Abstract
Strains formerly identified as Streptococcus bovis were allotted to two groups by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of whole-cell proteins. Strains from humans with infections, mostly patients with endocarditis, and strains from pigeons with septicemia clustered with the recently described species Streptococcus gallolyticus. The original S. bovis type strain and strains exclusively from ruminants formed the second cluster. The findings indicate that S. gallolyticus is more likely to be involved in human and animal infections than S. bovis. Growth characteristics and several biochemical reactions were found to be useful in the differentiation of S. gallolyticus from S. bovis.
The taxonomy of organisms designated Streptococcus bovis has a very complex history. S. bovis and Streptococcus equinus are listed as separate species in Bergey’s Manual of Systematic Bacteriology (10) but were reported to be subjective synonyms by Farrow et al. (8). The conclusions of the latter study were primarily based on DNA-DNA hybridization values. The specific epithet S. equinus has priority over S. bovis but is rarely used in human clinical bacteriology. In veterinary medicine both names are in use.
Recently the situation has become more complex by the description of two novel species for strains originally identified as S. bovis: Streptococcus caprinus (2) and Streptococcus gallolyticus (13). The epithet gallolyticus was derived from the ability of the strains to decarboxylate gallic acid. This characteristic and the tannase activities of the strains were characteristics of S. bovis-like strains originally detected in the feces of koalas. The epithet caprinus referred to animals that forage on the same types of foods. However, later work revealed the synonymy of these two species, but the specific epithet gallolyticus has nomenclatural priority (17).
In order to differentiate correctly putative S. bovis strains from those of human and animal origins, we undertook a study of strains identified as S. bovis by conventional and commercially available miniaturized identification techniques. The study included strains from domestic animals and from humans, including clinical and nonclinical strains, field strains, and collection strains.
MATERIALS AND METHODS
Strains.
Thirty-seven strains (Table 1) were investigated. Seven strains were isolated from human clinical samples, mainly from patients with endocarditis. Four strains were isolated from pigeons with septicemia. The exact origins of two collection strains, LMG 15051 (J. M. Sherman 38) and LMG 15052 (E. M. Barnes C101), were unknown. The remaining strains were cultured from the intestinal contents or from feces from the animal hosts listed in Table 1.
TABLE 1.
Strains examined
| Cluster | Straina | Other designation | Culture collectionb | Specimen type |
|---|---|---|---|---|
| I | Streptococcus bovis strains | |||
| LMG 8518T | NCFB 597T | NCFB | Cow dung | |
| LMG 14814 | Lille 3 | Wastewater | ||
| LMG 14852 | S329 | Goat intestine | ||
| LMG 14858 | S276 | Goat intestine | ||
| LMG 14864 | Ton 520 | Cow tonsils | ||
| LMG 14867 | RS20a | Cow feces | ||
| LMG 14868 | RS7b | Cow feces | ||
| LMG 14869 | RS19a | Cow feces | ||
| LMG 15048 | NCFB 1253 | NCFB | Cow rumen | |
| LMG 15052 | NCFB 2087 | Cow mastitis | ||
| LMG 15055 | NCFB 2128 | Cow dung | ||
| LMG 15062 | NCFB 2500 | Cow rumen | ||
| LMG 15064 | NCFB 2574 | Sheep rumen | ||
| LMG 15065 | NCFB 2596 | Cow vulval swab | ||
| II | Streptococcus gallolyticus strains | |||
| LMB 14618 | STR 600 | Pigeon septicemia | ||
| LMB 14620 | STR 603 | Pigeon septicemia | ||
| LMB 14621 | STR 106 | Pigeon abscess | ||
| LMB 14622 | STR 673 | Pigeon septicemia | ||
| LMB 14821 | STR 598 | Pigeon intestine | ||
| LMB 15049 | NCFB 2019 | NCFB | Cow mastitis | |
| LMB 15050 | NCFB 2079 | NCFB | ||
| LMB 15051 | NCFB 2080 (formerly a type strain) | NCFB | ||
| LMB 15053 | NCFB 2088 | NCFB | Cow mastitis | |
| LMB 15056 | NCFB 2134 | NCFB | Cow mastitis | |
| LMB 15063 | NCFB 2572 | NCFB | Cow rumen | |
| LMB 15120 | NCFB 2631 | NCFB | ||
| LMB 15456 | CCUG 19454 | CCUG | Human urine | |
| LMB 15462 | 67 | H. Verburgh | Human endocarditis | |
| LMB 15464 | 106 | H. Verburgh | Human endocarditis | |
| LMB 15477 | 260 | H. Verburgh | Human endocariditis | |
| LMB 15478 | 305 | H. Verburgh | Human endocariditis | |
| LMB 15479 | 332 | H. Verburgh | Human endocariditis | |
| LMB 15486 | 488 | H. Verburgh | Human endocariditis | |
| LMB 15572 | ACM 3969 | L. Blackall | Feral goat rumen | |
| LMB 15573 | ACM 3970 | L. Blackall | Feral goat rumen | |
| LMB 15582 | PDH 818 | Pigeon feces | ||
| LMB 16802T | CCUG 35224T | CCUG | Koala feces |
LMG indicates the LMG/BCCM culture collection at the Laboratorium voor Microbiologie Gent, Universiteit Gent, Ghent, Belgium.
