Abstract
Dynamic protein phosphorylation and dephosphorylation are essential regulatory mechanisms that ensure proper cellular signaling and biological functions. Deregulation of either reaction has been implicated in several human diseases. Here, we focus on the mechanisms that govern the specificity of the dephosphorylation reaction. Most cellular serine/threonine dephosphorylation is catalyzed by 13 highly conserved Phosphoprotein Phosphatase (PPP) catalytic subunits, which form hundreds of holoenzymes by binding to regulatory and scaffolding subunits. PPP holoenzymes recognize phosphorylation site consensus motifs and interact with short linear motifs (SLiMs) or structural elements distal to the phosphorylation site. We review recent advances in understanding the mechanisms of PPP site-specific dephosphorylation preference and substrate recruitment and highlight examples of their interplay in the regulation of cell division.
Keywords: Phosphoprotein Phosphatases (PPPs), protein phosphorylation, phosphorylation site consensus motifs, short linear motif (SLiM), kinetochore, mitosis
Protein Phosphorylation
Reversible post-translational modification (PTM) of proteins by phosphorylation is a critical regulatory event that controls most cellular processes by altering protein structure, stability, interactions, subcellular location, and/or activity. Deregulation of protein phosphorylation has been implicated in many human diseases, including cancer, diabetes, and neurodegenerative and cardiovascular diseases, among others.
Mass spectrometry-based proteomic and phosphoproteomic analyses have identified over 300,000 phosphorylation sites [1], with more than 50,000 distinct phosphorylation sites detected in a single cell line [2]. Indeed, protein phosphorylation is considered the most prevalent PTM, with at least 75% of detected proteins being phosphoproteins in a specific cell line and over 85% of detected proteins overall reported as phosphorylated [2,3]. Furthermore, many proteins are phosphorylated at more than one residue, allowing for signal integration, amplification, and scaled responses.
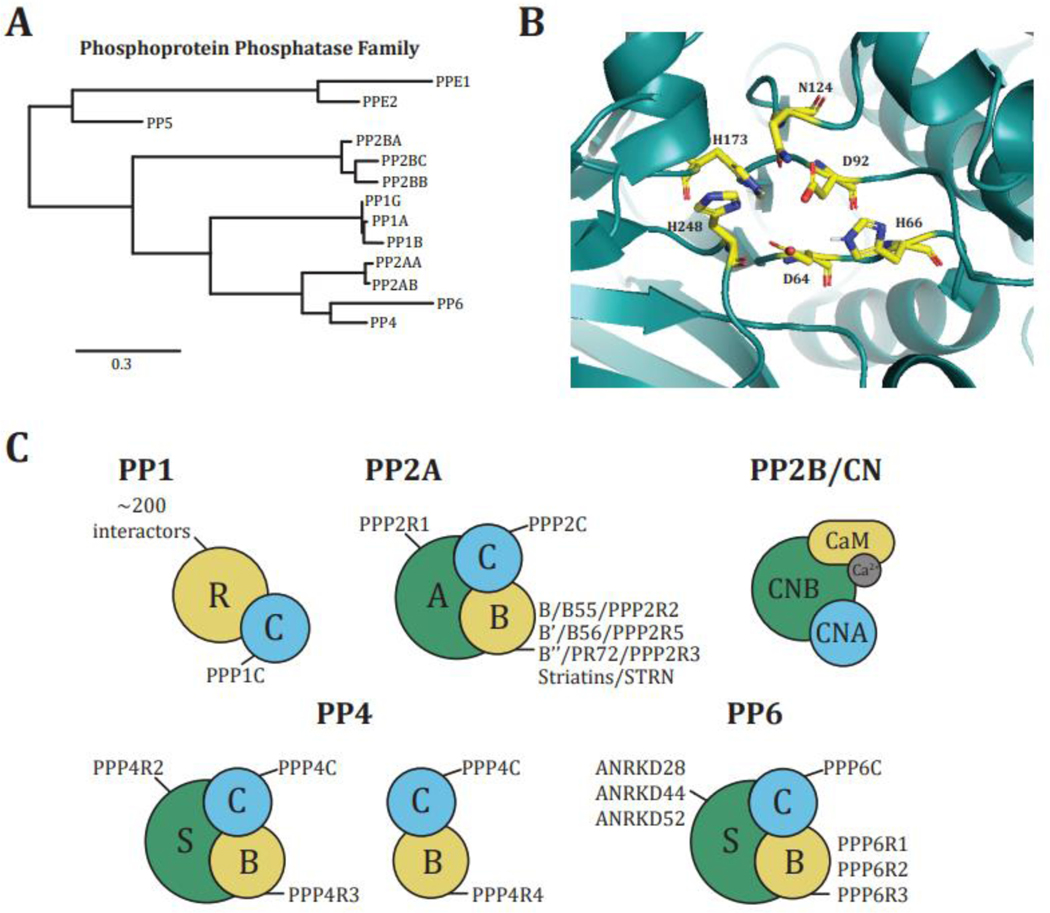
Phosphorylation is catalyzed by protein kinases that add a phosphoryl group to serine (S), threonine (T), and/or tyrosine (Y) residues and is removed by protein phosphatases that catalyze the hydrolysis of the phosphoester bond. There are 518 protein kinases encoded in the human genome, and about 400 are S/T kinases [4]. Conversely, 189 protein phosphatase catalytic subunits have been identified, with the 13 members of the Phosphoprotein Phosphatase (PPP) family responsible for 90% of the S/T dephosphorylation [5]. The PPP family consists of the catalytic subunits of protein phosphatase 1 (PP1), which has three isoforms (PPP1CA, PPP1CB, and PPP1CC), PP2A with two isoforms (PPP2CA, PPP2CB), PP2B/PP3/calcineurin (CN) with three isoforms (PPP3CA, PPP3CB, PPP3CC), PP4 (PPP4C), PP5 (PPP5C), PP6 (PPP6C) and the two isoforms of PP7 (PPEF1, PPEF2) (Box 1, Figure IA).
Text Box 1.
The PPP catalytic subunits are metalloenzymes with two bivalent metal ions (Fe2+, Mn2+, or Zn2+) located in the active site that catalyze phosphoester hydrolysis. The nature of the metal ions present in the active site depends on the source from which the enzyme was purified, the expression (recombinant or exogenous), and the purification strategy. However, for most PPPs, the specific metal ion has not been determined. The amino acid residues that coordinate these metal ions are conserved in all members of the family (Figure IB) [100]. Other active site residues allow for differential specificity of natural toxins, such as microcystin, fostriencin, and tautomycetin for certain PPPs, suggesting potential preferences for amino acids surrounding the dephosphorylation site [36,101,102]. However, substrate interactions beyond the active site are needed to achieve sufficient specificity to differentiate between substrates.
