Abstract
Background
HPV infection can cause cancer, and standard treatments often result in recurrence. The extent to which liquid biopsy using HPV circulating tumor DNA (HPV ctDNA) can be used as a promising marker for predicting recurrence in HPV‐related cancers remains to be validated. Here we conducted a systematic review and meta‐analysis to assess its effectiveness in predicting treatment response.
Methods
We conducted a systematic literature search of online databases, including PubMed, Embase, Scopus, and the Cochrane Library, up to December 2022. The goal was to identify survival studies that evaluated the potential of plasma HPV ctDNA at baseline and end‐of‐treatment (EoT) in predicting recurrence of related cancers. Hazard ratios were estimated directly from models or extracted from Kaplan–Meier plots.
Results
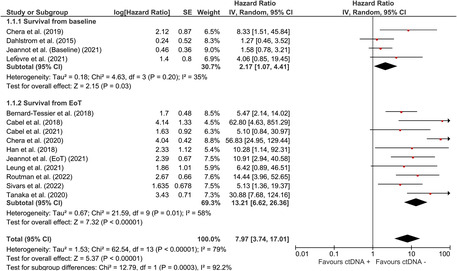
The pooled effect of HPV ctDNA presence on disease recurrence was estimated to be HR = 7.97 (95% CI: [3.74, 17.01]). Subgroup analysis showed that the risk of recurrence was HR = 2.17 (95% CI: [1.07, 4.41]) for baseline‐positive cases and HR = 13.21 (95% CI: [6.62, 26.36]) for EoT‐positive cases. Significant associations were also observed between recurrence of oropharyngeal squamous cell carcinoma (HR = 12.25 (95% CI: [2.62, 57.36])) and cervical cancer (HR = 4.60 (95% CI: [2.08, 10.17])) in plasma HPV ctDNA‐positive patients.
Conclusions
The study found that HPV ctDNA detection can predict the rate of relapse or recurrence after treatment, with post‐treatment measurement being more effective than baseline assessment. HPV ctDNA could be used as a surrogate or incorporated with other methods for detecting residual disease.
Keywords: circulating tumor DNA, HPV ctDNA, HPV‐related cancer, human papillomavirus, survival analysis
The study showed that detecting HPV ctDNA can predict the rate of relapse or recurrence after treatment, with post‐treatment measurement appearing to be superior to baseline assessment in recurrence prediction. This highlights the potential of HPV ctDNA as a surrogate or in combination with other methods for detecting residual disease.
1. INTRODUCTION
Among viruses that contribute to malignant transformation, the human papillomavirus (HPV) has attracted major attention. This virus is the culprit for about one‐third of infection‐related cancers and plays remarkable roles in different steps of cancer evolution. 1 Infections with mucosal HPV types can contribute to carcinomas originating from anogenital mucosa, and head and neck, mostly from the oropharynx. 2 , 3 , 4 These diverse types of cancer burdens attributed to HPV are potentially preventable by screening and vaccination programs. 5 Based on agents classified by the IARC Monographs, 14 high‐risk HPV types (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) are considered Groups 1 and 2A carcinogen. 6 Nearly 75% of all HPV‐associated squamous cell carcinomas and more than 90% of all adenocarcinomas are attributable to high‐risk subtypes 16, 18, 31, 33, and 45. 1 , 7 HPV16 is known to be the most common type of HPV that is associated with oropharyngeal cancer and alone is responsible for more than 90% of HPV‐positive oropharyngeal cancers, which is significantly higher than its prevalence in cervical cancer. 8
Nowadays in the setting of major HPV‐related cancers like cervical cancer (CC), head and neck squamous cell carcinoma (HNSCC), and anal squamous cell carcinoma (ASCC) the preferred standard‐of‐care therapeutic approach includes the combination of platinum‐based chemotherapy and radiation. 9 , 10 , 11 Although chemoradiotherapy (CRT) alone has shown benefits for many years, this method faces some considerable challenges like resistance and adverse events making reliable response prediction biomarkers to be identified essentially. 10 , 11 A significant proportion of patients with advanced HNSCC, cervical, vulval, and anal cancers undergoing CRT have a persistent or recurrent disease 12 ; which can be monitored by biomarkers obtained from blood‐based liquid biopsies. 13 , 14 This non‐invasive cost‐effective biopsy method can provide dynamic information on tumor development by giving biological data of heterogeneous tumor tissue and the metastatic tissues simultaneously. 15
HPV‐related tumor cells usually contain several copies of the HPV genome, as a single genome and multiple tandem head‐to‐tail repeats in humans, as well as in episomal form. 16 Small fragments of the double‐stranded HPV genome, as reported for genomic ctDNA, are released into the bloodstream by apoptosis, necrosis, pyroptosis, phagocytosis, cytoplasmic swelling (oncosis), or ferroptosis. 17 These fragments, referred to as HPV circulating tumor DNA (HPV ctDNA), are detected using ddPCR for HPV E7/E6 sequences. The detection of these fragments have higher specificity as a predictive biomarker for relapse compared to the detection of HPV integration sites. 18 Recent meta‐analyses have reported the diagnostic accuracy of HPV ctDNA in HPV‐associated cancers. 19 , 20 , 21 , 22 Gu et al. suggested that the detection of HPV ctDNA in patients with CC appears to be a highly specific but relatively sensitive diagnostic parameter. 21 Jensen et al. indicated that plasma HPV ctDNA is a promising tool for surveillance in patients with HPV‐related HNSCC. 22 Campo et al. concluded cell‐free human papillomavirus‐DNA (cfHPV‐DNA) represents a potential confirmatory and complementary assay for the positive results of positron emission tomography/computed tomography (PET/CT) scan. 20 In addition, Balachandra et al. and Galati et al. indicated that HPV16 E6 antibodies and circulating HPV‐16 DNA are valuable candidate blood‐based biomarkers of HPV‐associated cancers. 19 , 23 Both next‐generation sequencing (NGS) and digital droplet polymerase chain reaction (ddPCR) as high throughput techniques are major methods to study HPV ctDNA and have been demonstrated to be more accurate than quantitative PCR (qPCR). 13 The use of ddPCR, or a bead‐based assay such like the E7‐MPG, in liquid biopsy improves the diagnostic value of HPV ctDNA significantly. 21 , 23 , 24 There is a discrepancy in the data published on HPV ctDNA by OMIC techniques as a predictive biomarker in the surveillance of HPV‐related cancers. 25 , 26 This meta‐analysis aims to pool the recent publications to evaluate whether the detection of plasma HPV ctDNA can be used as a reliable tool for treatment response monitoring in HPV‐related cancers by considering the recurrence endpoints.
