Abstract
Respiratory syncytial (RS) viruses isolated over three epidemic periods in a children’s hospital in the United States were analyzed. The viruses (n = 174) were characterized as to major antigenic group (group A or B) by a PCR-based assay. Group A RS viruses were dominant the first 2 years, followed by a year with group B dominance (ratios of group A to group B viruses for epidemic periods, 56/4 for 1993–1994, 42/3 for 1994–1995, and 19/50 for 1995–1996). Genetic variability within the groups was assessed by restriction fragment analysis of PCR products; 79 isolates were also analyzed by nucleotide sequence determination of a variable region of the glycoprotein G gene. Among the group A RS virus isolates, this G-protein variable region had amino acid differences of as great as 38%. The G-protein amino acids of the group A viruses differed by up to 31% from the G-protein amino acids of a prototype (A2) group A virus. Among the group B RS virus G proteins, amino acid differences were as great as 14%. The G-protein amino acids of the group B viruses differed by up to 27% from the G-protein amino acids of a prototype (18537) group B virus. The group A and group B RS viruses demonstrated genetic variability between years and within individual years. Phylogenetic analysis revealed that there were multiple evolutionary lineages among both the group A and group B viruses. Among the recent group B isolates, variability was less than that seen for the group A viruses. However, comparisons to prototype strains revealed that the group B RS viruses may vary more extensively than was observed over the 3 years studied in the present investigation.
Respiratory syncytial (RS) virus is the most common viral cause of lower respiratory tract infection in infants and young children. Annual fall and winter epidemics occur in temperate regions (10). Two major antigenic groups, groups A and B, of RS virus were originally delineated by their reactivities with monoclonal antibodies (MAbs) (3, 19). Genetic diversity of the protein-G genes occurs within and between the two groups of RS virus (16, 22).
Epidemiologic studies conducted in the United States with MAbs to define the antigenic groups showed that there are three types of RS virus epidemics: those in which group A or group B viruses were dominant and those in which both groups circulate concurrently (2, 13, 14). Multiple lineages or strains of RS virus cocirculate (2, 6). The analysis of group A clinical isolates from successive epidemics in Birmingham, United Kingdom, showed that different lineages predominated in each epidemic and that not all lineages were present in every epidemic (7). Clinical isolates of group A RS virus collected in Uruguay and Spain have been analyzed. Viruses from separate phylogenetic branches were isolated during the same epidemic period, and very similar viruses were isolated in distant places and different years (12).
Several methods have been used to categorize RS virus clinical isolates as to group and to assess variability within the groups. In addition to antigenic characterization with MAbs, these methods include restriction endonuclease analysis of small hydrophobic and nucleocapsid RS virus protein genes (8), restriction endonuclease analysis of G-protein gene cDNA (5, 24), RNase protection analysis (11, 21), and nucleotide sequence analysis of the G-protein gene (5, 23).
In this study, we analyzed RS viruses from the Children’s Hospital of Alabama from three consecutive epidemic periods (1993 to 1996). The viruses were characterized as to group, and variability within the groups was assessed by restriction fragment analysis of amplified cDNAs and limited nucleotide sequencing of a variable region of the G-protein gene. Both group A and group B viruses were studied. The data presented here are the results of a molecular epidemiologic study of RS virus in a children’s hospital in the United States over three epidemic periods.
(This work was presented in part at the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., 17 to 20 September 1995.)
MATERIALS AND METHODS
Cells and viruses.
Clinical samples were submitted to the diagnostic virology laboratory of the Children’s Hospital of Alabama for respiratory viral cultures. Samples which grew RS virus were recultivated in our laboratory, and a cell lysate was prepared for reverse transcription-PCR (RT-PCR) as described previously (24).
RT-PCR.
RNA was extracted from infected cell lysates by a hot phenol extraction procedure. The extracted viral RNA was used as a template for cDNA synthesis. RT-PCR was performed with primers F164, G32, and G267 as described previously (24). Agarose gel electrophoresis allowed a group designation to be made on the basis of the size of the DNA fragment of the PCR product: 1.1 kb in length for group B virus and 0.9 kb for group A virus. We repeated the RT-PCR for the group A viruses with primer G10 (GCAAACATGTCCAAAAACAAG; complementary to bases 10 to 30 of the G-protein mRNA of the A2 virus) and F164 to yield a longer 1.1-kb group A DNA product.
