Abstract
The endoplasmic reticulum (ER) membrane protein complex (EMC) is essential for the insertion of a wide variety of transmembrane proteins into the plasma membrane across cell types. Each EMC is composed of Emc1–7, Emc10, and either Emc8 or Emc9. Recent human genetics studies have implicated variants in EMC genes as the basis for a group of human congenital diseases. The patient phenotypes are varied but appear to affect a subset of tissues more prominently than others. Namely, craniofacial development seems to be commonly affected. We previously developed an array of assays in Xenopus tropicalis to assess the effects of emc1 depletion on the neural crest, craniofacial cartilage, and neuromuscular function. We sought to extend this approach to additional EMC components identified in patients with congenital malformations. Through this approach we determine that EMC9 and EMC10 are important for neural crest development and the development of craniofacial structures. The phenotypes observed in patients and our Xenopus model were similar to EMC1 loss of function likely due to a similar mechanism of dysfunction in transmembrane protein topogenesis.
Keywords: Xenopus tropicalis, EMC, Neural Crest, Craniofacial Cartilage, CHD, NDD, Development
Introduction:
Membrane proteins must be inserted into lipid bilayers with specific topologies to carry out their functions. Many membrane proteins require the EMC in the process of proper membranous insertion (Chitwood, Juszkiewicz, Guna, Shao, & Hegde, 2018; Jonikas et al., 2009). The EMC is conserved from yeast to vertebrates and functions across kingdoms (Jonikas et al., 2009). Work in multiple vertebrates has shown a diverse array of transmembrane proteins such as acetylcholine receptors and rhodopsin that rely on the EMC (Richard, Boulin, Robert, Richmond, & Bessereau, 2013; Satoh, Ohba, Liu, Inagaki, & Satoh, 2015).
Multiple genetic studies in patients with congenital malformations have now suggested that variants in EMC subunits may be at the root of various congenital malformations (Abu-Safieh et al., 2013; Geetha et al., 2018; Haddad-Eid, Gur, Eid, Pilowsky-Peleg, & Straussberg, 2022; Harel et al., 2016; Homsy et al., 2015; Jin et al., 2017; Shao et al., 2021; Umair et al., 2020). Though we previously investigated the mechanism of pathogenesis for EMC1, we now sought to determine whether variants identified in genes encoding other EMC subunits might lead to disease in a similar manner. Such additional variants have been described for EMC9 and EMC10 (Haddad-Eid et al., 2022; Shao et al., 2021; Umair et al., 2020).
We sought to create models for EMC9 and EMC10 loss-of-function to improve our understanding of how dysfunction in different EMC subunits may lead to congenital disease. We based our approach on the assays we developed to assess mechanisms of dysfunction in emc1 knockout and knockdown models. In our Xenopus tropicalis model, emc9 and emc10 depletion recapitulated patient phenotypes including craniofacial malformations. The neural crest cell (NCC) lineage, which is critical for craniofacial development, appears to be affected in these phenotypes. NCCs delaminate from the neural plate border and migrate to diverse destinations within the developing embryo. NCCs then differentiate into a multitude of cell types, including chondrocytes that establish craniofacial cartilage (Martik & Bronner, 2017; Simoes-Costa & Bronner, 2015; Stuhlmiller & Garcia-Castro, 2012). Amongst several signaling pathways, WNTs have a demonstrated role in multiple steps of NCC function by regulating the expression of neural plate border and neural crest specifiers (Sutton, Kelsh, & Scholpp, 2021; Wu, Saint-Jeannet, & Klein, 2003).
Here we demonstrate that depletion of either emc9 or emc10 leads to NCC dysfunction via disruption of WNT signaling. This leads to abnormalities in craniofacial and neuromuscular development observed in our Xenopus model. As with depletion of emc1, depletion of these additional EMC subunits appears to cause depletion of the Fzd receptor that is a key component of WNT signal transduction. Downstream WNT deficits were evident via assaying nuclear pools of β-catenin that serves as a downstream effector of this pathway when translocated from the cytoplasm. Our results support that EMC9 and EMC10 dysfunction contribute to human disease via dysregulation of protein folding that affects NCC development as well as neuromuscular signaling.
Results:
Our previous work evaluated the effects of emc1 loss of function on development and uncovered significant deficits in transmembrane protein topogenesis and subsequent NCC function (Marquez et al., 2020). Because of the critical importance of the EMC in inserting transmembrane proteins, we wondered whether other variants in genes encoding EMC subunits might also contribute to early human disease. In our review of the literature, we focused on variants that were likely to disrupt protein function (protein truncations, frameshifts, or splicing disruptions) which included EMC9 and EMC10 (Table 1 and Figure 1), prompting us to focus on these EMC components (Jin et al., 2017; Shao et al., 2021; Umair et al., 2020). Patient phenotypes included congenital heart disease (CHD), neurodevelopmental delay (NDD), craniofacial dysmorphology (CFD), umbilical/inguinal hernias, renal anomalies, and limb abnormalities. CFD was the most common phenotype across reported patients with EMC gene variants, though dysmorphologies appeared heterogeneous and varied even amongst patients with the same genotype. Given the prevalence of CFDs in these patients with variants in EMC9/10, we sought to investigate the specification of NCCs given their importance in subsequent craniofacial development.