Although there are only 13 PPP catalytic subunits (Figure IA), a large number of scaffolding and regulatory subunits allow for the formation of hundreds of unique multimeric holoenzymes that carry out the majority (>90%) of cellular dephosphorylation (Figure IC). For example, the catalytic subunits of PP1 and CN form heterodimeric complexes with regulatory subunits, while PP2A, PP4, and PP6 form heterotrimers consisting of catalytic, regulatory, and scaffolding subunits [6,103]. The three PP1 catalytic subunits form holoenzymes with over 200 regulatory subunits to regulate a wide variety of cellular functions through subcellular localization, enzymatic regulation, and differential substrate dephosphorylation [53]. In contrast, CN is in an obligate heterodimer consisting of the catalytic calcineurin A subunit (CNA) and the regulatory calcineurin B (CNB) subunit. CN is the only calcium-dependent PPP [104]. CNA contains a calmodulin-binding domain through which it binds to Ca2+-bound calmodulin. Furthermore, CNB has two EF-hands, one with a high affinity for Ca2+, which constitutively binds one Ca2+ molecule, and another with a lower affinity, which is loaded with Ca2+ depending on the intracellular Ca2+ concentration [104]. PP2A forms heterotrimeric holoenzymes that consist of a scaffolding (A) subunit, a catalytic (C) subunit, and a regulatory (B) subunit [6,103]. There are four families of B subunits: B/B55/PPP2R2, B’/B56/PPP2R5, B’’/PR72/PPP2R3, and Striatins/STRN, each with several isoforms. Thus, through combinatorial assembly, there are over sixty unique PP2A holoenzymes, each likely to have unique functions and substrates.
Figure I: PPP conservation and structure.
A, PPP phylogenetic tree (Tree was created using www.phylogeny.fr [108]). B, PPP catalytic subunit active site. Conserved residues are shown (modified from PDB: 3V4Y [61]). C, PPP holoenzyme composition.
The conserved nature of the PPP catalytic subunits and the imbalance in the numbers of protein S/T kinases and PPPs led to early doubts about the selectivity of the dephosphorylation reaction and the labeling of PPPs as constitutively active housekeeping enzymes [6]. However, recent insights into PPP substrate recruitment and specificity, partly achieved through new mass spectrometry-based proteomic and phosphoproteomic approaches, have corrected this notion [7–17]. Here, we focus on PP1, PP2A, and CN holoenzymes (see Glossary), describing both their respective phosphorylation site consensus motif preferences and the distal short linear motifs (SLiMs) (see Glossary) they recognize in substrates and scaffolds that may impart further substrate specificity (Figure 1). To highlight the interplay between active site specificity and SLiM interactions on PPP substrate selection, we discuss some roles of PPPs in mitosis.
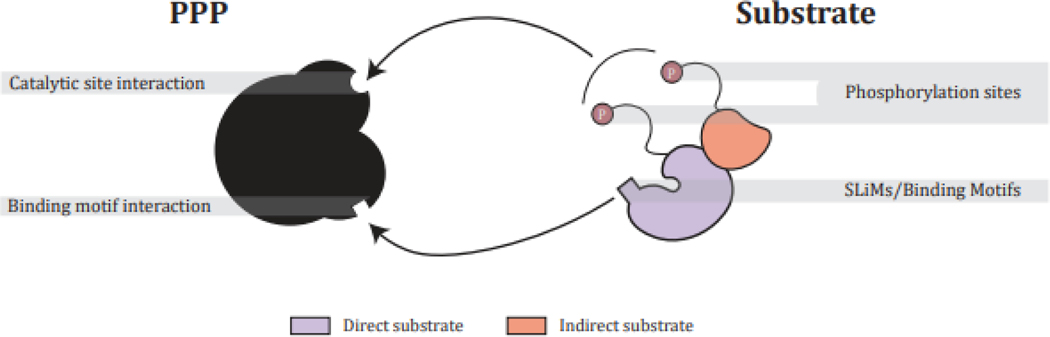
Figure 1: PPP – substrate interactions.
PPPs achieve substrate specificity through (1) catalytic site interactions with the phosphoacceptor residue and proximal amino acids and (2) SLiMs or structural elements distal to the phosphorylation site. Substrates are recruited directly or indirectly by motif containing scaffold proteins.
Clues from the opponents: phosphorylation site consensus motif preferences of kinases
The frequency of S:T:Y residues is 2.5:1.8:1 in the human proteome. However, experiments employing labeling of cells with 32P-orthophosphate followed by acid hydrolysis determined a phospho-amino acid distribution of pS:pT:pY of 184:15:1 [18]. This ratio was validated by mass spectrometry-based phosphoproteomic analyses (pS:pT:pY distribution of 162:31:1) [2]. In part, this discrepancy in amino acid versus phospho-amino acid distribution might lie in the frequencies of S and T residues in phosphorylated regions of the protein. The majority (more than 90%) of S/T phosphorylation occurs in intrinsically disordered regions, which are enriched for S compared to ordered regions [19–22]. In contrast, Ts are equally distributed between disordered and ordered protein regions [19,22]. Furthermore, while most S/T kinases preferentially phosphorylate S over T or do not distinguish between either residue, several PPP holoenzymes have a dephosphorylation preference for phospho-T (pT) over phospho-S (pS), tipping the overall phosphorylation balance towards pS [23–25]. Turk and colleagues recently demonstrated that the preference of a specific kinase for S or T phosphoacceptors is in part determined by the residue immediately downstream of the conserved Asp-Phe-Gly (DFG) motif, the DFG+1 residue. Large hydrophobic residues (phenylalanine, tryptophane, tyrosine) in the DFG+1 promote S phosphorylation, while smaller beta-branched residues (valine, isoleucine, threonine) promote T phosphorylation [25,26].
Phosphorylation site consensus motif preferences of PPPs
As noted above, several PPP holoenzymes preferentially dephosphorylate pT over pS (Figure 2A) [8,11,27–30]. For example, in vitro analysis of PP2A holoenzymes purified from rabbit skeletal muscle and the PP1 catalytic subunit purified from dog liver revealed a preference for the dephosphorylation of pT [31–35]. A comprehensive mass spectrometry-based analysis of dephosphorylation events of over 25,000 randomized synthetic phosphopeptides by purified PP1 and PP2A catalytic subunits confirmed their pT preference in vitro [9]. Further, depletion of PP2A substrate binding-regulatory subunit B55 or inhibition of PP2A-B55 activity preferentially stabilized T over S phosphorylation sites in cell lysates (Figure 2B) [11,29]. However, inhibition of PP2A-B56 dephosphorylation through substrate competition in cells equally affects pT and pS [29], suggesting differences in PPP2C dephosphorylations based on regulatory subunit binding.
