Abstract
The telomeric repeat-containing RNA, TERRA, associates with both telomeric DNA and telomeric proteins, often forming RNA:DNA hybrids (R-loops). TERRA is most abundant in cancer cells utilizing the alternative lengthening of telomeres (ALT) pathway for telomere maintenance, suggesting that persistent TERRA R-loops may contribute to activation of the ALT mechanism. Therefore, we sought to identify the enzyme(s) that regulate TERRA metabolism in mammalian cells. Here, we identify that the 5’-3’ exoribonuclease XRN2 regulates the stability of TERRA RNA. Moreover, while stabilization of TERRA alone was insufficient to drive ALT, depletion of XRN2 in ALT-positive cells led to a significant increase in TERRA R-loops and exacerbated ALT activity. Together, our findings highlight XRN2 as a key determinant of TERRA metabolism and telomere stability in cancer cells that rely on the ALT pathway.
Keywords: TERRA, Alternative Lengthening of Telomeres (ALT), Auxin-Inducible Degron (AID), XRN2, R-Loops
Graphical Abstract:
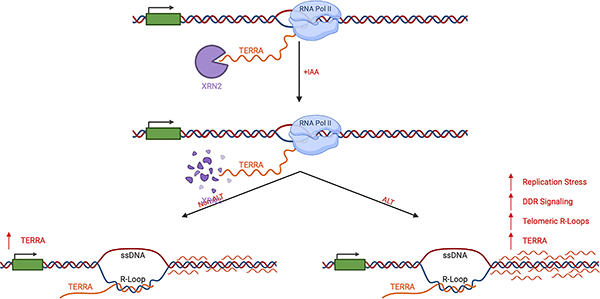
The 5’-3’ exoribonuclease XRN2 directly modulates telomere repeat-containing RNA (TERRA) stability in mammalian cells. XRN2 depletion led to TERRA stabilization and localization on chromatin. In cells utilizing the alternative lengthening of telomeres (ALT) pathway, XRN2 depletion stabilized R-loops and exacerbated ALT activity. Collectively, defects in TERRA metabolism alter telomere stability and drive telomere dysfunction in ALT cells.
Introduction
Telomeres are repetitive DNA elements that cap the ends of linear chromosomes and function as a molecular barrier to the human genome. In mammalian cells, telomeres are primarily composed of a series of (TTAGGG)n hexameric repeats terminating in a single-stranded overhang that loops back onto itself, invades the double-stranded region, and forms a structure known as the T-loop [1–3]. Progressive telomere shortening following each cellular division jeopardizes the formation of the T-loop [4–6]. Consequently, critically-short or dysfunctional telomeres are a significant source of genomic instability. To counter telomere dysfunction, cells rely on the multi-subunit protein complex shelterin, which not only facilitates T-loop formation but also shields telomere ends from recognition by the DNA damage response machinery [7]. In addition to shelterin, the telomeric cap has expanded to include the long non-coding telomere repeat-containing RNA, TERRA [8,9]. The discovery of TERRA suggested that there could be additional layers of regulation underlying telomere maintenance that have yet to be defined, highlighting the importance of characterizing TERRA function at telomeric DNA.
TERRA is expressed in several eukaryotes, including plants, yeast, fish, and mammals [8–11]. In mammalian cells, TERRA is transcribed from the C-rich strand of telomeric DNA by RNA polymerase II [8,9]. Transcription initiation is believed to occur within CpG island repeats located in the subtelomeric region on approximately half of all chromosome ends and proceeds distally toward the telomeric repeats [12]. The resulting TERRA transcript contains both subtelomeric sequence and G-rich (UUAGGG)n telomeric repeats, ranging in length from 200bp to 9kb [8,9]. TERRA is localized within the nucleus and has been demonstrated to associate with a number of telomeric proteins, including the shelterin components TRF1 and TRF2 [13]. Through these protein interactions, TERRA has been implicated in the regulation of telomere length maintenance, heterochromatin formation, and the DNA damage response [14–17].
In addition to the association with telomeric binding proteins, TERRA has also been shown to interact with the telomeric DNA itself, forming DNA:RNA hybrid structures known as R-loops [9,18–21]. R-loops are triple-stranded nucleic acid structures that arise as a byproduct of RNA transcription and can act as a molecular roadblock to the replication machinery leading to the formation of transcription-replication collisions (TRCs) [22–25]. TRCs can occur co-directionally, where the DNA replisome and RNA Pol II travel in the same direction on DNA, or TRCs can occur head-on, where the replication machinery and the transcriptional machinery travel towards one another. Co-directional TRCs can be resolved when the replisome overtakes the transcriptional machinery, leading to the displacement of the RNA from DNA. In contrast, head-on TRCs can impede the replication machinery, causing replication stress and driving the collapse of the replication fork into DNA double-strand breaks [22,26]. Thus, the formation of TERRA R-loops at telomere ends could increase the opportunity for co-directional and/or head-on TRCs within the telomeres and, ultimately, compromise telomere stability.
TERRA R-loops are most prevalent in cells that rely on the alternative lengthening of telomeres (ALT) pathway to promote telomere elongation [19]. Early evidence demonstrated that ALT telomeres displayed an increase in spontaneous DNA damage and telomere length heterogeneity, suggesting a reliance on recombination to promote telomere elongation [27,28]. Over the past decade, the mechanistic details have been further refined, demonstrating that DNA double-strand breaks can catalyze repair and, consequently, telomere elongation via a recombination process known as break-induced replication (BIR) [29–31]. BIR is a pathway that repairs one-ended double-strand breaks (DSBs), which arise when challenges to the replisome cause replication fork stalling. These telomeric DNA DSBs are clustered into nuclear bodies that contain the scaffolding protein promyelocytic leukemia protein (PML). These ALT-associated PML-bodies (APB) provide the framework for the recruitment of the DNA replication and recombination proteins required to initiate repair and, ultimately, promote the elongation of ALT telomeres [32–34]. Although it is unclear mechanistically what drives the DSBs required for BIR at ALT telomeres, chronic replication stress undoubtedly contributes to the process [35–37]. The enrichment of TERRA in ALT cells has led to the hypothesis that TERRA R-loops promote the replication stress associated with the activation of ALT. However, it remains unclear whether the increase in TERRA alone is sufficient to induce R-loops and, ultimately, drive ALT activity.
Here, we sought to identify the enzyme(s) that regulate TERRA stability in an effort to define additional factors that may catalyze ALT activity. These studies add to the growing body of literature surrounding TERRA by demonstrating that the 5’-3’ exoribonuclease XRN2 plays a critical role in TERRA degradation in mammalian cells. We show that rapid depletion of XRN2 leads to an increase in TERRA and the accumulation of TERRA transcripts on chromatin. While the increase in TERRA did not drive telomere dysfunction in non-ALT cells, we observed a significant increase in R-loop formation and DNA damage at telomeric DNA in cells with an active ALT mechanism. Our data suggest that the regulation of TERRA metabolism is a critical determinant of TERRA interaction at telomeres. Additionally, defects in TERRA metabolism can alter telomere stability and drive telomere dysfunction in the context of ALT.
Materials & Methods
Cell Lines and Cell Culture Conditions
All cell lines were submitted for Short Tandem Repeat (STR) analysis by ATCC, and certificates of authentication can be provided upon request. HCT116, U2OS, HeLa, HEK 293FT, Cal-72, and RPE cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). SaOS2 cells were cultured in RPMI 1640 media supplemented with 10% FBS and 1% P/S. MG-63 and NY cells were cultured in DMEM/F-12 supplemented with 5% FBS and 1% P/S. SJSA-1, HuO9, G-292, and NOS1 cells were cultured in RPMI 1640 media supplemented with 5% FBS, 1% sodium pyruvate (NaPy), and 1% P/S. Hu03N1 and Cal-78 cells were cultured in RPMI 1640 media supplemented with 10% FBS, 1% NaPy, and 1% P/S. hFOB 1.19 cells were cultured in DMEM/F-12 supplemented with 10% FBS, 0.3mg/mL G418, and 1% P/S. HOS cells were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% FBS and 1% P/S. IAA-inducible HCT116 cells were previously engineered, as outlined in Davidson et al. (2019) [41]. Likewise, the IAA-inducible SaOS2 cells were generated using the same pipeline. Briefly, cells were transfected with AAVS1 T2 CRISPR/Cas plasmid (based on pX330 and encoding gRNA and SpCas9) with pMK232 (CMV-OsTir1) and selected with puromycin to generate stable clones. Clones expressing OsTir1 were then transfected with pX330-based CRISPR/Cas plasmid (encoding gRNA and SpCas9) and a donor plasmid (Hygro-mAID pMK287) to integrate a 3xAID tag at the 3’ end of the XRN2 locus. To induce OsTir1 interaction with AID-tagged protein, cells were treated with 500μM IAA unless otherwise stated. Cell culture media and supplements were obtained from Gibco Invitrogen, and all plasticware came from Corning (Corning, NY). All cells were maintained at 37°C in a humidified incubator at 5% CO2.
Antibodies, Probes, Plasmids, and Primers
The following antibodies, probes, plasmids and primers were used where indicated: DIS3 (H-3, Santa Cruz); EXOSC10 (B-8, Santa Cruz); XRN2 (H-3, Santa Cruz); αTubulin (11H10, Cell Signaling); OsTir1 (MBL Int. Co.), αDNA-RNA Hybrid Antibody (S9.6, EMD Millipore); PML (PG-M3, Santa Cruz); γH2AX Ser139 (JBW301, EMD Millipore); pRPA32 S33 (Bethyl); pRPA32 S4/S8 (Bethyl); TRF2 (4A794, EMD Millipore); ATRX (H-300, Santa Cruz); Peroxidase-Conjugated Goat Anti-Mouse IgG (Jackson ImmunoResearch); Peroxidase-Conjugated Goat Anti-Rabbit IgG (Jackson ImmunoResearch); Alexa Fluor 488-Conjugated Donkey Anti-Mouse IgG (Jackson ImmunoResearch); Cy5-Conjugated Goat Anti-Rabbit IgG (Abcam); C-Rich Telomere/TERRA Probe (5’ - CCCTAACCCTAACCCTAACCCTAA - 3’); G-Rich Telomere Probe (5’ - TTAGGGTTAGGGTTAGGGTTAGGG - 3’); 28S Probe (5’ - AACGATCAGAGTAGTGGTATTTCACC - 3’); PNA Telomeric Probes (TelC-Cy3 and TelG-Cy3, PNA Bio Inc.); WT-XRN2 and D235A-XRN2 vectors for stable transfection were previously made by Eaton et al. (2018) [42] via insertion of the XRN2 coding sequence into a pSBbi-Puro Empty Backbone (Addgene #60523); shATRX Plasmid (MISSION shATRX #3 - NM_000489.3–2357s21c1); psPAX2 Plasmid (Addgene #12260); pMD2G Plasmid (Addgene #12259); TelBam3.4 Forward Primer (5’ - CAGAGTTCTCCTCAGGTCAGA - 3’); TelBam3.4 Reverse Primer (5’ - GGAATGCTGCTCCACCTTTA - 3’); RPPH1 Forward Primer (5’ - GTGCGTCCTGTCACTCCACT - 3’); RPPH1 Reverse Primer (5’ - TTCCAAGCTCCGGCAAAGGA - 3’).
