Abstract
Drug-induced liver injury (DILI) is a significant concern in drug development, often leading to drug withdrawal. Although many studies aim to identify biomarkers and gene/pathway signatures related to liver toxicity and aim to predict DILI compounds, this remains a challenge in drug discovery. With a strong development of high-content screening/imaging (HCS/HCI) for phenotypic screening, we explored the morphological cell perturbations induced by DILI compounds. In the first step, cell morphological signatures were associated with two datasets of DILI chemicals (DILIRank and eTox). The mechanisms of action were then analyzed for chemicals having transcriptomics data and sharing similar morphological perturbations. Signaling pathways associated with liver toxicity (cell cycle, cell growth, apoptosis, ...) were then captured, and a hypothetical relation between cell morphological perturbations and gene deregulation was illustrated within our analysis. Finally, using the cell morphological signatures, machine learning approaches were developed to predict chemicals with a potential risk of DILI. Some models showed relevant performance with validation set balanced accuracies between 0.645 and 0.739. Overall, our findings demonstrate the utility of combining HCI with transcriptomics data to identify the morphological and gene expression signatures related to DILI chemicals. Moreover, our protocol could be extended to other toxicity end points, offering a promising avenue for comprehensive toxicity assessment in drug discovery.
Introduction
The general success rate to bring a drug candidate to market is estimated at 8%.1 One of the most significant reasons for attrition is adverse clinical side effects and toxicity. Since drug-induced liver injury (DILI) is a major reason for the withdrawal of drugs,2 being able to detect molecules with the potential risk of DILI as early as possible during drug development is a crucial issue in drug discovery.
Many studies have been performed to identify biomarkers and gene/pathway signatures related to liver toxicity based on toxicogenomics studies3−9 or machine learning models.10−14 In all these studies, it is assumed that chemicals having a similar structure or inducing specific gene expression profiles to known DILI chemicals can be identified as putatively toxic based on shared compound structures or common mechanisms of response at the molecular level. Nevertheless, the challenge remains to be able to predict the potential toxicological profile of a drug in humans and to capture the underlying mechanistic events associated with this toxicity.
Recently, with the advances in automated fluorescence microscopy and image analysis pioneered by Anne Carpenter and her team, there was a reinvestment in high-content screening/imaging (HCS/HCI) and notably in cell morphology analysis provided by the Cell Painting assay.15 The Cell Painting assay uses a mixture of 6 fluorescent dyes to stain different compartments of a cell (nucleus, cytoplasm, plasma membrane, ...), which are then represented by morphological features (i.e., shape, intensity, and texture, among others).16 Such an assay originally uses the human osteosarcoma U2OS immortalized cell line and allows the investigation of the morphological perturbation profiles induced by large sets of chemicals to inform on phenotypic changes associated with different modes of action (MoAs).17,18
However, although HCS/HCI allows the observation of the morphological perturbation of a cell caused by a chemical, it does not provide direct readouts on the molecular mechanism that underlies the cell perturbation, and the results obtained need to be interpreted using dedicated specific approaches.19 The association of molecular and cellular features to be able to describe biological effects has started to be reported.20−23 Furthermore, Cell Painting data have been combined with transcriptomics data to assess the potential link between transcriptomic changes and the corresponding alterations in morphology.24−26 Overall, these studies highlighted that the relation between gene expression and morphology profiles is challenging, and the choice of cell line has a non-negligible effect on the predictive accuracy.27
In this context, the objective of this study was to analyze the morphological perturbations of cells induced by DILI chemicals that are part of the Cell Painting data released by the Broad Institute. Specific MoAs based on transcriptomics data for sets of DILI chemicals that share similar Cell Painting features were determined. Concerning the liver toxicity data, two datasets of chemicals were considered, one based only on human data (DILIrank)28 and a second one (eTox) which classifies DILI versus non-DILI chemicals combining human and rat information from clinical reports provided by pharmaceutical companies.12 Based on selected morphological cell features from the first analysis, machine learning approaches were developed on these 2 datasets to predict chemicals with a potential risk of DILI. Importantly, the outcome of this analysis showed that DILI chemicals sharing similar morphological perturbation profiles also have similar pathways associated with liver toxicity (cell cycle, RNA splicing, ...).
Materials and Methods
Steps of the complete analysis workflow of this study are summarized in Figure 1, and each step is detailed below.
Figure 1.
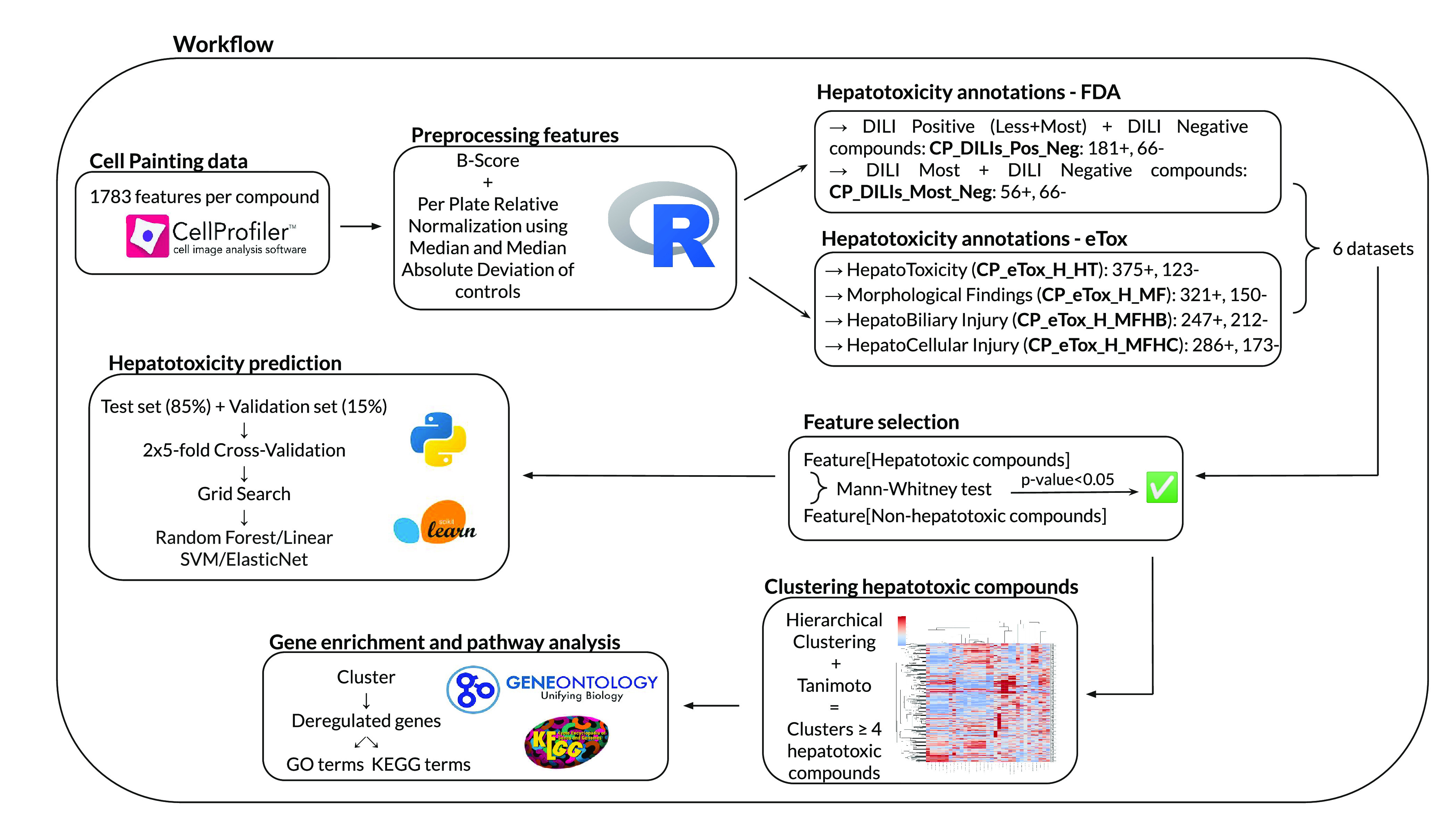
Schematic representation of the complete study workflow. Cell Painting data were preprocessed, 6 datasets related to 6 hepatotoxicity end points were created using annotations from the DILIrank and eTox databases, features significantly discriminative of hepatotoxicity were selected and used to build clusters of hepatotoxicity compounds before gene enrichment, and pathway analyses were performed on them. Finally, machine learning models were trained to test the predictive potential of Cell Painting features regarding DILI.
Cell Painting Data
We used a publicly available dataset provided by the Broad Institute, containing morphological features for 30 616 chemical components tested in the Cell Painting assay15,29 using the human osteosarcoma U2OS immortalized cell line. A total of 406 384-well plates were used, each containing 64 placed DMSO controls. Each compound had 4 to 12 replicates spread across different plates. The dataset provided 1783 features at a per-well level. Features were already extracted using CellProfiler image analysis software30 and were sorted at three levels:
The first is the compartment related to the measurement, including “Cell” in its entirety, “Cytoplasm”, and “Nuclei”.
The second is the category and type of measurement (10 categories are available: “AreaShape”, “Correlation”, “Granularity”, “Intensity”, “Location”, “Neighbors”, “RadialDistribution”, “Texture”, “Parent”, and “Number”).
Lastly, the third level is for cell compartments and organelles stained during the Cell Painting assay: “DNA” for the nucleus, “ER” for the endoplasmic reticulum, “RNA” for nucleolus and cytoplasmic RNA, “AGP” for F-actin cytoskeleton, Golgi, and plasma membrane, and “Mito” for mitochondria.
Preprocessing
Data Curation
We excluded 9 plates from the 406 plates included in the Broad Institute dataset because more than 20% of wells were without values. Plates with less than 20% of missing values (30 plates) are completed with “NA” values to be normalized and used in the study. Features without variance are also excluded from the dataset, bringing the number of features from 1783 to 1779.
Normalization
Normalization comprises a plate-layout effect correction and a feature transformation. R software (v4.0.4) is used to normalize the data. The plate-layout effect is corrected using a two-way median polish, also called B-Score.31 The B-Score is applied to each plate individually. We have checked that control wells are distributed uniformly across the plate, as this is required to apply the B-Score. The R package platetools (v0.1.5) already provides an implemented version of the B-Score method (Warchal Scott (Last updated: 2021, June), Tools and Plots for Multi-Well Plates. Retrieved from https://cran.r-project.org/web/packages/platetools/platetools.pdf). For each row of a plate, the row median is calculated. The median of the row medians, i.e. the row overall effect, is deduced. We subtracted the row median from the wells for all rows and then subtracted the overall effect from each row median and repeated all the last steps for columns. We added the column overall effect to the row overall effect. Finally, we repeated the loop until the global overall effect became unchanged.32
Z-Score relative normalization is commonly used in biology to put values of all features in the same range, which is frequently necessary for downstream analysis algorithms.33 Because it is based on mean and standard deviation, Z-Score may be very sensitive to outliers. We chose here a similar normalization called Median and Median Absolute Deviation (MMAD) that centers and scales data using median and median absolute deviation features with respect to a control population34 (i.e., DMSO).
