Abstract
Low-molecular-weight (LMW) thiols are an abundant class of cysteine-derived small molecules found in all forms of life that maintain reducing conditions within cells. While their contributions to cellular redox homeostasis are well established, LMW thiols can also mediate other aspects of cellular physiology, including intercellular interactions between microbial and host cells. Here we discuss emerging roles for these redox-active metabolites at the host–microbe interface. We begin by providing an overview of chemical and computational approaches to LMW-thiol discovery. Next, we highlight mechanisms of virulence regulation by LMW thiols in infected cells. Finally, we describe how microbial metabolism of these compounds may influence host physiology.
Keywords: Low-molecular-weight thiol, redox regulation, host–microbe interactions, metabolism
Introduction
Low-molecular-weight (LMW) thiols are a class of thiol-containing small molecules found in all three domains of life that maintain intracellular redox homeostasis [1]. The antioxidant properties of these compounds stem from the nucleophilicity and redox reactivity of the thiol group. LMW thiols can detoxify reactive oxygen species and other electrophilic agents, serve as essential cofactors for antioxidant enzymes, and regulate cell signaling via the covalent modification of protein thiols. The paradigmatic LMW thiol is glutathione (GSH), an abundant tripeptide derived from glutamate, cysteine, and glycine that is synthesized by eukaryotes and many Gram-negative bacteria (Figure 1a). Organisms lacking GSH employ alternative LMW thiols to maintain redox balance, such as mycothiol (MSH) in certain Actinobacteria, bacillithiol (BSH) in Bacilli, and trypanothione (TSH), a polyamine derivative of GSH found in trypanosomatids (Figure 1a) [2]. In addition, certain fungal and bacterial species produce the thione-containing antioxidant ergothioneine (EGT) (Figure 1a), which can accumulate in plant and animal tissues from dietary sources [3]. Although EGT exists primarily as a thione, it is routinely classified as a member of the LMW-thiol family because it exhibits partial thiolate character via resonance and can form a thiol-based tautomer [4–6]. The unique chemical features and protective properties of these small molecules have been the subject of several comprehensive reviews [1,2,7]; however, recent studies have uncovered unexpected roles for LMW thiols at the host–microbe interface. Beyond serving as antioxidant stewards of cellular health, LMW thiols are emerging as redox-active regulators of microbial virulence and key substrates in the production of diverse, bioactive metabolites. Here we describe recent technical advances in the discovery of microbial LMW thiols and highlight how the sulfur-based chemistry of these compounds mediates host–microbe interactions.
Figure 1. Chemical and bioinformatic approaches to LMW-thiol discovery.
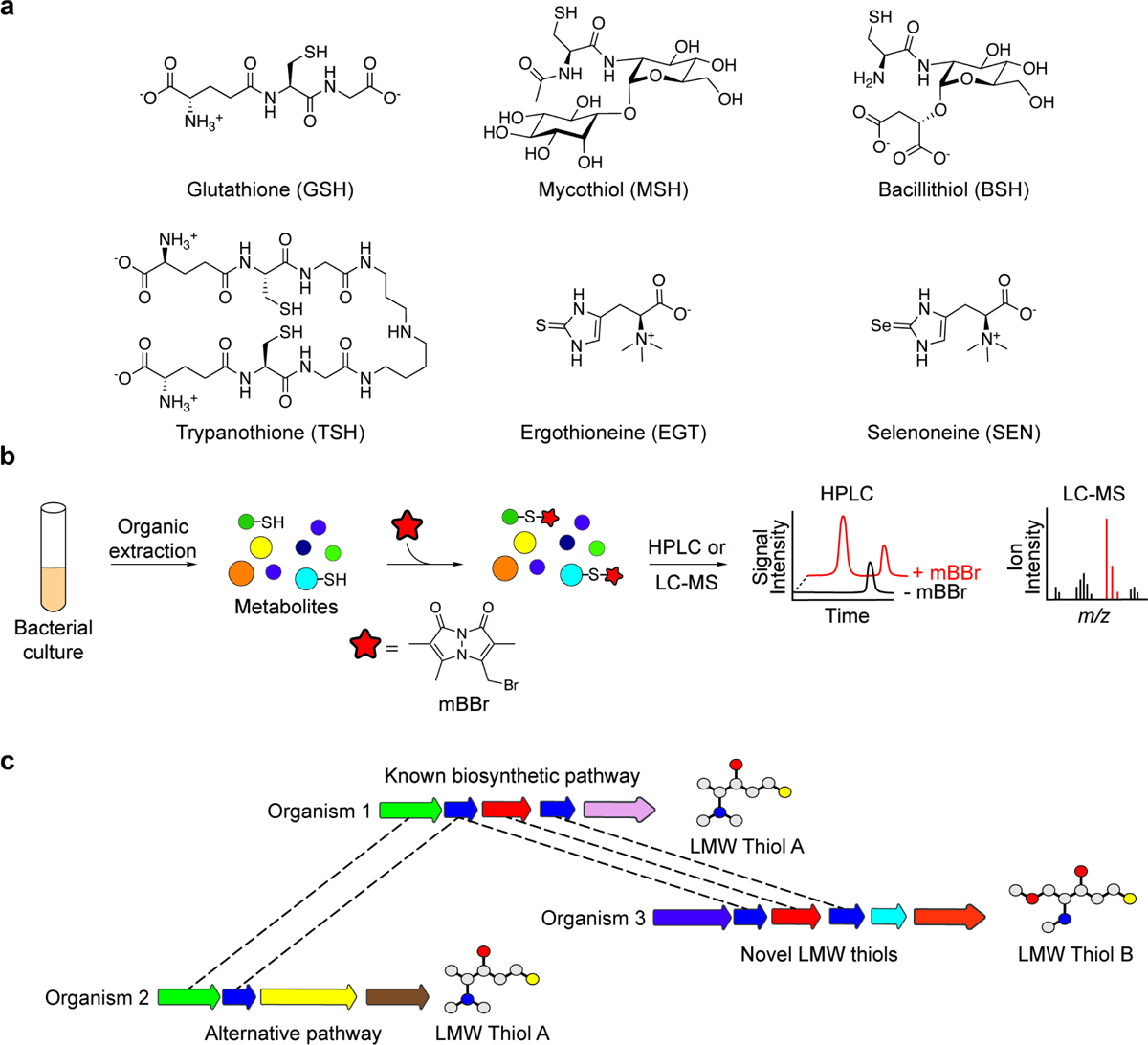
(a) Chemical structures of representative LMW thiols are shown, including glutathione (GSH), mycothiol (MSH), bacillithiol (BSH), trypanothione (TSH), ergothioneine (EGT), and selenoneine (SEN). (b) Organic extracts of bacterial cultures can be treated with the thiol-reactive probe monobromobimane (mBBr) to selectively derivatize thiol-containing metabolites in complex samples. Comparison of probe-labeled and unlabeled samples by HPLC–fluorescence analysis or LC–MS enables the identification and characterization of LMW thiols. (c) Bioinformatic analyses can facilitate the discovery of new biosynthetic pathways for known LMW thiols or LMW-thiol analogs. For example, parts of the LMW Thiol A biosynthetic pathway (organism 1) are conserved in organisms 2 and 3 (denoted by dashed lines), implying the production of related compounds.