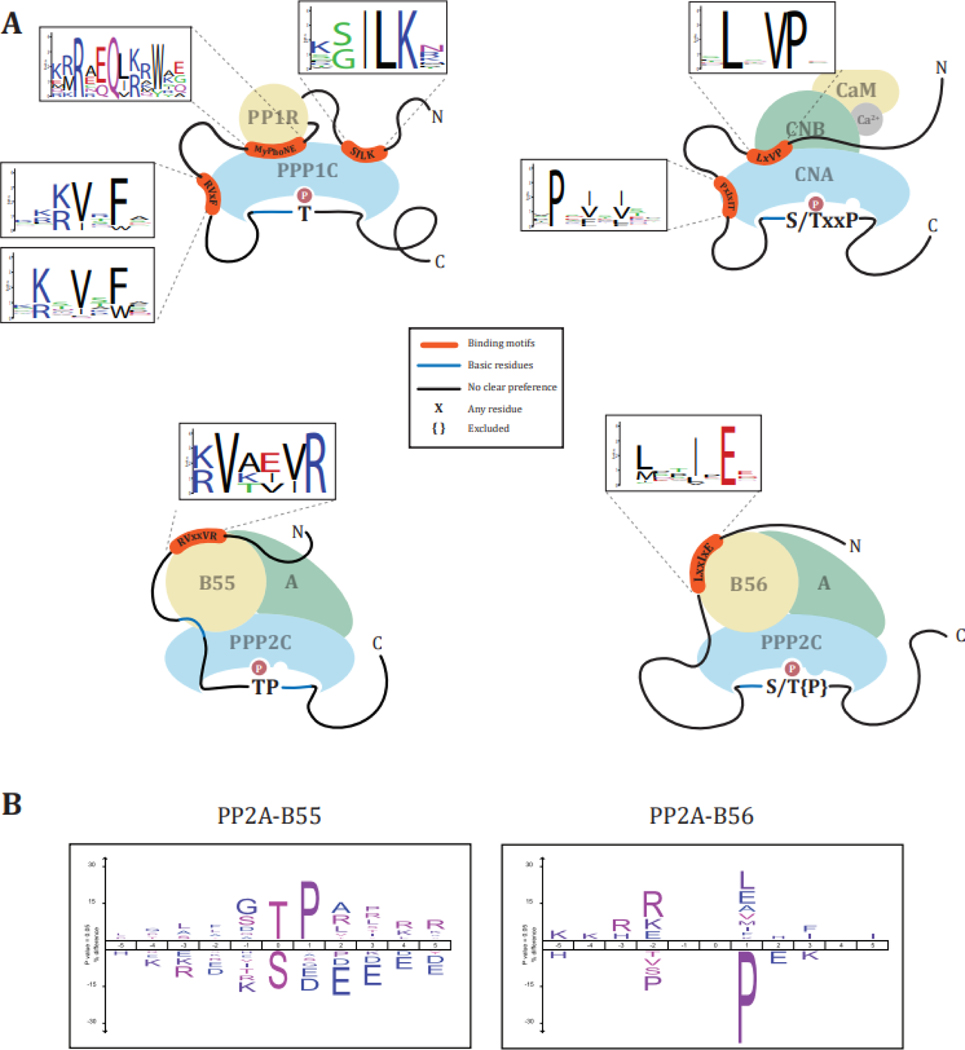
Figure 2: PPP phosphorylation site preferences and binding motifs.
(A) Active site dephosphorylation preferences and deselections for phosphoacceptor and surrounding amino acids and SLiMs determinants and degeneracy of PP1 [105,106], CN [107], PP2A-B55 [69], and PP2A-B56 [10]. (B) PP2A-B55 and -B56 preferences and deselections of phosphoacceptors and surrounding amino acids (modified from Kruse et al. [29]).
Beyond active site residues, an acidic, a hydrophobic, and a C-terminal groove on the surface of the PP1 and PP2A catalytic subunits promote interactions with residues surrounding the phosphorylation site [36–38]. Due to a more extensive acidic groove on the surface of PPP1C compared to PPP2C, PPP1C has a more pronounced preference for basophilic phosphorylation site consensus motifs than PPP2C [9].
For PP2A holoenzymes, acidic patches on the B55 and B56 regulatory subunits promote dephosphorylation of sites with basic residues amino (N)-terminal to the phosphorylation site [9,11,29,32,39]. In vitro, dephosphorylation of peptide substrates by PP2A-B55 could be increased by including N-terminal basic amino acids [32].
Peptides containing a pS and pT with proline in the +1 position (pSP/pTP) are generally poor substrates for PP1 [31,35] and PP2A [35], with PP2A-B55 dephosphorylating proline-directed phosphorylation site with higher efficiency than PP2A-B56, PP2A-PR72, or PP1 [29,31,35,40,41] (Figure 2A). In support of this, using siRNA-mediated depletion of B55 pathway components, Cundell et al. [11] demonstrated a preference of PP2A-B55 for proline-directed sites, specifically pTP (Figure 2B). Furthermore, they identified a bipartite polybasic motif N (position −16 to −11) and carboxyl (C)-terminal (position +2 to +6) from the phosphorylation site enriched in candidate PP2A-B55 substrates and demonstrated that mutation of these basic amino acids to alanines reduced the dephosphorylation rates [11]. Intriguingly, dephosphorylation of pSP/pTP by PP2A-B55 is conformation specific and only occurs for the trans, not cis, isomer [42]. Isomerization of pSP/pTP is catalyzed by the prolyl isomerase Pin1, adding a layer of regulation to the dephosphorylation reaction [43,44].
Using protein-based inhibitors, Kruse et al. [29] confirmed the pTP/pSP preference of PP2A-B55, and found that PP2A-B56 preferentially dephosphorylated sites with basic amino acids in the −2 and −3 position and deselected pTP/pSP (Figure 2B). Intriguingly, they found in mitotic cells a preference of PP2A-B56 for acidic residues in the −2 position N-terminal to the phosphorylation site, suggesting a potential role of PP2A-B56 in opposing Polo-like kinase 1 (Plk1) phosphorylation site in mitosis [20]. In vitro dephosphorylation assays confirmed these preferences and deselections of PP2A-B56α and revealed that acidic residues C-terminal to the phosphorylation site abrogated the ability of PP2A-B56α to dephosphorylate the site [29]. The differences in consensus motif preferences of PP2A-B55 and PP2A-B56 are surprising as both enzymes share the same catalytic subunit and point to the role of the respective regulatory subunits in influencing active site selectivity. As these differences are seen in vitro in assays using short phosphopeptides as substrates [29], substrate recruitment through the regulatory subunits can most likely be excluded as the cause.
In contrast to PP1 and PP2A-B55, CN shows less of a preference for T over S and efficiently dephosphorylates proline-directed phosphorylation sites (Figure 2A) [45]. Specifically, a proline in the +3 position (pS/pTxxP) promotes rapid dephosphorylation [46]. Basic amino acids N-terminal to the phosphorylation site increase, and acidic residues C-terminal to the phosphorylation site decrease, CN dephosphorylation ability [45]. Intriguingly, in a comparison of 28 peptides, a sequence containing a PP2B short linear motif (LxVP) N-terminal to the phosphorylation site dramatically increased PP2Bs dephosphorylation efficiency [45,47].
These in vitro and in cell analyses demonstrate that the nature of phosphorylatable residues (T or S) and the consensus motif of amino acids surrounding the phosphorylation sites are essential determinants of PPP specificity and fine-tune dephosphorylation kinetics. However, as for kinases, while consensus motifs allow the delineation of PPP-substrate relationships and provide a degree of specificity at the active site, motif degeneracy and redundancy limit their impact. Thus, additional mechanisms are needed for PPPs to specifically select dephosphorylation sites and substrates.
Substrate selection mechanisms beyond the active site: recognition and binding through docking motifs
An emerging theme in PPP biology is the recruitment of substrates directly or indirectly through specific docking elements, either short linear motifs (SLiMs) or structural motifs distal to the phosphorylation site (Table 1). SLiMs are stretches of four to ten amino acids located in intrinsically disordered regions (IDRs) [48–50]. It remains an outstanding question how broadly docking motif strength and distance to the phosphorylation site affects substrate dephosphorylation and PPPs’ phosphoacceptor and phosphorylation site motif preferences.
