 were related to one another, resulting in these types of relations: Dose-Boost, Dose-Date, Dose-Dose, Dose-FxFreq, Dose-FxNo, and Dose-Site. Of note, Dose-Dose relations are possible, linking 2 different Dose mentions in the same RT instance; for example, in the string “50 Gy in 2 Gy/fx,” “50 Gy” and “2 Gy” would be linked in a Dose-Dose relation.
Forty-eight RT instances, including 209 RT properties and 258 relations, were double-annotated by a radiation oncologist (DSB) and an experienced medical coder (DH). Interannotator agreement (IAA) was calculated for all properties and relations (Table E1). The remaining documents were annotated by a single anno-tator (DH or DSB). A sample annotation is in Fig. 1. We used the Anafora annotation tool22 to create the gold annotations.
Fig. 1.
Example of annotated radiation therapy (RT) text. Mock text segment describing RT illustrating the Dose-Site relationships between the bolded entities. Two adjacent RT instances are distinguished by wavy and dash underlines. The italicized entity “left breast” is a nonrelated anatomic site close to but not within the first RT instance.
Named entity recognition component
Named entity recognition (NER) focuses on the extraction of entity mentions of interest, which for RT are Dose, FxFreq, FxNo, Date, Site, and Boost. Our NER method builds on state-of-the-art (SOTA) transformer models (BioClinicalBert23,24 and RoBERTa25), which are deep neural networks based on stacked self-attention layers. For each entity type, we developed a model fine-tuned on the train split gold instances and optimized on the development split (for data distribution see Table 1). Optimization hyperparameters are in Table E2. For the Dose model, the full texts were used as input. For non-Dose models, as in our previous work,26 Dose entity mentions served as the anchors around which a ±53 token window was created to define the space for possibly relevant Boost, Date, Dose, FxFreq, FxNo, and Site mentions. The ±53 token window represents the 99th percentile of token span lengths between related Dose-property mentions in the same RT instances in the train and development gold annotation sets. The text within these windows was used as model input for the NER models other than Dose. We implemented these models using the Clinical NLP Transformers v0.3.0 (cnlpt)27 library (which includes BioClinicalBERT and RoBERTa), which adds abstraction on top of Hugging Face Transformers library28 for many clinical NLP research use cases.
Relation extraction component
The RT relations are defined between an anchor Dose mention and each of the other RT properties. Thus, the relations are Dose-Boost, Dose-Date, Dose-Dose, Dose-FxFreq, Dose-FxNo, and Dose-Site. Relation extraction models aim to identify these relations. In this component, relations were extracted and tested using only gold annotated entities in the text.
A candidate relation was generated between each anchor Dose mention and any RT Property mention within its ±53 text window. Demarcation of each Dose and other RT Property was done using an established previously described technique.29 Dose-Property relations within the text window were true positive if both entities were part of the same gold RT instance, otherwise a negative instance of a Dose-Property relation (None relation). Our model is a multilabel classifier built on top of a SOTA transformer neural model (RoBERTa) fine-tuned on the train split and optimized on the development split (for data distribution, see Table 1). Optimized hyperparameters are in Table E2. As with the NER component, we used the cnlpt library to facilitate our work.
End2end pipeline
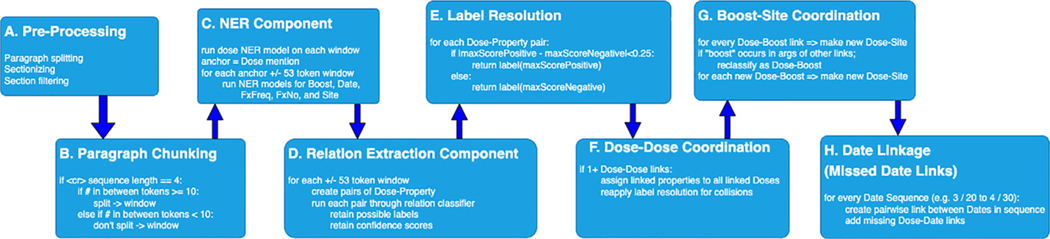
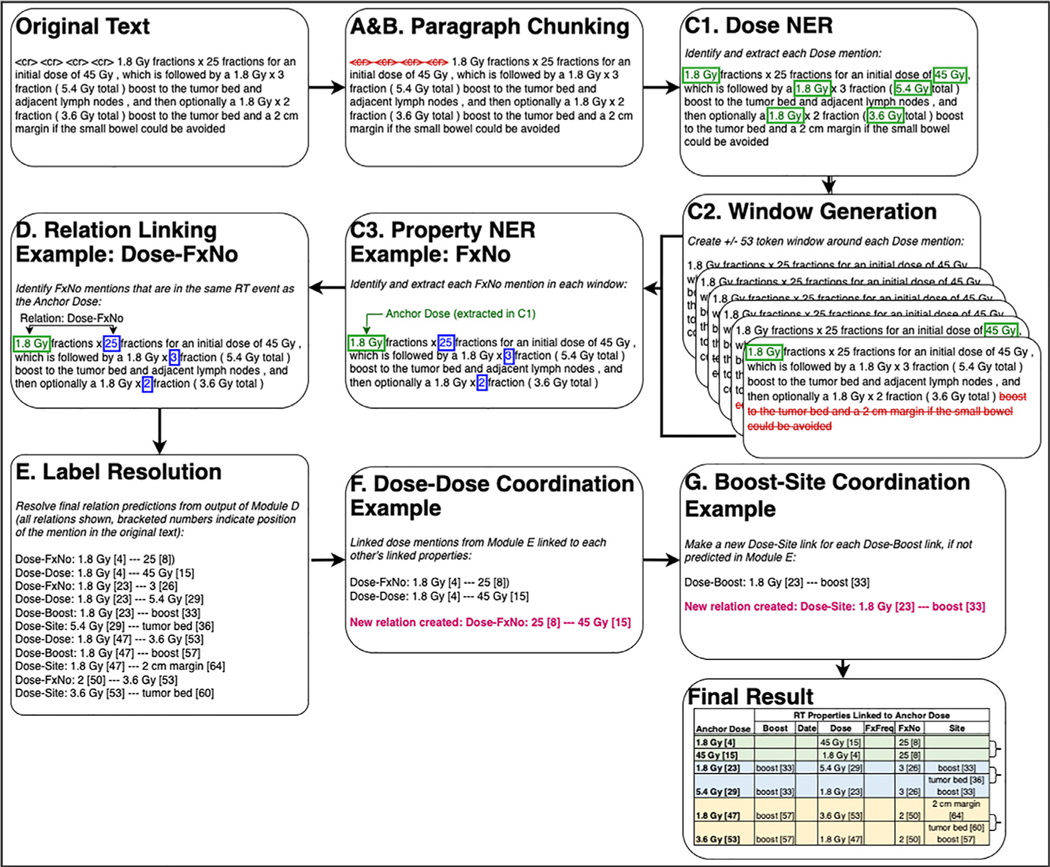
‘What matters in the end is the ability to extract relations from raw input text, which is what is addressed in the end2end pipeline. That is, the end2end pipeline performs complete processing from raw text to Dose-Property relations representing RT Instances. The end2end pipeline encompasses more than just the NER and Relation Extraction components linked together. As is presented in Fig. 2, there are several preprocessing steps and rules linking the neural network components into a full pipeline. These steps were developed based on iterative review of the development set output for each component and the full pipeline. Figure 3 presents an example text passed through each module. First, Module A performs some basic text preprocessing, including paragraph splitting, section splitting, and section