CCUG, Culture Collection University of Göteborg, Department of Clinical Bacteriology, Göteborg, Sweden; NCFB, the National Collection of Food Bacteria, Agricultural and Food Research Council, Institute of Food Research, Reading Laboratory, Reading, Berkshire, United Kingdom. Spaces left blank indicate our own culture collection.
Whole-cell protein analysis.
Whole-cell protein analysis, preparation of cellular protein extracts, polyacrylamide gel electrophoresis, densitometric analysis, registration of protein profiles, normalization of the densitometric traces and interpolation of the protein profiles, grouping of strains by the Pearson product moment correlation coefficient, and unweighted pair group method using arithmetic averages (UPGMA) cluster analysis were performed as described before (16, 20) with the GelCompar 4.0 software package (Applied Maths, Kortrijk, Belgium) (21).
Growth and biochemical activity.
We studied growth in brain heart infusion (Biolife, Milano, Italy) and Columbia agar (LAB M, Bury, United Kingdom) with 5% bovine blood and on the following selective media: Slanetz and Bartley agar (Oxoid, Basingstoke, United Kingdom), Edwards agar (Oxoid) with 5% bovine blood, bile esculin agar (Difco, Detroit, Mich.), and Rogosa SL agar (Difco). We also tested for clotting in litmus milk (Oxoid). Tannase was detected as described by Osawa and Walsh (14). Amylase activity was investigated by spot inoculating strains on Columbia agar without added blood. Reaction zones were recorded after we flooded the plates with Gram’s iodine following overnight incubation. Motility in semisolid motility-indole-ornithine (MIO) medium (Gibco, Paisley, United Kingdom) was sought. We also tested for urease activity in 0.01 M sodium phosphate buffer containing 0.2% urea and 0.05% phenol red (pH 6.5). Other enzymatic and carbohydrate reactions were determined with a BBL CRYSTAL gram-positive identification kit (Becton Dickinson, Cockeysville, Md.) and with API 20 Strep, API 50CH, and Rapid ID32 Strep galleries (BioMérieux, La Balme les Grottes, France). The reaction mixtures for all tests except the galleries, urease test, and certain comparative growth tests were incubated in air with 5% CO2. Rogosa SL agar was incubated anaerobically in H2 and CO2.
RESULTS
Whole-cell protein analysis.
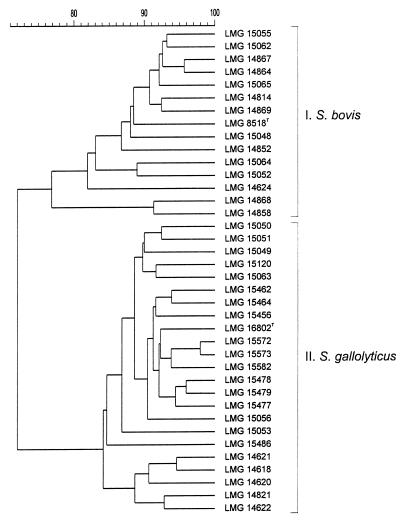
The strains were separated into two clusters (Fig. 1 and 2). Seven reference strains, all of ruminant origin, were comprised in cluster I: LMG 8518T (type strain of S. bovis), LMG 15048, LMG 15052, LMG 15055, LMG 15062, LMG 15064, and LMG 15065. Another strain of the 14 cluster I strains was isolated from wastewater of a dairy farm, one strain was obtained from the tonsils of a calf, and the remaining strains were from goat and cow intestines or feces. Cluster II comprised 23 strains, including 11 reference strains: LMG 15049, LMG 15050, LMG 15051, LMG 15053, LMG 15056, LMG 15063, LMG 15120, LMG 15456, and LMG 15572T (type strain of S. caprinus), LMG 15573, and LMG 16602T (type strain of S. gallolyticus). The other strains were from human infections and from intestinal contents (two strains) or septicemia specimens (four strains) from pigeons.
