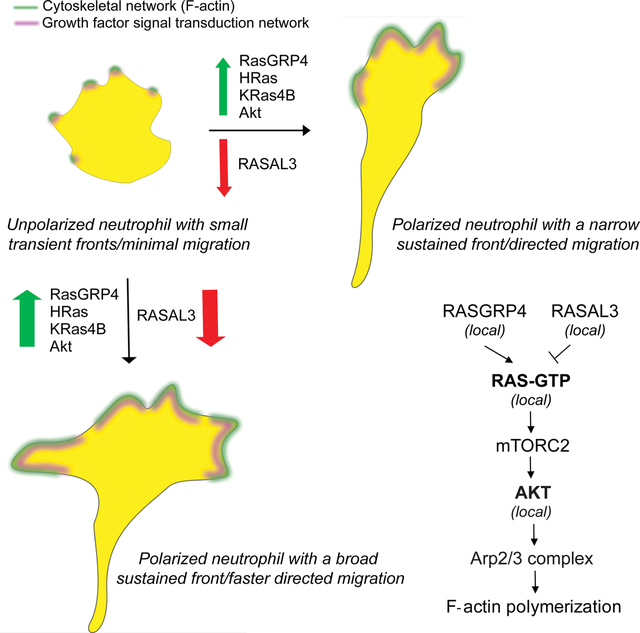
Summary
Ras signaling is typically associated with cell growth but not in direct regulation of motility or polarity. By optogenetically targeting different nodes in Ras/PI3K/Akt network in differentiated human HL-60 neutrophils, we abruptly altered protrusive activity, bypassing chemoattractant receptor/G-protein network. First, global recruitment of active KRas4B/HRas isoforms or RasGEF, RasGRP4, immediately increased spreading and random motility. Second, activating Ras at the cell rear generated new protrusions, reversed pre-existing polarity, and steered sustained migration in neutrophils or murine RAW264.7 macrophages. Third, recruiting RasGAP, RASAL3, to cell fronts extinguished protrusions and changed migration direction. Remarkably, persistent RASAL3 recruitment at stable fronts abrogated directed migration in three different chemoattractant gradients. Fourth, local recruitment of Ras-mTorC2 effector, Akt, in neutrophils or Dictyostelium amoebae generated new protrusions and rearranged pre-existing polarity. Overall, these optogenetic effects were mTorC2-dependent, but relatively independent of PI3K. Thus, receptor-independent, local activations of classical growth-control pathways directly control actin assembly, cell shape, and migration modes.
Keywords: immunity, biochemical excitability, cancer, leukocytes, development, metastasis
eTOC BLURB
Pal et al uncover that chemoattractant receptor-independent, local activation of the Ras-mTorC2-Akt growth factor signal transduction network is directly required for actin organization, polarity, and directed migration in immune cells. Moreover, localized network activity is necessary for immune cell chemotaxis and may be considered essential for immune response signaling.
Graphical Abstract:
Introduction
Neutrophils initiate the innate immune response by sensing chemical cues and rapidly migrating towards sites of tissue damage and infection1–3. This directed migration is coordinated by rearrangements of signal transduction proteins and lipids to the leading or trailing edges of the cell. This dictates the location and dynamics of cellular protrusions and contractions that move the cell4–11. Misdirected neutrophil migration is responsible for a wide array of autoimmune and inflammatory conditions in adults1,12,13. Discovering the key signal transduction events that direct cytoskeletal activity, cellular protrusions, and migratory behavior in cells of the immune system will facilitate development of next generation treatments14,15.
Multiple pathways have been implicated in migration in neutrophils and other mammalian cells; it is likely that each contributes to a larger network. Chemoattractant mediated receptor/G-protein stimulation generates PIP3 at the cell’s leading edge and local PI3K activation triggers Rac activation, actin polymerization, protrusion formation, and migration9,10,16–34. However, other studies suggest that PI3K/PIP3 signaling is not singularly essential for directed migration27,29,35–43. This is highlighted in observations of PI3K-independent pathways involving Gβγ-DOCK2-ELMO1 or Gβγ-PAK1-PIXα mediated Rac or Cdc42 activation in F-actin formation at the leading edge44–50. It is also unclear whether Akt, which is brought to the cell’s leading edge by PI3K/PIP3, is necessary for mammalian cell migration since it has been reported that Akt1 isoform is a negative regulator of motility whereas Akt2 promotes it42,51–54. Notably, G-protein coupled chemoattractant receptors also activate Ras, which in turn stimulates mTorC2 and Akt, and is important for cell growth, survival, and energy metabolism55–60.
Although it may control gene expression events involved in cell migration, it is unknown whether the Ras-mTorC2-Akt growth factor signaling axis is directly required for actin organization, polarity, or directed migration. Knockout or knockdown studies with modulators of Ras activity have not given a conclusive understanding of Ras proteins in mammalian chemotaxis56,61. Biochemical and genetic investigations in the established model organism, Dictyostelium discoideum, have shown that Ras is activated by G-protein coupled receptor stimulation but also spontaneously at protrusions, which has not been shown in migrating mammalian cells5,6,62–71. However, as multiple Ras isoforms are expressed in this amoeba it has been difficult to demonstrate that Ras activity is essential for migration and has prevented assignment of specific roles for each isoform by gene deletions70,72–74. To circumvent this problem, researchers conditionally expressed constitutively active Ras isoforms which produced phenotypes over many hours, however, allowing sufficient time for signaling networks to re-adjust through protein rearrangement or differential gene expression63,70,75–77. Although these studies implicated Ras in migration, they did not show that local activation of these growth control pathways on the membrane could bring about localized protrusions and directed motility. Moreover, these results were observed in free-living amoeba, and may or may not apply to migratory cells in humans.
In the past decade, optogenetic tools have been developed to locally perturb upstream receptor/G-protein networks or downstream cytoskeletal components, namely Rac, Cdc42, and RhoA, to induce polarity and persistent directed motility in different cellular systems18,78–88. Modifications of these optical approaches allowed us to address critical questions about growth factor pathways which could not be answered with previous methods. First, is Ras and its downstream effectors important for mammalian migration? Can activation of individual signaling components of growth factor networks on the cell membrane influence cytoskeletal arrangement and affect motility? Does activation of different growth signaling nodes, such as Ras or Akt, have similar or opposing effects on cellular protrusions and migration? Second, chemoattractant gradients can bias pre-existing front-back axis of the cell, but the occupied receptors activate a vast array of downstream signaling events80,89–94. Can localized activation of individual components of growth control pathways at the cell back override pre-existing polarity? Conversely, can localized inhibition of growth control pathway components at the cell front convert it to a back? Can such inhibition override the signal from chemoattractants? Using subcellular optogenetics, we abruptly and locally perturbed Ras and Akt activity, bypassing chemoattractant-sensing receptor/G-protein network. Our studies answer these questions and show that cell shape, actin assembly, and migration modes in mammalian cells are controlled by local, spontaneous activities of the Ras-mTorC2-Akt axis of classical growth-control pathways.
Results
For optical manipulation of signaling in migratory neutrophils, we engineered a blue light-inducible, cryptochrome-based dimerization system in differentiated human HL-60 neutrophil-like cells. This process was performed in a stepwise manner. First, the plasma membrane component CIBN, fused to a C-terminal CAAX motif, was expressed from an integrated lentiviral vector in wildtype neutrophils. Second, the F-actin polymerization biosensor, LifeAct tagged with an infrared fluorophore, was stably co-expressed to generate a dual-expressing cell line. Third, the cytosolic recruitable component, CRY2PHR-mcherry2, was stably introduced to these dual-expressing cells by transposon-based integration (Figure S1A). Illuminating the entire periphery of these ‘triple expressors’ with 488 nm light resulted in global recruitment of cytosolic CRY2PHR, fused with a protein of interest, to the plasma membrane (Figure S1B–D). The corresponding linescan shows that the CRY2PHR-mcherry2 intensity peak shifts from cytosol to plasma membrane when blue light is switched on, demonstrating membrane recruitment (Figure S1E). Similarly, once blue light was selectively shined on front or back of the cell, the CRY2PHR-fused protein of interest was locally recruited to the illuminated region of the membrane (Figure S1B–C). This system thus allowed us to spatio-temporally control activities of signaling components on the membrane in a tightly regulated fashion.
Global Ras activation increased spreading and motility in neutrophils
Using our optogenetic system, we investigated role of Ras activation on neutrophil morphology in absence of chemoattractant-mediated receptor stimulation. Within 30 seconds of turning on the 488 nm light, constitutively activated HRas G12V isoform lacking its C-terminal CAAX motif, HRas G12V ΔCAAX, was recruited uniformly on the cell membrane. In next 90 seconds, an increase in F-actin-rich protrusions, marked by LifeAct, was observed and cells started moving rapidly (Figure 1A, Video S1A). In a representative kymograph for these cells, narrow regions of LifeAct intensity existed in absence of blue light. However, once light was switched on, LifeAct intensity on the membrane increased substantially with a concomitant increase in cell area. In HRas-activated state, cells also demonstrated spreading and contraction periodically (Figure 1B). As illustrated in Figure 1C, there was an overall ~40% increase in cell area. Additionally, HRas activity on the membrane polarized cells and improved their migratory ability (Figure 1A). This led to 70% and 20% increase in migration speed (Figure 1D–F) and aspect ratio (proxy for polarity; Figure 1G), respectively, in recruited cells. As one control, we looked at cells in the same population which showed no detectable recruitment of HRas G12V ΔCAAX to the membrane, presumably due to low expression of CIBN-CAAX. These cells, even in presence of blue light, did not show any appreciable change in size of F-actin patches, protrusion shape, cell area, migration speed or polarity (Figure S1F–L). Thus, HRas-mediated signaling at the membrane improves spreading and migration of neutrophils.
Figure 1. Global recruitment of constitutively active Ras isoforms improves neutrophil polarity and migration.
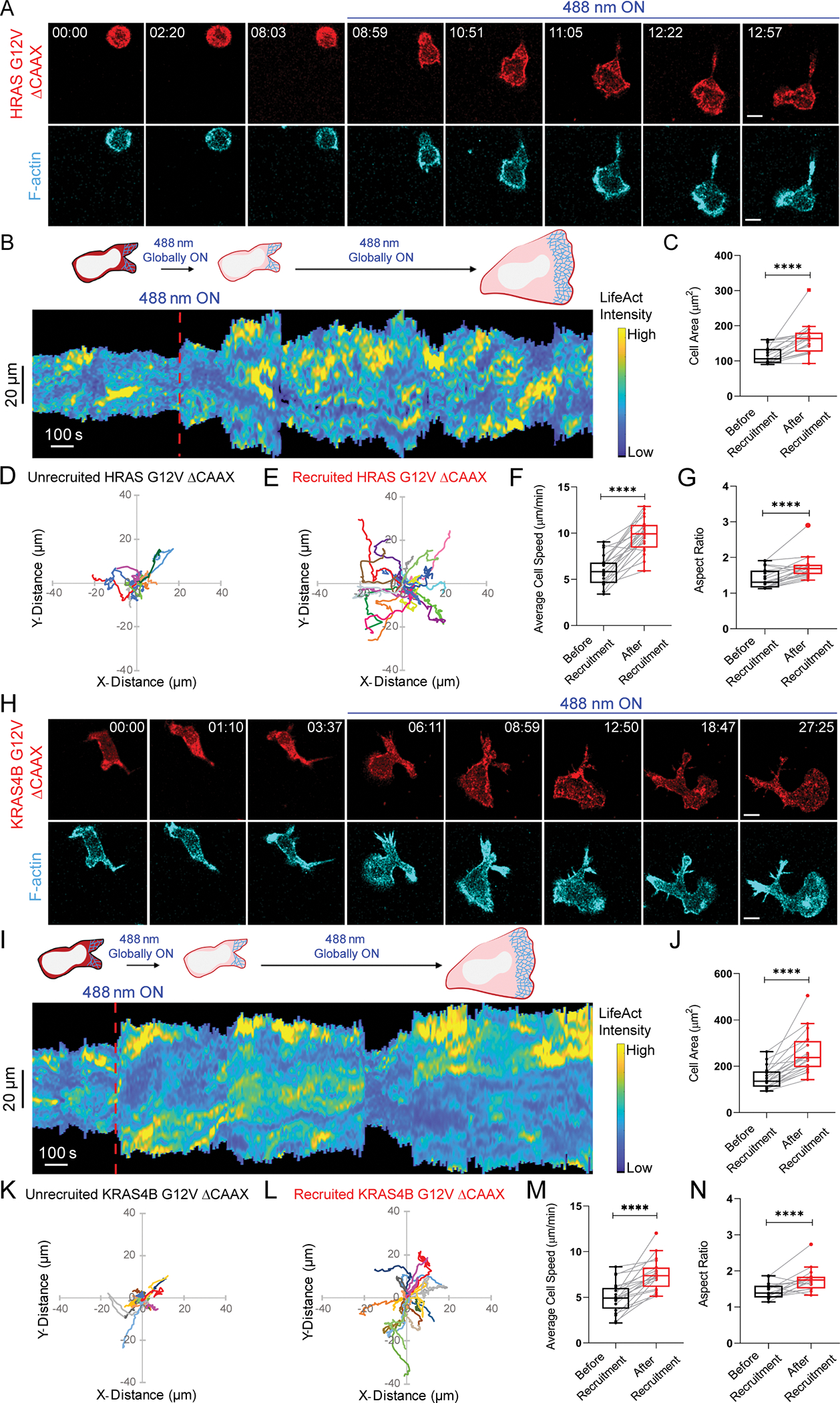
Confocal images of differentiated HL-60 neutrophil expressing (A) CRY2PHR-mcherry2-HRas G12V ΔCAAX or (H) CRY2PHR-mcherry2-KRas4B G12V ΔCAAX (red; upper panel) and LifeActmiRFP703 (cyan; lower panel), before or after 488 nm illumination. Time in min:sec format; scale bars 5 μm. (B, I) Representative kymographs of cortical LifeAct in cells of A and H. Cartoon depicts recruitment, F-actin polymerization, or cell shape status corresponding to the kymographs. Box-and-whisker plots of (C, J) cell area, (F, M) average speed, and (G, N) aspect ratio, before (black) and after (red) HRas G12V ΔCAAX or KRas4B G12V ΔCAAX recruitment. Centroid tracks of neutrophils showing random motility (D, K) before and (E, L) after HRas G12V ΔCAAX or KRas4B G12V ΔCAAX recruitment. Each track lasts at least 5 mins and was reset to same origin. nc=19 (HRas G12V ΔCAAX) or nc=20 (KRas4B G12V ΔCAAX) from atleast 3 independent experiments; asterisks indicate significant difference, ****P ≤ 0.0001 (Wilcoxon-Mann-Whitney rank sum test). See also Figures S1–S2.