Finally, features were concatenated at a compound level by grouping replicates using the median.
Hepatotoxicity Annotations
Hepatotoxicity annotations of the Cell Painting compounds were collected from two datasets:
DILIrank
The DILIrank dataset was provided by the Food and Drug Administration (FDA)28 and contains annotations for 1036 FDA-approved drugs according to their capacity to induce DILI in humans. Drugs were classified into 4 categories: “No-DILIconcern”, “Less-DILIconcern”, “Most-DILIconcern”, and “Ambiguous-DILIconcern”. 309 molecules from the DILIrank dataset have been tested on Cell Painting. Respectively, 62 compounds were annotated as Ambiguous-DILIconcern, 66 as No-DILIconcern, 125 as Less-DILIconcern, and 56 as Most-DILIconcern. The analysis on DILIrank was performed on 2 sets combining different categories: one dataset named CP_DILIs_most_neg, containing compounds marked as Most-DILIconcern and No-DILIconcern. The second dataset, named CP_DILIs_pos_neg, combined Less-DILIconcern and Most-DILIconcern (defined as Positive-DILIconcern) and No DILIconcern. Ambiguous compounds were excluded from this study.
eTox
eTox contains hepatotoxicity data for 3712 compounds from preclinical reports collected by pharmaceutical industries. It includes human and rodent findings, which were classified into different interlocked categories from general hepatotoxicity to more specific morphological and clinical findings.12 We focused our analysis on four of these end points: “Human Hepatotoxicity” (H_HT), “Human Morphological Findings” (H_MF), “Human Morphological Hepatobiliary Injury” (H_MFHB), and “Human Morphological Hepatocellular Injury” (H_MFHC), for which 498, 471, 459, and 459 compounds, respectively, were tested in Cell Painting. We built 4 datasets representing these 4 end points: CP_eTox_H_HT, CP_eTox_H_MF, CP_eTox_H_MFHB, and CP_eTox_H_MFHC.
Definitions of the 6 end point datasets’ names obtained from DILIrank and eTox are explained in Table 1. DILIrank and eTox compounds’ annotations are available in Table S1 in SI (Supporting Information).
Table 1. Term Definitions of End Point Datasets Created from DILIrank and eTox Databases.
| Dataset name | Definition |
|---|---|
| CP_DILIs_pos_neg | Compounds tested in the Cell Painting assay and annotated as Positive-DILIconcern (either Less-DILIconcern or Most-DILIconcern) or No-DILIconcern in the DILIrank database |
| CP_DILIs_most_neg | Compounds tested in the Cell Painting assay and annotated as Most-DILIconcern or No-DILIconcern in the DILIrank database |
| CP_eTox_H_HT | Compounds tested in the Cell Painting assay and annotated as positive or negative regarding the “Human Hepatotoxicity” (H_HT) end point in the eTox database |
| CP_eTox_H_MF | Compounds tested in the Cell Painting assay and annotated as positive or negative regarding the ‘Human Morphological Findings’ (H_MF) end point in the eTox database |
| CP_eTox_H_MFHB | Compounds tested in the Cell Painting assay and annotated as positive or negative regarding the ‘Human Morphological HepatoBiliary’ (H_MFHB) end point in the eTox database |
| CP_eTox_H_MFHC | Compounds tested in the Cell Painting assay and annotated as positive or negative regarding the ‘Human Morphological HepatoCellular’ (H_MFHC) end point in the eTox database |
Feature Selection
A dataset-specific feature selection was processed. For each of the six reduced datasets (2 from DILIrank and 4 from eTox), we excluded features without variance. For each feature, the means of the values of both the positive and negative compounds were calculated, and a Mann–Whitney test was then calculated to select features that showed a significant difference (p-value < 0.05) between hepatotoxic and non-hepatotoxic compounds. For each dataset, Fisher’s Exact Tests were performed to identify among the feature selection significantly enriched CellProfiler feature categories.
Clustering of Similar Hepatotoxic Compounds
Hierarchical clustered heatmaps (i.e., clustermaps) were calculated with the Python seaborn package (v0.11.2) based on features selected in the previous step. It allowed grouping of chemicals sharing a similar cell morphological profile. The scale was set up at 0 and calculated with minimum and maximum, respectively, set at the 25th and 75th percentiles.
As we were not interested in non-hepatotoxic compounds at this stage, only DILI compounds of each dataset were selected to build the related clustermaps. The procedure of clustering was repeated for each of the 6 datasets. First, linkage matrices were extracted from compound dendrograms of clustermaps and used to create clusters of DILI compounds sharing a similar cell morphological profile. Different clusters were formed according to the distance’s threshold considered (going from 1 to 0.1) with an interval of 0.1, i.e. an increasingly stringent threshold. Thus, we iterated through the initial clusters to split clusters that were initially too wide. To obtain final clusters, a threshold cluster size was defined as the mean size of the initial clusters at a threshold of 0.8. Initial clusters smaller than this size were kept as they were. For larger than the initial clusters’ mean size, a continuous Tanimoto coefficient was calculated on the features for each combination of compounds in cluster35 (eq 1). Contrary to the classical Tanimoto coefficient used with binary encoded chemical structures, the continuous Tanimoto coefficient is used with the continuous values collected from the morphological features. If the mean score of all coefficients calculated for the cluster was superior or equal to 0.7, i.e. 70% of similarities between compounds, the cluster was then kept as it was. Otherwise, it was split regarding the next smaller distance threshold. This loop had a smaller size than the threshold cluster size or until the smallest threshold distance of 0.1 was reached. Thereafter, only clusters with at least 4 compounds were considered for further analysis.
| 1 |
The scaling is different from one dataset compared to another one, as it is dependent on the set of features. The scaling was used only at this stage and not for the machine learning approaches.
Gene Enrichment and Pathway Analysis
Once the hepatotoxic chemicals were grouped by morphological features, we were interested to see if they also shared similar biological processes. To do so, both gene and pathway enrichments were performed for our clusters of DILI compounds using a list of differentially expressed genes, provided by the publicly available L1000 mRNA profiling assay36 (https://lincsproject.org/LINCS/tools/workflows/find-the-best-place-to-obtain-the-lincs-l1000-data). L1000 data for 19 811 chemicals were available at different levels of preprocessing, going from 0 for raw data to 5 for differential gene expression signatures. In order to extract a list of differentially expressed genes by our hepatotoxic compounds of interest, we used the L1000 data at level 4, i.e. “gene signatures computed using Z-scores relative to a population of controls”. All differentially expressed genes by DILI compounds belonging to the same cluster and tested after 6 h of treatment, in a HepG2 cell line, and at a concentration of 10 μM (the condition for which we have the highest number of chemicals with transcriptomics data) were collected. To get only the most perturbed genes, only those with a Z-score below −3.00 (downregulated) and above 3.00 (upregulated) were considered. Gene enrichment and pathway analysis were then performed to identify gene sets and biological pathways that could be disrupted by similar hepatotoxic compounds. The Gene Ontology37,38 (GO) was used for the study of gene enrichment whereas the KEGG database39−41 was used for pathways analysis. Among all the available GO terms, we focused only on terms dealing with biological processes (BPs) and cellular components (CCs).
Enrichment was performed using the R package clusterProfiler (v4.2.2).42 We selected significant GO terms and KEGG pathways with an adjusted (with a Bonferroni correction) p-value lower than 0.05. Heatmaps were applied to visualize pathways that were representative of clusters regarding their adjusted p-value and to study the similarity between clusters of hepatotoxic compounds in the same dataset. Clusters in which only one compound had gene expression data were excluded from the analysis.
As some clusters were not considered in the analysis because of a size smaller than 4 or the lack of gene expression data, the final cluster numbering used in the study, starting with 1, is not continuous.
Tanimoto Coefficient Calculation
The structural similarity between two molecules was calculated using the Tanimoto coefficient. To do so, we used the Open Babel program43 (The Open Babel Package, version 3.0.0, http://openbabel.org, accessed Oct 2022) and the MACCS fingerprints.
Comparison of Classification Models Based on DILIrank and eTox
With the objective of assessing the relevance of morphological features selected with the DILIrank and eTox datasets in the prediction of DILI compounds, computational models were performed using three machine learning models, Random Forest (RF), Linear Support-Vector-Machine (SVM), and ElasticNet, implemented in Python (v3.9.5). For each dataset, data were split into a training dataset (85%) and a validation dataset (15%), keeping the same proportion of hepatotoxic and non-hepatotoxic compounds in each dataset. For Linear SVM and ElasticNet, the datasets were standardized before prediction. Each model was developed using the scikit-learn package (v1.0.1). Stratified 2 times 5-fold cross-validation was included in the training procedure. For each model, we performed hyperparameter tuning with Grid Search optimization. The details of the parameter settings and model performance assessment are available in Table S2 in SI. Model efficiency was measured with balanced accuracy (BA), specificity (SP), and sensitivity (SN) metrics of which formulas are available in Table S3 in SI. The entire model-fitting and validation process was repeated on permuted datasets too. Parameter settings of models previously optimized and trained on real data were selected instead of a Grid Search optimization. Permutation tests were repeated 100 times. Finally, we compared the machine learning performance of ElasticNet models for hepatotoxicity prediction based on Cell Painting features with Recursive Feature Elimination (RFE) included in the loop of training. We used a RF algorithm as RFE estimator. Stratified 2 times 5-fold cross-validation was included in the training procedure.
Results
Overlapping between DILIrank and eTox Compounds and Annotations
To account for the different information provided by DILIrank and eTox, we created 6 different datasets corresponding to the different end points. DILIrank gave information on human hepatotoxicity, while eTox annotations were collected from preclinical studies on rodents. We used DILIrank annotations to create two datasets: CP_DILIs_pos_neg and CP_DILIs_most neg. CP_DILIs_pos_neg regrouped DILIrank non-hepatotoxic compounds, and all the compounds annotated as hepatotoxic regardless of the level of hepatotoxicity (Less-DILI and Most-DILI). CP_DILIs_most_neg included only the most hepatotoxic compounds (Most-DILI) in addition to the non-hepatotoxic compounds. The eTox datasets were nested and provided more specific information regarding the type of hepatotoxicity, going from general hepatotoxicity (CP_eTox_H_HT) to hepatotoxic morphological findings (CP_eTox_H_MF) that included morphological findings at both hepatobiliary and hepatocellular levels (respectively CP_eTox_H_MFHB and CP_eTox_H_MFHC).