Table 1:
Mutations in EMC9 and EMC10 in 18 individuals from ten families with congenital anomalies
| Subunit Gene | Coding Sequence Change | Amino Acid Change | Effect | gnomAD MAF | Source | Patients/Families | Zygosity | Phenotypes |
|---|---|---|---|---|---|---|---|---|
| EMC9 | C.307C>T | p.Arg103Ter | Stopgain | 1.59E-05 | Jin et al. 2017 | 1/1 | Het | CHD, CFD, NDD |
| EMC10 | c.736delG | p.Gly246fs | Frameshift | 0 | Jin et al. 2017 | 1/1 | Het | CHD, CFD |
| EMC10 | c.679–1 G>A | N/A | Splice Acceptor | 0 | Umair et al. 2020 | 2/1 | Hom | CFD, NDD |
| EMC10 | c.287delG | p.Gly96fs | Frameshift | 2.54E-05 | Shao et al. 2021 | 14/7 | Hom | CHD, CFD, NDD |
CFD, craniofacial defects; CHD, congenital heart disease; gnomAD, Genome Aggregation Database http://gnomad.broadinstitute.org; Het, heterozygous; Hom, homozygous; MAF, minor allele frequency; N/A, not applicable; NDD, neurodevelopmental delay
Figure 1:
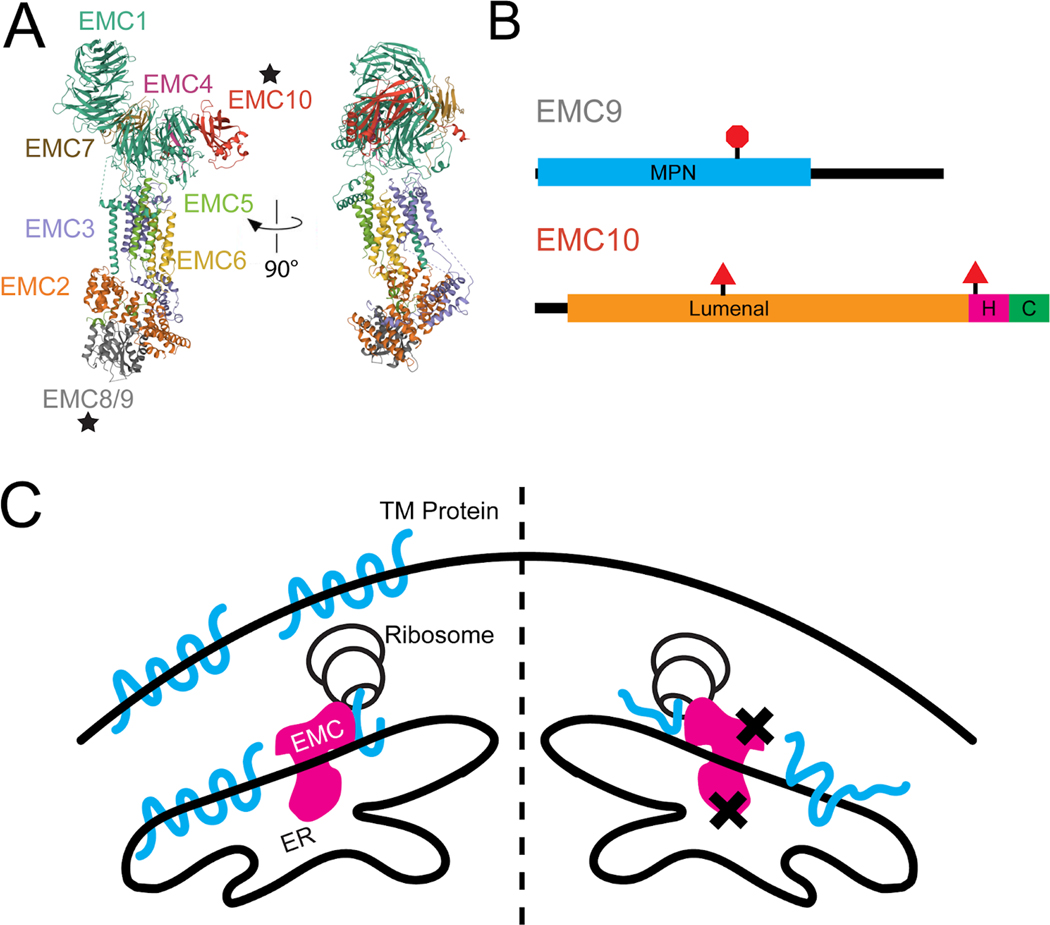
Loss of EMC9 or EMC10 is Predicted to Result in Abnormal Protein Folding. (A) Ribbon diagram of endoplasmic reticulum membrane protein complex. Stars indicate positions of EMC9 and EMC10. (B) Schematic diagram of EMC9 and EMC10 protein with domain annotations and locations and effects of variants. Red triangle indicates splice variant and red octagon indicates truncating variant. All of these variants would be predicted to result in loss of function and depletion of the protein. (C) Model of resultant protein misfolding due to EMC dysfunction predicted to result from loss of function variants in EMC9 and EMC10. Xs indicate losses of functional EMC9 or EMC10 in the EMC. C: cytoplasmic domain, ER: endoplasmic reticulum, H: helical transmembrane domain, MPN: Mpr1, Pad1 N-terminal domain, TM: transmembrane.
