Abstract
Background
In recent years, machine learning (ML) has had notable success in providing automated analyses of neuroimaging studies, and its role is likely to increase in the future. Thus, it is paramount for clinicians to understand these approaches, gain facility with interpreting ML results, and learn how to assess algorithm performance.
Objective
To provide an overview of ML, present its role in acute stroke imaging, discuss methods to evaluate algorithms, and then provide an assessment of existing approaches.
Methods
In this review, we give an overview of ML techniques commonly used in medical imaging analysis and methods to evaluate performance. We then review the literature for relevant publications. Searches were run in November 2021 in Ovid Medline and PubMed. Inclusion criteria included studies in English reporting use of artificial intelligence (AI), machine learning, or similar techniques in the setting of, and in applications for, acute ischemic stroke or mechanical thrombectomy. Articles that included image-level data with meaningful results and sound ML approaches were included in this discussion.
Results
Many publications on acute stroke imaging, including detection of large vessel occlusion, detection and quantification of intracranial hemorrhage and detection of infarct core, have been published using ML methods. Imaging inputs have included non-contrast head CT, CT angiograph and MRI, with a range of performances. We discuss and review several of the most relevant publications.
Conclusions
ML in acute ischemic stroke imaging has already made tremendous headway. Additional applications and further integration with clinical care is inevitable. Thus, facility with these approaches is critical for the neurointerventional clinician.
In the last decade, the role of neuroimaging in acute ischemic stroke (AIS) has increased considerably with the establishment of endovascular therapy (EVT). Now more than ever, there is a tremendous need for high-quality, universally available, and rapidly performed AIS imaging interpretations. In response to this need, machine learning (ML) approaches have been increasingly used, and the motivation for their use is clear—reducing even a few minutes of delay in treatment can improve clinical outcomes. These algorithms already play a central role in patient triage for AIS treatments, and in the coming years the number of applications making use of ML techniques is certain to grow. Thus, it is important for neurointerventional practitioners to understand the fundamentals of ML, including how ML works, how to evaluate existing algorithms, and reasons to implement them in their own practices. Building from there, an understanding of the capabilities of ML coupled with a knowledge of clinical needs would enable improvement of existing approaches or development of novel ones, with the potential for transformative changes in clinical practice.1
In this review, we provide an overview of ML, present its role in acute stroke imaging, discuss methods to evaluate algorithms, and then provide an assessment of existing approaches. We review the various algorithms currently in clinical use for AIS imaging and provide insights for future applications of ML in AIS beyond neuroimaging.
OVERVIEW OF MACHINE LEARNING IN STROKE IMAGING
Medical image computing (MIC) is the interdisciplinary research area that uses computational methods to analyze or transform medical images. In the last 20 years, more and more medical image computing pipelines have started to rely on ML algorithms to produce their output.2 Common MIC tasks include classification to automatically assign a label to an image (ie, the presence or absence of large vessel occlusion (LVO)) and semantic segmentation to assign a different label to each pixel of an image (ie, infarct core, penumbra, or healthy brain).
In stroke applications using data acquired from clinical scanners, brain/neck imaging pipelines involving ML algorithms typically have these initial preprocessing steps in common:
Field of view detection. The areas of interest, such as brain or neck, are detected and the remaining parts of the image are cropped out.
Skull stripping. For brain imaging application, the brain is isolated using segmentation algorithms.
Registration. The image is aligned to a common space, either with rigid or non-rigid approaches. In some cases, the registration is performed in the opposite direction, where an image template in a common space is aligned to the patient space. In other uncommon cases, this step can be skipped if the subsequent algorithm is insensitive to rotation, translation, scaling, and if it is not necessary to use an image atlas as part of the algorithm.
Currently, these preprocessing steps are often performed without ML approaches, and implemented in software packages such as FSL,3 Free-Surfer,4 Elastix,5 or ITK.6 However, active research has identified ML strategies to improve the speed and accuracy of these steps, especially for skull stripping7 and registration,8 which are the most time consuming and susceptible to errors. Automated image quality estimation is another step that can be added to the preprocessing pipeline in order to flag images that do not have the required characteristics for the subsequent ML algorithm.
After these preprocessing steps the actual ML model can be used. ML models have two phases: a training phase, and an inference phase. During training, the model attempts to learn the appropriate internal parameters that allow it to produce outputs as close to the ground truth as possible. During inference, the model parameters are fixed, and the model analyzes data unseen during the training phase. ML algorithms can be trained in a supervised, unsupervised, or semisupervised manner.8 Supervised training requires a predefined dataset with corresponding target variables (also known as ground truth) to learn from; examples are numerical variables describing modified Rankin Scale or manually drawn areas of infarct core on imaging. Unsupervised training is based on naturally occurring patterns in the dataset, without using any ground truth. These techniques are less commonly used in AIS applications. There are also semisupervised methodologies that can improve on a supervised model by adding unlabeled data able to give more information on the inner structure of the dataset. In stroke applications, supervised models are the most common, as they provide the best performance with the most immediate clinical application.