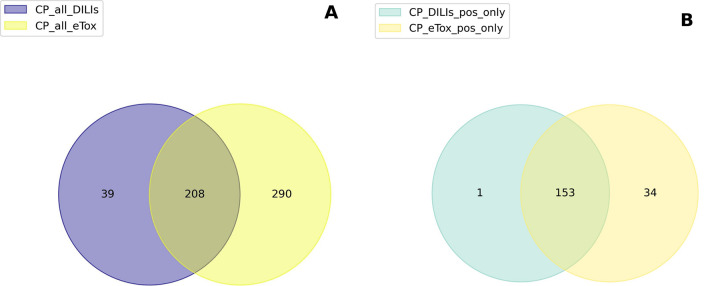
DILIrank and eTox contained annotations at all levels for 247 and 498 compounds, respectively, with 208 compounds in common between the two datasets (Figure 2A). We checked if DILIrank and eTox agreed on their annotations of hepatotoxicity. To do that, we compared the common positive compounds of CP_DILIs_pos_neg and CP_eTox_H_HT and found that out of 188 compounds, 153 were annotated as hepatotoxic in both DILIrank and eTox (Figure 2B). Only 1 compound was defined as hepatotoxic in DILIrank and non-hepatotoxic in eTox, compared to 34 compounds mentioned as hepatotoxic in eTox and non-hepatotoxic in DILIrank. The overall agreement in hepatotoxicity annotations between DILIrank and eTox tallies at 81%.
Figure 2.
Venn diagrams of compounds and annotations in common between the DILIrank and eTox datasets: (A) DILIrank compounds vs eTox compounds; (B) hepatotoxic DILIrank compounds vs hepatotoxic eTox compounds.
Identification of Morphological Features Associated with Hepatotoxicity
Cell Painting data initially provided 1783 features per compound. After removing features without variance, 1779 features remained. A high number of features can frequently lead to information redundancy, over-representation of some features, and noise, making the interpretation more challenging.44,45 Therefore, we performed feature selection in the workflow of Cell Painting data analysis to reduce the number of features by keeping only the most informative ones regarding the biological question of interest, in this case hepatotoxicity.
Feature selection, based on a Mann–Whitney test, was processed on each dataset separately to select significant features that discriminate between DILI and non-DILI compounds. Morphological features with a p-value lower than 0.05 were considered significant and selected for further investigation (Table 2). The list of selected features per dataset is detailed in Table S4 in SI.
Table 2. Number of Features per Dataset after Feature Selection with the Mann-Whitney Test.
| Number of DILI-Discriminative Selected Features | ||||||
|---|---|---|---|---|---|---|
| Dataset | CP_DILIs_pos_neg | CP_DILIs_most_neg | CP_eTox_H_HT | CP_eTox_H_MF | CP_eTox_H_MFHB | CP_eTox_H_MFHC |
| Number of DILI compounds | 181 | 56 | 375 | 321 | 247 | 286 |
| Number of nonDILI compounds | 66 | 66 | 123 | 150 | 212 | 173 |
| Number of selected features | 149 | 335 | 33 | 49 | 83 | 73 |
The number of selected features was more stringent for the eTox datasets (33 to 83 features) compared to the DILIrank datasets (149 and 335 features). Among the DILIrank datasets, the Mann–Whitney test selected the highest number of features with the CP DILIs_most_neg dataset (335 features). The inclusion of Less-DILIconcern compounds within the Most-DILIconcern compounds increased the number of features that discriminated significantly (p-value < 0.05) from the No-DILIconcern compounds. In contrast, the observation was different with the eTox datasets, although the number of compounds was larger. CP_eTox_H_HT, containing the less specific annotations of hepatotoxicity in the eTox classification, had the smallest number of selected features (33 features). On the contrary, CP_eTox_H_MF, CP_eTox_H_MFHB, and CP_eTox_H_MFHC, which contained hepatotoxicity annotations resulting from more specific observations at a morphological level, were less stringent in terms of feature selection (49 to 83 features).
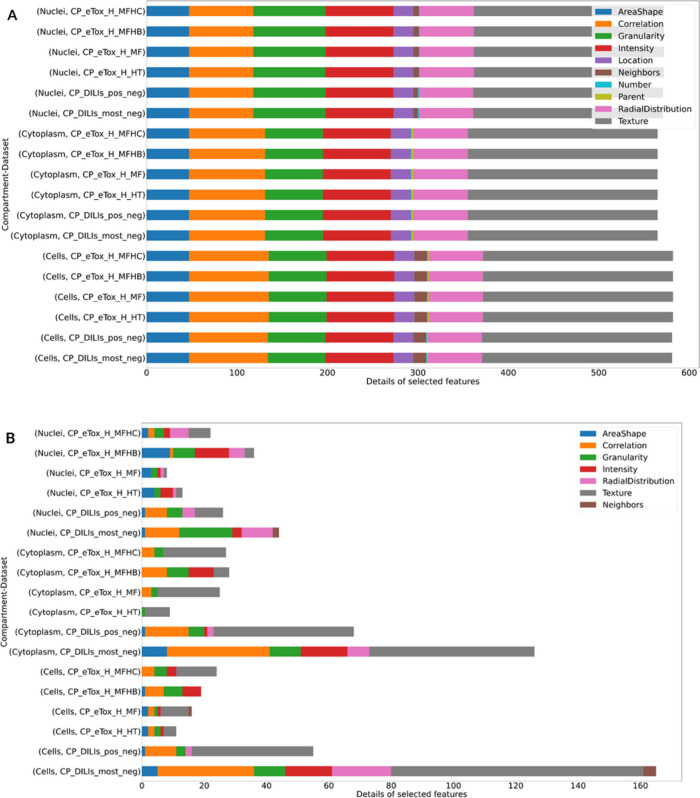
Regarding the analysis of cell morphology features, they were examined before and after feature selection in the function of the cell’s compartments, i.e. “Cell”, “Cytoplasm”, “Nuclei”, and the feature categories (Figure 3). Initially, it is clear that there is approximately the same number of features assigned to the three compartments (Figure 3A). Category proportion was almost the same for each compartment. Features belonging to the “Texture” category represented more than one-third of the total features whereas “Location”, “Neighbors”, “Number”, and “Parent” features were under-represented (0 to 4%) (Table S5 in SI). The remaining categories accounted for 8% to 15% of the features.
Figure 3.
Details of number of features per category in each compartment and dataset: (A) before feature selection and (B) after feature selection. The figures represent the number of features in each category of CellProfiler measurements and sorted the analysis by CellProfiler compartments and datasets. Percentages of feature categories in each compartment and dataset before and after feature selection are available in Tables S5 and S6 in SI, and the as ratio of percentages in the selected features in Table S7.
After feature selection and according to the dataset, the number of selected features per compartment was of comparable magnitude (Figure 3B). Among the measurement categories, only 7 categories out of 10 were represented after selection: “AreaShape”, “Correlation”, “Granularity”, “Intensity”, “RadialDistribution”, “Texture”, and “Neighbors” (Figure 3B). However, the “Location”, “Parent”, and “Number” categories were already under-represented before feature selection (Figure 3A). The “Texture”, “Correlation”, and “Granularity” categories were the most represented in the feature selection for both the DILIrank and eTox datasets, as confirmed by the ratio of their percentages in the selective features relative to the background percentage in the complete set of features before selection (Table S7). “Texture” measurements pertain to the general aspect of objects, i.e. the cell in its entirety, cytoplasm, and nuclei, and especially their roughness and smoothness. “Correlation” measurements represent the correlation between pixel intensities in different images. “Granularity” measurements are texture measurements that take into account the global image more than objects by trying to fit a granular spectrum to the image. “Texture” measurements also appeared to be more represented in “Cell” and “Cytoplasm” compartments, while “RadialDistibution” and “AreaShape” were more specific to the “Nuclei” compartment (Tables S6 and S7). The “RadialDistribution” category measures the spatial distribution of pixel intensities in cells, cytoplasm, and nuclei. “AreaShape” applies to the image area that is occupied by objects like the cell, cytoplasm, or nucleus and to the shape of these objects. However, Fisher’s Exact Tests revealed that “Texture” was the only significantly enriched feature category, in the CP_DILIs_pos_neg, CP_DILIs_most_neg, CP_eTox_H_MF, and CP_eTox_H_MFHC datasets for the “Cells” compartment and in every dataset except CP_eTox_H_MFHB for the “Cytoplasm” compartment (Table S8).
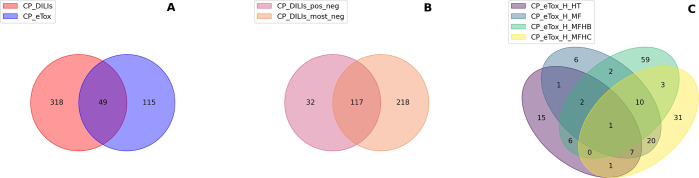
In the subsequent analysis, we examined the overlap of selected features across datasets. We first intersected features selected in at least one of the two DILIrank datasets and those that were selected in at least one of the eTox datasets, resulting in 49 common features (Figure 4A). Out of these 49 features, 8 were associated with the nucleus, 23 with the cytoplasm, and 18 with the cell. The majority of these common features (30) fell under the “Texture” category. We then compared the features selected in each individual DILIrank dataset. It appeared that CP_DILIs_pos_neg shared 78% of its selected features with CP_DILIs_most_neg (Figure 4B), with the majority being cell and cytoplasm features (respectively, 45% and 45%). The selected features of the categories “Texture”, “Granularity”, and “Correlation” were consistently in the majority. Conversely, we found less similarity among the selected features in the eTox datasets (Figure 4C). CP_eTox_H_MF and CP_eTox_H_MFHC exhibited the highest similarity, sharing 38 out the 49 selected features. Interestingly, only 1 feature, “Cytoplasm_Texture_Entropy_AGP_5_0″, was common to all the eTox datasets.
Figure 4.
Venn diagrams of DILI-discriminative features in common between datasets after feature selection: (A) merged DILIrank datasets vs merged eTox datasets; (B) CP_DILIs_pos_neg vs CP_DILIs_most_neg; (C) CP_eTox_H_HT vs CP_eTox_H_MF vs CP_eTox_H_MFHB vs CP_eTox_H_MFHC.
Clustering of Compounds and Features
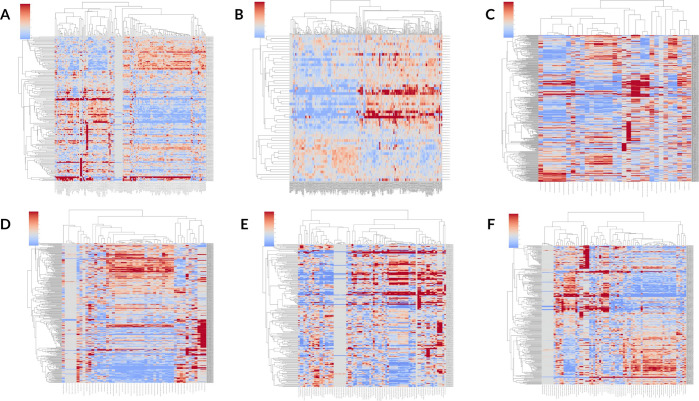
To investigate the relationship between Cell Painting features and hepatotoxicity indications from the DILI compounds of our datasets, hierarchical-clustered heatmaps (clustermaps) of DILI compounds were built based on the morphological features previously selected. Clustermaps are presented in Figure 5 with hepatotoxic compounds in rows and cell morphological features in columns.