Restriction endonuclease analysis.
The DNA fragments obtained by PCR were analyzed for their genetic variability. Three restriction endonuclease enzymes, PstI, RsaI, and HincII, were used for the digestion of the group A PCR products. Group B virus PCR products were cut with AluI, RsaI, and HincII enzymes. Digestion and analysis of the resulting products were done as described previously (24). The patterns obtained from the clinical samples were assigned lowercase letter designations. Thus, each virus had a restriction pattern designated with three lowercase letters, e.g., abc or pno (24).
Nucleotide sequence analysis.
Selected isolates were evaluated by nucleotide sequence determination. The cDNA PCR products were purified by agarose gel electrophoresis followed by DNA extraction (Qiagen, Chatsworth, Calif.). Sequencing reactions were carried out with a Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science Inc., Cleveland, Ohio). The primers were G714 (GCCAACCATCAACACCACC; complementary to bases 714 to 722 of the A2 G-protein mRNA sequence) (9) for group A viruses or G718 (CCAACCCTCAAGACCAC; complementary to bases 718 to 734 of the 8/60 G-protein mRNA sequence) for group B viruses. A small number of group A viruses with restriction pattern abc required primers G546 and F16 for sequencing. Primer G546 (CCCTGCAGCATATGCAGC) corresponded to bases 529 to 546 of the A2 G protein mRNA. Primer F16 (GAGGATTGGCAACTCC) was complementary to bases 16 to 31 of a group A RS virus (strain WV12342) F protein mRNA. DNA and deduced amino acid sequence analyses were performed with the Wisconsin Package, version 9.1, Genetics Computer Group (Madison, Wis.) sequence analysis programs. Phylogenetic analysis was performed by using the Clustal X (26), PAUP (25), and MacClade (17) programs.
The percent nucleotide changes resulting in amino acid changes (see Table 2) represents the total number of amino acid changes divided by the total number of nucleotide changes determined by MacClade analysis of the neighbor-joining trees (1). The amino acid and nucleotide changes were determined by calculating the number of changes observed in each branch of the tree as one counts from the root of the tree to each terminal node. Then, the sum of all these changes is calculated for each indicated region of the protein. The G-protein amino acids included in each region that were compared (see Table 2) were chosen from the aligned amino acid sequences as follows for the group A and B viruses (numbers were based on the published sequences for the G proteins of the A2 [group A] and 8/60 [group B] viruses): region 1, 1 to 66 for group A and 6 to 83 for group B (the first 5 amino acids were omitted for group B because information was not available for all of the isolates); region 2, 67 to 157 for group A and 84 to 152 for group B; region 3, 158 to 207 for group A and 153 to 221 for group B; region 4, 208 to 298 for group A and 222 to 292 for group B; and region 5, 246 to 298 for group A and 249 to 292 for group B.
TABLE 2.
Percent nucleotide changes resulting in amino acid changes
| Sequence (region no.) | % Changes
|
|
|---|---|---|
| Group A RS virus | Group B RS virus | |
| Full length sequences | ||
| G-protein regions | ||
| N terminus (1) | 29 | 15 |
| First variable region in ectodomain (2) | 59 | 54 |
| Central conserved region in ectodomain (3) | 13 | 15 |
| Second variable region in ectodomain, C terminus (4) | 64 | 67 |
| Complete G protein | 54 | 52 |
| Variable region analyzed in this study (5)b | 61 | 66 |
The methods used for the calculation of the comparisons made here and the regions used are described in Materials and Methods. The full-length G-protein gene sequences compared are listed in the legend to Fig. 3.
The sequences were from previously published sequences and the sequences of isolates examined in the present study.
Nucleotide sequence accession numbers.