To test whether these cytoskeletal changes are Ras isoform-specific, we expressed constitutively active KRas4B G12V ΔCAAX in neutrophils. Once blue laser was switched on and KRas4B membrane recruitment occurred, within 40 seconds, these cells formed broad F-actin-driven lamellipodium and showed increased motility (Figure 1H, Video S1B). Representative kymograph showed that, upon activating KRas4B, LifeAct intensity was tightly coordinated with cell area. During spreading and contracting stages of the cell, LifeAct intensity increased and decreased accordingly (Figure 1I). In non-recruiting cells of this population, narrow regions of LifeAct intensity were present stochastically, and motility was minimal (Figure S2A–B). Overall, KRas4B activation gave rise to 70%, 50%, and 20% increase in cell area, migration speed, and polarity, respectively (Figure 1J–N). In non-recruiting cells, these migration parameters remained unchanged over time (Figure S2C–G). Altogether, HRas activity was a stronger inducer of motility whereas KRas4B promoted more spreading.
Although Ras activation-mediated cytoskeletal changes and motility occurred without chemoattractant, we wanted to ensure that these phenotypes were caused solely by active Ras recruitment and not basal G-protein coupled receptor signaling95. To test this, we inhibited heterotrimeric G-protein activity in neutrophils using a combination of Gαi and Gβγ inhibitors, pertussis toxin (PTX) and gallein, respectively (Figure S2H–I)51,78,92,96–99. Dual inhibitor treatment prevented typical, uniform chemoattractant-induced burst of F-actin polymerization around the cell periphery and subsequent polarized migration, indicating that G-protein signaling was completely stalled (Figure S2I). However, within 30–40 seconds of KRas4B G12V ΔCAAX global recruitment in these inhibited cells, they polarized and migrated with broad fronts as observed previously (Figures 1H, S2J, Video S1B–C). Representative kymograph showed that narrow LifeAct patches were converted to broad ones, accompanied with increase in cell area once laser was switched on (Figure S2K). This suggests that spontaneous Ras activity on the membrane, independent of receptor-mediated G-protein signaling, is sufficient to activate neutrophil motility and polarity, and Ras should be included as an intermediary coupling G-protein activity to downstream signaling.
Since individual Ras isoforms could promote migration, we wondered whether concerted Ras activation by a RasGEF would have a similar or stronger effect. We selected RasGRP4 since it is expressed exclusively in myeloid cells and its dysregulation causes various diseases100,101. Its involvement in immune cell migration is yet unknown. Time-lapse imaging and kymograph analysis showed that, within 3 minutes of RasGRP4 membrane recruitment, narrow, transient pseudopods were converted to wide, sustained lamellipodium resulting in prolonged migration (Figure 2A–C, Video S1D). Altogether, RasGRP4 recruitment caused over 86% increase in cell area, accompanied with ~70% and ~25% rise in migration speed and polarity, respectively (Figure 2D–H). Neutrophils in the same population with cytosolic, non-recruiting RasGRP4 were not activated (Figure S3A–G). Also, no response was observed when CRY2PHR component, without being fused to either activated Ras or RasGEF, was recruited (Figure S3H–N, Video S1E).
Figure 2. RasGEF activation on neutrophil membrane induces polarity and migration.
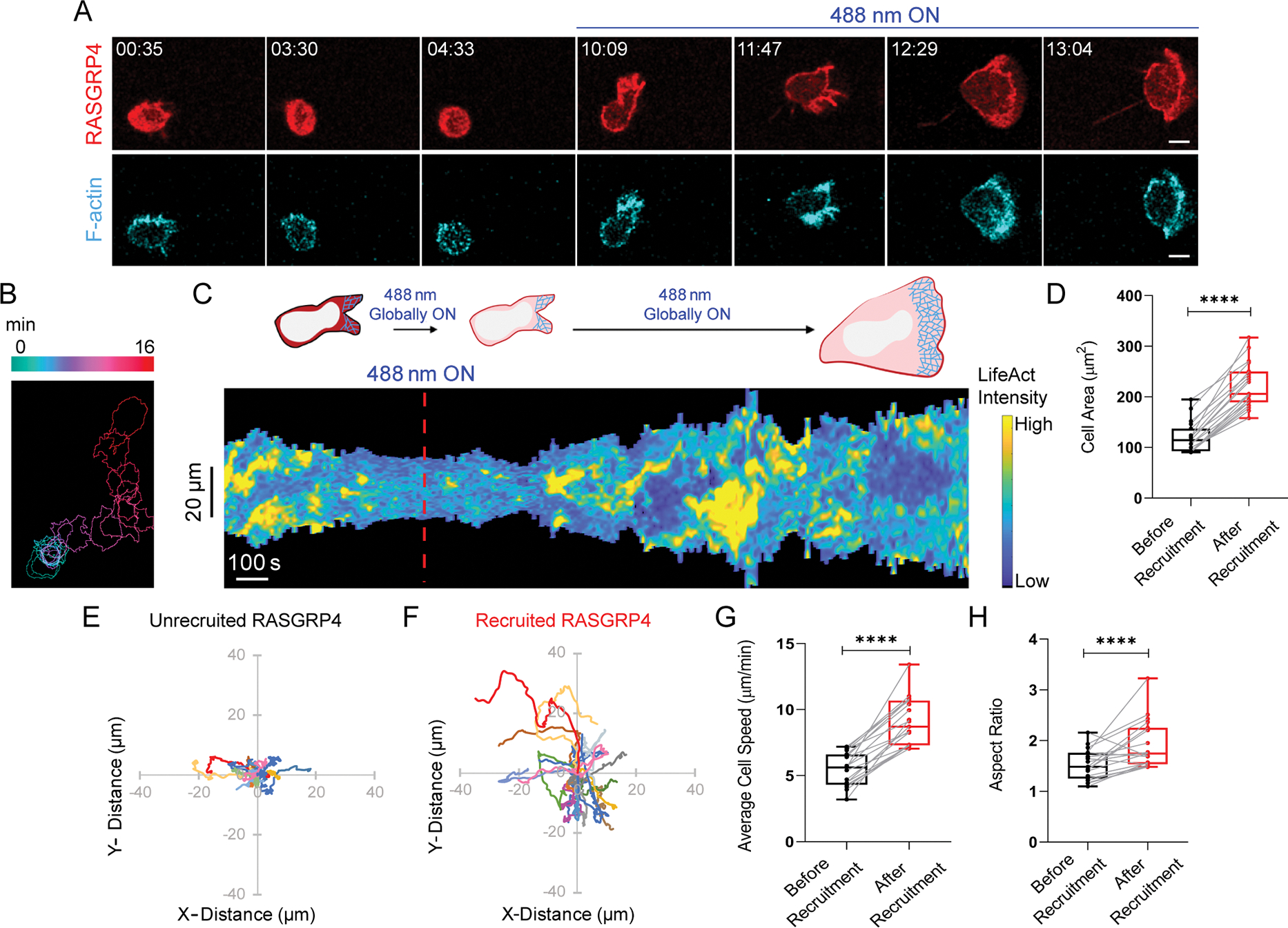
(A) Confocal images of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; upper panel) and LifeActmiRFP703 (cyan; lower panel), before and after 488 nm illumination. Time for mat and scale bars as in Figure 1. (B) Color-coded (at 1 min intervals) outlines of cell shown in A. (C) Representative kymograph of cortical LifeAct intensity in cell in panel A. Color map as in Figure 1. Cartoon depicts membrane recruitment, actin polymerization or cell shape status corresponding to kymograph. Box-and-whisker plots of (D) cell area, (G) average speed, and (H) aspect ratio, before (black) or after (red) RasGRP4 recruitment (nc=19). Statistical analysis as in Figure 1. Centroid tracks of neutrophils (nc=19) showing random motility before (E) or after (F) recruitment. Tracks presented as in Figure 1. See also Figure S3.
Next, we aimed to provide a mechanistic insight to actin assembly guiding Ras-driven lamellipodium formation. We assessed effects of blocking the actin nucleator, Arp2/3 complex, whose activation by Scar/WAVE causes lamellipodia formation in immune cells10,102,103. After treating recruitable RasGRP4-expressing neutrophils with Arp2/3 complex inhibitor, CK-666, they rounded up, actin polymerization on the cortex vanished, and migration was abrogated. Despite RasGRP4 recruitment, neutrophils did not display any F-actin-rich protrusions, spreading, motility, or polarity (Figure S4A–H). Altogether, Ras activity on the membrane leads to Arp2/3-mediated F-actin polymerization causing spreading and migration.
Local Ras activation or inhibition reverses pre-existing polarity
Activating Ras over the entire cell periphery led to formation of actin-rich lamellipodia, increased polarity, and persistent migration, but could local Ras activation at the cell rear generate new protrusions and re-organize polarity? To answer this, we recruited RasGRP4 to a quiescent back region in migrating neutrophils by intermittently applying 488 nm light near it, as indicated by dashed white box (Figure 3A, Video S2A). Immediately after RasGRP4 localized to the back, the front contracted, ruffles started diminishing, and cell slowed down. Simultaneously, several finger-like F-actin-rich structures appeared at the recruitment site, which gradually broadened into sustained protrusions. This forced the cell to move in the other direction by rearranging its front-rear axis (Figure 3A–E). This phenomenon was analyzed in the angular histogram which showed that probability of new protrusion generation was highest at or near RasGRP4 recruitment site (Figure 3B).
Figure 3. Localized Ras activation or inhibition rearranges front-rear polarity in neutrophils.
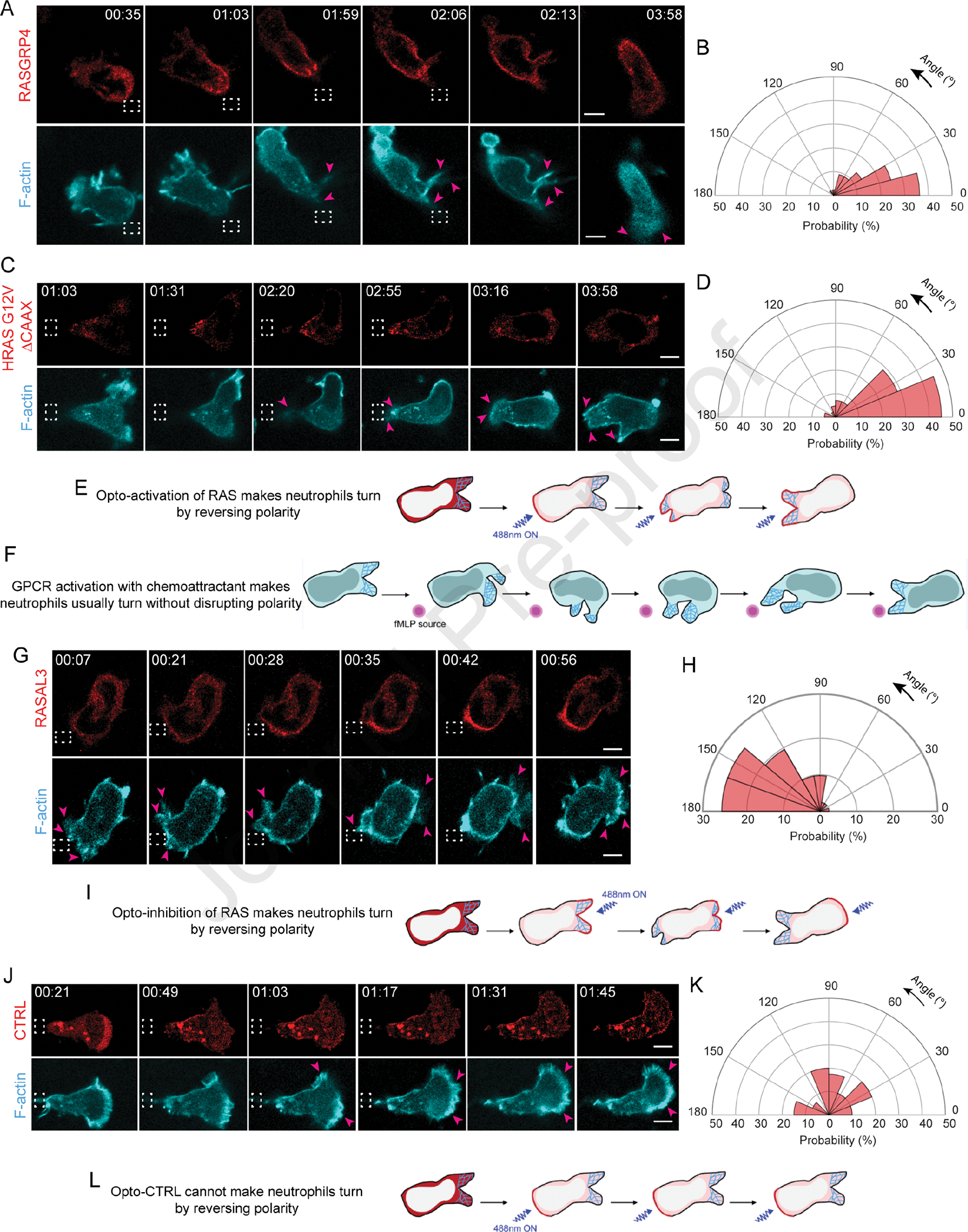
Time-lapse confocal images of differentiated HL-60 neutrophil expressing (A) CRY2PHR-mcherry2-RasGRP4, (C) CRY2PHR-mcherry2-HRas G12V ΔCAAX, (G) CRY2PHR-mcherry2-RASAL3, or (J) CRY2PHR-mcherry2-CTRL (red; upper panel) and LifeActmiRFP703 (cyan; lower panel). RasGRP4, HRas G12V ΔCAAX or CTRL was recruited to the cell back whereas RASAL3 was recruited to the front by local 488 nm illumination, as shown by dashed white boxes. For RasGRP4, HRas G12V ΔCAAX or CTRL, pink arrows highlight new protrusion formation at the opposite end of recruitment. For RASAL3, pink arrows highlight protrusion disappearance at recruitment site and appearance of new ones away from it. Time and scale bars as in Figure 1. (B, D) Polar histograms of RasGRP4 (nc=16 and np=44) or HRas G12V ΔCAAX (nc=23 and np=42) demonstrate higher probability of fresh protrusion formation near recruitment area. (H, K) Polar histogram for RASAL3 (nc=21 and np=41) shows higher probability of fresh protrusion formation away from recruitment area whereas for CTRL (nc=15 and np=40), new protrusion formation is not biased by its recruitment. (E, I, L) Cartoons depict Ras opto-activation or -inhibition at the cell back or front by RasGRP4/HRas G12V ΔCAAX or RASAL3 recruitment causes formation of new front or back, respectively. For opto-CTRL (control), its recruitment has no effect on reversing pre-existing polarity. (F) Cartoon showing cell making a ‘U-turn’ to reorient itself towards fMLP source without breaking pre-existing polarity. See also Figure S4.