Chemical and bioinformatic approaches to LMW-thiol discovery
The identification of LMW thiols in diverse microbes has leveraged the intrinsic nucleophilicity of the thiolate anion, which can be chemically derivatized to facilitate chromatographic separation and detection of thiol-containing metabolites in complex samples. The thiol-alkylating agent monobromobimane (mBBr) has been especially useful in this regard. When added to cell extracts, mBBr selectively reacts with thiol-containing metabolites to generate fluorescent bimane derivatives that are readily detectable by high-performance liquid chromatography (HPLC) (Figure 1b) [8]. Advances in whole-genome and metagenomic sequencing coupled with bioinformatic tools have further accelerated the characterization of novel LMW-thiol biosynthetic pathways. In this section, we provide an overview of reactivity-guided and sequence-based approaches to the discovery of prominent LMW thiols and their structural analogs.
Reactivity-guided detection of LMW thiols
One of the first microbial GSH analogs characterized was TSH, reported by Fairlamb and Cerami in 1985 [9]. After purifying the putative GSH reductase of Trypanosoma brucei from the blood of infected rats, the authors determined that an LMW, Trypanosomatid-derived factor was required to reconstitute the activity of the dialyzed enzyme. Enzyme activity was inhibited by treating pre-reduced LMW preparations with the thiol-reactive agent N-ethylmaleimide, suggesting the LMW factor contained a thiol group. Indeed, through subsequent structural analyses, the purified cofactor was identified as N1,N8-bis(glutathionyl)spermidine, or TSH (Figure 1a) [10], an LMW thiol now known to be essential for the pathogenic lifestyle of subtropical protozoan parasites including Trypanosoma and Leishmania species.
Fairlamb and Cerami relied on an inhibitor of thiol reactivity to gain insight into the chemical properties of TSH; thiol reactivity has since been widely harnessed for the unbiased identification of LMW thiols in other microbes. By using mBBr labeling and HPLC–fluorescence analysis to survey LMW thiols in multiple Streptomyces and Flavobacterium species, Newton et al. discovered MSH, a disaccharide of glucosamine and myo-inositol linked to N-acetylcysteine (Figure 1a) [11]. BSH was similarly identified via the analysis of LMW thiols in mBBr-treated extracts from Bacillus anthracis [12]. Although structurally related to MSH, BSH contains a malic acid in lieu of myo-inositol and a non-acetylated cysteine (Figure 1a) [13,14].
Based on the success of HPLC-based fluorometric approaches in detecting bimane adducts of LMW thiols, we recently coupled mBBr labeling with liquid chromatography–mass spectrometry (LC–MS) to detect microbial LMW thiols using a reactivity-guided metabolomics approach (Figure 1b). By profiling thiol-containing metabolites in the gastric pathogen Helicobacter pylori, we identified the human dietary antioxidant EGT, and subsequently determined that H. pylori imports EGT from the host environment using the ATP-binding cassette transporter EgtUV [15]. A complementary study demonstrated that EGT is also imported by the respiratory pathogen Streptococcus pneumoniae using a solute-binding domain that binds EGT with remarkable affinity (KD = 50 nM) [16]. Bioinformatic analyses revealed that egtUV is predominantly found in host-associated microorganisms and has likely spread throughout the gastrointestinal microbiome via horizontal gene transfer [15,16], suggesting microbial uptake of host EGT may be an important mechanism of microbial adaptation to the host.
Bioinformatic approaches to LMW-thiol discovery
The functional characterization of genes involved in LMW-thiol biosynthesis can lead to the discovery of orthologs that contribute to the production of structurally related compounds in other bacteria (Figure 1c). Likewise, the identification of enzymes that catalyze defined steps in LMW-thiol biosynthesis can be guided by the chemical structures of known pathway intermediates. These approaches proved instrumental in the characterization of four distinct, yet partially overlapping biosynthetic pathways for EGT. Seebeck reported the first pathway in Mycobacteria, which involves the trimethylation of histidine, followed by its oxidative sulfurization and subsequent conversion to EGT [17]. Further bioinformatic searches revealed three additional pathways in fungi [18], Cyanobacteria [19], and thermophilic bacteria and archaea [20], respectively, that employ alternative mechanisms and/or substrates for C–S bond formation. Likewise, characterization of the MSH biosynthetic pathway provided critical insight into the BSH pathway due to the structural similarities between these LMW thiols (Figure 1a, 1c) [14].
