Abstract
In this paper we demonstrate the advantages of a new molecular typing procedure, multilocus sequence typing, for the unambiguous characterization of penicillin-resistant pneumococci. The sequences of ∼450-bp fragments of seven housekeeping genes were determined for 74 penicillin-resistant Taiwanese isolates of Streptococcus pneumoniae (MIC of penicillin > 0.5 μg/ml). The combination of alleles at the seven loci defined an allelic profile for each strain, and a dendrogram, based on the pairwise mismatches in allelic profiles, grouped 86% of the isolates into one of three penicillin-resistant clones for which the MICs of penicillin were 1 to 2 μg/ml. Isolates within each clone had identical alleles at all seven loci or differed at only a single locus, and the fingerprints of their pbp1A, pbp2B, and pbp2X genes were uniform. Isolates of the Taiwan-19F clone and the Taiwan-23F clone were resistant to penicillin, tetracycline, and erythromycin but were susceptible to chloramphenicol. A second serotype 23F clone and serotype 19F variants of this clone were resistant to penicillin, tetracycline, chloramphenicol, and, in some cases, erythromycin. Comparisons of the allelic profiles of the three major clones with those of reference isolates of the known penicillin-resistant clones showed that the Taiwan-19F and Taiwan-23F clones were previously undescribed, whereas the second serotype 23F clone was indistinguishable from the Spanish multidrug-resistant serotype 23F clone. Single isolates of the Spanish penicillin-resistant serotype 9V clone and the Spanish multidrug-resistant serotype 6B clone were also identified in the collection.
Isolates of Streptococcus pneumoniae with intermediate-level penicillin resistance (MIC ≥ 0.12 μg/ml) or high-level resistance (MIC ≥ 2 μg/ml) have been reported from many countries in recent years (1, 6, 30). In an increasing number of countries a high proportion of pneumococcal isolates from carriers and patients with disease are penicillin resistant or multiply antibiotic resistant (1, 30).
Molecular characterization of penicillin-resistant pneumococci requires an assessment of the overall genetic relatedness of the isolates and the relatedness of their penicillin-binding protein (pbp) genes (6). Several different techniques, including multilocus enzyme electrophoresis (20, 21, 24, 26), pulsed-field gel electrophoresis (18, 23, 26), PCR with repetitive element primers (9, 19, 31), and restriction fragment end labeling (11), have been used to assess the genetic relatedness of isolates; analysis of pbp genes is normally carried out by high-resolution fingerprinting of the PCR-amplified pbp1A, pbp2B, and pbp2X genes (5). By these approaches, penicillin-resistant isolates that are identical (or very similar) in overall genotype and which have the same allelic forms of the three pbp genes can be assigned as members of the same clonal group.
These molecular typing studies have identified a number of clones of highly penicillin-resistant pneumococci, some of which have spread globally (6, 30). The most extensively studied clones are those that appear to have emerged within Spain (5, 7, 9, 11, 20, 24, 26). Predominant among these are the major Spanish multidrug-resistant serotype 6B (26), 14 (7), and 23F (20) clones (MMSp6B, MMSp14, and MMSp23F, respectively) and the major penicillin-resistant serotype 9V clone (MPSp9V) (5, 24). Variants of these clones that differ in serotype have also emerged (5, 9, 11, 22) by recombinational exchanges at the capsular biosynthetic locus (8), and some of these have become prevalent (e.g., the serotype 19F variant of the MMSp23F clone [7]). Other clones have been found to predominate in countries that have a high incidence of antibiotic-resistant pneumococci, e.g., Hungary (21), Slovakia (23), South Africa (25), and parts of the United States (18).
The currently used methods for assessing the overall relatedness of resistant isolates each have advantages and disadvantages but are generally adequate for a local epidemiology study, in which a laboratory typically wishes to identify clusters of identical isolates (clones) within a local population of penicillin-resistant pneumococci. However, these methods are less satisfactory when a laboratory wishes to establish whether an identified penicillin-resistant clone is unique to their locality or has been identified previously in other countries (global epidemiology). For global epidemiology, the currently used molecular typing methods produce data that are poorly portable between laboratories as the methods are predominantly based on comparisons of DNA fragment patterns on agarose gels.
We recently developed a procedure that provides a highly portable approach to global epidemiology, multilocus sequence typing (MLST) (17). In this procedure the sequences of ∼450-bp internal fragments of seven housekeeping genes are obtained for each bacterial isolate and the alleles at each of the seven loci define an allelic profile or sequence type. MLST therefore uses the well-established principles of multilocus enzyme electrophoresis but assigns alleles directly via DNA sequencing, rather than indirectly from the electrophoretic mobilities of their gene products. A great advantage of MLST is that sequence data are unambiguous and electronically portable, so that any laboratory can compare the allelic profiles of their isolates with those held in a central MLST database on the World Wide Web via the Internet. MLST was originally described and validated for meningococci (17) but has subsequently been developed and validated for pneumococci (10). In this paper we show how MLST can be used to identify clusters of related isolates (clones) among a population of penicillin-resistant pneumococci and to compare the resistant isolates with those found in other countries.