The nucleotide sequences corresponding to the amino acid sequences, presented in Fig. 1, of the viruses (the designations presented here differ from those in Fig. 1 by the addition of a prefix indicating the location [AL, Alabama] and month and year of isolation) were submitted to GenBank and given the indicated accession nos.: AL-12-93-179381, AF086868; AL-12-93-179522, AF086869; AL-1-94-180081, AF086870; AL-1-94-180893, AF086871; AL-1-94-181691, AF086872; AL-2-94-182473, AF086873; AL-2-94-182701, AF086874; AL-2-94-183221, AF086875; AL-3-94-184002, AF086876; AL-3-94-184431, AF086877; AL-4-94-185413, AF086878; AL-12-94-193563, AF086879; AL-12-94-193651, AF086880; AL-12-94-194522, AF086881; AL-11-94-194581, AF086882; AL-1-95-195462, AF086883; AL-2-95-195563, AF086884; AL-2-95-195901, AF086885; AL-3-95-196775, AF086886; AL-4-95-198921, AF086887; AL-10-95-203721, AF086888; AL-11-95-204664, AF086889; AL-11-95-204682, AF086890; AL-12-95-205342, AF086891; AL-12-95-205464, AF086892; AL-12-95-205865, AF086893; AL-1-96-206145, AF086894; AL-1-96-206344, AF086895; AL-1-96-206846, AF086896; AL-1-96-207347, AF086897; AL-2-96-207385, AF086898; and AL-11-93-177771, AF086899.
FIG. 1.
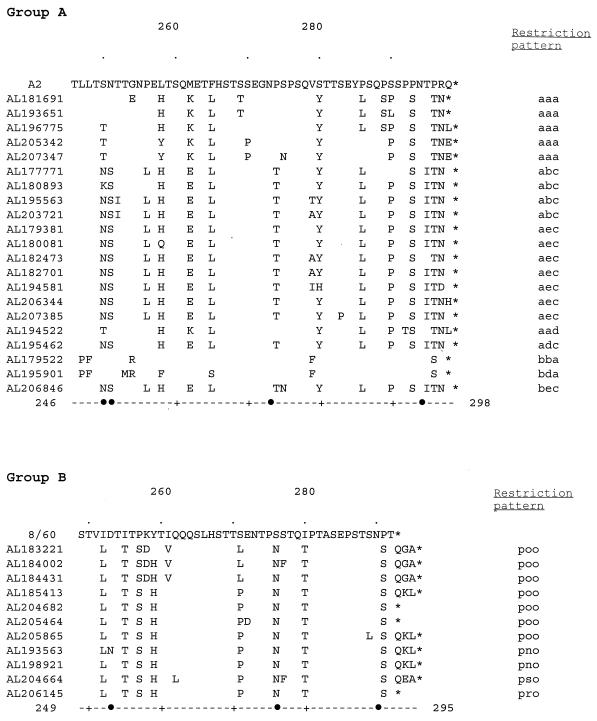
Alignment of the deduced amino acid sequences of G proteins of the group A (A) and group B (B) isolates from a children’s hospital. The sequences from amino acids 246 (group A) or 249 (group B) to the end of the G protein are shown in comparison with the sequence of prototype isolate A2 (A) or 8/60 (B). Differences in the sequences of the isolates from clinical samples in comparison to the sequences of the prototype isolates are shown. The name of each virus isolated in the Children’s Hospital of Alabama begins each line, and restriction patterns are shown at the end of the sequence. Symbols: •, potential N-linked glycosylation sites in any of the sequences; ∗, termination codons. The residues are indicated by numbers at the ends of the dashed lines at the bottom; dots above the sequences indicate 10-residue increments, with residues 260 and 280 being shown.
RESULTS
PCR analysis of intergroup RS virus variation and restriction fragment analysis of within-group variation.
RS viruses isolated over three annual epidemic periods (1993–1994, 1994–1995, and 1995–1996) at a children’s hospital in the United States were analyzed. The viruses were characterized as to group (group A or B) by a PCR-based assay. Genetic variability within the groups was initially assessed by restriction fragment analysis of the PCR products of 174 isolates (24). Some isolates (n = 79) were also analyzed by nucleotide sequence determination of a region of the glycoprotein-G gene (23).
Group A viruses were predominant during the first two epidemic periods, whereas the group B viruses dominated during the last period (Table 1). Restriction fragment analysis of the group A virus PCR products revealed additional genetic variability within the group, with four to seven restriction patterns observed during each period. Fewer group B viruses were available from the first two epidemic periods, and only one restriction pattern was observed; among the isolates from the last period, when more viruses were available, three group B restriction patterns were found. For the group A viruses the predominant restriction pattern was different among viruses from each of the three periods. Among the group B viruses the single pattern seen during the first period was also the dominant pattern during the third period.