2. MATERIALS AND METHODS
2.1. Search strategy and inclusion criteria
Online databases of PubMed, Embase, Scopus, and the Cochrane Library were systematically searched to find all relevant studies on HPV ctDNA and CC, HNSCC, and ASCC up to December 2022. A combination of subsequent MeSH terms, including “Human Papillomavirus Viruses”, “Circulating Tumor DNA”, “Cell‐Free Nucleic Acids”, “Uterine Cervical Neoplasms”, “Anus Neoplasms”, “Head and Neck Neoplasms”, “Disease‐Free Survival”, “Progression‐Free Survival”, “Minimal Residual Disease”, “Survival Analysis” and “Kaplan–Meier Estimate” were used to search the literature. Moreover, search terms that had no available MeSH terms included “HPV ctDNA”, “Relapse‐Free Survival”, “Recurrence‐Free Survival”, “Locoregional Relapse‐Free Survival”, “Distant Metastasis‐Free Survival (DMFS)”, “Liquid Biopsy Analysis”, “Prognostic Value”, “Prognostic Factor”, “Prognostic Indicator”, “Prognostic and Predictive Biomarkers”, “Recurrence Risk”, “Predictive for Outcome”. The flow diagram of the meta‐analysis and exclusion criteria are shown in Figure 1. The flow diagram was prepared based on The Preferred Reporting Items for Systematic reviews and Meta‐Analyses (PRISMA) 2020 statement guideline. 27
FIGURE 1.
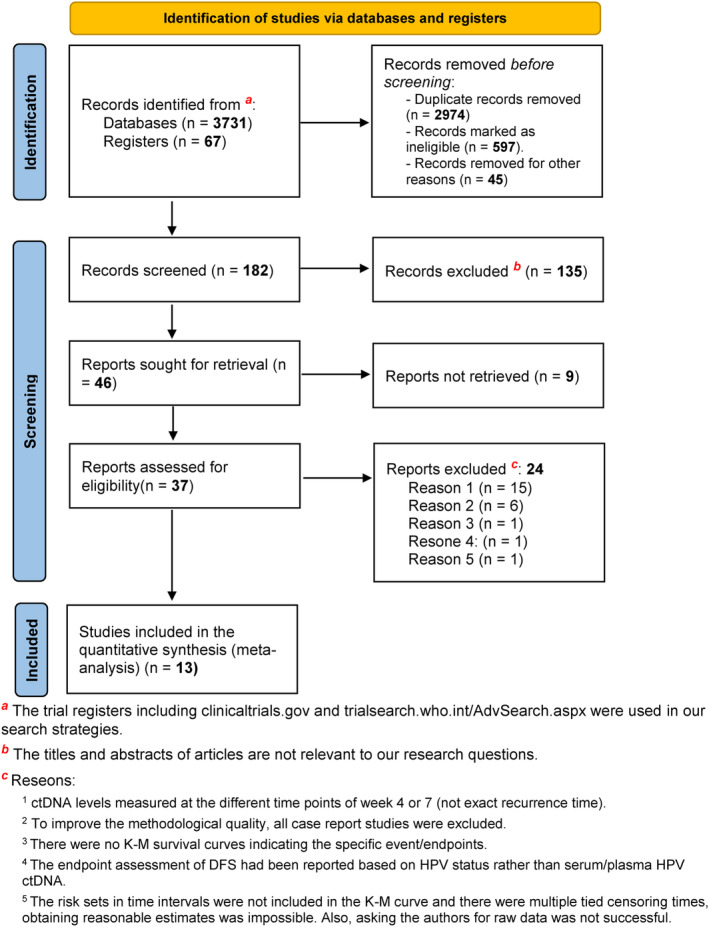
PRISMA flow diagram of the study. In this study, we used the “PRISMA 2020 flow diagram” template which included searches of only databases and registers.