Table 1:
SLiM and structural interaction motifs
| PPP | Motif | Definition | Ref |
|---|---|---|---|
| PP1 | RVxFa | [RK]-x(0|1)-[VI]-{FIMYDP}-[FW] | [51–54] |
| SILK | [GS]-I-L-[KR]-{DE} | [54,60,63] | |
| ΦΦ | hydrophobic amino acids | [61,62] | |
| KiR | F-D-x-x-L-P-[PA]-N-[ST]-P-L-[RK]-[RK]-G-x-[ST]-P [62] | ||
| MyPhoNE | R{P}[DEQ]Q[VIL]([RK]{P}|{P}[RK])[YW] | [52] | |
| PP2A-B55 | B55 motif | [RK]-[VIL]-x-x-[VILM]-[RK] | [69] |
| PP2A-B56 | LxxIxE | [LMFYWIC]-x-x-[IVLWC]-x- E | [10,66,67,75] |
| CN | PxIxIT | P-x-[FILV]-x-[FILV] | [72,74,75] |
| LxVP | [NQDESRTH]-[YRTDFILV]-L-x-[VPL]-[PK] | [47,73,75] | |
x is any residue, [ ] residues allowed, { } residues excluded, ( | ) either or
Structural studies revealed a hydrophobic groove on the PP1 catalytic subunit that binds to over 200 regulatory proteins by recognizing a SLiM with the consensus sequence RVxF (Figure 2A) [51–55]. These regulatory proteins can be scaffolding proteins that target PPP1C to specific subcellular locations or recruit substrates to PPP1C, direct substrates, or competitive inhibitors [53,56–59]. RVxF motifs often co-occur with a second SLiM such as the SILK, KiR, or ΦΦ motif, or the structural myosin phosphatase N-terminal element (MyPhoNE) motif (Figure 2A) [52,54,55,60–63]. Regulatory proteins are often unstructured in their free form, and binding to PPP1C induces folding. For instance, the PP1 regulatory subunit Spinophilin is unstructured in its unbound form, but upon interaction with the PPP1C folds and binds to a large surface of the catalytic subunit, including the RVxF and C-terminal binding grooves [64].
In the case of PP2A, the regulatory subunits of the holoenzymes recognize substrates through SLiMs. The first SLiM recognized by a PP2A holoenzyme was identified for the B56 regulatory subunits. A highly conserved binding pocket in a HEAT repeat helix found in all isoforms of B56 binds to a LxxIxE motif found in substrates (Figure 2A) [10,65–67]. In a subset of LxxIxE-containing proteins, the interaction is enhanced by a dynamic charge-charge interaction between a positively charged motif which interacts with a negatively charged groove on B56 [68].
For the PP2A-B55 holoenzyme, two substrate interaction motifs have been identified. A conserved acidic pocket on the B55 regulatory subunit binds to basic residues N-terminal of the phosphorylation sites [11,39]. Additionally, investigations into the mode of interaction of the PP2A-B55 substrates p107, Tau, and MAP2 identified a common sequence motif [RK]-[VIL]-x-x-[VILM]-[RK] (Figure 2A) [69]. A follow-up study on the interaction of PP2A-B55 and its substrate FAM122A revealed that this motif is not a SLiM but a structural motif in the form of a short α-helix that binds to the B55 regulatory subunit [70].
While no SLiM motif has been defined for the PR72 families of PP2A regulatory subunits, structural analysis has revealed specific binding sites for the recruitment of substrates. PR70, a member of the PR72 family, interacts with its substrate Cdc6 via a region near the PP2A holoenzyme active site with a specific peptide sequence in Cdc6 (49KALPLSPRKRLGDDNLCNTPHLPPCSPPKQGKKENGPPHSHT90) [71].
CN recognizes two SLiMs: the PxIxIT motif, which binds to a binding pocket on CNA close to the active site, and the LxVP motif, which binds to a pocket at the interface of CNA and CNB (Table 1; Figure 2A) [47,72–75]. CN is inhibited by the fungal-derived immunosuppressants cyclosporin A (CSA) and FK506, which are employed to prevent rejection in post-organ transplantation. Intriguingly, both molecules in complex with their respective endogenous protein receptors (cyclophilin for CSA, FKBP for FK506) inhibit CN by blocking binding to the LxVP [47].
Regulation of SLiM-mediated interactions
The affinity of the PPP-substrate/scaffold interaction and dephosphorylation rate are determined by multiple factors and can be modulated. SLiMs are degenerate in their sequence, and the amino acid composition can strongly influence their affinity [10,54]. For instance, a proline at the fourth position in the RVxF motif greatly decreases its binding capability to PP1, whereas a tryptophan residue instead of phenylalanine in the fourth position enhances the binding affinity (Figure 2A) [55]. For the B56 SLiM LxxIxE, acidic residues C-terminal to the core motif increase its binding affinity to B56 (Figure 2A) [10]. Surprisingly, for CN, the binding affinity is not correlated with the amino acid composition of the LxVP SLiM but with the on-rate, which can vary by ~100-fold between LxVP sequences [73]. Furthermore, the multiple SLiMs recognized by the PP1 catalytic subunit and the CN holoenzyme increase the specificity and affinity for their specific substrates through avidity (Figure 2A). The binding sites on the PP1 catalytic subunit can theoretically generate more than 4,000 unique combinations of binding motifs, explaining the broad range of biological functions, specific interactions, and catalytic activities connected to PP1 [52,53,60,64,76,77]. Comparable to kinases, larger interaction surfaces through multiple SLiMs or structural motifs increase the stringency of the dephosphorylation reaction while reducing the number of substrates [78].
Besides composition, SLiM-based interactions are modulated by PTMs, specifically phosphorylation. For PP1, the x position in the PP1 RVxF motif is frequently an S or T (Figure 2A), and phosphorylation of this residue reduces the affinity of the interaction [62,79,80]. Conversely, phosphorylation of an S or T residue in any of the x positions of the PP2A-B56 LxxIxE motif or residues directly adjacent to it increases the interaction with B56 [10,65]. For example, Cdk1 phosphorylation of the Repo-man LxxIxE motif increases its binding to PP2A-B56, while the phosphorylation of its RVxF motif decreases its binding to PP1 to dynamically coordinate dephosphorylation of its substrates during mitotic progression [81]. Less is known about the phosphorylation-dependent modulation of the affinity of the CN recognized SLiMs PxIxIT and LxVP and the effects that phosphorylation may have on their interaction with CN. However, an overrepresentation of T residues at the sixth position of the PxIxIT motif points to phosphorylation as a potential regulatory mechanism (Figure 2A) [75].
In addition to the direct motif-dependent recruitment of substrates, proteins containing these docking motifs frequently function as scaffolds for indirect substrate recruitment (Figure 1) [9,11,29,80,82]. The LxxIxE SLiM-containing protein BubR1 recruits PP2A-B56 to the kinetochore to dephosphorylate Aurora kinase B substrates [80], and the Ebola virus nucleoprotein recruits PP2A-B56 via a LxxIxE-motif to dephosphorylate the viral transcription factor VP30 [82]. Furthermore, a phosphoproteomic-based global analysis of the PP2A-B56 substrates revealed that only 5% of identified PP2A-B56-dependent phosphorylation sites are located on proteins with known or predicted LxxIxE-motif, while 75% were found on proteins known to interact with LxxIxE-motif-containing proteins [29]. Thus, the indirect recruitment of substrates through SLiM-containing proteins as scaffolds appear to be an important regulatory mechanism for the recruitment of PP2A-B56 substrates. It will be interesting to see as more PPP SLiM sequences are discovered, and the substrates of these phosphatase holoenzymes are identified if they share this indirect recruitment mechanism. With PPPs being responsible for most of the tens of thousands of S/T phosphorylation sites found in a cell, it seems plausible that indirect recruitment is necessary to accomplish this task.