RNA Dot Blot
Total RNA was extracted from the three biological replicates using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Twenty micrograms of RNA were diluted in 1:1 water to formamide solution with 1 mM EDTA. Samples were heated to 65°C before being transferred onto a Hybond N+ membrane using a Bio-Dot SF microfiltration apparatus (BioRad). Membranes were crosslinked at 254 nm at 150 mJ/cm². Membranes were then incubated in ULTRAhyb hybridization buffer (Ambion) overnight at 50°C with a DNA probe complementary to the TERRA sequence (CCCTAA)4. DNA probes were end-labeled using a DIG oligonucleotide 3’-end labeling kit (Roche) according to the manufacturer’s instructions. The following day, the membrane was washed twice with 2xSSC containing 0.1% SDS at room temperature for 5 minutes each and twice with 0.5xSSC containing 0.1% SDS at 50°C for 15 minutes each before development. The membrane was developed using Sigma-Aldrich Anti-Digoxigenin-AP (Roche #11093274910) and Sigma-Aldrich chemiluminescent substrate CDP-Star (Roche #11685627001) with the DIG Wash and Block Buffer Set (Roche) following manufacturer’s instructions. Images were captured and visualized using a BioRad ChemiDoc XRS+ imaging system. TERRA was quantified by densitometry using Image Lab 6.1 software.
Western Blot
Cells were collected by trypsinization and washed with ice-cold 1xPBS. Samples were then lysed in 2x Sample Buffer and sonicated in a water bath at 4°C for 10 minutes (20-second pulse-on, 30-second pulse-off at 100% amplitude) before denaturing at 95°C for 15 minutes. Samples were separated by SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked in TBS-T (1xTBS with 0.1% Tween-20) containing 5% milk for 1 hour and then incubated overnight at 4°C in primary antibody. Following overnight incubation, membranes were washed three times for 5 minutes each in TBS-T before incubation in TBS-T containing 5% milk with peroxidase-conjugated secondary antibodies for 90 minutes at room temperature. The membrane was then again washed three times for 5 minutes each in TBS-T before protein detection using enhanced chemiluminescence reagents from BioRad and Thermo Fisher.
Lentiviral Infection
HEK 293-FT cells were seeded at a density of 5×105 cells per well and transfected using Fugene 6 Transfection Reagent (Promega #E2691). Standard lentiviral packaging plasmids pMD2.G (0.5μg) and psPAX2 (2.5μg) were incubated with 2μg of target plasmid DNA with 12μL Fugene 6 diluted in Opti-MEM overnight according to the manufacturer’s instructions. Transfection media was removed the next day and replaced with fresh media before allowing the cells to proliferate 48hr. The supernatant was then collected and filtered using a 0.45μm filter (Corning #431220). Filtered viral supernatant was then used to infect target cells directly. Cells were seeded at a density of 5×105 cells per well one day prior to infection. Cell culture media was removed and replaced with media containing lentivirus and 0.8μg/mL polybrene and then incubated overnight. The next day, the lentivirus media was removed and replaced with fresh media. After another 24hr, selection media containing 0.2μg/mL puromycin (Gibco Invitrogen) was added, and the cells were allowed to grow for 72hr. Fresh non-selection media was then replaced, and the cells were allowed to proliferate for 7 days before being processed for downstream analysis.
cDNA Overexpression
All cDNA plasmid overexpression was transfected using Fugene 6 Transfection Reagent (Promega #E2691). Cells for transfection were plated 24 hours prior to Fugene transfection. cDNA was mixed with Fugene 6 in Opti-MEM according to the manufacturer’s instructions and incubated for 30 minutes at room temperature before being added to cell culture media overnight. Fresh media was replaced the next day, and cells were given an additional 48 hours to ensure abundant plasmid expression prior to downstream applications.
Immunofluorescence (IF)
Prior to staining, cells were grown on Cell-Tak (Corning)-coated glass coverslips. Cells were then rinsed with 1xPBS before pre-extraction using cytobuffer (100mM NaCl, 300mM Sucrose, 3mM MgCl2, 10mM PIPES pH 7.0, and 0.1% Triton X-100) for 7 minutes at 4°C. Cells were then rinsed with 1xPBS before being fixed in 4% paraformaldehyde for 10 minutes at room temperature. Cells were again rinsed with 1xPBS, then permeabilized with 0.5% NP-40 in 1xPBS for 10 min at room temperature. Cells were rinsed one last time with 1xPBS before incubating in PBG blocking buffer (0.5% BSA, 0.2% Fish Gelatin in 1xPBS) for 1 hour at room temperature. Cells were then incubated overnight at 4°C in a humidified chamber with primary antibody in PBG blocking buffer at a concentration of 1:500 unless otherwise specified. The following day, coverslips were washed three times with 1xPBS for 5 minutes each at room temperature before incubation in secondary antibodies diluted in PBG buffer for 1 hour at room temperature. The coverslips were washed twice with 1xPBS for 5 minutes each at room temperature and once with 1xPBS containing DAPI for 10 minutes at room temperature. Coverslips were then mounted on glass microscope slides with vectashield mounting medium and analyzed using a Zeiss LSM 710 confocal microscope.
Combined IF-DNA Fluorescence in situ Hybridization (DNA FISH)
For combined IF-DNA FISH, cells were processed in the same manner as the IF protocol; however, prior to the DAPI counterstain, coverslips were fixed with 4% paraformaldehyde for 10 minutes at room temperature. Coverslips were then digested with 200μg/mL RNaseA diluted in 2xSSC for 30 minutes at 37°C. The coverslips were then dehydrated with a series of ethanol washes (70%, 85%, and 100%) for 2 minutes each at room temperature, before being allowed to air dry at 37°C. Dehydrated coverslips were then mounted on glass slides with 10nM of a fluorophore-coupled PNA Telomeric Probe in hybridization buffer (70% Formamide, 0.25% Roche Blocking Reagent, 10mM Tris pH 7.5, 4.1mM Na2HPO4, 1.25mM MgCl2, and 0.45mM Citric Acid, Dextran Sulfate) at a concentration of 1:500, unless otherwise specified. Coverslips were denatured at 85°C for 3 minutes before being incubated overnight at 37°C in a humidified chamber. The next day, coverslips were washed 3 times for 5 minutes with 2xSSC/formamide (1:1) at 37°C, 3 times for 5 minutes with 2xSSC at 37°C, and a final wash in 2xSSC containing DAPI for 10 minutes at room temperature. Coverslips were then mounted on glass microscope slides with vectashield mounting medium and analyzed using a Zeiss LSM 710 confocal microscope.
Combined IF-RNA Fluorescence in situ Hybridization (RNA FISH)
For combined IF-RNA FISH, prior to staining, cells were pre-extraction using cytobuffer (100mM NaCl, 300mM Sucrose, 3mM MgCl2, 10mM PIPES pH 7.0, and 0.1% Triton X-100) supplemented with 10mM Ribonucleoside-Vanadyl Complex (New England Biolabs) for 7 minutes at 4°C. Cells were then processed in the same manner as the IF protocol; however, prior to the DAPI counterstain, coverslips were fixed with 4% paraformaldehyde for 10 minutes at room temperature. The coverslips were then dehydrated with a series of ethanol washes (70%, 85%, and 100%) for 2 minutes each at room temperature before being allowed to air dry at 37°C. Dehydrated coverslips were then mounted on glass slides with 10nM of a fluorophore-coupled PNA Telomeric Probe in hybridization buffer (70% Formamide, 0.25% Roche Blocking Reagent, 10mM Tris pH 7.5, 4.1mM Na2HPO4, 1.25mM MgCl2, and 0.45mM Citric Acid, Dextran Sulfate) supplemented with 10mM Ribonucleoside-Vanadyl Complex at a concentration of 1:500 and incubated overnight at 37°C in a humidified chamber. The next day, coverslips were washed 3 times for 5 minutes with 2xSSC/formamide (1:1) at 37°C, 3 times for 5 minutes with 2xSSC at 37°C, and a final wash in 2xSSC containing DAPI for 10 minutes at room temperature. Coverslips were then mounted on glass microscope slides with vectashield mounting medium and analyzed using a Zeiss LSM 710 confocal microscope.
DNA-RNA Immunoprecipitation (DRIP)
DRIP was performed as in Sanz and Chédin (2019) [71] with some modifications to allow for telomeric DRIP blotting and DRIP-qPCR. Briefly, an approximately 80% confluent 10cm plate of cells was collected by trypsinization and washed with ice-cold 1xPBS before being resuspended in TE buffer supplemented with SDS and proteinase K and rotated overnight at 37°C. High-density Maxtract Phase-Lock Gel Tubes (Qiagen) were used in conjunction with phenol/chloroform/isoamyl alcohol (25:24:1) to isolate chromatin, which was then centrifuged, and the top aqueous phase was then precipitated in 100% ethanol supplemented with 3M sodium acetate (pH 5.2). Precipitated nucleic acids were then spooled out of the mixture and washed with 85% ethanol before allowing it to air dry. Isolated chromatin was resuspended in TE buffer before sonication in a water bath at 4°C for 90 minutes (20-second pulse-on, 30-second pulse-off at 100% amplitude) to obtain fragments of approximately 200bp. Nucleic acid fragments were then purified using phenol/chloroform/isoamyl alcohol (25:24:1) extraction with Phase-Lock Gel Light Tubes (QuantaBio) and precipitated at −20°C in 100% ethanol supplemented with 3M sodium acetate (pH 5.2) and glycogen. Precipitated chromatin was centrifuged to form pellets and washed with 85% ethanol before allowing them to air dry. Purified chromatin was then resuspended in TE buffer for DNA-RNA immunoprecipitation. 20μg of purified chromatin was incubated overnight at 4°C in TE buffer supplemented with 10xDRIP Binding Buffer (100mM Sodium Phosphate pH 7.0, 1.4M NaCl, and 0.5% Triton X-100) and 20μg αDNA-RNA Hybrid Antibody S9.6 while inverting. When performing DRIP-qPCR, 20μg of purified chromatin was immunoprecipitated with 5μg αDNA-RNA Hybrid Antibody S9.6. Five percent of the sample was saved as input for DRIP-qPCR and 0.5 percent of the sample was saved as input for DRIP dot blot, and inputs were treated with 100U RNase H in 1x RNase H Buffer per reaction overnight at 37°C (New England Biosystems). Protein A/Protein G Dynabeads (1:1) were washed in 10xDRIP Binding Buffer before antibody-incubated chromatin was added and incubated for 2hr at 4°C while rotating. Dynabead-bound DNA-RNA hybrids were then washed with 1xDRIP Binding Buffer before being eluted in DRIP Elution Buffer (50mM Tris pH 8.0, 10mM EDTA pH 8.0, and 0.5% SDS) supplemented with proteinase K at 55°C for 1 hour with shaking. Eluted DNA-RNA hybrids were then purified using Phase-Lock Gel Light Tubes (QuantaBio) in conjunction with phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated at −20°C in 100% ethanol supplemented with 3M sodium acetate (pH 5.2) and glycogen. Precipitated DNA-RNA hybrids were then centrifuged to form pellets and washed with 85% ethanol before allowing them to air dry and resuspending the purified DNA-RNA hybrids in TE buffer before downstream applications (see below).