FIG. 1.
Dendrogram derived from the unweighted pair group average linkages of correlation coefficients (expressed for convenience as percentages of value) between whole-cell protein patterns of all of the strains examined.
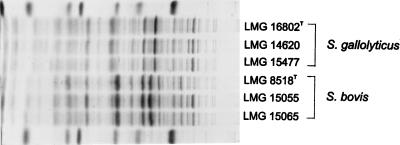
FIG. 2.
Electrophoretic protein profiles of a selection of S. bovis and S. gallolyticus strains. Strains LMG 16802T, LMG 14620, and LMG 15477 were isolated from a feral goat, a pigeon septicemia specimen, and human endocarditis specimens, respectively. The molecular weight markers used (indicated in the top and bottom lanes) are (from left to right) as follows: lysozyme, 14,500; trypsin inhibitor, 20,100; trypsinogen, 24,000; carbonic anhydrase, 29,000; glyceraldehyde-3-phosphate dehydrogenase, 36,000; egg albumin, 45,000; and bovine albumin, 66,000.
Morphology and growth characteristics.
All strains were gram-positive coccobacilli. The cells were smaller than those commonly seen with typical pyogenic streptococci and enterococci. All produced homogeneous growth after 1 day of incubation. Growth at 42°C was similar or slightly better than at 37°C and definitely better than at 30°C. No or very slight and delayed growth was seen at 25°C. Incubation in air supplemented with 5% CO2 increased growth of all strains. The strains were not pigmented and not motile. Only two cluster II strains were alpha-hemolytic, while all strains of cluster I produced small zones of alpha-hemolysis without greening.
Resistance and growth on selective media.
No growth occurred in 6.5% NaCl broth. All except one cluster II strain grew and caused blackening on esculin bile medium. All grew on Edwards medium and all lysed esculin in this medium. All cluster I strains produced browning and blackening on Edwards medium, while none of the cluster II strains caused similar discolorations. On Slanetz and Bartley agar, cluster I colonies tended to be whitish while cluster II strains were slightly pink. All cluster I strains except one produced extremely watery or mucoid colonies on Rogosa SL agar due to dextran formation from the sucrose present in this medium. Only one of the cluster II strains had watery colonies on Rogosa agar.
Biochemical activity.
All strains were positive in the Voges-Proskauer reaction test and in tests for alanyl-phenylalanyl proline arylamidase, α-galactosidase, β-glucosidase, leucine arylamidase, 4-methylumbelliferyl (4MU)-β-d-glucoside, l-valine-7-amino-methylcoumarin (AMC) l-phenylalanine-AMC, 4MU-α-d-glycoside, l-pyroglutamic acid-AMC, l-tryptophan-AMC, l-arginine-AMC, and l-isoleucine-AMC. All strains produced acid from amygdalin, cellobiose, esculin, d-fructose, N-acetylglucosamine, methyl-β-d-glucopyranoside, galactose, d-glucose, lactose, maltose, mannose, saccharose, and salicin, and all except one S. gallolyticus strain utilized β-gentiobiose, glycogen, maltotriose, d-raffinose, and starch. All except one S. gallolyticus and one S. bovis strain each produced acid from melibiose. All strains hydrolyzed p-nitrophenyl-β-d-glucoside, proline- and leucine-p-nitroanilide, o-nitrophenyl-β-d-galactoside, and p-nitrophenyl-α-d-galactoside.