The phosphorylation site consensus motif, the affinity of the PPP for the respective SLiM sequence or structural motif in the substrate or scaffold, the distance between the phosphorylation site and the SLiM motif, and direct/indirect recruitment mechanisms cooperatively contribute to the dephosphorylation reaction efficiency, resulting in specific dephosphorylation kinetics depending on the combination of the contributing factors. For example, with increasing affinity of the LxxIxE SLiM in FoxO3, more PP2A-B56 is recruited, resulting in a proportional decrease in phosphorylation occupancy of two sites on FoxO3 [29]. Conversely, although proline-directed phosphorylation motifs are not preferred substrates of PP2A-B56, shortening the distance between the phosphorylation site and the SLiM motif can increase dephosphorylation efficiency, even for proline-containing phosphorylation motifs [29]. Indeed, the metalloprotease Adam17 contains a LxxIxE-motif at position 759–764 (MDTIQE) and is dephosphorylated at the proline-directed T735 and basophilic S808 by PP2A-B56 [29]. Intriguingly, decreasing the affinity of the SLiM results in ~2-fold and ~7-fold increase in the phosphorylation occupancy of pT735 and pS808, respectively, supporting the notion that SLiM affinity and phosphorylation site consensus motif combinatorially contribute to the dephosphorylation rate.
Regulation of mitotic progression through PPP phosphorylation site preferences and SLiMs
To illustrate the biological impact of the interplay of PPP phosphorylation site preferences and motif-based recruitment and its regulation, we will discuss examples of the collaboration and counteraction of PPP and kinase activities during cell division.
Entry into mitosis and early mitotic processes such as chromosome condensation, disassembly of organelles, formation of the mitotic spindle, and chromosome alignment are driven by dynamic phosphorylation [83–86]. Activation of mitotic kinases, most prominently Cyclin-dependent kinase 1 (Cdk1), Aurora kinases A and B, and Polo-like kinase 1 (Plk1) results in an overall increase in phosphorylation [83–86]. However, activation of kinases alone is not sufficient to achieve the phosphorylation occupancy necessary for mitotic progression but requires the inhibition of specific phosphatases (Figure 3A) [86–88].
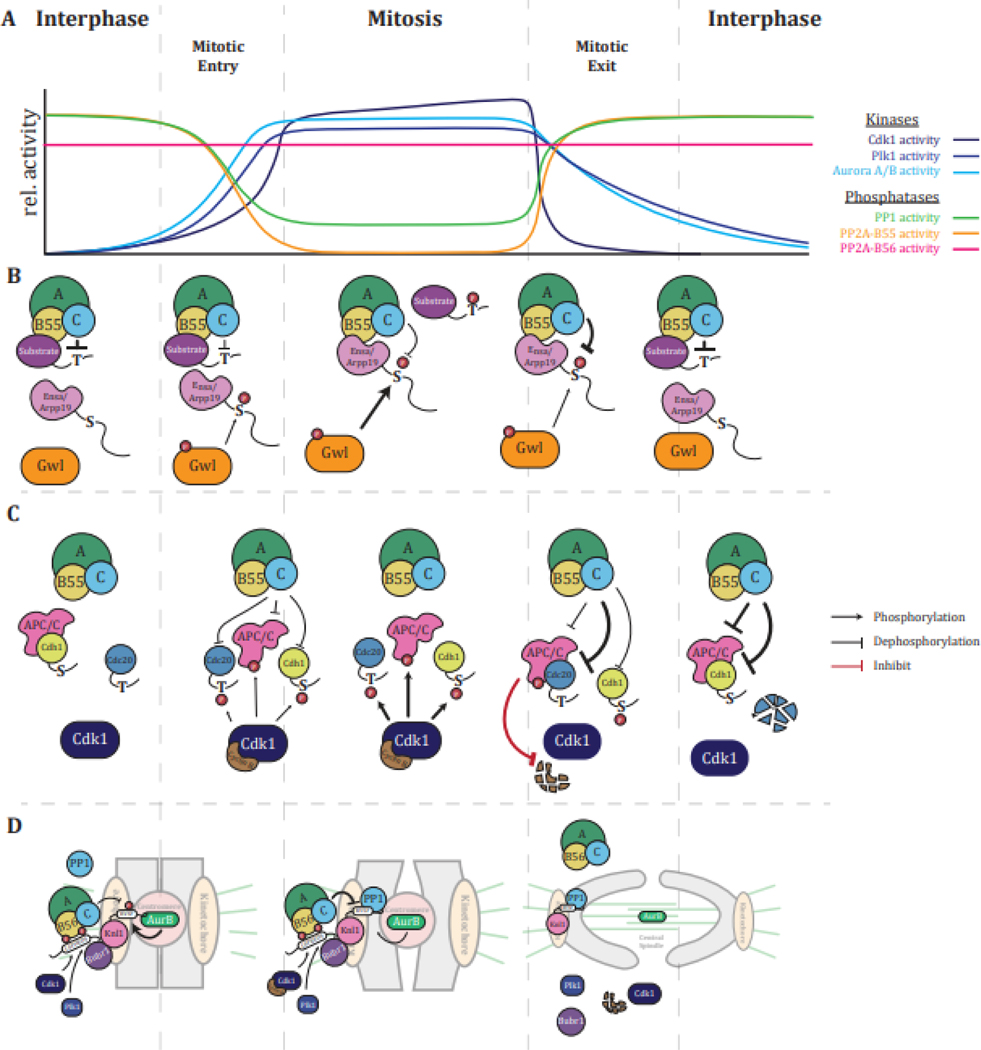
Figure 3: Regulation of mitotic progression by dephosphorylation.
(A) Scheme of relative activities of mitotic kinases and PPPs between cell cycles (adapted from Nasa and Kettenbach [83]). (B) Mechanism of regulation of PP2A-B55 holoenzyme activity by Ensa/Arpp19 during mitotic entry and exit. (C) Orderly reactivation of APC/C by Cdc20 and Cdh1 upon mitotic exit based on PP2A-B55 holoenzyme phosphorylation site preference for pT over pS. (D) Phosphorylation-dependent regulation of PPP SLiM-based substrate recruitment by PP2A-B56 and PP1 and its effects on the regulation of microtubule attachment and spindle assembly checkpoint signaling at the kinetochore.
Cdk1 preferentially phosphorylates proline-directed motifs [21,23,89], and PP2A-B55 is the main PPP that dephosphorylates these motifs [11,29,31,35,40,41]. To achieve high phosphorylation occupancy of Cdk1 substrates, PP2A-B55 is inhibited from late G2 until the onset of anaphase [84,86,87,90]. Inhibition of PP2A-B55 is triggered by the Cdk1-dependent phosphorylation of the PP2A catalytic subunit and the activation of Greatwall kinase, which in turn phosphorylates Arpp19 (cAMP regulated phosphoprotein 19) on S62 and Ensa (endosulfine alpha) on S67 at a conserved amino acid sequence FDSGD [87,88,90,91]. Phosphorylated Arpp19/Ensa inhibit PP2A-B55 through competition with PP2A-B55 substrates [87,88,91]. Arpp19/Ensa bind PP2A-B55 tightly, but the FDpSGD sequence is a poor consensus motif for PP2A-B55, and dephosphorylation occurs slowly with a Kcat of ~0.02s−1 compared to a Kcat of 21–25s−1 of proline-directed phosphorylation sites (Figure 3B) [92].
PP2A-B55 and PP2A-B56 share the same catalytic subunit. However, their distinct phosphorylation site consensus motif preferences are key for the differential regulation of their activities in mitosis [29]. While PP2A-B55 is inhibited, PP2A-B56 remains active throughout mitosis and dynamically opposes the kinases Aurora B on basophilic and Plk1 on acidophilic phosphorylation sites to ensure proper chromosome alignment and segregation and progression through mitosis [10,29,83,84,86] (Figure 3A).
