Abstract
Background:
Short structural variants (SSVs) including indels are common in the human genome and impact disease risk. The role of SSVs in late onset Alzheimer’s disease (LOAD) has been understudied. Here we developed a bioinformatics pipeline of SSVs within LOAD-GWAS regions to prioritize regulatory SSVs based on the strength of their predicted effect on transcription factor (TFs) binding sites.
Methods:
The pipeline utilized publicly available functional genomics data sources including candidate cis-regulatory elements (cCREs) from ENCODE and single-nucleus (sn)RNA-seq data from LOAD patient samples.
Results:
We catalogued 1581 SSVs in candidate cis-regulatory elements (cCRE) in LOAD-GWAS regions that disrupted 737 TF sites. That included SSVs that disrupt the binding of RUNX3, SPI1 and SMAD3, within the APOE-TOMM40, SPI1 and MS4A6A LOAD-regions.
Conclusions:
The pipeline developed here prioritized noncoding SSVs in cCREs and characterized their putative effects on TF binding. The approach integrates multi-omics datasets for validation experiments using disease models.
1. NARRATIVE
1.1. Contextual background
Structural variants (SVs) are common in the human genome and were implicated in human traits and diseases including neurodegenerative diseases such as Late Onset Alzheimer’s Disease (LOAD) [1, 2]. The broadest class of structural variation includes deletions, duplications, large copy number variants, insertions, inversions, tandem repeats and translocations. While single nucleotide polymorphisms (SNPs) are limited to a single nucleotide base change and are primarily bi-allelic, SVs can be multi-allelic and repeated multiple times within the genome. Short structural variants (SSVs) are a subclass of SVs and include the same types of variation but are shorter in length, covering variation less than 50 bp[3]. A subset of SSVs include simple sequence/tandem repeats (SSRs/STRs); these are also known as microsatellites or short tandem repeats, including homopolymer stretches and indels. These variants are abundant in the human genome and one of the most polymorphic classes characterized by multiple alleles. Many SSVs have functional roles through which they mediate disease causality [4–6]. However, SSVs were not included in disease GWAS and expression quantitative trait (eQTL) studies mainly due to technological limitation in high throughput studies. Recent studies have suggested that SSVs are involved in many complex diseases and contribute to variation in gene expression in humans[4–7]. A structural variant (SV) map of 2504 human genomes showed that SVs are enriched in haplotypes identified by GWAS [8]. Other studies elucidated the role of short tandem repeats[4, 5] and extendable repeats[6] in human disease. It has been suggested that the effect of SVs on human diseases and traits is mediated via dysregulation of gene transcription[9–14], splicing[15], and translation. However, the roles of noncoding SSVs in human complex diseases, including LOAD, and specifically the mechanisms whereby SSVs regulate gene expression and exert their pathogenic effects, have yet to be discovered.
Examples of SSVs involved in neurodegenerative disorders include the Rep1 repeat site in the SNCA gene, involved in Parkinson’s disease[16–18] and the c9orf72 hexanucleotide repeat that is associated with frontotemporal dementia (FTD) and ALS[19, 20]. Recent research on ALS-linked genes have highlighted polymorphic STRs and SSVs that could influence disease risk and progression[21, 22] and showed that SSVs may account for some of the missing heritability for ALS[22]. For example, an indel variant within intron 5 of the SQSTM1 gene[23] and a poly-T repeat in the SCAF4 gene were associated with ALS [24]. Of note, the indel variant in the SQSTM1 gene was found to influence transcript levels and was significantly associated with familial ALS, especially in patients who had a superoxide 1 (SOD1) mutation [23]. Although the role of SSVs in LOAD has been understudied, examples such as the impact of the intronic poly-T variant in the TOMM40 gene and the impact of haplotypes in the APOE-TOMM40 region with LOAD risk and age of onset[25–31] suggest that systematic exploration of SSVs within LOAD risk regions will advance the understanding of the genetic architecture of LOAD.
Tagging-SNPs associated with LOAD risk are mainly located in noncoding regions that contain cCREs, suggesting that the actual causal variants have a regulatory role in gene expression. However, the regulated gene might be distal from the cCRE and interact via chromatin looping[32]. Gene-cCRE interactions are further complex as one cCRE may regulate more than a single gene and one gene can be regulated by several cCREs [33]. Thus, identification of regulatory variants and their target genes within LOAD associated regions has been a challenge in the field of LOAD genetics. Previously we developed a bioinformatics pipeline that characterizes and prioritizes candidate regulatory SNPs in cCREs within LOAD-GWAS regions[34]. Here we extend the bioinformatics pipeline to include SSVs, and developed a framework to catalogue a filtered set of candidate regulatory SSVs that have a predicted strong effect on TF binding. The outcomes prioritized candidate SSVs and transcription factors (TFs) for in vitro and/or in vivo validation in cellular models or animal models, respectively.
1.2. Study approach and findings
We aimed to catalogue candidate functional SSVs in LOAD-GWAS regions that impact transcriptional regulation by examination of their effects on TF binding affinities. Towards this goal we utilized public bioinformatics databases and specifically, recently available single nucleus (sn)RNA-seq data from LOAD brains and age-matched controls. In this paper, we present four example SSVs and their regulatory impact on specific TFs (Table 1) and provide data for all of the SSVs in LOAD-GWAS regions in the Supplemental Material. The example SSVs were chosen to optimize two different criteria: minimize distance between the SSV and the GWAS-SNP and/or to maximize the difference in the TF binding scores for the reference and alternate alleles.
TABLE 1.
Impact of specific SSVs on TF binding and relationship with tagging GWAS SNP for four example LOAD GWAS loci.
| SSV/Transcription Factor | ||||||||
|---|---|---|---|---|---|---|---|---|
| Transcription factor | Gene | Criteria | Binding loss or gain | Allele difference | MotifbreakR p value | RS number | Chr | Start |
| RUNX1 | SPI1 | large difference in allele score | - | −2.10 | 0.011 | rs4647710 | 11 | 47,216,593 |
| SMAD2 | APOE | distance to GWAS SNP | - | −1.54 | 0.010 | rs546328656 | 19 | 44,903,706 |
| FOXD3 | MS4A6A | distance to GWAS SNP | - | −0.99 | 0.003 | rs575848267 | 11 | 60,183,623 |
| TAL1 | FERMT2 | large difference in allele score | - | −2.99 | 0.020 | rs1277882435 | 14 | 52,951,884 |
| SSV/Transcription Factor | ||||||||
|---|---|---|---|---|---|---|---|---|
| Transcription factor | Gene | End | Width | Strand | REF | ALT | Variant type | motifPos |
| RUNX1 | SPI1 | 47,216,596 | 4 | + | TGTG | TG | Deletion | c(−2| 6) |
| SMAD2 | APOE | 44,903,720 | 15 | + | TCTCAAGTGTGTCTG | - | Deletion | c(9| 3) |
| FOXD3 | MS4A6A | 60,183,638 | 16 | + | (GTTT)4 | (GTTT)3 | Deletion | c(3| 11) |
| TAL1 | FERMT2 | 52,951,903 | 20 | + | AGATGATGCTCTTGCCTGAG | AG | Deletion | c(−3| 4) |
| SSV/Transcription Factor | GWAS | ||||||
|---|---|---|---|---|---|---|---|
| Transcription factor | Gene | SNP | major/minor allele | SNP location | GWAS SNP to variant (bp) | LD r2 | LD D’ |
| RUNX1 | SPI1 | rs3740688 | T/G | 47,358,789 | 142,197 | 0.0100 | 1.00 |
| SMAD2 | APOE | rs429358 | T/C | 44,908,684 | 4,979 | 0.0020 | 0.41 |
| FOXD3 | MS4A6A | rs7933202 | A/C | 60,169,453 | 14,169 | 0.0002 | 1.00 |
| TAL1 | FERMT2 | rs17125924 | A/G | 52,924,962 | 26,921 | 0.0006 | 0.43 |
The present study builds on our prior work where we developed a bioinformatics strategy to identify candidate LOAD causal genes in LOAD-GWAS regions based on evidence for 3D interactions between promoters in genes and active enhancer elements[35] and subsequent work to predict the impact of enhancer SNPs within LOAD-GWAS regions on transcription factor (TF) binding sites with the goal to catalogue and prioritize candidate LOAD functional SNPs[34]. The current study presents two major innovations and advancements in moving forward the prior work. First, the bioinformatics analysis specifically addresses SSVs compared to SNPs. Second, the bioinformatics pipeline integrates recent sources of data including single nucleus RNA sequencing (snRNA-seq) data from LOAD brains and age and sex matched controls and data from the expanded encyclopaedia of DNA elements (ENCODE) on cCREs. The code for the bioinformatics pipeline is publicly available and is extensible to cover both SNPs and SSVs.