All strains tested were negative in tests for arginine dihydrolase, β-mannosidase, alkaline phosphatase, pyrrolidonyl arylamidase, glycyltryptophan arylamidase, hippurate, N-acetyl-β-glucosaminidase, 4MU-phosphate, 4MU-β-d-glucuronide, p-nitrophenylphosphate, and p-nitrophenyl-α-d-maltoside. None of the strains produced acid from adonitol, d- and l-arabitol, d- and l-arabinose, cyclodextrine, dulcitol, erythritol, d- and l-fucose, α-methyl-d-glucoside, gluconate, 2- and 5-ketogluconate, glycerol, inositol, d-lyxose, melezitose, ribose, sorbitol, l-sorbose, tagatose (except one S. gallolyticus strain), xylitol, d- and l-xylose, and β-methylxyloside.
Varied and differentiating reactions are listed in Table 2.
TABLE 2.
Reactions differing between S. bovis and S. gallolyticus strains and species
| Characteristic | No. of strains
|
|
|---|---|---|
| S. bovis(n = 14) | S. gallolyticus(n = 23) | |
| Alpha-hemolytic | 14 | 2 |
| Browns and blackens on Edwards medium | 14 | 0 |
| Forms watery colonies on Rogosa SL agar | 13 | 1 |
| Clots of litmus milk | 14 | 0 |
| Produces tannase | 0 | 18 |
| Amylase plate test positive | 14 | 18 |
| Produces β-galactosidase | 0 | 4 |
| Produces β-glucuronidase | 0 | 4 |
| Urease negativea | 12 | 15 |
| Hydrolyses: | ||
| 4MU-N-acetyl-β-d-glucosaminidine | 0 | 18b |
| p-Nitrophenyl-β-d-cellobioside | 9 | 23 |
| Produces acid from: | ||
| d-arabinose | 5 | 0 |
| Arbutine | 8 | 23 |
| Mannitol | 0 | 19 |
| Inulin | 10 | 16 |
| Methyl-β-d-glucoside | 3c | 0 |
| Pullulan | 0 | 18 |
| Rhamnose | 0 | 7c |
| Trehalose | 10 | 20 |
| d-Turanose | 2 | 0 |
Negative in BBL CRYSTAL and Rapid ID32 Strep galleries.
Weak reactions were noted in the remaining five S. gallolyticus strains, while no trace of reaction was seen with the S. bovis strains.
Weak and/or delayed reactions.
DISCUSSION
The suitability of highly standardized one-dimensional whole-organism protein electrophoresis for the differentiation of streptococcal species has been demonstrated in several previous studies (18, 19, 20). In the present study this technique allowed the subdivision of strains routinely identified as S. bovis into two distinct clusters.
These clusters contain several strains which have been examined by DNA-DNA hybridization. Two cluster I strains, the S. bovis type strain and LMG 15055, have been shown by Osawa et al. (13) to be closely related and to belong to a DNA-DNA homology group which differs from that of S. gallolyticus. The same strains as well as another cluster I reference strain, LMG 15062, were studied by Farrow et al. (8) and allotted to DNA-DNA homology group 1. Cluster II comprised eight strains identified by Osawa et al. (13) as S. gallolyticus by DNA-DNA hybridization experiments. They include the type strain, five reference strains consigned to homology group 2 by Farrow et al. (LMG 15049, LMG 15053, LMG 15056, LMG 15063, and LMG 15120), and two strains described as S. caprinus (2). Two additional DNA-DNA homology group 2 strains also belonged to this cluster: LMG 15050 and LMG 15051. These results support the synonymy among S. gallolyticus, S. caprinus, and the strains in DNA-DNA homology group 2 of Farrow et al. (8).
Data reported in the literature do not allow easy and reliable differentiation of S. gallolyticus from S. bovis by conventional and miniaturized methods. Farrow et al. (8) found only the test for acid production from mannitol to be a useful differentiating tool among a wide array of biochemical tests. Their DNA-DNA homology group 1 strains were mannitol negative as were similar strains studied by others (2, 13) and the strains of our cluster I (Table 2). However, the differentiating value of this reaction is limited: results obtained in the present work as well as data reported in the literature (8, 13) demonstrate that not all S. gallolyticus strains produce acid from this carbohydrate. In the description of S. caprinus (2) several differentiating reactions that reflect the presence or absence of a trait have been tabulated, but these results are at variance with results obtained in other studies (8, 13) and in our work. The use of special laboratory-prepared media to detect tannase and gallate decarboxylase has been advocated as an additional possibility for differentiation (14, 15); the majority of the S. gallolyticus strains tested were positive in the first test and all were positive in the second test. Tests for certain more easily observable characteristics can be useful alternatives to these tests, as described here (Table 2). Hemolysis, browning and blackening on esculin-containing Edwards agar, and clotting of litmus milk are particularly valuable differentiating characteristics. However, exceptions in individual characteristics may occur. In order to obtain reliable identifications, results of these tests have to be combined with each other and to be supplemented with results from enzymatic and carbohydrate breakdown tests readily available in commercial microtest systems. Most S. gallolyticus strains belong to so-called S. bovis biotype I and a few belong to S. bovis biotype II2 (3). This subdivision is used in the API identification galleries. The majority of the cluster I strains are identified as S. bovis biotype I in these systems. Hydrolysis of 4MU-N-acetyl-β-d-glucosaminide is a very useful differentiating test incorporated in the BBL CRYSTAL gram-positive identification kit (Table 2).