Since RasGRP4 may activate numerous Ras proteins, we next asked whether local activation of a specific Ras isoform could elicit a similar response at the cortex. To examine this, we transiently recruited HRas G12V ΔCAAX to the back of migrating neutrophils. This arrested protrusion formation at the front, and caused a concomitant increase in F-actin polymerization at the recruitment site. As a result, the cell changed its direction of migration, reversing its pre-existing polarity (Figure 3C–E, Video S2B). The corresponding angular histogram demonstrated that new protrusion formation could be biased towards activated HRas G12V recruitment site (Figure 3D). When stimulated by chemoattractant from behind, neutrophils usually maintain their pre-existing polarity and make a U-turn towards the new source; they can only switch polarity if gradient is extraordinarily steep and perfectly positioned (Figure 3F)90. Apparently, local Ras activation is equivalent to a very steep chemoattractant gradient.
We next examined effects of inhibiting Ras activity on cytoskeletal assembly and protrusions, which might be expected to stop migration. We studied a RasGAP, RASAL3, which was recently characterized for its immunomodulatory functions in neutrophils. It is possible that these effects on immune response were due to inhibition of migration, but this was not assessed104. For our experiment, we transiently recruited RASAL3 to the leading front by applying light periodically near the existing protrusions, as shown by dashed white box (Figure 3G, Video S2C). Upon doing so, protrusions were restricted and eventually collapsed in on themselves. Thus, a new back formed at the recruitment site, whereas a new F-actin rich front emerged on the other end of the cell. The angular histogram analysis agreed with our observation that all new protrusions formed away from the RASAL3 recruitment site (Figure 3H). A similar observation was made when RASAL3 was locally recruited in activated macrophages (Figure S4I, Video S2D). Therefore, through its RasGAP function, RASAL3 changed cell’s direction by reversing its pre-existing polarity (Figure 3G, I).
We next enquired whether dynamically recruiting RASAL3 over the entire cell periphery would shut down all protrusive activity. For this experiment, we selected chemoattractant-activated macrophage possessing F-actin driven protrusions around its periphery, as shown by pink arrows. Once we applied blue light continuously around the perimeter, RASAL3 was recruited globally causing cell shrinkage and protrusions vanishing instantaneously (Figure S4I, Video S2D). We could reproduce this phenotype in non-polarized neutrophil having protrusions around its perimeter. Upon applying blue laser continuously around its periphery, RASAL3 was globally recruited causing protrusions to disappear completely. This was accompanied by a concomitant decrease in cell area (Figure S4J–K, Video S2E).
Locally recruiting CRY2PHR alone did not bias new protrusion formation nor affect front-rear axis of the cell (Figure 3J–L, Video S2F). This suggested that reversing polarity, either to create new protrusions at the back or inhibit existing protrusions at the front, is due to localized Ras activation or inhibition, and not due to cryptochrome recruitment or light irradiation. Altogether, transient Ras activation is sufficient and necessary to orchestrate cytoskeletal arrangement and generate protrusions.
Local Ras signaling is necessary for directionally persistent migration
These results prompted us to ask whether local Ras activation could direct persistent migration in neutrophils. To test this, Ras was stably and locally activated on the membrane by persistently recruiting RasGRP4 in cells without chemoattractants. First, when we applied laser near the cell rear, as highlighted by dashed white boxes, it reoriented itself and moved towards the light source. Next, when we shifted the light source further in the same direction, the cell followed obediently. Again, once we shifted the light near the cell back, it readjusted its pre-existing polarity, created a new front, and made its way to the light. We observed this directionally-sensitive migration with all subsequent changes in light source location (Figure 4A, Video S3A). Across the population, neutrophils persistently migrated over several cell lengths towards the light source irrespective of the direction of optical input, as demonstrated by their directional index of 0.46±0.26 (Figure 4B–C). Even in quiescent macrophages, which have low basal motility, locally activating Ras directed movement towards the light, albeit slowly (Figure S4L–M, Video S3B). Therefore, local Ras signaling is sufficient for directed motility in immune cells.
Figure 4. Local Ras signaling is necessary for directionally persistent migration.
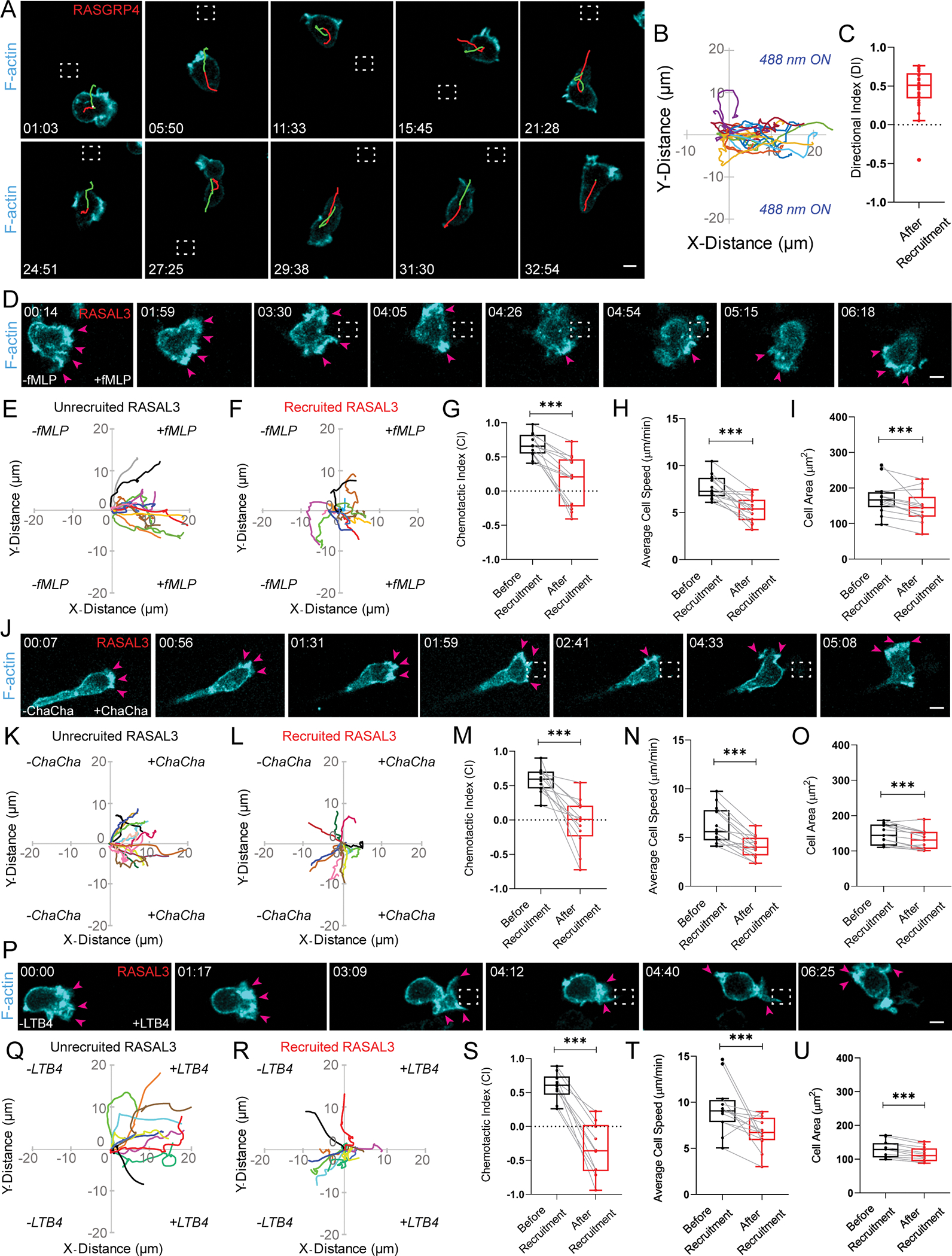
(A) Confocal images of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 and LifeActmiRFP703 (cyan). RasGRP4 was recruited by locally applying 488 nm light (dashed white box), which was repositioned continuously as the cell migrated. Shown in green and red are the centroid locations of the cell during the 15 previous (red) and future (green) frames (truncated at the first and final frames). Time and scale bars as in Figure 1. (B) Cell trajectories (nt=25, nc=10) after laser was applied. The trajectory was rotated so that, in the new coordinates, moving along the x-direction corresponds to movement toward the laser location. Tracks lasted atleast 70 secs and were reset to same origin. (C) Box and whisker plot of directional index (DI) for trajectories in B. Sign rank statistical test against the zero DI gives p< 3e-5 significance. (D, J, P) Confocal images of HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) chemotaxing in gradients of N-formyl-met-leu-phe (fMLP), C5a receptor agonist (ChaCha peptide), or Leukotriene B4 (LTB4), respectively. Local 488 nm illumination was applied continuously near front protrusions (dashed white boxes). Pink arrows highlight protrusion disappearance at illumination site and appearance of new ones away from it. Time and scale bars as in Figure 1. (E-F, K-L, Q-R) Neutrophil centroid tracks showing directed motility towards fMLP (nc=13), ChaCha peptide (nc=12) or LTB4 (nc=10), respectively, before and after RASAL3 membrane recruitment. Tracks last atleast 2 mins and were reset to same origin. Box-and-whisker plots of (G, M, S) chemotactic indices, (H, N, T) average cell speed and (I, O, U) cell area before (black) or after (red) RASAL3 recruitment in fMLP (nc=13), ChaCha peptide (nc=12), and LTB4 (nc=10) gradients, respectively; atleast 3 experiments. Statistical analysis as in Figure 1. See also Figure S4.