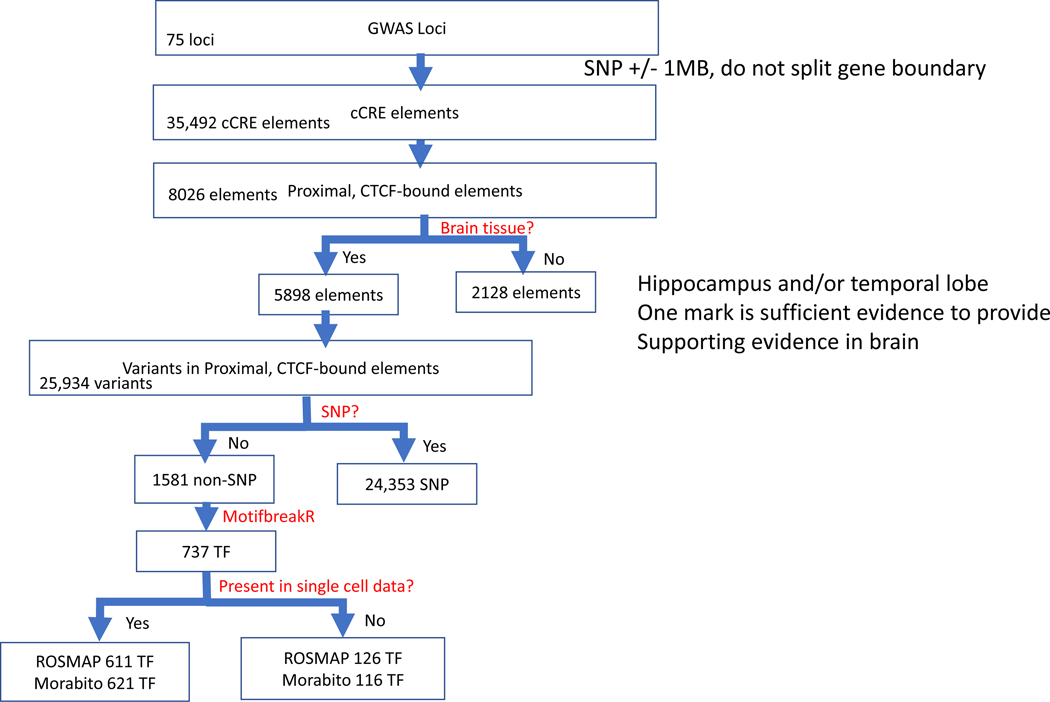
The overall strategy for the study is shown in Figure 1. Effectively, each step in the pipeline filters prioritized SSVs to proceed from the GWAS loci (broad range, ±1 Mb around the tagging SNP) to pinpoint candidate regulatory SSVs that impact binding affinities of LOAD relevant TFs and their linked genes. The version of the pipeline used for this study is focused on CCCTC-transcription binding factors (CTCF). CCCTC-binding factors are a family of highly conserved zinc finger proteins. The choice of CTCF binding sites is based on their ability to bind at chromatin domain boundaries, at enhancers and gene promoters, and inside gene bodies and their involvement in chromatin loops formation[36]. In addition, CTCF TFs can attract many other TFs to chromatin, including tissue-specific transcriptional activators and repressors[36]. The developed pipeline is built stepwise and is based on evidence of genomic attributes: (1) location of the SSV in a genomic locus, identified as associated with LOAD risk from large, consortium GWAS, (2) presence of the SSV in a proximal, CTCF-bound candidate cis-regulatory element, (3) evidence for at least one epigenetic mark in the hippocampus and/or temporal lobe, (4) evidence that the SSV disrupts TF binding, either gain or loss of binding sites, (5) evidence of the TF expression in snRNA-seq dataset, (6) identification of the TF as a differentially expressed gene (DEG) in LOAD. Each of these criteria could be relaxed or strengthened, depending on the number of SSV/TF hypotheses for further investigations. The number and size of specific GWAS loci could be expanded or reduced, the cCRE search could be expanded to more distal enhancer-like signatures and not restricted to CTCF, epigenetic marks could be expanded to more brain regions and types of marks, different stringency levels can be set for impact on TF binding, forthcoming larger snRNA-seq datasets could be used.
Figure 1. Schematic of the bioinformatics pipeline.
Flowchart illustrating the analytical scheme used to progress from SSVs located in candidate cis-regulatory element within LOAD GWAS regions to a filtered set of SSVs that have a predictive regulatory effect on transcription factor binding in LOAD-disease relevant tissues with supporting data from snRNA-seq data.
1.2.1. The utilization of the bioinformatics pipeline: Identification of candidate LOAD functional SSVs and the interacting TFs.
We demonstrated the utility of the bioinformatics pipeline with detailed examples for the SPI1 LOAD GWAS region interaction with the RUNX1 TF (Fig. 2) and for the MS4A2 LOAD GWAS region interaction with the FOXA3 TF (Fig. 3). We present these examples together since biologically, these genes are involved with immune system processes and microglial activation and function. We discuss our results with SSVs and TF binding in context with prior studies of these LOAD GWAS regions. We also present consolidated results for two additional regions to further illustrate the utility and range of applications of the bioinformatics pipeline: (1) APOE genetic locus and the SMAD2 TF (Supplemental Fig. 1), (2) FERMT2 genetic locus and TAL1 TF (Supplemental Fig. 2). These examples of the deletion type demonstrated the strongest evidence for SSV and TF pairs in LOAD regions from the entire catalogue examined in this study. Both the APOE and SPI1 loci have strong literature precedent and GWAS results supporting their involvement in LOAD and recent work suggested mechanistic role in the microglia[32, 37–40]. Relevant to our paper, several transcriptomic studies including snRNA-seq demonstrated that APOE and SPI1 are LOAD-DEGs upregulated in microglia[29, 41]. Here we showed specific deletions potentially modify TF binding which may in turn affect their expression in LOAD. APOE exemplified an SSV that is in close proximity to the GWAS tagging SNP, and SPI1 provides an example for the robust change in TF binding affinities between the alternative SSV alleles.
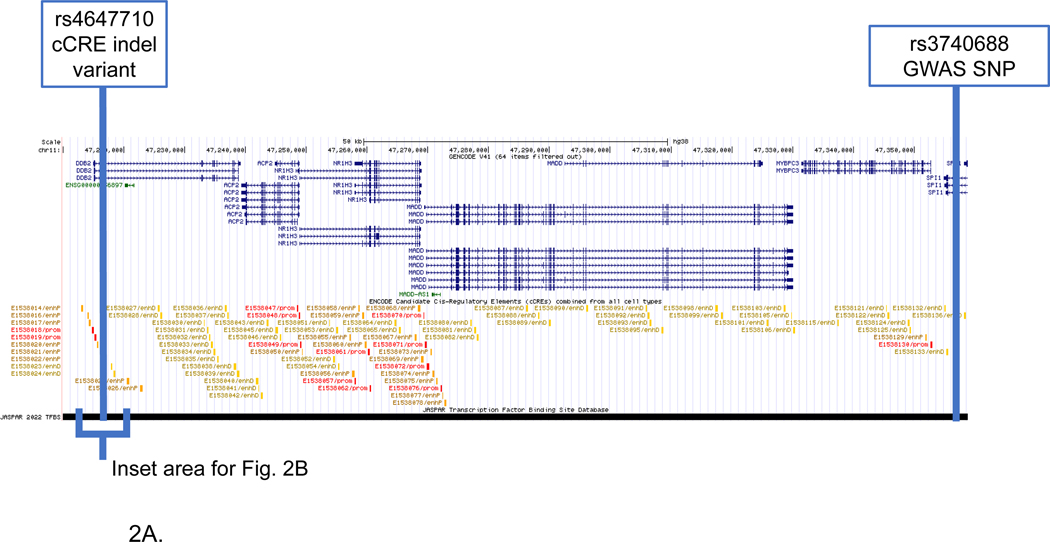
Figure 2. SPI1 LOAD GWAS locus.
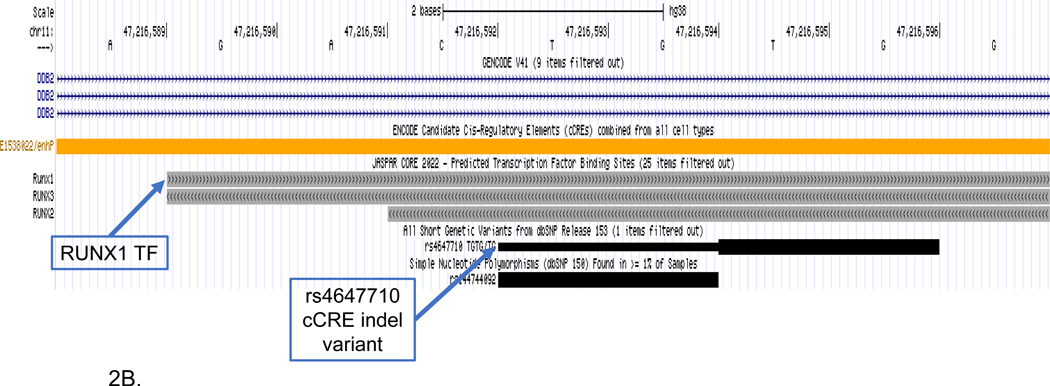
Genome browser view of SSV deletion variant (rs4647710) disruption of RUNX transcription factors. Tracks include (upper to lower): gene structure (GENCODE V41); candidate cCCREs (ENCODE) TFs from the JASPAR core collection; and SSVs from short genetic variants (dbSNP153). The deletion disrupts the RUNX1, RUNX2 and RUNX3 TFs. (A) Displayed genomic region includes the GWAS SNP (rs3740688) and the deletion variant (Rs4647710) located in the enhancer element (E1538022). (B) Inset that shows detail surrounding the enhancer SSV (Rs4647710).
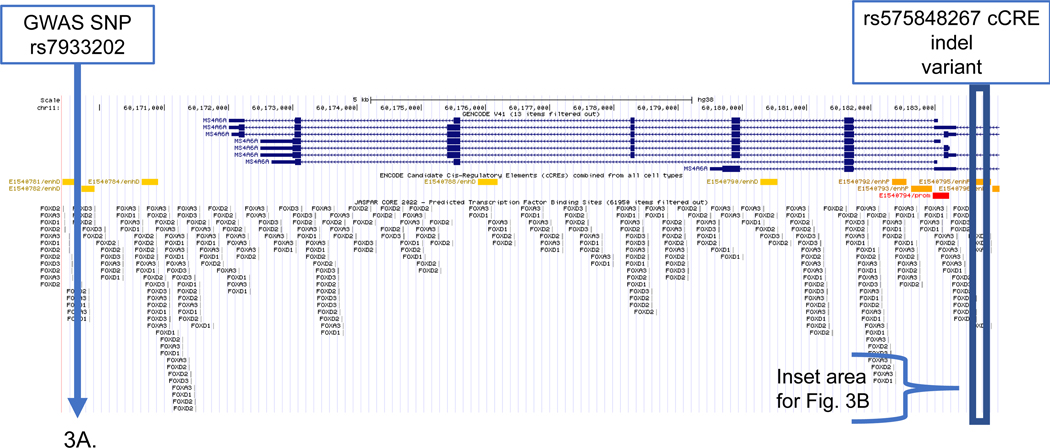
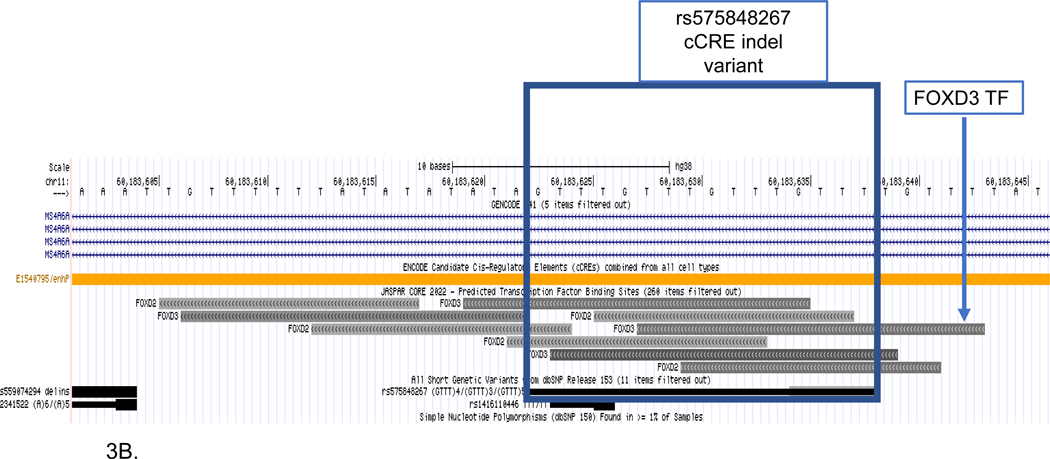
Figure 3. MS4A6A LOAD GWAS locus.