Even after the preprocessing steps previously described, the input data might still need to be further prepped. This process is called feature engineering, in which the relevant features (as determined by an expert with knowledge of the field) are extracted from the dataset.9 An example may be a threshold based on Hounsfield units when looking for acute hemorrhage. This approach works well when the dataset is small or the pattern on the data to be learned is well defined. However, feature engineering with imaging data typically involves a lot of trial and error and needs to be performed by researchers with significant experience in image processing. Another approach is feature learning, wherein the algorithm can learn to extract these features automatically. Doing so greatly speeds up development time and allows the creation of general-purpose ML algorithms that can work with many data sources and MIC tasks. The main drawbacks of this approach are that it requires much larger data-sets, and the internal representation is not always fully predictable, creating concerns of interpretability.
Deep learning ML algorithms are, by far, the most common type of feature learning ML approaches. They are essentially neural networks composed of hierarchical layers, where the lower layers are designed to learn compact data representation, and layers higher in the hierarchy are designed to use this higher-level data representation to learn a set of parameters enabling the final algorithm decision. In stroke imaging, the most common layers used to learn a compact data representation are called convolutional layers, and these types of model are called deep convolutional neural networks.10
MEASURING THE PERFORMANCE OF ML IMAGING ALGORITHMS
As ML algorithms are increasingly incorporated into routine practice, it has become increasingly important for clinicians to understand how these algorithms are designed, and how they can be evaluated. In the same way that clinicians should have facility in interpreting p-values, ORs, and other statistical methods used in clinical trials so that they may make an informed decision on the relevance of the trial results to their practice, the same is true for methods for evaluating ML algorithms. In this section, we will introduce some of the commonly used metrics for imaging-based ML methods, including ones that are already being used in the cerebrovascular literature for topic-relevant tasks. In the next section, we will describe some of those commercially used or published algorithms and then understand their performance using these metrics.
DICHOTOMIZED OUTPUTS (CLASSIFICATION TASKS)
One of the common ML tasks is that of classification. An input is given to an algorithm, and it is tasked with determining into which of two categories the data belong. In the cerebrovascular space, the most common example is that of LVO detection—there is either an LVO (ie, ‘yes’) or not (ie, ‘no’). This type of output can also be used in other tasks, including dichotomized measures of infarct core or penumbra (ie, above or below a certain volume threshold).
Classification performance can be measured with receiver-operator curves (ROCs) and further simplified to a single scalar number as the area under the curve (AUC) metric. In many instances, however, a related curve called precision-recall (PR) may be preferable to ROC. Unlike ROC, PR will be more robust in cases of significant imbalance between the two classes yes vs no), such as when the event is very rare. As an example, consider an algorithm screening all non-contrast head CT scans to detect LVO. The frequency of LVO will probably be low in this dataset, and an ROC may overestimate model performance in this setting. PR curves are created by plotting precision (true positives/(true positives+false positives) versus recall (ie, sensitivity), and can similarly be evaluated using an AUC metric. Like ROC, the closer to 1.0, the better the performance, and an AUC of 0.5 indicates a performance equivalent to random guessing.
IMAGE OUTPUTS (INCLUDING SEGMENTATION)
Another common output for imaging-based ML methods in cerebrovascular disease is an image output or segmented volume output. Examples of such algorithms include those that analyze CT or MRI imaging and output the volume of infarcted tissue. Quantifying the performance of such methods can be slightly more challenging. Here, we will demonstrate why, and then describe the calculation for one of the common metrics called the dice similarity coefficient (DSC, also known as the F1 score).11
One of the simplest methods to measure performance might be to calculate the percentage of pixels that the algorithm annotated correctly. The formula for this measurement would be:
However, as shown in figure 1, this approach fails in most situations. Suppose the ground truth is given in figure 1A and the hypothetical algorithm output is given in figure 1B. The black boxes in figure 1 measure 200×200 pixels, and figure 1A has three red squares, each measuring 25×25 pixels. Thus, there are 1875 red pixels out of the total 40 000. A segmentation algorithm that fails to identify any red squares, as in figure 1B, would give a pixel accuracy of over 95%! Thus, a metric that considers not just the areas identified correctly, but the total regions of the two images is needed.
Figure 1.