Our results show that localized Ras activation can steer chemoattractant-independent motility, but is it necessary for chemotaxis? We answered this by optically recruiting RASAL3 to the fronts of neutrophils migrating in gradients of different classes of chemoattractants37,105–108. In stable gradients of ‘end-target’ chemoattractants, such as N-formyl-met-leu-phe (fMLP) and C5a receptor agonist (ChaCha peptide), cells migrated persistently towards the attractant source in the right reservoir of microfluidic chamber-slide, as underlined by their chemotactic indices (CIs) of 0.66±0.17 and 0.58±0.18, respectively. However, when laser was applied continuously at the front of chemotaxing cells, as shown by dashed white boxes, their F-actin protrusions slowly diminished, they slowed down, and eventually formed new protrusions away from the direction of chemoattractant source. These stark changes were reflected in their CI of 0.17±0.35 or −0.04±0.35 in fMLP or ChaCha peptide gradient, respectively (Figure 4D–G, J–M, Video S3C–D). Overall, local Ras inhibition resulted in 30% or 35% reduction in average cell speed and 13% or 10% decrease in cell area in populations exposed to fMLP or ChaCha gradient, respectively (Figure 4H–I, N–O). Next, we tested an ‘intermediary’ chemoattractant, such as Leukotriene B4 (LTB4), and observed a similar stalling of directed migration after recruiting RASAL3 to the front. This caused a significant drop of CI from 0.59±0.21 to −0.35±0.4, and 30% or 12% reduction in cell speed or area, respectively (Figure 4P–U, Video S3E). Altogether, localized Ras activity is necessary for neutrophil chemotaxis and may be considered essential for immune response signaling.
mTorC2 is more important than PI3K for Ras activation of polarity and migration
Our next aim was to delineate Ras downstream signaling in neutrophils. We first examined the role of PI3K/PIP3 pathway using a combination of optogenetic and pharmacological inhibition approaches. Although PI3K is a direct effector of Ras and an important regulator of polarization and migration in many cells, role of Ras-activated PI3K in neutrophil motility is unknown22,41,42,51. We selected the PI3Kγ inhibitor, AS605240, which depleted PIP3 as indicated by signal loss of its biosensor, PH-Akt, on the neutrophil membrane (Figure S5A)109,110. Upon inhibitor treatment, RasGRP4-expressing cells lost polarity and migration was largely diminished (Figure 5A–B, Video S3F). Kymograph showed a few spontaneous, narrow patches of LifeAct in PI3K-inhibited cells (Figure 5C). Within 2 minutes of RasGRP4 recruitment, PI3K-inhibited cells regained polarity, developed F-actin rich fronts, and migrated (Figure 5B, Video S3F). Upon recruitment, LifeAct patches broadened and increased in number along with widening of cell area (Figure 5C). Overall, Ras activation caused an increase of 50%, 38%, and 64% in cell area, polarity, and migration speed, respectively, in PI3K-inhibited population (Figure 5D–H). We observed a similar recovery in polarity and migration in pan-PI3K inhibitor, LY294002-treated neutrophils after RasGRP4 recruitment (Figure S5B–E, Video S3G)111. In non-recruiting cells where RasGRP4 recruitment was undetectable, there was no recovery from PI3K inhibition (Figure S5F–L). We independently assessed opto-RasGRP4-induced Akt phosphorylation at Thr308 and Ser473 positions as an indirect readout of Ras activation in PI3K-inhibited population. Basal phosphorylation at both crucial residues disappeared with inhibitor treatment, but recovered to untreated levels upon RasGRP4 recruitment, indicating Ras activates downstream Akt even when PI3K is strongly suppressed (Figures 5I, S6A–C). These suggest that while PI3K activity may be crucial for spontaneous motility, it is not as important for optically-triggered migration.
Figure 5. mTorC2 is more important than PI3K for Ras activation of polarity and migration.
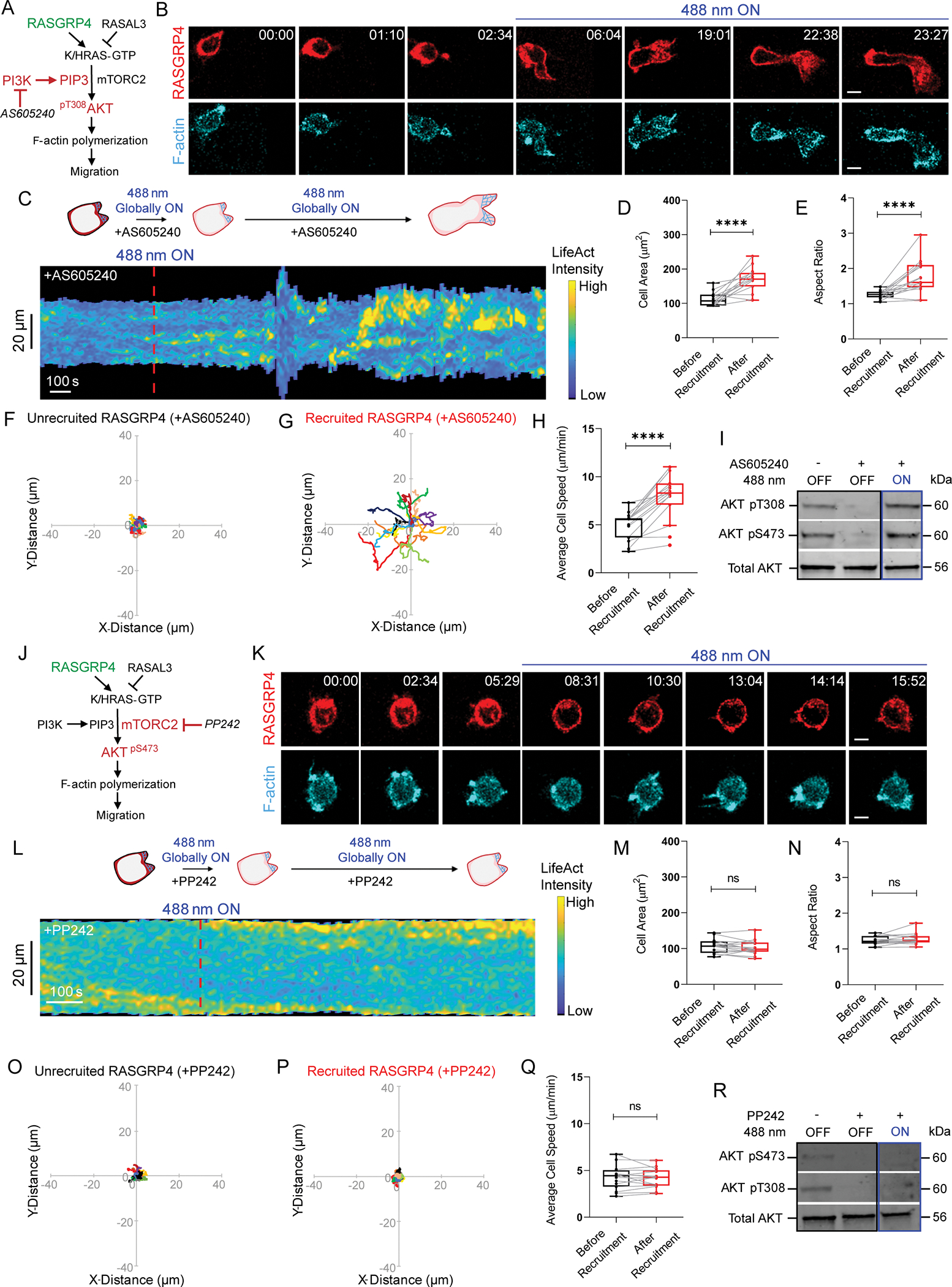
Strategy for testing effects of (A) PI3Kγ inhibitor, AS605240, or (J) mTor inhibitor, PP242 on RasGRP4-mediated F-actin polymerization and migration. Confocal images of (B) AS605240- or (K) PP242-treated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; upper panel) and LifeActmiRFP703 (cyan; lower panel), before or after global 488 nm illumination. Time and scale bars as in Figure 1. (C, L) Representative kymographs of cortical LifeAct intensity in AS605240- or PP242-treated RasGRP4-expressing neutrophil of panel B or K, before and after 488 nm illumination. Cartoon depicts recruitment, F-actin polymerization or cell shape status corresponding to the kymographs. Box-and-whisker plots of (D, M) cell area, (E, N) aspect ratio and (H, Q) average speed, before (black) and after (red) RasGRP4 recruitment in AS605240- or PP242-treated neutrophils, respectively. nc=15 from atleast 3 independent experiments. Statistical analyses as in Figure 1. Centroid tracks of AS605240- or PP242-treated neutrophils (nc=15) showing random motility before (F, O) or after (G, P) RasGRP4 recruitment, respectively. Tracks presented as in Figure 1. (I, R) Representative western blots demonstrating phospho-Akt levels (Thr308 and Ser473; 60kDa) in untreated/unrecruited, AS605240- or PP242-treated/unrecruited or AS605240- and PP242-treated/RasGRP4-recruited whole cell lysates. Total Akt (56kDa) was used as loading control. For each condition, results were triplicated (n=3). See also Figures S5–S6.
To test whether Ras regulates mTorC2 signaling in neutrophil polarization and migration, we treated RasGRP4-expressing cells with mTor inhibitor, PP242 (Figure 5J)112. Inhibited cells rounded up and ceased motility quickly, and we did not observe any recovery upon recruiting RasGRP4 (Figure 5K–Q, Video S3H). The mTorC1 inhibitor, rapamycin, did not affect neutrophil shape or size, suggesting that PP242-induced cytoskeletal changes were due to mTorC2 inhibition, and not mTorC1 (Figure S6D–E). Additionally, Akt phosphorylation at both Thr308 and Ser473 failed to recover with RasGRP4 recruitment in PP242-treated population (Figures 5R, S6F–H), suggesting Ras activates Akt via mTorC2 complex.
Ras-mTorC2 effector, Akt, activates cellular protrusions and migration
We next asked whether Akt activity directly affects cellular protrusive activity. There are two Akt isoforms in neutrophils, Akt1 and Akt2. Despite several studies, there is no clear consensus as to whether the effect of each on motility is positive or negative53,54. To resolve this dilemma, we transiently recruited Akt1 to the quiescent back of a moving cell. As a result, front protrusions started retracting, and a broad new protrusion soon emerged at the site of recruitment (Figure 6A–C, E, Video S4A). This phenomenon was observed consistently across the population, as highlighted in the angular histogram (Figure 6D). To test if the polarity reversing ability of Akt1 was due to its kinase activity, we similarly recruited a Akt kinase-dead mutant, Akt1T308A, to the cell rear113. Akt1T308A mutant could not bias fresh protrusion formation at its recruitment site and polarity remained unaffected, as confirmed by its angular histogram (Figure S6I–M, Video S4B).
Figure 6. Spontaneous Akt1 activation rearranges polarity and promotes migration even in presence of PI3K inhibitors.
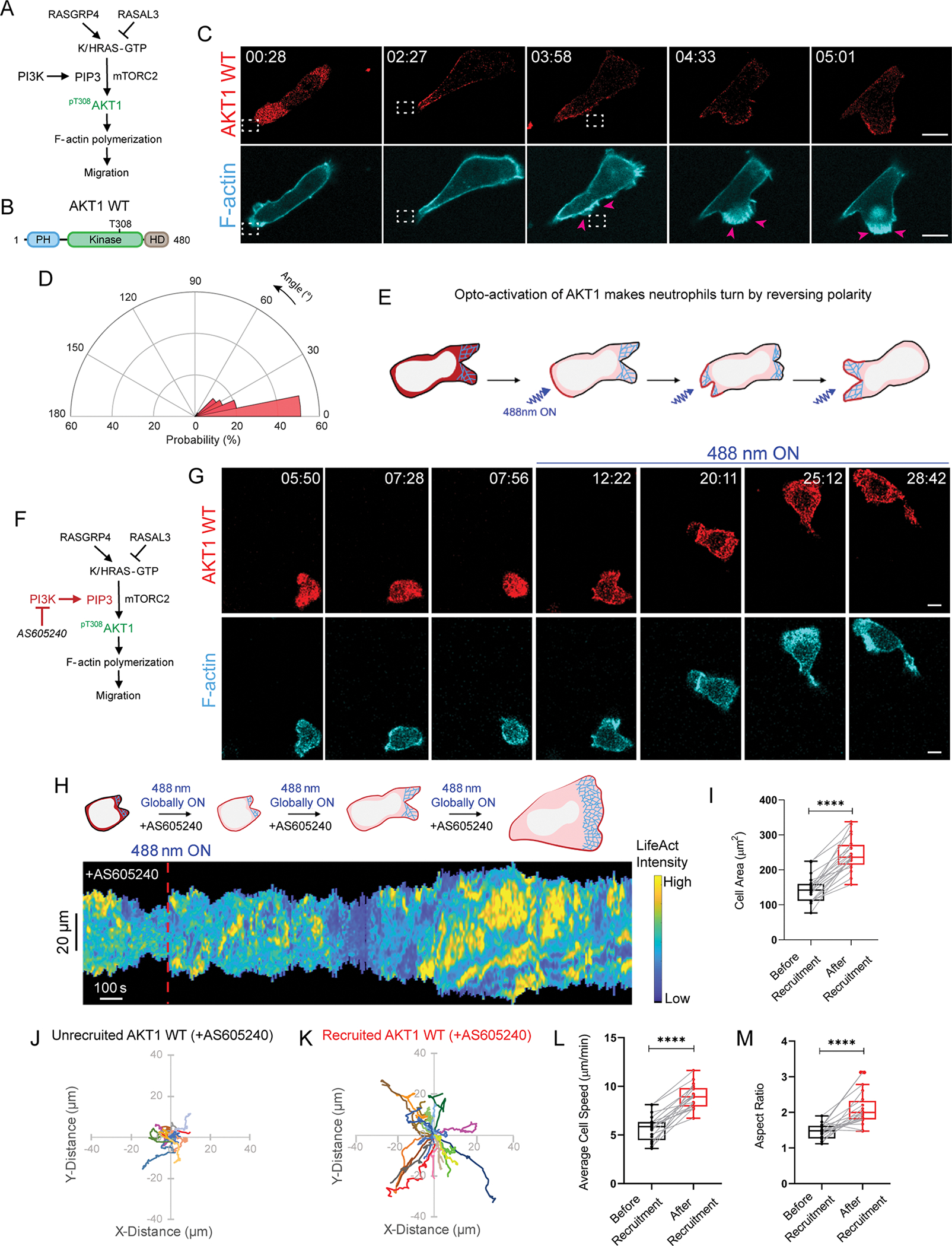
Strategy for testing if Akt1 opto-activation promotes F-actin polymerization and migration in absence (A) or presence (F) of PI3Kγ inhibitor, AS605240. (B) Cartoon of Akt1 WT protein sequence with PH, kinase, and hypervariable domains, highlighting T308 phosphorylation as critical for kinase activity. (C, G) Time-lapse confocal images of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-Akt1 WT (red; upper panel) and LifeActmiRFP703 (cyan; lower panel). Akt1 WT was recruited (C) exclusively to cell back by applying 488 nm laser near it (dashed white box) or (G) all over cell boundary by global illumination. In C, pink arrows highlight Akt1 WT-induced new protrusion formation. Time and scale bars as in Figure 1. (D) Polar histogram demonstrates higher probability of fresh protrusion formation near recruitment area; nc=19 and np=49. (E) Cartoon illustrating Akt1 opto-activation at the back makes neutrophils turn by reversing polarity. (H) Representative kymograph of cortical LifeAct intensity in AS605240-treated Akt1 WT-expressing neutrophil in panel G, before and after 488 nm laser was turned on. Color map as in Figure 1. Cartoon depicts recruitment, F-actin polymerization or cell shape status corresponding to kymograph. Box-and-whisker plots of (I) cell area, (L) average speed, and (M) aspect ratio, before (black) or after (red) global Akt1 WT recruitment. Statistical analyses as in Figure 1. Centroid tracks of neutrophils showing random motility before (J) or after (K) recruitment. Tracks presented as in Figure 1. nc=20 from atleast 3 independent experiments. See also Figures S6–S8.