Genome browser view of SSV deletion variant (rs575848267) disruption of the FOXA3 transcription factor. Tracks include (upper to lower): gene structure (GENCODE V41); candidate cCCREs (ENCODE) TFs from the JASPAR core collection; and SSVs from short genetic variants (dbSNP153). The deletion disrupts the FOXA3 TF. (A) Displayed genomic region includes the GWAS SNP (rs7933202) and the deletion variant (rs575848267) located in the enhancer element (E1540795). (B) Inset that shows detail surrounding the enhancer deletion SSV (rs575848267).
1.2.2. Characterization of SSV impact on TF binding sites and prioritization of SSV/TF pairs
Table 1 provides a summary of SSVs effects on TF binding affinity for each of the four examples described above. Each row shows the results for a specific SSV/TF pair and indicates the corresponding GWAS locus and tagging SNP. The program MotifbreakR was used to calculate the relative entropy for the reference and alternative alleles of the SSV. The table reports whether the alternate deletion-allele results in a loss or gain of binding function and the binding affinity difference. Two additional criteria were used to select SSV/TF pairs for progression in the analytical pipeline: (1) the magnitude of difference in TF binding scores between the reference and alternate alleles of the SSV or (2) proximity to the GWAS SNP.
These two criteria are guidelines that can be used to select SSVs for validation studies, using genome editing approaches such as CRISPR/Cas experiments. However, the criteria are not mutually exclusive, thus, in a scenario where a strong impact on TF binding is the main criteria the selection would be based on the largest allele differences and therefore down weighting distance between the GWAS SNP and the SSV. Based on the MotifbreakR classifications for the effect size of the allele differences, values greater than 1.6 are considered very strong effects, greater than 0.7 are strong effects, less than 0.7 are weak effects. The distance between the GWAS SNP and the SSV that disrupts the TF is effectively a proxy for the genetic association between the variants or linkage disequilibrium (LD). The GWAS SNP effectively tags a region of the genome and the specific causal variants and/or gene may be proximal to the GWAS SNP or may be distal. In this study, we purposely expand the region around the GWAS SNP to include potential TFs that may be associated with LOAD but are not in strong LD with the GWAS SNP. We report the LD (D’ and r2) in addition to the distance between the GWAS SNP and the SSVs in the results tables. The correlation between distance (Kb) and LD has been extensively examined in human genetics with some general principles. For haplotypes, the swept radius, 1/ɛ, estimates the distance in kb at which association falls to approximately 1/3 of For disease haplotypes, the swept radius is estimated between 300 and 500 kb and for random haplotypes it is somewhat smaller than 300 kb[42]. More recently, LD at larger distances (1 centimorgan or approximately 1Mb), has been characterized at the chromosome-wide level[43] and epistasis at distances of long-range LD (>0.25 cM) has been reported for complex diseases including Alzheimer’s disease[44]. Noteworthy, the LD is also influenced by factors including allele frequencies of the variants and is often ancestry-specific.
1.2.3. SPI1 locus
Figure 2A visualizes the extended genomic region for the SPI1 gene including the RUNX1 TF binding site, GWAS tagging-SNP and location of the 2 bp deletion (rs4647710) in the E1538022 cCRE. Figure 2B shows the magnified region surrounding the deletion variant and the RUNX1 TF. The deletion is in predicted TFs of RUNX1, RUNX2 and RUNX3. We present this SSV-TF pair as an example for a relatively large (−2.10) difference in RUNX1 binding scores between the SSV alleles. The region was defined using the GWAS tagging-SNP in the SPI1 gene (rs3740688) as an anchor. However, the candidate SSV is distal (140kb) from the tagging-SNP and the LD between them was reduced (r2=0.01). The deletion is located in predicted cCREs for brain hippocampus middle, temporal cortex and prefrontal cortex (orange color blocks). Furthermore, epigenetic evidence for the E1538022 cCRE supports a regulatory role in brain tissue relevant to LOAD. There is a high signal for H3K4me3m, a histone mark that usually indicates an active gene promoter in hippocampus, temporal lobe and mid frontal cortex (Supplemental Table S1) and a high signal for H3K27ac, a marker for an active enhancer that activates transcription in the same brain tissues (Supplemental Table S1).
SPI1 encodes PU.1, a transcription factor that is critical for myeloid cell development and function[45]. Previous studies suggested that PU.1 may regulate the expression of multiple LOAD associated genes in myeloid lineage cells mainly microglia [45]. Overexpression and down regulation of PU.1 levels in mouse microglial cells affected the expression of mouse orthologs of several LOAD risk genes and the phagocytic activity. GWAS have shown that the SPI1 locus is associated with LOAD[46]. Fine-mapping approaches and integrative multi genomic analysis have nominated SPI1 as the most likely gene mediating the CELF1/SPI1 locus association with LOAD risk[32, 45].
The SPI1 genomic region demonstrated the complexity of the gene regulation network associated with LOAD. Previously, we identified a SNP, rs116371174 located in a predicted active enhancer adjacent to the SPI1 gene for three brain tissues, frontal cortex, temporal cortex and hippocampus [34]. Like the SSV deletion, this SNP disrupts the binding site of the TF RUNX3 and is also adjacent to the PU.1 binding site. Although the SNP was not predicted to affect PU.1 binding, the proximity of the two TF binding sites suggested a possible interaction between the TFs in this region that may have a biological consequence. Moreover, it was shown that the candidate LOAD SNPs within the myeloid cCRE modulated expression of several genes, suggesting regional network regulation of several genes with plausible contribution to LOAD risk [32].
1.2.4. MS4A6A locus
In the example of the SSV deletion within the MS4A6A locus, the GWAS SNP and the candidate SSV were relatively close (approximately 14kb), which was the main factor driving the selection of this candidate SSV/TF pair as a candidate for further evaluation. On the other hand, the SSV deletion showed a moderate (−0.99) difference in the predicted allele scores for the binding of the FOXA3 TF. Figure 3 shows the annotated genomic MS4A6A locus from the bioinformatics analysis.
The MS4A gene cluster encodes a family of cellular membrane spanning proteins with a similar protein sequence and with similar predicted topological structure, which have roles in signal transduction and regulation of cell activation and are highly expressed in microglia[39]. The MS4 family genes are involved as chemosensory receptors[39]. These genes are expressed in microglia and regulate cell activation[47]. A recent LOAD GWAS study reported significant association signal within 1.0 kb windows at MS4A6A and MS4A4A for CpG-related SNPs (CGSes) and identified a strong negative dosage effect of the CGSes on LOAD risk[48]. The window containing the MS4A4A CGSes was found to be associated with increased DNA methylation in brain and blood[48]. LOAD-associated SNPs within the MS4A locus are associated with lower MS4A4A and MS4A6A expression in myeloid cells, which confers a protective effect on LOAD[32, 38]. An example includes the functional SNP (rs636317) that was validated in hiPSC-derived microglia[38], It was shown that MS4As were highly expressed in microglia and peripheral immune cells[32], and suggested that these proteins function as lipid receptors and may interact with TREM2 [32, 49].
Several studies examined shared genetic etiology between LOAD and neuropsychiatric diseases including post-traumatic stress disorder (PTSD)[50] and major depressive disorder (MDD) [51]. These studies found that among the most significant shared loci were genes mapped on chromosome 11 from the MS4A gene family[50, 51]. We found an enrichment for SNPs in the MS4A gene family for LOAD across increasingly stringent levels of significance with PTSD GWAS association, e.g. statistical association for SNPs in the MS4A locus conditional with an association with PTSD or (LOAD|PTSD) in two independent cohorts and also found a modest enrichment for the reverse conditional association (PTSD|LOAD) for the SNPs in this locus[50]. African-American samples showed moderate enrichment for (LOAD|PTSD), however, no FDR-significant associations were reported[50]. Similar conditional associations between SNPs in the MS4A locus were reported for LOAD and major depressive disorder (MDD)[51]. Moreover, for the LOAD and MDD pleiotropy study, significant associations were observed between four SNPs clustered at the MS4A locus and mRNA levels of MS4A6A gene in whole blood and with proxy SNPs for mRNA levels of MS4A6A in monocytes[51]. A recent study identified two SNPs in the MS4A locus that were modifiers of AD risk, one protective (rs1582763) and one risk (rs6591561)[52]. This study reported a chemokine microglial subpopulation that is altered in individuals who carry the MS4A variants[52]. Moreover, the study identified MS4A4 as the major regulator and provided a mechanistic explanation for the AD variants in the MS4A locus[52]. Our study is complementary to this work in terms of providing details on SSV variants in this region and potential TF targets.
1.2.5. TF cell-type specific expression in LOAD
The results of the LOAD-associated differential expression analysis for the genes and TFs considered in this study are summarized in Figure 4 and Supplemental Table S2. We used two independent snRNA-seq datasets and reported results that showed FDR-significance for LOAD associated differential expression by both datasets (Table 2). The replication by two independent snRNA-seq datasets provided a strong statistical rigor of the developed pipeline. An FDR-corrected p value ≤ 0.1 for differential expression by LOAD diagnosis compared with controls and an absolute value of log2(fold change) ≥ 0.2 was used to determine a differentially expressed gene (DEG).