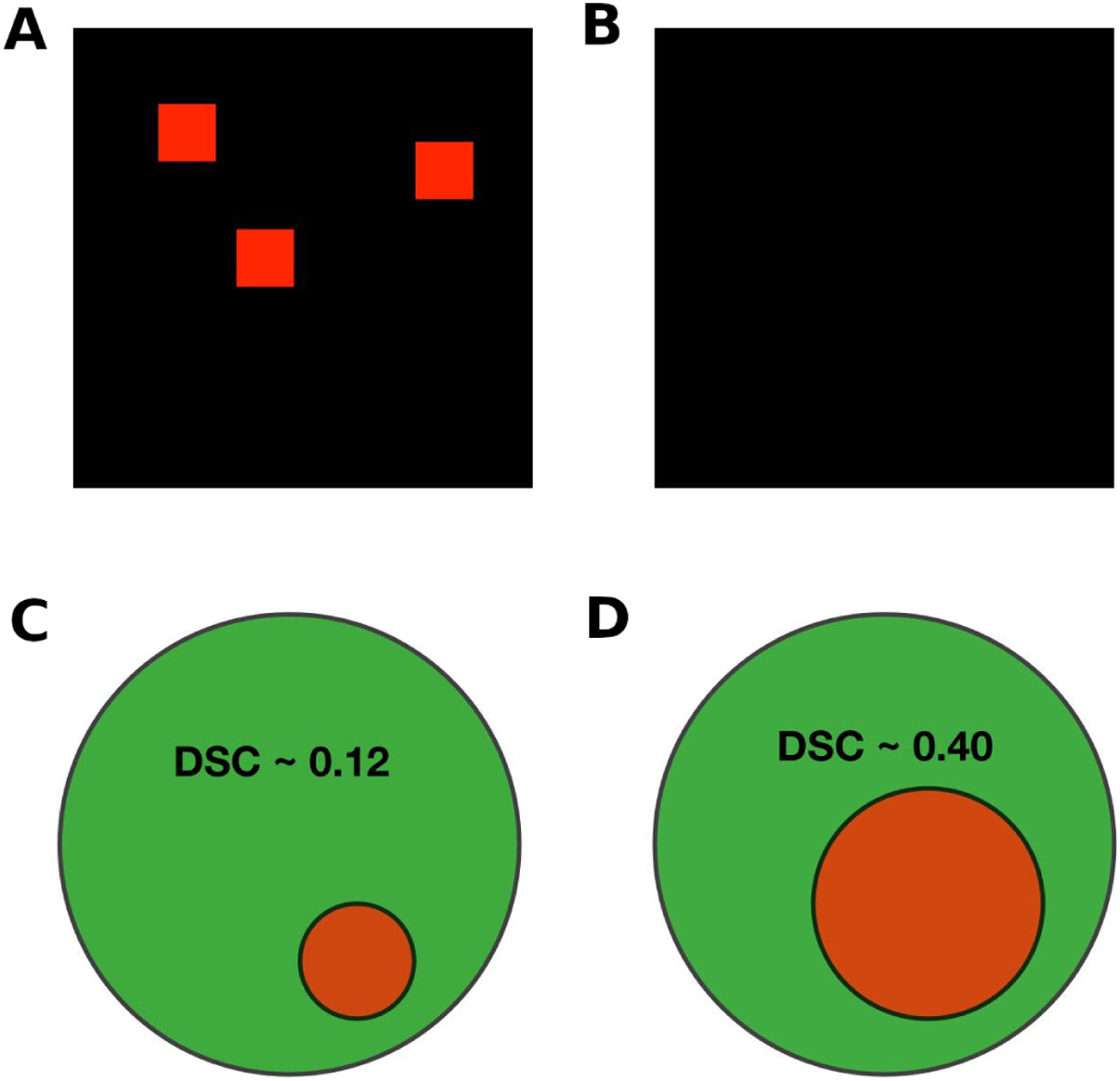
Examples of segmentation tasks and results. (A) A sample ‘ground truth’ image; (B) an example algorithm output; (C) and (D) examples of segmentation results. The dice similarity coefficients (DSCs) are given for two sample outputs, in which the green circles represent a ‘ground truth’ and the red circles the sample algorithm outputs.
The DSC serves such a role, and is calculated by the following formula:
This measurement is more robust to class imbalances unlike pixel accuracy, which suffered from this problem in the example illustrated above. Using the example of figure 1, the DSC would be a much more modest 48% (an average of 95% for the black color and 0% for the red color).
It is worth having an intuitive sense of segmentation accuracy as measured by the DSC, and examples are given in figure 1C,D. A clinically relevant correlate would be an infarct core that is represented by the green circle, and the red circle is an algorithm’s estimation of the infarct core. As shown in the figure, a DSC value of 0.4, which is the range of performance of many algorithms we discuss in the next section, may not ‘look’ like an impressive segmentation result.