Related bioinformatic approaches led to the discovery of a biosynthetic pathway for the selenium-containing analog of EGT, selenoneine (SEN) (Figure 1a, c) [21]. To identify selenometabolite biosynthetic pathways, Kayrouz et al. searched for biosynthetic gene clusters (BGCs) encoding homologs of the selenophosphate synthetase SelD, which generates the selenium donor required for the production of selenocysteine. An operon encoding the SelD homolog SenC, the putative glycosyltransferase SenB, and SenA, a homolog of the EGT C–S bond-forming enzyme EgtB, was identified in actinomycetes and β-proteobacteria. When reconstituted in vitro, these enzymes were shown to synthesize SEN. Although little is known about its physiological function, a recent study indicates that SEN is imported by human cells and can mediate the detoxification of methylmercury in zebrafish embryos [22]. Future studies are warranted to examine whether SEN produced by gut bacteria can influence host physiology.
Altogether, these studies highlight the utility of computational approaches in characterizing the assembly of LMW thiols and structurally related metabolites. Modern computational algorithms for identifying BGCs in complex datasets [23–25], including soil and fecal metagenomes [26], will likely create new opportunities for LMW-thiol discovery in untapped microbial communities. Additionally, we anticipate that reactivity-guided metabolomic approaches will be widely applicable to the discovery and characterization of bioactive small molecules beyond LMW thiols (e.g., biogenic amines, small-molecule electrophiles) in diverse biological settings.
LMW thiols as mediators of bacterial virulence
LMW thiols can serve as environmental signals, cofactors, and covalent modifiers that modulate bacterial virulence during infection [27,28]. Certain pathogens exploit the reducing activity of host-derived GSH to activate virulence-associated transcriptional programs, whereas the covalent modification of proteins with LMW thiols can regulate molecular mechanisms of pathogenesis at the post-translational level. To date, most evidence of virulence regulation has been drawn from studies of GSH; however, analogous mechanisms of virulence control are likely to emerge for other LMW thiols. Here, we describe how the chemical properties of LMW thiols can influence virulence pathways in infected cells.
Mechanisms of virulence activation by GSH
Two seminal studies established that host-derived GSH can regulate virulence gene expression in intracellular pathogens (Figure 2, Virulence). Through a genetic screen of the foodborne pathogen Listeria monocytogenes in infected macrophages, Reniere et al. discovered that bacterial GSH enhances the expression of virulence genes regulated by the transcription factor PrfA [29]. Notably, in the absence of bacterial GSH biosynthesis, virulence gene expression could still be induced by GSH from the host cell cytosol, suggesting that both bacterial and host-derived GSH contribute to PrfA activation. Indeed, purified PrfA was found to bind to GSH in vitro, and a subsequent structural analysis of PrfA in complex with GSH and DNA confirmed that GSH regulates PrfA activity through an allosteric mechanism [30]. Altogether, these findings demonstrate that GSH serves as an environmental cue that triggers L. monocytogenes virulence programs in infected cells.
Figure 2. Representative functions of LMW thiols.
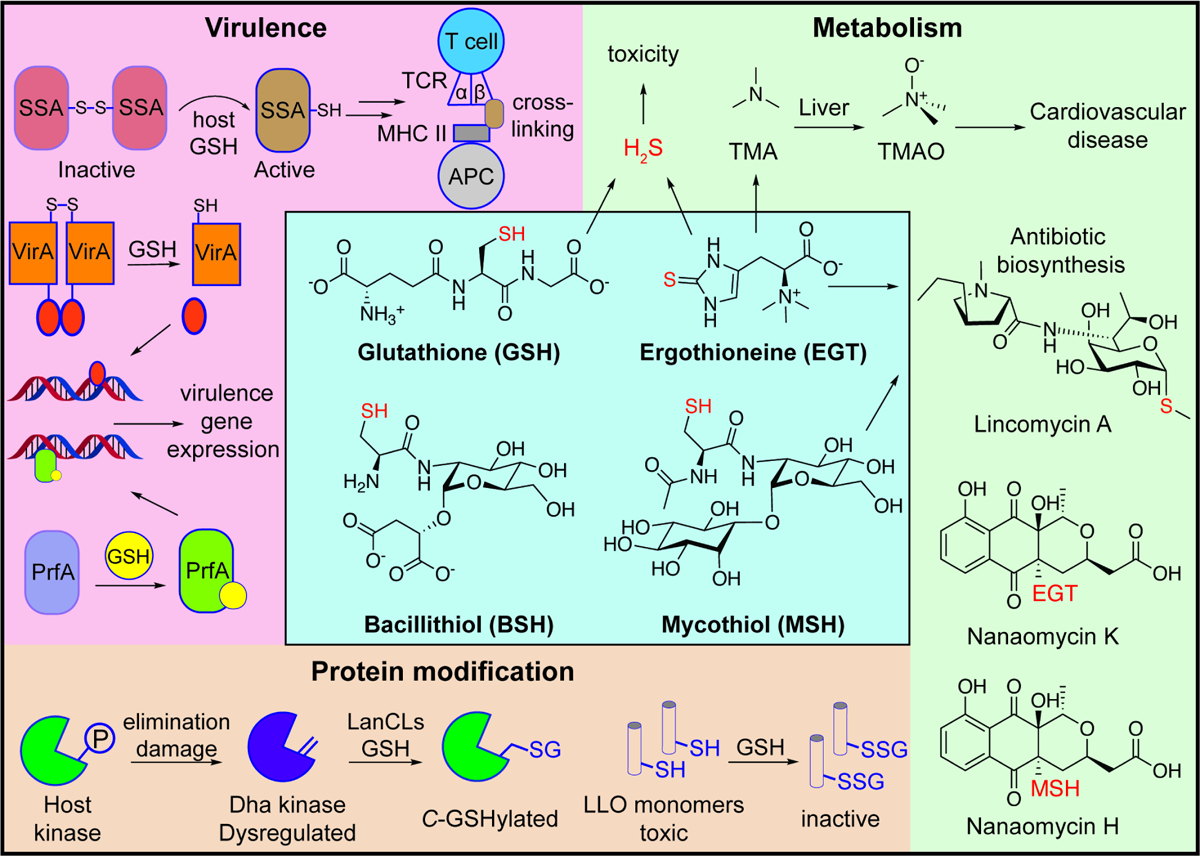
The chemical structures of GSH, EGT, BSH, and MSH are shown, with their representative functions depicted along the periphery. Virulence, top: GSH reduces and activates the S. pyogenes streptococcal superantigen SSA, which can then crosslink major histocompatibility complex (MHC) class II proteins on antigen-presenting cells (APC) to the β-chains of T-cell receptors (TCR). Virulence, bottom: GSH reduction of the B. pseudomallei histidine sensor kinase VirA generates activated monomers that induce virulence gene expression. Allosteric binding of GSH to the L. monocytogenes transcription factor PrfA promotes virulence gene expression. Protein modification: elimination of phosphate groups (P) from host kinases generates electrophilic dehydroalanine (Dha) sites that dysregulate protein signaling. LanCL enzymes can modify these sites via C-glutathionylation. S-glutathionylation of L. monocytogenes LLO monomers inhibits their pore-forming activity. Metabolism, top: microbial metabolism of GSH and EGT can produce the toxic compound H2S. Catabolism of EGT also generates trimethylamine (TMA), which undergoes oxidation in the liver to the cardiovascular disease-associated compound trimethylamine N-oxide (TMAO). Metabolism, bottom: EGT and MSH participate in the formation of microbial secondary metabolites including the antibiotic lincomycin A and nanaomycin K & H.