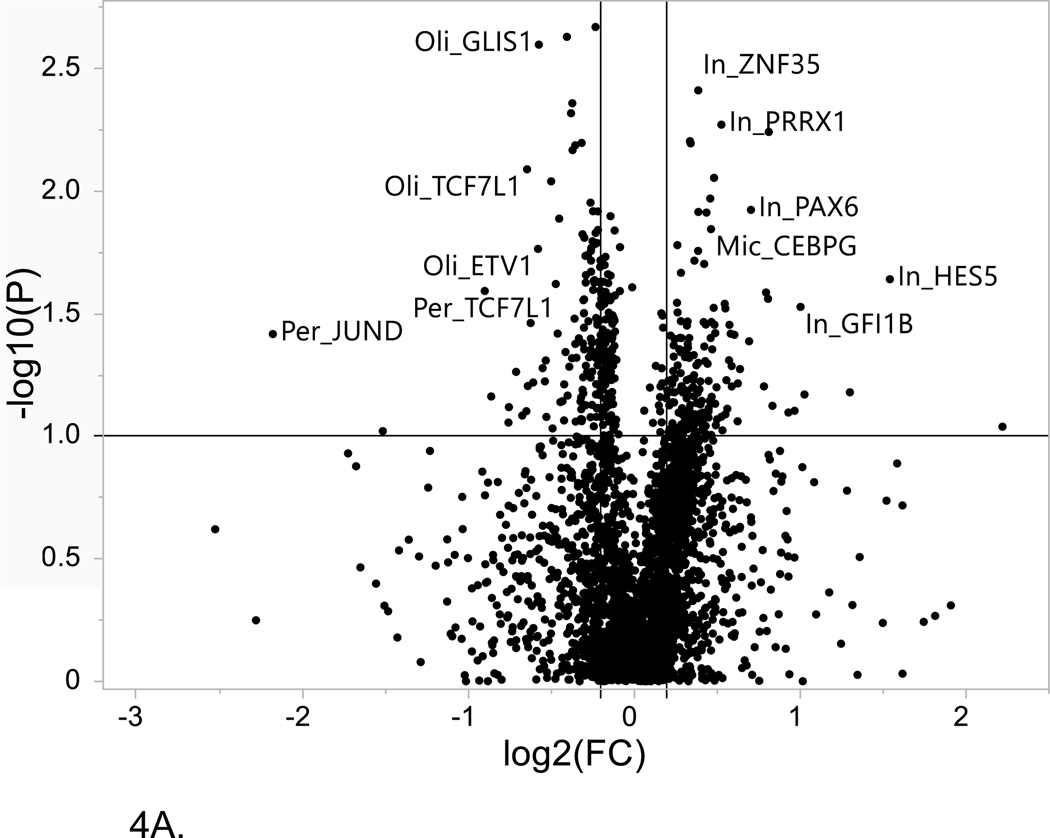
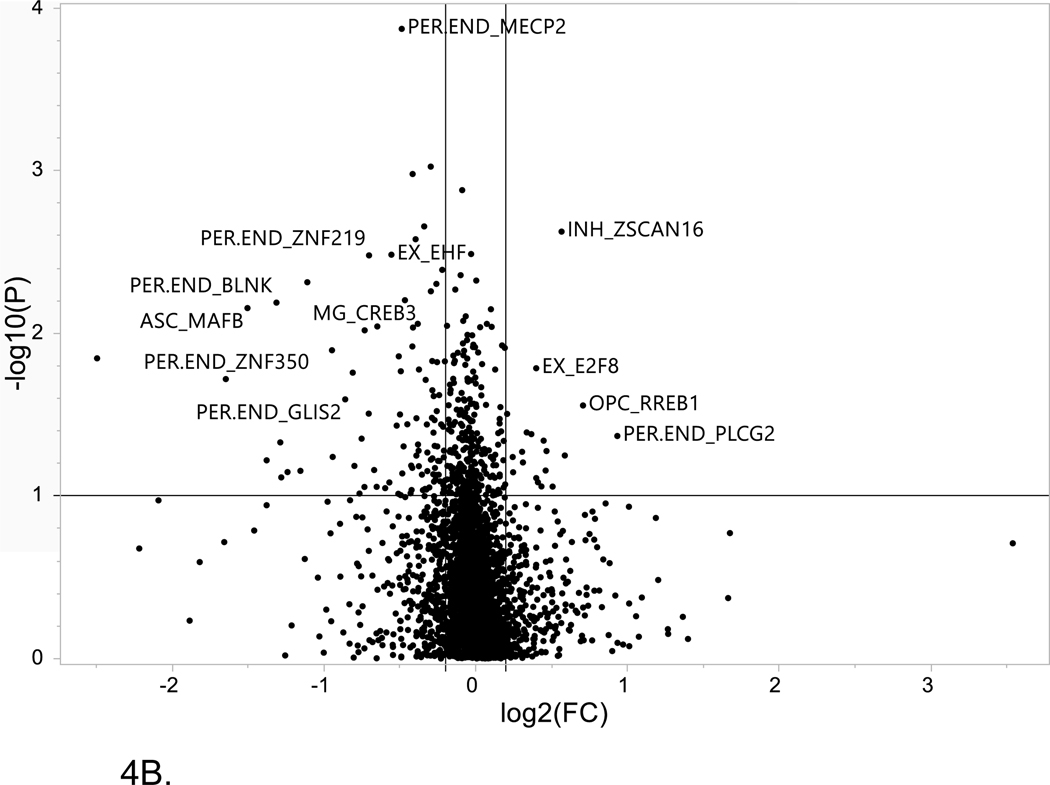
Figure 4. Volcano plot of single cell LOAD data.
This plot is based on the fold change differences in the snRNA-seq data for the genes and transcription factors identified in the bioinformatics analysis for all of the LOAD GWAS regions. The fold change is calculated as the difference between late-stage AD (LOAD) and age-matched cognitively healthy controls (74–90+years old). The plot shows the relationship between fold change (log2 fold change (FC); horizontal axis) and FDR-adjusted statistical significance (−log10 (p value); vertical axis), respectively. The horizontal line corresponds to a cut-off of FDR P value ≤ 0.1, vertical lines correspond to log2 fold change thresholds of ± 0.2. (A) ROSMAP data analysis. Cell type abbreviations: Oli-oligodendrocytes, In-inhibitory neurons, Mic-microglia, Per-pericytes (B) Sample analysis from Morabito et al. data[40] Cell type abbreviations: INH-inhibitory neurons, EX-excitatory neurons, ASC-astrocytes, MG-microglia, PER-END-pericytes/endothelial cells, OPC-oligodendrocyte progenitor cells.
TABLE 2.
Single-cell differential expression results for genes and transcription factors identified by the bioinformatics pipeline.
| Cell type | Gene | log2(FC) ROSMAP | log2(FC) Morabito | FDR p value ROSMAP | FDR p value Morabito |
|---|---|---|---|---|---|
| Astrocytes | APOE | 0.35 | −0.30 | 0.082 | 0.060 |
| Astrocytes | IRF7 | 0.60 | −0.22 | 0.039 | 0.038 |
| Astrocytes | PPARG | 0.26 | −0.42 | 0.051 | 0.001 |
| Astrocytes | TBX2 | 0.78 | −0.20 | 0.063 | 0.015 |
| Astrocytes | TP63 | 0.83 | 0.59 | 0.075 | 0.057 |
| Astrocytes | VAX2 | 0.53 | 0.46 | 0.095 | 0.070 |
| Excitatory neurons | GLIS3 | 0.31 | −0.21 | 0.087 | 0.089 |
| Pericytes | ATF4 | −0.86 | −0.42 | 0.069 | 0.067 |
| Pericytes | JUND | −2.18 | −0.27 | 0.038 | 0.068 |
Six of the 9 gene/TFs were DEGs in astrocytes for both snRNA-seq datasets. Notably, both APOE and PPARG are found to be differentially expressed in astrocytes between LOAD cases and controls. Numerous studies have considered the role of PPARG in the development of Alzheimer’s disease[53–55] and indeed PPARG agonists have been evaluated as potential repurposed drugs to delay the onset of LOAD[56, 57]. IRF7 is a TF that is a master regulator of type I interferons after activation by pathogen recognition receptors and was reported to be associated with decreased levels in LOAD brains relative to controls; supporting a hypothesis that the innate immune system is impaired in LOAD. Studies have supported the role of IRF7 in activation of interferon pathways as part of the neuroinflammatory response to brain amyloidosis and showed an association of IRF7 expression with neuritic plaque burden, clinical dementia rating and Braak score[58] and described IRF7 regulation of type-I IFN-mediated immune suppression in AD and tau-associated neurodegenerative diseases including LOAD[59]. Although these gene/TFs show a statistically-significant log2 fold change ≥ 0.2, for 5 of the 9 comparisons between the datasets, there is a different direction of effect. Further studies with larger samples sizes are needed to confirm the direction of change in expression levels with respect to clinically-relevant phenotypes..
1.3. Limitations, future studies and disease implications
Regulation of gene expression is complex and controlled by several pre- and post- transcriptional mechanisms. In this work we focused on the affinities of TFs for their corresponding binding sites which represents only one facet of the gene regulation process. Furthermore, we investigated the relationships between a particular SSV and TF, while it is likely that multiple genetic variants and TFs contribute in concert to changes in the transcription of key genes in LOAD. Another limitation is related to the parameters of our bioinformatics pipeline. While we considered the impact of changing parameters on the number of candidates SSVs and TFs, specific sensitivity to parameters including cCRE type needs to be determined. Finally, although the results presented were for cases with a reference and an alternate allele, the extension to SSVs offers the opportunity to evaluate multi-allelic variants. Currently, the MotifbreakR algorithm is limited to the evaluation of bi-allelic variants, however, our bioinformatics pipeline is easily extended to multi-allelic variants by designating one allele as the reference and repeatedly running the MotifbreakR algorithm. Future work will expand the analysis for the LOAD GWAS regions beyond biallelic, short insertions, deletions and indels to larger classes of SSVs. These SSVs are often polymorphicMotifbreakR. Finally, the effect of different ancestral genetic backgrounds should be included in future analysis, notably as results for GWAS for cohorts of individuals from diverse and under-represented ancestries are completed.
The current study developed a bioinformatics pipeline that uses genomic attributes to catalogue and prioritize candidate SSV and TF pairs. The pipeline employed in silico tools and existing datasets to generate evidence and assess each SSV/TF pair. Next steps will be essential to test experimentally the specific alleles of the SSVs using gene editing techniques such as CRISPR/Cas technologies. Initial studies would focus on generating isogenic hiPSC lines for the candidate SSVs identified by our bioinformatics analysis and evaluatiom of their cis regulatory effects in the respective brain cell-type by differentiation into microglia, astrocytes and neurons. The advantage of using isogenic hiPSC-derived models is that only the SSV is modified on the same genomic background allowing direct evaluation of the SSV function. These studies would confirm the effect of the SSV on gene transcription, the linked target gene, and the TF that mediate the effect. Follow up analyses could use 3D models including organoids and co-cultures to further examine impact on downstream cellular mechanisms, neurodegeneration and other disease perturbations, The knowledge gained by the combined computational and experimental approach will provide the foundation for the development of new emerging actionable therapeutic targets for prevention and/or treatment of LOAD. Also, as more single cell multi-omics datasets from LOAD became available we could apply the bioinformatics pipeline to study in depth cell-type and subtype LOAD-associated changes in gene regulation driven by cis-trans interactions. Future work will consider the complexity of gene regulation networks including, more distal enhancers governed by SNPs and SSV and crosstalk between cCREs and gene promoters.
2. CONSOLIDATED METHODS AND RESULTS
2.1. Consolidated Methods
2.1.1. Bioinformatics analysis
The schematic for the bioinformatic pipeline is shown in Figure 1 with the corresponding number of elements identified at each step provided.