An important aspect of our study concerns host specificity. All strains belonging to S. bovis (cluster I) were isolated from ruminants. The S. gallolyticus strains studied originated mostly from humans and pigeons, but others were from cattle, goats, and a koala. In the study of Osawa et al. (13) S. gallolyticus was identified from humans and a variety of domestic and wild animals while strains showing high DNA-DNA homology values with strain NCDO 597 (S. bovis LMG 8518T in our study) were from cattle and horses.
The frequent association of S. gallolyticus with pathological conditions is noteworthy. All seven human lesion strains included in the study turned out to be S. gallolyticus. An earlier study (11) had already demonstrated the low DNA-DNA homology between the S. bovis type strain ATCC 33317T (LMG 8518T) from cow dung and 10 strains from human infections which were designated at that time as S. bovis and S. bovis variants (7). This association of S. gallolyticus with lesion origin is also evident among the animal strains. Strains identified to date as S. bovis are frequent causes of septicemia and related conditions in pigeons (4, 6) and relatively rare causes of mastitis in cattle (12). All pigeon lesion strains included in our study turned out to be S. gallolyticus (cluster II), and all of the four S. gallolyticus reference strains with known animal host sources in the study of Osawa et al. (13) originated from bovine mastitis specimens while none of their seven reference strains showing high DNA-DNA homology with strain NCDO 597 (cluster I, LMG 8518T) originated from lesions. The association of milk-coagulating and mannitol-positive S. bovis strains, presumably S. gallolyticus (Table 2), with mastitis in cows has already been documented in the older literature (9). However, the association of S. gallolyticus with pathogenicity is far from absolute, as has been demonstrated convincingly for pigeons. These bacteria are normal components of the intestinal floras of pigeons in many lofts, and certain serotypes are more virulent than others (5). It should be noted that one S. gallolyticus strain used in the present study had been isolated from bovine rumen and that one S. bovis strain was from a bovine mastitis specimen. The latter strain was of low pathogenicity in experimental infections (9).
Certain unresolved taxonomical problems concerning S. bovis- and S. equinus-like strains remain to be elucidated. A human strain named MG Fras (3) remained unclassified in the DNA-DNA hybridization study of Osawa et al. (13). This and certain other human clinical strains have been shown to belong to a DNA homology group (3) containing strains of biotype II1. These differed from strains which are presumably S. gallolyticus. Another problem concerns S. equinus. In an earlier DNA-DNA homology study (8), certain strains identified as S. equinus, among which was the type strain, belonged to the same homology group as strains identified as S. bovis, which in our study were also identified as S. bovis (cluster I). S. alactolyticus, an apparently exclusively animal-associated species, is clearly a distinct taxon, although phylogenic affinity with the S. bovis-S. equinus group is evident (1).
ACKNOWLEDGMENTS
P.V. and K.K. are indebted to the Fund for Scientific Research—Flanders (Brussels, Belgium) for research and personal grants, respectively, and for positions as postdoctoral fellows. M.V. was awarded a grant by the Flemish Institute for the Advancement of Research in Industry. Our research was also supported by the Prime Minister’s Services—Federal Office for Scientific, Technical and Cultural Affairs, Brussels, Belgium.
We are grateful to all depositors of the strains examined.