To test if protrusion-promoting effects of Akt are isoform-specific, we recruited Akt2 over the entire cell perimeter. After 6 minutes of recruitment, non-polarized, motility-incompetent neutrophils developed F-actin at the front, polarized, and eventually migrated with broad lamellipodium (Figure S7A, Video S5A). Representative kymograph showed a gradual increase in LifeAct intensity along with widening of cell area (Figure S7B). Akt2 activation caused 83%, 47%, and 21% improvement in cell area, migration speed, and aspect ratio, respectively (Figure S7C–G). No such phenotypic change was observed when CRY2PHR, without being fused to Akt2, was recruited (Figure S3H–N, Video S1E). Additionally, Akt2 recruitment induced 3-fold increase in Ras activation, suggesting Akt may have positive feedback to Ras in migrating neutrophils (Figure S7H–J).
Are Akt-mediated cytoskeletal effects conserved across cell types and species? To answer this, we globally recruited PKBA (Akt1 homolog) on the Dictyostelium membrane, using iLID system114,115. Within a minute, rounded, quiescent cells polarized and migrated efficiently (Figure S7K, Video S5B). This phenomenon did not occur when control SsrA binding partner (SsbB) lacking PKBA was globally recruited, suggesting PKBA activation was responsible, and not light irradiation or SspB recruitment (Figure S7L, Video S5C). We proved that both Akt isoforms positively regulate protrusion generation and migration in an evolutionary conserved manner.
Akt function overcomes PI3K inhibitors
It is well established that PI3K leads to phosphorylation of Akt kinase to promote proliferation, survival, and metabolism, in response to extracellular signals116. We checked if that holds true in neutrophil polarization and migration. We treated Akt1-expressing neutrophils with PI3Kγ inhibitor, AS605240, which removed polarity, depleted PIP3, and strongly suppressed motility. Interestingly, within 10 minutes of Akt1 recruitment, cells re-polarized, formed sustained lamellipodium, and migrated (Figure 6F–G, S5A, Video S5D). Kymograph showed slightly delayed increase in F-actin polymerization and cell spreading after switching on laser (Figure 6H). Box and whisker plots showed 70%, 54%, and 47% increment in cell area, speed, and polarity, respectively (Figure 6I–M). Non-recruiting cells in same population were unable to polarize, spread, or move (Figure S8A–G). Upon treatment with pan-PI3K inhibitor, LY294002, we observed a similar recovery in neutrophil polarity, actin polymerization, and migration after Akt1 recruitment (Figure S8H–K, Video S5E). Thus, PI3K-inhibited neutrophils could be activated only when detectable Akt1 accumulated on the membrane. We next checked whether kinase activity of Akt1 was responsible for these cytoskeletal changes. For this, we globally recruited kinase-dead mutant, Akt1T308A, in PI3K-inhibited cells, which could not rescue actin polymerization, cell spread, motility or polarity (Figure S8L–N, Video S5F). Kymograph, bar plot and cell track analyses confirmed our observations (Figure S8O–T). Altogether, we confirmed that Akt phosphorylation and activation can occur even in presence of PI3K inhibitors.
Discussion
Our studies show that local activities of growth factor signaling cascades on the plasma membrane directly regulate immune cell polarity and migration. Our optogenetic approaches rule out secondary effects of long-term adaptation and assess immediate effects of localized activations. In absence of chemoattractant-mediated receptor stimulation, global increase in Ras activity promoted F-actin-driven lamellipodium formation, stable polarization, and persistent motility in neutrophils. Moreover, strategic activation of Ras alone at inactive, back regions of the cell membrane could generate fresh protrusions thereby reversing pre-existing polarity. Additionally, stably localized Ras activity caused persistent, directional migration in these cells. Although similar effects can be brought about by local G-protein coupled receptor or cytoskeletal activation, our approach showed that localized activity of single components of Ras-mTorC2-Akt growth factor network could directly alter cell behavior. Moreover, our methods allowed us to locally inhibit Ras activity at the front, which extinguished existing protrusions and created a new cell rear. Importantly, local Ras inhibition at the fronts of chemotaxing cells abrogated directed migration in gradients of different classes of attractants, suggesting Ras activity is necessary for chemotaxis. Ras-mediated cytoskeletal effects worked primarily through mTorC2 complex. Consistently, spontaneously activating the Ras/mTorC2 effector, Akt, could reverse polarity and induce actin polymerization-driven protrusions and migration even in presence of PI3K inhibitors. Our results suggest that previously reported pathways for directed migration are likely part of a larger network, and compel us to include Ras-mTorC2-Akt growth factor pathway as a central part of the mammalian chemotactic network (Figure 7).
Figure 7. Proposed model of local growth factor network activation-mediated actin polymerization and migration.
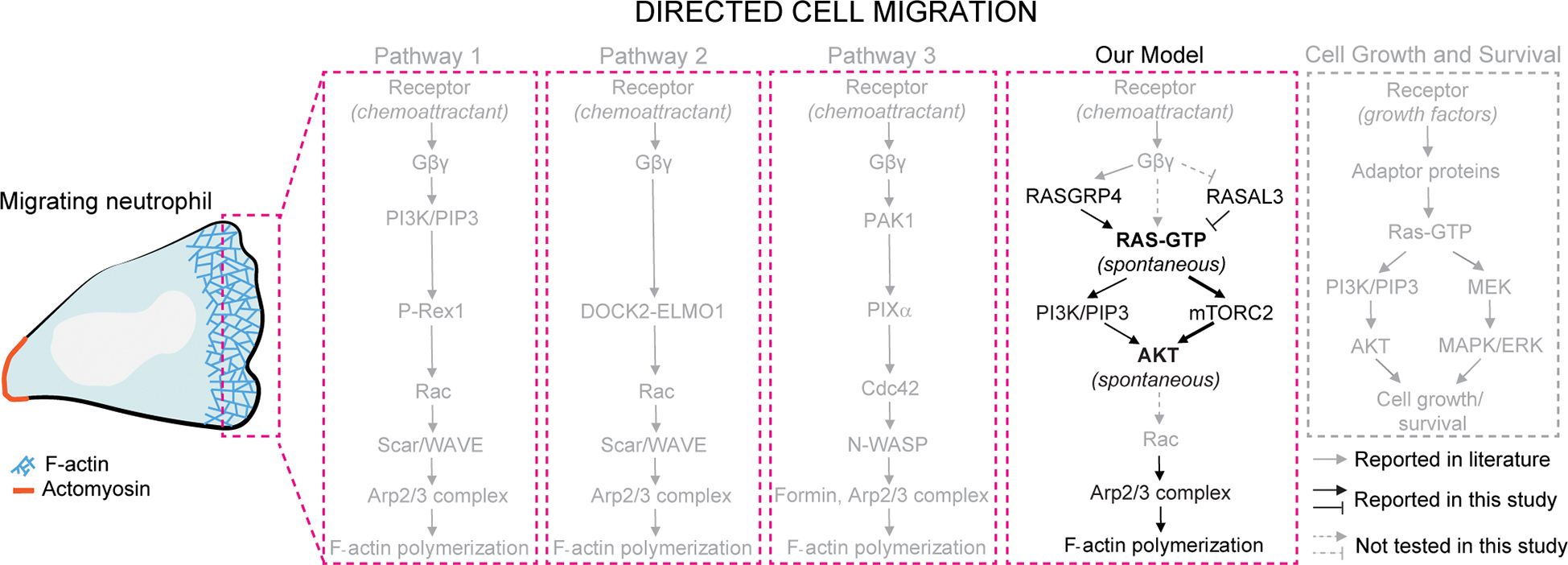
The prevalent pathways in directed cell migration are illustrated in grey within pink dotted boxes. In pathway 1, chemoattractant-mediated G-protein coupled receptor stimulation activates PI3K/PIP3 which, through Rac and Scar/WAVE, initiates Arp2/3 complex-mediated actin polymerization and migration. Pathways 2 and 3 are PI3K-independent ones where receptor-Gβγ-DOCK2-ELMO1 or receptor-Gβγ-PAK1-PIXα mediated Rac or Cdc42 activation causes F-actin formation at the leading edge. In our illustration, we have not included feedback loops in these pathways. Our model demonstrates that chemoattractant/G-protein network-independent localized Ras activity is modulated by RasGRP4 and RASAL3. Ras locally activates mTorC2, and not PI3K, to activate Akt which triggers Arp2/3-based actin polymerization, protrusion formation, and directed migration even in absence of chemoattractant gradients. Moreover, spontaneous Akt activation directs cytoskeletal organization and migration even when PI3K signaling is strongly suppressed. We did not test whether Ras is activated by Gβγ or Akt triggers Rac (grey dashed arrows). Altogether, Ras-mTorC2-Akt growth factor network is integrated into mammalian chemotactic network.
In this study, we optogenetically actuated nodes in signaling networks that are conventionally associated with cell growth and survival as opposed to cytoskeletal organization or motility. Recently, optically perturbing receptor/G-protein signaling or downstream cytoskeletal activities of Rac, Cdc42, and RhoA, helped steer actin-driven migration. These studies highlighted that local activation or inactivation of single components can enhance existing polarity or reverse it and re-establish migration in a new direction18,78–88. We show that activation of single components of growth control pathways is not only sufficient for directed migration but necessary for chemoattractant-directed motility. These results likely explain why components of these signaling cascades have been reported as important for immune response. Altogether, these studies suggest that traditional concepts of “upstream” and “downstream” pathway components are an outdated and simplistic view, and that single component activation at multiple nodes can trigger global networks through extensive crosstalk and feedbacks.
Prioritizing response between different groups of chemoattractants allows neutrophils to transition between guidance cues and perform their physiological functions. ‘End-target’ attractants, such as fMLP and C5a, are pathogen- or injury-derived and compel neutrophils to focus their cytotoxic functions on their targets. ‘Intermediary’ chemoattractants, such as LTB4 and IL8, are secreted from those cells and guide more neutrophils out of vasculature and towards the pathogen or injury site. Although reports propose that PI3K-Akt or p38 MAPK signaling is needed for migratory response to intermediary or end-target signal, respectively, molecular mechanisms underlying this prioritization are unclear105–108,117. However, both chemoattractant types stimulate receptor to activate Rac, Cdc42 and RhoA and polymerize actin118,119. Our local RASAL3 recruitment results suggest that productive Ras signaling at fronts of chemotaxing cells in gradients is essential for both types of attractants. We conclude that the two signaling pathways, mediating decision-making, are fundamentally similar.
Previous studies have attributed polarity in neutrophils to various factors. A compelling argument was made for membrane tension as an inhibitor of cytoskeletal activation120–122. Our results are consistent with a membrane or cortical tension scheme since eliciting a protrusion at the back with RasGRP4 is accompanied by a collapse of the front, and conversely, causing a collapse of a front with RASAL3 leads to protrusion formation in the erstwhile back. Other experiments suggest that cell cortex and cytoskeleton integrity, but not necessarily its dynamics, is essential for polarity94. We do not directly test this model since our experiments always involved dynamic cytoskeletal changes. However, using our optical tools, role of the cytoskeleton could be tested in neutrophils immobilized with a cocktail of cytoskeleton-stalling inhibitors. Would optical localization of a single component, such as Ras, recruit other components in the signaling cascade to the same place, or would localization of the second component be also subjected to the polarity axis? Interesting questions like these could be answered using our system.
Some of the most notable regulators of mammalian chemotaxis are PI3K/PIP3 and mTorC251,122–125. PIP3 is localized at leading edge of migrating cells but its precise role in chemotaxis has been debated22,29,35–43. In neutrophils, recruitment of PI3K and focal production of PIP3, elicits a local protrusion27,51. In cells lacking PTEN or Ship1, which have elevated levels of PIP3, protrusion formation is drastically altered126–128. Furthermore, PIP3 was required for leading edge protrusion formation elicited by local optogenetic activation of Rac18. In our studies, PIP3 was required for basal motility but was largely dispensable for Ras-mediated protrusion formation. On the other hand, inhibitors of mTor, but not mTorC1, completely blocked basal and RasGEF-induced motility, suggesting that mTorC2 was critical for both. In Dictyostelium, TorC2 is important but not essential for chemotaxis4,63,129,130. Using optogenetic approaches to combine perturbations and expansion to many more components will help unravel networks.