2.1.2. Sample demographics
Sample demographics, as described in the primary studies that provided data for each of the bioinformatics analysis steps including the original GWAS study, evidence for brain tissue and single cell RNAseq data are provided in Table 3. All individuals in Stage 1 of the original AD GWAS were of European ancestry[46]. For the single cell RNAseq data, individuals were also primarily of European ancestry, Sufficiently powered GWAS from multi-ancestry populations will be essential to develop a clear understanding of the genetics of AD, to provide the detailed annotations of SSVs and TFs and to elucidate which genomic properties apply generally across ancestries and which are unique.
TABLE 3.
Sample demographics.
| GWAS or proxy-LOAD cases | GWAS controls | Brain tissue evidence | ROSMAP single cell cases | ROSMAP single cell controls | Morabito single cell cases | Morabito single cell controls | |
|---|---|---|---|---|---|---|---|
| N | 111,326 | 677,663 | 9 | 24 | 24 | 12 | 8 |
| Age mean (SD) | 76.8 (6.6) | 67.6 (10.9) | 77.7 (3.5) | 85.7 (4.2) | 85.6 (4.3) | 87.7 (3.6) | 81.6 (5.7) |
| Age of AD onset mean (SD) | 78.1 (4.8) | - | - | - | - | - | - |
| females % | 63 | 54 | 33 | 50 | 50 | 50 | 38 |
| APOE e4 allele frequency % | 30 | 14 | - | 32 | 14 | 33 | 0 |
2.1.3. Cataloguing SSVs in LOAD defined cCREs
For the current study, the recent LOAD GWAS data reported by Bellenguez et al.[46] defined the LOAD-associated loci. The approach for identifying active enhancers in LOAD GWAS regions is described in detail in Lutz et al.[35]. In brief, the region tagged by each LOAD-SNP was initially defined by anchoring the center of the region on the GWAS SNP and extending 500kb in each direction to cover a 1Mb locus. The GWAS SNP effectively tags a region of the genome, the specific SSVs may or may not be in LD with the GWAS SNP, however, the bioinformatics analysis is designed to find SSVs that cause a disruption (gain or loss) in specific TFs in the genomic region. Using a 1Mb range is a conservative boundary based on studies to predict the range of linkage disequilibrium (LD) for mapping disease genes[42]. Genes on the boundary of the 1Mb region were examined and the locus extended to cover the full length of the gene if the boundary intersects within a gene. Alternative methods for definition of LOAD GWAS-associated regions to search for active enhancers include using LD blocks and/or selection of specific enhancer types (e.g. proximal, distal or promoter).
The extended GWAS regions were used to search the ENCODE Registry (GENCODE V24) of candidate cis-regulatory elements (cCREs)[60] for proximal, CTCF-bound cCREs. This registry includes 926,535 cCREs for 839 cell types. We downloaded all human cCREs using the ENCODE Screen tool (https://screen.encodeproject.org/) and tested for evidence (designated as high expression) of at least one epigenetic mark (H3K4me3, H3K27ac) in specific brain tissues relevant to LOAD (hippocampus, temporal lobe, mid frontal lobe, astrocytes in the hippocampus, astrocytes in the cerebellum). From 34,492 cCRE elements, 8026 are filtered as proximal, CTCF-bound and 5898 had at least one epigenetic mark providing supporting evidence for a role in the brain.
SSVs located within the enhancers were catalogued using the UCSC Table Browser[61, 62] to load data for the Thousand Genomes Project, Phase 3. This dataset contains 73 million single nucleotide variants (SNVs) and 5 million short insertions/deletions (indels) produced by the International Genome Sample Resource (IGSR) from sequence data generated by the 1000 Genomes Project in its Phase 3 sequencing of 2,504 genomes from 16 populations. Importantly, this is an extensive source of indel annotation where the variant genotypes are phased with a designated reference and alternate alleles which is the information required for analysis of transcription factor binding. There were 1581 SSVs identified in the 5898 cCRE elements.
2.1.4. Predicting TF binding sites affected by SSVs in LOAD cCREs
Prediction of TF binding sites was completed for the 1581 SSVs. The software package/algorithm MotifbreakR[63] was used to estimate or predict whether the sequence surrounding a SSV matches to specific TF binding sites, and how one allele of the SSV relative to the other affects the strength of the TF binding site (gain or loss of the TF binding affinity). MotifbreakR can predict effects for novel or previously described variants in public databases. For our study, we utilized the information content (ic) algorithm and position weight matrices from Homer, HOCOMOCO, Factorbook and ENCODE.
Each SSV from the catalogue we generated for LOAD-GWAS enhancers filtered by the prior steps was evaluated for the potential to disrupt/gain TF binding sites using a predicted p value < 1× 10-4. The choice of the 1× 10−4 threshold is a first level filtering parameter recommended in the MotifbreakR user manual[64] for the p value for the position weight matrix match to the sequence. All p values for the sequence match will be at this level or lower with the final p values for the reference and alternate allele scores reported after a second step of p value calculation for the resulting set of TFs. The SSVs were evaluated for impact on specific TF binding with calculation of a permutation p value, score for impact on binding and assessment of loss or gain of a binding site based on the MotifbreakR calculations. There were 737 TFs identified by the MotifbreakR analysis.
2.1.5. Evaluation of candidate TFs and their binding sites in snRNA-seq data for LOAD and control brain samples.
The candidate TFs from the bioinformatics analysis were evaluated in single cell RNA-seq (snRNA-seq) data from LOAD and control brain samples to interrogate expression in specific brain cell subtypes (astrocytes, microglia, excitatory neurons, inhibitory neurons, oligodendrocytes, oligodendrocyte progenitor cells and pericytes/endothelial cells).
To provide a replication framework, two snRNA-seq datasets were used to evaluate expression of the candidate TFs and corresponding genes.
The first sample was obtained from a public dataset made available by the Swarup Lab (https://swaruplab.bio.uci.edu/singlenucleiAD/), that is described in detail in Morabito et al.[40]. The data was downloaded from Synapse (Synapse ID: syn22079621). In short, single nuclei suspensions were isolated from ~ 50mg frozen human prefrontal cortex, as described in [40, 65]. Nuclei were FACS sorted with DAPI (NucBlue Fixed Cell ReadyProbe Reagent, Cat#R37606, Thermo) before running on the 10x ChromiumTM Single Cell 3’ v3 platform. cDNA library quantification and quality were assessed as in bulk RNA-seq. Libraries were sequenced using Illumina Novaseq 6000 S4 platform at the New York Genome Centre, using 100bp paired-end sequencing. Detailed demographic and technical information on the samples is provided in Morabito et al.[40]. There were 20 samples analyzed, 12 AD, 8 control samples, age matched across diagnoses (Table 3).
The second sample was taken from a subset of the Religious Orders Study and Rush Memory and Aging Project (ROSMAP) dataset [14,22–24]. ROS has enlisted nuns and brothers since 1994. MAP has recruited individuals from the northern Illinois region since 1997. Both studies were run by the same investigators using similar data collection techniques. Thus, the results from both were comparable. The ROSMAP snRNA-seq dataset is described in detail in Mathys et al.[66]. The data was downloaded from Synapse (Synapse ID: syn16780177). In short, single nuclei were isolated from the prefrontal cortex (Brodmann area 10) and profiled by RNA-sequencing using the DroNc-seq protocol[67], modified to work on the 10x Genomics Chromium platform. This approach uses droplet technology for high-throughput, massively parallel single-nucleus RNA-sequencing, and is suited for profiling cells from tissues that cannot be easily dissociated (human brain, for example), or from samples that have been previously frozen. Detailed information on the samples and the snRNA-seq analysis are provided in Mathys et al.[66]. There were 48 samples analyzed, 24 clinically-diagnosed AD, 24 controls, age- and sex-matched across diagnoses (Table 3).
Differential expression analysis to calculate log2(Fold Change) and FDR-adjusted P values for the genes and TFs identified from the bioinformatics analysis were calculated using the approach described in the Seurat program[68].
2.1.6. Genome version and coordinates
All genomic data and coordinates are based on the December 2013 version of the genome: hg38, GRCh38.
2.2. Consolidated Results
Results for two additional examples, APOE and FERMT2 loci were reported in this section. Table 1 reports the detailed bioinformatics analysis for these loci and full analysis results were reported for all of the LOAD-GWAS loci in supplemental table S3.
2.2.1. APOE Locus
The association between APOE ε2/3/4, determined by two coding SNPs, and LOAD risk and age of onset have been well established. In recent years accumulating evidence has suggested that specific haplotypes across the APOE locus have regulatory roles that also contribute to LOAD risk and pathogenesis. We identified an SSV deletion variant (rs546328656) that disrupts the SMAD2 TF (Supplemental Figure 1). The SSV is positioned in a genomic region proximal to both APOE and its nearby TOMM40 gene. The distance between the SSV deletion and the APOE ε4 coding SNP (rs429358) is approximately 5Kb, thus, in high LD with variants across the APOE and TOMM40 region. This example illustrated the criteria of proximity between the LOAD SNPs (APOE isoforms defining SNPs) and the SSV as an important factor for prioritizing candidate SSV/ TF pairs for further evaluation.
2.2.2. FERMT2 locus
The identified SSV deletion within the FERMT2 locus showed a large difference (−2.99) in the predicted binding allele scores for the TAL1 TF. Thus, this is another example where the magnitude of difference in TF binding scores was the main factor driving selection of the candidate SSV/ TF pair. The annotated genomic locus plot for FERMT2 is provided in Supplemental Figure 2.
2.2.3. Other LOAD GWAS loci
Supplemental Table S3 shows the complete set of results for all of the LOAD GWAS loci. We found 1581 unique SSVs in the LOAD GWAS regions and 737 TFs are identified by the MotifbreakR analysis. Overall the bioinformatics pipeline identified 7587 combinations of SSVs and TFs pairs; 5934 showed strong effects on TF binding affinities and 1653 showed weak effects. For three of the 75 LOAD GWAS loci (EPHA1, PTK2B and CLU) the pipeline did not identify any SSV/TF pair.