TABLE 1.
Categorization of the group A and B RS viruses by their restriction patterns and distributions over 3 epidemic years
| Group and restriction patterna | No. of viruses isolated in the following epidemic years:
|
||
|---|---|---|---|
| 1993–1994 | 1994–1995 | 1995–1996 | |
| Group A | 56 | 42 | 19 |
| abc (48, 14, 4) | 44 | 1 | 3 |
| aec (18, 8, 7) | 7 | 1 | 2 |
| bba (40, 18, 1) | 4 | 33 | 3 |
| aaa (14, 9, 5) | 1 | 3 | 10 |
| aad (1) | 1 | ||
| adc (1) | 1 | ||
| bda (1) | 1 | ||
| bec (1) | 1 | ||
| Group B | 4 | 3 | 50 |
| pno (3, 3, 2) | 3 | ||
| poo (46, 16, 7) | 4 | 42 | |
| pso (7, 7, 1) | 7 | ||
| pro (1) | 1 | ||
See Materials and Methods for the categorization of viruses by restriction pattern. The numbers in parentheses are total number of viruses in restriction pattern group, number of sequences determined, number of unique amino acid sequences within restriction pattern group.
Nucleotide sequence analysis.
The restriction fragment analysis revealed that there was within-group genetic variability. To more accurately define the extent of genetic variability within the individual groups, nucleotide sequences were determined for part of the G-protein genes for selected isolates. At least one virus from each restriction pattern group from each period was analyzed, and for restriction pattern groups with multiple isolates, additional viruses were tested (Table 1). The region for which sequences were determined is one of two variable regions of the protein and is C terminal to the central conserved region of the G protein. The nucleotides determined corresponded to bases 750 to 918 in the prototype group A virus (A2 strain) mRNA for the group A viruses (28). For the group B viruses the nucleotides determined corresponded to bases 760 to 921 in the prototype group B virus (8/60 strain) mRNA (22). Within each restriction pattern group for which more than one isolate was analyzed, nucleotide sequence variability was noted for all except one group. All 18 group A RS viruses with restriction pattern bba had identical nucleotide sequences over the three epidemic periods.
Amino acid sequence analysis.
For each restriction pattern group, one example of each unique amino acid sequence is shown (Fig. 1). Deduced amino acid sequences for group A RS virus were 52 or 53 residues, which would result in G-protein lengths of 297 or 298 amino acids (assuming normal reading frame usage for the full protein). All but two of the group A amino acid sequences included recognition signals for potential N-linked sugar addition at either residue 250 or residue 251, some of the isolates had an additional potential N-linked sugar site at residue 273, and all of the isolates except for the prototype A2 strain had a potential N-linked sugar site at residue 294. All of the group B isolates, but not the prototype 8/60 strain, had potential N-linked sugar sites at residues 276 and 290. Deduced amino acid sequences for group B RS virus were from 43 to 46 residues, which would result in G-protein lengths of from 291 to 294 amino acids (assuming normal reading frame usage for the full protein).
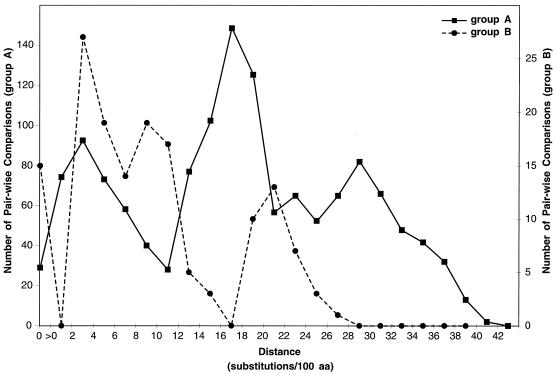
The extent of amino acid variation was assessed by pairwise comparisons within each antigenic group. These results were summarized graphically (Fig. 2). Among the group A isolates from the children’s hospital, the greatest extent of amino acid differences was 38%. Compared to the sequence of the prototype A2 isolate (isolated in Australia in 1962), the sequences of the group A isolates from the children’s hospital differed by as much as 31%. The sequences of the group A isolates from the children’s hospital were also compared to published sequences of the group A virus G protein available through GenBank (for a list of the viral sequences which were compared, see Fig. 3); differences of up to 40.4% were observed for the region examined here.