Figure 5.
Clustermaps based on DILI-discriminative features of DILI-compounds of each dataset only: (A) CP_DILIs_pos_neg; (B) CP_DILIs_most_neg; (C) CP_eTox_H_HT; (D) CP_eTox_H_MF; (E) CP_eTox_H_MFHB; (F) CP_eTox_H_MFHC. Each clustermap is related to a dataset. Only hepatotoxic compounds of each dataset were clustered. Features are clustered in columns, and compounds are clustered in rows. A spectrum of color is centered on zero (gray) from dark blue (negative value) to dark red (positive value) and calculated with minimum and maximum, respectively, set at the 25th and 75th percentiles of feature values.
Interestingly, the features were not grouped either by CellProfiler compartments or categories after clustering. In addition, some features exhibited minimal variation across the compounds, as depicted by the gray areas in Figure 5A and D–F. In fact, variations of these features were linked to only a few extreme compounds, i.e., features with extreme values.
For each dataset, clusters of compounds could be visible in the dendrogram as well as feature patterns (red for compounds having a higher morphological feature value than the mean and blue for compounds having a lower morphological feature value than the mean). It can be hypothesized that compounds of the same cluster, i.e., compounds with close values on the same morphological features, have similar effects on cells, i.e., associated in our case with hepatotoxic effects. Since DILI can stem from different biological pathways, this could explain why DILI compounds were not all grouped together in the clustermap. To further investigate the mechanistic link between liver toxicity and clusters of DILI compounds, we extracted clusters of similar hepatotoxic compounds using the dendrogram performed on the compound side (on the left of the clustermap) (see Materials and Methods) and we performed enrichment analysis for chemicals having in vitro gene deregulations in the HepG2 cell line under the same conditions (when available).
From the dendrogram, we obtained 15 clusters for CP_DILIs_pos_neg, 6 for CP_DILIs_most_neg, 32 for CP_eTox_H_HT, 34 for CP_eTox_H_MF, 22 for CP_eTox_H_MFHB, and 28 for CP_eTox_H_MFHC, with at least 4 chemicals (Table S9). The cluster sizes ranged from 4 to 20 chemicals. Further details of the number of clusters and their mean size are provided in Table S10 in SI.
Hierarchical clustering on Cell Painting features allowed us to gather compounds that did not necessarily share structural similarities or similar pharmacological properties but that had similar morphological signatures on cells presumably associated with hepatotoxicity. For example, cluster 26 extracted from the CP_DILIs_pos_neg dataset contained 4 compounds: Paclitaxel, Mebendazole, Etoposide, and Amlodipine. Although Paclitaxel and Etoposide are known chemotherapeutic agents,46,47 they are not structurally similar, as they have a Tanimoto coefficient of 0.49 (calculated with Open Babel). More generally, none of the compounds of cluster 26 was structurally similar to another. Mebendazole and Amlodipine are also used for different pharmacological purposes, as they are anthelmintic and antihypertensive agents, respectively.48,49 However, all 4 compounds were associated with liver toxicity in the DILIrank database and may have similar MoAs for the induction of hepatotoxicity.
Enrichment and Pathway Analysis
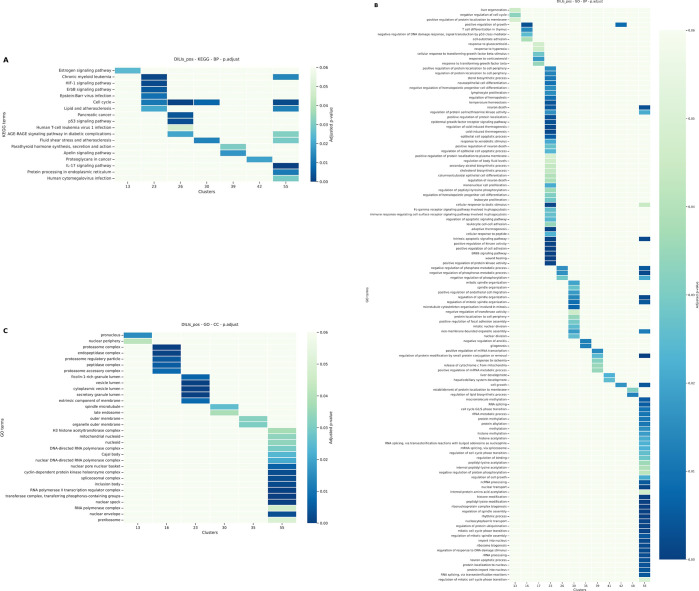
Based on the hypothesis that chemicals having similar DILI-discriminative morphological features may have similar mechanisms of action, we collected transcriptomics data from compounds tested in L1000 and we performed enrichment analysis with DILI chemicals grouped in the same clusters. As our biological organ is the liver, chemicals tested on HepG2 cells at a concentration of 10 μM and after 6 h of treatment were considered. We used the GO and the KEGG databases and focused on terms dealing with BP and CC. To visualize the results of the pathway analysis, a heatmap per enrichment type and per dataset was built. Results for the CP_DILIs_pos_neg dataset are presented and discussed in Figure 6).
Figure 6.
Heatmaps of pathways significantly deregulated by clusters of hepatotoxic compounds from the CP_DILIs_pos_neg dataset. A. KEGG pathway analysis. B. GO BP pathway analysis. C. GO CC pathway analysis. Color scale is related to p−value. P−values equal or above 0.05 are in light green.
Based on the hierarchical clustering on the CP_DILIs_pos_neg dataset, 15 clusters were further investigated for gene enrichment and pathways analysis. Interestingly, the KEGG enrichment analysis (Figure 6) revealed that the cell cycle was significantly impacted by DILI chemicals in clusters 13, 26, 30, and 55. Among the genes that contribute to cell cycle modulation, SMC1A and WEE1 are significantly deregulated by these 4 clusters. SMC1A is a gene belonging to the structural maintenance of the chromosome family. It is implicated in DNA damage repair and plays a role during the mitosis stage of the cell cycle.50 WEE1 regulates the entry in mitosis too and its deregulation is known to be associated with several types of cancer, including hepatocellular carcinoma.51 These 2 genes, expressed in HepG2, are also expressed in U2OS, as reported by the Human Protein Atlas (https://www.proteinatlas.org).
Subpathways of the cell cycle such as the p53-dependent apoptotic pathway or the mitotic cell cycle phase also appeared in our enrichment analysis (Figure 6). To ensure that the deregulation of the cell cycle was not biased by the cytotoxicity of the compounds in clusters 13, 26, 30, and 55, we examined the cytotoxic median concentrations of available compounds using the CompTox Chemistry Dashboard52 (https://comptox.epa.gov/dashboard/, v2.2). Except for Mebendazole, compounds in clusters 13, 26, 30, and 55 (Metoprolol, Omeprazole, Clomipramine, Tamoxifen, Progesterone, Lovastatin, and Simvastatin) had a cytotoxic median concentration higher than the 10 μM concentration used in the enrichment analysis.
Compounds belonging to cluster 55 exhibited differentially expressed genes associated with the RNA splicing (Figure 6), as exemplified by CASC3.53 GO enrichment analysis at the cellular component level corroborated this implication, as spliceosome was also mentioned for this cluster (Figure 6). RNA alternative splicing is a process that transforms pre-messenger RNAs into messenger RNAs.
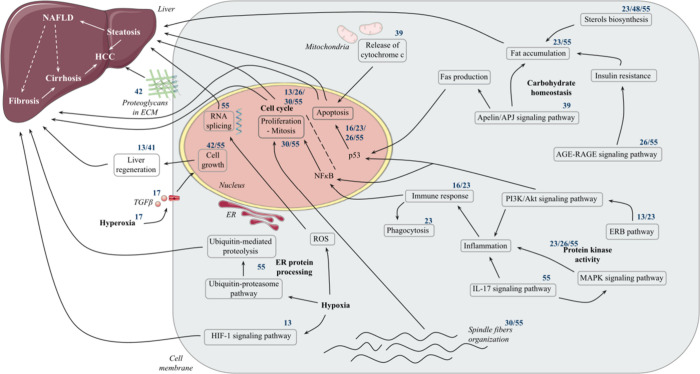
Figure 7 summarizes and illustrates the relationships between all clusters/pathways and cellular compartments associated with morphological perturbations.
Figure 7.
Schematic representation of pathway analysis results for clusters of the CP_DILIs_pos_neg dataset and their link to different hepatic pathologies. Cluster numbers are specified in superscript above the involved pathway or hepatic pathology. Clusters 6, 27, 31, 35, and 48 are not included in the figure due to the lack of informative pathway analysis results. ECM, extracellular matrix; ER, endoplasmic reticulum; HCC, hepatocellular carcinoma; NAFLD, nonalcoholic fatty liver disease. The figure was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
The analysis of the remaining pathways for the CP_DILIs_pos_neg dataset, involving pathways related to the immune response and kinase activity, the carbohydrate homeostasis, the endoplasmic reticulum (ER) protein processing, the hypoxia-inducible factor 1 (HIF-1) signaling pathway, response to hyperoxia, proteoglycans in cancer, spindle fibers’ disorganization, and the release of cytochrome c from mitochondria, is described in SI. A similar exercise has also been performed with the 5 other datasets and can be found in SI. The gene enrichment results of each cluster are compiled in Table S9 in SI, including the ensemble of compounds, the number of both down-regulated and up-regulated genes, the number of KEGG pathways perturbed by the cluster after pathway analysis, and the number of both BP and CC terms from the GO affected by the cluster of chemicals.
Hepatotoxicity Prediction
While the selection of hepatotoxicity-related Cell Painting features allowed us to determine clusters of compounds both with similar morphological profiles and known to be associated with DILI, the pathway analysis resulting from these clusters highlighted potential common mechanisms for liver injury. In a second step, we hypothesized that selected Cell Painting features for DILI compounds could enable discrimination between DILI vs non-DILI chemicals and we developed machine learning models to evaluate their capability to predict potential new hepatotoxic compounds.