Traditionally, growth factors activate PI3K which leads to phosphorylation of activation loop of Akt in almost all cell types42,131. Here, we showed that Akt activation may take place even when PI3K signaling is strongly suppressed. When PI3K was pharmacologically inhibited in neutrophils, polarity and migration were drastically improved by optically activating Akt on the membrane. Here, Akt may either be phosphorylated by PDKs independently of PIP3, or residual PIP3 in PI3K-inhibited cells may be sufficient in activating recruited Akt132. Thus, productive PI3K signaling may not be as important for functional Akt activation.
Despite being activated through similar mechanisms, it has been reported that Akt isoforms perform different roles in growth and metabolism133,134. In migration, it has been suggested that Akt1 and Akt2 serve opposing functions135–139. Even with the same isoform, opposing functions have been implied in different experiments115,140. We suggest that these differences might be attributable to selective genetic compensation, which can occur in knockout and overexpression studies, or to different assays in different cell types. Using our optogenetic tools to bypass such experimental artefacts, we demonstrated that, irrespective of isoform or cell type, spontaneous Akt activation promotes cell polarization and motility.
Although neutrophils’ intimate involvement in diseases make them ideal therapeutic targets, measures are required to delimitate their harmful side-effects from beneficial responses. Current neutrophil-based therapeutics primarily target cell surface chemokine receptors141. Since chemokine receptors activate multiple downstream signaling pathways, pharmacologically targeting them have several off-target effects. Our optogenetic study in model leukemic neutrophils showed that individually targeting activities of these signaling components have strong effects on motility and polarity. We provided proof-of-concept that targeting specific signaling nodes affects neutrophil behavior, but with greater control.
Limitations of the study
Although we discovered mechanisms of directed migration in mammalian cells, the study has several limitations. First, migration involves a network of molecular interactions. The experiments were limited to the study of just a few components which were chosen based on our hypotheses. Our experiments demonstrate that local activation of growth factor pathway components cause immediate, local protrusions to form. Our diagrams suggest that growth factor pathway feeds into previously proposed events including PI3K and Rac activation, leading to cytoskeletal rearrangements. We cannot rule out that activation of growth factor pathway modulates cytoskeletal activity differently. Second, we speculate that our perturbations set in motion the coordinated activation of many components controlling polarity and migration. However, due to technical limitations, we could monitor only a few. A powerful approach would be to monitor numerous biosensors during optogenetic perturbations. Perhaps all act in concert or only certain ones are involved.
STAR Methods
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter N. Devreotes (pnd@jhmi.edu)
Materials Availability
All unique reagents generated in this study are available from the lead contact without restriction.
Data and Code Availability
All data are provided in the main or supplementary text. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Female human neutrophil-like HL-60 cell line was a kind gift from Dr. Orion Weiner (UCSF). Cells were cultured in RPMI medium 1640 containing L-glutamine and 25 mM HEPES (Gibco; 22400–089). This medium was supplemented with 15% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific; 16140071) and 1% penicillin-streptomycin (Thermo Fisher Scientific; 15140122). For subculturing, cells were split at a density of 0.15 million cells/ml and passaged every 3 days. Stable lines were maintained similarly, in presence of selection antibiotics. For differentiating, 1.3% DMSO was added to cells at a density of 0.15 million cells/ml, followed by incubation for 5–7 days, before experimentation142. This myeloid leukemia cell line, upon differentiation, serves as an effective model to investigate human neutrophils61,143. Stable lines of HL-60 cells were similarly differentiated, and selection antibiotics were removed for experimentation.
Male mouse monocyte/macrophage RAW 264.7 cell line was obtained from Dr. N. Gautam (Washington University School of Medicine). Cell line was maintained in DMEM medium containing 4.5 g/L glucose, sodium pyruvate, and sodium bicarbonate (Sigma-Aldrich; D6429). Culture medium was supplemented with 10% FBS and 1% penicillin-streptomycin. At ~90% confluency, cells were harvested by scraping and subcultured at a split ratio of 1:4144.
Female human embryonic kidney HEK293T cell line was grown in DMEM medium containing 4.5 g/L glucose and sodium pyruvate (Gibco; 10569–010). Medium was supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were maintained according to ATCC guidelines.
All mammalian cells were maintained under humidified conditions at 37°C and 5% CO2, and experiments were done with cells at low passage.
Wild type Dictyostelium discoideum cells of the axenic strain AX2 were used in this study. Cells were subcultured, either in suspension or on tissue culture dishes, in HL-5 medium at 22°C. Stable cell lines, generated by electroporation, were grown in presence of hygromycin B and G418 sulphate. All experiments were carried out within 2 months of thawing the cells from frozen stocks.
METHOD DETAILS
Preparation of reagents and inhibitors
Fibronectin (Sigma-Aldrich; F4759–2MG) was dissolved in 2 ml sterile water, followed by dilution in 8 ml PBS to make a stock solution of 200 μg/ml. N-Formyl-Met-Leu-Phe (fMLP, Sigma-Aldrich; 47729) was dissolved in DMSO (Sigma Aldrich; D2650) to make stock solution of 10 mM. FKP-(D-Cha)-Cha-r (ChaCha peptide, Anaspec; 65121) was dissolved in PBS to prepare a 2.5 mM stock solution. Leukotriene B4 (LTB4, Cayman Chemical; 20110) was purchased as a ready-made stock solution of 297 μM in ethanol. AS605240 (Sigma-Aldrich; A0233), LY294002 (Invitrogen; PHZ1144) or PP242 (EMD Millipore; 475988) was dissolved in DMSO to make a stock solution of 20 mM, 50 mM or 20 mM, respectively. Pertussis toxin (PTX, Sigma-Aldrich; 516560–50UG) was reconstituted in 500 μl sterile water to prepare a 100 μg/ml stock. Gallein (Sigma-Aldrich; 371708–5MG) or CK-666 (EMD Millipore; 182515) was dissolved in DMSO to make 20 mM or 50 mM stock solution, respectively. Janelia Fluor 646 HaloTag (Promega Corporation; GA1120) was dissolved in DMSO to make a 200 μM stock. 2.5 mg/ml puromycin (Sigma-Aldrich; P8833) or 10 mg/ml blasticidine S (Sigma-Aldrich; 15205) stock was prepared in sterile water. Hygromycin B (Thermo Fisher Scientific; 10687010) or G418 sulphate (Thermo Fisher Scientific; 10131035) was available as ready-made 50 mg/ml stock solution. All stocks were aliquoted and stored at −20°C. According to experimental requirements, further dilutions were made in PBS or culture medium before adding to cells.
Cloning
All DNA oligonucleotides were ordered from Sigma-Aldrich. For lentiviral constructs, CIBN-CAAX (Addgene #79574) or LifeActmiRFP703 (Addgene #79993) ORF was subcloned into pLJM1-eGFP plasmid (Addgene #19319), in place of the eGFP gene, using primer sets P1/P2 or P3/P4 respectively (Table S1). We procured PiggyBac™ transposon system from Dr. Sean Collins (UC Davis) which consists of a transposon and a transposase expression plasmid145. CRY2PHR-mcherry2 gene (Addgene #26866) was first sub-cloned into the transposon plasmid using primers P5/P6. Next, at the C-terminal of CRY2PHR-mcherry2 gene in the transposon plasmid, we introduced HRas G12V ΔCAAX (Addgene #18666), KRas4B G12V ΔCAAX (Addgene #9052), RasGRP4 (Addgene #70533), RASAL3 (Addgene #70521), Akt1 (Addgene #86631), Akt1T308A (Addgene #49189), or Akt2 (Addgene #86593) gene using primer sets P7/P8, P9/P10, P11/P12, P13/P14, P15/P16, P17/P18, or P19/P20, respectively (Table S1). pFUW2-RFP-PH-Akt construct was developed previously in our lab146.
For non-viral constructs, RASAL3 (Addgene #70521) or RasGRP4 (Addgene #70533) was subcloned into pCRY2PHR–mCherryN1 plasmid (Addgene #26866), at the C-terminal of CRY2PHR-mcherry2 gene, using primers P21/22 or P23/24 (Table S1). pEGFPN1-Lifeact-7Alinker-Halo7 plasmid was a kind gift from Dr. Akihiro Kusumi (Okinawa Institute of Science and Technology Graduate University)147.
For Dictyostelium constructs, CAAX-deleted Venus-iLID (Addgene #60411) fused to plasma membrane targeting N150 fragment or tgRFPt-SSPB R73Q (Addgene #60416) gene was subcloned in pDM358 (dictyBase #534) or pCV5 (dictyBase #23) plasmid using AgeI/SbfI or AgeI/BamHI restriction digestion, respectively. Next, at the C-terminal of tgRFPt-SSPB R73Q gene in pCV5, we introduced PKBA gene using primer sets P25/26 (Table S1).
All constructs were verified by diagnostic restriction digestion and Sanger sequencing (JHMI Synthesis and Sequencing Facility).
Stable cell line construction
Stable expression lines in HL-60 cells were generated by a combination of 3rd-generation lentiviral- and PiggyBac™ transposon-integration based approaches78,148. Virus was prepared in HEK293T cells grown to ~80% confluency in 10 cm cell culture dish (Greiner Bio-One; 664160). For each reaction, a mixture of 3.32 μg pMD2.G (Addgene plasmid #12259), 2 μg pMDLg/pRRE (Addgene plasmid #12251), 4.64 μg pRSV-Rev (Addgene plasmid #12253), and 10 μg pLJM1 construct with gene of interest (CIBN-CAAX or LifeActmiRFP703) or pFUW2-RFP-PH-Akt construct were transfected using Lipofectamine 3000® as per manufacturer’s instructions (Invitrogen; L3000–008)149. After 96 hours, virus containing culture medium was harvested at 3000 rpm for 20 mins at 4°C. In a 6-well plate (Greiner Bio-One; 657160), entire viral medium was added to 4×106 HL-60 cells (seeded at a density of 0.25×106 cells/mL) in presence of 10 μg/mL polybrene (Sigma; TR1003). After 24 hours, viral medium was removed, and cells were introduced to a mix of fresh and conditioned (mixed) culture medium. For selecting LifeActmiRFP703 or RFP-PH-Akt expressors, infected cells were sorted after 5 days, and subsequently, grown to confluency. For selecting CIBN-CAAX-expressors, infected cells were allowed to recover for 24 hours, and subsequently, incubated with 1 μg/mL puromycin in 24-well cell culture plate (Greiner Bio-One; 662160) for 4–5 days. Resistant cells were grown to confluency in puromycin.
For transposon integration in HL-60 cell line, 5 μg transposon plasmid, containing CRY2PHR-mcherry2 fused to a gene of interest, was co-electroporated with an equal amount of transposase expression plasmid into two million cells using Neon™ transfection kit (Invitrogen; MPK10025B). Cells and DNA mix were resuspended in buffer ‘R’ before electroporation in 100 μl pipettes at 1350 volts for 35 ms in Neon™ electroporation system (Invitrogen; MPK5000). Cells were resuspended in mixed culture medium in a 6-well plate and allowed to recover for 24 hours. Post-recovery, transfected cells were selected in presence of 10 μg/mL blasticidine S for 5–6 days. Once blasticidine S was removed, resistant cells were transferred to 48-well cell culture plate (Sarstedt; 83.3923), and further grown over 3–4 weeks into stable cell lines. Stable cells were maintained throughout in Blasticidine S.
Stable lines in Dictyostelium cells were generated by electroporation as described previously130,150. Briefly, 5×106 cells were harvested, washed and resuspended in 100 μl chilled H-50 buffer. Next, 2 μg each of N150-Venus-iLID/pDM358 and tgRFPt-SSPB R73Q-CTRL/pCV5 (control) or tgRFPt-SSPB R73Q-PKBA/pCV5 plasmids were mixed with cell suspension, and transferred to chilled 0.1 cm-gap cuvette (Bio-Rad, 1652089). Transfection took place over two rounds of electroporation at 850 V and 25 μF with an interval of 5 secs (Bio-Rad Gene Pulser Xcell Electroporation Systems, 1652660). Electroporated cells were incubated on ice for 10 mins, and subsequently transferred to HL-5 culture medium, supplemented with heat-killed Klebsiella aerogenes (lab stock), in 10 cm cell culture dish. On the following day, 50 μg/ml hygromycin B and 20 μg/ml G418 sulphate were added to cells and selected over 3–4 weeks.
Transient transfection
RAW 264.7 macrophage-like cells were transiently transfected by nucleofection using Amaxa cell line kit V (Lonza; VACA-1003) as previously described130,144. Briefly, 3×106 cells were harvested and added to 100 μl supplemented Nucleofector Solution V containing 0.7 μg CIBN-CAAX (Addgene #79574), 1.1 μg CRY2PHRmcherry2-RASAL3 or RasGRP4, and 0.7 μg pEGFPN1-Lifeact-7Alinker-Halo7151. Cell and DNA were mixed gently, transferred to a Lonza cuvette and electroporated with Amaxa Nucleofector II device (Lonza; LOAAB-1001) using program ‘D-32’. After a single pulse, cells were transferred carefully to 0.5 ml pre-warmed culture medium and incubated at 37°C and 5% CO2 for 10 mins. Subsequently, 2×105 cells were transferred to each well of 8-well chambered coverglass (LAB-TEK; 155409) and left in the incubator for 1 hour. Next, 0.5 ml warm culture medium was added to each sample and incubated for 4 hours before imaging.