filtering. The sections filtered out of the text in Module A are listed in Appendix E2 and were chosen because they rarely contain text describing RT events. Module B further refines paragraphs into text chunks. It allows for RT events in tabular format to be discovered by allowing text fragments separated by newlines to be combined as a chunk if each fragment is <10 tokens; otherwise, every fragment that is >10 tokens and between newlines becomes a separate chunk. This chunk is then passed to Module C, which runs the NER neural models described previously to extract anchor Dose mentions, then defines a window within the chunk of text within ±53 tokens from each anchor. The NER neural models for the other properties (Boost, Date, FxFreq, FxNo, Site) are then run on this window, which is passed to Module D. Module D runs the relation extraction neural model described previously on Dose-Property candidate pairs to assign the relation label and confidence score. Module E finalizes the relation label assignment based on a confidence score function. In Module F, for each Dose-Dose pair, every non-Dose property linked to one Dose is linked to the other dose, if such a link between the other Dose and that non-Dose property was not already predicted by the neural model. In Module G, because “boost” was also defined as a Site in the annotation guidelines, every “boost” occurrence in a Dose-Boost pair is also classified as a Dose-Site pair, and vice versa, if not already classified as such. In Module H, symbolic rules are used to determine whether system-identified Date instances are in a range, and where one of these Date mentions in a range is linked to a Dose mention, we link the other Date mention to that same Dose mention. For example, in “3/20/20 to 4/30/20,” where the system identified both “3/20/20” as “4/20/20” as a Date in Module C, this module will link “4/30/20” to the same Dose mentions as “3/20/2020” and vice versa. Taken together, the end2end pipeline is a hybrid one, combining supervised deep learning approaches with symbolic and lexical rules.
Fig. 2.
End2end pipeline processing flow. Visualization of each module within the fully automated end2end radiation therapy event extraction pipeline, which combined both rules and neural models. Each module carries out specific tasks of the text passed from the preceding module. Modules C and D implement the neural models. The remaining modules carry out rules-based processing.
Fig. 3.
Sample text and its output from each end2end pipeline processing flow component. The text in the boxes shows how the text data are transformed at each module of the end2end pipeline, starting from the original input text and end at the final data extracted by the pipeline. The letters correspond to the modules in Fig. 2 and are summarized as follows: (1) basic text preprocessing (Modules A and B), (2) NER to identify Dose mentions (Module C1), (3) extraction of ±53 token windows around each Dose mention (Module C2), (4) NER to identify all other radiation therapy (RT) properties in a window, (5) relation extraction to link the anchor Dose for that window with the other identified properties (Modules D and E), and (6) postprocessing rules (Modules F and G). The Final Result table shows the final data that are extracted after the text passes through all modules; each row in the Final Result table shows the RT properties that describe the same RT event as the anchor Dose. Curly brackets on the right of the Final Result table indicate the 3 distinct RT events extracted from the sample text, defined by anchor doses being related to each other in a Dose-Dose relation. Please note that Module H, which implements riles to handle date ranges, is not shows in this example.
Evaluation
Standard metrics of precision (P), recall (R), and F1 score are used for evaluation against the gold standard labels of the test split (for data distribution see Table 1):
| (1) |
| (2) |
| (3) |
where TP is true positive count, FP is false positive count, and FN is false negative count. We report results using both the instance-level approach (A) where all TPs, FPs, and FNs from all documents are counted and P, R, and F1 are computed on the total counts, and with the document-level approach (B) where TPs, FPs and FNs per document are counted, and P, R and F1 score are computed per document, and then averaged across all documents. Instance-level (A) gives a global view of the evaluation, while document-level (B) is a macro result capturing what happens on average per document.