FIG. 2.
Pairwise distances among group A (A) and group B (B) RS virus G proteins. All possible pairwise comparisons were made among amino acid sequences from isolates at the children’s hospital and published human RS virus G-protein sequences available through GenBank for the region of the G protein analyzed in this report. See Fig. 3 for a list of the published sequences which were used here. The distances represent the number of substitutions/100 amino acids (aa) for each pairwise comparison. No correction was used for multiple substitutions at single sites. The figure plots the number of pairwise sequence comparisons that have distance measurements within each indicated range. Sequence distances for group A and group B viruses are shown separately by using different scales for the number of sequence comparisons.
FIG. 3.
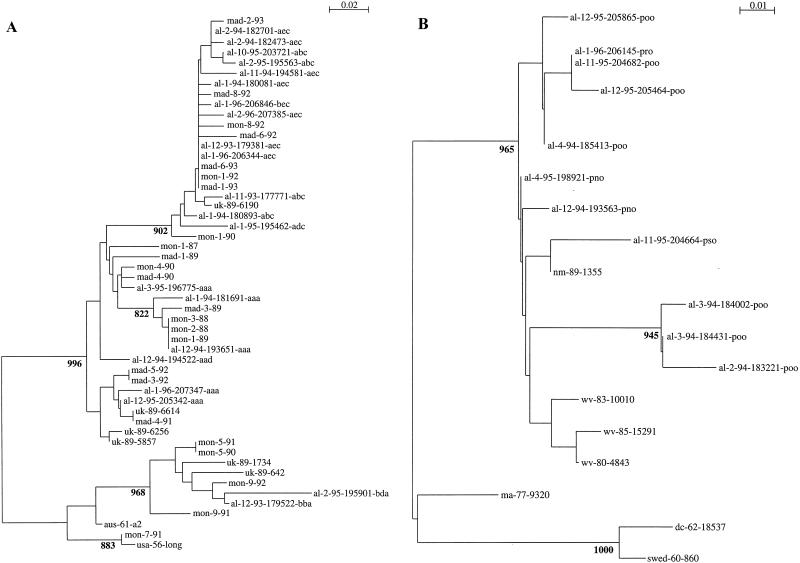
Group A (A) and B (B) RS virus G-protein phylogenetic relationships. Partial G-protein sequences from RS viruses isolated at the Children’s Hospital of Alabama were compared to published G-protein sequences available through GenBank. Viruses are identified by the geographic location (from the United States, al, Alabama; wv, West Virginia [23]; dc, District of Columbia [16], ma, Massachusetts [23]; md, Maryland [16], and nm, New Mexico [23]; from the United Kingdom, uk [5]; from Spain, mad, Madrid; from Uruguay, mon, Montevideo [12]; from Sweden, swed [22]; and from Australia, aus [28]), year or month and year of isolation, and, for isolates from the United States and the United Kingdom a number designation. For isolates from Alabama a three-letter designation that describes the restriction pattern observed after restriction endonuclease digestion of the PCR DNA products follows the designations described above. A single sequence was used for each unique nucleotide sequence within each restriction fragment pattern among the isolates described here (Table 1). The nucleotide sequence alignments of either group A or group B sequences were used to create the neighbor-joining trees displayed in the figure. The scales represent either 0.02 (group A) or 0.01 (group B) substitutions per base per indicated horizontal distance. The numbers present at some of the internal nodes of the trees represent the number of bootstrap replicates of 1,000 that display the indicated sequence groupings. Only significant bootstrap replicate numbers with values of greater than 800 are shown.
Among the group B RS virus isolates from the children’s hospital, the greatest extent of amino acid differences was 14%. Compared to the sequence of the prototype 8/60 group B isolate (isolated in Sweden in 1960), the sequences of the group B isolates from the children’s hospital differed by up to 25%, and compared to the sequence of the prototype 18537 group B isolate (isolated in Washington, D.C., in 1962), the sequences of the group B isolates from the children’s hospital differed by up to 27%. The sequences of the group B isolates from the children’s hospital were also compared to published sequences of the G protein from viruses isolated from 1977 to 1989 (for a list of the viral sequences which were compared, see Fig. 3); the differences were less than those seen for the 8/60 and 18537 viruses.