Microscopy and FACS
Two microscopes were used for time-lapse imaging: 1) Zeiss LSM780-FCS single-point, laser scanning confocal microscope (Zeiss Axio Observer with 780-Quasar confocal module) and 2) Zeiss LSM800 GaAsP single-point, laser scanning confocal microscope with a wide-field camera. In these microscopes, argon laser (488 nm excitation) was used for GFP or Venus visualization, solid-state laser (561 nm excitation) was used for mcherry2 or RFP, and diode laser (633 nm excitation) was used for miRFP703 or Janelia Fluor 646 HaloTag. 40X/1.30 Plan-Neofluar oil objective was used, along with digital zoom. Microscopes were equipped with temperature-controlled chamber held at 5% CO2 and 37°C for imaging mammalian cells. Zeiss 780 and 800 were operated by ZEN Black and ZEN Blue software, respectively.
For high-speed sorting, we used two instruments: 1) BD FACSAria IIu cell sorter and 2) SH800S cell sorter (Sony). We used 561 nm excitation laser to sort for RFP expression, and 633 nm excitation to sort miRFP703 expressors. Briefly, cells were harvested, resuspended in sorting buffer (1x PBS, Ca2+/Mg2+ free; 0.9% heat-inactivated FBS; 2% penicillin-streptomycin) at a density of 15×106 cells/ml, and sorted using 100 μm microfluidic sorting chip. High expressors (top 1%-10%) were collected in fresh culture medium (containing 2% penicillin-streptomycin) and grown to confluency.
Optogenetics
All optogenetic experiments with differentiated HL-60 cells, except for chemotaxis assays, were done in absence of any chemoattractant. On day of the experiment, differentiated neutrophils were allowed to attach on chambered coverglass, coated with fibronectin at a density of ~35 μg/cm2, for 40 mins. Next, depending on experimental requirements, attached cells were treated with inhibitors (20 μM AS605240, 50 μM LY294002, 20 μM PP242, 20 μM gallein, or 100 μM CK-666) for 10 mins before imaging. For pertussis toxin (PTX) treatment, differentiated cells were incubated with 1 μg/ml PTX for 20 hours before imaging. For global recruitment experiments on the Zeiss LSM780 microscope, 488 nm excitation laser was switched on after imaging for at least 300 secs. Photoactivation and image acquisition were done once every 7 secs for single plane imaging. Laser intensity during image capture was at a low level (laser power of 1.7% or ~0.05 mW at the objective) so that protein recruitment over the entire cell periphery was maintained throughout without inducing light damage. For local recruitment studies on the Zeiss LSM800 microscope, a small region of interest was drawn (shown as dashed white box in images or solid yellow box in videos), which was irradiated with 488 nm laser (power of 5% or ~0.21 mW at the objective) in multiple iteration. For optically-driven migration experiments, regions of interest were repositioned as neutrophils or macrophages migrated, to maintain an approximately constant distance between them and the cell boundary. In chemotaxis assays, regions of interest were positioned continuously close to F-actin rich fronts of migrating cells. 488 nm excitation laser was switched on after imaging for at least 120 secs.
We discovered that pre-treatment of differentiated, migration-competent HL-60 cells with heat-killed Klebsiella aerogenes vastly improved efficiency of CRY2-CIBN optogenetic system. Briefly, we treated 107 differentiated cells, grown on 10 cm cell culture dish, with 13 μg/ml heat killed Klebsiella aerogenes (lab stock) for 7 hours. Dead bacteria were rinsed off along with non-adherent neutrophils on fibronectin-coated chambered coverglass, and all imaging was completed within 5 hours.
Transiently transfected RAW 264.7 cells were transferred to 450 μL pre-warmed HBSS buffer supplemented with 1 g/L glucose, and subsequently stained with 5 nM Janelia Fluor 646 HaloTag for 15 mins. During imaging, cells were activated with 10 μM C5a-receptor agonist FKP-(D-Cha)-Cha-r. Local recruitment studies were performed similarly to HL-60 cells.
Optogenetic experiments with Dictyostelium cells were done in the absence of any chemoattractant. Cells were allowed to attach on chambered coverglass for 30 mins before imaging. For recruitment experiments, 488 nm excitation laser was switched on after imaging for at least 250 secs. Photoactivation and image acquisition were done once every 5–10 secs for single plane imaging. Very low laser intensity (10–20% of what was used for mammalian optogenetics) was sufficient to stably recruit SspB over the cell perimeter without inducing light damage.
2D Chemotaxis assay
All chemotaxis assays using chemotaxis μ-slides (Ibidi; 80322) were performed as described previously152,153. Briefly, 2×106 differentiated HL-60 cells, pre-treated with heat-killed bacteria, were harvested, and resuspended in 100 μl fresh culture medium. 20 μl cell suspension was loaded in the collagen-coated channel and cells were allowed to adhere for 1 hour. Subsequently, each reservoir was filled with 55 μl fresh medium. On the Zeiss LSM800 microscope, 30 μl 10 nM freshly-made chemoattractant solution (fMLP, ChaCha peptide or LTB4) was added to the right reservoir and imaging was started immediately. Photoactivation and image acquisition of chemotaxing cells was done for upto 2 hours, as described in the ‘optogenetics’ section.
Akt phosphorylation assay
This assay was performed as described previously with minor modifications27. Briefly, 107 differentiated and pre-treated opto-RasGRP4 expressing HL-60 cells were harvested and resuspended in 1 mL culture medium. Next, cell suspension was treated with 20 μM AS605240 or PP242 for 10 mins. The inhibitor-treated sample was split equally into two wells of a 6-well plate; one well was illuminated for 10 mins using a 470 nm LED with a current output of 900 mA (Lightspeed Technologies) while the other well was not. An untreated and unilluminated control was maintained throughout the experiment. Finally, cells from all samples were harvested, resuspended in 225 μl 1× SDS sample buffer, and boiled for 5 mins for western blot analysis.
Ras activation assay
This assay was done using Ras activation assay biochem kit (Cytoskeleton, Inc.; BK008S) as per the manufacturer’s instructions. Briefly, 4×107 differentiated and pre-treated opto-Akt2 expressing HL-60 cells were harvested, resuspended in 500 μl culture medium and illuminated with 470 nm LED for 10 mins as described previously. An unilluminated control was maintained throughout the experiment. Next, samples were placed on ice, media was aspirated, and washed with chilled 1×PBS. Cells were then resuspended in 1 ml ice-cold lysis buffer supplemented with 1× protease inhibitor cocktail and lysed for 10 mins on ice. Immediately, lysates were clarified at 10,000×g, 4°C for 5 mins. 100 μl of each supernatant (input) was removed for western blot quantitation of total Ras. Remaining 900 μl from each supernatant was incubated with 100 μg (30 μl) agarose beads conjugated with RBD (active Ras binding domain of human Raf1) at 4°C for 1.5 hours. The Raf-RBD beads were pelleted by centrifugation at 3500×g at 4°C for 1 min, washed once with 500 μl wash buffer, and finally resuspended in 30 μl 2× SDS sample buffer. Beads and inputs were boiled for 5 mins for subsequent western blot analysis.
Immunoblotting
For western blots, protein samples equivalent to at least 106 cells or 30 μl beads, were loaded per well of pre-cast 4–15% polyacrylamide gels (Bio-Rad; 5671085) at 100 V for 1.5 hours. Gels were transferred onto PVDF membranes (Bio-Rad; 162–0262) for 10 mins using a Trans-Blot Turbo semi-dry transfer apparatus (Bio-Rad; 1704150EDU). Membranes were blocked using Intercept Blocking buffer (Li-Cor; 927–60001) for 1 hour. Akt phosphorylation at Thr308 or Ser473 (~60 kDa) was detected with rabbit anti-phospho-AktT308 (Cell Signaling; 13038) or anti-phospho-AktS473 (Cell Signaling; 4060) antibody, respectively. Total Akt (~56 kDa) was detected with rabbit anti-pan Akt antibody (Cell Signaling; 9272). Total GST tagged Raf1-RBD protein bound to beads was detected with rabbit anti-GST antibody (EMD Millipore; AB3282). These primary antibodies were used at a dilution of 1:1000. Total Ras (~20 kDa) was detected by mouse anti-pan Ras antibody (Cytoskeleton Inc.; AESA02) at a dilution of 1:250. After overnight primary antibody incubation at 4°C, blots were probed with goat anti-rabbit IRDye 680RD- or goat anti-mouse IRDye 800CW-conjugated secondary antibody (1:10,000 dilution; Li-Cor; 925–68071 or 925–32210) for 1 hour. Near-infrared signal from the blots was detected via Odyssey CLx imaging system (Li-Cor).
QUANTIFICATION AND STATISTICAL ANALYSIS
Image Analysis
Images were analyzed on Fiji/ImageJ 1.52i (NIH) and MATLAB 2019a (MathWorks, MA, USA) as described previously130. Results were plotted using GraphPad Prism 8 (GraphPad software, CA, USA) and Microsoft Excel (Microsoft, WA, USA).
Linescan intensity profile
Linescans were generated in Fiji/ImageJ 1.52i software. A straight line segment (width of 12 pixels) was drawn on the red channel, using the line tool option, across the cell. Using the “Plot Profile” option, we obtained the average intensity value along that line for the red channel. Values were normalized and graphed in Microsoft Excel.
Kymographs
Cell segmentation was done against the background using a custom code written in MATLAB 2019b, after standard image processing steps were carried out. Membrane kymographs were generated from segmented cells as described earlier154. We used a linear color map for normalized intensities, where blue indicated lowest intensity and yellow denoted highest.
Cell migration analysis
Analysis was performed by segmenting neutrophil cells in Fiji/ImageJ 1.52i software. For this, image stack was thresholded using the ‘Threshold’ option. ‘Calculate threshold for each image’ box was unchecked, and range was not reset. Next, cell masks were created by size-based thresholding using the ‘Analyzed particles’ option. To optimize binarized masks, ‘Fill holes’, ‘Dilate’, and ‘Erode’ were done several times. For creating temporal color-coded cell outlines, ‘Outline’ was applied on binarized masks, followed by ‘Temporal-Color Code’ option. Next, ‘Centroid’ and ‘Shape descriptors’ boxes were checked in ‘Set Measurements’ option under ‘Analyze’ tab. This provided us with values for centroid coordinates and aspect ratio. Mean and SEM from replicates of aspect ratio values were determined and plotted in GraphPad Prism 8. The starting point for centroid values was set to zero for each track, and these new coordinates were plotted in Microsoft Excel to generate migration tracks. Before- and after-recruitment tracks for a cell have the same color-code to aid comparison. Velocity was calculated by computing displacement between two consecutive frames. Displacement was then divided by time interval to obtain speed for each cell. These speed values were then time-averaged over all frames to produce data points for cell speed which were plotted as box-and-whisker graphs in GraphPad Prism 8.
Directional analysis of cells in Fig. 4A and B was done with semi-automatic segmentation code in MATLAB.
Chemotactic and Directional Indices
For cells moving in a chemoattractant gradient aligned along the -axis, the chemotactic index (CI) was computed using the formula:
which corresponds to the net displacement along the -axis, divided by the total displacement along the trajectory. For cells responding to a light stimulus, a similar formula was obtained. However, because the laser light is not aligned to any particular axis, we rotated the trajectories so that the displacement along the rotated x-axis is towards the location of the laser source. Specifically, suppose that when the laser light is applied at location the cell centroid is in position . We compute the rotation angle: . We then rotate each cell centroid position to for each frame during the run according to:
The rotated coordinates where then used to compute the directional index (DI):
Local protrusion formation analysis
The region of recruitment in the red channel was marked using the ‘segmented line’ tool in ImageJ software. Next, ‘Fit spline’ and ‘Straighten’, two custom-written macros, were sequentially used to determine the midpoint of the recruited region. The centroid was estimated with help of another ImageJ macro. Holding centroid as vertex, the angle between the midpoint of recruitment region and new protrusion was determined using the ‘angle’ tool. In MATLAB, these values were plotted with the help of ‘polarhistogram’ command. The minimum number of bins for each plot was determined by Sturges’ formula. For each histogram, at least 40 fresh protrusions were considered.
Statistical Analysis
Unless otherwise mentioned, all statistical analyses were performed by paired or unpaired 2-tailed non-parametric tests on GraphPad Prism 8 and Microsoft Excel. For Fig. 4C, the signrank test in MATLAB was used. All results are expressed as mean ± SD from at least 3 independent experiments. ns denotes P>0.05, * denotes P ≤ 0.05, ** denotes P ≤ 0.01, *** denotes P ≤ 0.001, **** denotes P ≤ 0.0001.