The NER component was evaluated against the gold labels in the test split, which was our held-out data never seen in model training and optimization. The relation extraction component, which represents the output of the hybrid end2end system, was evaluated under 2 settings. Under Setting 1, the input is the gold entities, thus simulating a perfect NER system. Under Setting 2, the input is system-generated entities, thus presenting the real-world application scenario. Setting 1 is represented in Tables 3 and 4 under “Input: gold entities all” and Setting 2 under “end2end.”
Table 3.
Results on the test split for relation extraction: Instance level
| Input: gold entities all* |
End2end: all* |
End2end: HemOnc |
End2end: EMR |
End2end: NAACCR abstracts |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relation | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 |
|
| |||||||||||||||
| None† | 0.97 | 0.97 | 0.97 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
| Dose-Boost (IAA 0.67) | 0.92 | 0.85 | 0.88 | 0.92 | 1.00 | 0.96 | 0.75 | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Dose-Date (IAA 1) | 0.91 | 0.88 | 0.89 | 0.83 | 0.63 | 0.72 | NA‡ | NA‡ | NA‡ | 0.70 | 0.68 | 0.69 | 0.94 | 0.61 | 0.74 |
| Dose-Dose (IAA 0.94) | 0.78 | 0.76 | 0.77 | 0.84 | 0.60 | 0.70 | 0.87 | 0.65 | 0.74 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| Dose-FxFreq (IAA 1) | 0.67 | 0.75 | 0.71 | 0.69 | 0.86 | 0.77 | 0.75 | 0.86 | 0.80 | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ |
| Dose-FxNo (IAA 0.99) | 0.92 | 0.91 | 0.92 | 0.94 | 0.80 | 0.87 | 0.95 | 0.84 | 0.89 | 0.93 | 0.75 | 0.83 | 0.94 | 0.83 | 0.88 |
| Dose-Site (IAA 0.9) | 0.86 | 0.87 | 0.86 | 0.78 | 0.55 | 0.65 | 0.76 | 0.49 | 0.60 | 0.75 | 0.47 | 0.58 | 0.84 | 0.78 | 0.81 |
| Average | 0.86 | 0.86 | 0.86 | 0.86 | 0.78 | 0.81 | 0.85 | 0.81 | 0.81 | 0.73 | 0.65 | 0.68 | 0.98 | 0.87 | 0.90 |
Abbreviations: EMR = electronic medical record; FxFreq = fraction frequency; FxNo = fraction number; IAA = interannotator agreement; NAACCR = National American Association of Central Cancer Registries; P = precision/PPV; R = recall/sensitivity; site = treatment site.
All = HemOnc + EMR + NAACCR abstracts.
Indicates pairs of entities that are not linked.
Indicates no instance.
Table 4.
Results on the test split for relation extraction: Document level
| Input: gold entities all* |
End2end: all* |
End2end: HemOnc |
End2end: EMR |
End2end: NAACCR abstracts |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relation | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 |
|
| |||||||||||||||
| None† | 0.81 | 0.89 | 0.83 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
| Dose-Boost | 0.86 | 0.83 | 0.84 | 0.97 | 1.00 | 0.98 | 0.78 | 1.00 | 0.86 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Dose-Date | 0.91 | 0.85 | 0.87 | 0.69 | 0.66 | 0.65 | NA‡ | NA‡ | NA‡ | 0.59 | 0.63 | 0.58 | 0.77 | 0.69 | 0.72 |
| Dose-Dose | 0.47 | 0.39 | 0.41 | 0.31 | 0.28 | 0.29 | 0.87 | 0.65 | 0.74 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| Dose-FxFreq | 0.95 | 0.86 | 0.90 | 0.75 | 0.86 | 0.80 | 0.75 | 0.86 | 0.80 | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ | NA‡ |
| Dose-FxNo | 0.95 | 0.92 | 0.93 | 0.92 | 0.86 | 0.88 | 0.95 | 0.84 | 0.89 | 0.92 | 0.86 | 0.88 | 0.92 | 0.87 | 0.88 |
| Dose-Site | 0.91 | 0.90 | 0.90 | 0.68 | 0.63 | 0.64 | 0.76 | 0.49 | 0.60 | 0.55 | 0.49 | 0.51 | 0.92 | 0.89 | 0.88 |
| Average | 0.84 | 0.81 | 0.81 | 0.76 | 0.75 | 0.75 | 0.85 | 0.81 | 0.81 | 0.68 | 0.66 | 0.66 | 0.93 | 0.91 | 0.91 |
Abbreviations: EMR = electronic medical record; FxFreq = fraction frequency; FxNo = fraction number; IAA = interannotator agreement; NAACCR = National American Association of Central Cancer Registries; P = precision/PPV; R = recall/sensitivity; site = treatment site.
All = HemOnc + EMR + NAACCR abstracts.
Indicates pairs of entities that are not linked.
Indicates no instance.
Furthermore, to explore methods performance on the combination of the 3 data sets and on each of them separately, evaluation was done on the combined data and on each data set.
Manual error analysis was carried out on the test set to identify error modes of the end2end pipeline (Table 5).
Table 5.