Phylogenetic analysis.
Phylogenetic comparisons of the C-terminal variable region among the isolates described here and the published sequences for RS virus G-protein genes available through GenBank were performed (Fig. 3). When group A and group B viral sequences were analyzed together, two lineages separating the group A and B viruses were evident. To facilitate comparisons, the group A and group B sequences were analyzed separately. Among the group A viruses, two broad lineages and multiple sublineages were seen (Fig. 3A). Viruses from Alabama appeared in both group A lineages. Isolates from Birmingham, United Kingdom; Montevideo, Uruguay; and Madrid, Spain, also appeared in both lineages (5, 12, 23).
Among the group B viruses, multiple lineages were also evident (Fig. 3B). The number of previously described group B viruses available for comparison was more limited than the number of group A viruses available for comparison. The three oldest isolates, 8/60, 18537, and 9320, were on a branch separate from the more recent isolates.
Selection for change.
Earlier studies have shown that there are conserved and variable regions of the G protein. In addition, a high proportion of the nucleotide changes (∼50%, as calculated for the entire protein) have been observed to result in amino acid changes, suggesting that there may be a selective pressure for change (5, 15, 16, 23). The percent nucleotide changes which resulted in amino acid changes was calculated (Table 2). Among the group A viruses the proportions of nucleotide changes resulting in amino acid changes were 13% for the conserved central region and 61% for the second variable region analyzed here. The group B viruses had similar results. If nucleotide changes are occurring randomly, 24% of the encoded amino acids will change (1). Thus, for both the group A and group B viruses, these results were compatible with there being a positive selection for change in the variable regions of the G protein and conservation of sequences in the central region. These data were in agreement with those from earlier analyses of group A RS virus G-protein variability (12).
DISCUSSION
Genetic variability was analyzed for RS viruses isolated at a children’s hospital in the United States over a 3-year period. Viruses were characterized as to antigenic group (group A or B), and within-group variability was assessed by restriction fragment analysis and nucleotide sequence determination.
Although viruses of both antigenic groups were present each year, the group A viruses were dominant for 2 years, followed by a year in which group B isolates predominated. Similar variability has been described in other studies, and overall, the group A viruses are more frequently identified than the group B viruses (2, 7, 14, 20). In a study performed in Finland, alternating group A and B virus dominance has been observed (27). Infections with a virus of one antigenic group increase the likelihood that if a reinfection occurs it will be with a virus of the opposite antigenic group (18, 27).
In addition to the differences in the viral groups isolated each year, restriction pattern analysis showed that the group A viruses which were dominant during the first two epidemic periods were genetically distinct from year to year. Nucleotide and deduced amino acid sequences were determined for isolates from each restriction pattern group. Additional genetic variability was found among isolates within most but not all restriction fragment groups. A previous study suggested that there was less variability among the group B viruses than among the group A viruses (23). The data presented here confirm this observation for isolates from a period spanning three epidemics at a single location (Fig. 2). However, the group B viruses have the potential for additional variability, as evidenced by comparisons to the prototype strain. Whether the analysis of additional group B isolates would reveal even greater differences remains to be determined. The extent to which the individual viruses vary locally and over time could influence the pattern of reinfections with different viral strains. The group A viruses may vary more extensively than the group B viruses. We postulate that this might play a role in the predominance of group A viruses over group B viruses in many studies of RS virus epidemiology.
Human antibody responses to linear epitopes of the variable region of the group A RS virus G protein analyzed here have been assessed. The reactivity of human antibodies with synthetic peptides varied with the infecting RS virus group. In addition, all of the defined linear epitopes included potential N-linked glycosylation sites in some of the RS virus isolates. It was suggested that the modulation of glycosylation sites might be a mechanism for evasion of the host immune response by RS virus (4). Interestingly, for the viruses studied here, variation was noted among potential N-linked glycosylation sites described in the study mentioned above (4) (Fig. 1). The variable reactivities of human antibodies against peptides from this region of the G protein suggest the potential importance of antigenic changes in this region.