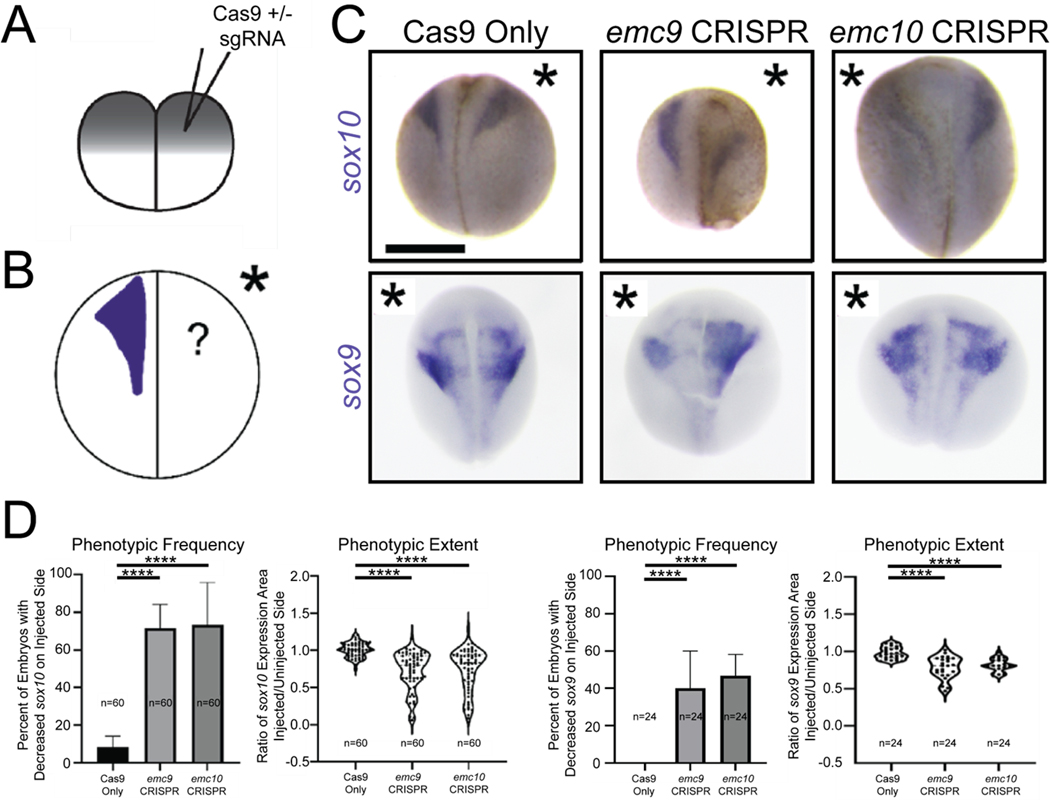
After neurulation, NCCs delaminate from the neural plate border and undergo an extensive pattern of migration in response to environmental cues. Once they arrive at their destinations, they can differentiate into an extraordinary array of cell types including chondrocytes that establish craniofacial structure. To assess NCC development, we employed whole mount in situ hybridization (WISH) to evaluate sox10 as a NCC marker that defines NCC specification and migration and an earlier NCC marker sox9. For these experiments, we exploited an advantage of Xenopus. Due to holoblastic cleavages, injection of one cell at the two-cell stage (Figure 2A) can lead to targeting of either the left or right side of the embryo, providing the unaffected side as an internal control that can be easily identified by co-injecting fluorescent tracers. Thus, we depleted emc subunit genes unilaterally via CRISPR/Cas9 injection in developing embryos. We confirmed targeting of the appropriate locus by our sgRNAs through inference of CRISPR edits analysis(Conant et al., 2022; Hsiau et al., 2019) (Figure S1). We then examined both sox9 and sox10 expression patterns (Figure 2C, D). Indeed, these markers were reduced in affected tissue when emc9 or emc10 was depleted (Figure 2C, D). This phenotype was consistent with multiple additional guide RNAs targeting emc9 or emc10 (Figure S2) Importantly, reduction in these markers was not due to excess apoptotic cell death based on terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) analysis (Figure S3).
Figure 2:
Depletion of emc9 or emc10 in Xenopus results in defective Neural Crest Development. (A) Schematic of injection after holoblastic cleavage at the 2-cell stage (B) Schematic of experimental set up to assess sox9 and sox10 expression via WISH (C) Representative images of WISH for sox10 and sox9 demonstrates decreased expression at stage 20 in tissues depleted of emc9 or emc10 (injected halves of embryos indicated by asterisks) (D) Quantitation of sox10 and sox9 expression in tissues depleted of emc9 or emc10 (n = 60 per condition over 3 replicates for sox10 n = 24 per condition over 3 replicates for sox9). Scale bar: 500 μm. Statistical tests carried out as chi-squared tests with Yates correction for frequency of phenotype and two-tailed t-tests for expression area ratios. **** indicates p<0.0001. Bars indicate mean and SD of replicate values for frequency and SD of individual values for area of expression.
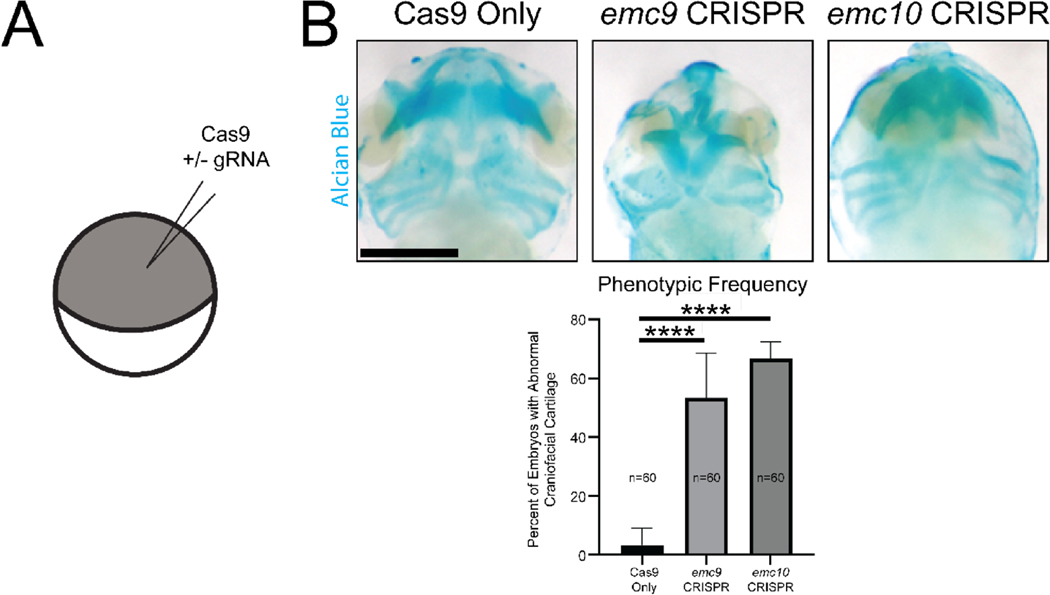
We further assessed subsequent development of craniofacial cartilage and cardiac morphology. Here we employed one cell CRISPR injected embryos to more easily evaluate the overall appearance of craniofacial morphology (Figure 3A). We noted a dysmorphology in craniofacial cartilages for both emc9 and emc10 depletion (Figure 3B). This appeared to impact rostral craniofacial cartilage to a greater extent than more caudal craniofacial cartilage (Figure S4). In addition, we observed decreased caliber of the cardiac outflow tract in emc9 and emc10 depleted (Figure S5). From these studies, we conclude that NCC, craniofacial cartilage, and cardiac development are altered in emc9 and emc10 depleted embryonic tissue.