Error modes with example text segments
| Examples | ||||
|---|---|---|---|---|
|
| ||||
| Error mode | No. | Text | Gold labels | Predicted labels |
|
| ||||
| FN: Missed atypical and/or RT-specific descriptions | 1 | “He ... received 6250 cGy to the presacral nodule and 45 Gy in 25 fractions to a larger field.” | Dose-Site: “6250 cGy”-“presacral nodule” Dose-FxNo: “6250 cGy”-“25 fractions” Dose-Site: “45 Gy”-“larger field” Dose-FxNo: “45 Gy”-“25 fractions” |
Dose-Site: “6250 cGy”-“presacral nodule” Dose-Site: “45 Gy”-“presacral nodule” Dose-FxNo: “45 Gy”-“25 fractions” |
| 2 | “The prescription dose delivered to PTV3 was 54 Gy in 28 fractions” |
Dose-Site: “54 Gy”-“PTV3” Dose-FxNo: “54 Gy”-“28 fractions” |
Dose-FxNo: “54 Gy”-“28 fractions” | |
| 3 | “Dr. University of Pittsburgh delivered 12.5 Gy to two sites, using 5-cm applicators. One was in the common iliac region, and the second site was in the presacral region.” | Dose-Site: “12.5 Gy”-“two sites” Dose-Site: “12.5 Gy”-”common iliac region” Dose-Site: “12.5 Gy”-“presacral region” |
Dose-FxNo: “12.5 Gy”-“two” | |
| 4 | “Synchronously, PTV1 and PTV2 received 45 and 50.4 Gy, respectively.” |
Dose-Site: “45”-“PTV1”
Dose-Site: “50.4 Gy”-“PTV2” |
Dose-Dose: “45 and”-“50.4 Gy” | |
| FN: Missed relations in tabular text format | 5 | “Left breast 6 MV IMRT 50.4 Gy January 19, 2021 February 25, 2021 28” | Dose-Site: “50.4 Gy”-“Left breast” Dose-Date: “50.4 Gy”-“January 19, 2021” Dose-Date: “50.4 Gy”-“February 25, 2021” Dose-FxNo: “50.4 Gy”-“28” |
Dose-Site: “50.4 Gy”-“Left breast” Dose-Date: “50.4 Gy”-“January 19, 2021” Dose-Date: “50.4 Gy”-“February 25, 2021” |
| FN: Missed relations between entities with intervening text/crossing sentence boundaries | 6 | “Dr. University of Pittsburgh delivered 12.5 Gy to two sites, using 5-cm applicators. One was in the common iliac region, and the second site was in the presacral region.” | Dose-Site: “12.5 Gy”-“two sites” Dose-Site: “12.5 Gy”-“common iliac region” Dose-Site: “12.5 Gy”-“presacral region” |
Dose-FxNo: “12.5 Gy”-“two” |
| 7 | “I discussed the potential role for palliative radiation therapy to the lumbar spine to help reduce some of her radicular symptoms. This could be done in a short course (8 Gy in one fraction) or a modified palliative course (20 Gy in five fractions) being the most common treatment scheduled we use here.” | Dose-Site: “S Gy”-“lumbar spine” Dose-FxNo: “8 Gy”-“one fraction” Dose-Site: “20 Gy”-“lumbar spine” Dose-FxNo: “20 Gy”-“five fractions” |
Dose-Site: “8 Gy”-“lumbar spine” Dose-FxNo: “8 Gy”-“one fraction” Dose-FxNo: “20 Gy”-“five fractions” |
|
| FN: Missed relations because NER model did not tag dose mention | 8 | “08/26/19 through 09/23/19 RT breast 42.56 16 fractions” |
Dose-Date: “08/26/19”-“42.56” Dose-Date: “09/23/19”-“42.56” Dose-FxNo: “42.56”-“16 fractions”
Dose-Site: “42.56”-“RT breast” |
None |
| FN and FP: Relations incorrectly tagged when RT phases of the same total course described together | 9 | “Synchronously, PTV1 and PTV2 received 45 and 50.4 Gy, respectively.” | Dose-Site: “45”-“PTV1” Dose-Site: “50.4 Gy”-“PTV2” |
Dose-Dose: “45 and-50.4 Gy |
| 10 | “He . . . received 6250 cGy to the presacral nodule and 45 Gy in 25 fractions to a larger field.” | Dose-Site: “6250 cGy”-“presacral nodule” Dose-FxNo: “6250 cGy”-“25 fractions” Dose-Site: “45 Gy”-“larger field” Dose-FxNo: “45 Gy”-“25 fractions” |
Dose-Site: “6250 cGy”-“presacral nodule” Dose-Site: “45 Gy”-“presacral nodule” Dose-FxNo: “45 Gy”-“25 fractions” |
|
| 11 | “He . . . received 6250 cGy to the presacral nodule and 45 Gy in 25 fractions to a larger field.” | Dose-Site: “6250 cGy”-“presacral nodule” Dose-FxNo: “6250 cGy”-“25 fractions” Dose-Site: “45 Gy”-“larger field” Dose-FxNo: “45 Gy”-“25 fractions” |
Dose-Site: “6250 cGy”-“presacral nodule” Dose-Site: “45 Gy”-“presacral nodule” Dose-FxNo: “45 Gy”-”25 fractions” |
|
| FP: Nearby medication details mistaken for radiation details | 12 | “50.4 Gy in 2S fractions would be indicated. We will use a fluoropyrimidine as a radiation sensitizer. I have discussed with Mr. University of Pittsburgh that this could either be done with infusional 5-FU or capecitabine. He opted to take oral medication. We will, therefore, start capecitabine S25 mg/m2 twice-daily for five days per week.” | Dose-FxNo: “50.4 Gy”-“2S fractions” | Dose-FxNo: “50.4 Gy”-“28” Dose-FxFreq: “825”-“twice-daily” Dose-FxFreq: “825”-“five days per week” |
| Human annotation error | 13 | “69-year-old White female with a history of T4, N2, colorectal cancer treated with preoperative radiation (25 Gy in 5 fractions)” |
Dose-Site: “25 Gy”-“colorectal cancer” Dose-FxNo: “25 Gy”-“5 fractions” |
Dose-FxNo: “25 Gy”-“5 fractions” |
| 14 | “She has completed 30 Gy in 15 fractions for recurrent colon cancer in the pelvis.” | Dose-FxNo: “30 Gy”-“ 15 fractions” Dose-Site: “30 Gy”-“colon cancer” Dose-Site: “30 Gy”-“pelvis” |
Dose-FxNo: “30 Gy”-“ 15 fractions” Dose-Site: “30 Gy”-“pelvis” |
|
| 15 | “He underwent APR on June 20, 2014, as well as resection of the liver metastases and 12.5 Gy intraoperative radiation therapy delivered to the common iliac and presacral regions.” | Dose-Site: “12.5 Gy”-“common iliac” Dose-Site: “12.5 Gy”-“presacral regions” |
Dose-Date: “12.5 Gy”-“June 20, 2014” Dose-Site: “12.5 Gy”-“common iliac” Dose-Site: “12.5 Gy”-“presacral regions” |
|
| 16 | “We would treat the patient with a six-week course of radiation and use a total dose of 50.4 Gy in 28 fractions with a field reduction after 45 Gy.” | Dose-FxNo: “50.4 Gy”-“2S fractions” Dose-Site: “50.4 Gy”-“field” Dose-Dose: “50.4 Gy”-“45 Gy” Dose-Dose: “45 Gy”-”28 fractions” |
Dose-FxNo: “50.4 Gy-28 fractions Dose-Dose: “45 Gy-28 fractions | |
Abbreviations: APR = University of Pittsburgh; FN = false negative; FP = false positive; FxFreq = fraction frequency; FxNo = fraction number; RT = radiation therapy; site = treatment site.