For telomeric DRIP dot blot, samples were denatured at 65°C for 15 minutes before being transferred to a Hybond N+ membrane, and telomeric DNA was analyzed using DIG-labeled (TTAGGG4) as previously described above. Images were captured and visualized using a BioRad ChemiDoc XRS+ imaging system. Telomeric DRIP signals were quantified using Image Lab 6.1 software.
For telomeric DRIP-qPCR analysis, 2μL of isolated IP DNA-RNA hybrids and their inputs were prepared for qPCR reaction. PowerUp SYBR Green Master Mix and 10μM of TelBam3.4 or RPPH1 primers (see above) were used for qPCR reactions according to the manufacturer’s instructions with samples in technical triplicate. qPCR was performed on a StepOne qPCR machine using the following program: 50°C 2 min, 95°C 2 min, then 40 cycles (95°C 15s, 60°C 15s, 72°C 1 min), followed by a standard melt curve. Data were analyzed by both the ΔΔCT and percent input method.
Statistical Analysis
All results are representative of at least three biological replicates. All statistical analysis was performed using GraphPad Prism 9 software. Two-tailed unpaired Student’s t-tests and analysis of variance (ANOVA) were used for statistical analysis. Results were considered statistically significant if p<0.05.
Results
The Catalytic Activity of XRN2 Regulates TERRA Stability in Mammalian Cells
To define the factors that regulate TERRA metabolism, we asked which enzymes were responsible for the degradation of TERRA in mammalian cells. Given that TERRA is primarily localized within the nucleus, we narrowed our focus to enzymes that regulate RNA degradation within the nuclear compartment, including DIS3, EXOSC10, and XRN2. DIS3 (also known as Rrp44) and EXOSC10 (also known as Rrp6) function as the catalytically-active components of the nuclear RNA exosome complex and ensure the degradation of a wide range of RNA transcripts [38,39]. XRN2 is an exoribonuclease that plays a crucial role in RNA transcription termination and the subsequent 5’-3’ degradation of RNA transcripts [40–42]. We predicted that if these factors were involved in the degradation of TERRA in mammalian cells, then the loss of any one factor would lead to the stabilization of the TERRA transcript. To modulate the expression of these essential enzymes, we induced rapid degradation of the proteins DIS3, EXOSC10, or XRN2 using a previously-established auxin-inducible degron (AID) system [41].
In this system, HCT116 cells were engineered to stably express the plant E3 ubiquitin ligase, OsTir1, and also an AID tag at the C-terminus of the endogenous DIS3, EXOSC10, or XRN2 gene loci. When auxin, or 3-Indoleacetic acid (IAA), is added to the cell culture media, AID-tagged proteins are rapidly degraded by the proteasome [41]. Here, we have used HCT116 cells expressing OsTir1 alone as a control (HCT116OsTir1), or HCT116 cells expressing OsTir1 and AID-tagged DIS3, EXOSC10, or AID-XRN2 (HCT116AID-DIS3, HCT116AID-EXOSC10, or HCT116AID-XRN2). To assess the effects of rapid depletion of DIS3, EXOSC10, or XRN2 on the stability of TERRA, we added IAA to cell culture media and analyzed protein expression by western blot at 0, 1, and 6 hours following IAA treatment. In these cells, we saw complete protein degradation in as little as 1 hour (Figure 1A), and this degradation was maintained as long as IAA remained in the cell culture media. To determine which of the tagged nucleases regulated TERRA stability, we induced degradation of DIS3, EXOSC10, or XRN2 with IAA and analyzed TERRA by dot blot using TERRA-specific probes (Figures 1B and C). Although TERRA remained unchanged following acute depletion of DIS3 and EXOSC10, depletion of XRN2 led to a 3.5-fold increase in TERRA at the 6-hour time point. This observed increase in TERRA stability was maintained over time (Figures 1D and E), leading to an approximately 5.5-fold increase in TERRA by 48 hours following IAA treatment.
Figure 1. The catalytic activity of XRN2 regulates TERRA stability in mammalian cells.
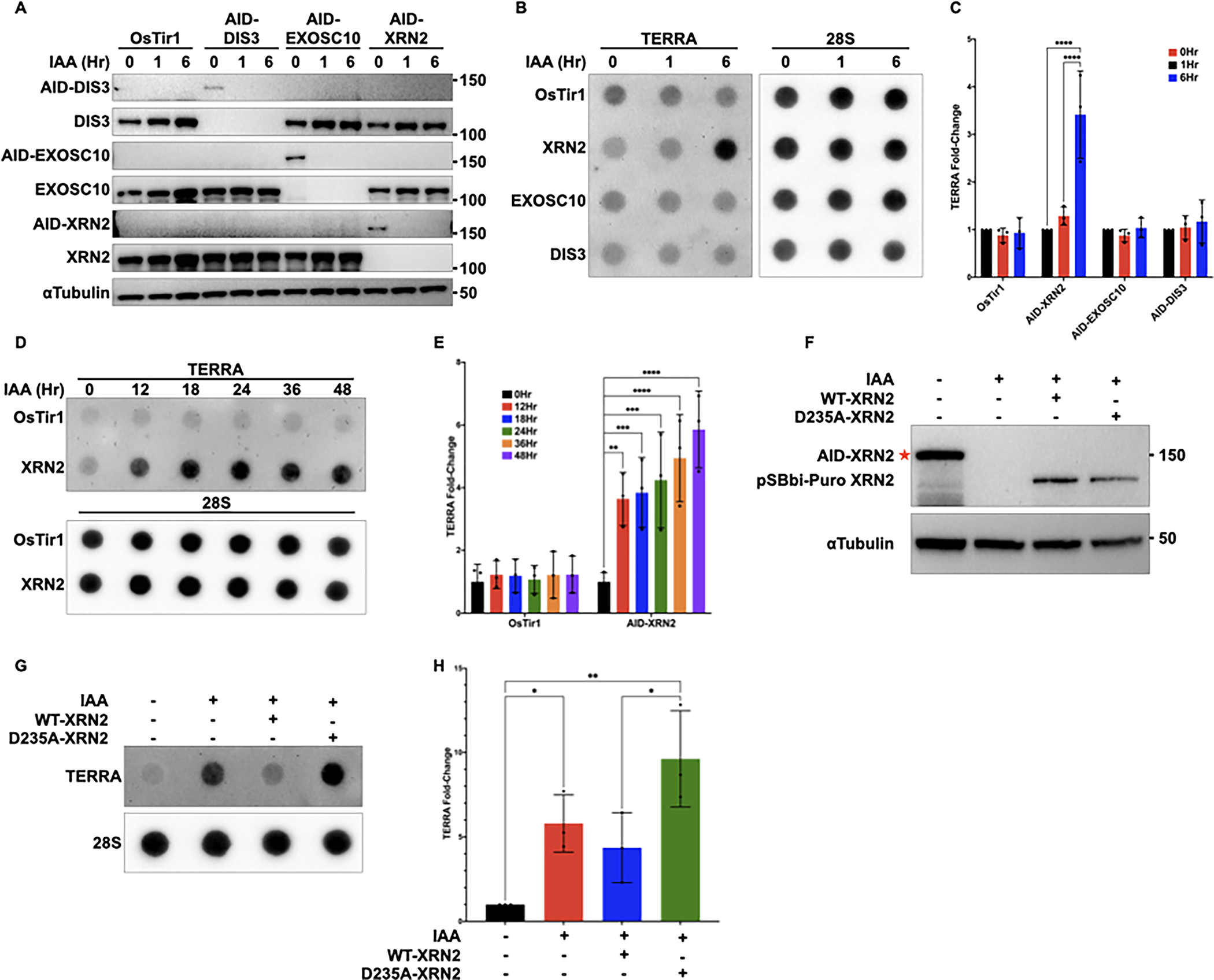
(A) Western blot of DIS3, EXOSC10, XRN2, or αTubulin in HCT116OsTir1, HCT116AID-DIS3, HCT116AID-EXOSC10, and HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 1 or 6 hours. All proteins were analyzed using the indicated antibodies. AID-tagged proteins run at a higher molecular weight and are shown as separate blots (B) RNA dot blot analysis of TERRA in HCT116OsTir1, HCT116AID-DIS3, HCT116AID-EXOSC10, and HCT116AID-XRN2 cells using DIG-labeled probes for both TERRA and 28S as a loading control. Cells were either left untreated or treated with 500μM IAA for 1 or 6 hours (C) Quantification of dot blot data shown in (B). The data are presented as the mean ± SD of three independent experiments (n=3) analyzed by two-way ANOVA followed by Tukey’s test for multiple comparisons (D) RNA dot blot analysis of TERRA in HCT116OsTir1 and HCT116AID-XRN2 cells treated with 500μM IAA at the indicated time points. TERRA and 28S were detected using DIG-labeled probes (E) Quantification of dot blot data shown in (D). Data are presented as mean ± SD of three independent experiments (n=3) analyzed by two-way ANOVA followed by Dunnett’s test for multiple comparisons (F) Western blot analysis of XRN2 in HCT116AID-XRN2 cells overexpressing WT-XRN2 or D235A-XRN2 from a pSBbi-Puro background plasmid. Cells were treated with 500μM IAA for 24 hours and transfected with WT-XRN2 or D235A-XRN2, and proteins were detected using the indicated antibodies. The red star indicates AID-tagged XRN2, which runs at a higher molecular weight than exogenous WT or D235A-XRN2 (G) RNA dot blot analysis of TERRA in HCT116AID-XRN2 cells overexpressing WT-XRN2 or D235A-XRN2. Cells were treated with 500μM IAA for 24 hours and transfected with WT-XRN2 or D235A-XRN2 (H) Quantification of dot blot data shown in (G) (I) Representative Northern blot analysis of HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 24 or 72 hours. TERRA transcripts were detected using a 32P-labeled (CCCTAA)4 probe and total RNA was stained with ethidium bromide and hybridized with a probe to U3 as controls. Data in (C), (E), and (H) are presented as mean ± SD of three independent experiments (n=3) analyzed by one-way ANOVA followed by Dunnett’s test for multiple comparisons. For all data *p≤0.0332, **p≤0.0021, ***p≤0.0002, and ****p≤0.0001
To ensure that the observed increase in TERRA was a direct result of XRN2 loss, we performed a rescue experiment by exogenously expressing either WT-XRN2 or a catalytically inactive D235A-XRN2 in the IAA-treated HCT116AID-XRN2 cells. Following IAA treatment, we confirmed the depletion of the endogenous XRN2 proteins and re-expression of either WT-XRN2 or D235A-XRN2 protein by western blot (Figure 1F). Following RNA extraction and dot blot analysis, we found that expression of exogenous WT-XRN2 rescued TERRA to within untreated levels in HCT116AID-XRN2 cells, while catalytically-inactive D235A-XRN2 did not rescue TERRA levels (Figure 1G and H). [40,43]Taken together, our results suggest that the 5’-3’ exoribonuclease XRN2 mediates the degradation of TERRA in mammalian cells.