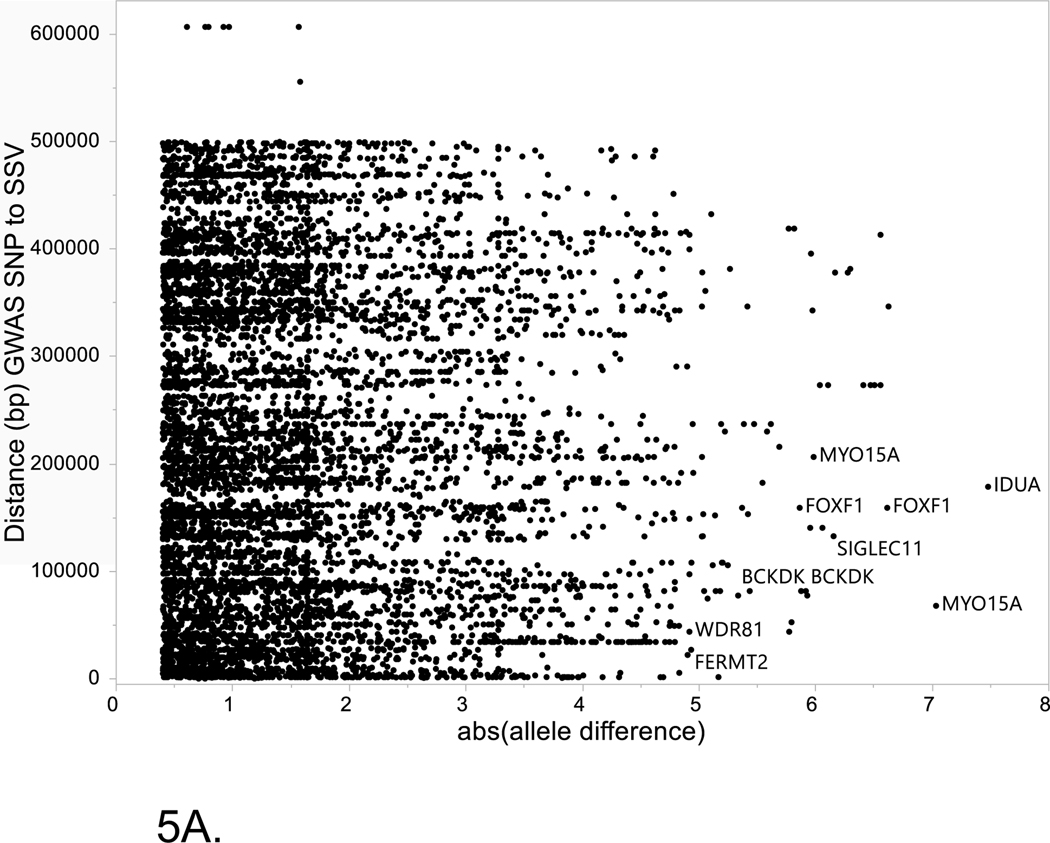
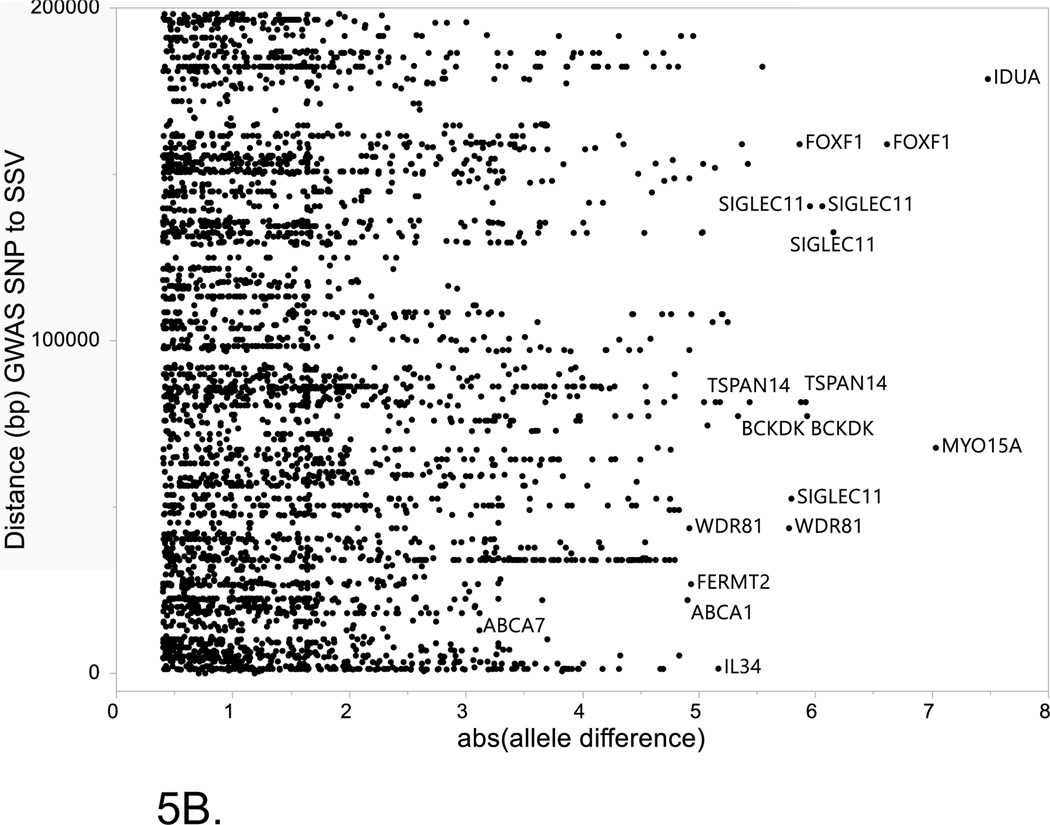
The two major alternative evidence supporting the prioritization of SSV/TFs pairs within LOAD risk loci are: (1) close proximity between the SSV and TF binding site to the GWAS tagging SNP, (2) the magnitude of the differences in binding potential for the alternative alleles of the SSV. Figure 5 shows the relationship between these criteria for all of the SSVs listed in Supplemental Table S3. Of note, allele differences greater than 3 were found for several SSVs in the ABCA7 locus and these also showed close proximity to the GWAS SNP. In contrast, many of the entries in Supplemental Table S3 were distal to the GWAS SNP by over 100k, thus, unlikely in LD with the GWAS SNP.
Figure 5. Relationship between absolute value of the allele difference for the reference and alternate alleles of each SSV and the distance (bp) between the GWAS SNP and the SSV.
Horizontal axis is the absolute value between the reference allele and the alternate allele for disruption of TF binding as estimate by MotifbreakR. Vertical axis is the distance (bp) between the GWAS SNP and the start of the SSV. (A) Entire set of data for all SSVs analyzed. (B) Inset that is restricted to data where the distance between the GWAS SNP and SSV is ≤ 200Kb.
2.2.4. Bioinformatics pipeline computational and performance details
The bioinformatics pipeline is comprised of a number of steps which involve downloading data from public repositories using bioinformatics software (UCSC Table Browser, ENCODE Screen Tool, Synapse for snRNA-seq data and performing the operations detailed in Figure 1 and in the Consolidated Methods section for each level of analysis. The most computationally-intensive steps are the MotifbreakR analysis and the single cell analysis using the Seurat program; both of these analyses were run using R version 4.1. Statistical analysis for reporting and summarizing the results was done using JMP (version 16.2.0, SAS Institute, Cary NC) and SAS (version 9.4, SAS Institute Cary). A visual basic macro is available on the public resource site that maps SSVs from the MotifbreakR output to the input GWAS genomic regions. Overall elapsed time from starting with a list of loci to completion of the MotifbreakR analysis, prior to single cell analysis, is about 30 minutes with time dependent primarily on length of time to download the specific data needed for each step and formatting of the files for each part of the analysis.
The two major computational steps, MotifbreakR analysis and single cell analysis (Seurat) were run on two computational platforms, a Windows 10 PC, intel i-7 6600U 2.6 Ghz processor with 15Gb RAM and on a local linux compute cluster with 1300 nodes, 30,000 VCPUs and 200Tb RAM. The run times for MotifbreakR strongly depends on the number of input SSVs. For 10, 20 and 40 SSVs, the run times were 2, 5 and 8 minutes on the Windows PC. MotifbreakR run times were similar for the linux compute cluster, although the 40 SSV run was slightly faster at 6 minutes. The MotifbreakR program utilizes the R package TFMsc2pv[69] to compute p values for scores for the reference and alternate alleles based on the specific position weight matrices. This step can be a very memory and time intensive process if the algorithm doesn’t converge rapidly. We set the “granularity” parameter to improve the speed of computation without sacrificing accuracy of the p values. We used a value of 1×10−4 as recommended in the MotifbreakR user manual[64] as a compromise between computational time and p value accuracy. We used the information criteria as the method for scoring probabilities for the different positions in the sequence; for this approach, the position-specific scoring matrix is created using a scoring method that directly weights the score by the importance of the position within the match sequence in contrast to a simple sum of the log probabilities for each position within the sequence.
The run times for the Seurat, single cell analysis are typical for single cell experiments, however, since a subset of the full dataset was used to limit the analysis to the specific genes and TFs of interest identified by the bioinformatics pipeline analysis, run times were shorter than for the entire set of genes. For the ROSMAP dataset (48 samples), run times on the linux cluster were approximately 6 minutes while on the PC run times were on order of 12 minutes.
To show the impact of the number of genomic regions that are used as the primary input for the bioinformatics pipeline, Supplemental Table 4 shows the number of elements (cCREs, SSV variants, TFs) for 24 and 75 regions respectively. The approximately 3-fold increase in number of regions results in approximately a 2-fold increase in number of TFs identified as disrupted by the SSV variants while the number of elements at different steps ranges from 2-fold to 7-fold with the greatest increase for the number of cCREs. Other aspects of the bioinformatics study design included decisions on the inclusion of SNPs in addition to SSVs, more distal cCRE types and extensions to include testing in other types of brain tissue.
Selection criteria at several steps will have a significant impact on the number of elements (cCRES, SSVs, TFs) considered at each step. Moreover, the bioinformatics pipeline can be used to consider both SNPs and SSVs in the same analysis for a set of genomic regions. For both SNPs and SSVs, minor allele frequency can be used as an effective filtering step. Supplemental Table 5 shows the number of different types of variants (SNPs, SSVs) at two MAF levels: MAF > 0.01 and unfiltered (e.g. includes rare variation). This Table shows the number of variants by the two levels of MAF filtering across all of the AD GWAS regions and for the APOE-TOMM40 region (chr19:44,892,743–44,909,743). The increase of number of variants varies greatly by type, there are approximately 50 times as many SNPs when the MAF > 0.01 restriction is removed for both the AD GWAS regions and the APOE-TOMM40 region. For the AD GWAS regions, removing the MAF > 0.01 restriction results in about 8 times as many insertions and 16 times as many deletions. The decision to use a filter based on MAF largely centers on whether rare variation is to be included and the threshold for detection of common variation (e.g. greater than 1% or 5%). Rare variants near the APOE region have been reported to be associated with CSF and neuroimaging biomarkers of Alzheimer’s disease and were hypothesized to account for some of the missing heritability of Alzheimer’s disease not explained by the common variation assessed by GWAS[70]. Analysis of rare variation from whole-genome sequencing datasets also revealed two novel Alzheimer’s disease-associated genes: DTNB and DLG2[71].