Similarly, the intracellular pathogen Burkholderia pseudomallei activates expression of the type VI secretion system 1 (T6SS1) following exposure to GSH in the host cytosol [31]. After invading host cells, B. pseudomallei is initially contained in endocytic vacuoles or phagosomes, but subsequently escapes into the cytosol where it forms actin protrusions that mediate intercellular transmission [32]. Expression of B. pseudomallei T6SS1, which is required for cell-to-cell spread, is controlled by the sensor histidine kinase VirA. VirA is maintained as a disulfide-linked dimer in its inactive form; however, following vacuolar escape of B. pseudomallei, host cytosolic GSH reduces the disulfide bonds in VirA and generates activated monomers that promote T6SS1 expression (Figure 2, Virulence) [31].
The Gram-positive bacterium Streptococcus pyogenes, which causes wide-ranging infections including strep throat and necrotizing fasciitis [33], deploys the pore-forming toxin streptolysin O to drive efflux of host GSH into the extracellular space [34]. Extracellular GSH can then activate the streptococcal superantigen SSA by reducing a surface-exposed cysteine residue, Cys26, which normally maintains SSA as an inactive, disulfide-linked dimer. SSA Cys26 forms mixed disulfide bonds with the β-chains of T-cell receptors (TCR) and facilitates crosslinking with major histocompatibility complex (MHC) class II molecules of antigen-presenting cells (APC), thereby inducing severe T-cell responses and uncontrolled cytokine release (Figure 2, Virulence) [35]. In addition, S. pyogenes uses the GSH-binding protein GshT to import host-derived GSH, which enhances S. pyogenes resistance to oxidative stress and the expression of multiple virulence factors [36]. As such, S. pyogenes uses the reducing activity of host GSH both to provoke inflammation and shield itself from collateral oxidative damage.
Post-translational regulation of virulence pathways
LMW thiols can also tune bacterial virulence through the post-translational modification of proteins at defined sites. For example, LMW thiols can form mixed disulfide bonds with cysteine residues in a process termed S-thiolation, which can alter protein function or binding interactions [1]. The L. monocytogenes pore-forming toxin listeriolysin O (LLO) is activated by the reduction of a single cysteine residue (Cys484) within the toxin’s membrane-binding domain. Portman et al. found that S-glutathionylation of Cys484 reduces binding of purified LLO to red blood cells by 35-fold relative to unmodified LLO and decreases cell lysis by 1000-fold (Figure 2, Protein modification) [37]. The authors suggest that S-glutathionylation may serve to temporarily inhibit toxin activity and enable the spatiotemporal activation of LLO by host oxidoreductases. Further studies are needed to confirm whether Cys484 of LLO undergoes S-glutathionylation and/or other oxidative modifications in infected cells.
GSH can also post-translationally modify protein dehydroamino acid sites through C-glutathionylation, which is catalyzed by bacterial LanC or eukaryotic LanC-like (LanCL) enzymes [38,39]. In mammals, dehydroamino acids are typically formed via the enzymatic or spontaneous elimination of phosphate groups from phosphoserine or phosphothreonine side chains [40], which are often found in the activation loops of mammalian kinases. Human LanCLs add GSH to dehydroamino acids (Figure 2, Protein modification) [38], thereby eliminating electrophilic sites from the proteome that can dysregulate cell signaling or promote the formation of deleterious protein crosslinks [41]. Notably, certain pathogenic bacteria, including Salmonella and Shigella species, secrete phosphothreonine lyases into host cells that catalyze the formation of dehydroamino acids within host kinases [42,43]. C-glutathionylation of these sites may constitute a strategy of host defense in infected cells, though this remains to be tested. LanCLs can also catalyze C-glutathionylation of dehydroamino acids in cytolysins produced by Enterococcus faecalis [38]; however, it is unknown whether this modification inhibits cytolysin toxicity during infection.