MATERIALS AND METHODS
Pneumococcal isolates.
The 74 penicillin-resistant isolates were recovered from patients in six hospitals in Taiwan between 1993 and 1997 (Table 1). Of the isolates, 33 were from Taichung Veterans General Hospital (TCVGH), 26 were from Mackay Memorial Hospital (MMH), 9 were from Taipei Veterans General Hospital (TPVGH), and 2 each were from Hsinchu Hospital (HCH), Cathay General Hospital (CGH), and Kaohsiung Veterans General Hospital (KSVGH). A total of 19 isolates (26%) were from blood, cerebrospinal fluid (CSF), or pleural effusion; 26 isolates (35%) were from sputum; and the remainder were mostly from pus, the nasopharynx, or the middle ear. The isolates from TCVGH and MMH represented all of the penicillin-resistant isolates for which MICs were >0.5 μg/ml obtained, predominantly from patients with disease, during 1993 to 1997 and the first half of 1997, respectively. Those from the other hospitals were a random selection of similar isolates provided to us by C.-P. Fung. Serotyping was carried out by the quellung reaction with antisera from the Statens Serum Institut, Copenhagen, Denmark. MICs were determined by the E test (AB Biodisk, Solna, Sweden).
TABLE 1.
Properties of 74 Taiwanese penicillin-resistant pneumococcia
| Strain | Allele no.
|
Serotype | Hospital | Source | Yr | MIC (μg/ml) ofb:
|
Allele pattern
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | PEN | ERY | TET | CM | pbp1A | pbp2B | pbp2X | |||||
| TW1 | 15 | 16 | 19 | 15 | 6 | 20 | 1 | 19F | TCVGH | Blood | 1993 | 2 | 1.5 | 24 | 1 | A | A | A |
| TW53 | 15 | 16 | 19 | 15 | 6 | 20 | 1 | 19F | MMH | Ear | 1996 | 1 | 2 | 12 | 1.5 | A | A | A |
| TW55 | 15 | 16 | 19 | 15 | 6 | 20 | 1 | 19F | MMH | Pus | 1996 | 1 | 2 | 12 | 2 | A | A | A |
| TW56 | 15 | 16 | 19 | 15 | 6 | 20 | 1 | 19F | TPVGH | Throat | 1996 | 0.75 | 8 | 16 | 2 | A | A | A |
| TW64 | 15 | 16 | 19 | 15 | 6 | 20 | 1 | 19F | TPVGH | Sputum | 1996 | 1.5 | 3 | 32 | 2 | A | A | A |
| TW2 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Unknown | 1993 | 2 | 4 | 16 | 2 | A | A | A |
| TW6 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1995 | 1 | 4 | 12 | 0.75 | A | A | A |
| TW8 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1996 | 1 | 4 | 24 | 1.5 | A | A | A |
| TW10 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Sputum | 1996 | 1 | 3 | 16 | 1.5 | A | A | A |
| TW12 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Pus | 1997 | 1.5 | 2 | 32 | 2 | A | A | A |
| TW13 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1997 | 1 | 3 | 24 | 1.5 | A | A | A |
| TW14 | 15 | 16 | 19 | 22 | 6 | 20 | 26 | 19F | TCVGH | Sputum | 1997 | 1.5 | 2 | 12 | 1.5 | A | A | A |
| TW15 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Unknown | 1997 | 1 | 2 | 32 | 2 | A | A | A |
| TW18 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Sputum | 1997 | 1 | 4 | 24 | 2 | A | A | A |
| TW20 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1997 | 1 | 4 | 16 | 2 | A | A | A |
| TW26 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Nose | 1997 | 1 | 4 | 12 | 2 | A | A | A |
| TW30 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Unknown | 1997 | 1 | 6 | 16 | 1.5 | A | A | A |
| TW31 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | CSF | 1997 | 2 | 3 | 32 | 0.75 | A | A | A |
| TW32 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Sputum | 1997 | 1 | 3 | 32 | 1 | A | A | A |
| TW33 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Sputum | 1997 | 1.5 | 6 | 16 | 1 | A | A | A |
| TW34 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Sputum | 1997 | 1 | 3 | 32 | 1.5 | A | A | A |
| TW35 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Pus | 1997 | 1 | 4 | 32 | 0.75 | A | A | A |
| TW36 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Sputum | 1997 | 1.5 | 4 | 32 | 1 | A | A | A |
| TW37 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Nose | 1997 | 1.5 | 4 | 32 | 1 | A | A | A |
| TW38 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | MMH | Unknown | 1997 | 1 | 3 | 24 | 0.75 | A | A | A |
| TW60 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TPVGH | Sputum | 1996 | 2 | 3 | 32 | 2 | A | A | A |
| TW71 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1993 | 2 | 3 | 24 | 0.5 | A | A | A |