The public resource (https://github.com/NCTrailRunner/Alzheimer-SSV-TF-analysis) provides datasets for each step of the bioinformatics pipeline for investigators to use any aspect of the datasets for further analysis and for the design of follow up bioinformatics or experimental studies.
Supplementary Material
Systematic review:
The authors reviewed the literature using Pubmed, meeting abstracts and presentations, selected computational tools and downloaded publicly available datasets. Short Structural Variants (SSVs), including indels are common multi- allelic variants in the human genome and impact substantially human traits and diseases. The role of indels in brain diseases including late onset Alzheimer’s disease (LOAD) has been understudied. Previously we developed a bioinformatics pipeline that characterizes and prioritizes candidate regulatory SNPs in enhancers located in LOAD-GWAS regions. Here we extend the pipeline to the analysis of SSVs. The developed bioinformatics pipeline progresses from SSVs located in LOAD-GWAS regions to a filtered set of candidate regulatory SSVs that have a predicted strong effect on transcription factor (TFs) binding sites.
Interpretation:
This study provides an analytical framework to catalogue and prioritize noncoding indel variants in candidate cis-regulatory elements (cCREs) located in LOAD-GWAS loci and characterize their putative effects on TF binding sites. This study extended prior work with cCRE within LOAD-GWAS regions to include SSVs and to rank the top candidate SSV/TF pairs for validation experiments. The bioinformatics pipeline was utilized to characterize several LOAD-GWAS loci including SPI1 and APOE.
Future directions:
Future work will focus on testing experimentally the effect of the different SSV alleles using gene editing techniques such as CRISPR/Cas technologies. Initial studies would focus on generating isogenic hiPSC lines for the candidate SSVs identified by our bioinformatics pipeline and evaluate their cis regulatory effects in the respective brain cell-type by differentiation into microglia, astrocytes and neurons. These studies would confirm the effect of the SSV on gene transcription, the linked target gene, and the TF that mediate the effect. Follow up analyses could use 3D models including organoids and co-cultures to further examine impact on downstream cellular mechanisms, neurodegeneration and other disease perturbations. This knowledge would be essential for the development of new therapeutic targets for prevention and treatment of LOAD. Future computational biology work will consider the complexity of gene regulation networks including, more distal enhancers governed by SNPs and SSV and crosstalk between cCREs and gene promoters.
Acknowledgements
We thank all the participants of ROS and MAP studies. These studies were funded by the National Institute on Aging: P30AG010161 ADCC, P30AG072975 ADRC, R01AG015819 RISK, R01AG017917 MAP, U01AG46152 AMP-AD Pipeline I, U01AG61356 AMP-AD Pipeline II.
Grant Funding:
This work was funded in part by the National Institutes of Health/National Institute of Neurological Disorders and Stroke – NIH/NIA (R01 AG057522-01 to O.C.-F. and RF1 AG077695 to O.C.-F). The funding source did not have a role in the design of the study, collection of data, analysis, interpretation of the data or writing of the manuscript.
Grant funding for the past 36 months. O.C.-F: 1. SRA 2020 Chiba-Falek, Kantor (MPI) 08/26/2020 – 02/25/2024 Seelos Therapeutics, 2. 22-AAIIA-953269, Alzheimer’s Association Ornit Chiba-Falek (PI), 3. 1RF1-NS113548-01A1 Chiba-Falek (PI) National Institutes of Health 4. R56-AG062344 Chiba-Falek (subaward PI) Wang, 5. 1R56-AG062302 Lutz, Chiba-Falek, Luo Williamson National Institutes of Health/NIA, 6. 1R01-AG057522 Chiba-Falek, Lutz (MPI) National Institutes of Health/NIH. 7. RF1 AG077695, Chiba-Falek (PI) National Institutes of Health, 8. R41AG077992 Chiba-Falek (PI) National Institutes of Health, 9. MJFF-021362, Chiba-Falek, Kantor (MPI) Michael J. Fox Foundation for Parkinson’s Research.
Michael W. Lutz: 1. R01AG057522 Chiba-Falek (PI) National Institutes of Health, 2. RF1-AG057895 Lutz (co-PI) National Institutes of Health, 3. R01-AG066184 Badea (PI) National Institutes of Health, 4. R01-AG064803-02 Luo (PI) National Institutes of Health, 5. P01-AG031719 Vaupel (PI) National Institutes of Health, 6. R01-ES024288 Plassman (PI) National Institutes of Health, 7. R56-AG062302 Lutz (PI) National Institutes of Health.
Conflict of Interest and Disclosure Statement
Ornit Chiba-Falek: Dr. Chiba-Falek is a consultant to Seelos Therapeutics and co-founder at CLAIRIgene. Dr. Chiba-Falek reports filing patent applications: PCT/US2019/028786 entitled “Downregulation of SNCA Expression by Targeted Editing of DNA-Methylation”; PCT/US2021/054475 “Composition of Matter and Methods for treating Alzheimer’s disease”; PCT/US2022/78260 “Compositions and Methods relating to Epigenetic Modulation”
Michael W. Lutz: Dr. Lutz received consulting fees and travel expenses to attend scientific conferences from Zinfandel Pharmaceuticals.
ABBREVIATIONS
- AD
Alzheimer’s Disease
- LOAD
Late-onset Alzheimer’s Disease
- GWAS
Genome-Wide Association Study
- cCRE
Candidate cis-regulatory element
- SNP
Single Nucleotide Polymorphism
- SV
Structural Variant
- SSV
Short structural variant
- TF
Transcription Factor
- UCSC
University of California, Santa Cruz
- ENCODE
Encyclopedia of DNA elements
- LD
Linkage Disequilibrium
- SnRNA-seq
Single Nucleus RNA sequencing
- SnATAC-seq
Single Nucleus Assays for Transposase Accessible Chromatin using sequencing
- bp, Kb, Mb
base pair, kilobases, megabases
Footnotes
DECLARATIONS
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Data and computer code availability:
All computer code and example scripts are available on Github at the following link (https://github.com/NCTrailRunner/Alzheimer-SSV-TF-analysis). Tables containing the genomic regions and annotations described in the bioinformatics analysis of this paper are also available through this public resource.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Publicly available bioinformatics software and data sources were used for all analyses. Software:
MotifbreakR: https://www.bioconductor.org/packages/release/bioc/html/MotifbreakR.html
UCSC Table Browser: https://genome.ucsc.edu/cgi-bin/hgTables
ENCODE SCREEN tool: https://screen.encodeproject.org/
Databases:
dbSNP v153: https://www.ncbi.nlm.nih.gov/snp/
TF binding motifs from MotifDb: https://bioconductor.org/packages/release/bioc/html/MotifDb.html
1000 Genomes Phase 3 release: https://www.internationalgenome.org/category/phase-3/
Single nucleus (sn)RNA-seq data from the Swarup lab for AD samples and controls: https://www.synapse.org/#!Synapse:syn22079621/wiki/603535
snRNA-seq data from ROSMAP for AD samples and controls: The single-nucleus RNA-Sequencing data is available at Synapse (https://www.synapse.org/#!Synapse:syn18485175). The DOI for this dataset is: 10.7303/syn18485175. The DOI for the ROSMAP metadata is: 10.7303/syn3157322.
REFERENCES
- 1.Jakubosky D, D’Antonio M, Bonder MJ, Smail C, Donovan MKR, Young Greenwald WW, et al. Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nat Commun. 2020;11(1):2927. PMCID: PMC7286898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–51. [DOI] [PubMed] [Google Scholar]
- 3.Saul R, Lutz MW, Burns DK, Roses AD, Chiba-Falek O. The SSV Evaluation System: A Tool to Prioritize Short Structural Variants for Studies of Possible Regulatory and Causal Variants. Hum Mutat. 2016;37(9):877–83. PMCID: PMC4983215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, et al. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 2016;48(1):22–9. PMCID: PMC4909355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willems T, Gymrek M, Highnam G, Genomes Project C, Mittelman D, Erlich Y. The landscape of human STR variation. Genome Res. 2014;24(11):1894–904. PMCID: PMC4216929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447(7147):932–40. [DOI] [PubMed] [Google Scholar]
- 7.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nature reviews Genetics. 2005;6(10):729–42. [DOI] [PubMed] [Google Scholar]
- 8.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akai J, Kimura A, Hata RI. Transcriptional regulation of the human type I collagen alpha2 (COL1A2) gene by the combination of two dinucleotide repeats. Gene. 1999;239(1):65–73. [DOI] [PubMed] [Google Scholar]
- 10.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10(26):3101–9. [DOI] [PubMed] [Google Scholar]
- 11.Okladnova O, Syagailo YV, Tranitz M, Stober G, Riederer P, Mossner R, et al. A promoter-associated polymorphic repeat modulates PAX-6 expression in human brain. Biochem Biophys Res Commun. 1998;248(2):402–5. [DOI] [PubMed] [Google Scholar]
- 12.Peters DG, Kassam A, St Jean PL, Yonas H, Ferrell RE. Functional polymorphism in the matrix metalloproteinase-9 promoter as a potential risk factor for intracranial aneurysm. Stroke. 1999;30(12):2612–6. [DOI] [PubMed] [Google Scholar]
- 13.Searle S, Blackwell JM. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. Journal of medical genetics. 1999;36(4):295–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, et al. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455(1–2):70–4. [DOI] [PubMed] [Google Scholar]
- 15.Hefferon TW, Groman JD, Yurk CE, Cutting GR. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci U S A. 2004;101(10):3504–9. PMCID: PMC373492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, Orrison BM, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18(17):3274–85. PMCID: PMC2722989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiba-Falek O, Touchman JW, Nussbaum RL. Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum Genet. 2003;113(5):426–31. [DOI] [PubMed] [Google Scholar]