Phylogenetic analysis showed that the group A and group B viruses were placed in multiple lineages. Most of the major phylogenetic branches included viruses which were isolated in this study, reflecting the great diversity of RS viruses which may be present in a community over a period of 3 years. However, among the group A viruses it was also clear that viruses isolated at different times and from different places could be very similar to the viruses isolated at our children’s hospital. Thus, viruses from the United Kingdom, Spain, and Uruguay which were isolated several years before the viruses studied here were isolated could be grouped phylogenetically with these viruses. The observation that very similar viruses are isolated at different times and from geographically distant sites demonstrates that the virus is capable of worldwide spread (9, 12).
The study presented here was designed to assess variability in a detailed manner over a limited period of time. Thus, the study was not intended to address the issue of change over time. For the group A RS viruses, there appears to be an accumulation of change over time (9). The oldest group B isolates, 8/60, 18537, and 9320 (isolated in 1960, 1962, and 1977, respectively), were in a separate lineage from the more recent group B isolates. This observation is also compatible with the accumulation of changes among the group B RS viruses over time. However, until a greater number of more chronologically and geographically disperse isolates have been examined, this issue remains speculative for the group B RS viruses.
These results confirm the variability among group A RS virus isolates which has been demonstrated previously (9, 12). The data also show that variability occurs among group B RS viruses, although to a lesser extent than among the group A viruses. Among both the group A and group B RS viruses, a high percentage (>60%) of nucleotide changes resulted in amino acid coding changes in the variable region of the G protein studied here. These data indicate that the group B RS virus G proteins, while less variable in this study, are no more likely to have synonymous mutations than are the group A RS virus G proteins. The high percentage of nucleotide changes which resulted in amino acid coding changes suggested that there may be a selective advantage to G-protein changes (1, 5, 23). One possible advantage would be that such changes result in an escape from the host immune response. This might contribute to the ability of RS viruses to establish infections throughout life. Longitudinal community-based studies of RS virus variability will be necessary to define precisely the contribution of antigenic variation to RS virus reinfections.
ACKNOWLEDGMENTS
Support for this study was received from Public Health Service grant AI33425 (to W.M.S.). Support for nucleotide sequence analysis was provided by the Center for AIDS Research (PO AI27767) and the X-Ray Crystallography Core Facility (CA13148) at the University of Alabama. Support for the synthesis of oligonucleotides was provided though NCI grant CA13148 to the University of Alabama Comprehensive Cancer Center.
We thank Kimberly Grantham Edwards and Margaret Amsler for technical assistance, Dana Pinson for secretarial support, and the members of the Diagnostic Virology Laboratory for assistance.
REFERENCES
- 1.Air G M, Gibbs A J, Laver W, Webster R G. Evolutionary changes in influenza B are not primarily governed by antibody selection. Proc Natl Acad Sci USA. 1990;87:3884–3888. doi: 10.1073/pnas.87.10.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson L J, Hendry R M, Pierik L T, Tsou C, McIntosh K. Multicenter study of strains of respiratory syncytial virus. J Infect Dis. 1991;163:687–692. doi: 10.1093/infdis/163.4.687. [DOI] [PubMed] [Google Scholar]
- 3.Anderson L J, Hierholzer J C, Tsou C, Hendry R M, Fernie B F, Stone Y, McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 4.Cane P A. Analysis of linear epitopes recognised by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J Med Virol. 1997;51:297–304. doi: 10.1002/(sici)1096-9071(199704)51:4<297::aid-jmv7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Cane P A, Matthews D A, Pringle C R. Identification of variable domains of the attachment (G) protein of subgroup A respiratory syncytial viruses. J Gen Virol. 1991;72:2091–2096. doi: 10.1099/0022-1317-72-9-2091. [DOI] [PubMed] [Google Scholar]
- 6.Cane P A, Matthews D A, Pringle C R. Analysis of relatedness of subgroup A respiratory syncytial viruses isolated worldwide. Virus Res. 1992;25:15–22. doi: 10.1016/0168-1702(92)90096-r. [DOI] [PubMed] [Google Scholar]