| TW78 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | CSF | 1996 | 2 | 3 | 12 | 2 | A | A | A |
| TW19 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 19F | TCVGH | Blood | 1997 | 2 | 3 | 16 | 3 | A | A | B |
| TW5 | 15 | 16 | 19 | 15 | 30 | 20 | 39 | 19F | TCVGH | Blood | 1994 | 1 | 4 | 12 | 2 | A | C | A |
| TW4 | 15 | 29 | 4 | 21 | 9 | 1 | 14 | 23F | TCVGH | Sputum | 1994 | 1 | >256 | 24 | 4 | A | C | B |
| TW11 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Unknown | 1997 | 1.5 | >256 | 24 | 3 | A | C | B |
| TW16 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Sputum | 1997 | 1 | >256 | 48 | 4 | A | C | B |
| TW17 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Blood | 1997 | 1 | >256 | 24 | 3 | A | C | B |
| TW23 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Sputum | 1997 | 1.5 | >256 | 24 | 2 | A | C | B |
| TW24 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Sputum | 1997 | 2 | >256 | 24 | 3 | A | C | B |
| TW27 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Sputum | 1997 | 1 | >256 | 32 | 3 | A | C | B |
| TW29 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | TCVGH | Sputum | 1997 | 1.5 | >256 | 32 | 2 | A | C | B |
| TW48 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | MMH | Pus | 1997 | 1 | >256 | 48 | 1 | A | C | B |
| TW49 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | MMH | Pus | 1997 | 1 | >256 | 48 | 1.5 | A | C | B |
| TW54 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | MMH | Pus | 1996 | 1 | >256 | 24 | 2 | A | C | B |
| TW65 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | HCH | Blood | 1996 | 0.75 | >256 | 48 | 2 | A | C | B |
| TW66 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 23F | HCH | Sputum | 1996 | 2 | >256 | 64 | 2 | A | C | B |
| TW80 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 19F | TCVGH | CSF | 1996 | 0.75 | >256 | 32 | 3 | A | C | B |
| TW47 | 15 | 29 | 4 | 21 | 30 | 1 | 1 | 23F | MMH | Sputum | 1997 | 2 | 0.06 | 0.75 | 1.5 | A | A | B |
| TW7 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | TCVGH | Nose | 1996 | 1 | 2 | 16 | 12 | B | B | B |
| TW9 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | TCVGH | Sputum | 1996 | 1 | 4 | 24 | 24 | B | B | B |
| TW58 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | TPVGH | Throat | 1996 | 0.75 | 1 | 48 | 24 | B | B | B |
| TW61 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | TPVGH | Throat | 1996 | 1 | 3 | 24 | 16 | B | B | B |
| TW62 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | TPVGH | Pleural fluid | 1996 | 1 | 2 | 12 | 24 | B | B | B |
| TW63 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | TPVGH | Urine | 1996 | 1 | 2 | 32 | 16 | B | B | B |
| TW67 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | CGH | Ear | 1996 | 1 | 3 | 16 | 24 | B | B | B |
| TW68 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | CGH | Eye | 1996 | 1 | 4 | 16 | 16 | B | B | B |
| TW69 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | KSVGH | Sputum | 1997 | 1 | >256 | 24 | 24 | B | B | B |
| TW70 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | KSVGH | Blood | 1996 | 1 | 4 | 16 | 24 | B | B | B |
| TW21 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | TCVGH | Nose | 1997 | 1.5 | >256 | 64 | 16 | B | B | B |
| TW28 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | TCVGH | Sputum | 1997 | 1 | 1 | 12 | 24 | B | B | B |
| TW40 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | MMH | Sputum | 1997 | 1 | 4 | 12 | 24 | B | B | B |
| TW41 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | MMH | Blood | 1997 | 1 | 0.047 | 16 | 24 | B | B | B |
| TW45 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | MMH | Unknown | 1997 | 1 | >256 | 16 | 24 | B | B | B |
| TW46 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | MMH | Unknown | 1997 | 1 | >256 | 16 | 24 | B | B | B |
| MMSp23F | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | R | S/R | R | R | B | B | B | |||
| TW39 | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | MMH | Pus | 1997 | 1 | >256 | 64 | 16 | B | B | B |
| TW42 | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | MMH | Sputum | 1997 | 1 | >256 | 32 | 16 | B | B | B |