Text in boldface highlights FN or FP errors due to the specified error mode. Examples have been deidentified.
The code to train the neural models and process text in the pipeline is released at https://github.com/Machine-Learning-for-Medical-Language/radiotherapy_end2end. We have provided formatted training and evaluation data from HemOnc.org, along with instructions, for reproducibility. The neural models trained on the full data set cannot be released to protect patient privacy.
Results
Results from the NER models are in Table 2. The Dose NER model achieves the best instance-level performance of 0.96 F1, followed by the Boost and FxNo models, each achieving 0.94 F1. The lowest instance-level performance is 0.67 F1 for the Site model. NER model performance exceeds the IAA for Boost and Fraction Number.
Table 2.
Results on the test split for named entity recognition
| A: Instance level |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All* |
HemOnc |
EMR |
NAACCR abstracts |
|||||||||
| Category | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 |
|
| ||||||||||||
| Boost (IAA 0.80) | 0.98 | 0.90 | 0.94 | 1.00 | 0.86 | 0.93 | 1.00 | 1.00 | 1.00 | 0.94 | 0.94 | 0.94 |
| Date (IAA 1) | 0.81 | 0.96 | 0.88 | NA | NA | NA | 0.93 | 0.88 | 0.90 | 0.82 | 1.00 | 0.90 |
| Dose (0.99) | 0.94 | 0.98 | 0.96 | 0.96 | 0.97 | 0.97 | 0.86 | 0.98 | 0.92 | 1.00 | 1.00 | 1.00 |
| Fraction frequency (IAA 1) | 0.88 | 0.89 | 0.88 | 0.89 | 0.91 | 0.90 | NA | NA | NA | NA | NA | NA |
| Fraction number (IAA 0.83) | 0.94 | 0.94 | 0.94 | 0.95 | 0.96 | 0.96 | 0.90 | 0.90 | 0.90 | 0.92 | 0.88 | 0.90 |
| Site (IAA 1) | 0.70 | 0.64 | 0.67 | 0.69 | 0.63 | 0.66 | 0.67 | 0.58 | 0.62 | 0.80 | 0.74 | 0.77 |
| Average | 0.88 | 0.89 | 0.88 | 0.90 | 0.87 | 0.88 | 0.87 | 0.87 | 0.87 | 0.90 | 0.91 | 0.90 |
| B: Document level | ||||||||||||
| All* | HemOnc | EMR | NAACCR abstracts | |||||||||
| Category | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 |
| Boost (IAA 0.80) | 0.89 | 0.90 | 0.89 | 1.00 | 0.86 | 0.93 | 1.00 | 1.00 | 1.00 | 1.00 | 0.86 | 0.93 |
| Date (IAA 1) | 0.85 | 0.90 | 0.86 | NA | NA | NA | 0.75 | 0.78 | 0.76 | 0.93 | 1.00 | 0.95 |
| Dose (0.99) | 0.95 | 0.99 | 0.96 | 0.96 | 0.97 | 0.97 | 0.90 | 0.99 | 0.93 | 1.00 | 1.00 | 1.00 |
| Fraction frequency (IAA 1) | 0.30 | 0.30 | 0.30 | 0.89 | 0.91 | 0.90 | NA | NA | NA | NA | NA | NA |
| Fraction number (IAA 0.83) | 0.94 | 0.93 | 0.93 | 0.95 | 0.96 | 0.96 | 0.92 | 0.94 | 0.92 | 0.96 | 0.93 | 0.94 |
| Site (IAA 1) | 0.68 | 0.68 | 0.66 | 0.69 | 0.63 | 0.66 | 0.59 | 0.61 | 0.58 | 0.85 | 0.80 | 0.82 |
| Average | 0.77 | 0.78 | 0.77 | 0.90 | 0.87 | 0.88 | 0.83 | 0.86 | 0.84 | 0.95 | 0.92 | 0.93 |
Abbreviations: EMR = electronic medical record; IAA = interannotator agreement; NAACCR = National American Association of Central Cancer Registries; P = precision/PPV; R = recall/sensitivity.
All = HemOnc + EMR + NAACCR abstracts.