2.2. Data extraction
The following information was obtained from each study: first author, year of publication, cancer type, HPV ctDNA detection/quantitation methods, number of patients in the time‐to‐event analysis, treatment strategies, and recurrent event endpoints data. Chemotherapy with radiation therapy (CRT), chemotherapy only, and radiation therapy only were chosen among other standard treatment regimens. To assess the efficacy of treatment strategies on recurrent event endpoints, we assumed that the widely used endpoints for tumor assessments—progression‐free survival (PFS), disease‐free survival (DFS), recurrence‐free survival (RFS), and event‐free survival (EFS)—represent the same concept, based on studies documenting their interchangeability. The Food and Drug Administration (FDA) defines DFS and PFS as below for approval of cancer drugs and biologics; DFS is defined as the time from randomization or treatment until disease recurrence or death from any cause. PFS is defined as the time from randomization or treatment until objective tumor progression or death, whichever occurs first. 28 DFS also known as relapse‐free survival (RFS) is the time from treatment to the relapse (local, regional, and distant).
Based on the assumptions of these documents, we conclude that DFS, PFS, RFS, and EFS are the time elapsed between the date of initiation or end of treatment and the date of cancer relapse or tumor recurrence. All endpoints are estimated by the Kaplan–Meier (K–M) plots and survival curves. Plasma/serum HPV ctDNA values of pre‐therapeutic (baseline), end‐of‐treatment (EoT), and recurrence at follow‐up were used for estimations of these endpoints across studies.
We use two types of analysis for time‐to‐event data: one for the positivity of HPV ctDNA at EoT and the second for HPV ctDNA value at baseline. To have homogeneous Kaplan–Meier (K–M) data, all reported values were converted to hazard ratio (HR). To minimize underestimation of HPV ctDNA detection, documents that used ddPCR and HPV sequencing (HPV‐seq) methods were chosen for final analysis.
Titles and abstracts of the papers were carefully reviewed, as articles with similar keywords but content unrelated to the purpose, articles without an appropriate treatment strategy, unclear HPV DNA detection methods, case report studies, and papers assessing only the primary efficacy endpoint of overall survival (OS), and paper having HPV ctDNA value just in recurrence time were excluded from the study.
2.3. Statistical analysis
To conduct a meta‐analysis on survival studies, we needed estimates on hazard ratios comparing the groups and their standard errors (SE). However, survival studies report different information and most of the time, required data for meta‐analysis are not directly reported. We retrieved data on HR and SE directly or by back‐calculation from confidence intervals (CI) for those studies that had reported this information. For those that had reported the K–M curve along with risk sets in groups at specific time intervals, we estimated the required information by using approximation methods available in the literature. 29 Some studies neither report HR estimates directly nor represent the risk sets on K–M curves, but show censoring times on the K–M curve by symbols. For these studies, we used GetData Graph Digitizer software (getdata‐graph‐digitizer.com) to extract original data from the K–M curves and then used the Cox PH model to estimate HRs and corresponding SEs. As indicated by heterogeneity indices, a random‐effects model was used to calculate pooled HR and corresponding CI. Pooled estimates and related plots were obtained by Review Manager software (RevMan) version 5.4. 30
3. RESULTS
3.1. Study selection
The initial search, according to the detailed list of the search terms yielded the detection of 3798 records (Figure 1). After the duplicates are identified, 597 publications were assessed for systematic search by filtering to further narrow down these results. Then, 183 records were screened in the title and abstract. Forty‐seven records remained eligible for full‐text assessment. After full‐text assessments, 13 studies meeting the search criteria were eligible for final review. An overview of the studies having eligible time‐to‐event analysis data and their main clinical characteristics are shown in Tables 1 and 2.
TABLE 1.
Overview of the studies stating time‐to‐event analysis in HPV‐related cancer.
| No. | Authors and Year | Country | Tumor type | HPV ctDNA quantification | Cases No. in time‐to‐event analysis | Surrogate endpoint assessment (pre‐post‐treatment marker) | Medical treatment for outcome measurement |
|---|---|---|---|---|---|---|---|
| 1 | Leung et al. (2021) 31 | Canada | LACC | HPV‐seq | 16 | PFS: EOT detectable HPV ctDNA | CRT |
| 2 | Jeannot et al. (2021) 18 | France | CC | ddPCR | 40 | PFS: EOT detectable HPV ctDNA | CRT, Neo‐chemotherapy |
| PFS: Baseline detectable HPV ctDNA | |||||||
| 3 | Cabel et al. (2021) 32 | France | LACC | ddPCR | 14 | DFS: EOT detectable HPV ctDNA | CRT |
| 4 | Lefèvre et al. (2021) 33 | Denmark | SCCA | ddPCR | 45 | DFS: Baseline high and low pHPV | CRT |
| 5 | Tanaka et al. (2020) 34 | Japan | HNSCC | ddPCR | 35 | EFS: EOT detectable HPV ctDNA 16 | Radiotherapy with or without chemotherapy |
| 6 | Chera et al. (2020) 35 | USA | OPSCC | ddPCR | 115 | RFS: EOT detectable HPV ctDNA | CRT |
| 7 | Chera et al. (2019) 36 | USA | OPSCC | ddPCR | 48 | RDFS a : Baseline favorable and unfavorable clearance HPV ctDNA16 | CRT |
| 8 | Bernard‐Tessier et al. (2018) 37 | France | SCCA | ddPCR | 36 | PFS: EOT detectable HPV ctDNA | Chemotherapy |
| 9 | Cabel et al. (2018) 38 | France | ASCC [locally advanced] | ddPCR | 18 | DFS: EOT detectable HPV ctDNA | CRT |
| 10 | Dahlstrom et al. (2015) 26 | USA | OPSCC [OPC] | qPCR | 99 | PFS: Baseline detectable circulating HPV DNA | Chemotherapy and Radiotherapy b |
| 11 | Han et al. (2018) 39 | Canada | LACC | ddPCR | 19 | PFS: EOT detectable HPV ctDNA | CRT |
| 12 | Routman et al. (2022) 40 | USA | OPSCC | ddPCR | 159 | RFS: EOT detectable HPV ctDNA | Adjuvant radiation therapy |
| 13 | Sivars et al. (2022) 41 | Sweden | LACC | ddPCR | 18 | PFS: EOT detectable HPV ctDNA | CRT and EBRT |
Abbreviations: CC: cervical cancer; CRT: chemoradiotherapy; DFS: disease‐free survival; EBRT: external beam radiation therapy; EFS, event‐free survival; HNSCC: squamous cell carcinoma of the head and neck; LACC: locally advanced cervical cancer; OPSCC: oropharyngeal squamous cell carcinoma; PFS: progression‐free survival; RDFS: regional disease‐free survival; RFS: recurrence‐free survival; SCCA: Squamous cell carcinoma of the anus; SCCA: squamous cell carcinomas of the anal canal [Anal squamous cell carcinoma (SCCA)].