| TW43 | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | MMH | Pus | 1997 | 1.5 | >256 | 12 | 12 | B | B | B |
| TW44 | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | MMH | Ear | 1997 | 1 | >256 | 48 | 12 | B | B | B |
| TW77 | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | TCVGH | CSF | 1995 | 2 | >256 | 24 | 24 | B | B | B |
| TW52 | 10 | 13 | 6 | 1 | 6 | 38 | 1 | 23F | MMH | Sputum | 1996 | 1 | >256 | 16 | 1.5 | NDc | G | B |
| TW59 | 10 | 13 | 6 | 1 | 6 | 38 | 1 | 23F | TPVGH | Sputum | 1996 | 2 | >256 | 48 | 2 | ND | G | B |
| TW25 | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | TCVGH | Sputum | 1997 | 1 | >256 | 64 | 32 | B | F | C |
| MMSp6B | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | R | S/R | R | R | B | F | C | |||
| TW50 | 7 | 11 | 10 | 1 | 6 | 1 | 1 | 9V | MMH | Pus | 1997 | 1.5 | >256 | 16 | 0.75 | B | B | B |
| MPSp9V | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | R | S/R | S | S | B | B | B | |||
| TW51 | 2 | 8 | 7 | 4 | 6 | 1 | 1 | 14 | MMH | Blood | 1997 | 1.5 | >256 | 12 | 2 | B | D | C |
| TW57 | 2 | 13 | 9 | 15 | 6 | 19 | 42 | 6B | TPVGH | Ear | 1996 | 0.75 | 4 | 48 | 2 | B | C | B |
| TW72 | 10 | 13 | 19 | 16 | 6 | 37 | 31 | 23F | TCVGH | Blood | 1993 | 1.5 | >256 | 16 | 24 | C | H | D |
| TW22 | 8 | 11 | 10 | 1 | 6 | 36 | 38 | 9V | TCVGH | Sputum | 1997 | 1.5 | >256 | 1.5 | 2 | B | E | B |
Alleles of housekeeping genes, pbp genes, and serotypes that are different from those normally found in the major clones are underlined. The allelic profiles and pbp alleles of reference isolates of the major Spanish clones (MMSp23F, MMSp6B, and MPSp9V) are shown.
Abbreviations: PEN, penicillin; ERY, erythromycin; TET, tetracycline; CM, chloramphenicol; R, resistant; S, susceptible; S/R, susceptible or resistant.
ND, not determined.
The MLST database included the allelic profiles of 306 pneumococcal isolates, obtained predominantly from patients with invasive disease (10), and examples of each of the major clones of penicillin- and multidrug-resistant isolates. These included isolates of the MMSp23F (SP264), MMSp6B (SP681), MMSp14 (VH14), and MPSp9V (SP665) clones (7); the highly cephalosporin-resistant serotype 23F clone from the United States (CS111) (18); the serotype 14 clone from Slovakia (CZ29044) (23); and previously undescribed serotype 14 and 23F clones from Uruguay and Poland (4a).
Genetic relatedness of isolates.
The nucleotide sequences of ∼450-bp internal regions from the aroE, ddl, gdh, gki, recP, spi, and xpt genes were amplified by PCR using the primers described previously (10). The gene fragments were sequenced on both strands, by using the same primers, on an ABI 377 Prism automated sequencer with dRhodamine-labeled terminators (PE Applied Biosystems). For each gene, the sequences were compared with each other and with those in our pneumococcal MLST database (http://mlst.zoo.ox.ac.uk) by using the Macintosh computer program Sequence Output (http://epunix.biols.susx.ac.uk/biols/Biochem/Molbiol/). Sequences were assigned as known alleles if they were identical to alleles in our database or as new alleles if they differed in sequence from any of the known alleles. No weighting was given to the degree of sequence divergence between alleles, since, in the absence of knowledge of the proportion of allelic changes that are due to recombination rather than mutation, we cannot say that alleles differing at many sites are any more distantly related than those that differ at a single site. The alleles at each of the seven loci defined the allelic profile, or sequence type, and the relatedness of isolates was determined by constructing a tree by the unweighted pair group method with arithmetic means (UPGMA) from the matrix of pairwise differences between the allelic profiles by using Statistica software (StatSoft, Tulsa, Okla.), as described elsewhere (10, 17).
Analysis of pbp genes.
Fingerprints of the pbp1A, pbp2B, and pbp2X genes were obtained as described previously (5), except that DNA fragments were detected on polyacrylamide gels by staining with ethidium bromide rather than by end labeling and autoradiography. For each pbp gene, isolates that produced identical patterns of DNA fragments were assigned the same allele number (7).