EXAMPLES OF ML ALGORITHMS CURRENTLY IN USE
ML has already made significant inroads in AIS care, and here we review some of the most relevant published works. The literature was searched by two authors. Search strategies were created using a combination of keywords and standardized index terms, which included ‘artificial intelligence’, ‘automated’, ‘machine learning’, ‘deep learning’, ‘stroke’, ‘thrombectomy’, ‘large vessel occlusion’. Searches were run in November 2021 in Ovid Medline and PubMed. Inclusion criteria were studies in English reporting a use of AI, machine learning, or similar techniques in the setting of, or in applications for, AIS or mechanical thrombectomy. Articles that included image-level data with meaningful results and sound ML approaches were included in this discussion.
Given the extreme time-sensitivity of AIS, it is not surprising that one of the first applications of ML in clinical medicine was for rapid triage and identification of possible EVT candidates. Figure 2 depicts the workflow enhancement that such algorithms could produce. Viz.AI created one of the earliest such applications, employing a convolutional neural network method for detection of LVO from CT angiography (CTA) images.12 The performance of this algorithm was evaluated in a recent single-center study. A total of 1167 CTA scans were included, from which 75 were confirmed LVO cases (M1 (n=47), carotid terminus (n=28)), and the algorithm detected the occlusions in a robust fashion (AUC of 0.91).13
Figure 2.
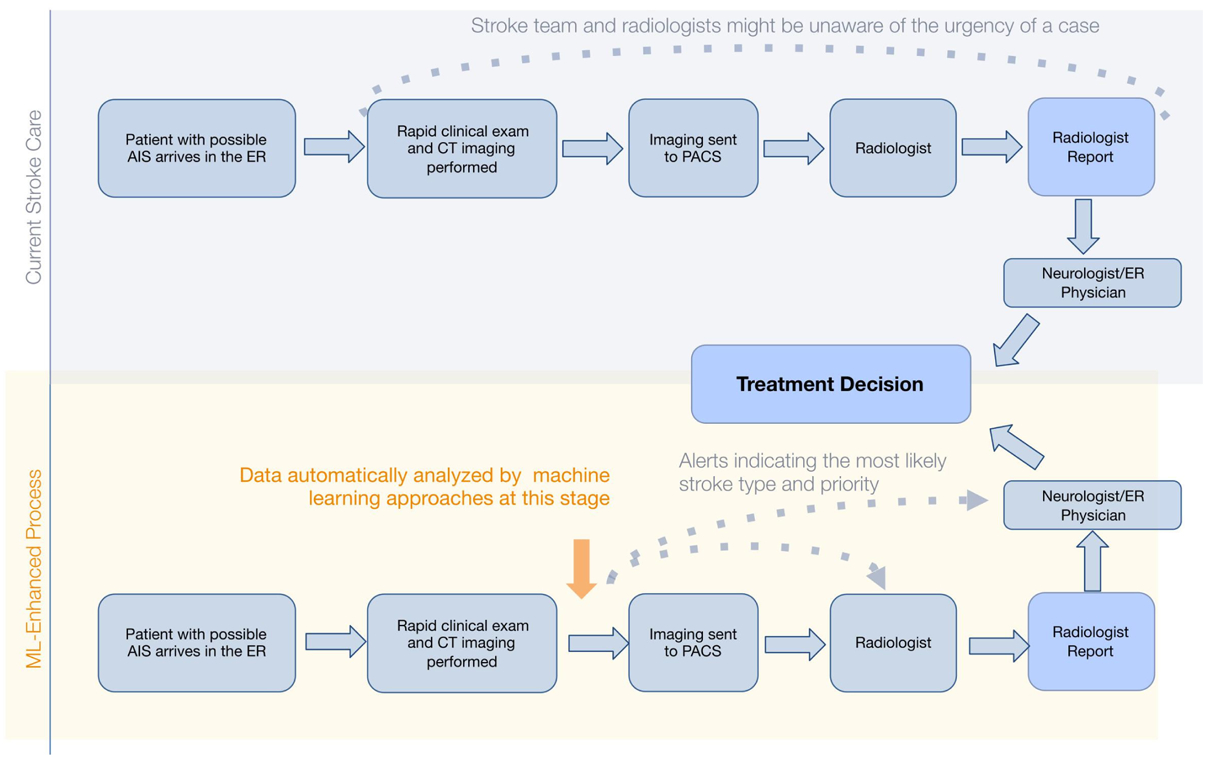
Process map demonstrating the benefit of automated detection of large vessel occlusion in acute ischemic stroke triage workflows. AIS, acute ischemic stroke; ER, emergency room; ML, machine learning; PACS, picture archiving and communication system.