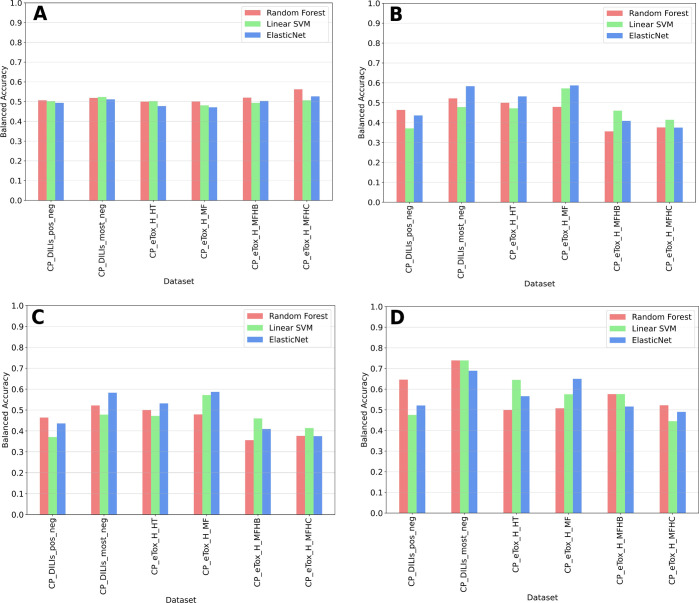
We compared three different machine learning algorithms: RF, Linear SVM, and ElasticNet. Each model was trained on each dataset, either with the original 1779 Cell Painting features (i.e. “complete” feature set) or with only features that were selected for each dataset after the feature selection step (i.e. “reduced” feature set). We built a bar chart of BAs obtained after cross-validation runs and on the validation set for each model and dataset (Figure 8). Exact BA, SP, and SN scores across cross-validation runs for training and validation sets can be found in Table S11 in SI. From a technical point of view, no single machine learning algorithm stood out from the others. Regarding the complete set of cell morphological features, the highest BA in the validation set reached 0.583 and 0.587 for the CP_DILIs_most_neg and CP_eTox_H_MF datasets, respectively, both using the ElasticNet model (Figure 8A–B), but with cross-validation mean BAs of 0.511 and 0.471, respectively. With the selected features containing only hepatotoxicity-discriminating features, cross-validation mean BAs between 0.567 and 0.603 and validation set BAs between 0.646 and 0.739 were obtained for the CP_DILIs_pos_neg and CP_DILIs_most_neg datasets (Figure 8C–D, Table S11). For eTox, the mean BAs were 0.555 and 0.541 across cross-validation and 0.645 and 0.65 with the validation set for the CP_eTox_H_HT and CP_eTox_H_MF datasets, respectively (Figure 8C–D, Table S11). None of the models succeeded in giving correct performances for the CP_eTox_H_MFHB and CP_eTox_H_MFHC datasets. Performances were slightly better for DILIrank datasets than for eTox ones, which could be explained by the higher number of selected features for the DILIrank datasets. Finally, we performed permutation tests for each combination of model and dataset. For combinations of model and dataset associated with BAs above 0.6 for their given validation set (RF-CP_DILIs_pos_neg, RF/Linear SVM/ElasticNet-CP_DILIs_most_neg, Linear SVM-CP_eTox_H_HT, and ElasticNet-CP_eTox_H_MFHC), less than 2% of the permutation tests succeeded in reaching higher BAs (Figure S6, Table S12). As ElasticNet models were the more stable across all datasets, we also conducted a supplementary analysis with a RFE step within the cross-validation loop, but we did not observe improved performances (Table S13).
Figure 8.
Comparison of balanced accuracies obtained before and after feature selection for the 6 datasets, during cross-validation and on the validation set, with Random Forest, Linear SVM, and ElasticNet: (A) Mean cross-validation BAs obtained with models trained on datasets with all Cell Painting features; (B) BAs obtained on the validation set with models trained on datasets with all Cell Painting features; (C) mean cross-validation BAs with models trained on datasets after feature selection; (D) BAs obtained on the validation set with models trained on datasets after feature selection.
Looking at SP and SN scores, models based on selected features exhibited more balanced values (around 0.6) compared to models using the full list of Cell Painting features, which showed either high SP (prediction of truly nonhepatotoxic chemicals) and low SN, or high SN (prediction of truly hepatotoxic chemicals) and low SP (Table S11). In general, the selection of Cell Painting features related to DILI/non-DILI chemicals improved the stability of the models and slightly enhanced performance for the DILIrank datasets and the eTox_H_HT and eTox_H_MFHC datasets. SP and SN were the best for the CP_DILIs_most_net dataset, using either Random Forest or Linear SVM, with respective values of 0.78 and 0.70 on the validation set (Table S11).
Discussion
High-Throughput Phenotypic Profiling (HTPP) and High-Throughput Transcriptomics (HTTr) are currently highly considered in the assessment of chemical-induced toxicity, including for DILI.54 DILI is characterized by a variety of mechanisms that can lead to adverse outcomes in the human liver. Thus, it makes it challenging to assess a global hepatotoxic risk without focusing on a particular mechanism or without studying compounds on an individual basis.
The objective of our study was to assess DILI using a combination of image-based information from Cell Painting and gene expression data. Cell Painting is an HCS assay that allows the observation of cell morphology perturbations induced by a compound. However, it provides limited information on the potential mechanism of action at the molecular level and essentially informs on phenotypic changes. In this regard, combining image-based information with transcriptomics data is an interesting solution to explore. For example, Way et al. recently demonstrated how Cell Painting and the L1000 mRNA profiling assay were highly complementary in terms of capturing the MoA.55
Based on the assumption that chemicals having similar DILI-specific morphological features might have similar MoAs, we exploited 2 datasets of chemicals with DILI information (DILIrank and eTox). First, we selected features of interest for our phenotypic end point associated with DILI categories. It has been shown that Cell Painting can benefit from supervised feature selection and that a smaller size of features is more informative.56 To do the feature selection, we used a Mann–Whitney test, which allowed us to propose sufficient morphology features that allow discrimination between DILI versus non-DILI chemicals. Notably, we noticed that the feature selection was larger for DILIrank compared to eTox. One explanation could be that eTox annotations came from preclinical studies carried out on rodents and suggested to cause DILI in humans whereas DILIrank annotations are observations coming from clinical studies. As the Cell Painting assay was carried out on human cells, it may be more challenging to link human in vitro observations with a rodent in vivo annotation. Interestingly, we also noted that the features perturbed by the two sets of DILI chemicals were relatively different (only 49 morphological features in common between both datasets). This highlights that there is not a unique morphological cell signature associated with DILIs and that their mechanisms of action are multiple. It also suggests that the mechanisms of liver toxicity could be different from human to rodents.
Nevertheless, by clustering DILI chemicals sharing a similar morphological profile and by analyzing transcriptomics data for these chemicals, we managed to identify potential significant functional signaling pathways, reported in the literature, which can lead to hepatic injury. For example, previous studies have demonstrated that inhibition of the cell cycle with fewer cell divisions is associated with liver inflammation and fibrosis,57 while other studies showed evidence of RNA splicing as a precursor of liver disease development at an early stage, as steatosis, and at a later stage, as HCC.58 Additionally, the transcription factor and tumor suppressor p53, expressed in the hepatic tissue and in U2OS, has been implicated in liver disease development.59,60 Some pathways were found across different clusters, indicating that although the chemicals were not categorized to the same cluster (thus having different morphological profiles), they shared some common gene deregulation profiles. For instance, both Metoprolol and Quinidine were clustered separately (respectively, clusters 13 and 41) but both disrupted liver regeneration (Figure 7). Other pathways were unique to specific clusters, suggesting that these chemicals have a distinct mechanism of action. This is the case for chemicals such as the Cetirizine (cluster 42), which would impact essential proteoglycans in cancer, opposite to Amitriptyline (cluster 17), which would rather act on spindle fibers’ organization during mitosis (Figure 7). Cluster analysis showed that compounds of the same cluster do not necessarily share structural or pharmacological properties but still cause DILIs and affect cells in similar ways. We repeated our gene enrichment and pathway analysis with randomly created clusters of chemicals, of the same size as the mean size of Cell Painting features-based clusters (Table S14). Pathways that showed up were similar to the ones highlighted with the Cell Painting features-based clusters (Figures S7–S12), which was expected, as the same sets of hepatotoxic compounds were used to create both types of clusters. In addition, the analysis revealed a diverse list of deregulated pathways, with a weaker significance, demonstrating a higher scattering and false positive associations between randomly created clusters and deregulated pathways. Although supplemental studies would be required to confirm an association between the hepatotoxic MoA highlighted here and each compound, our protocol provides hypotheses to explore regarding Cell Painting features-based clusters and their utility in studying chemical toxicity.
Although exploring DILI mechanisms through gene expression analysis already allowed the identification of some adverse outcome pathways (AOPs) occurring in the liver,7,61,62 linking biological pathways with some cell morphological features through the use of Cell Painting remains a challenge. Indeed, contrary to features from other software such as cellXpress,63 CellProfiler does not allow the ability to easily connect features to precise cellular components or proteins. It is notable that the biological pathways highlighted in our study could not always be associated with a unique cell compartment. For example, signaling pathways can take place in both cytoplasm and nuclei but could also be interpreted as the cell in CellProfiler features. Although the complete feature names provided by CellProfiler contain the name of the dye used to stain the corresponding compartment (mitochondria, nucleus, ER, ...), the precise parameter measured by the feature remains vague and difficult to interpret, thereby preventing a clear association between cell morphological features and biological pathways. Thus, a dedicated analysis of the selected Cell Painting features is necessary to establish a more precise linkage with the specific pathways highlighted in our analysis. Integrating a quantitative metric to associate morphological perturbations observed in the Cell Painting assay with disrupted biological pathways would also improve our study. However, our approach of combining Cell Painting and gene expression data allowed us to study compound modes of action in an untargeted way when studies tend to focus on predicting precise targets and MoAs.64,27
Another limitation in this analysis is that the transcriptomics data were generated using the hepatic cell line HepG2, whereas the Cell Painting assay was carried out using the human osteosarcoma U2OS immortalized cell line. Although 76.7% of HepG2 genes that were differentially expressed during the enrichment were also known to be expressed in the U2OS cell lines (https://www.proteinatlas.org/) (representing 2293 genes out of 2990 differentially expressed genes in HepG2 in the enrichment analysis), it would have been preferable to use Cell Painting and transcriptomics data derived from the same cell type. Some studies adapting the Cell Painting protocol to various cell lines are ongoing, which should bring more confidence to such analysis.65,66
In a second step, we aimed to evaluate the relevance of our morphological feature selection in the development of a predictive DILI model that could be used to assess the potential of a compound to induce DILI based on its Cell Painting profile. So far, Cell Painting data have been used for cytotoxicity or for DILI prediction67,68 and concentration-response modeling approaches to estimate drug bioactivity69−71 thresholds. With the improvement of machine learning strategies observed these past years, many studies have shown that in silico models can be trained to predict the toxicity outcomes of hundreds of chemicals using image-based profiling.45 Consequently, Cell Painting has emerged as a promising tool in drug discovery.72,73 Previous studies that employed Cell Painting data for toxicity prediction achieved correct performances by integrating morphological features with for example structural information or gene expression,18 or by employing deep learning approaches.74 For instance in their study, Chavan et al. processed RF models on HCI data to predict hepatotoxicity end points, resulting in BA scores of 0.59 during cross-validation and 0.58 on their validation set.68 Here, targeting the same end point, we achieved similar predictive performances during cross-validation and BAs up to 0.739 on the validation set. The next step could be to combine both Cell Painting and gene expression data in a deep learning model for enhanced predictive capabilities.