Nucleotide sequence accession numbers.
The nucleotide sequences described in the present work have been assigned GenBank accession no. AJ233886 and AJ233896.
RESULTS
Properties of penicillin-resistant isolates.
All 74 strains were resistant to >0.5 μg of penicillin/ml, and all but one of them (TW47) were multidrug resistant (Table 1). Resistance to erythromycin (MIC ≥ 1 μg/ml), tetracycline (MIC ≥ 8 μg/ml), and chloramphenicol was found in 97, 97, and 31% of the isolates, respectively.
Analysis of relatedness of isolates.
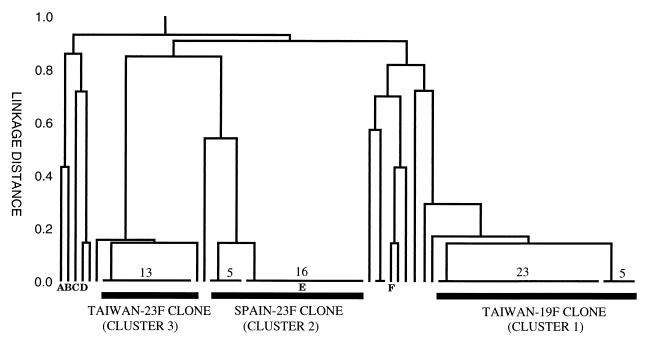
The sequences of the seven housekeeping gene fragments were obtained from all 74 resistant isolates, and a dendrogram was constructed from the matrix of pairwise differences in their allelic profiles (Fig. 1). Sixteen different allelic profiles were distinguished among the 74 isolates, and, at a genetic distance of 0.2, there were three major clusters of isolates, which together included 65 of the isolates. The largest cluster (cluster 1) included 29 serotype 19F isolates, of which 28 had indistinguishable pbp1A, pbp2B, and pbp2X gene fingerprints (pattern A-A-A). All of these 29 isolates had the same MLST allelic profile (15-16-19-15-6-20-26) or differed from this profile at a single locus, and they were defined as the Taiwan-19F clone (Table 1). TW19 had the normal allelic profile but differed in having a variant pbp2X gene. Five of the six single-locus variants were identical and differed from the typical allelic profile at the ddl locus. The other single locus variant (TW14) differed at the recP locus. One other serotype 19F isolate (TW5) clustered with the Taiwan-19F clone, but it appeared to be less closely related as it had a variant pbp2B gene and also differed from the normal allelic profile at two of the seven housekeeping loci. All 29 isolates of the Taiwan-19F clone (and TW5) were resistant to penicillin (MICs, 0.75 to 2 μg/ml), tetracycline (MICs, 12 to 32 μg/ml), and erythromycin (MICs, 1.5 to 8 μg/ml). They were all susceptible to chloramphenicol (MICs < 4 μg/ml).
FIG. 1.
Relatedness of penicillin-resistant pneumococci from Taiwan. The positions on the dendrogram of reference isolates of the Slovakian serotype 14 (A), MMSp14 (B), U.S. serotype 23F (C), MMSp6B (D), MMSp23F (E), and MPSp9V (F) clones are shown. The numbers of isolates of each of the major sequence types are shown.
Cluster 2 contained 21 isolates which were either serotype 23F (16 isolates) or serotype 19F (5 isolates) (Table 1). Sixteen of these isolates had the same allelic profile (4-4-2-4-4-1-1), and the other five differed from this profile at only a single locus (spi). All 21 isolates had indistinguishable pbp1A, pbp2B, and pbp2X gene fingerprints (pattern B-B-B), and all were resistant to penicillin (MICs, 0.75 to 2 μg/ml), tetracycline (MICs, 12 to 64 μg/ml), and chloramphenicol (MICs, 12 to 24 μg/ml). Of 21 isolates, 20 were resistant to erythromycin (MICs ≥ 1 μg/ml), and 4 isolates with the typical allelic profile and all 5 isolates of the single-locus variant were highly resistant (MIC > 256 μg/ml).
Cluster 3 contained 15 closely related isolates, which were all serotype 23F, except strain TW80, which was serotype 19F. Fourteen isolates gave indistinguishable pbp1A, pbp2B, and pbp2X gene fingerprints (pattern A-C-B); had the same allelic profile (15-29-4-21-30-1-14), except TW4, which differed at the spi locus; and were defined as the Taiwan-23F clone (Table 1). All of these isolates were resistant to penicillin (MICs, 0.75 to 2 μg/ml), erythromycin (MICs > 256 μg/ml), and tetracycline (MICs, 24 to 64 μg/ml) but were susceptible to chloramphenicol (MICs ≤ 4 μg/ml). The other strain in this cluster (TW47) was considered a variant of the Taiwan-23F clone as it differed from the typical allelic profile at only a single locus (ddl), but it had a variant pbp2B gene fingerprint and was susceptible to erythromycin and tetracycline (Table 1).