Stabilization of TERRA Does Not Induce Replication Stress
TERRA is localized within the nucleus and associates with telomeric chromatin. Therefore, we asked whether the increase in TERRA following XRN2 depletion also led to an increase in the accumulation of TERRA on chromatin. To evaluate the localization of TERRA following XRN2 depletion, we used a combination of immunofluorescence (IF) and RNA fluorescence in situ hybridization (RNA FISH). Notably, we found a significant increase in TERRA localization on chromatin by IF-FISH following IAA-mediated depletion of XRN2 (Figures 2A and B). Given the propensity for TERRA to form telomeric R-loops and the role of XRN2 in the resolution of R-loops throughout the cell, we asked whether XRN2 depletion led to an increase in TERRA on chromatin in the form of R-loops. Previous studies have demonstrated that TERRA R-loops can form within the CpG island repeats of the TERRA promoter or the telomeric repeats themselves [19–21,44]. Therefore, we isolated R-loops using DNA-RNA immunoprecipitation (DRIP) and quantified the abundance of these structures within the predicted promoter region of TERRA (TelBam3.4) by qPCR or within the telomeric repeats themselves by dot blot and asked whether the depletion of XRN2 might lead to the accumulation of TERRA R-loops at these sites. As a positive control, we also analyzed R-loop formation at the RPPH1 loci which demonstrates an accumulation of R-loops following XRN2 loss [45] and treated a subset of lysates with RNaseH to degrade R-loops as a negative control [46]. Following DRIP and amplification by qPCR, we detected a significant increase in RPPH1 R-loops in HCT116AID-XRN2 for control samples following IAA treatment, and R-loop signal was reduced following treatment with RNaseH (Figure 2C). However, we did not observe any significant increase in R-loops at the predicted TERRA promoter TelBam3.4 in HCT116AID-XRN2 cells following IAA treatment (Figure 2D). In addition, although R-loops were detected within the telomeric repeats of HCT116AID-XRN2 cells and control ALT-positive U2OS cells by dot blot, we failed to detect a significant increase in R-loops within the telomeric repeats of HCT116AID-XRN2 cells following IAA treatment (Figures 2E and F). Taken together, our data suggest that increasing TERRA alone is insufficient to drive R-loop formation at telomeres.
Figure 2. Stabilization of TERRA does not induce replication stress.
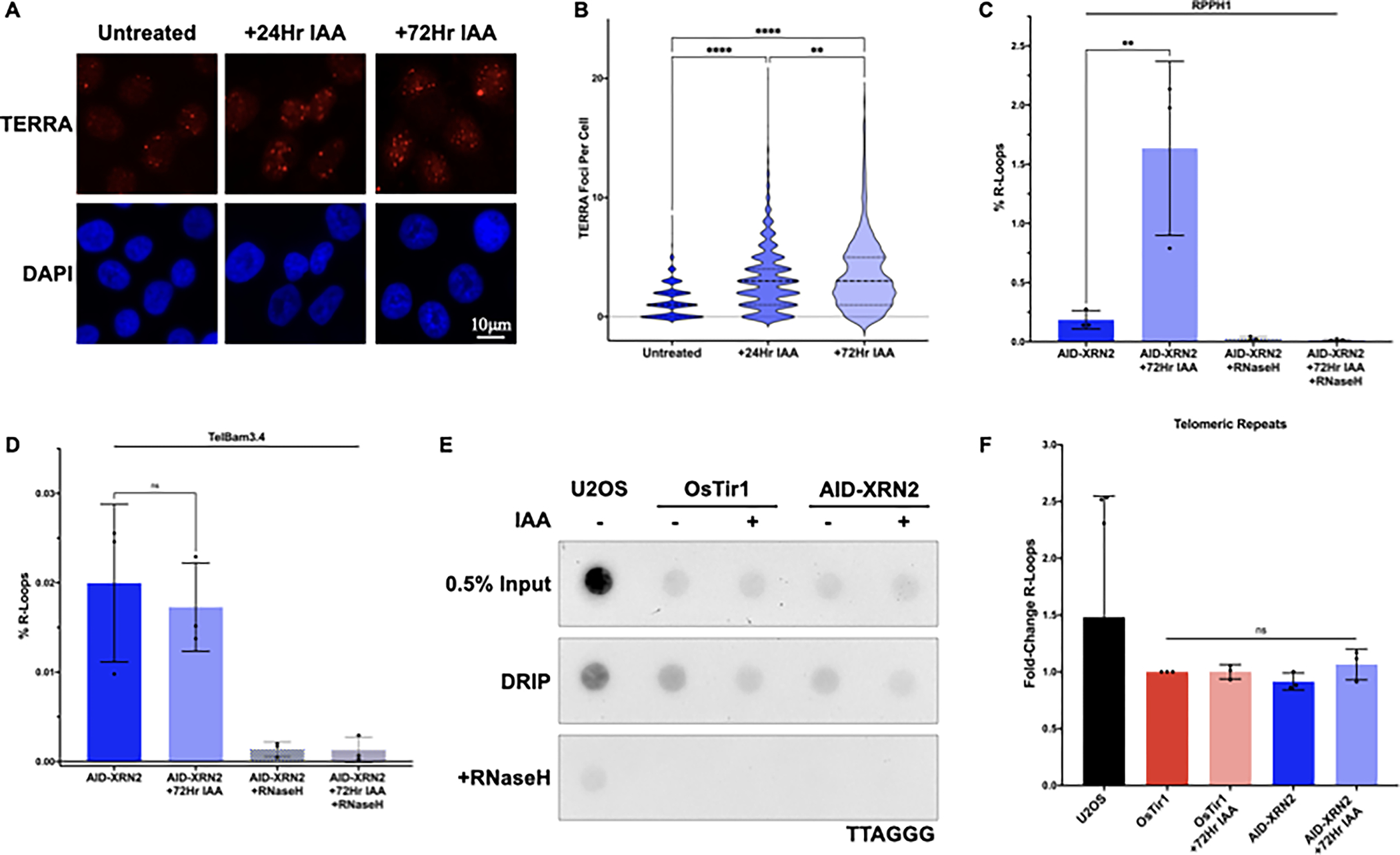
(A) RNA Fluorescence in situ Hybridization (RNA FISH) of HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 24 or 72 hours. TERRA was detected using a TelC-Cy3 PNA telomeric probe. Representative nuclei are shown with DAPI staining, scale bar ~10 μm (B) Quantification of RNA FISH data shown in (A) (C) Quantification of DRIP-qPCR from HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Sequence-specific primers for RPPH1 were used as a positive control for R-loops and RNaseH treatment as a negative control for R-loops (D) Quantification of DRIP-qPCR from HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. The predicted TERRA promoter TelBam3.4 was amplified using sequence-specific primers with RNaseH treatment as a negative control for R-loops (E) Telomeric DRIP blot of U2OS and HCT116AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Telomeric signal was detected following DRIP using a DIG-labeled probe for TTAGGG. 0.5% inputs are shown, and RNaseH treatment was used as a negative control (F) Quantification of telomeric DRIP blot data shown in (E), data are presented as fold-change of R-loops at telomeres. All data are presented as the means ± SD of three independent experiments (n=3), except in (F), where (n=6) for U2OS. RNA FISH results represent quantifications of at least n=300 cells for each condition. Data were analyzed by one-way ANOVA followed by Tukey’s test for multiple comparisons (B-D) or Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 test for multiple comparisons (F). For all data ns = p≤0.1234, **p≤0.0021, and ****p≤0.0001
Given that TERRA stabilization alone could not drive R-loop formation, we considered whether additional factors might cooperate with increased TERRA to incite R-loop formation at telomeres. TERRA has been shown to interact with proteins that function at telomeric DNA, including the chromatin-remodeling enzyme ATRX. ATRX binds the histone chaperone DAXX, and together this complex functions to deposit the histone variant H3.3 within heterochromatic regions of the genome, including centromeric and telomeric DNA. ATRX has also been shown to interact with repeat-containing RNAs (such as TERRA) through its RNA binding region and suppress R-loop formation by preventing RNA hybridization with complementary DNA [47]. Therefore, defects in ATRX function may not only disrupt heterochromatic formation but could also promote R-loop formation through a more open chromatin environment, consequently leading to an increase in replication stress. It is, therefore, not surprising that ATRX is often mutated in ALT-positive tumors [48]. To determine whether loss of ATRX cooperated with loss of XRN2 to induce TERRA R-loops at telomeric DNA, we knocked down ATRX using shRNA in the HCT116AID-XRN2 cells (HCT116AID-XRN2 + shATRX) (Figure 3A) and evaluated the effect of IAA treatment on the stabilization of TERRA and, ultimately, the formation of telomeric R-loops. As anticipated, depletion of XRN2 alone and in combination with ATRX knockdown led to a significant increase in TERRA by dot blot. Notably, knockdown of ATRX alone in HCT116AID-XRN2 cells in the absence of IAA treatment also led to an increase in TERRA, as has been previously described [36,49] (Figures 3B and C). Next, we asked whether ATRX knockdown generated the replication stress that, when coupled with XRN2 loss, would drive R-loop formation at telomeres. However, similar to our initial results, we did not observe any significant accumulation of R-loops within the promoter nor within the telomeric repeats themselves following IAA treatment in HCT116AID-XRN2 + shATRX cells (Figures 3D–F). Thus, these results suggest that IAA-mediated TERRA stabilization is insufficient to drive telomeric R-loop formation at telomeric DNA even when combined with the loss of ATRX.
Figure 3. Loss of ATRX and increased TERRA do not cooperate to induce replication stress in non-ALT cells.