Altogether, protein glutathionylation is an emerging mechanism of virulence regulation that warrants further study. It is likely that protein S-thiolation by other microbial LMW thiols regulates virulence pathways during infection in a similar manner. Indeed, oxidative stress induces extensive protein S-mycothiolation and S-trypanothionylation in Mycobacterium smegmatis and T. brucei [44,45], respectively, suggesting these modifications may tune cellular physiology in the host environment. Chemical proteomic methods [46–48] hold promise for uncovering pathways regulated by LMW thiols in the context of infection.
Microbial metabolism of LMW thiols
LMW thiols represent a rich source of nutrients for microbes in the host environment. Several microbes encode dedicated enzymes for the catabolism of GSH and EGT, which are maintained at millimolar levels within host cells [7,49]. LMW thiols can also serve as substrates for the microbial biosynthesis of secondary metabolites with diverse bioactivities [50]. Here, we describe metabolic transactions driven by LMW thiols that shape host–microbe interactions.
Bioactive metabolites derived from microbial catabolism of LMW thiols
Many microbes can use extracellular GSH as a nutrient source. The first step in GSH degradation is typically catalyzed by the enzyme γ-glutamyl transpeptidase (GGT), which hydrolyzes GSH to glutamate and cysteinylglycine [51]. GSH degradation supports microbial metabolism but can also drive the production of toxic byproducts with deleterious consequences for the host. For example, the oral pathogen Treponema denticola uses GGT and a cysteinylglycinase to extract cysteine from environmental GSH [52,53]. Cysteine is further metabolized by the cysteine desulfurase cystalysin, generating ammonia, pyruvate, and the toxic volatile thiol, hydrogen sulfide (H2S) (Figure 2, Metabolism), which drives host cell apoptosis and is associated with periodontal pathologies [53].
Similarly, many microbes degrade EGT, which is abundant in the human diet [54]. Microbial catabolism of EGT can generate bioactive byproducts including trimethylamine (TMA) and H2S (Figure 2, Metabolism) [55–57]. In humans, TMA undergoes oxidation by a liver monooxygenase to generate trimethylamine N-oxide (TMAO), a metabolite associated with cardiovascular disease that can activate the unfolded protein stress response [58,59]. We showed that EGT catabolic activity is widespread in human fecal samples [15], suggesting that microbial uptake and breakdown of EGT may contribute to TMAO and H2S production in vivo. Notably, high EGT levels have been associated with health benefits in humans [60], whereas depleted levels have been associated with disease risk [61]. These findings raise the intriguing possibility that microbial metabolism could influence the bioavailability of dietary EGT and restrict its protective effects in humans.
Secondary metabolites derived from LMW thiols
LMW thiols have recently been implicated in the biosynthesis of secondary metabolites. For example, MSH and EGT are required for the biosynthesis of the antibiotic lincomycin A by Streptomyces lincolnensis (Figure 2, Metabolism) [62]. EGT serves as a temporary carrier of the lincosamide core before undergoing thiol-exchange with MSH, which acts as the ultimate sulfur donor. Numerous other natural products containing EGT- or MSH-derived moieties have been identified, including nanaomycin H [63], nanaomycin K [64], trichothioneic acid [65], the mycothiogranaticins [66], N-acetyl-cysteinylated streptophenazines [67], clithioneine [68], JBIR-73 [69], and spithioneine A and B [70] (Figure 2, Metabolism). Several of these compounds are likely produced via conjugation of EGT (e.g., spithioneine A & B, nanaomycin K) or MSH (e.g., mycothiogranaticins, nanaomycin H) with natural product precursor scaffolds containing reactive groups such as epoxides [71,72]. Whether these LMW-thiol conjugates form spontaneously or via specific enzymatic pathways is unknown. Some of these compounds have notable biological activities; for example nanaomycin K displays anti-tumor activity [73], and N-acetyl-cysteinylated streptophenazines, likely derived from mycothiolated streptophenazines, display broad-spectrum antibacterial activity [67]. Mass spectrometry-based approaches such as isotope-tracing metabolomics may further advance the discovery of LMW-thiol-derived metabolites that exhibit important bioactivities.
Conclusion
Although LMW thiols are principally known for their protective antioxidant properties, the biological roles of these ubiquitous small molecules are proving far more complex. Host-associated LMW thiols can activate virulence mechanisms in invading pathogens and serve as substrates for microbial metabolism, fueling the production of bioactive metabolites that may be harmful to the host. While the contrasting effects of LMW thiols on host biology during infection have been most extensively parsed for GSH, we anticipate similar roles will be uncovered for other LMW thiols at the host–microbe interface. These compounds likely regulate other fundamental aspects of cell biology and physiology that await discovery. Additional functions of LMW thiols may be unveiled through systematic identification of interacting proteins or LMW-thiol-dependent gene expression patterns in host and microbial cells. Advances in metabolomic and bioinformatic pipelines for small-molecule discovery will also continue to expand the known catalogue of alternative LMW thiols and their derivative metabolites, creating new opportunities to explore the unique redox biology and therapeutic potential of these functionally versatile compounds.