Because time to treatment is such a vital driver of outcomes in AIS, the Centers for Medicare and Medicaid Services recently approved a new technology add-on payment for LVO detection algorithms. Prior publications have demonstrated that these algorithms reduce treatment times for patients presenting at, or transferring to, endovascular-capable centers.14 15 In a health econometrics study, accelerating thrombectomy by an average of 10 min across the USA would save an estimated $249 million annually.16 By subsidizing the cost of implementing LVO-detection ML algorithms, the new technology add-on payment enables implementation of the software at high-volume centers, and also at lower-volume centers, where the need may be even greater.17
ML algorithms for segmentation have also been applied extensively to identify infarct core from cross-sectional imaging acquisition, particularly non-contrast head CT (NCHCT) and CT perfusion (CTP). The ISLES challenge invited teams to analyze CTP images using MRI DWI as a ground truth. As segmentation algorithms, they were evaluated using DSC. Peak performance of the convolutional neural networks developed for this challenge approached a DSC of 0.51 (±0.31).18 Many CTP postprocessing algorithms, including RAPID (IschemaView), produce segmentation maps of regions predicted to be infarct core and penumbra; however, the performance of these segmentation algorithms is not readily available. In the ISLES challenge, a pure threshold-based approach, which RAPID uses, obtained a DSC of 0.34 (±0.29) compared with DWI ground truth.
Lee et al tested several ML methodologies analyzing MRI DWI and fluid-attenuated inversion recovery (FLAIR) images to identify patients presenting with unknown onset time who might qualify for thrombolysis.19 They used logistic regression, support vector machine, and random forest algorithms against human readings of DWI–FLAIR mismatch. All three ML methods were able to identify patients with AIS within the time window with superior sensitivity (>70%) and comparable specificity (>85%) compared with human readings (48.5% sensitivity and 91.3% specificity).
Because infarct core determinations play such an important role in treatment eligibility for revascularization procedures for LVO AIS, many groups have developed algorithms to determine infarct core using imaging modalities more readily available than CTP and DWI.20 Our group used CTA to define infarct core, given the relative ubiquity of this technique relative to CTP and emergent DWI. The algorithm, named DeepSymNet, was trained against CTP-RAPID outputs as the gold standard. Using a dichotomous outcome, with cut-off points set as ≤30 mL and ≤50 mL for infarct core size, the algorithm performed with AUC 0.88 and 0.90, respectively. Its performance was robust across the full range of infarct sizes and time windows of presentation relative to imaging. In a subsequent study, an updated version of DeepSymNet was compared against CTP-RAPID with DWI as the ground truth. The ML model, using CTA images alone, was equivalent to, if not superior to, CTP-RAPID.21 Figure 3 provides a visual overview of the training and possible implementation of this approach.
Figure 3.
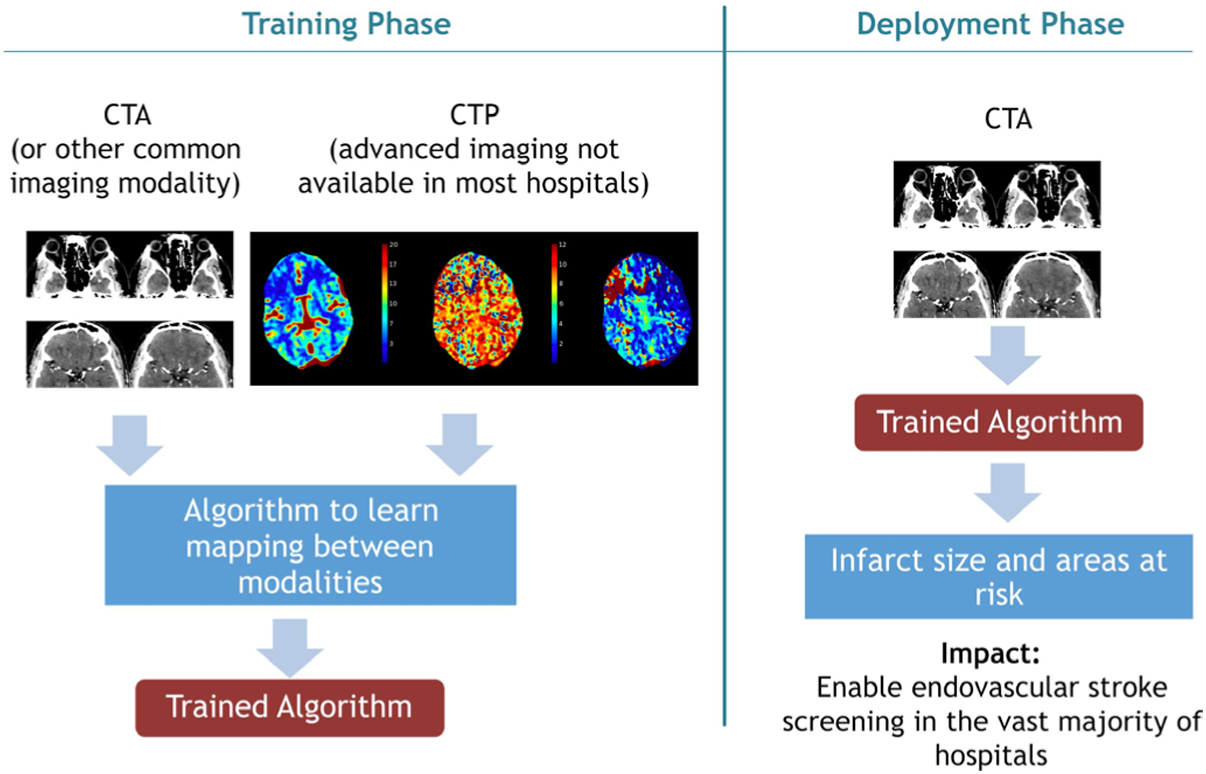
Schematic detailing development and deployment of a novel machine learning algorithm detecting infarct core from CT angiography (CTA), by training on CT perfusion (CTP).