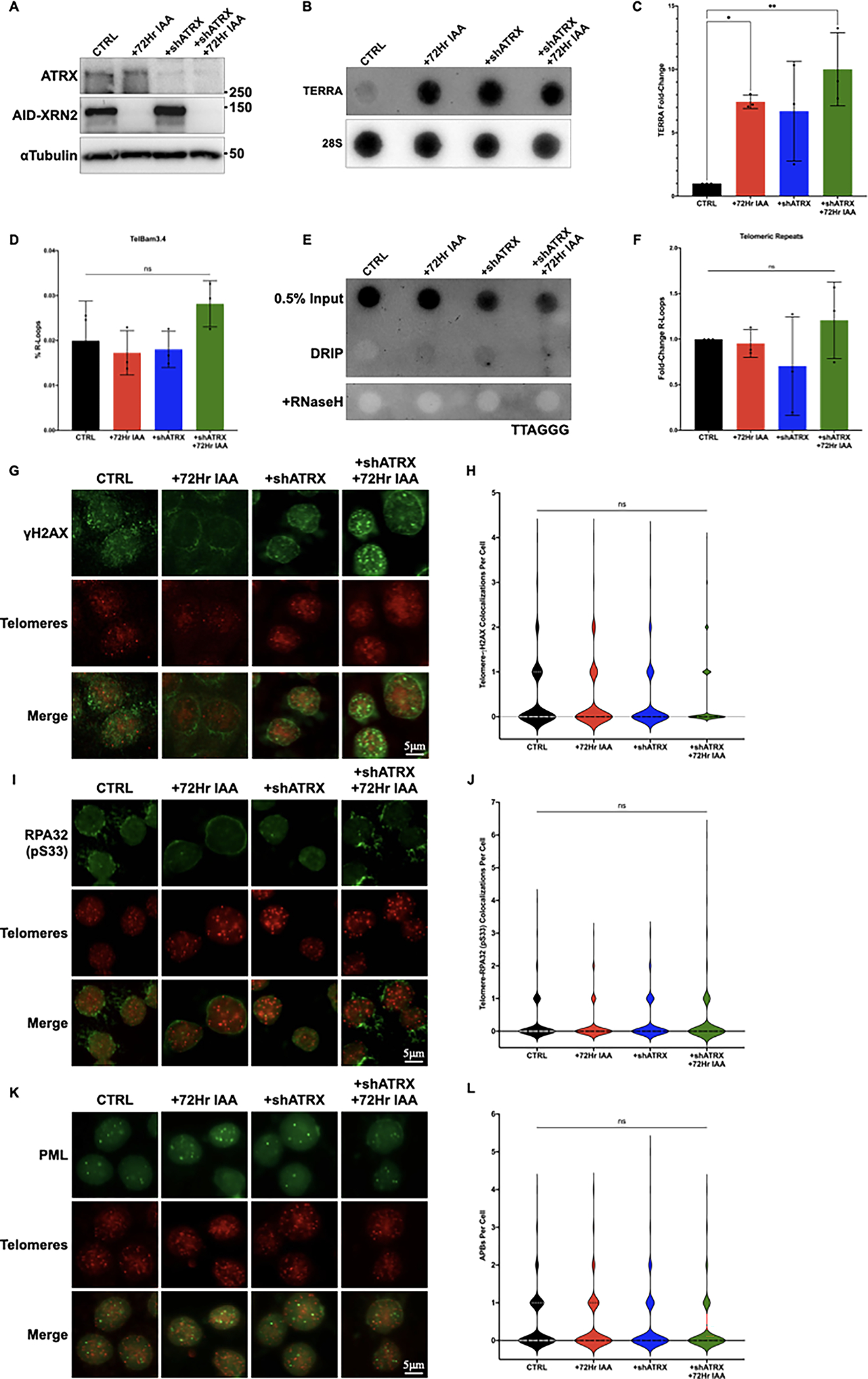
(A) Western blot of ATRX, XRN2, or αTubulin in HCT116AID-XRN2 cells. Cells with and without expression of shATRX were either left untreated or treated with 500μM IAA for 72 hours. All proteins were analyzed using the indicated antibodies (B) RNA dot blot analysis of TERRA in HCT116AID-XRN2 cells either left untreated, stably expressing shATRX, treated with 500μM IAA for 72 hours, or combined shATRX and IAA treatment. DIG-labeled probes for both TERRA and 28S as a loading control are shown (C) Quantification of dot blot data shown in (B) (D) Quantification of DRIP-qPCR from the predicted TERRA promoter TelBam3.4 in HCT116AID-XRN2 cells. Cells with and without expression of shATRX were either left untreated or treated with 500μM IAA for 72 hours (E) Telomeric DRIP blot of HCT116AID-XRN2 cells either left untreated, stably expressing shATRX, treated with 500μM IAA for 72 hours, or combined shATRX and IAA treatment. Telomeric signal was detected following DRIP using a DIG-labeled telomeric (TTAGGG) probe. 0.5% inputs are shown, and RNaseH treatment was used as a negative control (F) Quantification of telomeric DRIP blot data shown in (E), data are presented as fold-change of R-loops at telomeres (G) Combined immunofluorescence (IF) and DNA Fluorescence in situ Hybridization (DNA FISH) of HCT116AID-XRN2 cells. Cells with and without expression of shATRX were either left untreated or treated with 500μM IAA for 72 hours. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and γH2AX was detected with the indicated antibody. Representative nuclei are shown, scale bar = 5 μm (H) Quantification of IF-DNA FISH data shown in (G) (I) Combined IF-DNA FISH of HCT116AID-XRN2 cells either left untreated, stably expressing shATRX, treated with 500μM IAA for 72 hours, or combined shATRX and IAA treatment. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and phosphorylated RPA32 Ser33 was detected using the indicated antibody. Representative nuclei are shown, scale bar = 5 μm (J) Quantification of IF-DNA FISH data shown in (I) (K) Combined IF-DNA FISH of HCT116AID-XRN2 cells either left untreated, stably expressing shATRX, treated with 500μM IAA for 72 hours, or combined shATRX and IAA treatment. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and PML was detected using the indicated antibody. Representative nuclei are shown, scale bar = 5 μm (L) Quantification of IF-DNA FISH data shown in (K). All figure quantifications are presented as the means ± SD of three independent experiments (n=3). IF-DNA FISH results represent quantifications of at least n=300 cells for each condition. Data were analyzed by one-way ANOVA followed by Tukey’s test for multiple comparisons, except in (F), which was analyzed by Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 test for multiple comparisons. For all data ns = p≤0.1234, *p≤0.0332, and **p≤0.0021
Conceivably, the analysis of R-loops isolated from total genomic DNA may not provide the sensitivity needed to detect initial, early increases in TERRA R-loops that may be indicative of telomere dysfunction. If co-depletion of ATRX and XRN2 does indeed promote TERRA R-loops, we may be able to detect an increase in replication stress and genome instability within individual cells. Therefore, initially, we analyzed the accumulation of the DNA damage marker γH2AX at telomeric DNA in HCT116AID-XRN2 +/− shATRX cells following IAA treatment using a combined IF-FISH approach. However, we did not detect a significant increase in γH2AX foci at telomeres in the IAA-treated HCT116AID-XRN2 + shATRX cells (Figures 3G and H). Given that ATRX not only functions to inhibit replication stress but is commonly mutated in the progression towards ALT, we hypothesized that the depletion of ATRX in the context of increased TERRA might drive the replication stress typically associated with ALT. Therefore, we analyzed typical markers of replication stress by IF-FISH, including the phosphorylation of RPA32 at serine 33 (RPA32 pS33) and the formation of APBs. However, similar to our analysis of γH2AX, we failed to detect significant changes in RPA32 pS33 at telomeric DNA, nor did we detect changes in APBs in the IAA-treated HCT116AID-XRN2 +/− shATRX cells (Figures 3I–L). Collectively, we have shown that IAA-mediated stabilization of TERRA does not increase telomeric R-loops nor induce the replication stress associated with the activation of ALT, even in the context of ATRX loss. These results demonstrate that the precise relationship between TERRA and the emergence of ALT phenotypes in telomerase-positive mammalian cells is complex and that TERRA alone may be insufficient to trigger the ALT mechanism.
TERRA is Significantly Increased in Cells That Rely on the ALT Pathway
Although we observed a significant increase in TERRA in HCT116AID-XRN2 cells treated with IAA, the increase in TERRA may not be sufficient to drive TRCs and induce replication stress in our system. Therefore, we analyzed TERRA across a panel of telomerase and ALT-positive cell lines and osteosarcoma tumors. To determine ALT status, we analyzed APB formation in our collection of primary human tumor samples by combined IF-DNA FISH (Figures 4B and C). As has been previously described, samples with greater than 0.5% of cells positive for APBs were considered to be ALT-positive [50]. The prevalence of ALT across all cancers is approximately 10%; however, the prevalence of ALT in osteosarcoma is estimated to be over 50%, making osteosarcoma an attractive tumor type for the analysis of TERRA [51,52]. The rarity of osteosarcoma makes a comprehensive analysis of ALT in primary tumors difficult. However, research in our lab suggests that ALT is prevalent in approximately 88% of pediatric osteosarcoma [53,54]. Following dot blot analysis, we demonstrated a significant increase in TERRA in ALT-positive samples compared to ALT-negative samples (Figures 4A and D). Interestingly, the relative increase in TERRA in ALT-positive samples was not uniform, as some samples demonstrated a 35–50-fold increase in TERRA while others maintained TERRA at levels comparable to TERRA in ALT-negative samples (Figures 4D).
Figure 4. TERRA is significantly increased in cells that rely on the ALT pathway.