RDFS: was measured from the first CRT dose and defined as the time to detect or develop persistent or recurrent disease in the cervical lymph nodes.
During and after treatment, patients underwent regular clinical and radiologic examinations with a team of specialists in head and neck cancer, including surgeons, radiation oncologists, and medical oncologists.
TABLE 2.
Main outcomes of the included studies.
| No | Study type | Study type | Sample | HPV ctDNA monitoring outcome |
|---|---|---|---|---|
| 1 | Leung et al. | Prospective clinical trial | Plasma | Detectable plasma HPV ctDNA at EOT timepoint was associated with shorter PFS |
| 2 | Jeannot et al. | Prospective cohort (the multicenter trial) | Serum | Significant association between HPV ctDNA at EOT and PFS was obtained |
| Baseline HPV ctDNA levels was not associated with PFS | ||||
| 3 | Cabel et al. | Prospective study | Serum/Plasma | HPV‐ctDNA detection at the end of CRT and/or during follow‐up was associated with shorter DFS |
| 4 | Lefèvre et al. | Prospective study | Plasma | High versus low median pretreatments plasma HPV levels had different DFS |
| 5 | Tanaka et al. | a Not specified | Plasma | No detectable ctHPV16DNA was associated with longer EFS |
| 6 | Chera et al. | Prospective biomarker clinical trial | Plasma | Actuarial 2‐year RFS rates between undetectable HPV ctDNA at all surveillance time points vs patients with at least one abnormal HPV ctDNA level were 30% for patients with an abnormal HPV ctDNA valus during post‐treatment surveillance and 100% for patients with negative HPV ctDNA value |
| 7 | Chera et al. | Prospective biomarker study | Plasma | Favorable clearance of ctHPV16DNA was associated with 100% RDFS |
| 8 | Bernard‐Tessier et al. | Multicenter prospective single‐arm trial | Serum | Detectable HPV ctDNA at the end of chemotherapy was associated with poorer PFS |
| 9 | Cabel et al. | Prospective study | Plasma/Serum | HPV ctDNA detection after CRT was strongly associated with shorter DFS |
| 10 | Dahlstrom et al. | Prospective cohort study | Plasma/serum | Negative and positive HPV ctDNA levels at the pretreatment timepoint were not significantly associated with longer PFS |
| 11 | Han et al. | Prospective multicenter study | Plasma | Patients with undetectable HPV ctDNA at CRT completion time had significantly longer PFS than those with detectable levels. |
| 12 | Routman et al. | Prospective study or part of clinical trials | Serum | Detectable HPV ctDNA level after surgical treatment was significantly associated with shorter RFS |
| 13 | Leung et al. | Not specified | Plasma | Patients with detectable HPV ctDNA at EOT timepoint had significantly shorter PFS compared to those with negative HPV ctDNA values |
The patients were newly diagnosed with HPV16‐related HNSCC and underwent radiotherapy with or without chemotherapy.
3.2. Variability in study design and quality assessment
Of 13 studies included in the meta‐analysis, 5 studies were related to cervical, 3 to anal, and 6 to head and neck cancers. Both squamous cell carcinoma (SCCA) and anal squamous cell carcinoma (ASCC) are considered neoplasms of the anal canal. In the case of HNSCC, most of the records were HPV‐positive oropharyngeal squamous cell carcinoma (OPSCC). Only one study indicated HPV16‐related HNSCC patients treated by radiotherapy with or without chemotherapy.