- 18.Afek A, Tagliafierro L, Glenn OC, Lukatsky DB, Gordan R, Chiba-Falek O. Toward deciphering the mechanistic role of variations in the Rep1 repeat site in the transcription regulation of SNCA gene. Neurogenetics. 2018;19(3):135–44. PMCID: PMC6054541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. PMCID: PMC3202986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. PMCID: PMC3200438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theunissen F, Flynn LL, Anderton RS, Akkari PA. Short structural variants as informative genetic markers for ALS disease risk and progression. BMC Med. 2022;20(1):11. PMCID: PMC8762977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theunissen F, Flynn LL, Anderton RS, Mastaglia F, Pytte J, Jiang L, et al. Structural Variants May Be a Source of Missing Heritability in sALS. Front Neurosci. 2020;14:47. PMCID: PMC7005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pytte J, Anderton RS, Flynn LL, Theunissen F, Jiang L, Pitout I, et al. Association of a structural variant within the SQSTM1 gene with amyotrophic lateral sclerosis. Neurol Genet. 2020;6(2):e406. PMCID: PMC7061286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pytte J, Flynn LL, Anderton RS, Mastaglia FL, Theunissen F, James I, et al. Disease-modifying effects of an SCAF4 structural variant in a predominantly SOD1 ALS cohort. Neurol Genet. 2020;6(4):e470. PMCID: PMC7357414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deters KD, Mormino EC, Yu L, Lutz MW, Bennett DA, Barnes LL. TOMM40-APOE haplotypes are associated with cognitive decline in non-demented Blacks. Alzheimers Dement. 2021;17(8):1287–96. PMCID: PMC8855738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuytemans K, Lipkin Vasquez M, Wang L, Van Booven D, Griswold AJ, Rajabli F, et al. Identifying differential regulatory control of APOE varepsilon4 on African versus European haplotypes as potential therapeutic targets. Alzheimers Dement. 2022. PMCID: PMC9250552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulminski AM, Philipp I, Shu L, Culminskaya I. Definitive roles of TOMM40-APOE-APOC1 variants in the Alzheimer’s risk. Neurobiol Aging. 2022;110:122–31. PMCID: PMC8758518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulminski AM, Philipp I, Loika Y, He L, Culminskaya I. Haplotype architecture of the Alzheimer’s risk in the APOE region via co-skewness. Alzheimers Dement (Amst). 2020;12(1):e12129. PMCID: PMC7656174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L, Loika Y, Kulminski AM. Allele-specific analysis reveals exon- and cell-type-specific regulatory effects of Alzheimer’s disease-associated genetic variants. Transl Psychiatry. 2022;12(1):163. PMCID: PMC9016079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiba-Falek O, Gottschalk WK, Lutz MW. The effects of the TOMM40 poly-T alleles on Alzheimer’s disease phenotypes. Alzheimers Dement. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L, Lutz MW, Wilson RS, Burns DK, Roses AD, Saunders AM, et al. APOE epsilon4-TOMM40 ‘523 haplotypes and the risk of Alzheimer’s disease in older Caucasian and African Americans. PLoS One. 2017;12(7):e0180356. PMCID: PMC5495438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero-Molina C, Garretti F, Andrews SJ, Marcora E, Goate AM. Microglial efferocytosis: Diving into the Alzheimer’s disease gene pool. Neuron. 2022;110(21):3513–33. [DOI] [PubMed] [Google Scholar]
- 33.Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, et al. A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet. 2019;51(10):1442–9. PMCID: PMC6778519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutz MW, Chiba-Falek O. Bioinformatics pipeline to guide late-onset Alzheimer’s disease (LOAD) post-GWAS studies: Prioritizing transcription regulatory variants within LOAD-associated regions. Alzheimers Dement (N Y). 2022;8(1):e12244. PMCID: PMC8864953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutz MW, Sprague D, Chiba-Falek O. Bioinformatics strategy to advance the interpretation of Alzheimer’s disease GWAS discoveries: The roads from association to causation. Alzheimers Dement. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holwerda SJ, de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120369. PMCID: PMC3682731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimenova AA, Herbinet M, Gupta I, Machlovi SI, Bowles KR, Marcora E, et al. Alzheimer’s-associated PU.1 expression levels regulate microglial inflammatory response. Neurobiol Dis. 2021;148:105217. PMCID: PMC7808757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novikova G, Kapoor M, Tcw J, Abud EM, Efthymiou AG, Chen SX, et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat Commun. 2021;12(1):1610. PMCID: PMC7955030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimenova AA, Raj T, Goate AM. Untangling Genetic Risk for Alzheimer’s Disease. Biol Psychiatry. 2018;83(4):300–10. PMCID: PMC5699970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morabito S, Miyoshi E, Michael N, Shahin S, Martini AC, Head E, et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat Genet. 2021;53(8):1143–55. PMCID: PMC8766217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan K, Friedman BA, Etxeberria A, Huntley MA, van der Brug MP, Foreman O, et al. Alzheimer’s Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation. Cell Rep. 2020;31(13):107843. PMCID: PMC7422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ott J. Predicting the range of linkage disequilibrium. Proc Natl Acad Sci U S A. 2000;97(1):2–3. PMCID: PMC33508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koch E, Ristroph M, Kirkpatrick M. Long range linkage disequilibrium across the human genome. PLoS One. 2013;8(12):e80754. PMCID: PMC3861250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singhal P, Veturi Y, Dudek SM, Lucas A, Frase A, van Steen K, et al. Evidence of epistasis in regions of long-range linkage disequilibrium across five complex diseases in the UK Biobank and eMERGE datasets. Am J Hum Genet. 2023;110(4):575–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017;20(8):1052–61. PMCID: PMC5759334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet. 2022;54(4):412–36. PMCID: PMC9005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eon Kuek L, Leffler M, Mackay GA, Hulett MD. The MS4A family: counting past 1, 2 and 3. Immunol Cell Biol. 2016;94(1):11–23. [DOI] [PubMed] [Google Scholar]
- 48.Ma Y, Jun GR, Chung J, Zhang X, Kunkle BW, Naj AC, et al. CpG-related SNPs in the MS4A region have a dose-dependent effect on risk of late-onset Alzheimer disease. Aging Cell. 2019:e12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greer PL, Bear DM, Lassance JM, Bloom ML, Tsukahara T, Pashkovski SL, et al. A Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction. Cell. 2016;165(7):1734–48. PMCID: PMC4912422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lutz MW, Luo S, Williamson DE, Chiba-Falek O. Shared genetic etiology underlying late-onset Alzheimer’s disease and posttraumatic stress syndrome. Alzheimers Dement. 2020;16(9):1280–92. PMCID: PMC7769164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz MW, Sprague D, Barrera J, Chiba-Falek O. Shared genetic etiology underlying Alzheimer’s disease and major depressive disorder. Transl Psychiatry. 2020;10(1):88. PMCID: PMC7062839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You SF, Brase L, Filipello F, Iyer AK, Del-Aguila J, He J, et al. MS4A4A modifies the risk of Alzheimer disease by regulating lipid metabolism and immune response in a unique microglia state. medRxiv. 2023. PMCID: PMC9934804. [Google Scholar]
- 53.Gu L, Ju Y, Hu M, Zheng M, Li Q, Zhang X. Research progress of PPARgamma regulation of cholesterol and inflammation in Alzheimer’s disease. Metab Brain Dis. 2023;38(3):839–54. [DOI] [PubMed] [Google Scholar]
- 54.Khan MA, Alam Q, Haque A, Ashafaq M, Khan MJ, Ashraf GM, et al. Current Progress on Peroxisome Proliferator-activated Receptor Gamma Agonist as an Emerging Therapeutic Approach for the Treatment of Alzheimer’s Disease: An Update. Curr Neuropharmacol. 2019;17(3):232–46. PMCID: PMC6425074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian S, Gottschalk WK, Kim SY, Roses AD, Chiba-Falek O. The effects of PPARgamma on the regulation of the TOMM40-APOE-C1 genes cluster. Biochim Biophys Acta Mol Basis Dis. 2017;1863(3):810–6. PMCID: PMC5285471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saunders AM, Burns DK, Gottschalk WK. Reassessment of Pioglitazone for Alzheimer’s Disease. Front Neurosci. 2021;15:666958. PMCID: PMC8243371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong H, Geng R, Zhang Y, Ding J, Liu M, Deng S, et al. Effects of Peroxisome Proliferator-Activated Receptor-Gamma Agonists on Cognitive Function: A Systematic Review and Meta-Analysis. Biomedicines. 2023;11(2). PMCID: PMC9953157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy ER, Wang B, Wan YW, Chiu G, Cole A, Yin Z, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest. 2020;130(4):1912–30. PMCID: PMC7108898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanford SAI, McEwan WA. Type-I Interferons in Alzheimer’s Disease and Other Tauopathies. Front Cell Neurosci. 2022;16:949340. PMCID: PMC9334774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium EP, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710. PMCID: PMC7410828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31(1):51–4. PMCID: PMC165576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–6. PMCID: PMC308837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31(23):3847–9. PMCID: PMC4653394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coetzee SG, Coetzee GA, Hazelett DG. motifbreakR: an Introduction. 2023. [updated 2023; cited]; Available from: https://bioconductor.org/packages/release/bioc/vignettes/motifbreakR/inst/doc/motifbreakR-vignette.html.
- 65.Habib N, McCabe C, Medina S, Varshavsky M, Kitsberg D, Dvir-Szternfeld R, et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat Neurosci. 2020;23(6):701–6. PMCID: PMC9262034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570(7761):332–7. PMCID: PMC6865822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14(10):955–8. PMCID: PMC5623139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–87 e29. PMCID: PMC8238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Touzet H, Varre JS. Efficient and accurate P-value computation for Position Weight Matrices. Algorithms Mol Biol. 2007;2:15. PMCID: PMC2238751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nho K, Kim S, Horgusluoglu E, Risacher SL, Shen L, Kim D, et al. Association analysis of rare variants near the APOE region with CSF and neuroimaging biomarkers of Alzheimer’s disease. BMC Med Genomics. 2017;10(Suppl 1):29. PMCID: PMC5461522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prokopenko D, Lee S, Hecker J, Mullin K, Morgan S, Katsumata Y, et al. Region-based analysis of rare genomic variants in whole-genome sequencing datasets reveal two novel Alzheimer’s disease-associated genes: DTNB and DLG2. Mol Psychiatry. 2022;27(4):1963–9. PMCID: PMC9126808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available bioinformatics software and data sources were used for all analyses. Software:
MotifbreakR: https://www.bioconductor.org/packages/release/bioc/html/MotifbreakR.html
UCSC Table Browser: https://genome.ucsc.edu/cgi-bin/hgTables
ENCODE SCREEN tool: https://screen.encodeproject.org/
Databases:
dbSNP v153: https://www.ncbi.nlm.nih.gov/snp/
TF binding motifs from MotifDb: https://bioconductor.org/packages/release/bioc/html/MotifDb.html
1000 Genomes Phase 3 release: https://www.internationalgenome.org/category/phase-3/
Single nucleus (sn)RNA-seq data from the Swarup lab for AD samples and controls: https://www.synapse.org/#!Synapse:syn22079621/wiki/603535
snRNA-seq data from ROSMAP for AD samples and controls: The single-nucleus RNA-Sequencing data is available at Synapse (https://www.synapse.org/#!Synapse:syn18485175). The DOI for this dataset is: 10.7303/syn18485175. The DOI for the ROSMAP metadata is: 10.7303/syn3157322.