In mitotic exit, the dephosphorylation preference of PP2A-B55 for pT over pS sites is a crucial factor in the ordering of events. Exit from mitosis requires the activation of the Anaphase-Promoting Complex/Cyclosome (APC/C) through dephosphorylation and binding of its activators Cdc20 in metaphase and Cdh1 in late mitosis [93]. The order of APC/C activation is encoded in phosphorylation sites on Cdc20 and Cdh1. While Cdc20 is phosphorylated by Cdk1 on TP, Cdh1 is phosphorylated on SP [8]. Thus, the preference of PP2A-B55 for pTP over pSP results in the dephosphorylation of Cdc20 before Cdh1 [8]. Similarly, rapid dephosphorylation of a pTP site on the chromosomal passenger complex protein INCENP (Inner centromere protein) by PP2A-B55 is required for proper mitotic and meiotic transitions, and mutation to an SP impedes cell cycle progression (Figure 3C) [8,94].
PTMs of SLiM motifs further contribute to the regulation of substrate dephosphorylation throughout mitosis. In mitosis, PP2A-B56, several PP1 holoenzymes, Plk1, Cdk1, and Aurora kinase B are enriched at kinetochores and centromeres, and their coordinated actions regulate chromosome congression [84]. The PP2A-B56 SLiM LxxIxE can contain S/T residues at the x-position or adjacent to it, and the phosphorylation of these residues increases their affinity for PP2A-B56 [10,65]. PP2A-B56 is recruited to kinetochores by binding to the LxxIxE motif in the protein BUBR1 (Bub1-related 1), which has the sequence LSPIIE [80,95]. Cdk1 phosphorylates the S within the LSPIIE motif, while Plk1 phosphorylates an adjacent S, and both events result in increased PP2A-B56 binding [80,95]. One of the kinetochore substrates of PP2A-B56 is the protein KNL1, which recruits PP1 catalytic subunits via RVxF, SILK, and ΦΦ motifs [96,97]. Phosphorylation of the RVxF motif by Aurora kinase B on kinetochores not under tension due to lack of or incorrect microtubule attachments inhibits PP1 catalytic subunit binding [79,96,98]. PP2A-B56 opposes Aurora B and dephosphorylates the KNL1 RVxF motif, promoting PP1 catalytic subunit recruitment (Figure 3D). Thus, at the kinetochore, Cdk1, Plk1, PP2A-B56, and PP1-Knl1 cooperate to counteract Aurora kinase B activity and ensure correct chromosome alignment [80,95].
These examples demonstrate how phosphorylation site preference of kinases and PPPs, and PPP binding motifs, are crucial to the regulation of complex biological processes such as cell division. Through these mechanisms, specificity of dephosphorylation reactions that coordinate mitotic events is achieved to ensure the faithful inheritance of genomic and cellular content. Furthermore, PPPs, their phosphorylation site preferences, and their motif-based recruitment mechanisms, are all highly conserved [6]. For instance, as in human cells, in the budding yeast Saccharomyces cerevisiae, PP2A-B55 (PP2ACdc55) preferentially dephosphorylates pTP over pSP sites to counteract Cdk activity and order cell cycle progression [28]. Similarly, in the sea star Patiria miniata, the preference of PP2A-B55 for pTP over pSP dephosphorylation is crucial for the proper coordination of meiosis I-meiosis II transition [94]. In Caenorhabditis elegans, Bub1 targets PP2A-B56 to chromosomes via an LxxIxE motif; phosphorylation of this motif increases its affinity for B56 [99]. Thus, it is likely that these mechanisms are broadly applied throughout evolution to control phosphorylation signaling in cell cycle control and beyond.
Concluding Remarks
Our understanding of substrate targeting mechanisms utilized by PPPs has greatly increased since the days when PPPs were described as constitutively active housekeeping enzymes. Global mass spectrometry-based phosphoproteomic analyses of PPP substrates have lent support to the early observation of preferential consensus motif dephosphorylation based on the study of a limited number of peptides in vitro and greatly increased our knowledge of PPP substrates. The discovery of distinct SLiM recruitment mechanisms for several PPPs allows for the prediction and identification of direct substrates and scaffolds of indirect substrate recruitment and provides new insights into the complexity of PPP signaling. Combined, phosphorylation site census motif and SLiM-based recruitment mechanisms will enable the elucidation of intricate inter- and counteractions of protein kinases and PPPs in the regulation of cellular signaling (see Outstanding questions).
Outstanding Questions.
How specific are PPP-substrate relationships? Are phosphorylation sites exclusively dephosphorylated by a specific PPP, or is there redundancy?
Do isoforms and splice variants of PPP subunits affect phosphorylation site preferences and substrate recruitment?
Does phosphoacceptor preference influence which phosphorylation site consensus motifs are dephosphorylated?
Does the location of the SLiM relative to the phosphorylation site determine dephosphorylation kinetics?
Does the distance between the SLiM and phosphorylation sites on direct and indirect substrates or the affinity of SLiM for the PPP impact the type of phosphorylation site consensus motifs that are dephosphorylated?
Are the mechanisms of substrate recognition through phosphorylation site preferences and SLiMs conserved in other PPP family members?
Are SLiM affinities modified by post-translational modifications other than phosphorylation?
What regulatory mechanisms govern the dephosphorylation of sites that are part of or directly adjacent to SLiMs? Is it autodephosphorylation or phosphatase crosstalk?
How do regulatory subunits influence the phosphorylation site motif preferences of the catalytic subunit?
Highlights.
Global mass spectrometry-based phosphoproteomic analyses have revealed preferences of PPPs for distinct phosphorylation site consensus motifs.
Phosphoacceptor and phosphorylation site preferences determine dephosphorylation kinetics and provide temporal control of signaling events.
Identification of a growing number of PPP holoenzyme-specific SLiMs has revealed specific PPP-substrate/scaffold interactions.
SLiM-containing proteins are direct PPP substrates or function as scaffolds for indirect substrate recruitment.
Phosphorylation site preferences, SLiM affinity, and distance between the phosphorylation site and SLiM combinatorially contribute to the dephosphorylation kinetics.
Recent studies demonstrated PPP phosphorylation site preferences and modulation of SLiM affinity in biological contexts, specifically mitosis.
Acknowledgments
This work was supported by NIH grant R35GM119455. We thank Scott Gerber for critically reading the manuscript, Paul Robustelli for discussing IDRs, Wolfgang Peti and Rebecca Page for discussing PP1 inhibitory subunits, and Jakob Nilsson for discussing the role of PPPs substrate specificity. The authors apologize to colleagues whose important contributions to the field were not cited due to space limitations.