Accurate segmentation of infarct core from NCHCT would be of tremendous benefit, and a recent publication presented an algorithm to that end, using MRI as the ground truth. Although a more formal comparison of segmented volumes using a metric such as DSC was not provided, the correlation appeared strong.22 This approach has also been used by commercial ventures, including Brainomix (Oxford, UK) with their eASPECTS software. This algorithm was recently shown to correlate relatively well with human expert reads of Alberta Stroke Program Early CT (ASPECT) scores, and the best among three algorithms that were tested.23 Another recent publication demonstrated reasonable success for early ischemic changes in posterior circulation ASPECTS using NCHCT.24
One of the most exciting possibilities for ML in AIS is extending the usefulness of imaging beyond what we have traditionally considered. In a recent study, an algorithm named Methinks was able to detect LVO from NCHCT alone with reasonable accuracy. The MethinksLVO (Methinks, Barcelona, Spain) and MethinksLVO+ (in which National Institutes of Health Stroke Scale score was included in the model) performed with AUC of 0.87 and 0.91, respectively.25 This approach could be useful in accelerating stroke care, by avoiding the need for CTA. Another study trained an algorithm using as the input data the region on NCHCT where a middles cerebral artery occlusion was harbored. The algorithm was able to learn the likelihood of successful aspiration thrombectomy (AUC 0.88), and even predict the number of EVT passes that would be required for substantial reperfusion (Pearson correlation coefficient of 0.73).26
APPLICATIONS OF MACHINE LEARNING FOR THE CLINICIAN
Workflow optimization
ML models hold great promise in optimizing daily clinical work-flow. During image acquisition, ML models can help reduce scan times27 and improve image quality of both cross-sectional and digital subtraction angiography studies.28 29 ML models have been shown to facilitate neuroimaging triaging through early smartphone app notifications of critical findings such as LVOs or quantification of ischemic penumbra/core to the treatment team (examples of software packages include RapidAi or Viz. ai).14 30 Similarly, automated detection of intracranial hemorrhage (ICH) from NCHCT can help with screening for thrombolytic eligibility, reducing the risk of errors, and accelerating patient triage.31
Automated detection of ICH can be paired with flagging on the diagnostic reading list, a feature available on commercial software packages such as Aidoc (Aidoc, Tel Aviv, Israel), Rapid ICH or Viz ICH (Viz.Ai), helping prioritization of the radiologist’s workflow. Ginat showed how ML-aided flagging of positive cases is associated with a significantly shorter view and read delay than unflagged cases.32
Clinical trial screening
ML has also been proposed for use in clinical trial screening. Current applications focus on automated enrollment and patient screening. The ENRICH trial is one of the first studies on hemorrhagic stroke to use automated ICH detection software (Viz ICH) and a ML automated recruiting software (Viz Recruit) to actively enroll potential patients, with a significant increase in screening and enrollment rates.33
Moreover, ML offers the potential to maximize the success and efficiency of clinical trials. Trials.ai is an application where investigators can upload their protocols, evaluate, and adjust for potential pitfalls identified through natural language processing algorithms. This can simulate the trial before the protocol is even fully developed, helping to optimize planning and design.