Figure 3:
Depletion of emc9 or emc10 in Xenopus results in Craniofacial Cartilage Abnormalities. (A) Schematic of injection at the 1-cell stage (B) Representative images and quantitation of Alcian Blue staining of craniofacial cartilage demonstrates abnormalities at stage 45 in tissues depleted of emc9 or emc10 (n = 60 per condition over 3 replicates). Scale bar: 250 μm. Statistical tests carried out as chi-squared tests with Yates correction. **** indicates p<0.0001. Bars indicate SD of replicate values.
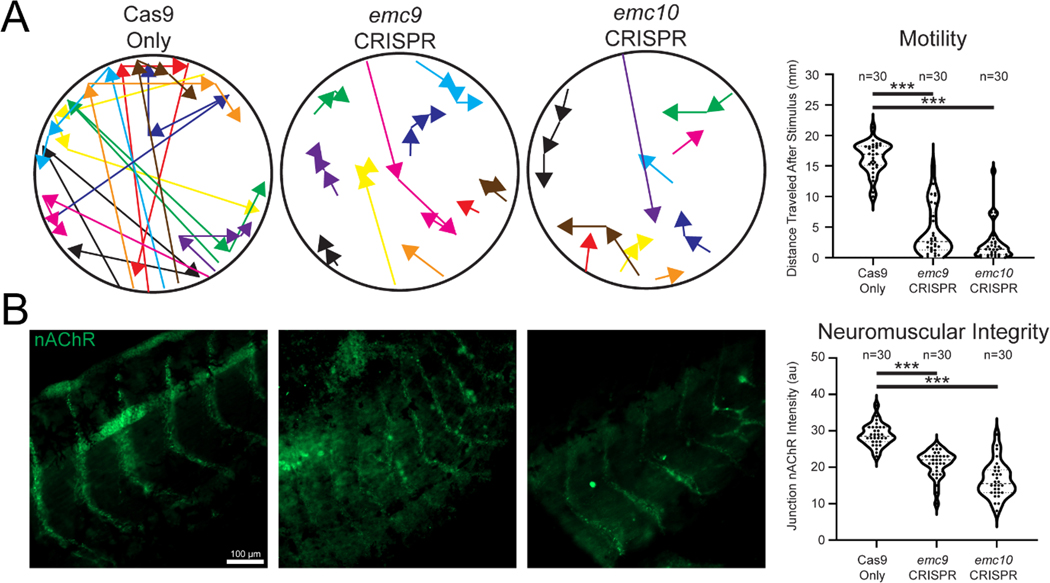
As the transmembrane nicotinic acetylcholine receptor (nAChR) has been shown to rely on EMC mediated insertion (Richard et al., 2013) and is crucial in neuromuscular signaling, we next assayed movement in tadpoles depleted of emc9 or emc10. We measured the distance tadpoles moved after stimulation. emc9 or emc10 deficient embryos moved considerably shorter distances than their stage-matched siblings in the control group (Figure 4A). To test whether this might indeed be due to nAChR abnormalities, we performed immunofluorescence for nAChR in tadpole tails, examining the locations where muscle contraction impulses were generated (Figure 4B). In control embryos, the nAChR signal was a sharp arc across the somitic muscle, whereas in emc9 or emc10 depleted embryos, the nAChR signal appeared weaker and more discontiguous. From these results, we conclude that properly localized nAChR is reduced in tadpoles depleted of EMC subunits.
Figure 4:
Depletion of emc9 or emc10 in Xenopus affects embryo motility and neuromuscular acetylcholine receptor patterning. (A) Sample traces and measurement of control (n = 30), emc9 depleted (n = 30), and emc10 depleted tadpole movement over 10 seconds after stimulation (different colors differentiate distinct tadpoles) over 3 replicates. (B) Labeling of tail neuromuscular acetylcholine receptor distribution reveals sparse signals in (n = 30) emc9 and (n = 30) emc10 depleted junctions.