Conclusions
In this study, we aimed to associate mechanisms of liver toxicity with groups of chemicals with similar phenotypic profiles based on the assumption that chemicals having similar morphological patterns might hold similar mechanisms of action too. Through our computational methodology, we succeeded in associating clusters of chemicals with biological pathways such as RNA splicing or deregulation of the cell cycle, which are known to be related to hepatotoxicity. Furthermore, based on the cell morphology feature selection from our analysis, we developed machine learning models that can contribute to the prediction of potential DILI chemicals. Such approaches could be applied for other biological/toxicological end points providing there are enough Cell Painting and transcriptomics data to exploit.
Glossary
Abbreviations
- AOP
adverse outcome pathway
- BA
balanced accuracy
- BP
biological process
- CC
cellular component
- DILI
drug-induced liver injury
- ECM
extracellular matrix
- ER
endoplasmic reticulum
- FDA
Food and Drug Administration
- GO
Gene Ontology
- HCC
hepatocellular carcinoma
- HCI
high-content imaging
- HCS
high-content screening
- HIF-1
hypoxia-inducible factor 1
- H_HT
human hepatotoxicity
- H_MF
human morphological findings
- H_MFHB
human hepatobiliary injury
- H_MFHC
human hepatocellular injury
- HTPP
high-throughput phenotypic profiling
- HTTr
high-throughput transcriptomics
- MMAD
median and median absolute deviation
- MoA
mode of action
- NAFLD
nonalcoholic fatty liver disease
- RF
Random Forest
- RFE
recursive feature elimination
- SN
sensitivity
- SP
specificity
- SVM
Support Vector Machine.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.2c00381.
Details of model parameters; metrics formulas; lists of selected features after feature selection; percentages of feature categories in each compartment and dataset before and after feature selection; ratio in percentage in the selected features categories; Fisher’s Exact Tests between the number of features before and after selection for each dataset; Cell Painting features-based and random clustering details and gene enrichment results; details of the number of Cell Painting features-based clusters and their mean size; additional pathway analysis of the CP_DILIs_pos_neg dataset; heatmaps of pathways significantly deregulated by Cell Painting features-based and randomly created clusters of hepatotoxic compounds from the CP_DILIs_most_neg, CP_eTox_H_HT, CP_eTox_H_MF, CP_eTox_H_MFHB, and CP_eTox_H_MFHC datasets, and respective pathway analysis; comparison of machine learning models’ performances for hepatotoxicity prediction based on Cell Painting features; histograms of permutation test results and percentage of balanced accuracies higher than original scores; performances of ElasticNet models including recursive feature elimination in the training loop (PDF)
DILIrank and eTox compounds’ annotations (XLSX)
Author Contributions
Conceived and designed the experiments: OT, DR, VL. Performed the experiments: VL, NC. Wrote and reviewed the manuscript: All. CRediT: Vanille Lejal data curation, formal analysis, methodology, visualization, writing-original draft; Natacha Cerisier formal analysis, writing-review & editing; David Rouquié conceptualization, funding acquisition, writing-review & editing; Olivier Taboureau conceptualization, funding acquisition, supervision, writing-original draft.
This work was supported by the project RISK-HUNT3R: RISK assessment of chemicals integrating HUman centric Next generation Testing strategies promoting the 3Rs. RISK-HUNT3R has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 964537. This work is also supported by Bayer Crop Science through GIE AIFOR PhD Cifre funding (2021-0755).
The authors declare no competing financial interest.
Supplementary Material
References
- Paul S. M.; Mytelka D. S.; Dunwiddie C. T.; Persinger C. C.; Munos B. H.; Lindborg S. R.; Schacht A. L. How to Improve R&D Productivity: The Pharmaceutical Industry’s Grand Challenge. Nat. Rev. Drug Discovery 2010, 9 (3), 203–214. 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- Siramshetty V. B.; Nickel J.; Omieczynski C.; Gohlke B.-O.; Drwal M. N.; Preissner R. WITHDRAWN—a Resource for Withdrawn and Discontinued Drugs. Nucleic Acids Res. 2016, 44 (D1), D1080–D1086. 10.1093/nar/gkv1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Roberts R.; Tong W.; Liu Z. Tox-GAN: An Artificial Intelligence Approach Alternative to Animal Studies—A Case Study With Toxicogenomics. Toxicol. Sci. 2022, 186 (2), 242–259. 10.1093/toxsci/kfab157. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Chen M.; Tong W. Is Toxicogenomics a More Reliable and Sensitive Biomarker than Conventional Indicators from Rats To Predict Drug-Induced Liver Injury in Humans?. Chem. Res. Toxicol. 2012, 25 (1), 122–129. 10.1021/tx200320e. [DOI] [PubMed] [Google Scholar]
- Zhang J. D.; Berntenis N.; Roth A.; Ebeling M. Data Mining Reveals a Network of Early-Response Genes as a Consensus Signature of Drug-Induced in Vitro and in Vivo Toxicity. Pharmacogenomics J. 2014, 14 (3), 208–216. 10.1038/tpj.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauschke V. M. Toxicogenomics of Drug Induced Liver Injury – from Mechanistic Understanding to Early Prediction. Drug Metab. Rev. 2021, 53 (2), 245–252. 10.1080/03602532.2021.1894571. [DOI] [PubMed] [Google Scholar]
- Aguayo-Orozco A.; Bois F. Y.; Brunak S.; Taboureau O. Analysis of Time-Series Gene Expression Data to Explore Mechanisms of Chemical-Induced Hepatic Steatosis Toxicity. Front. Genet. 2018, 9, 396. 10.3389/fgene.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegaro G.; Kunnen S. J.; Trairatphisan P.; Grosdidier S.; Niemeijer M.; den Hollander W.; Guney E.; Piñero Gonzalez J.; Furlong L.; Webster Y. W.; Saez-Rodriguez J.; Sutherland J. J.; Mollon J.; Stevens J. L.; van de Water B. The Human Hepatocyte TXG-MAPr: Gene Co-Expression Network Modules to Support Mechanism-Based Risk Assessment. Arch. Toxicol. 2021, 95 (12), 3745–3775. 10.1007/s00204-021-03141-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrach J.; Mueller S. O.; Hewitt P. Development of an in Vitro Liver Toxicity Prediction Model Based on Longer Term Primary Rat Hepatocyte Culture. Toxicol. Lett. 2011, 206 (2), 189–196. 10.1016/j.toxlet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Aguayo-Orozco A.; Brunak S.; Taboureau O. Extrapolation of Drug Induced Liver Injury Responses from Cancer Cell Lines Using Machine Learning Approaches. Comput. Toxicol. 2021, 17, 100147 10.1016/j.comtox.2020.100147. [DOI] [Google Scholar]
- Liu A.; Walter M.; Wright P.; Bartosik A.; Dolciami D.; Elbasir A.; Yang H.; Bender A. Prediction and Mechanistic Analysis of Drug-Induced Liver Injury (DILI) Based on Chemical Structure. Biol. Direct 2021, 16 (1), 6. 10.1186/s13062-020-00285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulliner D.; Schmidt F.; Stolte M.; Spirkl H.-P.; Czich A.; Amberg A. Computational Models for Human and Animal Hepatotoxicity with a Global Application Scope. Chem. Res. Toxicol. 2016, 29 (5), 757–767. 10.1021/acs.chemrestox.5b00465. [DOI] [PubMed] [Google Scholar]
- Kotsampasakou E.; Montanari F.; Ecker G. F. Predicting Drug-Induced Liver Injury: The Importance of Data Curation. Toxicology 2017, 389, 139–145. 10.1016/j.tox.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Kelly R.; Fang H.; Ding D.; Tong W. Comparative Analysis of Predictive Models for Nongenotoxic Hepatocarcinogenicity Using Both Toxicogenomics and Quantitative Structure–Activity Relationships. Chem. Res. Toxicol. 2011, 24 (7), 1062–1070. 10.1021/tx2000637. [DOI] [PubMed] [Google Scholar]
- Bray M.-A.; Gustafsdottir S. M.; Rohban M. H.; Singh S.; Ljosa V.; Sokolnicki K. L.; Bittker J. A.; Bodycombe N. E.; Dančík V.; Hasaka T. P.; Hon C. S.; Kemp M. M.; Li K.; Walpita D.; Wawer M. J.; Golub T. R.; Schreiber S. L.; Clemons P. A.; Shamji A. F.; Carpenter A. E.. A Dataset of Images and Morphological Profiles of 30 000 Small-Molecule Treatments Using the Cell Painting Assay. GigaScience 2017, 6 ( (12), ), giw014. 10.1093/gigascience/giw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M.-A.; Singh S.; Han H.; Davis C. T.; Borgeson B.; Hartland C.; Kost-Alimova M.; Gustafsdottir S. M.; Gibson C. C.; Carpenter A. E. Cell Painting, a High-Content Image-Based Assay for Morphological Profiling Using Multiplexed Fluorescent Dyes. Nat. Protoc. 2016, 11 (9), 1757–1774. 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerisier N.; Dafniet B.; Badel A.; Taboureau O. Linking Chemicals, Genes and Morphological Perturbations to Diseases. Toxicol. Appl. Pharmacol. 2023, 461, 116407 10.1016/j.taap.2023.116407. [DOI] [PubMed] [Google Scholar]
- Seal S.; Carreras-Puigvert J.; Trapotsi M.-A.; Yang H.; Spjuth O.; Bender A. Integrating Cell Morphology with Gene Expression and Chemical Structure to Aid Mitochondrial Toxicity Detection. Commun. Biol. 2022, 5 (1), 858. 10.1038/s42003-022-03763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder J.; Sedrani R.; Wiesmann C. The Discovery of First-in-Class Drugs: Origins and Evolution. Nat. Rev. Drug Discovery 2014, 13 (8), 577–587. 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- Hofmarcher M.; Rumetshofer E.; Clevert D.-A.; Hochreiter S.; Klambauer G. Accurate Prediction of Biological Assays with High-Throughput Microscopy Images and Convolutional Networks. J. Chem. Inf. Model. 2019, 59 (3), 1163–1171. 10.1021/acs.jcim.8b00670. [DOI] [PubMed] [Google Scholar]
- Simm J.; Klambauer G.; Arany A.; Steijaert M.; Wegner J. K.; Gustin E.; Chupakhin V.; Chong Y. T.; Vialard J.; Buijnsters P.; Velter I.; Vapirev A.; Singh S.; Carpenter A. E.; Wuyts R.; Hochreiter S.; Moreau Y.; Ceulemans H. Repurposing High-Throughput Image Assays Enables Biological Activity Prediction for Drug Discovery. Cell Chem. Biol. 2018, 25 (5), 611–618. 10.1016/j.chembiol.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapotsi M.-A.; Mervin L. H.; Afzal A. M.; Sturm N.; Engkvist O.; Barrett I. P.; Bender A. Comparison of Chemical Structure and Cell Morphology Information for Multitask Bioactivity Predictions. J. Chem. Inf. Model. 2021, 61 (3), 1444–1456. 10.1021/acs.jcim.0c00864. [DOI] [PubMed] [Google Scholar]
- Cox M. J.; Jaensch S.; Van de Waeter J.; Cougnaud L.; Seynaeve D.; Benalla S.; Koo S. J.; Van Den Wyngaert I.; Neefs J.-M.; Malkov D.; Bittremieux M.; Steemans M.; Peeters P. J.; Wegner J. K.; Ceulemans H.; Gustin E.; Chong Y. T.; Göhlmann H. W. H. Tales of 1,008 Small Molecules: Phenomic Profiling through Live-Cell Imaging in a Panel of Reporter Cell Lines. Sci. Rep. 2020, 10 (1), 13262. 10.1038/s41598-020-69354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassiri I.; McCall M. N. Systematic Exploration of Cell Morphological Phenotypes Associated with a Transcriptomic Query. Nucleic Acids Res. 2018, 46 (19), e116–e116. 10.1093/nar/gky626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi M.; Caicedo J. C.; Cimini B. A.; Carpenter A. E.; Singh S.. High-Dimensional Gene Expression and Morphology Profiles of Cells across 28,000 Genetic and Chemical Perturbations. Nat. Methods 2022, 19, 1550. 10.1038/s41592-022-01667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohban M. H.; Abbasi H. S.; Singh S.; Carpenter A. E. Capturing Single-Cell Heterogeneity via Data Fusion Improves Image-Based Profiling. Nat. Commun. 2019, 10 (1), 2082. 10.1038/s41467-019-10154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapins M.; Spjuth O.. Evaluation of Gene Expression and Phenotypic Profiling Data as Quantitative Descriptors for Predicting Drug Targets and Mechanisms of Action. bioRxiv 2019. 10.1101/580654 (accessed 2023-03-10). [DOI] [Google Scholar]
- Chen M.; Suzuki A.; Thakkar S.; Yu K.; Hu C.; Tong W. DILIrank: The Largest Reference Drug List Ranked by the Risk for Developing Drug-Induced Liver Injury in Humans. Drug Discovery Today 2016, 21 (4), 648–653. 10.1016/j.drudis.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Cell Painting dataset provided by the Broad Institute, http://gigadb.org/dataset/100351 (accessed 2021-10-18).