- 7.Cane P A, Matthews D A, Pringle C R. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J Clin Microbiol. 1994;32:1–4. doi: 10.1128/jcm.32.1.1-4.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cane P A, Pringle C R. Respiratory syncytial virus heterogeneity during an epidemic: analysis by limited nucleotide sequencing (SH gene) and restriction mapping (N gene) J Gen Virol. 1991;72:349–357. doi: 10.1099/0022-1317-72-2-349. [DOI] [PubMed] [Google Scholar]
- 9.Cane P A, Pringle C R. Evolution of subgroup A respiratory syncytial virus: evidence for progressive accumulation of amino acid changes in the attachment protein. J Virol. 1995;69:2918–2925. doi: 10.1128/jvi.69.5.2918-2925.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1313–1351. [Google Scholar]
- 11.Cristina J, Lopez J A, Albo C, Garcia-Barreno B, Garcia J, Melero J A, Portela A. Analysis of genetic variability in human respiratory syncytial virus by the RNase A mismatch cleavage method: subtype divergence and heterogeneity. Virology. 1990;174:126–134. doi: 10.1016/0042-6822(90)90061-u. [DOI] [PubMed] [Google Scholar]
- 12.Garcia O, Martin M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Brena P, Martinez I, Garcia-Barreno B, Melero J A. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol. 1994;68:5448–5459. doi: 10.1128/jvi.68.9.5448-5459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall C B, Walsh E E, Schnabel K C, Long C E, McConnochie K M, Hildreth S W, Anderson L J. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–1290. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- 14.Hendry R M, Pierik L T, McIntosh K. Prevalence of respiratory syncytial virus subgroups over six consecutive outbreaks: 1981–1987. J Infect Dis. 1989;160:185–190. doi: 10.1093/infdis/160.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Johnson P R, Collins P L. The 1B (NS2), 1C (NS1) and N proteins of human respiratory syncytial virus (RSV) of antigenic subgroups A and B: sequence conservation and divergence within RSV genomic RNA. J Gen Virol. 1989;70:1539–1547. doi: 10.1099/0022-1317-70-6-1539. [DOI] [PubMed] [Google Scholar]
- 16.Johnson P R, Spriggs M K, Olmsted R A, Collins P L. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci USA. 1987;84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddison W P, Maddison D R. MacClade. Analysis of phylogeny and character evoluation, 33. Sunderland, Mass: Sinauer Associates, Inc.; 1992. [Google Scholar]
- 18.Mufson M A, Belshe R B, Orvell C, Norrby E. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J Clin Microbiol. 1987;25:1536–1539. doi: 10.1128/jcm.25.8.1535-1539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mufson M A, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- 20.Storch G A, Park C S. Monoclonal antibodies demonstrate heterogeneity in the G glycoprotein of prototype strains and clinical isolates of respiratory syncytial virus. J Med Virol. 1987;22:345–356. doi: 10.1002/jmv.1890220407. [DOI] [PubMed] [Google Scholar]
- 21.Storch G A, Park C S, Dohner D E. RNA fingerprinting of respiratory syncytial virus using ribonuclease protection. Application to molecular epidemiology. J Clin Invest. 1989;83:1894–1902. doi: 10.1172/JCI114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullender W M, Anderson K, Wertz G W. The respiratory syncytial virus subgroup B attachment glycoprotein: analysis of sequence, expression from a recombinant vector, and evaluation as an immunogen against homologous and heterologous subgroup virus challenge. Virology. 1990;178:195–203. doi: 10.1016/0042-6822(90)90394-7. [DOI] [PubMed] [Google Scholar]
- 23.Sullender W M, Anderson L J, Mufson M A, Wertz G W. Genetic diversity of the attachment protein of subgroup B respiratory syncytial viruses. J Virol. 1991;65:5425–5434. doi: 10.1128/jvi.65.10.5425-5434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullender W M, Sun L, Anderson L J. Analysis of respiratory syncytial virus genetic variability with amplified cDNAs. J Clin Microbiol. 1993;31:1224–1231. doi: 10.1128/jcm.31.5.1224-1231.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford D L. PAUP: phylogenetic analysis using parsimony, 3.1. Champaign: Illinois Natural History Survey; 1993. [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waris M. Pattern of respiratory syncytial virus epidemics in Finland: two-year cycles with alternating prevalence of groups A and B. J Infect Dis. 1991;163:464–469. doi: 10.1093/infdis/163.3.464. [DOI] [PubMed] [Google Scholar]
- 28.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]