Acknowledgments
We gratefully acknowledge support from National Institutes of Health (NIH) Predoctoral Training Grant T32 GM067543 (DGD), NIH R35 GM137952 (SKH), an Arnold and Mabel Beckman Young Investigator Award (SKH), and a Conquer Cancer Now Award from the Concern Foundation (SKH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
References
- 1.Ulrich K, Jakob U: The role of thiols in antioxidant systems. Free Radic Biol Med 2019, 140:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahey RC: Glutathione analogs in prokaryotes. Biochim Biophys Acta 2013, 1830:3182–3198. [DOI] [PubMed] [Google Scholar]
- 3.Halliwell B, Cheah IK, Tang RMY: Ergothioneine – a diet-derived antioxidant with therapeutic potential. FEBS Lett 2018, 592:3357–3366. [DOI] [PubMed] [Google Scholar]
- 4.Hartman PE: Ergothioneine as antioxidant. Methods Enzymol 1990, 186:310–318. [DOI] [PubMed] [Google Scholar]
- 5.Sugihara A, Uemura K, Matsuura Y, Tanaka N, Ashida T, Kakudo M: The crystal structure of l-ergothioneine dihydrate, C9H15N3O2S. 2H2O. Acta Crystallogr B Struct Cryst Cryst Chem 1976, 32:181–185. [Google Scholar]
- 6.Cumming BM, Chinta KC, Reddy VP, Steyn AJC: Role of Ergothioneine in Microbial Physiology and Pathogenesis. Antioxid Redox Signal 2018, 28:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hand CE, Honek JF: Biological Chemistry of Naturally Occurring Thiols of Microbial and Marine Origin. J Nat Prod 2005, 68:293–308. [DOI] [PubMed] [Google Scholar]
- 8.Fahey RC, Newton GL: Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol 1987, 143:85–96. [DOI] [PubMed] [Google Scholar]
- 9.Fairlamb AH, Cerami A: Identification of a novel, thiol-containing co-factor essential for glutathione reductase enzyme activity in trypanosomatids. Mol Biochem Parasitol 1985, 14:187–198. [DOI] [PubMed] [Google Scholar]
- 10.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A: Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science 1985, 227:1485–1487. [DOI] [PubMed] [Google Scholar]
- 11.Newton GL, Fahey RC, Cohen G, Aharonowitz Y: Low-molecular-weight thiols in streptomycetes and their potential role as antioxidants. J Bacteriol 1993, 175:2734–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicely NI, Parsonage D, Paige C, Newton GL, Fahey RC, Leonardi R, Jackowski S, Mallett TC, Claiborne A: Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology. Biochemistry 2007, 46:3234–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, Fahey RC: Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 2009, 5:625–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, Helmann JD: Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci USA 2010, 107:6482–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumitrescu DG, Gordon EM, Kovalyova Y, Seminara AB, Duncan-Lowey B, Forster ER, Zhou W, Booth CJ, Shen A, Kranzusch PJ, et al. : A microbial transporter of the dietary antioxidant ergothioneine. Cell 2022, 185:4526–4540.e4518. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes the application of reactivity-guided metabolomics to the discovery of the LMW thiol EGT and its corresponding transporter in Helicobacter pylori and other host-associated bacteria.
- 16.Zhang Y, Gonzalez-Gutierrez G, Legg KA, Walsh BJC, Pis Diez CM, Edmonds KA, Giedroc DP: Discovery and structure of a widespread bacterial ABC transporter specific for ergothioneine. Nat Commun 2022, 13:7586. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper also reports the discovery of a bacterial transporter of EGT in S. pneumoniae and provides a detailed biophysical characterization of EGT-binding specificity.
- 17.Seebeck FP: In Vitro Reconstitution of Mycobacterial Ergothioneine Biosynthesis. J Am Chem Soc 2010, 132:6632–6633. [DOI] [PubMed] [Google Scholar]
- 18.Bello MH, Barrera-Perez V, Morin D, Epstein L: The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet Biol 2012, 49:160–172. [DOI] [PubMed] [Google Scholar]
- 19.Burn R, Misson L, Meury M, Seebeck FP: Anaerobic Origin of Ergothioneine. Angew Chem Int Ed Engl 2017, 56:12508–12511. [DOI] [PubMed] [Google Scholar]
- 20.Beliaeva MA, Seebeck FP: Discovery and Characterization of the Metallopterin-Dependent Ergothioneine Synthase from Caldithrix abyssi. JACS Au 2022, 2:2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayrouz CM, Huang J, Hauser N, Seyedsayamdost MR: Biosynthesis of selenium-containing small molecules in diverse microorganisms. Nature 2022, 610:199–204. [DOI] [PubMed] [Google Scholar]; ** This paper uses genome-mining approaches to uncover a targeted pathway for the production of the selenium-containing EGT analog selenoneine.
- 22.Yamashita M, Yamashita Y, Suzuki T, Kani Y, Mizusawa N, Imamura S, Takemoto K, Hara T, Hossain MA, Yabu T, et al. : Selenoneine, a novel selenium-containing compound, mediates detoxification mechanisms against methylmercury accumulation and toxicity in zebrafish embryo. Mar Biotechnol (NY) 2013, 15:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R: antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 2011, 39:W339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, et al. : Minimum Information about a Biosynthetic Gene cluster. Nat Chem Biol 2015, 11:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medema MH, Fischbach MA: Computational approaches to natural product discovery. Nat Chem Biol 2015, 11:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimoto Y, Camacho FR, Wang S, Chankhamjon P, Odabas A, Biswas A, Jeffrey PD, Donia MS: A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science 2019, 366:eaax9176. [DOI] [PubMed] [Google Scholar]
- 27.Ku JWK, Gan Y-H: Modulation of bacterial virulence and fitness by host glutathione. Curr Opin Microbiol 2019, 47:8–13. [DOI] [PubMed] [Google Scholar]
- 28.Ku JWK, Gan Y-H: New roles for glutathione: Modulators of bacterial virulence and pathogenesis. Redox Biol 2021, 44:102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA: Glutathione activates virulence gene expression of an intracellular pathogen. Nature 2015, 517:170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This is one of the first papers describing how host cell-derived GSH can drive virulence gene expression in enteric pathogens.