Future applications
The potential clinical applications of AI in acute neuroimaging go far beyond the image analysis tasks that are currently implemented; ML algorithms are far from being fully integrated into clinical settings. Future investigations should focus on providing on-the-fly critical clinical information regarding outcomes and risk of potential complications.34–37 These patient-specific predictions can support the time-sensitive decision-making required in acute stroke interventions. We envision individualized predicted feasibility and benefits of reperfusion, risk of hemorrhagic complications, and even 90-day forecasted modified Rankin Scale scores, which can be presented to the neurointerventionalist before the procedure as decision support. Detection and volumetric analysis of chronic subdural hematomas will also help to identify potential candidates for middle meningeal artery embolization.38 39
CONCLUSIONS
Neurointerventional physicians are fortunate to be in a field that has actively embraced ML. It is hard to think of another area of medicine that has incorporated ML algorithms so seamlessly into daily clinical practice. Financial incentives to hospitals through programs like new technology add-on payment have probably played a substantial role. The workflow enhancements and improvement of clinical outcomes are already apparent, within only a few years of software availability. Looking to the future, the expansion of ML is inevitable. In the near future, ML algorithms will probably play a larger role in identification of hemorrhagic disease and treatment determinations. Algorithms will become more sophisticated, and probably include other imaging sources including clinical data to provide predictions on intermediate variables (such as infarct core, hemorrhage size) and also outcome variables (such as likelihood of improvement with or without treatment). Real-time interpretation of imaging is also a possibility. And as neurointervention itself becomes more sophisticated with brain–computer interfaces, optical coherence tomographic imaging, and other new techniques arriving on the scene, ML will undoubtedly play an increasingly prominent role.
Supplementary Material
Funding
SAS and LG report funding from the National Institutes of Health (R01NS121154).
Footnotes
Competing interests PK is on the editorial board of JNIS.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Additional supplemental material for this paper are available online. To view these files, please visit the journal online (http://dx.doi.org/10.1136/neurintsurg-2021-018142).
REFERENCES
- 1.Obermeyer Z, Lee TH. Lost in thought - the limits of the human mind and the future of medicine. N Engl J Med 2017;377:1209–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronneberger O, Fischer P. Brox T. U-Net: Convolutional networks for biomedical image segmentation. Arxiv 2015. [Google Scholar]
- 3.Jenkinson M, Beckmann CF, Behrens TEJ, et al. FSL. Neuroimage 2012;62:782–90. [DOI] [PubMed] [Google Scholar]
- 4.Fischl B FreeSurfer. Neuroimage 2012;62:774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein S, Staring M, Murphy K, et al. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196–205. [DOI] [PubMed] [Google Scholar]
- 6.McCormick M, Liu X, Jomier J, et al. ITK: enabling reproducible research and open science. Front Neuroinform 2014;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleesiek J, Urban G, Hubert A, et al. Deep MRI brain extraction: a 3D convolutional neural network for skull stripping. Neuroimage 2016;129:460–9. [DOI] [PubMed] [Google Scholar]
- 8.de Vos BD, Berendsen FF, Viergever MA, et al. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal 2019;52:128–43. [DOI] [PubMed] [Google Scholar]
- 9.Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 2013;35:1798–828. [DOI] [PubMed] [Google Scholar]
- 10.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 11.Tiu E Metrics to evaluate your semantic segmentation model https://towardsdatascience.com/metrics-to-evaluate-your-semantic-segmentation-model-6bcb99639aa2 [Accessed 20 May 2022].