Instance-level results for the relation model when input with the gold entities (Setting 1 from Evaluation) are in Table 3. Results range from 0.71 F1 (Dose-FxFreq) to 0.92 F1 (Dose-FxNo). Results for our hybrid end2end pipeline (Setting 2 from Evaluation) range from 0.65 F1 (Dose-Site) to 0.96 F1 (Dose-Boost) excluding None. Dose-Dose EMR has only 4 instances which the hybrid system missed. Of note is that in both settings, the Dose-Boost results exceed the IAA of 0.67. Per–data set results show that the end2end pipeline performs best on the text from the NAACCR abstracts where the average F1 for the end2end system exceeds the average F1 when the input is gold entities.
Table 4 presents document-level results which exhibit the same trend as the instance-level results. The worst result for the end2end system is Dose-Dose (0.29 F1) compared with 0.41 F1 with gold entities input, which reflects the performance for that type of relation on EMR text. Of note, the results for HemOnc using both instance-level and document-level evaluations are the same because all instances were compiled in one document.
We identified several error modes during error analysis, which are summarized in Table 5 along with representative de-identified examples, including: False negatives due to atypical and/or RT-specific language:
The most common error mode arose from atypical language that was not well-represented in the training set (eg, “common iliac region” or “presacral region” in example 3), and/or RT-specific jargon. The latter scenario was particularly common for descriptions of RT Sites that describe an RT field or volume (eg, “field” in example 1 and “PTV” in example 4). By contrast, false negatives were less common when Site mentions were true anatomic sites.
False negatives due to missed relation in tabular text format: Some, but not all, relations were missed when the text was formatted in a tabular format, as shown in example 5.
False negatives due to missed relations between distant but related RT properties: Relations were more frequently missed if there was significant intervening text between 2 related properties, especially when the intervening text or when the relation crossed sentence boundaries (examples 6 and 7).
False negatives due to a missed Dose mention: By design, if all Dose mentions in an RT instance were missed, no relations were identified (example 8). Although this occurred rarely given the excellent performance of the Dose NER model (0.96 F1), it led to several false negative relation predictions. This demonstrates that even the few errors resulting from almost perfect performance on one task propagate with an increasing effect on downstream tasks.
False negatives and positives due to relations incorrectly predicted when multiple RT phases of the same total course are described: Because a total RT course is often delivered in multiple sequential or concurrent phases, adjacent or overlapping RT instances were common. In some cases, this led to false positives (examples 9, 10) or negatives (example 11). Situations in which this error mode may commonly arise include descriptions of simultaneous integrated boosts and field-in-field techniques.
False positives due to medication dosage and administration frequencies incorrectly predicted to be RT Dose and FxFreq mentions: When a mention of medication dose and/fraction frequency occurred near an RT instance, it was sometimes incorrectly predicted to be an RT Dose or FxFreq mention (example 12). However, there were other instances where the pipeline correctly distinguished between mentions describing a medication event and mentions describing an RT event.
Discussion
Our hybrid end2end system identifies RT instances and their most important properties. The foundation of our system are 2 deep learning—based components: (1) NER component that carries out NER to identify and extract RT properties and (2) relation extraction component to synthesize the properties into coherent treatment events. The averaged instance-level result for the end-to-end system is F1 of 0.81 and is as high as 0.90 for the free text in the NAACCR abstracts. We identified error modes that inform situations in which system predictions should be interpreted with caution by the end-user.
Site NER performance had an important downstream effect on the ultimate performance of the end2end system, as evidenced by the discrepancy between Dose-Site relation extraction using gold entity input versus end2end input (instance-level F1 0.86 vs 0.65, respectively). Error analysis revealed that FNs for treatment site extraction were common when they were described using RT-specific descriptions of relative regions, volumes, and fields. This is likely due to underrepresentation of this type of language in the data used for both pretraining and fine-tuning the neural models. This is supported by the fact that Dose-Site performance was much better among the NAACCR abstract data set, which only had one such description of an RT volume overall and none in the test set. In the future, this limitation could be remedied by incorporating RT-specific lexicons into the system.30
The system’s Dose NER performance was near-perfect (instance-level F1 0.96), supporting our approach of using Dose as the anchor entity to identify RT properties that are part of the same RT event. However, Dose-Dose relation extraction was more challenging (instance-level F1 0.70). This may be because many Dose-Dose relations require discriminating between close and semantically similar Dose mentions that are part of separate RT phases but the same overall RT course—a challenging task even for humans. Of note, it is difficult to fully understand the performance of Dose-Dose extraction in NAACCR abstracts, as there was only 1 Dose-Dose instance in this data set.
Temporal information extraction is especially important for understanding patients’ cancer trajectories but is a notoriously challenging NLP task.2 Given this, our system achieved excellent instance-level Date NER performance of 0.88 and an instance-level Dose-Date F1 of 0.72.
The excellent hybrid end-to-end system performance of 0.90 F1 in the NAACCR abstract data set (consisting of free text from clinical notes copy–pasted into XML elements) may be especially effective for real-world evidence generation. Many cancer registries, including SEER9 and National Cancer Database,10 include only limited details about patients’ initial RT courses, and have documented several limitations to the completeness of these manually abstracted data.11 As patients with cancer are increasingly receiving multiple courses of RT and new combinations of RT with systemic therapies, there is an urgent need for such real-world evidence to understand survivorship and safety. Our methods hold the potential to address this need by improving the accuracy and efficiency of RT event abstraction for populating and enhancing cancer registry records.