Glossary
- Holoenzyme
a biochemically active enzyme complex formed by a combination of an enzyme and its coenzyme
- Centromere
a region of a chromosome where sister chromatids are held together, and the kinetochore assembles
- Chromosome congression
a process of aligning chromosomes on the metaphase plate
- Kinetochore
a large protein complex that assembles on the centromere and is the attachment site for spindle microtubules
- Short linear motif (SLiM)
a stretch of four to ten amino acids with three to five residues that mediate protein-protein interactions. SLiMs are degenerate in sequence and often located in intrinsically disordered regions of the protein
- Intrinsically disordered regions (IDR)
a stretch of amino acids in a protein not likely to support the formation of three-dimensional folded structures
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hornbeck P. v. et al. (2019) 15 years of PhosphoSitePlus ® : Integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res 47, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma K et al. (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep 8, 1583–1594 [DOI] [PubMed] [Google Scholar]
- 3.Ochoa D et al. (2020) The functional landscape of the human phosphoproteome. Nat Biotechnol 38, 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning G et al. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 5.Chen MJ et al. (2017) Genomics and evolution of protein phosphatases. Sci Signal 10, eaag1796 [DOI] [PubMed] [Google Scholar]
- 6.Brautigan DL (2013) Protein Ser/Thr phosphatases--the ugly ducklings of cell signalling. FEBS J 280, 324–345 [DOI] [PubMed] [Google Scholar]
- 7.Davey NE et al. (2022) ProP-PD for proteome-wide motif-mediated interaction discovery. Trends Biochem Sci 47, 547–548 [DOI] [PubMed] [Google Scholar]
- 8.Hein JB et al. (2017) Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat Cell Biol DOI: 10.1038/ncb3634 [DOI] [PubMed] [Google Scholar]
- 9.Hoermann B et al. (2020) Dissecting the sequence determinants for dephosphorylation by the catalytic subunits of phosphatases PP1 and PP2A. Nat Commun 11, 3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertz EPT et al. (2016) A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol Cell 63, 686–695 [DOI] [PubMed] [Google Scholar]
- 11.Cundell MJ et al. (2016) A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J Cell Biol 214, 539–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swingle MR and Honkanen RE (2019) Inhibitors of Serine/Threonine Protein Phosphatases: Biochemical and Structural Studies Provide Insight for Further Development. Curr Med Chem 26, 2634–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvanska DH and Nilsson J (2020) Specificity determinants of phosphoprotein phosphatases controlling kinetochore functions. Essays Biochem 64, 325–336 [DOI] [PubMed] [Google Scholar]
- 14.Fowle H et al. (2019) PP2A holoenzymes, substrate specificity driving cellular functions and deregulation in cancer. Adv Cancer Res 144, 55–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seok SH (2021) Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life (Basel) 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amin P et al. (2022) PP2A-B55: substrates and regulators in the control of cellular functions. Oncogene 41 [DOI] [PubMed] [Google Scholar]
- 17.Kokot T and Köhn M (2022) Emerging insights into serine/threonine-specific phosphoprotein phosphatase function and selectivity. J Cell Sci 135 [DOI] [PubMed] [Google Scholar]
- 18.Hunter T and Sefton BM (1980) Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A 77, 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iakoucheva LM et al. (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kettenbach AN et al. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal 4, rs5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt LJ et al. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uversky VN (2013) The alphabet of intrinsic disorder: II. Various roles of glutamic acid in ordered and intrinsically disordered proteins. Intrinsically Disord Proteins 1, e24684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller ML et al. (2008) Linear Motif Atlas for Phosphorylation-Dependent Signaling. Sci Signal 1, ra2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kettenbach AN et al. (2012) Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem Biol 19, 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JL et al. (2023) An atlas of substrate specificities for the human serine/threonine kinome. Nature 613, 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C et al. (2014) Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol Cell 53, 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinna LA et al. (1976) Preferential dephosphorylation of protein bound phosphorylthreonine and phosphorylserine residues by cytosol and mitochondrial “casein phosphatases”. Biochem Biophys Res Commun 70, 1308–1315 [DOI] [PubMed] [Google Scholar]
- 28.Godfrey M et al. (2017) PP2A(Cdc55) Phosphatase Imposes Ordered Cell-Cycle Phosphorylation by Opposing Threonine Phosphorylation. Mol Cell 65, 393–402.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruse T et al. (2020) Mechanisms of site‐specific dephosphorylation and kinase opposition imposed by PP2A regulatory subunits. EMBO J 39, e103695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueki Y et al. (2019) A Consensus Binding Motif for the PP4 Protein Phosphatase. Mol Cell 76, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.AGOSTINIS P et al. (1992) Specificity of the polycation‐stimulated (type‐2A) and ATP, Mg-dependent (type‐1) protein phosphatases toward substrates phosphorylated by P34cdc2 kinase. Eur J Biochem DOI: 10.1111/j.1432-1033.1992.tb16774.x [DOI] [PubMed] [Google Scholar]
- 32.Agostinis P et al. (1990) Synthetic peptides as model substrates for the study of the specificity of the polycation-stimulated protein phosphatases. Eur J Biochem 189, 235–241 [DOI] [PubMed] [Google Scholar]
- 33.Deana AD and Pinna LA (1988) Identification of pseudo “phosphothreonyl-specific” protein phosphatase T with a fraction of polycation-stimulated protein phosphatase 2A. Biochim Biophys Acta 968, 179–185 [DOI] [PubMed] [Google Scholar]
- 34.Deana AD et al. (1982) Dephosphorylation of synthetic phosphopeptides by protein phosphatase-T, a phosphothreonyl protein phosphatase. J Biol Chem 257, 8565–8568 [PubMed] [Google Scholar]
- 35.Agostinis P et al. (1987) Dephosphorylation of phosphoproteins and synthetic phosphopeptides. Study of the specificity of the polycation-stimulated and MgATP-dependent phosphorylase phosphatases. J Biol Chem 262, 1060–1064 [PubMed] [Google Scholar]
- 36.Goldberg J et al. (1995) Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376, 745–753 [DOI] [PubMed] [Google Scholar]
- 37.Kita A et al. (2002) Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1. Structure 10, 715–724 [DOI] [PubMed] [Google Scholar]
- 38.Xing Y et al. (2006) Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins. Cell 127, 341–353 [DOI] [PubMed] [Google Scholar]
- 39.Xu Y et al. (2008) Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sola MM et al. (1991) p34cdc2 phosphorylation sites in histone H1 are dephosphorylated by protein phosphatase 2A1. Biochim Biophys Acta 1094, 211–216 [DOI] [PubMed] [Google Scholar]
- 41.Ferrigno P et al. (1993) Protein phosphatase 2A1 is the major enzyme in vertebrate cell extracts that dephosphorylates several physiological substrates for cyclin-dependent protein kinases. Mol Biol Cell 4, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou XZ et al. (2000) Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell 6, 873–883 [DOI] [PubMed] [Google Scholar]
- 43.Schutkowski M et al. (1998) Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry 37, 5566–5575 [DOI] [PubMed] [Google Scholar]
- 44.Yaffe MB et al. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957–1960 [DOI] [PubMed] [Google Scholar]
- 45.Donella-Deana A et al. (1994) Dephosphorylation of phosphopeptides by calcineurin (protein phosphatase 2B). Eur J Biochem 219, 109–117 [DOI] [PubMed] [Google Scholar]
- 46.Hendus-Altenburger R et al. (2019) Molecular basis for the binding and selective dephosphorylation of Na(+)/H(+) exchanger 1 by calcineurin. Nat Commun 10, 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grigoriu S et al. (2013) The Molecular Mechanism of Substrate Engagement and Immunosuppressant Inhibition of Calcineurin. PLoS Biol 11, 1001492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davey NE et al. (2015) Short linear motifs - ex nihilo evolution of protein regulation. Cell Commun Signal 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tompa P et al. (2014) A million peptide motifs for the molecular biologist. Mol Cell 55, 161–169 [DOI] [PubMed] [Google Scholar]
- 50.van Roey K and Davey NE (2015) Motif co-regulation and co-operativity are common mechanisms in transcriptional, post-transcriptional and post-translational regulation. Cell Commun Signal 13, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egloff MP et al. (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16, 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terrak M et al. (2004) Structural basis of protein phosphatase 1 regulation. Nature 429, 780–784 [DOI] [PubMed] [Google Scholar]