Two highly penicillin-resistant serotype 23F isolates (MICs, 1 to 2 μg/ml) from different hospitals had identical allelic profiles (10-13-6-1-38-1) and identical pbp2B and pbp2X gene fingerprints (their pbp1A genes could not be amplified with our normal primers). Both isolates were resistant to erythromycin and tetracycline but were susceptible to chloramphenicol. The other six penicillin-resistant isolates each had unique allelic profiles.
Relationship of Taiwanese penicillin-resistant isolates to those from other countries.
The allelic profiles of the 74 Taiwanese isolates were compared with those in our MLST database, which included isolates of the major Spanish penicillin-resistant and multidrug-resistant clones (MPSp9V, MMSp6B, MMSp14, MMSp15F, and MMSp23F), isolates of the highly cephalosporin-resistant serotype 23F U.S. clone (18), and the highly penicillin-resistant serotype 14 clone from Slovakia (23), as well as yet-undescribed resistant clones from South America and Eastern Europe and individual resistant isolates from a number of other countries. The Taiwan-19F and Taiwan-23F clones were not closely related to any of the previously characterized penicillin-resistant clones. However, two high-level penicillin-resistant serotype 19F isolates in our MLST database, recovered from blood cultures from patients with pneumonia in the same London hospital in 1995 (4 months apart), had the same allelic profile as the majority of the isolates of the Taiwan-19F clone (15-16-19-15-6-20-26).
The typical allelic profile of the serotype 19F and 23F isolates within cluster 2 was identical to that of the MMSp23F clone (4-4-2-4-4-1-1). As expected, the antibiotic resistance profile of these Taiwanese isolates and their pbp1A, pbp2B, and pbp2X gene fingerprints corresponded to those of a reference isolate of the MMSp23F clone (7).
One serotype 6B isolate (TW25) was assigned by MLST to the MMSp6B clonal complex, and one serotype 9V isolate (TW50) was assigned to the MPSp9V clonal complex. In both cases the isolates had the characteristic pbp1A, pbp2B, and pbp2X gene fingerprints of the corresponding Spanish clones (7) and differed from their typical allelic profiles at only a single locus (Table 1). The antibiotic resistance profile of the Taiwanese isolate of the MMSp6B clone was typical of this clone (26). This single locus variant of the MMSp6B clone has also been found in Thailand (32). The Taiwanese isolate of the MPSp9V clone was resistant to penicillin and tetracycline, whereas members of this clonal complex are normally tetracycline susceptible. The four other penicillin-resistant isolates, which each had unique allelic profiles, were not closely related to strains in our MLST database.
DISCUSSION
Penicillin-resistant pneumococci have increased dramatically in frequency in the Far East, and several countries, including South Korea, Japan, and Hong Kong, have recently reported resistance rates of >40% (1, 4, 14–16, 27). The incidence of antibiotic-resistant pneumococci in Taiwanese hospitals has also increased greatly during the 1990s. In Taipei (northern Taiwan), <10% of isolates from children with pneumococcal disease were penicillin resistant in 1991 and 1992 (13), and in southern Taiwan, 12% of clinical isolates from all age groups were penicillin resistant from 1990 to 1993 (12). However, in 1994 and 1995, 45% of isolates from children in Taipei were penicillin-resistant (13), and 71% of isolates from children in Kaohsiung were penicillin resistant from 1995 to 1997, with 58% of these isolates exhibiting high-level resistance (3). Resistance to erythromycin and tetracycline is also particularly prevalent in Taiwan (>60% of isolates [2, 3, 12]), and almost all penicillin-resistant isolates are resistant to at least two other classes of antibiotic (3, 12). In the present study, almost all of the penicillin-resistant isolates were resistant to tetracycline (MIC ≥ 8 μg/ml), and erythromycin (MIC ≥ 1 μg/ml), with 40% of all isolates being highly erythromycin resistant (MICs > 256 μg/ml). Resistance to chloramphenicol was 31% but, with one exception, was restricted to isolates of the MMSp23F and MMSp6B clones that have been imported into Taiwan.