- 12.Murray NM, Unberath M, Hager GD, et al. Artificial intelligence to diagnose ischemic stroke and identify large vessel occlusions: a systematic review. J Neurointerv Surg 2020;12:156–64. [DOI] [PubMed] [Google Scholar]
- 13.Yahav-Dovrat A, Saban M, Merhav G, et al. Evaluation of artificial Intelligence-powered identification of large-vessel occlusions in a comprehensive stroke center. AJNR Am J Neuroradiol 2021;42:247–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan AE, Ringheanu VM, Rabah RR, et al. Early experience utilizing artificial intelligence shows significant reduction in transfer times and length of stay in a hub and spoke model. Interv Neuroradiol 2020;26:615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morey JR, Zhang X, Yaeger KA, et al. Real-world experience with artificial Intelligence-based triage in transferred large vessel occlusion stroke patients. Cerebrovasc Dis 2021;50:450–5. [DOI] [PubMed] [Google Scholar]
- 16.Kunz WG, Hunink MG, Almekhlafi MA, et al. Public health and cost consequences of time delays to thrombectomy for acute ischemic stroke. Neurology 2020;95:e2465–75. [DOI] [PubMed] [Google Scholar]
- 17.Hassan AE. New Technology Add-on Payment (NTAP) for Viz LVO: a win for stroke care. J Neurointerv Surg 2021;13:406–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakim A, Christensen S, Winzeck S, et al. Predicting infarct core from computed tomography perfusion in acute ischemia with machine learning: lessons from the Isles challenge. Stroke 2021;52:2328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Lee E-J, Ham S, et al. Machine learning approach to identify stroke within 4.5 hours. Stroke 2020;51:860–6. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Lee S, Abdelkhaleq R, et al. Utilization and availability of advanced imaging in patients with acute ischemic stroke. Circ Cardiovasc Qual Outcomes 2021;14:e006989. [DOI] [PubMed] [Google Scholar]
- 21.Abdelkhaleq R, Kim Y, Khose S, et al. Automated prediction of final infarct volume in patients with large-vessel occlusion acute ischemic stroke. Neurosurg Focus 2021;51:E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu W, Kuang H, Teleg E, et al. Machine learning for detecting early infarction in acute stroke with non-contrast-enhanced CT. Radiology 2020;294:638–44. [DOI] [PubMed] [Google Scholar]
- 23.Hoelter P, Muehlen I, Goelitz P, et al. Automated ASPECT scoring in acute ischemic stroke: comparison of three software tools. Neuroradiology 2020;62:1231–8. [DOI] [PubMed] [Google Scholar]
- 24.Kniep HC, Sporns PB, Broocks G, et al. Posterior circulation stroke: machine learning-based detection of early ischemic changes in acute non-contrast CT scans. J Neurol 2020;267:2632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olive-Gadea M, Crespo C, Granes C, et al. Deep learning based software to identify large vessel occlusion on noncontrast computed tomography. Stroke 2020;51:3133–7. [DOI] [PubMed] [Google Scholar]
- 26.Hofmeister J, Bernava G, Rosi A, et al. Clot-based radiomics predict a mechanical thrombectomy strategy for successful recanalization in acute ischemic stroke. Stroke 2020;51:2488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 2018;392:2388–96. [DOI] [PubMed] [Google Scholar]
- 28.Ueda D, Katayama Y, Yamamoto A, et al. Deep learning–based angiogram generation model for cerebral angiography without misregistration artifacts. Radiology 2021;299:203692. [DOI] [PubMed] [Google Scholar]
- 29.McKinley R, Hung F, Wiest R, et al. A machine learning approach to perfusion imaging with dynamic susceptibility contrast MR. Front Neurol 2018;9:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morey JR, Fiano E, Yaeger KA. Impact of Viz LVO on time-to-treatment and clinical outcomes in large vessel occlusion stroke patients presenting to primary stroke centers. Medrxiv. [Google Scholar]
- 31.Austein F, Wodarg F, Jürgensen N, et al. Automated versus manual imaging assessment of early ischemic changes in acute stroke: comparison of two software packages and expert consensus. Eur Radiol 2019;29:6285–92. [DOI] [PubMed] [Google Scholar]
- 32.Ginat D Implementation of machine learning software on the radiology worklist decreases scan view delay for the detection of intracranial hemorrhage on CT. Brain Sci 2021;11:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987;40:373–83. [DOI] [PubMed] [Google Scholar]
- 34.Jiang B, Zhu G, Xie Y, et al. Prediction of clinical outcome in patients with large-vessel acute ischemic stroke: performance of machine learning versus SPAN-100. AJNR Am J Neuroradiol 2021;42:240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo J, Yoon JG, Park H, et al. Machine learning-based model for prediction of outcomes in acute stroke. Stroke 2019;50:1263–5. [DOI] [PubMed] [Google Scholar]
- 36.Brugnara G, Neuberger U, Mahmutoglu MA, et al. Multimodal predictive modeling of endovascular treatment outcome for acute ischemic stroke using machine-learning. Stroke 2020;51:3541–51. [DOI] [PubMed] [Google Scholar]
- 37.Nishi H, Oishi N, Ishii A, et al. Predicting clinical outcomes of large vessel occlusion before mechanical thrombectomy using machine learning. Stroke 2019;50:2379–88. [DOI] [PubMed] [Google Scholar]
- 38.Farzaneh N, Williamson CA, Jiang C, et al. Automated segmentation and severity analysis of subdural hematoma for patients with traumatic brain injuries. Diagnostics 2020;10:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellogg RT, Vargas J, Barros G, et al. Segmentation of chronic subdural hematomas using 3D convolutional neural networks. World Neurosurg 2021;148:e58–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