Despite the promising results, this system is not ready for clinical implementation, at least not in the fully automated setting. Near-perfect performance and more extensive evaluation on diverse EMR data sets is needed for implementation without a human check,19 and end-user testing is needed before implementation as clinical decision support to demonstrate whether the system can improve the efficiency and accuracy of EMR review in the human-in-the-loop setting. However, in the future, such systems may have clinical value to support quality assurance, for example, as a silent additional screening for prior RT to supplement in a semiautomated fashion, but not replace, manual EMR review and patient interviews, and as a method to identify inconsistencies that may point to inaccurate documentation of RT in the EMR.
To our knowledge, this is the first NLP system for automated RT event extraction, and the first to extract comprehensive cancer treatment events from clinical free text in general. To date, most clinical NLP work for cancer phenotyping has focused on identifying baseline cancer characteristics and general treatment procedures without associated treatment details.2,3,8,31–33 For example, several NLP methods exist to identify mentions of RT procedures, such as “radiation” and “radiotherapy,” which complement our methods but do not provide the information about when, how, and where the patient was treated that is needed for research and clinical decision-making.34,35 Si and Roberts proposed a frame-based system that can capture cancer treatment details, but performance at this level is not reported.36 Treatment event extraction in the general medical domain is also limited and centered on medication events, which tend to be described in a more predictable fashion than RT events.37–39
Limitations of our work include the relatively small document data sets, especially the EMR data set, used to develop the system, which may limit the performance and generalizability. However, this is in part offset by the fact that the models were developed on a hybrid data set reflecting primary clinical text from multiple institutions, expert curations, and bibliometric extractions. The EMR data set contained a diverse set of note types written by a variety of providers, not only radiation oncologists. We acknowledge that RT treatment summary notes likely have the most accurate RT information among clinical documents, and in an ideal world would be the most accurate document from which to extract RT event information. We also acknowledge that structured data present in the treatment planning system, when available, is the most accurate source of RT information. However, patients often do not have all of their cancer treatment at the same institution, and in our fragmented health care system, electronic prescriptions and comprehensive treatment summary notes are not always available to all abstractors and clinicians. Instead, we aimed to develop a proof-of-concept system that can handle a variety of note types when these primary sources of RT information are not available. By design, our system will only extract RT events if they include at least 1 Dose mention, but we found that most substantive RT descriptions include Dose. The NER modules can still be used to facilitate identification of RT descriptions that do not include Dose. In addition, the system does not distinguish between planned and delivered RT events; the annotation guidelines detail how to annotate RT properties for these attributes, and this is an area of future work. Moreover, patient-level summarization is not supported by our system but is a focus of future work.
Conclusion
We developed the first NLP system that automatically extracts RT events from clinical texts. Our system’s performance is relatively high in the end2end evaluation setting. These methods serve as proof-of-concept that NLP can support real-world data collection for research and, in the future, could play a role in clinical care via automatic oncology history summarization and quality assurance from the EMR.
Supplementary Material
Acknowledgments—
We thank Dr Ramakanth Kavuluru for excellent feedback on the manuscript draft. We thank Jong Cheol Jeong and Isaac Hands for their assistance with acquiring the North American Association of Central Cancer Registries reports and University of Kentucky clinical notes.
The work was supported by National Institutes of Health grants UH3CA243120 (E.G., S.F., D.H., E.B.D., H.H., J.L.W., G.K.S.), U24CA248010 (E.G., S.F., H.H., J.L.W., G.K.S.), R01LM010090 (T.M., G.K.S.), R01LM013486 (T.M., G.K.S.), 5R01GM11435 (T.M., G.K.S.), U24CA265879 (J.L.W.), HHSN261201800013I/HHSN26100001 (E.B.D.), and P30 CA177558 (E.B.D.).
Disclosures:
X J.L.W. received funding from Brown Physicians Inc outside the submitted work; is a consultant for Westat, Melax Tech, Roche, and Flatiron all outside the submitted work; and is the deputy editor of HemOnc.org and cofounder of HemOnc.org LLC. D.S.B. is an associate editor of the Radiation Oncology section of HemOnc.org.
Footnotes
The data underlying this article cannot be shared owing to the privacy of individuals whose clinical text data were used in the study.
Although we cannot release the data or the fully trained system to protect identifiable health information, we publicly release the code for training and evaluating the system, as well as a sample training data set from HemOnc.org to facilitate implementation. The code is available at: https://github.com/Machine-Learning-for-Medical-Language/radiotherapy_end2end.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ijrobp.2023.03.055.
References
- 1.Bitterman DS, Miller TA, Mak RH, Savova GK. Clinical natural language processing for radiation oncology: A review and practical primer. Int J Radiat Oncol Biol Phys 2021;110:641–655. [DOI] [PubMed] [Google Scholar]
- 2.Savova GK, Danciu I, Alamudun F, et al. Use of natural language processing to extract clinical cancer phenotypes from electronic medical records. Cancer Res 2019;79:5463–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yim W-W, Yetisgen M, Harris WP, Kwan SW. Natural language processing in oncology: A review. JAMA Oncol 2016;2:797–804. [DOI] [PubMed] [Google Scholar]
- 4.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer 2005;104:1129–1137. [DOI] [PubMed] [Google Scholar]
- 5.Smith BD, Haffy BG, Wilson LD, et al. The future of radiation oncology in the United States from 2010 to 2020: Will supply keep pace with demand? J Clin Oncol 2010;28:5160–5165. [DOI] [PubMed] [Google Scholar]
- 6.Matuszak MM, Fuller CD, Yock TI, et al. Performance/outcomes data and physician process challenges for practical big data efforts in radiation oncology. Med Phys 2018;45:e811–e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huynh E, Hosny A, Guthier C, et al. Artificial intelligence in radiation oncology. Nat Rev Clin Oncol 2020;17:771–781. [DOI] [PubMed] [Google Scholar]
- 8.Warner JL, Anick P, Hong P, Xue N. Natural language processing and the oncologic history: Is there a match? J Oncol Pract 2011;7:e15–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) Program. Available at: https://seer.cancer.gov/. Accessed September 1, 2022.