- Carpenter A. E.; Jones T. R.; Lamprecht M. R.; Clarke C.; Kang I.; Friman O.; Guertin D. A.; Chang J.; Lindquist R. A.; Moffat J.; Golland P.; Sabatini D. M. CellProfiler: Image Analysis Software for Identifying and Quantifying Cell Phenotypes. Genome Biol. 2006, 7 (10), R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteller F.; Tukey J. W.. Data Analysis and Regression: A Second Course in Statistics; Addison-Wesley Publishing Company: University of Michigan, 1977. [Google Scholar]
- Ajoge I.; Adam M. B.; Anwar F.; Sadiq A. Y. Median Polish with Covariate on Before and After Data. IOSR J. Math. 2016, 12 (3), 64–73. [Google Scholar]
- Birmingham A.; Selfors L. M.; Forster T.; Wrobel D.; Kennedy C. J.; Shanks E.; Santoyo-Lopez J.; Dunican D. J.; Long A.; Kelleher D.; Smith Q.; Beijersbergen R. L.; Ghazal P.; Shamu C. E. Statistical Methods for Analysis of High-Throughput RNA Interference Screens. Nat. Methods 2009, 6 (8), 569–575. 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N.; Zhang X. D.; Kreamer A.; Locco L.; Kuan P.-F.; Bartz S.; Linsley P. S.; Ferrer M.; Strulovici B. Median Absolute Deviation to Improve Hit Selection for Genome-Scale RNAi Screens. SLAS Discovery 2008, 13 (2), 149–158. 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- Bajusz D.; Rácz A.; Héberger K. Why Is Tanimoto Index an Appropriate Choice for Fingerprint-Based Similarity Calculations?. J. Cheminformatics 2015, 7 (1), 20. 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.; Narayan R.; Corsello S. M.; Peck D. D.; Natoli T. E.; Lu X.; Gould J.; Davis J. F.; Tubelli A. A.; Asiedu J. K.; Lahr D. L.; Hirschman J. E.; Liu Z.; Donahue M.; Julian B.; Khan M.; Wadden D.; Smith I. C.; Lam D.; Liberzon A.; Toder C.; Bagul M.; Orzechowski M.; Enache O. M.; Piccioni F.; Johnson S. A.; Lyons N. J.; Berger A. H.; Shamji A. F.; Brooks A. N.; Vrcic A.; Flynn C.; Rosains J.; Takeda D. Y.; Hu R.; Davison D.; Lamb J.; Ardlie K.; Hogstrom L.; Greenside P.; Gray N. S.; Clemons P. A.; Silver S.; Wu X.; Zhao W.-N.; Read-Button W.; Wu X.; Haggarty S. J.; Ronco L. V.; Boehm J. S.; Schreiber S. L.; Doench J. G.; Bittker J. A.; Root D. E.; Wong B.; Golub T. R. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171 (6), 1437–1452.e17. 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M.; Ball C. A.; Blake J. A.; Botstein D.; Butler H.; Cherry J. M.; Davis A. P.; Dolinski K.; Dwight S. S.; Eppig J. T.; Harris M. A.; Hill D. P.; Issel-Tarver L.; Kasarskis A.; Lewis S.; Matese J. C.; Richardson J. E.; Ringwald M.; Rubin G. M.; Sherlock G. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25 (1), 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S.; Douglass E.; Good B. M.; Unni D. R.; Harris N. L.; Mungall C. J.; Basu S.; Chisholm R. L.; Dodson R. J.; Hartline E.; Fey P.; Thomas P. D.; Albou L.-P.; Ebert D.; Kesling M. J.; Mi H.; Muruganujan A.; Huang X.; Mushayahama T.; LaBonte S. A.; Siegele D. A.; Antonazzo G.; Attrill H.; Brown N. H.; Garapati P.; Marygold S. J.; Trovisco V.; dos Santos G.; Falls K.; Tabone C.; Zhou P.; Goodman J. L.; Strelets V. B.; Thurmond J.; Garmiri P.; Ishtiaq R.; Rodríguez-López M.; Acencio M. L.; Kuiper M.; Lægreid A.; Logie C.; Lovering R. C.; Kramarz B.; Saverimuttu S. C. C.; Pinheiro S. M.; Gunn H.; Su R.; Thurlow K. E.; Chibucos M.; Giglio M.; Nadendla S.; Munro J.; Jackson R.; Duesbury M. J.; Del-Toro N.; Meldal B. H. M.; Paneerselvam K.; Perfetto L.; Porras P.; Orchard S.; Shrivastava A.; Chang H.-Y.; Finn R. D.; Mitchell A. L.; Rawlings N. D.; Richardson L.; Sangrador-Vegas A.; Blake J. A.; Christie K. R.; Dolan M. E.; Drabkin H. J.; Hill D. P.; Ni L.; Sitnikov D. M.; Harris M. A.; Oliver S. G.; Rutherford K.; Wood V.; Hayles J.; Bähler J.; Bolton E. R.; De Pons J. L.; Dwinell M. R.; Hayman G. T.; Kaldunski M. L.; Kwitek A. E.; Laulederkind S. J. F.; Plasterer C.; Tutaj M. A.; Vedi M.; Wang S.-J.; D’Eustachio P.; Matthews L.; Balhoff J. P.; Aleksander S. A.; Alexander M. J.; Cherry J. M.; Engel S. R.; Gondwe F.; Karra K.; Miyasato S. R.; Nash R. S.; Simison M.; Skrzypek M. S.; Weng S.; Wong E. D.; Feuermann M.; Gaudet P.; Morgat A.; Bakker E.; Berardini T. Z.; Reiser L.; Subramaniam S.; Huala E.; Arighi C. N.; Auchincloss A.; Axelsen K.; Argoud-Puy G.; Bateman A.; Blatter M.-C.; Boutet E.; Bowler E.; Breuza L.; Bridge A.; Britto R.; Bye-A-Jee H.; Casas C. C.; Coudert E.; Denny P.; Estreicher A.; Famiglietti M. L.; Georghiou G.; Gos A.; Gruaz-Gumowski N.; Hatton-Ellis E.; Hulo C.; Ignatchenko A.; Jungo F.; Laiho K.; Le Mercier P.; Lieberherr D.; Lock A.; Lussi Y.; MacDougall A.; Magrane M.; Martin M. J.; Masson P.; Natale D. A.; Hyka-Nouspikel N.; Orchard S.; Pedruzzi I.; Pourcel L.; Poux S.; Pundir S.; Rivoire C.; Speretta E.; Sundaram S.; Tyagi N.; Warner K.; Zaru R.; Wu C. H.; Diehl A. D.; Chan J. N.; Grove C.; Lee R. Y. N.; Muller H.-M.; Raciti D.; Van Auken K.; Sternberg P. W.; Berriman M.; Paulini M.; Howe K.; Gao S.; Wright A.; Stein L.; Howe D. G.; Toro S.; Westerfield M.; Jaiswal P.; Cooper L.; Elser J. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49 (D1), D325–D334. 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28 (1), 27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28 (11), 1947–1951. 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Sato Y.; Kawashima M.; Ishiguro-Watanabe M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2023, 51, D587–D592. 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.; Hu E.; Xu S.; Chen M.; Guo P.; Dai Z.; Feng T.; Zhou L.; Tang W.; Zhan L.; Fu X.; Liu S.; Bo X.; Yu G. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. The. Innovation 2021, 2 (3), 100141 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An Open Chemical Toolbox. J. Cheminformatics 2011, 3 (1), 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo J. C.; Cooper S.; Heigwer F.; Warchal S.; Qiu P.; Molnar C.; Vasilevich A. S.; Barry J. D.; Bansal H. S.; Kraus O.; Wawer M.; Paavolainen L.; Herrmann M. D.; Rohban M.; Hung J.; Hennig H.; Concannon J.; Smith I.; Clemons P. A.; Singh S.; Rees P.; Horvath P.; Linington R. G.; Carpenter A. E. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14 (9), 849–863. 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S. N.Image-Based Profiling for Drug Discovery: Due for a Machine-Learning Upgrade? Nat. Rev. Drug Discovery 2020, 20, 145–159 10.1038/s41573-020-00117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehate C.; Jain S.; Saneja A.; Khare V.; Alam N.; Dubey R.; Gupta P. Paclitaxel Formulations: Challenges and Novel Delivery Options. CDD 2014, 11 (6), 666–686. 10.2174/1567201811666140609154949. [DOI] [PubMed] [Google Scholar]
- Hande K. R. Etoposide: Four Decades of Development of a Topoisomerase II Inhibitor. Eur. J. Cancer 1998, 34 (10), 1514–1521. 10.1016/S0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- Guerini; Triggiani; Maddalo; Bonù; Frassine; Baiguini; Alghisi; Tomasini; Borghetti; Pasinetti; Bresciani; Magrini; Buglione Mebendazole as a Candidate for Drug Repurposing in Oncology: An Extensive Review of Current Literature. Cancers 2019, 11 (9), 1284. 10.3390/cancers11091284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H.; DiNicolantonio J. J.; O’Keefe J. H.; Lavie C. J. Amlodipine in Hypertension: A First-Line Agent with Efficacy for Improving Blood Pressure and Patient Outcomes. Open Heart 2016, 3 (2), e000473 10.1136/openhrt-2016-000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A. The Multiple Facets of the SMC1A Gene. Gene 2020, 743, 144612 10.1016/j.gene.2020.144612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Wu J.; Wang S.; Luo X.; Du Y.; Huang D.; Gu D.; Zhang F. PKMYT1 Promoted the Growth and Motility of Hepatocellular Carcinoma Cells by Activating Beta-Catenin/TCF Signaling. Exp. Cell Res. 2017, 358 (2), 209–216. 10.1016/j.yexcr.2017.06.014. [DOI] [PubMed] [Google Scholar]