We chose to use MLST to identify clusters of penicillin-resistant pneumococci from Taiwan that are closely related in their overall genotype. MLST provides a powerful method for the characterization of penicillin-resistant pneumococci as it has a very high discriminatory power, which results from the presence of a large number of alleles at each of the seven housekeeping loci (10). The average number of alleles per locus for the ∼500 pneumococcal isolates currently in the MLST database is >30, providing the ability to distinguish >307 (22 billion) genotypes. The expected frequency at which any allelic profile will occur by chance can be estimated from the products of the frequencies of each allele in the pneumococcal population (10). For example, on the basis of allele frequency data from our pneumococcal MLST database, the allelic profiles of the three major penicillin-resistant clones in Taiwan would each be expected to occur by chance at a frequency of <10−10. The high discriminatory power therefore makes it very unlikely that unrelated isolates will by chance have identical, or even similar, allelic profiles (10).
In this study isolates that had the same allelic profile or that differed at one locus were considered to be members of the same clonal complex. According to this criterion there were three major multidrug-resistant clones among the 74 penicillin-resistant isolates from Taiwanese hospitals. This clustering of isolates by MLST was validated by the demonstration that (with minor exceptions) isolates within each cluster had the same pbp1A, pbp2B, and pbp2X gene fingerprints and serotypes. One isolate of the Taiwan-23F clone and five isolates of the MMSp23F clone were serotype 19F, and these presumably represent serotype variants of these clones. Serotype 19F variants of the MMSp23F clone are commonly encountered in Spain (7) and have also been reported in other countries, including South Korea (19) and Thailand (11).
The Taiwan-19F clone was the most prevalent (39% of isolates) and has been present in Taiwan since at least 1993. The Taiwan-23F clone (19% of isolates) has been present since 1994. Both of these clones were recovered from three hospitals in Taiwan, and they appear to differ from previously described penicillin-resistant clones. This suggests that they may have emerged within the Far East, where the incidence of antibiotic-resistant pneumococci is now very high. The Taiwan-19F clone appears to have already spread intercontinentally, as two penicillin-resistant serotype 19F isolates from the United Kingdom that were indistinguishable from this clone were identified in our MLST database (10). All of the isolates of the Taiwan-19F and Taiwan-23F clones were resistant to erythromycin, although the MICs for the isolates of the latter clone were >256 μg/ml, presumably due to the presence of the ermAM genes, which encode an rRNA methylase, whereas the MICs for the former clone were 1.5 to 8 μg/ml and may result from drug efflux due to mefE (28).
The 21 isolates within cluster 2 were unambiguously identified as members of the MMSp23F clonal complex by MLST combined with pbp gene fingerprinting. Most of these isolates had an allelic profile identical to those of isolates typical of the MMSp23F clonal complex, but there was also a distinctive highly erythromycin-resistant single-locus variant of this clone with a different allele of the spi gene within Taiwan. The latter isolates are clearly members of the MMSp23F clonal complex, as they have the characteristic pbp1A, pbp2B, and pbp2X gene fingerprints and serotype of this clone. Isolates of the MMSp23F clone in Taiwan were, with one exception, resistant to erythromycin, although some of them had high-level resistance characteristic of ermAM and others had the lower levels of resistance characteristic of drug efflux (28). The prevalence of the MMSp23F clone in Taiwan is not surprising since in recent years it has become established globally and has been previously reported to be present in the Far East (11, 19, 29). We also identified single isolates of both the MMSp6B clone and the MPSp9V clone within Taiwan. In each case, the isolates differed at a single locus from the typical allelic profile of these clones, but they were clearly members of the clones as they had the characteristic pbp gene fingerprint patterns and serotypes.
More than 90% of the 74 penicillin-resistant isolates from Taiwan were serotype 19F or 23F, and a similar predominance of these two serotypes is found among highly penicillin-resistant isolates from other countries in the Far East (4, 14, 16, 27). Comparisons of strains from these countries with those described here are needed to determine the contribution of the Taiwan-19F and Taiwan-23F clones and serotype 19F and 23F isolates of the MMSp23F clone to the high prevalence of multidrug-resistant pneumococci in the Far East.
This paper demonstrates that MLST provides an unambiguous method for identifying clones within populations of penicillin-resistant pneumococci and is particularly suitable for comparing isolates or clones with those described previously. Characterization of a clone of interest by MLST is straightforward and only requires that the seven housekeeping gene fragments be amplified by PCR and sequenced on both strands, by using a single primer for each direction. The sequences can then be submitted electronically to our MLST Web site, where they can be compared with those in the database and an allelic profile can be assigned. The allelic profile of a penicillin-resistant clone can then be compared with the allelic profiles in the database, which includes those of each of the known penicillin-resistant clones, making it simple to ascertain whether the clone of interest is a known or a previously undocumented penicillin-resistant clone, without the need to obtain reference strains. For definitive characterization, MLST or any other method for comparing the overall relatedness of penicillin-resistant isolates should be combined with an analysis of the pbp genes since penicillin resistance may emerge independently on more than one occasion in closely related lineages.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust. B.G.S. is a Wellcome Trust Principal Research Fellow. Z.-Y.S. was supported by the TCHVGH.