- 30.Hall M, Grundström C, Begum A, Lindberg MJ, Sauer UH, Almqvist F, Johansson J, Sauer-Eriksson AE: Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc Natl Acad Sci USA 2016, 113:14733–14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong J, Chen Y, Gan Y-H: Host Cytosolic Glutathione Sensing by a Membrane Histidine Kinase Activates the Type VI Secretion System in an Intracellular Bacterium. Cell Host Microbe 2015, 18:38–48. [DOI] [PubMed] [Google Scholar]
- 32.Stevens MP, Galyov EE: Exploitation of host cells by Burkholderia pseudomallei. Int J Med Microbiol 2004, 293:549–555. [DOI] [PubMed] [Google Scholar]
- 33.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V: Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev 2014, 27:264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouwer S, Barnett TC, Ly D, Kasper KJ, De Oliveira DMP, Rivera-Hernandez T, Cork AJ, McIntyre L, Jespersen MG, Richter J, et al. : Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat Commun 2020, 11:5018. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper describes a mechanism wherein S. pyogenes secretes streptolysin O to promote efflux of host GSH, which then activates the superantigen SSA to drive inflammation in the host.
- 35.Reda KB, Kapur V, Mollick JA, Lamphear JG, Musser JM, Rich RR: Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Streptococcus pyogenes. Infect Immun 1994, 62:1867–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brouwer S, Jespersen MG, Ong CY, De Oliveira DMP, Keller B, Cork AJ, Djoko KY, Davies MR, Walker MJ: Streptococcus pyogenes Hijacks Host Glutathione for Growth and Innate Immune Evasion. mBio 2022, 13:e0067622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portman JL, Huang Q, Reniere ML, Iavarone AT, Portnoy DA: Activity of the Pore-Forming Virulence Factor Listeriolysin O Is Reversibly Inhibited by Naturally Occurring S-Glutathionylation. Infect Immun 2017, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai K-Y, Galan SRG, Zeng Y, Zhou TH, He C, Raj R, Riedl J, Liu S, Chooi KP, Garg N, et al. : LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome. Cell 2021, 184:2680–2695.e2626. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper demonstrates that host LanCL enzymes catalyze the C-glutathionylation of dehydroamino-acid sites in host cell kinases and in cytolysin peptides from E. faecalis.
- 39.Repka LM, Chekan JR, Nair SK, van der Donk WA: Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem Rev 2017, 117:5457–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Lyons B, Truscott RJW, Schey KL: Human protein aging: modification and crosslinking through dehydroalanine and dehydrobutyrine intermediates. Aging Cell 2014, 13:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linetsky M, Hill JMW, LeGrand RD, Hu F: Dehydroalanine crosslinks in human lens. Exp Eye Res 2004, 79:499–512. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou J-M, Shao F: The Phosphothreonine Lyase Activity of a Bacterial Type III Effector Family. Science 2007, 315:1000–1003. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, Chen S, Wang D-C, Shao F: Structural Insights into the Enzymatic Mechanism of the Pathogenic MAPK Phosphothreonine Lyase. Mol Cell 2007, 28:899–913. [DOI] [PubMed] [Google Scholar]
- 44.Ulrich K, Finkenzeller C, Merker S, Rojas F, Matthews K, Ruppert T, Krauth-Siegel RL: Stress-Induced Protein S-Glutathionylation and S-Trypanothionylation in African Trypanosomes-A Quantitative Redox Proteome and Thiol Analysis. Antioxid Redox Signal 2017, 27:517–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hillion M, Bernhardt J, Busche T, Rossius M, Maaß S, Becher D, Rawat M, Wirtz M, Hell R, Rückert C, et al. : Monitoring global protein thiol-oxidation and protein S-mycothiolation in Mycobacterium smegmatis under hypochlorite stress. Sci Rep 2017, 7:1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulsen CE, Carroll KS: Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem Rev 2013, 113:4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weerapana E, Speers AE, Cravatt BF: Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc 2007, 2:1414–1425. [DOI] [PubMed] [Google Scholar]
- 48.Samarasinghe KTG, Munkanatta Godage DNP, VanHecke GC, Ahn Y-H: Metabolic Synthesis of Clickable Glutathione for Chemoselective Detection of Glutathionylation. J Am Chem Soc 2014, 136:11566–11569. [DOI] [PubMed] [Google Scholar]
- 49.Forman HJ, Zhang H, Rinna A: Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med 2009, 30:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M, Zhao Q, Liu W: The versatile low-molecular-weight thiols: Beyond cell protection. Bioessays 2015, 37:1262–1267. [DOI] [PubMed] [Google Scholar]
- 51.Meister A, Tate SS: Glutathione and Related γ-Glutamyl Compounds: Biosynthesis and Utilization. Annu Rev Biochem 1976, 45:559–604. [DOI] [PubMed] [Google Scholar]
- 52.Chu L, Dong Z, Xu X, Cochran DL, Ebersole JL: Role of glutathione metabolism of Treponema denticola in bacterial growth and virulence expression. Infect Immun 2002, 70:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu L, Wu Y, Xu X, Phillips L, Kolodrubetz D: Glutathione catabolism by Treponema denticola impacts its pathogenic potential. Anaerobe 2020, 62:102170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halliwell B, Tang RMY, Cheah IK: Diet-Derived Antioxidants: The Special Case of Ergothioneine. Annu Rev Food Sci Technol 2023, 14:323–345. [DOI] [PubMed] [Google Scholar]
- 55.Beliaeva MA, Leisinger F, Seebeck FP: In Vitro Reconstitution of a Five-Step Pathway for Bacterial Ergothioneine Catabolism. ACS Chem Biol 2021, 16:397–403. [DOI] [PubMed] [Google Scholar]; * This paper reports a complete microbial pathway for EGT catabolism and describes the corresponding bioactive byproducts.