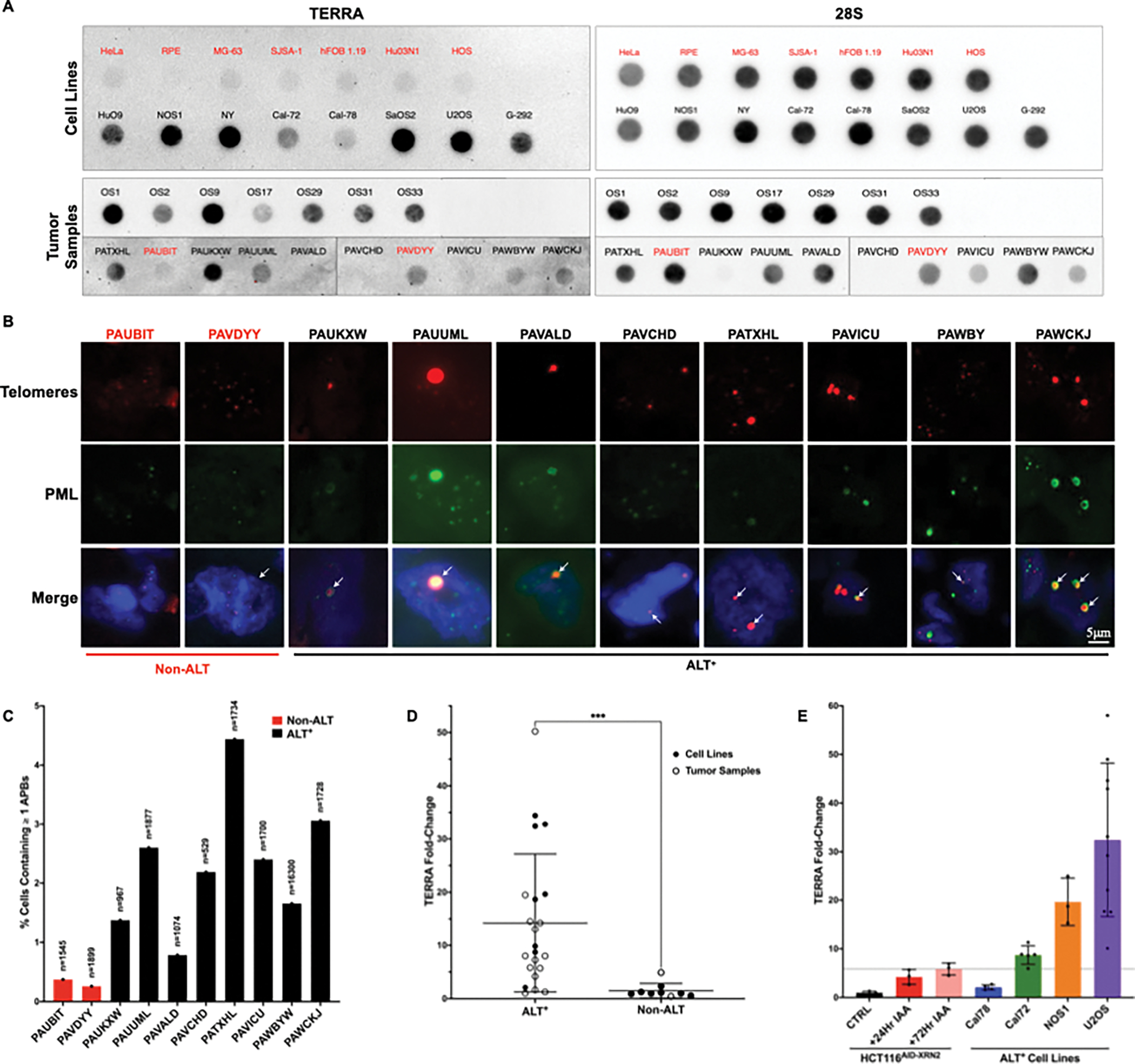
(A) RNA dot blot analysis of TERRA across a panel of cell lines and tumor samples using DIG-labeled probes for both TERRA and 28S as a loading control. Samples identified as ALT-positive are labeled in black, while non-ALT samples are identified in red (B) Combined IF-DNA FISH in patient samples. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and PML was detected using an antibody. Representative nuclei are shown with white arrows indicating colocalization events, scale bar = 5 μm. Samples identified as ALT-positive are labeled in black, while non-ALT samples are identified in red (C) Quantification of Telomere-PML colocalization events from images shown in (B). Graphed data represent the percent of total cells counted containing at least one APB with a minimum of n=950, except for human patient sample PAVCHD where n=529 due to limited sample availability (D) Quantification of dot blot data shown in (A). Cell line samples are marked with solid black circles, while tumor samples are marked with hollow black circles. Graphed data represent the mean value of each cell line or tumor sample with a minimum of n=3, except for human patient samples where n=1 due to limited sample availability. ALT versus non-ALT data is presented as the mean ± SD of all sample data and was analyzed by a two-tailed unpaired t-test followed by Welch’s correction for uneven SD, ***p≤0.0021 (E) Quantification of dot blot data shown in (A) and (1E). The data presented are the mean ± SD of at least three independent experiments (n=3). The dotted line represents the mean level of stabilized TERRA in HCT116AID-XRN2 cells following 500μM IAA treatment for 72 hours
Given that TERRA expression varies across samples, we asked whether the increase in TERRA in the IAA-treated HCT116AID-XRN2 cells matched the TERRA quantified in our ALT-positive samples. Using dot blot analysis, we found that TERRA in our IAA-treated HCT116AID-XRN2 cells is within the range of TERRA expression in other ALT-positive samples (Figure 4E). Although the expression of TERRA in our HCT116AID-XRN2 cells was comparable to the expression of TERRA in some of our ALT-positive samples (Figures 4A and D), the increase in TERRA in the HCT116AID-XRN2 cells failed to induce ALT phenotypes (Figures 3G–L). This suggests that the increase in TERRA alone in HCT116AID-XRN2 cells was insufficient to induce telomere dysfunction or invoke ALT phenotypes in the timeframe of these experiments.
Stabilization of TERRA Exacerbates ALT Phenotypes
Although increasing TERRA alone in non-ALT cells did not create an environment permissive for the activation of ALT, we asked whether stabilization of TERRA in an ALT-positive environment would exacerbate ALT phenotypes. To assess this, we created an ALT-positive SaOS2 cell line containing an AID tag at the C-terminus of the XRN2 loci (SaOS2AID-XRN2). Many osteosarcoma cell lines, including SaOS2, have undergone a whole-genome doubling event making it difficult to tag all XRN2 alleles within the cell. However, using PCR analysis, we confirmed that the majority of the XRN2 alleles were engineered to contain the AID tag at the C-terminus (Figure 5A, Top). Moreover, we detect an 85% reduction in XRN2 protein expression by western blot in SaOS2AID-XRN2 cells treated with IAA (Figure 5A, Bottom). Using this system, we evaluated TERRA in SaOS2AID-XRN2 cells following XRN2 depletion. Similar to our results in HCT116AID-XRN2 cells, we demonstrate a significant increase in TERRA in SaOS2AID-XRN2 cells following IAA-mediated loss of XRN2 (Figures 5B and C).
Figure 5. Stabilization of TERRA exacerbates ALT phenotypes.
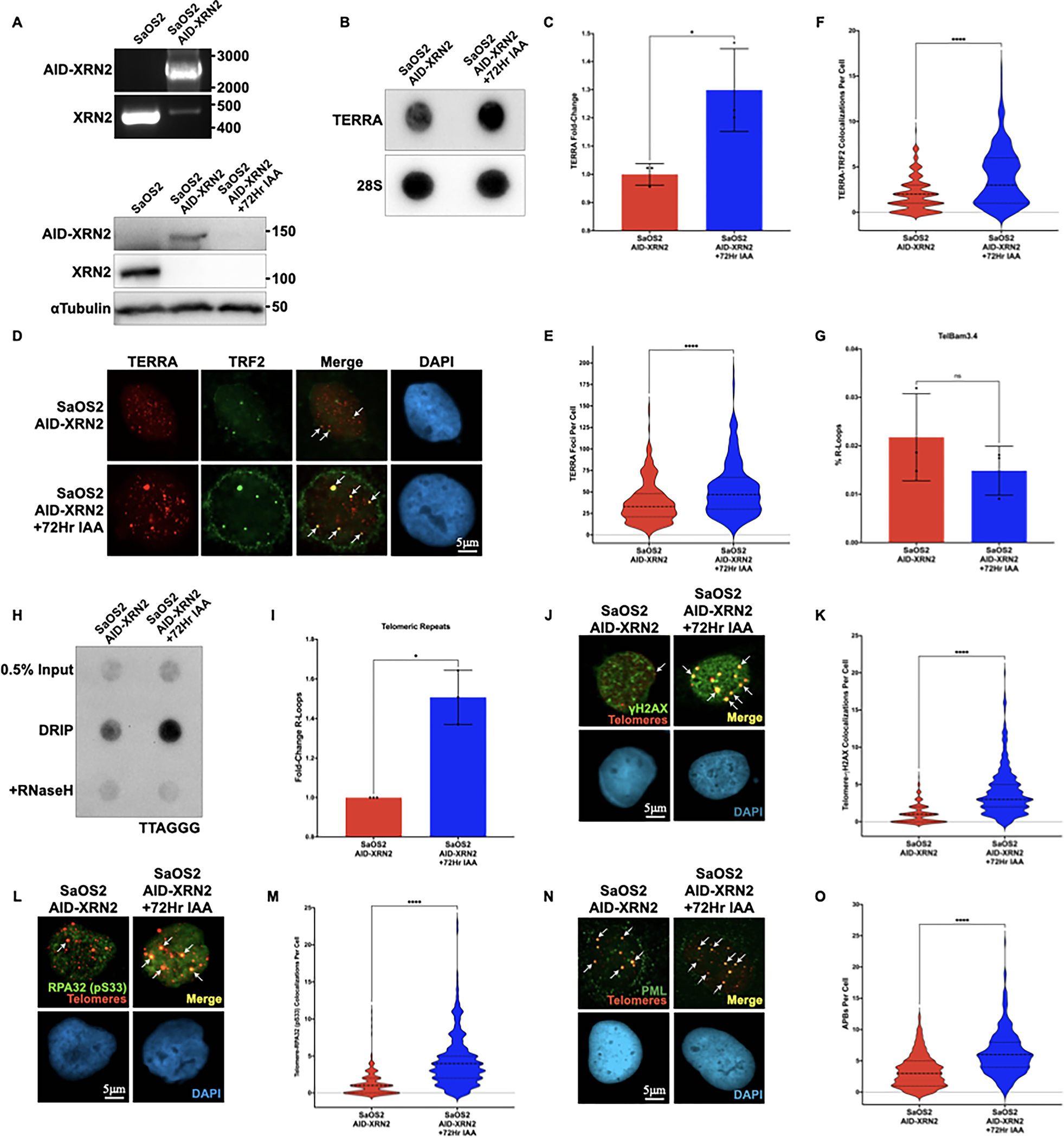
(A, Top) PCR analysis of the XRN2 locus in parental SaOS2 and SaOS2 cells integrated with an AID tag (SaOS2AID-XRN2). The wildtype XRN2 locus is approximately 450 bp in length, while Hygro-tagged AID-XRN2 is approximately 2500 bp in length (A, Bottom) Western blot of XRN2 or αTubulin in SaOS2 and SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. All proteins were analyzed using the indicated antibodies. AID-tagged proteins run at a higher molecular weight (B) RNA dot blot analysis of TERRA in SaOS2AID-XRN2 cells using DIG-labeled probes for both TERRA and 28S as a loading control. Cells were either left untreated or treated with 500μM IAA for 72 hours (C) Quantification of dot blot data shown in (B) (D) Combined IF-RNA FISH analysis of SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. TERRA was detected using a TelC-Cy3 PNA telomeric probe and TRF2 was detected with the indicated antibody. Representative nuclei are shown with DAPI staining, scale bar = 5 μm (E) Quantification of TERRA RNA FISH data shown in (D) (F) Quantification of IF-RNA FISH data shown in (D) (G) Quantification of DRIP-qPCR from the predicted TERRA promoter TelBam3.4 in SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours (H) Telomeric DRIP blot of SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Telomeric signal was detected following αDNA-RNA hybrid immunoprecipitation using a DIG-labeled probe for TTAGGG. 0.5% inputs are shown, and RNaseH treatment was used as a negative control (I) Quantification of telomeric DRIP blot data shown in (H), data are presented as fold-change of R-loops at telomeres (J) Combined IF-DNA FISH of SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and γH2AX was detected using the indicated antibody. Representative nuclei with DAPI staining are shown with white arrows indicating colocalization events, scale bar = 5 μm (K) Quantification of IF-DNA FISH data shown in (J) (L) Combined IF-DNA FISH of SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and phosphorylated RPA32 Ser33 was detected using the indicated antibody. Representative nuclei with DAPI staining are shown with white arrows indicating colonization events, scale bar = 5 μm (M) Quantification of IF-DNA FISH data shown in (L) (N) Combined IF-DNA FISH of SaOS2AID-XRN2 cells either left untreated or treated with 500μM IAA for 72 hours. Telomeres were detected using a TelC-Cy3 PNA telomeric probe, and PML was detected using the indicated antibody. Representative nuclei with DAPI staining are shown with white arrows indicating colonization events, scale bar = 5 μm (O) Quantification of IF-DNA FISH data shown in (N). All data are presented as the mean ± SD of at least three independent experiments (n=3) with at least n=300 cells per condition for FISH images. Data were analyzed by a two-tailed unpaired t-test, except in (I), which was analyzed by Welch’s two-tailed t-test. For all data ns = p≤0.1234, *p≤0.0332, **p≤0.0021, ***p≤0.0002, and ****p≤0.0001
Given that we successfully stabilized TERRA in the context of ALT, we asked whether there was a corresponding increase in TERRA, specifically on chromatin. As we had observed in HCT116 AID-XRN2 cells, following IAA treatment, we detected a significant increase in TERRA on chromatin by RNA FISH in SaOS2AID-XRN2 cells (Figures 5D and E). Next, we asked whether these TERRA transcripts specifically colocalized to telomeric DNA using IF-RNA FISH. Using the shelterin component TFR2 as a marker for telomere [13], we demonstrate a significant increase in colocalization events between TERRA and TRF2 in SaOS2AID-XRN2 following treatment with IAA (Figures 5D and F). To determine whether this increase in TERRA at telomeres corresponded to an increase in R-loops at telomeres in ALT, we performed DRIP analysis at both the TelBam3.4 promoter and the telomeric repeats themselves. While we did not detect an increase in R-loops within the TelBam3.4 promoter (Figure 5G), we did see a significant increase in R-loops at the telomeric repeats in SaOS2AID-XRN2 following treatment with IAA (Figures 5H and I).