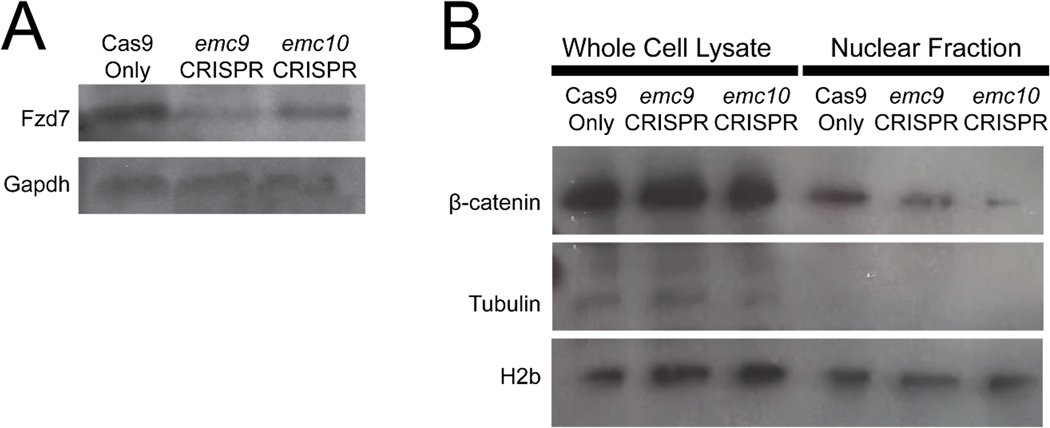
As we had previously observed a disruption in WNT signaling in emc1 dysfunction phenotypes, we assayed the levels of transmembrane WNT components. With both emc9 and emc10 depletion we observed a decrease in Fzd7 levels consistent with disruption of Fzd folding (Figure 5A). While this was suggestive of WNT dysfunction on the transmembrane protein level, we sought to assess the downstream effect on WNT signaling that involves nuclear import of β-catenin to enact transcriptional changes. Levels of nuclear β-catenin were decreased in embryos depleted of emc9 and emc10 (Figure 5B).
Figure 5:
Depletion of emc9 or emc10 in Xenopus affects WNT signaling via depletion of transmembrane protein levels. (A) Immunoblotting for Fzd7 showed a decrease in Fzd7 in pooled (n = 30 per stage per condition) emc9 depleted and emc10 depleted tadpoles as compared with control tadpoles at stage 45. (B) Immunoblotting for β-catenin showed a marked decrease in nuclear β-catenin in pooled (n = 30 per stage per condition) emc9 depleted and emc10 depleted tadpoles as compared with control tadpoles at stage 45.
Discussion:
As the EMC is both evolutionarily conserved and utilized in all cell types, we believed that dysfunction in different subunits would likely result in similar phenotypes via closely related mechanisms. Yet, recent work assessing multiple subunits of another large complex, the nuclear pore complex, uncovered distinct phenotypes and mechanisms leading to congenital disease (Braun et al., 2018; Braun et al., 2016; Chen et al., 2019; Del Viso et al., 2016; Marquez, Bhattacharya, Lusk, & Khokha, 2021; Miyake et al., 2015; Muir et al., 2020). Thus, modeling loss-of-function phenotypes of subunits in a large complex has proven a fruitful endeavor.
The observed phenotypes in patients with likely damaging variants of EMC9 and EMC10 closely resembled those of patients with EMC1 variants. Yet, the inheritance pattern and phenotype correlation among variants in EMC9 and EMC10 appears distinct. Heterozygous loss-of-function variants in EMC1 resulted in CHD while homozygous putative loss-of-function variants resulted in a syndromic NDD phenotype. The heterozygous putative loss-of-function variant in EMC9 may contribute to both CHD and NDD phenotypes while both heterozygous and homozygous putative loss-of-function variants in EMC10 appear to contribute to either CHD or NDD or both (Table 1). Larger clinical cohorts of patients with EMC variants will be essential to fully decipher these phenotype-genotype correlations.
Given these observations, we concluded that modeling loss-of-function in EMC9 and EMC10 in Xenopus would be useful in further determining the pathogenicity of loss-of-function in these genes. By assessing NCC development in emc9 and emc10 depleted tissues through multiple NCC markers, we observed mis-patterning which indicates developmental dysfunction within the NCC lineage. Later assessing craniofacial cartilage, we uncovered dysmorphology indicative of impaired craniofacial cartilage establishment downstream of impacts on NCC development. Cardiac patterning was also impaired in these embryos which may also be a result of NCC dysfunction as cardiac NCCs populate various areas of the developing heart (Figure S5). Our investigation of neurological dysfunction further supported the role of EMC subunit dependence for proper neuromuscular signaling. It therefore appears that emc9 and emc10 loss-of-function affects many of the same tissues as emc1 loss of function, including NCC derivatives. Indeed, given the overlap of the observed phenotype in our now multiple EMC subunit loss-of-function gene models, it appeared likely that dysfunction leads to congenital disease via a similar deficiency in protein folding (Figure 1C).
Though NCC development appears affected in emc9 and emc10 depleted tissues, the broader mechanism of disrupted protein folding likely contributes to pathogenesis through effects on tissues beyond NCC derivatives. Visualization of abnormal nAChR at tail neuromuscular junctions in Xenopus following emc9 or emc10 depletion suggests this broader pathogenic impact involves neuromuscular development that would be largely independent of NCC development and WNT signaling.
This disrupted protein localization of nAChR and decreases in Fzd7 further support the role of transmembrane protein misfolding as the basis for EMC subunit loss of function resulting in phenotypes observed in our Xenopus model and patients. In particular, it appears that the downstream diminution of WNT signaling likely plays a role in these phenotypes. We observed both loss of transmembrane components of WNT signaling as well as evidence of disrupted downstream WNT signaling.