- 56.Muramatsu H, Matsuo H, Okada N, Ueda M, Yamamoto H, Kato S, Nagata S: Characterization of ergothionase from Burkholderia sp. HME13 and its application to enzymatic quantification of ergothioneine. Appl Microbiol Biotechnol 2013, 97:5389–5400. [DOI] [PubMed] [Google Scholar]; * This paper reports the identification of ergothionase, which eliminates TMA from EGT.
- 57.Muramatsu H, Maguchi H, Harada T, Kashiwagi T, Kim C-S, Kato S-i, Nagata S: Identification of the gene encoding 3-(5-oxo-2-thioxoimidazolidin-4-yl) propionic acid desulfhydrase in Burkholderia sp. HME13. Biosci Biotechnol Biochem 2020, 85:626–629. [DOI] [PubMed] [Google Scholar]
- 58.Chen S, Henderson A, Petriello MC, Romano KA, Gearing M, Miao J, Schell M, Sandoval-Espinola WJ, Tao J, Sha B, et al. : Trimethylamine N-Oxide Binds and Activates PERK to Promote Metabolic Dysfunction. Cell Metab 2019, 30:1141–1151.e1145. [DOI] [PubMed] [Google Scholar]
- 59.Fennema D, Phillips IR, Shephard EA: Trimethylamine and Trimethylamine N-Oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-Mediated Host-Microbiome Metabolic Axis Implicated in Health and Disease. Drug Metab Dispos 2016, 44:1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith E, Ottosson F, Hellstrand S, Ericson U, Orho-Melander M, Fernandez C, Melander O: Ergothioneine is associated with reduced mortality and decreased risk of cardiovascular disease. Heart 2020, 106:691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheah IK, Halliwell B: Ergothioneine, recent developments. Redox Biol 2021, 42:101868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Q, Wang M, Xu D, Zhang Q, Liu W: Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature 2015, 518:115–119. [DOI] [PubMed] [Google Scholar]; ** This paper describes an unprecendented role for EGT and MSH in mediating the biosynthesis of the antibiotic lincomycin A.
- 63.Nakashima T, Kimura T, Miyano R, Matsuo H, Hirose T, Kimishima A, Nonaka K, Iwatsuki M, Nakanishi J, Takahashi Y, et al. : Nanaomycin H: A new nanaomycin analog. J Biosci Bioeng 2017, 123:765–770. [DOI] [PubMed] [Google Scholar]
- 64.Matsuo H, Nakanishi J, Noguchi Y, Kitagawa K, Shigemura K, Sunazuka T, Takahashi Y, Ōmura S, Nakashima T: Nanaomycin K, a new epithelial-mesenchymal transition inhibitor produced by the actinomycete “Streptomyces rosa subsp. notoensis” OS-3966. J Biosci Bioeng 2020, 129:291–295. [DOI] [PubMed] [Google Scholar]
- 65.Miyano R, Matsuo H, Mokudai T, Noguchi Y, Higo M, Nonaka K, Niwano Y, Sunazuka T, Shiomi K, Takahashi Y, et al. : Trichothioneic acid, a new antioxidant compound produced by the fungal strain Trichoderma virens FKI-7573. J Biosci Bioeng 2020, 129:508–513. [DOI] [PubMed] [Google Scholar]
- 66.Deng MR, Li Y, Luo X, Zheng XL, Chen Y, Zhang YL, Zhang W, Zhou H, Zhu H: Discovery of Mycothiogranaticins from Streptomyces vietnamensis GIMV4.0001 and the Regulatory Effect of Mycothiol on the Granaticin Biosynthesis. Front Chem 2021, 9:802279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vind K, Maffioli S, Fernandez Ciruelos B, Waschulin V, Brunati C, Simone M, Sosio M, Donadio S: N-Acetyl-Cysteinylated Streptophenazines from Streptomyces. J Nat Prod 2022, 85:1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konno K, Shirahama H, Matsumoto T: Clithioneine, an amino acid betaine from Clitocybe acromelalga. Phytochemistry 1984, 23:1003–1006. [Google Scholar]
- 69.Motohashi K, Nagai A, Takagi M, Shin-ya K: Two novel benzastatin derivatives, JBIR-67 and JBIR-73, isolated from Streptomyces sp. RI18. J Antibiot 2011, 64:281–283. [DOI] [PubMed] [Google Scholar]
- 70.Fu P, MacMillan JB: Spithioneines A and B, Two New Bohemamine Derivatives Possessing Ergothioneine Moiety from a Marine-Derived Streptomyces spinoverrucosus. Org Lett 2015, 17:3046–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasai M, Shirahata K, Ishii S, Mineura K, Marumo H, Tanaka H, Omura S: Structure of nanaomycin E, a new nanaomycin. J Antibiot (Tokyo) 1979, 32:442–445. [DOI] [PubMed] [Google Scholar]
- 72.Doyle TW, Nettleton DE, Balitz DM, Moseley JE, Grulich RE, McCabe T, Clardy J: Isolation and structure of bohemamine (1aβ,2α,6aβ,6bβ)-3-methyl-N-(1a,6,6a,6b-tetrahydro-2,6a-dimethyl-6-oxo-2H-oxireno[a]pyrrolizin-4-yl)-2-butenamide. J Org Chem 1980, 45:1324–1326. [Google Scholar]
- 73.Kitagawa K, Shigemura K, Ishii A, Nakashima T, Matsuo H, Takahashi Y, Omura S, Nakanishi J, Fujisawa M: Nanaomycin K inhibited epithelial mesenchymal transition and tumor growth in bladder cancer cells in vitro and in vivo. Sci Rep 2021, 11:9217. [DOI] [PMC free article] [PubMed] [Google Scholar]