- 10.American College of Surgeons. National Cancer Database. Available at: https://www.facs.org/quality-programs/cancer/ncdb. Accessed September 1, 2022.
- 11.National Cancer Institute. SEER treatment data limitations (November 2021. submission). Available at: https://seer.cancer.gov/data-software/documentation/seerstat/nov2021/treatment-limitations-nov2021.html. Accessed September 1, 2022.
- 12.Rule A, Bedrick S, Chiang MF, Hribar MR. Length and redundancy of outpatient progress notes across a decade at an academic medical center. JAMA Netw Open 2021;4 e2115334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurafsky D, Martin JH. Speech and Language Processing3 Hoboken, NJ: Prentice Hall; 2014. [Google Scholar]
- 14.Eisenstein J Introduction to Natural Language Processing. Cambridge, MA: MIT Press; 2019. [Google Scholar]
- 15.Hayman JA, Dekker A, Feng M, et al. Minimum data elements for radiation oncology: An American Society for Radiation Oncology Consensus Paper. Pract Radiat Oncol 2019;9:395–401. [DOI] [PubMed] [Google Scholar]
- 16.HemOnc.org. Available at: http://hemonc.org. Accessed November 18, 2022.
- 17.Warner JL, Cowan AJ, Hall AC, Yang PC. HemOnc.org: A collaborative online knowledge platform for oncology professionals. J Oncol Pract 2015;11:e336–e350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bethard S, Derczynksi L, Savovam GK et al. , SemEval-2015 Task 6: Clinical TempEval. In: Proceedings of the 9th International Workshop on Semantic Evaluation, Association for Computational Linguistics. 2015: 806–814. [Google Scholar]
- 19.Bitterman DS, Aerts HJWL, Mak RH. Approaching autonomy in medical artificial intelligence. Lancet Digit Health 2020;2:e447–e449. [DOI] [PubMed] [Google Scholar]
- 20.North American Association of Central Cancer Registries (NAACCR). Available at: http://datadictionary.naaccr.org. Accessed September 1, 2022.
- 21.RTAnnot: Guidelines for text-level annotation of radiotherapy treatment detail. (Github). Available at: https://github.com/RTParse/RTAnnot. Accessed September 1, 2022.
- 22.Chen W-T, Styler W Anafora: A web-based general purpose annotation tool. In: Proceedings of the 2013 NAACL HLT Demonstration Session 14–19. Atlanta, GA: Association for Computational Linguistics; 2013. [PMC free article] [PubMed] [Google Scholar]
- 23.Alsentzer E, Murphy JR, Boag W, et al. , Publicly available clinical BERT embeddings. Available at: https://arxiv.org/abs/1904.03323. Accessed February 15, 2022.
- 24.Hugging Face. emilyalsentzer/Bio_ClinicalBERT. Available at: https://huggingface.co/emilyalsentzer/Bio_ClinicalBERT?text=The+goal+of+life+is+%5BMASK. Accessed February 15, 2022.
- 25.Liu Y, Ott M, Goyal N, et al. RoBERTa: A robustly optimized BERT pretraining approach. Available at: https://arxiv.org/abs/1907.11692. Accessed December 1, 2021.
- 26.Bitterman Danielle, Miller Timothy, Harris David, Lin Chen, Finan Sean, Warner Jeremy, Mak Raymond, and Savova Guergana. Extracting Relations between Radiotherapy Treatment Details. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, Online. Association for Computational Linguistics. 2020: 194–200. [Google Scholar]
- 27.GitHub. cnlp_transformers: Transformers for Clinical NLP. Available at: https://github.com/Machine-Learning-for-Medical-Language/cnlp_transformers. Accessed September 1, 2022.
- 28.Hugging Face. Transformers. Available at: https://huggingface.co/docs/transformers/index. Accessed December 1, 2021.
- 29.Dmitriy Dligach, Miller Timothy, Lin Chen, Steven Bethard. and Savova Guergana. Neural Temporal Relation Extraction. Valencia, Spain: Association for Computational Linguistics; 2017:746–751. [Google Scholar]
- 30.Mayo CS, Moran JM, Bosch W, et al. American Association of Physicists in Medicine Task Group 263: Standardizing nomenclatures in radiation oncology. Int J Radiat Oncol Biol Phys 2018;100:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Fu S, Wen A, et al. Assessment of electronic health record for cancer research and patient care through a scoping review of cancer natural language processing. JCO Clin Cancer Inform 2022;6 e2200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng J, Banerjee I, Henry AS, et al. Natural language processing to identify cancer treatments with electronic medical records. JCO Clin Cancer Inform 2021;5:379–393. [DOI] [PubMed] [Google Scholar]
- 33.Datta S, Bernstam EV, Roberts K. A frame semantic overview of NLP-based information extraction for cancer-related EHR notes. J Biomed Inform 2019;100 103301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apache cTAKES. Examples. Available at: https://ctakes.apache.org/examples.html. Accessed July 1, 2019.
- 35.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si Y, Roberts K. A frame-based NLP system for cancer-related information extraction. AMIA Annu Symp Proc 2018;2018:1524–1533. [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Liu F, Antieau L, Cao Y, Yu H. Lancet: A high precision medication event extraction system for clinical text. J Am Med Inform Assoc 2010;17:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Q, Ji Z, Li Z, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc 2020;27:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller T, Geva A, Dligach D. Extracting adverse drug event information with minimal engineering. Proc Conf 2019;2019:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.