Our understanding of the EMC has flourished amidst recent studies on the structure, molecular function, and determination of the client proteins of this crucial complex (Chitwood et al., 2018; Guna, Volkmar, Christianson, & Hegde, 2018; Miller-Vedam et al., 2020; O’Donnell et al., 2020; Pleiner et al., 2020; Shurtleff et al., 2018; Tian et al., 2019). While the phenotypes observed in our Xenopus emc9 and emc10 loss-of-function models resembled those of emc1 loss of function this is somewhat surprising given the relative importance of EMC subunits. Studies of EMC function have identified similar structures and possibly even a similar evolutionary origin for EMC8 and EMC9 (Bai, You, Feng, Kovach, & Li, 2020; O’Donnell et al., 2020; Pleiner et al., 2020; Tian et al., 2019; Wideman, 2015). Given this potential redundancy between EMC8 and EMC9 we expected a potentially less severe phenotype in our emc9 loss-of-function model. As this was not apparent in our studies, it may point to a distinct role for EMC9 during the window of development that we interrogated.
As additional variants in EMC subunit genes are identified, we may begin to better understand a critical question about the role these proteins play in essential aspects of cell biology. Thus far, only putatively damaging variants in EMC1, EMC9, and EMC10 have been observed in patients. These genes encode portions of the EMC that are not considered “core” components. EMC2, EMC5, and EMC6, the core components of the EMC are required not only for proper folding of client EMC proteins but also for the establishment of the EMC itself (Volkmar et al., 2019). As no disease-causing variants have been observed in the genes encoding these subunits, perhaps this indicates that we have observed EMC1, EMC9, and EMC10 variants due to a greater tolerance for mutation albeit causing severe disease.
In summary, our results indicate that disruption of additional subunits of the EMC complex support a model of human disease in part via dysfunction in the neural crest and derived tissues stemming from transmembrane protein misfolding. This conclusion is readily evident in our Xenopus model and extends the implications of our previous work on the pathogenesis of dysfunctional EMC disease. Indeed, EMC subunit related diseases may be best understood as a category of developmental foldopathies. Future therapeutic approaches could aim to alleviate the excess of misfolded proteins as a strategy to ameliorate disease burden, given the difficulty of targeting WNT signaling itself. These mechanistic insights may serve to improve treatment and optimize care for future patients with EMC variants.
Methods:
Xenopus Husbandry
Xenopus tropicalis were housed and cared for according to established protocols approved by Yale IACUC. We induced ovulation and collected embryos by in vitro fertilization as previously described (Lane & Khokha, 2021). Embryos were raised in 1/9xMR + gentamycin. Staging of Xenopus tadpoles was performed as previously cataloged (Nieuwkoop, Faber, & Hubrecht-Laboratorium (Embryologisch Instituut), 1967).
Xenopus CRISPR Manipulations
CRISPR/Cas9-mediated genome editing in Xenopus tropicalis embryos was used as previously described (Bhattacharya, Marfo, Li, Lane, & Khokha, 2015). CRISPR sgRNAs targeting exon 2 of emc9 and exon 1 of emc10 were designed to generate F0 knockout embryos (emc9: 5’-GGATTGGGATACAGTCACTC-3’, emc10: 5’-GGCTGCCGGTTGTTTAGTT-3’). For targeted loss of function experiments 200 pg sgRNA along with 0.8 ng Cas9 (CP03, PNA Bio) in a 1 nl volume were injected into one cell of a two-cell stage embryo or 400 pg sgRNA along with 1.6 ng Cas9 (CP03, PNA Bio) in a 2 nl volume were injected into a one-cell stage embryo.
Inference of CRISPR Edits Analysis
We amplified the sequence adjacent to the CRISPR cut site using a polymerase chain reaction with Phusion HighFidelity DNA Polymerase (NEB). Resultant products were sequenced and analyzed via Synthego browser based platform (ice.synthego.com).
WISH
WISH was carried out as previously described (Henriquez, Cross, Vial, & Maccioni, 1995). Briefly Xenopus embryos were fixed in 4% paraformaldehyde and dehydrated through washes in methanol. Embryos were then rehydrated in PBS with 0.1% tween-20. Embryos were then hybridized with digoxigenin labeled RNA probes complementary to target genes. Embryos were then washed and blocked prior to incubation with anti-DIG-Fab fragments (Roche) overnight at 4 degrees. BM purple (Sigma) was used to visualize expression prior to post-fixation in 4% paraformaldehyde with 0.1% glutaraldehyde.
TUNEL Staining
TUNEL staining was carried out as previously described (Hensey C & Gautier J. 1998) on embryos that were injected in 1 at the 1-cell stage.
Whole Mount Alcian Blue Cartilage Staining
Stage 45 embryos were fixed in 100% ethanol for 48 hrs at room temperature and then washed briefly in acid alcohol (1.2% HCl in 70% EtOH). A 0.25% Alcian blue solution in acid alcohol was used to stain the embryos over 48 hrs at room temperature. Specimens were then washed in acid alcohol several times, rehydrated in water and bleached for 2 hours in 1.2% hydrogen peroxide under a bright (2500 lux) light. They were then washed several times in 2% KOH and left rocking overnight in 10% glycerol in 2% KOH. Samples were processed through 20%, 40%, 60, and 80% glycerol in 2% KOH. Scoring of cartilage morphology was carried out by measuring ventral regions of cartilage representing meckel, ceratohyal, and branchial gill cartilage in FIJI. Cartilage was scored as abnormal if it was absent or less than 75% of the size of average region size of regions in Cas9 only injected embryos.
Whole Mount Cardiac Outflow Tract Evaluation
Stage 45 embryo outflow tracts were measured immediately proximal to the ventricle. DIstance was measured perpendicular to the direction of the outflow tract in FIJI.
Motility Assay
Motility was assessed as previously described (Marquez et al., 2020). Briefly, stage 45 tadpoles were placed into separate wells of a 48-well culture dish and allowed to reach a resting state for 5 minutes. Tadpoles were then gently prodded at the rostral most aspect of the tail using a capillary pipette tip. Video of tadpole movement was captured over 30 seconds after this stimulation using an AccuScope Excelis camera mounted on a Nikon SMZ 745T stereomicroscope. Videos were analyzed using Kinovea 0.8.15 software (https://www.kinovea.org/). Analysis consisted of marking a center point of the tadpole head in each frame of a 30-second video and plotting this on a circular map. Average motion was determined via converting marked tracking points to vectors and measuring the sum of these vectors over the 30-second recorded time.