In the included studies, different surrogate endpoint assessments including PFS, DFS, EFS, RFS, and RDFS (Regional Disease‐Free Survival) indicated the same idea and concept for treatment response assessment, thanks to the same meaning, all of them were considered as an endpoint assessment in the final analysis. Of 13 studies, 11 studies used EoT [at end of treatment/post‐treatment], 3 studies used baseline, and one study reported both EoT and baseline presence/absence HPV ctDNA for estimating disease relapse/recurrence prediction. Two studies used HPV ctDNA 16 levels in response assessment and outcome. Since the HPV16 ctDNA biomarker is highly specific and sensitive to the detection HPV positive cancer; we considered these studies in the final analysis too. Twelve studies used high‐throughput assays of ddPCR and HPV sequencing for HPV ctDNA quantitation. As shown in Table 1, among 13 studies, nine studies used EoT detectable HPV ctDNA as the predictor of recurrence, two studies used baseline detectable HPV ctDNA, one study used both EoT and baseline detectable HPV ctDNA, one study used baseline favorable and unfavorable clearance HPV ctDNA. Quality assessment of the studies based on the QUADAS‐2 indicated that the variations mentioned above meet our selection criteria and the overall risk of bias was low. The 13 included studies fulfilled the criterion for the items of patient selection, medical treatment for outcome measurement, endpoint assessment, HPV ctDNA quantification, and tumor.
3.3. HPV ctDNA meta‐analysis
A total of 662 patients were included according to time‐to‐event analysis data from 13 HPV‐related studies. The pooled estimate of the effect of HPV ctDNA presence on disease recurrence in HPV‐related cancers was HR = 7.97 (95% CI: [3.74, 17.01]) (Figure 2). Subgroup analysis was carried out to find the source of heterogeneity. As indicated by reduced heterogeneity in subgroups, the estimates from baseline and EoT studies were basically different. The estimated risk of cancer recurrence for HPV ctDNA positivity at baseline and EoT timepoints were HR = 2.17 (95% CI: [1.07, 4.41]) and HR = 13.21 (95% CI: [6.62, 26.36]), respectively (Figure 2). The pooled estimate for OPSCC (Figure 3) was HR = 12.25 (95% CI: [2.62, 57.36]) for HPV ctDNA positive cases. As shown in Figure 4, the pooled effect for CC was estimated as HR = 4.60 (95% CI: [2.08, 10.170]) for HPV ctDNA+ cases.
FIGURE 2.
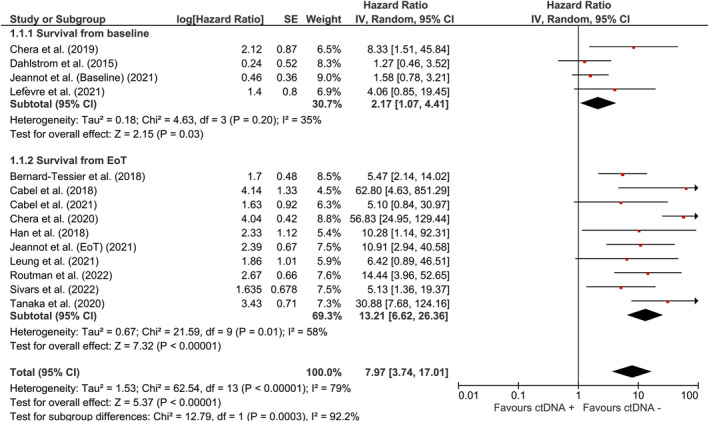
Risk of subsequent HPV‐related cancer recurrence for HPV ctDNA positive cases at baseline and EoT timepoints.
FIGURE 3.
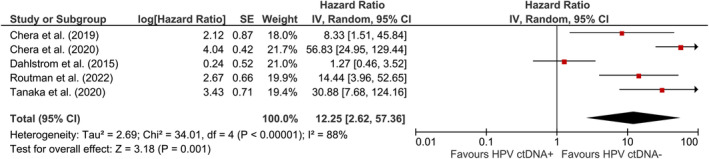
Risk of subsequent OPSCC recurrence in HPV ctDNA positive cases.
FIGURE 4.
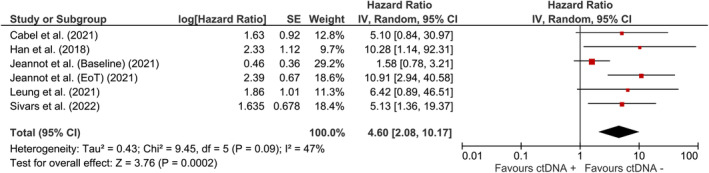
The risk of subsequent cervical cancer for HPV ctDNA positive cases.
3.4. Sensitivity analysis results
For the case that all studies included in the analysis (Figure 2), after removing Chera et al. (2020), I 2 was reduced from 79%, 35%, and 58% to 62%, 35%, and 0% for total, baseline, and EoT studies, respectively. Removal of the other studies showed negligible effect on heterogeneity (<2% reduction in I 2). For OPSCC incidence (Figure 3), after the removal of Dahstrom et al. (2015), I 2 dropped from 88% to 47%. None of these studies had an appreciable impact on the pooled estimate of the studies with respect to direction or significance.