We hypothesized that the accumulation of R-loops within the telomeric repeats in ALT cells could lead to increased replication stress at telomeres and, consequently, exacerbate ALT phenotypes. Using a combined IF-FISH approach, we observed a significant increase in the DNA damage marker γH2AX (Figures 5J and K) at telomeric DNA in SaOS2AID-XRN2 cells treated with IAA. Additionally, we observed a significant increase in RPA32 pS33 foci at telomeric DNA in SaOS2AID-XRN2 cells treated with IAA (Figures 5L and M) and the accumulation of APBs (Figures 5N and O), highlighting an increase in replication stress. Taken together, these results demonstrate that in the context of ALT, TERRA stabilization increases R-loops specifically within the telomeric repeats, driving replication stress at telomeres and, ultimately, exacerbating ALT phenotypes.
Discussion
Since the identification of TERRA more than a decade ago, there has been a growing interest in defining how TERRA functions within the cell to maintain telomere stability. The metabolism of TERRA within the cell will undoubtedly influence overall TERRA function. Therefore, we asked whether defining the enzymes responsible for TERRA stability could help us to better understand TERRA function at telomeric DNA. Here, we demonstrate that the 5’-3’ nuclear exoribonuclease XRN2 plays a crucial role in regulating TERRA degradation in mammalian cancer cells. Specifically, we show that the rapid depletion of XRN2 leads to increased TERRA transcript stability. These transcripts are localized within the nucleus and specifically associate with chromatin, providing an opportunity to explore how defects in TERRA metabolism might contribute to telomere dysfunction.
XRN2 is a nuclear 5’-3’ exoribonuclease that functions in the regulation of RNA metabolism [40]. XRN2 is one of the human orthologs (along with XRN1) of the S. cerevisiae protein Rat1p, which has been shown to regulate TERRA stability in yeast [10]. We have demonstrated that XRN2 plays a similar role in TERRA metabolism in mammalian cells, as depletion of XRN2 led to increased TERRA stability. XRN2 functions to degrade a diverse array of nuclear RNA substrates through its exoribonuclease activity. In mammalian cells, RNA can be protected from exonucleolytic degradation by a 7-methylguanosine (m7G) cap a the 5’ end of the transcript. However, XRN2 is known to interact with the decapping proteins DCP1a, DCP2, and EDC3, which function to remove the 5’ cap and facilitate XRN2-mediated degradation. Alternatively, XRN2 may gain access to the 5’ terminus of an RNA during 3’ end processing. Cleavage downstream of a polyadenylation signal (PAS) generates an uncapped downstream cleavage product that can be bound and degraded by XRN2. If XRN2 processivity exceeds the processivity of the RNA Pol II, XRN2 can overtake the polymerase to induce transcription termination [43,55]. Although TERRA does not contain a canonical PAS motif, TERRA does have a 5’ m7G cap suggesting that TERRA could undergo decapping prior to degradation by XRN2 and that this process contributes to the overall stability of TERRA. XRN2 not only functions to degrade RNA but also functions to facilitate the termination of RNA Polymerase II transcripts [40,43]. Therefore, it is possible that following depletion of XRN2, the observed increase in TERRA may not simply reflect a change in abundance but an increase in TERRA length and should be explored in future studies.
XRN2 not only functions to regulate RNA degradation and transcription termination but also interacts with enzymes associated with the transcriptional machinery, including the DNA:RNA helicase Senataxin (SETX) and the endonuclease RNaseH1, to resolve R-loops. Like XRN2, SETX and RNaseH1 have been demonstrated to contribute to the resolution of R-loops throughout the genome [56–59]. RNaseH1, SETX, and XRN2 associate with both telomeric DNA and TERRA [19,44,60] and have been suggested to inhibit replication stress at telomeric DNA through the resolution of TERRA R-loops. Enzymatically, RNaseH1 can cleave the RNA strand within an R-loop but cannot degrade RNA or the R-loop itself [59]. Therefore, following RNaseH1 cleavage, SETX may function to unwind the newly cleaved strand creating a free 5’ terminus that can be bound by XRN2 and, ultimately, drives the resolution of R-loops. Previous studies have demonstrated that XRN2 recruitment was substantially reduced, and TERRA was increased at R-loops in SETX-deficient cells [44,57]. In conjunction with the data we present here, it seems likely that there is a coordinated effort by RNaseH1, SETX, and XRN2 in mediating the resolution of TERRA R-loops in mammalian cells.
Depletion of XRN2 in HCT116AID-XRN2 cells did not lead to an increase in TERRA R-loops or drive replication stress at telomeric DNA, suggesting there could be additional complexities between transcription and replication dynamics at telomeres that have not yet been considered. Similar to the directionality of TERRA transcription, telomere replication is most often initiated within the subtelomeric region, and DNA synthesis progresses distally through the telomere end. When initiated within the CpG island promoter, TERRA transcription occurs approximately 1 kb upstream from the TTAGGG repeats, although more recent studies have identified promoters on other chromosome ends as far as 5–10 kb upstream [12,14,61]. The first telomeric origins of replication were identified in the subtelomeric DNA approximately 65–115 kb upstream from the repeats [62]. Thus, the bulk of telomere replication initiation events occurs upstream of TERRA initiation events suggesting that most TRC events would be co-directional in nature. As a result, TERRA R-loops and RNA Pol II are likely dissociated from the C-strand by the advancing replication machinery. Thus, one possible explanation for our inability to detect TERRA R-loops or replication stress in HCT116AID-XRN2 cells could be that any TERRA R-loops that arise are resolved by the passage of the replisome.
Single-molecule analysis of replicating DNA demonstrated that the induction of replication stress within telomeric DNA drives new replication initiation events within the repeats themselves [63]. Thus, increasing replication stress at telomeres may create an environment primed for head-on TRC events where the replisome traveling upstream from the origin would collide head-on with RNA Pol II transcribing TERRA on the C-rich strand traveling downstream. ATRX prevents replication stress by resolving G-quadruplex structures ahead of the replication fork [64]. Therefore, we reasoned that loss of ATRX could not only prime the telomeric chromatin landscape for the accumulation of R-loops, but may actually promote TRC events and trigger activation of the ALT pathway. However, as with the depletion of XRN2 alone, co-depletion of ATRX and XRN2 did not drive R-loop formation, replication stress, nor induce ALT phenotypes. Given these results, we considered the possibility that the kinetics of the system could limit the sheer number of TRC events. To increase the probability of inducing TRC events, we shifted our analysis to cells that rely on the ALT mechanism for telomere maintenance. ALT cells have both an increase in replication stress at telomeric DNA and elevated levels of TERRA transcript, leading to the hypothesis that TERRA R-loops may trigger TRC events and, ultimately, exacerbate ALT activity. Here, we demonstrate that by further increasing TERRA by inhibiting TERRA degradation, we can stimulate R-loop formation, replication stress, and ALT activity. These results are consistent with previous studies that have demonstrated that inhibition of TERRA transcription decreased replication stress and suppressed ALT activity [65,66], whereas inducing TERRA transcription increased ALT phenotypes and provoked ALT activity [65,66]. Taken together, our data suggest that increasing TERRA abundance alone may not be sufficient to induce ALT de novo but that in the context of ALT, increasing TERRA can exacerbate telomere dysfunction and provoke ALT activity.
Our studies continue to highlight the importance of cellular context in defining TERRA function in telomere maintenance. Previous studies have demonstrated that TERRA R-loops preferentially accumulate at short telomeres independent of cell cycle control and that persistent R-loops at short telomeres drive activation of the DNA damage response, facilitate the recruitment of recombination factors, and promote homology-directed repair to avoid premature senescence [67–70]. Although telomere length in HCT116 cells is approximately 5 kb in length, telomeres in ALT cells, such as SaOS2, can range in length from 1 kb up to 50 kb. This raises the possibility that the length of the telomeres of HCT116AID-XRN2 cells may not support TERRA R-loop formation and homology-directed repair following AID-mediated loss of XRN2. ALT-positive cells, in contrast, are more likely to possess a proportion of short telomeres that support the stabilization of TERRA R-loops that may, in turn, stimulate ALT activity. This is consistent with our observed increase in R-loops at telomeric repeats and the corresponding increase in ALT phenotypes, including γH2AX, RPA32 pS33, and APBs at telomeres following the IAA-mediated stabilization of TERRA in SaOS2AID-XRN2 cells. Continuing to gain a deeper understanding of the precise relationship between TERRA and the DNA damage response is critical to defining the mechanisms that regulate telomere stability and promote the acquisition of replicative immortality.