REFERENCES
- 1.Bentley R W, Leigh J E, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- 2.Brooker J D, O’Donovan L A, Skene I, Clarke K, Blackall L, Muslera P. Streptococcus caprinus sp. nov., a tannin-resistant ruminal bacterium from feral goats. Lett Appl Microbiol. 1994;18:313–318. [Google Scholar]
- 3.Coyckendall A L, Gustafson K B. Deoxyribonucleic acid hybridizations among strains of Streptococcus salivarius and Streptococcus bovis. Int J Syst Bacteriol. 1985;35:274–280. [Google Scholar]
- 4.De Herdt P, Haesebrouck H, Devriese L A, Ducatelle R. Biochemical and antigenic properties of Streptococcus bovis isolated from pigeons. J Clin Microbiol. 1992;30:2432–2434. doi: 10.1128/jcm.30.9.2432-2434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Herdt P, Haesebrouck F, Ducatelle R, De Groote B, Devriese L A. Streptococcus bovis infections in pigeons: virulence of different serotypes. Vet Microbiol. 1994;41:321–332. doi: 10.1016/0378-1135(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 6.Devriese L A, Uyttebroek E, Gevaert D, Van De Kerckhove P, Ceyssens K. Streptococcus bovis infections in pigeons. Avian Pathol. 1990;19:429–434. doi: 10.1080/03079459008418697. [DOI] [PubMed] [Google Scholar]
- 7.Facklam R R. Recognition of group D streptococcal strains of human origin by biochemical and physiological tests. Appl Microbiol. 1972;23:1131–1139. doi: 10.1128/am.23.6.1131-1139.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrow J A E, Kruze J, Phillips B A, Bramley A J, Collins M D. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp. nov. Syst Appl Microbiol. 1984;5:467–482. [Google Scholar]
- 9.Garvie E I, Bramley A J. Streptococcus bovis—an approach to its classification and its importance as a cause of bovine mastitis. J Appl Bacteriol. 1979;46:557–566. doi: 10.1111/j.1365-2672.1979.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardie J M. Other streptococci. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1068–1071. [Google Scholar]
- 11.Knight R G, Shlaes D M. Physiological characteristics and deoxyribonucleic acid relatedness of human isolates of Streptococcus bovis and Streptococcus bovis (var.) Int J Syst Bacteriol. 1985;35:357–361. [Google Scholar]
- 12.McDonald T J, McDonald J S. Streptococci isolated from bovine mammary tract infections. Am J Vet Res. 1976;37:377–381. [PubMed] [Google Scholar]
- 13.Osawa R, Fujisawa T, Sly L I. Streptococcus gallolyticus sp. nov.; gallate degrading organisms formerly assigned to Streptococcus bovis. Syst Appl Microbiol. 1995;18:74–78. [Google Scholar]
- 14.Osawa R, Walsh T P. A visual reading method for detection of bacterial tannase. Appl Environ Microbiol. 1993;59:1251–1252. doi: 10.1128/aem.59.4.1251-1252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osawa R, Walsh T P, Cork S J. Metabolism of tannin-protein complex by facultatively anaerobic bacteria isolated from koala feces. Biodegradation. 1993;4:91–99. [Google Scholar]
- 16.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O’Donnell A G, editors. Modern microbial methods. Chemical methods in bacterial systematics. Chichester, United Kingdom: J. Wiley and Sons; 1994. pp. 493–521. [Google Scholar]
- 17.Sly L I, Cahill M M, Osawa R, Fujisawa T. The tannin-degrading species Streptococcus gallolyticus and Streptococcus caprinus are subjective synonyms. Int J Syst Bacteriol. 1997;47:893–894. doi: 10.1099/00207713-47-3-893. [DOI] [PubMed] [Google Scholar]
- 18.Vandamme P, Pot B, Falsen E, Kersters K, Devriese L A. Taxonomic study of Lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–781. doi: 10.1099/00207713-46-3-774. [DOI] [PubMed] [Google Scholar]
- 19.Vandamme P, Devriese L A, Pot B, Kersters K, Melin P. Streptococcus difficile is a nonhemolytic group B, type Ib streptococcus. Int J Syst Bacteriol. 1997;47:81–85. doi: 10.1099/00207713-47-1-81. [DOI] [PubMed] [Google Scholar]
- 20.Vandamme P, Torck U, Falsen E, Pot B, Goossens H, Kersters K. Whole-cell protein electrophoretic analysis of viridans streptococci: evidence for heterogeneity among Streptococcus mitis biovars. Int J Syst Bacteriol. 1998;48:117–125. doi: 10.1099/00207713-48-1-117. [DOI] [PubMed] [Google Scholar]
- 21.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]