4. DISCUSSION
HPV ctDNA is linked to tumor burden, disease stage, and metastasis. 34 Previous studies have shown that detectible EoT and baseline HPV ctDNA values correlate well with tumor load and therefore HPV ctDNA dynamics can be used as a surrogate of treatment response. 18 Here, by combining results from multiple data sets, we showed that measurement of residual disease following concurrent use of chemotherapy and radiation—chemoradiotherapy (CRT)—and neoadjuvant therapy accurately well predict survival in HPV‐associated cancers. Some evidence indicates that the cytotoxic effects of chemotherapy or radiation therapy seem to increase cell‐free DNA levels due to cellular senescence. 42 , 43 Moreover, other studies have indicated that levels of HPV ctDNA drop significantly in patients with (CC) and LACC during CRT. 32 , 39 This means that the sensitivity of HPV ctDNA detection at the end of CRT is not perfect in predicting recurrence. However, our pooled analysis demonstrated that measuring plasma/serum HPV ctDNA levels at the end of treatment is more effective in predicting recurrence compared to measuring levels at baseline. According to survival data, HPV ctDNA clearance after CRT and/or neoadjuvant therapy can outperform tumor response to treatment. Although further studies are needed, HPV ctDNA postulates as an early surrogate of recurrence prediction being a promising endpoint assessment in the neoadjuvant and CRT setting; those administrations are widely used for oropharynx, cervical, and anal canal cancers. 44 , 45 , 46 Here, we showed that the predictive performance of HPV ctDNA for OPSCC was better than CC. It means that more focus is needed for HPV ctDNA in the decision‐making of postradiotherapy management of oropharyngeal cancer.
PET/CT is often used for long‐term follow‐up goals, treatment planning, and accurate prediction of treatment response in patients receiving radiation therapy (RT). Moreover, PET/CT is a routine tool for response evaluation after RT and follow‐up imaging for cancers with a high negative predictive value. Some unresolved issues, such as optimal uptaking of the probe, optimal cut‐off values (optimal threshold values that can be used to determine whether a tumor is responding to radiation therapy), and the optimal timing of PET/CT during RT in predicting the radiotherapy response of different tumors have diverse results and are still under research. 47 RT response evaluation using PET/CT is currently undertaken 10 to 12 weeks post CRT and can be postponed to minimize false‐positive results; like in the presence of inflammations, as the most common cause of false‐positive PET/CT findings post‐chemotherapy and radiotherapy that need to be is resolved. 48 Delaying PET/CT treatment response assessment would help decrease the frequency of false‐positive as well as false‐negative results. However, delaying PET‐CT treatment response assessment may lead to delayed salvage therapy and a poorer prognosis. 34 However, PET/CT imaging is not merely a solution after radiotherapy and can be complemented with HPV ctDNA measurement to empower decision‐making in the radiation oncology field. 34 We propose that positivity for HPV ctDNA at that end‐of‐treatment timepoint can outperform PET‐CT in predicting treatment failure after radiotherapy with or without chemotherapy in patients with HPV‐associated tumors, especially in HPV‐positive HNSCC tumors.
According to the risk ratio we found, patients with fast HPV ctDNA clearance and low recurrence risk after treatment could be considered for adaption of CRT with less‐intensive therapy in the last weeks of CRT, to avoid harmful side effects and unnecessary medications. Repeated time‐points may be needed to better predict relapse. As reported previously, two consecutively positive plasma HPV ctDNA detection in HNSCC patients during post‐treatment surveillance brought high (above 90%) positive and negative predictive values indicating its potential in facilitating the decision for salvage therapy. 35 Although several patients have achieved complete clearance of HPV ctDNA only at the end of CRT and did not subsequently experience relapse, so positive HPV ctDNA detection during CRT did not appear to be a reliable predictor of relapse. 32 This suggests that HPV ctDNA detection at the end of treatment timepoint has a superiority for earlier timepoints.
Question: If a tumor is predicted to have a good response to RT and/or CRT, it seems to be worth performing CRT or RT only as a treatment modality. Otherwise, if a tumor is predicted to be radio‐resistant, how we can predict the radiosensitivity of tumors using HPV ctDNA measurement remained to be evaluated.
4.1. Limitations
Since anal, penile, vaginal, and vulvar cancers are relatively rare, the potential role of HPV ctDNA in monitoring these cancers has yet to be fully evaluated. We were also unable to analyze cancer surveillance data according to tumor stage. No data were available regarding detectible HPV ctDNA at several time points, especially in mid‐treatment or weeks after treatment for real‐time ctDNA monitoring. Also, there were no qualitative attributes regarding HPV DNA fragments and HPV types in most of the studies and their implementation in clinical practice. Included studies had no relevant information regarding the immunohistochemical detection of p16INK4a (p16) as a reliable marker for active HPV infection.
5. CONCLUSIONS
In conclusion, ultrasensitive detection of HPV ctDNA can predict relapse or recurrence rate after treatment. Based on the obtained data, the end‐of‐treatment measurement of HPV ctDNA has superiority over baseline measurement in treatment monitoring and response assessment of HPV‐associated cancers. Also, OPSCC patients can ultimately benefit from HPV ctDNA measurement; so it is essential to incorporate HPV ctDNA measurement into the decision‐making of postradiotherapy management in OPSCC treatment settings to avoid unnecessary invasive procedures. Although the end‐of‐treatment timepoint sensitivity exceeded relapse prediction, however, a positive result might not guarantee subsequent recurrence prediction and could necessitate repeat testing or complemented with PET/CT scan. Future studies are needed for later time points measurements to guide real‐time treatment decision‐making of HPV‐associated cancers.