We thank Chang-Phone Fung for providing strains and Derrick Crook and the Oxford Vaccine Group for assistance with serotyping.
REFERENCES
- 1.Baquero F. Pneumococcal resistance to β-lactam antibiotics: a global geographic overview. Microb Drug Resist. 1995;1:115–120. doi: 10.1089/mdr.1995.1.115. [DOI] [PubMed] [Google Scholar]
- 2.Chang S C, Chen Y C, Luh K T, Hsieh W C. Macrolide resistance of common bacteria isolated from Taiwan. Diagn Microbiol Infect Dis. 1995;23:147–154. doi: 10.1016/0732-8893(95)00197-2. [DOI] [PubMed] [Google Scholar]
- 3.Chiou C C C, Liu Y C, Huang T S, Hwang W K, Wang J H, Lin H H, Yen M Y, Hsieh K S. Extremely high prevalence of nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae among children in Kaohsiung, Taiwan. J Clin Microbiol. 1998;36:1933–1937. doi: 10.1128/jcm.36.7.1933-1937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong Y, Lee K, Kwon O H, Henrichsen J. High incidence of multidrug-resistant Streptococcus pneumoniae in South Korea. Eur J Clin Microbiol Infect Dis. 1995;14:528–531. doi: 10.1007/BF02113433. [DOI] [PubMed] [Google Scholar]
- 4a.Coffey, T. J., and B. G. Spratt. Unpublished data.
- 5.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal gene transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Dowson C G, Daniels M, Spratt B G. Genetics and molecular biology of β-lactam-resistant pneumococci. Microb Drug Res. 1995;1:25–30. doi: 10.1089/mdr.1995.1.29. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Berrón S, Daniels M, Garcia-Leoni E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 9.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berrón S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Res. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 10.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 11.Hermans P W, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 12.Hsueh P R, Chen H M, Lu Y C, Wu J J. Antimicrobial resistance and serotype distribution of Streptococcus pneumoniae strains isolated in southern Taiwan. J Formos Med Assoc. 1996;95:29–36. [PubMed] [Google Scholar]
- 13.Huang F Y, Chiu N C, Liu S C. Penicillin-resistant pneumococcal infections in children. J Formos Med Assoc. 1997;96:414–418. [PubMed] [Google Scholar]
- 14.Kam K M, Luey K Y, Fung S M, Yiu P P, Harden T J, Cheung M M. Emergence of multiple-antibiotic-resistant Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1995;39:2667–2670. doi: 10.1128/aac.39.12.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S N, Kim S W, Choi I H, Pyo S N, Rhee D K. High incidence of multidrug-resistant Streptococcus pneumoniae in South Korea. Microb Drug Resist. 1996;2:401–406. doi: 10.1089/mdr.1996.2.401. [DOI] [PubMed] [Google Scholar]
- 16.Lyon D J, Scheel O, Fung K S, Cheng A F, Henrichsen J. Rapid emergence of penicillin-resistant pneumococci in Hong Kong. Scand J Infect Dis. 1996;28:375–376. doi: 10.3109/00365549609037922. [DOI] [PubMed] [Google Scholar]
- 17.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGee L, Klugman K P, Friedland D, Lee H J. Spread of the Spanish multi-resistant serotype 23F clone of Streptococcus pneumoniae to Seoul, Korea. Microb Drug Resist. 1997;3:253–257. doi: 10.1089/mdr.1997.3.253. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz R, Coffey T C, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 22.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 23.Sa Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 24.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 26.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980’s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 27.Song J H, Yang J W, Peck K R, Kim S, Lee N Y, Jacobs M R, Appelbaum P C, Pai C H. Spread of multidrug-resistant Streptococcus pneumoniae in South Korea. Clin Infect Dis. 1997;25:747–749. doi: 10.1086/516945. [DOI] [PubMed] [Google Scholar]
- 28.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarasi A, Chong Y, Lee K, Tomasz A. Spread of the serotype 23F multidrug-resistant Streptococcus pneumoniae clone to South Korea. Microb Drug Resist. 1997;3:105–109. doi: 10.1089/mdr.1997.3.105. [DOI] [PubMed] [Google Scholar]
- 30.Tomasz A. Antibiotic resistance in Streptococcus pneumoniae. Clin Infect Dis. 1997;24:S85–S88. doi: 10.1093/clinids/24.supplement_1.s85. [DOI] [PubMed] [Google Scholar]
- 31.Versalovic J, Kapur V, Mason E O, Shah V, Keouth T, Lupski J R, Musser J M. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, J., and B. G. Spratt. Unpublished data.