Immunofluorescence and Microscopy
Xenopus tails from stage 45 embryos were first fixed in 4% PFA for 30 minutes followed by brief permeabilization in 0.1% tween-20 in PBS. Samples were mounted in Prolong Glass (Thermo Fisher Scientific). Immunostained tails were imaged on a Zeiss Observer outfitted with optical interference (Apotome) microscopy. Antibodies are listed in Table S1. Fluorescent images were processed and analyzed with FIJI. nAChR signals were compared via selecting 100μm x 50μm regions encompassing neuromuscular bands in Xenopus tail samples. Comparisons were made between similar regions of the most proximal 500μm of Xenopus tail (measured from the junction of tadpole tail and body. Whole-mounted, craniofacial cartilage, WISH, and TUNEL stained embryos were imaged with a Canon EOS 5d digital camera mounted on a Zeiss discovery V8 stereomicroscope.
Immunoblot Analysis
Pooled embryos were lysed in RIPA buffer and immunoblots were performed with Bolt 4%–12% Bis-Tris plus gels and running buffer (Thermo Fisher Scientific) using standard methods. Antibodies are listed in Table S1. Nuclear fractionation was performed using centrifugation at 720 g for 5 min and collecting the nuclear pellet which was then washed with 500 μL of fractionation buffer (HEPES (pH 7.4) 20mM, KCl 10mM, MgCl2 2mM, EDTA 1mM, EGTA 1mM, DTT 1mM). The pellet was then dispersed via pipetting and centrifuged again at 720 g for 10 min discarding the supernatant and resuspending the pellet in TBS with 0.1% SDS.
Supplementary Material
Acknowledgements:
We thank Michael Slocum for animal husbandry. JM was supported by the Yale MSTP NIH T32GM07205 Training Grant, the Yale Predoctoral Program in Cellular and Molecular Biology NIH T32GM007223 Training Grant, and the Paul and Daisy Soros Fellowship for New Americans. This work was supported by the NIH R01HD081379 grant to MKK.
References:
- Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, . . . Alkuraya FS. (2013). Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res, 23(2), 236–247. doi: 10.1101/gr.144105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, You Q, Feng X, Kovach A, & Li H. (2020). Structure of the ER membrane complex, a transmembrane-domain insertase. Nature, 584(7821), 475–478. doi: 10.1038/s41586-020-2389-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, & Khokha MK. (2015). CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol, 408(2), 196–204. doi: 10.1016/j.ydbio.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Lovric S, Schapiro D, Schneider R, Marquez J, Asif M, . . . Hildebrandt F. (2018). Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest, 128(10), 4313–4328. doi: 10.1172/JCI98688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, . . . Hildebrandt F(2016). Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet, 48(4), 457–465. doi: 10.1038/ng.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Y, Yang S, Shi Z, Zeng W, Lu Z, & Zhou X. (2019). Bi-Allelic Mutations in NUP205 and NUP210 Are Associated With Abnormal Cardiac Left-Right Patterning. Circ Genom Precis Med, 12(7), e002492. doi: 10.1161/CIRCGEN.119.002492 [DOI] [PubMed] [Google Scholar]
- Chitwood PJ, Juszkiewicz S, Guna A, Shao S, & Hegde RS. (2018). EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell, 175(6), 1507–1519 e1516. doi: 10.1016/j.cell.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant D, Hsiau T, Rossi N, Oki J, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, Stoner R. (2022). Inference of CRISPR Edits from sanger trace data. CRISPR J 5(1), 123–130. 10.1089/crispr.2021.0113 [DOI] [PubMed] [Google Scholar]
- Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, . . . Khokha MK. (2016). Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia. Dev Cell, 38(5), 478–492. doi: 10.1016/j.devcel.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha TS, Lingappa L, Jain AR, Govindan H, Mandloi N, Murugan S, . . . Vedam R(2018). A novel splice variant in EMC1 is associated with cerebellar atrophy, visual impairment, psychomotor retardation with epilepsy. Mol Genet Genomic Med, 6(2), 282–287. doi: 10.1002/mgg3.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Volkmar N, Christianson JC, & Hegde RS. (2018). The ER membrane protein complex is a transmembrane domain insertase. Science, 359(6374), 470–473. doi: 10.1126/science.aao3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad-Eid E, Gur N, Eid S, Pilowsky-Peleg T, & Straussberg R. (2022). The phenotype of homozygous EMC10 variant: A new syndrome with intellectual disability and language impairment. Eur J Paediatr Neurol, 37, 56–61. doi: 10.1016/j.ejpn.2022.01.012 [DOI] [PubMed] [Google Scholar]
- Harel T, Yesil G, Bayram Y, Coban-Akdemir Z, Charng WL, Karaca E, . . . Lupski JR. (2016). Monoallelic and Biallelic Variants in EMC1 Identified in Individuals with Global Developmental Delay, Hypotonia, Scoliosis, and Cerebellar Atrophy. Am J Hum Genet, 98(3), 562–570. doi: 10.1016/j.ajhg.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez JP, Cross D, Vial C, & Maccioni RB. (1995). Subpopulations of tau interact with microtubules and actin filaments in various cell types. Cell Biochem Funct, 13(4), 239–250. doi: 10.1002/cbf.290130404 [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. (1998). Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol, 203(1):36–48. doi: 10.1006/dbio.1998.9028 [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, . . . Chung WK. (2015). De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science, 350(6265), 1262–1266. doi: 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, Stoner R. (2019). Inference of CRISPR Edits from sanger trace data. bioRxiv, 251082. 10.1101/251082 [DOI] [PubMed]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, . . . Brueckner M. (2017). Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet, 49(11), 1593–1601. doi: 10.1038/ng.3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, . . . Schuldiner M. (2009). Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science, 323(5922), 1693–1697. doi: 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M, & Khokha MK. (2021). Obtaining Xenopus tropicalis Embryos by In Vitro Fertilization. Cold Spring Harb Protoc. doi: 10.1101/pdb.prot106351 [DOI] [PubMed]