AUTHOR CONTRIBUTIONS
Abbas Karimi: Conceptualization (equal); data curation (equal); investigation (equal); methodology (equal); project administration (equal); supervision (equal); writing – original draft (equal); writing – review and editing (equal). Tohid JafariKoshki: Data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); validation (equal); writing – review and editing (equal). Mojtaba Zehtabi: Data curation (equal); investigation (equal); writing – review and editing (equal). Farzaneh Kargar: Investigation (equal). Tarik Gheit: Conceptualization (equal); investigation (equal); supervision (equal); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no conflicts of interest.
DISCLAIMER
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
ACKNOWLEDGMENTS
The authors thank Dr. Mohammad Haidari (Urmia University of Medical Sciences) for his advice on data extraction.
Karimi A, Jafari‐Koshki T, Zehtabi M, Kargar F, Gheit T. Predictive impact of human papillomavirus circulating tumor DNA in treatment response monitoring of HPV‐associated cancers; a meta‐analysis on recurrent event endpoints. Cancer Med. 2023;12:17592‐17602. doi: 10.1002/cam4.6377
Contributor Information
Abbas Karimi, Email: karimia@tbzmed.ac.ir.
Tarik Gheit, Email: gheitt@iarc.who.int.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from corresponding authors but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. However, data are available from the authors upon reasonable request and with permission of corresponding authors.
REFERENCES
- 1. Araldi RP, Sant'Ana TA, Módolo DG, et al. The human papillomavirus (HPV)‐related cancer biology: an overview. Biomed Pharmacother. 2018;106:1537‐1556. [DOI] [PubMed] [Google Scholar]
- 2. Bansal A, Singh MP, Rai B. Human papillomavirus‐associated cancers: a growing global problem. Int J Appl Basic Med Res. 2016;6(2):84‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brennan S, Baird AM, O'Regan E, Sheils O. The role of human papilloma virus in dictating outcomes in head and neck squamous cell carcinoma. Front Mol Biosci. 2021;8:677900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karimi A, Mohebbi E, McKay‐Chopin S, et al. Human papillomavirus and risk of head and neck squamous cell carcinoma in Iran. Microbiol Spectr. 2022;10(4):e0011722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. St Laurent J, Luckett R, Feldman S. HPV vaccination and the effects on rates of HPV‐related cancers. Curr Probl Cancer. 2018;42(5):493‐506. [DOI] [PubMed] [Google Scholar]
- 6. Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12‐19. [DOI] [PubMed] [Google Scholar]
- 7. De Vincenzo R, Ricci C, Conte C, Scambia G. HPV vaccine cross‐protection: highlights on additional clinical benefit. Gynecol Oncol. 2013;130(3):642‐651. [DOI] [PubMed] [Google Scholar]
- 8. Chaturvedi AK, Anderson WF, Lortet‐Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31(36):4550‐4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruiz FJ, Inkman M, Rashmi R, et al. HPV transcript expression affects cervical cancer response to chemoradiation. JCI Insight. 2021;6(16):e138734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patel U, Pandey M, Kannan S, et al. Prognostic and predictive significance of nuclear HIF1α expression in locally advanced HNSCC patients treated with chemoradiation with or without nimotuzumab. Br J Cancer. 2020;123(12):1757‐1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin D, Rödel F, Balermpas P, Rödel C, Fokas E. The immune microenvironment and HPV in anal cancer: rationale to complement chemoradiation with immunotherapy. Biochim Biophys Acta Rev Cancer. 2017;1868(1):221‐230. [DOI] [PubMed] [Google Scholar]
- 12. Sacks D, Baxter B, Campbell BCV, et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. 2018;13(6):612‐632. [DOI] [PubMed] [Google Scholar]
- 13. Cafforio P, Palmirotta R, Lovero D, et al. Liquid biopsy in cervical cancer: hopes and pitfalls. Cancers (Basel). 2021;13(16):3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmirotta R, Lovero D, Cafforio P, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1758835918794630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112‐123. [DOI] [PubMed] [Google Scholar]
- 16. McBride AA, Warburton A. The role of integration in oncogenic progression of HPV‐associated cancers. PLoS Pathog. 2017;13(4):e1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sánchez‐Herrero E, Serna‐Blasco R, Robado de Lope L, González‐Rumayor V, Romero A, Provencio M. Circulating tumor DNA as a cancer biomarker: an overview of biological features and factors that may impact on ctDNA analysis. FrontOncologia. 2022;12:943253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jeannot E, Latouche A, Bonneau C, et al. Circulating HPV DNA as a marker for early detection of relapse in patients with cervical cancer. Clin Cancer Res. 2021;27(21):5869‐5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balachandra S, Kusin SB, Lee R, et al. Blood‐based biomarkers of human papillomavirus‐associated cancers: a systematic review and meta‐analysis. Cancer. 2021;127(6):850‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campo F, Zocchi J, Moretto S, et al. Cell‐free human papillomavirus‐DNA for monitoring treatment response of head and neck squamous cell carcinoma: systematic review and meta‐analysis. Laryngoscope. 2022;132(3):560‐568. [DOI] [PubMed] [Google Scholar]
- 21. Gu Y, Wan C, Qiu J, Cui Y, Jiang T, Zhuang Z. Circulating HPV cDNA in the blood as a reliable biomarker for cervical cancer: a meta‐analysis. PloS One. 2020;15(2):e0224001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jensen KK, Grønhøj C, Jensen DH, von Buchwald C. Circulating human papillomavirus DNA as a surveillance tool in head and neck squamous cell carcinoma: a systematic review and meta‐analysis. Clin Otolaryngol. 2018;43(5):1242‐1249. [DOI] [PubMed] [Google Scholar]
- 23. Galati L, Combes JD, Le Calvez‐Kelm F, et al. Detection of circulating HPV16 DNA as a biomarker for cervical cancer by a bead‐based HPV genotyping assay. Microbiol Spectr. 2022;10(2):e0148021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheung TH, Yim SF, Yu MY, et al. Liquid biopsy of HPV DNA in cervical cancer. J Clin Virol. 2019;114:32‐36. [DOI] [PubMed] [Google Scholar]
- 25. Veyer D, Wack M, Mandavit M, et al. HPV circulating tumoral DNA quantification by droplet‐based digital PCR: a promising predictive and prognostic biomarker for HPV‐associated oropharyngeal cancers. Int J Cancer. 2020;147(4):1222‐1227. [DOI] [PubMed] [Google Scholar]
- 26. Dahlstrom KR, Li G, Hussey CS, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer. 2015;121(19):3455‐3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Available from: https://www.fda.gov/media/71195/download.