- Williams A. J.; Grulke C. M.; Edwards J.; McEachran A. D.; Mansouri K.; Baker N. C.; Patlewicz G.; Shah I.; Wambaugh J. F.; Judson R. S.; Richard A. M. The CompTox Chemistry Dashboard: A Community Data Resource for Environmental Chemistry. J. Cheminform 2017, 9 (1), 61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbracht J. V.; Boehm V.; Britto-Borges T.; Kallabis S.; Wiederstein J. L.; Ciriello S.; Aschemeier D. U.; Krüger M.; Frese C. K.; Altmüller J.; Dieterich C.; Gehring N. H. CASC3 Promotes Transcriptome-Wide Activation of Nonsense-Mediated Decay by the Exon Junction Complex. Nucleic Acids Res. 2020, 48 (15), 8626–8644. 10.1093/nar/gkaa564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I.; Antonijevic T.; Chambers B.; Harrill J.; Thomas R. Estimating Hepatotoxic Doses Using High-Content Imaging in Primary Hepatocytes. Toxicol. Sci. 2021, kfab091 10.1093/toxsci/kfab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way G. P.; Natoli T.; Adeboye A.; Litichevskiy L.; Yang A.; Lu X.; Caicedo J. C.; Cimini B. A.; Karhohs K.; Logan D. J.; Rohban M. H.; Kost-Alimova M.; Hartland K.; Bornholdt M.; Chandrasekaran S. N.; Haghighi M.; Weisbart E.; Singh S.; Subramanian A.; Carpenter A. E. Morphology and Gene Expression Profiling Provide Complementary Information for Mapping Cell State. Cell Syst. 2022, 13, 911–923.e9. 10.1016/j.cels.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegismund D.; Fassler M.; Heyse S.; Steigele S. Benchmarking Feature Selection Methods for Compressing Image Information in High-Content Screening. SLAS Technol. 2022, 27 (1), 85–93. 10.1016/j.slast.2021.10.015. [DOI] [PubMed] [Google Scholar]
- Dewhurst M. R.; Ow J. R.; Zafer G.; van Hul N. K. M.; Wollmann H.; Bisteau X.; Brough D.; Choi H.; Kaldis P. Loss of Hepatocyte Cell Division Leads to Liver Inflammation and Fibrosis. PLOS Genet. 2020, 16 (11), e1009084 10.1371/journal.pgen.1009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. Hyperoxia Accelerates Progression of Hepatic Fibrosis by Up-Regulation of Transforming Growth Factor-β Expression. WJG 2014, 20 (11), 3011. 10.3748/wjg.v20.i11.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic J.; Galhuber M.; Schulz T.; Schupp M.; Prokesch A. P53 as a Dichotomous Regulator of Liver Disease: The Dose Makes the Medicine. Int. J. Mol. Sci. 2018, 19 (3), 921. 10.3390/ijms19030921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi M. E. Apoptosis: A Mechanism of Acute and Chronic Liver Injury. Gut 2005, 54 (7), 1024–1033. 10.1136/gut.2004.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang S. A.; Oseini A.; Feaver R. E.; Cole B. K.; Asgharpour A.; Vincent R.; Siddiqui M.; Lawson M. J.; Day N. C.; Taylor J. M.; Wamhoff B. R.; Mirshahi F.; Contos M. J.; Idowu M.; Sanyal A. J. Gene Expression Predicts Histological Severity and Reveals Distinct Molecular Profiles of Nonalcoholic Fatty Liver Disease. Sci. Rep. 2019, 9 (1), 12541. 10.1038/s41598-019-48746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Wang X.; Zhuang L. Gene Expression Profiling Reveals Key Genes and Pathways Related to the Development of Non-Alcoholic Fatty Liver Disease. Ann. Hepatol. 2016, 15 (2), 190–199. [DOI] [PubMed] [Google Scholar]
- Hussain F.; Basu S.; Heng J. J. H.; Loo L.-H.; Zink D. Predicting Direct Hepatocyte Toxicity in Humans by Combining High-Throughput Imaging of HepaRG Cells and Machine Learning-Based Phenotypic Profiling. Arch. Toxicol. 2020, 94 (8), 2749–2767. 10.1007/s00204-020-02778-3. [DOI] [PubMed] [Google Scholar]
- Rose F.; Basu S.; Rexhepaj E.; Chauchereau A.; Del Nery E.; Genovesio A. Compound Functional Prediction Using Multiple Unrelated Morphological Profiling Assays. SLAS Technology 2018, 23 (3), 243–251. 10.1177/2472630317740831. [DOI] [PubMed] [Google Scholar]
- Willis C.; Nyffeler J.; Harrill J. Phenotypic Profiling of Reference Chemicals across Biologically Diverse Cell Types Using the Cell Painting Assay. SLAS Discovery Adv. Sci. Drug Discovery 2020, 25 (7), 755–769. 10.1177/2472555220928004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S. N.; Ackerman J.; Alix E.; Ando D. M.; Arevalo J.; Bennion M.; Boisseau N.; Borowa A.; Boyd J. D.; Brino L.; Byrne P. J.; Ceulemans H.; Ch’ng C.; Cimini B. A.; Clevert D.-A.; Deflaux N.; Doench J. G.; Dorval T.; Doyonnas R.; Dragone V.; Engkvist O.; Faloon P. W.; Fritchman B.; Fuchs F.; Garg S.; Gilbert T. J.; Glazer D.; Gnutt D.; Goodale A.; Grignard J.; Guenther J.; Han Y.; Hanifehlou Z.; Hariharan S.; Hernandez D.; Horman S. R.; Hormel G.; Huntley M.; Icke I.; Iida M.; Jacob C. B.; Jaensch S.; Khetan J.; Kost-Alimova M.; Krawiec T.; Kuhn D.; Lardeau C.-H.; Lembke A.; Lin F.; Little K. D.; Lofstrom K. R.; Lotfi S.; Logan D. J.; Luo Y.; Madoux F.; Marin Zapata P. A.; Marion B. A.; Martin G.; McCarthy N. J.; Mervin L.; Miller L.; Mohamed H.; Monteverde T.; Mouchet E.; Nicke B.; Ogier A.; Ong A.-L.; Osterland M.; Otrocka M.; Peeters P. J.; Pilling J.; Prechtl S.; Qian C.; Rataj K.; Root D. E.; Sakata S. K.; Scrace S.; Shimizu H.; Simon D.; Sommer P.; Spruiell C.; Sumia I.; Swalley S. E.; Terauchi H.; Thibaudeau A.; Unruh A.; Van De Waeter J.; Van Dyck M.; Van Staden C.; Warchoł M.; Weisbart E.; Weiss A.; Wiest-Daessle N.; Williams G.; Yu S.; Zapiec B.; Żyła M.; Singh S.; Carpenter A. E.. JUMP Cell Painting Dataset: Morphological Impact of 136,000 Chemical and Genetic Perturbations. bioRxiv, 2023. 10.1101/2023.03.23.534023 (accessed 2023-06-29). [DOI] [Google Scholar]
- Seal S.; Yang H.; Vollmers L.; Bender A. Comparison of Cellular Morphological Descriptors and Molecular Fingerprints for the Prediction of Cytotoxicity- and Proliferation-Related Assays. Chem. Res. Toxicol. 2021, 34 (2), 422–437. 10.1021/acs.chemrestox.0c00303. [DOI] [PubMed] [Google Scholar]
- Chavan S.; Scherbak N.; Engwall M.; Repsilber D. Predicting Chemical-Induced Liver Toxicity Using High-Content Imaging Phenotypes and Chemical Descriptors: A Random Forest Approach. Chem. Res. Toxicol. 2020, 33 (9), 2261–2275. 10.1021/acs.chemrestox.9b00459. [DOI] [PubMed] [Google Scholar]
- Nyffeler J.; Haggard D. E.; Willis C.; Setzer R. W.; Judson R.; Paul-Friedman K.; Everett L. J.; Harrill J. A. Comparison of Approaches for Determining Bioactivity Hits from High-Dimensional Profiling Data. SLAS Discovery 2021, 26 (2), 292–308. 10.1177/2472555220950245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I.; Setzer R. W.; Jack J.; Houck K. A.; Judson R. S.; Knudsen T. B.; Liu J.; Martin M. T.; Reif D. M.; Richard A. M.; Thomas R. S.; Crofton K. M.; Dix D. J.; Kavlock R. J. Using ToxCastTM Data to Reconstruct Dynamic Cell State Trajectories and Estimate Toxicological Points of Departure. Environ. Health Perspect. 2016, 124 (7), 910–919. 10.1289/ehp.1409029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyffeler J.; Willis C.; Harris F. R.; Foster M. J.; Chambers B.; Culbreth M.; Brockway R. E.; Davidson-Fritz S.; Dawson D.; Shah I.; Friedman K. P.; Chang D.; Everett L. J.; Wambaugh J. F.; Patlewicz G.; Harrill J. A. Application of Cell Painting for Chemical Hazard Evaluation in Support of Screening-Level Chemical Assessments. Toxicol. Appl. Pharmacol. 2023, 468, 116513 10.1016/j.taap.2023.116513. [DOI] [PubMed] [Google Scholar]
- Caicedo J. C.; Singh S.; Carpenter A. E. Applications in Image-Based Profiling of Perturbations. Curr. Opin. Biotechnol. 2016, 39, 134–142. 10.1016/j.copbio.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Scheeder C.; Heigwer F.; Boutros M. Machine Learning and Image-Based Profiling in Drug Discovery. Curr. Opin. Syst. Biol. 2018, 10, 43–52. 10.1016/j.coisb.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.; Harrison P. J.; Sreenivasan A. P.; Carreras-Puigvert J.; Spjuth O. Combining Molecular and Cell Painting Image Data for Mechanism of Action Prediction. Artificial Intelligence in the Life Sciences 2023, 3, 100060 10.1016/j.ailsci.2023.100060. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.