- Marquez J, Bhattacharya D, Lusk CP, & Khokha MK. (2021). Nucleoporin NUP205 plays a critical role in cilia and congenital disease. Dev Biol, 469, 46–53. doi: 10.1016/j.ydbio.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez J, Criscione J, Charney RM, Prasad MS, Hwang WY, Mis EK, . . . Khokha MK. (2020). Disrupted ER membrane protein complex-mediated topogenesis drives congenital neural crest defects. J Clin Invest, 130(2), 813–826. doi: 10.1172/JCI129308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik ML, & Bronner ME. (2017). Regulatory Logic Underlying Diversification of the Neural Crest. Trends Genet, 33(10), 715–727. doi: 10.1016/j.tig.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Vedam LE, Brauning B, Popova KD, Schirle Oakdale NT, Bonnar JL, Prabu JR, . . . Weissman JS. (2020). Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. Elife, 9. doi: 10.7554/eLife.62611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Tsukaguchi H, Koshimizu E, Shono A, Matsunaga S, Shiina M, . . . Matsumoto N. (2015). Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am J Hum Genet, 97(4), 555–566. doi: 10.1016/j.ajhg.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir AM, Cohen JL, Sheppard SE, Guttipatti P, Lo TY, Weed N, . . . Mefford HC. (2020). Bi-allelic Loss-of-Function Variants in NUP188 Cause a Recognizable Syndrome Characterized by Neurologic, Ocular, and Cardiac Abnormalities. Am J Hum Genet, 106(5), 623–631. doi: 10.1016/j.ajhg.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, & Hubrecht-Laboratorium (Embryologisch Instituut). (1967). Normal table of Xenopus laevis (Daudin); a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis (2d ed.). Amsterdam,: North-Holland Pub. Co. [Google Scholar]
- O’Donnell JP, Phillips BP, Yagita Y, Juszkiewicz S, Wagner A, Malinverni D, . . . Hegde RS. (2020). The architecture of EMC reveals a path for membrane protein insertion. Elife, 9. doi: 10.7554/eLife.57887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiner T, Tomaleri GP, Januszyk K, Inglis AJ, Hazu M, & Voorhees RM. (2020). Structural basis for membrane insertion by the human ER membrane protein complex. Science, 369(6502), 433–436. doi: 10.1126/science.abb5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Boulin T, Robert VJ, Richmond JE, & Bessereau JL. (2013). Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc Natl Acad Sci U S A, 110(11), E1055–1063. doi: 10.1073/pnas.1216154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Ohba A, Liu Z, Inagaki T, & Satoh AK. (2015). dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. Elife, 4. doi: 10.7554/eLife.06306 [DOI] [PMC free article] [PubMed]
- Shao DD, Straussberg R, Ahmed H, Khan A, Tian S, Hill RS, . . . Walsh CA. (2021). A recurrent, homozygous EMC10 frameshift variant is associated with a syndrome of developmental delay with variable seizures and dysmorphic features. Genet Med, 23(6), 1158–1162. doi: 10.1038/s41436-021-01097-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Itzhak DN, Hussmann JA, Schirle Oakdale NT, Costa EA, Jonikas M, . . . Weissman JS. (2018). The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife, 7. doi: 10.7554/eLife.37018 [DOI] [PMC free article] [PubMed]
- Simoes-Costa M, & Bronner ME. (2015). Establishing neural crest identity: a gene regulatory recipe. Development, 142(2), 242–257. doi: 10.1242/dev.105445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, & Garcia-Castro MI. (2012). Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci, 69(22), 3715–3737. doi: 10.1007/s00018-012-0991-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, Kelsh RN, & Scholpp S. (2021). Review: The Role of Wnt/beta-Catenin Signaling in Neural Crest Development in Zebrafish. Front Cell Dev Biol, 9, 782445. doi: 10.3389/fcell.2021.782445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Wu Q, Zhou B, Choi MY, Ding B, Yang W, & Dong M. (2019). Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex. Cell Rep, 28(10), 2517–2526 e2515. doi: 10.1016/j.celrep.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umair M, Ballow M, Asiri A, Alyafee Y, Al Tuwaijri A, Alhamoudi KM, . . . Alfadhel M. (2020). EMC10 homozygous variant identified in a family with global developmental delay, mild intellectual disability, and speech delay. Clin Genet, 98(6), 555–561. doi: 10.1111/cge.13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmar N, Thezenas ML, Louie SM, Juszkiewicz S, Nomura DK, Hegde RS, . . . Christianson JC. (2019). The ER membrane protein complex promotes biogenesis of sterol-related enzymes maintaining cholesterol homeostasis. J Cell Sci, 132(2). doi: 10.1242/jcs.223453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman JG. (2015). The ubiquitous and ancient ER membrane protein complex (EMC): tether or not? F1000Res, 4, 624. doi: 10.12688/f1000research.6944.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Saint-Jeannet JP, & Klein PS. (2003). Wnt-frizzled signaling in neural crest formation. Trends Neurosci, 26(1), 40–45. doi: 10.1016/s0166-2236(02)00011-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.