Acknowledgments
We thank Dr. Steven West for providing the HCT116-AID cell lines used in this study. We thank Alysia Bryll for her classification of a subset of the human tumor samples. We would also like to thank all Flynn and Ganem lab members for helpful discussions and suggestions. Tumor samples were provided by the Children’s Oncology Group (grant #AOST14B1-Q) and also obtained courtesy of Dr. Peter Houghton, UT Health San Antonio. RLF is supported by the National Center for Advancing Translational Sciences, the NIH, the Edward Mallinckrodt Junior Foundation Award, and through Boston University Clinical and Translational Science Institute (BU-CTSI) grant #1UL1TR001430. The findings of this work were supported by NIH grants R01CA214880, R01CA201446, R35GM146769.
Abbreviations:
- TERRA
Telomeric Repeat-Containing RNA
- ALT
Alternative Lengthening of Telomeres
- AID
Auxin Inducible Degron
- IAA
Auxin
- TRCs
Transcription-Replication Collision
- XRN2
5’-3’ exoribonuclease 2
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare
Data Availability
The data supporting the findings of this study are included within the article. Raw data supporting this study’s findings are available from the corresponding author, upon reasonable request.
References
- 1.T de L, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM & Varmus HE (1990) Structure and variability of human chromosome ends. Mol Cell Biol 10, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H & de Lange T (1999) Mammalian Telomeres End in a Large Duplex Loop. Cell 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 3.Doksani Y, Wu JY, De Lange T & Zhuang X (2013) XSuper-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB & Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 5.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK & Allshire RC (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868. [DOI] [PubMed] [Google Scholar]
- 6.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB & Bacchetti S (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palm W & de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42, 301–334. [DOI] [PubMed] [Google Scholar]
- 8.Schoeftner S & Blasco M a (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10, 228–236. [DOI] [PubMed] [Google Scholar]
- 9.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E & Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. [DOI] [PubMed] [Google Scholar]
- 10.Luke B, Panza A, Redon S, Iglesias N, Li Z & Lingner J (2008) The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32, 465–477. [DOI] [PubMed] [Google Scholar]
- 11.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W & Riha K (2010) siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet 6, e1000986–e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E & Azzalin CM (2009) CpG-island promoters drive transcription of human telomeres. RNA 15, 2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López de Silanes I, Stagno d’Alcontres M & Blasco M a (2010) TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat Commun 1, 33. [DOI] [PubMed] [Google Scholar]
- 14.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J & Lingner J (2014) Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 5, 5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnoult N, Van Beneden A & Decottignies A (2012) Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat Struct Mol Biol 19, 948–956. [DOI] [PubMed] [Google Scholar]
- 16.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J & Zou L (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Z, Norseen J, Wiedmer A, Riethman H & Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porro A, Feuerhahn S, Reichenbach P & Lingner J (2010) Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 30, 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T & Azzalin CM (2014) RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5, 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K & Luke B (2013) Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20, 1199–1205. [DOI] [PubMed] [Google Scholar]
- 21.Balk B, Dees M, Bender K & Luke B (2014) The differential processing of telomeres in response to increased telomeric transcription and RNA-DNA hybrid accumulation. RNA Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamperl S, Bocek MJ, Saldivar JC, Swigut T & Cimprich KA (2017) Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reaban ME, Lebowitz J & Griffin JA (1994) Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem 269, 21850–21857. [PubMed] [Google Scholar]
- 24.Reaban ME & Griffin JA (1990) Induction of RNA-stabilized DMA conformers by transcription of an immunoglobulin switch region. Nature 348, 342–344. [DOI] [PubMed] [Google Scholar]
- 25.Daniels GA & Lieber MR (1995) RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res 23, 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Pereira JM & Aguilera A (2015) R loops: new modulators of genome dynamics and function. Nat Rev Genet 16, 583–597. [DOI] [PubMed] [Google Scholar]
- 27.Bryan TM, Englezou a, Gupta J, Bacchetti S & Reddel RR(1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett H a, Neumann A a& Reddel RR(2009) Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol 16, 1244–1251. [DOI] [PubMed] [Google Scholar]
- 29.Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR & Greenberg RA (2016) Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobinoff AP, Allen JAM, Neumann AA, Yang SF, Walsh ME, Henson JD, Reddel RR & Pickett HA (2017) BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36, 2907 LP–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roumelioti F-M, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD & Gagos S (2016) Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 17, 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR & Reddel RR (1999) Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59, 4175–4179. [PubMed] [Google Scholar]
- 33.Cho NW, Dilley RL, Lampson M a& Greenberg R a(2014) Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 159, 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fasching CL, Neumann A a., Muntoni A, Yeager TR& Reddel RR(2007) DNA damage induces alternative lengthening of telomeres (ALT)-associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res 67, 7072–7077. [DOI] [PubMed] [Google Scholar]
- 35.O’Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G & Karlseder J (2014) Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol 21, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suvà ML, Benes CH, Haber DA, Boussin FD & Zou L (2015) Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science (1979) 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox KE, Maréchal A & Flynn RL (2016) SMARCAL1 Resolves Replication Stress at ALT Telomeres. Cell Rep 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Januszyk K, Liu Q & Lima CD (2011) Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA 17, 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A & Jensen TH (2010) The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29, 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West S, Gromak N & Proudfoot NJ (2004) Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432, 522–525. [DOI] [PubMed] [Google Scholar]
- 41.Davidson L, Francis L, Cordiner RA, Eaton JD, Estell C, Macias S, Cáceres JF & West S (2019) Rapid Depletion of DIS3, EXOSC10, or XRN2 Reveals the Immediate Impact of Exoribonucleolysis on Nuclear RNA Metabolism and Transcriptional Control. Cell Rep 26, 2779–2791.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eaton JD, Davidson L, Bauer DL V, Natsume T, Kanemaki MT & West S (2018) Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Dev 32, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eaton JD, Francis L, Davidson L & West S (2020) A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes. Genes Dev 34, 132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vohhodina J, Goehring LJ, Liu B, Kong Q, Botchkarev VV, Huynh M, Liu Z, Abderazzaq FO, Clark AP, Ficarro SB, Marto JA, Hatchi E & Livingston DM (2021) BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat Commun 12, 3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson J-Y & Richard S (2019) Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J 38, e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller W & Crouch R (1972) Degradation of DNA RNA Hybrids by Ribonuclease H and DNA Polymerases of Cellular and Viral Origin. Proceedings of the National Academy of Sciences 69, 3360–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan Q, Wulfridge P, Doherty J, Fernandez-Luna JL, Real PJ, Tang H-Y & Sarma K (2022) Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat Commun 13, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, McLendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SKN, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N & Meeker AK (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldberg AD, Banaszynski L a, Noh K-M,Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee Y-L, Boydston E a, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D & Allis CD (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AYM, Pickett H a & Reddel RR(2009) DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 27, 1181–1185. [DOI] [PubMed] [Google Scholar]
- 51.Heaphy CM, Subhawong AP, Hong S-M, Goggins MG, Montgomery E a, Gabrielson E, Netto GJ, Epstein JI, Lotan TL, Westra WH, Shih I-M, Iacobuzio-Donahue C a, Maitra A, Li QK, Eberhart CG, Taube JM, Rakheja D, Kurman RJ, Wu TC, Roden RB, Argani P, De Marzo AM, Terracciano L, Torbenson M & Meeker AK (2011) Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol 179, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henson JD, Hannay JA, Mccarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, Robinson BG, Learoyd DL, Stalley PD, Bonar SF, Yu D, Pollock RE & Reddel RR (2005) A Robust Assay for Alternative Lengthening of Telomeres in Tumors Shows the Significance of Alternative Lengthening of Telomeres in Sarcomas and Astrocytomas., 217–225. [PubMed] [Google Scholar]
- 53.Panier S, Maric M, Hewitt G, Mason-Osann E, Gali H, Dai A, Labadorf A, Guervilly J-H, Ruis P, Segura-Bayona S, Belan O, Marzec P, Gaillard P-HL, Flynn RL & Boulton SJ (2019) SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol Cell 76, 27–43.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mason-Osann E, Dai A, Floro J, Lock YJ, Reiss M, Gali H, Matschulat A, Labadorf A & Flynn RL (2018) Identification of a novel gene fusion in ALT positive osteosarcoma. Oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Connelly S & Manley JL (1988) A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2, 440–452. [DOI] [PubMed] [Google Scholar]
- 56.Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL & Boothman DA (2016) XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet 12, e1006107–e1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skourti-Stathaki K, Proudfoot NJ & Gromak N (2011) Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol Cell 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Groh M, Albulescu LO, Cristini A & Gromak N (2017) Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J Mol Biol 429, 3181–3195. [DOI] [PubMed] [Google Scholar]
- 59.Aguilera A & García-Muse T (2012) R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol Cell 46, 115–124. [DOI] [PubMed] [Google Scholar]
- 60.De Amicis A, Piane M, Ferrari F, Fanciulli M, Delia D & Chessa L (2011) Role of senataxin in DNA damage and telomeric stability. DNA Repair (Amst) 10, 199–209. [DOI] [PubMed] [Google Scholar]
- 61.Feretzaki M, Renck Nunes P & Lingner J (2019) Expression and differential regulation of human TERRA at several chromosome ends. RNA 25, 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG & Schildkraut CL (2012) Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J Cell Biol 197, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drosopoulos WC, Deng Z, Twayana S, Kosiyatrakul ST, Vladimirova O, Lieberman PM & Schildkraut CL (2020) TRF2 Mediates Replication Initiation within Human Telomeres to Prevent Telomere Dysfunction. Cell Rep 33, 108379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teng Y-C, Sundaresan A, O’Hara R, Gant VU, Li M, Martire S, Warshaw JN, Basu A & Banaszynski LA (2021) ATRX promotes heterochromatin formation to protect cells from G-quadruplex DNA-mediated stress. Nat Commun 12, 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva B, Arora R, Bione S & Azzalin CM (2021) TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat Commun 12, 3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silva B, Arora R & Azzalin CM (2022) The alternative lengthening of telomeres mechanism jeopardizes telomere integrity if not properly restricted. Proceedings of the National Academy of Sciences 119, e2208669119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L & Lingner J (2020) RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT & Luke B (2017) Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 170, 72–85.e14. [DOI] [PubMed] [Google Scholar]
- 69.Fernandes RV, Feretzaki M & Lingner J (2021) The makings of TERRA R-loops at chromosome ends. Cell Cycle 20, 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Misino S, Busch A, Wagner CB, Bento F & Luke B (2022) TERRA increases at short telomeres in yeast survivors and regulates survivor associated senescence (SAS). Nucleic Acids Res 50, 12829–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanz LA & Chédin F (2019) High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 14, 1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are included within the article. Raw data supporting this study’s findings are available from the corresponding author, upon reasonable request.