- 53.Heroes E et al. (2013) The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J 280, 584–595 [DOI] [PubMed] [Google Scholar]
- 54.Wakula P et al. (2003) Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. Journal of Biological Chemistry DOI: 10.1074/jbc.M300175200 [DOI] [PubMed] [Google Scholar]
- 55.Hendrickx A et al. (2009) Docking Motif-Guided Mapping of the Interactome of Protein Phosphatase-1. Chem Biol 16, 365–371 [DOI] [PubMed] [Google Scholar]
- 56.Lemaire S and Bollen M (2020) Protein phosphatase-1: dual activity regulation by Inhibitor-2. Biochem Soc Trans 48, 2229–2240 [DOI] [PubMed] [Google Scholar]
- 57.Kwon YG et al. (1997) Characterization of the interaction between DARPP-32 and protein phosphatase 1 (PP-1): DARPP-32 peptides antagonize the interaction of PP-1 with binding proteins. Proc Natl Acad Sci U S A 94, 3536–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava G et al. (2023) Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein–protein interaction. Nature Communications 2023 14:1 14, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao X et al. (2022) Protein phosphatase 1: life-course regulation by SDS22 and Inhibitor-3. FEBS J 289, 3072–3085 [DOI] [PubMed] [Google Scholar]
- 60.Hurley TD et al. (2007) Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem 282, 28874–28883 [DOI] [PubMed] [Google Scholar]
- 61.O’Connell N et al. (2012) The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure 20, 1746–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar GS et al. (2016) The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. Elife 5, e16539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang H Bin et al. (1999) Characterization of the Inhibition of Protein Phosphatase-1 by DARPP-32 and Inhibitor-2. Journal of Biological Chemistry 274, 7870–7878 [DOI] [PubMed] [Google Scholar]
- 64.Ragusa MJ et al. (2010) Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nature Publishing Group 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X et al. (2016) Expanding the PP2A Interactome by Defining a B56-Specific SLiM. Structure 24, 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu C-G et al. (2017) PP2A-B′ holoenzyme substrate recognition, regulation and role in cytokinesis. Cell Discov 3, 17027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J et al. (2016) Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein Cell 7, 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X et al. (2020) A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment. Elife DOI: 10.7554/eLife.55966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fowle H et al. (2021) PP2A/B55α substrate recruitment as defined by the retinoblastoma-related protein p107. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wasserman JS et al. FAM122A ensures cell cycle interphase progression and checkpoint control as a SLiM-2 dependent substrate-competitive inhibitor to the B55/PP2A phosphatase 3 4 5. DOI: 10.1101/2023.03.06.531310 [DOI] [Google Scholar]
- 71.Li Y et al. (2022) Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition. Elife 11, e79736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H et al. (2007) Structure of Calcineurin in Complex with PVIVIT Peptide: Portrait of a Low-affinity Signalling Interaction. J Mol Biol DOI: 10.1016/j.jmb.2007.04.032 [DOI] [PubMed] [Google Scholar]
- 73.Brauer BL et al. (2019) Leveraging New Definitions of the LxVP SLiM to Discover Novel Calcineurin Regulators and Substrates. ACS Chem Biol 14, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kissinger CR et al. (1995) Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 378, 641–644 [DOI] [PubMed] [Google Scholar]
- 75.Wigington CP et al. (2020) Systematic Discovery of Short Linear Motifs Decodes Calcineurin Phosphatase Signaling. Mol Cell 79, 342–358.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eto M et al. (2007) Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor. Structure 15, 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dancheck B et al. (2011) Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry 50, 1238–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller CJ and Turk BE (2018) Homing in: Mechanisms of Substrate Targeting by Protein Kinases. Trends Biochem Sci 43, 380–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nasa I et al. (2018) Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci Signal 11, eaai8669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suijkerbuijk SJE et al. (2012) Integration of Kinase and Phosphatase Activities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev Cell 23, 745–755 [DOI] [PubMed] [Google Scholar]
- 81.Qian J et al. (2015) Cdk1 orders mitotic events through coordination of a chromosome-associated phosphatase switch. Nat Commun 6, 10215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kruse T et al. (2018) The Ebola Virus Nucleoprotein Recruits the Host PP2A-B56 Phosphatase to Activate Transcriptional Support Activity of VP30. Mol Cell DOI: 10.1016/j.molcel.2017.11.034 [DOI] [PubMed] [Google Scholar]
- 83.Nasa I and Kettenbach AN (2018) Coordination of Protein Kinase and Phosphoprotein Phosphatase Activities in Mitosis. Front Cell Dev Biol 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nilsson J (2019) Protein phosphatases in the regulation of mitosis. Journal of Cell Biology 218, 395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barr FA et al. (2011) Protein phosphatases and the regulation of mitosis. J Cell Sci 124, 2323–2334 [DOI] [PubMed] [Google Scholar]
- 86.Mochida S et al. (2016) Two Bistable Switches Govern M Phase Entry. Current Biology 26, 3361–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mochida S et al. (2010) Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science (1979) 330, 1670–1673 [DOI] [PubMed] [Google Scholar]
- 88.Gharbi-Ayachi A et al. (2010) The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science (1979) 330, 1673–1677 [DOI] [PubMed] [Google Scholar]
- 89.Petrone A et al. (2016) Identification of Candidate Cyclin-dependent kinase 1 (Cdk1) Substrates in Mitosis by Quantitative Phosphoproteomics. Mol Cell Proteomics 15, 2448–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nasa I et al. (2020) Quantitative kinase and phosphatase profiling reveal that CDK1 phosphorylates PP2Ac to promote mitotic entry. Sci Signal 13, eaba7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vigneron S et al. (2009) Greatwall maintains mitosis through regulation of PP2A. EMBO J 28, 2786–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams BC et al. (2014) Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A-B55 heterotrimers. Elife 3, e01695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pines J (2011) Cubism and the cell cycle: the many faces of the APC/C. Nature Reviews Molecular Cell Biology 2011 12:7 12, 427–438 [DOI] [PubMed] [Google Scholar]
- 94.Swartz SZ et al. (2021) Selective dephosphorylation by PP2A-B55 directs the meiosis I-meiosis II transition in oocytes. Elife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kruse T et al. (2013) Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J Cell Sci 126, 1086–1092 [DOI] [PubMed] [Google Scholar]
- 96.Bajaj R et al. (2018) KNL1 Binding to PP1 and Microtubules Is Mutually Exclusive. Structure 26, 1327–1336.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu D et al. (2010) Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. Journal of Cell Biology 188, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nijenhuis W et al. (2014) Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol 16, 1257–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bel Borja L et al. (2020) BUB-1 targets PP2A:B56 to regulate chromosome congression during meiosis I in C. elegans oocytes. Elife 9, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shi Y (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 101.Choy MS et al. (2017) PP1:Tautomycetin Complex Reveals a Path toward the Development of PP1-Specific Inhibitors. J Am Chem Soc 139, 17703–17706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swingle M et al. (2007) Small-molecule inhibitors of ser/thr protein phosphatases: Specificity, use and common forms of abuse. Methods in Molecular Biology 365, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brautigan DL and Shenolikar S (2018) Protein Serine/Threonine Phosphatases: Keys to Unlocking Regulators and Substrates. Annu Rev Biochem 87, 921–964 [DOI] [PubMed] [Google Scholar]
- 104.Ulengin-Talkish I and Cyert MS (2023) A cellular atlas of calcineurin signaling. Biochim Biophys Acta Mol Cell Res 1870, 119366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.KVxF PP1 motif repository[Online]. Available: http://slim.icr.ac.uk/motifs/pp1/index.php?page=instances. [Accessed: 30-Mar-2023]
- 106.Kumar M et al. (2022) The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Res 50, D497–D508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cn docking motif repository[Online]. Available: http://slim.icr.ac.uk/motifs/calcineurin/index.php. [Accessed: 30-Mar-2023]
- 108.Dereeper A et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36 [DOI] [PMC free article] [PubMed] [Google Scholar]