- 29. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time‐to‐event data into meta‐analysis. Trials. 2007;8(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The Cochrane collaboration. Review Manager (RevMan) Version 5.4. 2020.
- 31. Leung E, Han K, Zou J, et al. HPV sequencing facilitates ultrasensitive detection of HPV circulating tumor DNA. Clin Cancer Res. 2021;27(21):5857‐5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cabel L, Bonneau C, Bernard‐Tessier A, et al. HPV ctDNA detection of high‐risk HPV types during chemoradiotherapy for locally advanced cervical cancer. ESMO Open. 2021;6(3):100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lefèvre AC, Pallisgaard N, Kronborg C, Wind KL, Krag SRP, Spindler KG. The clinical value of measuring circulating HPV DNA during chemo‐radiotherapy in squamous cell carcinoma of the anus. Cancers (Basel). 2021;13(10):2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tanaka H, Takemoto N, Horie M, et al. Circulating tumor HPV DNA complements PET‐CT in guiding management after radiotherapy in HPV‐related squamous cell carcinoma of the head and neck. Int J Cancer. 2021;148(4):995‐1005. [DOI] [PubMed] [Google Scholar]
- 35. Chera BS, Kumar S, Shen C, et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV‐associated oropharyngeal cancer. J Clin Oncol. 2020;38(10):1050‐1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chera BS, Kumar S, Beaty BT, et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV‐associated oropharyngeal cancer. Clin Cancer Res. 2019;25(15):4682‐4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bernard‐Tessier A, Jeannot E, Guenat D, et al. Clinical validity of HPV circulating tumor DNA in advanced anal carcinoma: an ancillary study to the epitopes‐HPV02 trial. Clin Cancer Res. 2019;25(7):2109‐2115. [DOI] [PubMed] [Google Scholar]
- 38. Cabel L, Jeannot E, Bieche I, et al. Prognostic impact of residual HPV ctDNA detection after chemoradiotherapy for anal squamous cell carcinoma. Clin Cancer Res. 2018;24(22):5767‐5771. [DOI] [PubMed] [Google Scholar]
- 39. Han K, Leung E, Barbera L, et al. Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precis Oncol. 2018;2:1‐8. [DOI] [PubMed] [Google Scholar]
- 40. Routman DM, Kumar S, Chera BS, et al. Detectable postoperative circulating tumor human papillomavirus DNA and association with recurrence in patients with HPV‐associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2022;113(3):530‐538. [DOI] [PubMed] [Google Scholar]
- 41. Sivars L, Hellman K, Crona Guterstam Y, et al. Circulating cell‐free tumor human papillomavirus DNA is a promising biomarker in cervical cancer. Gynecol Oncol. 2022;167(1):107‐114. [DOI] [PubMed] [Google Scholar]
- 42. Lo YM, Leung SF, Chan LY, et al. Kinetics of plasma Epstein‐Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res. 2000;60(9):2351‐2355. [PubMed] [Google Scholar]
- 43. Mair R, Mouliere F, Smith CG, et al. Measurement of plasma cell‐free mitochondrial tumor DNA improves detection of glioblastoma in patient‐derived Orthotopic xenograft models. Cancer Res. 2019;79(1):220‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pessia B, Romano L, Giuliani A, Lazzarin G, Carlei F, Schietroma M. Squamous cell anal cancer: management and therapeutic options. Ann Med Surg (Lond). 2020;55:36‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wotman M, Ghaly M, Massaro L, et al. Improving post‐CRT neck assessment in patients with HPV‐associated OPSCC (review). Mol Clin Oncol. 2020;13(4):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Musunuru HB, Pifer PM, Mohindra P, Albuquerque K, Beriwal S. Advances in management of locally advanced cervical cancer. Indian J Med Res. 2021;154(2):248‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shen LF, Zhou SH, Yu Q. Predicting response to radiotherapy in tumors with PET/CT: when and how? Transl Cancer Res. 2020;9(4):2972‐2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Long NM, Smith CS. Causes and imaging features of false positives and false negatives on F‐PET/CT in oncologic imaging. Insights Imaging. 2011;2(6):679‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from corresponding authors but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. However, data are available from the authors upon reasonable request and with permission of corresponding authors.