Supplementary Material
Video S1. Global Ras activation promotes F-actin-driven lamellipodia formation, cell spreading, polarity, and random motility. Related to Figures 1 and 2. (A) Global recruitment of HRas G12V increases random motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-HRas G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘08:10’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Global recruitment of KRas4B G12V promotes random motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-KRas4B G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:01’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Recruitment of KRas4B G12V recruitment in presence of G-protein inhibition. Time-lapse confocal microscopy of pertussis toxin- and gallein-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-KRas4B G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with pertussis toxin (PTX, Gαi inhibitor) and gallein (Gβγ inhibitor) for 20 hrs and 10 mins, respectively, before imaging, and kept in presence of both inhibitors throughout imaging. Pink arrows in both panels denote representative cell. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘03:23’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (D) Global recruitment of RasGRP4 improves spread and motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:47’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (E) Global recruitment of empty CTRL does not affect migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-CTRL(control, red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:08’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Video S2. Local Ras activation or inhibition rearranges pre-existing polarity. Related to Figure 3. (A) Local recruitment of RasGRP4 at the cell rear reverses polarity and makes new protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of RasGRP4 recruitment is marked by solid yellow box in the blue panel. Top right corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local recruitment of HRas G12V at the back reverses polarity and creates new protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-HRas G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. HRas G12V ΔCAAX was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of HRas G12V ΔCAAX recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Local recruitment of RASAL3 at the cell front abrogated existing protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited exclusively to the front of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar: 5 μm. (D) RASAL3 recruitment inhibits protrusions in activated macrophages. Time-lapse confocal microscopy of FKP-(D-Cha)-Cha-r-treated RAW264.7 macrophage expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeAct-Halo (cyan; right panel). LifeAct-Halo was visualized by standard Janelia Fluor 646 HaloTag labeling protocol. The membrane anchor, untagged CIBN-CAAX, was expressed. Imaging was started within a minute of adding the chemoattractant, FKP-(D-Cha)-Cha-r, to macrophages. RASAL3 was recruited to protrusions by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (E) RASAL3 recruitment silences protrusions in neutrophils. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 is recruited to protrusions by transiently applying 488 nm laser around the cell perimeter, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Top left corner shows time in min:sec format. Scale bar: 5 μm. (F) Local recruitment of CTRL to the rear does not reverse polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-CTRL (control, red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. CTRL was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. No formation of new protrusions at the site of CTRL recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 7 frames per second.
Video S3. Local Ras activation directs migration in absence or presence of chemoattractant gradients, primarily through mTorC2 complex. Related to Figures 4 and 5. (A) Local RasGRP4 recruitment steers persistent neutrophil migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 and LifeActmiRFP703 (cyan). The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited locally by continuously applying 488 nm laser near the cell, as denoted by the solid yellow box in the cyan panel. The box was repositioned as the neutrophil migrated, to maintain an approximately constant distance between the box and cell boundary. Top left corner shows time in min:sec format. Neutrophil was not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local RasGRP4 recruitment directs migration in macrophages. Time-lapse confocal microscopy of quiescent RAW264.7 macrophage expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel). The right panel is DIC. The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited to one side of the cell by consistently applying 488 nm laser near it, as denoted by the solid yellow box in the red panel. Appearance of protrusions at the RasGRP4 recruitment site, and subsequent migration, is marked by solid yellow box in the DIC channel. Top right corner shows time in min:sec format. Macrophages were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Local RASAL3 recruitment stops neutrophil chemotaxis in fMLP gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the N-formyl-met-leu-phe (fMLP) source in the right chamber. This is denoted as ‘−/+fMLP’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (D) Local RASAL3 recruitment stops neutrophil chemotaxis in C5a agonist gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the C5a receptor agonist (ChaCha peptide) source in the right chamber. This is denoted as ‘−/+ChaCha’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (E) Local RASAL3 recruitment stops neutrophil chemotaxis in LTB4 gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the Leukotriene B4 (LTB4) source in the right chamber. This is denoted as ‘−/+LTB4’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (F) Global RasGRP4 recruitment bypasses PI3Kγ inhibition in neutrophils. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging as denoted by ‘+AS605240’ on top of the video. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘03:09’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (G) Global RasGRP4 recruitment bypasses pan-PI3K inhibition in neutrophils. Time-lapse confocal microscopy of LY294002-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Cells were pre-treated with LY294002 (pan-PI3K inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:54’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (H) Global RasGRP4 recruitment cannot overcome mTorC2 inhibition. Time-lapse confocal microscopy of PP242-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with PP242 (mTor inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘06:11’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Video S4. Local Akt activation directly promotes F-actin rich protrusion formation and polarity rearrangement. Related to Figure 6. (A) Local recruitment of Akt1 to the cell rear generates protrusions and reverses pre-existing polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. AKT1 WT was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of AKT1 WT recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local recruitment of inactive Akt1T308A to the cell rear does not reverse pre-existing polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 T308A (control, red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. AKT1 T308A was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. No formation of new protrusions at the site of AKT1 T308A recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 7 frames per second.
Video S5. Global Akt activation promotes random migration in an evolutionary conserved manner even when PI3K signaling is strongly suppressed. Related to Figure 6. (A) Global recruitment of Akt2 polarizes neutrophil migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT2 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:26’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Global PKBA recruitment polarizes Dictyostelium migration. Time-lapse confocal microscopy of vegetative Dictyostelium expressing tgRFPt-SSPB R73Q-PKBA (red; left panel), before or after 488 nm laser was switched on globally. Right panel is DIC. The membrane anchor, Venus-iLID-CAAX, was expressed but not shown here. Pink and green arrows in both panels denote cells of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:05’ once ‘488 nm ON’ appears at the top of the video. Dictyostelium cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Global CTRL recruitment does not polarize Dictyostelium. Time-lapse confocal microscopy of vegetative Dictyostelium expressing tgRFPt-SSPB R73Q-CTRL (control, red; left panel), before or after 488 nm laser was switched on globally. Right panel is DIC. The membrane anchor, Venus-iLID-CAAX, was expressed but not shown here. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:10’ once ‘488 nm ON’ appears at the top of the video. Dictyostelium cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (D) Global Akt1 recruitment overcomes PI3Kγ inhibitor. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘08:24’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (E) Global Akt1 recruitment overcomes pan-PI3K inhibitor. Time-lapse confocal microscopy of LY294002-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Cells were pre-treated with LY294002 (pan-PI3K inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:26’ once ‘488 nm ON’ appears at the top of the video. Cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (F) Inactive Akt1T308A recruitment cannot overcome PI3Kγ inhibitor. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 T308A (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:50’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal phospho-Akt (Thr308) (D25E6) XP® | Cell Signaling Technology | Cat #13038S; RRID: AB_2629447 |
| Rabbit monoclonal phospho-Akt (Ser473) (D9E) XP® | Cell Signaling Technology | Cat #4060S; RRID: AB_2315049 |
| Rabbit polyclonal Akt | Cell Signaling Technology | Cat #9272S; RRID: AB_329827 |
| Mouse monoclonal p21 Ras | Cytoskeleton Inc. | Cat #AESA02 |
| Rabbit polyclonal GST | EMD Millipore | Cat # AB3282; RRID: AB_91439 |
| Goat anti-rabbit IRDye 680RD-conjugated secondary | Li-Cor | Cat #925-68071; RRID: AB_2721181 |
| Goat anti-mouse IRDye 800CW-conjugated secondary | Li-Cor | Cat #925-32210; RRID: AB_2687825 |
| Bacterial and virus strains | ||
| CIBN-CAAX expressing 3rd generation Lentivirus | this paper | N/A |
| LifeActmiRFP703 expressing 3rd generation Lentivirus | this paper | N/A |
| Klebsiella aerogenes | Lab stock | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI medium 1640 | Gibco | Cat #22400-089 |
| Heat-inactivated fetal bovine serum | Thermo Fisher Scientific | Cat#16140071 |
| Penicillin-streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| DMSO | Sigma-Aldrich | Cat # D2650 |
| DMEM medium | Sigma-Aldrich | Cat #D6429 |
| DMEM medium | Gibco | Cat #10569-010 |
| HL-5 medium | Lab stock | N/A |
| Fibronectin | Sigma-Aldrich | Cat #F4759-2MG |
| N-Formyl-Met-Leu-Phe | Sigma-Aldrich | Cat #47729 |
| FKP-(D-Cha)-Cha-r | Anaspec | Cat #65121 |
| Leukotriene B4 | Cayman Chemical | Cat #20110 |
| AS605240 | Sigma-Aldrich | Cat #A0233 |
| LY294002 | Invitrogen | Cat #PHZ1144 |
| PP242 | EMD Millipore | Cat #475988 |
| Rapamycin | Cayman Chemical | Cat #13346 |
| Pertussis toxin | Sigma-Aldrich | Cat #516560-50UG |
| Gallein | Sigma-Aldrich | Cat #371708-5MG |
| CK-666 | EMD Millipore | Cat #182515 |
| Janelia Fluor 646 HaloTag | Promega Corporation | Cat # GA1120 |
| Puromycin | Sigma-Aldrich | Cat # P8833 |
| Blasticidine S | Sigma-Aldrich | Cat #15205 |
| Hygromycin B | Thermo Fisher Scientific | Cat#10687010 |
| G418 sulphate | Thermo Fisher Scientific | Cat #10131035 |
| Lipofectamine 3000 | Invitrogen | Cat #L3000-008 |
| Polybrene | Sigma-Aldrich | Cat #TR1003 |
| Protease inhibitor cocktail | Roche | Cat#11836170001 |
| Pre-cast 4–15% polyacrylamide gel | Bio-Rad | Cat#5671085 |
| Intercept blocking buffer | Li-Cor | Cat #927-60001 |
| Critical commercial assays | ||
| Neon transfection kit | Invitrogen | Cat #MPK10025B |
| Amaxa cell line kit V | Lonza | Cat #VACA-1003 |
| Ras pull-down activation assay biochem kit | Cytoskeleton, Inc. | Cat #BK008S |
| Experimental models: Cell lines | ||
| Female human neutrophil-like HL-60 cell line | Millius et al142,143 | N/A |
| Male mouse monocyte/macrophage RAW 264.7 cell line | Meshik et al144 | N/A |
| Female human embryonic kidney HEK293T cell line | Lab stock | N/A |
| Dictyostelium discoideum cells of the axenic strain AX2 | Lab stock | N/A |
| Oligonucleotides | ||
| Primers (P1-P26) for cloning, See Table S1 | This paper | N/A |
| Recombinant DNA | ||
| CIBN-CAAX/pLJM1 | This paper | Addgene Plasmid #201749 |
| LifeActmiRFP703/pLJM1 | This paper | Addgene Plasmid #201750 |
| SC0085 transposase expression plasmid | Yusa et al145 | N/A |
| pFUW2-RFP-PH-Akt | Zhan et al146 | N/A |
| pEGFPN1-Lifeact-7Alinker-Halo7 | Shirai et al147 | N/A |
| pMD2.G | Didier Trono Lab (EPFL, Switzerland) (unpublished) | Addgene Plasmid #12259 |
| pMDLg/pRRE | Dull et al149 | Addgene Plasmid #12251 |
| pRSV-Rev | Dull et al149 | Addgene Plasmid #12253 |
| CRY2PHR-mcherry2-CTRL/SC0030 | This paper | Addgene Plasmid #201751 |
| CRY2PHR-mcherry2-HRas G12V ΔCAAX/SC0030 | This paper | Addgene Plasmid #201752 |
| CRY2PHR-mcherry2-KRas4B G12V ΔCAAX/SC0030 | This paper | Addgene Plasmid #201753 |
| CRY2PHR-mcherry2-RasGRP4/SC0030 | This paper | Addgene Plasmid #201754 |
| CRY2PHR-mcherry2-RASAL3/SC0030 | This paper | Addgene Plasmid #201755 |
| CRY2PHR-mcherry2-Akt1/SC0030 | This paper | Addgene Plasmid #201756 |
| CRY2PHR-mcherry2-Akt1T308A/SC0030 | This paper | Addgene Plasmid #201757 |
| CRY2PHR-mcherry2-Akt2/SC0030 | This paper | Addgene Plasmid #201758 |
| pCIBN-CAAX | Idevall-Hagren et al151 | Addgene Plasmid #79574 |
| pCRY2PHR-mcherry2-RasGRP4 | This paper | Addgene Plasmid #201759 |
| pCRY2PHR-mcherry2-RASAL3 | This paper | Addgene Plasmid #201760 |
| N150-Venus-iLID/pDM358 | This paper | Addgene Plasmid #201763 |
| tgRFPt-SSPB R73Q-CTRL/pCV5 | This paper | Addgene Plasmid #201761 |
| tgRFPt-SSPB R73Q-PKBA/pCV5 | This paper | Addgene Plasmid #201762 |
| Software and algorithms | ||
| Fiji (ImageJ) Ver 1.52i | N/A | http://imagej.nih.gov/ij |
| GraphPad Prism Ver 8.00 | N/A | https://graphpad.com |
| MATLAB Ver R2019a | N/A | https://mathworks.com |
| Other | ||
| Neon electroporation system | Invitrogen | Cat #MPK5000 |
| 0.1 cm-gap cuvette | Bio-Rad | Cat#1652089 |
| Gene Pulser Xcell Electroporation Systems | Bio-Rad | Cat#1652660 |
| Amaxa Nucleofector II device | Lonza | Cat #LOAAB-1001 |
| HPLS-45 470 nm LED controller system | LightSpeed Technologies | Cat #HPLS-R1000-3DExt |
| 10 cm cell culture dish | Greiner Bio-One | Cat#664160 |
| 6-well cell culture plate | Greiner Bio-One | Cat#657160 |
| 24-well cell culture plate | Greiner Bio-One | Cat#662160 |
| 48-well cell culture plate | Sarstedt | Cat #83.3923 |
| 8-well chambered coverglass | LAB-TEK | Cat#155409 |
| Chemotaxis μ-slides | Ibidi | Cat #80322 |
| PVDF membrane | Bio-Rad | Cat #162-0262 |
| Trans-Blot Turbo semi-dry transfer system | Bio-Rad | Cat #1704150EDU |
HIGHLIGHTS.
Global Ras activation directly increases random migration bypassing chemoattractants
Local Ras activity reverses polarity and steers migration via mTorC2-Akt activation
Local Ras inhibition blocks directed migration toward three different chemoattractants
Direct Akt activation improves polarity and migration, independently of PI3K activity
Acknowledgements
We thank Peter Devreotes, Pablo Iglesias, Douglas Robinson, and Miho Iijima labs (School of Medicine and Whiting School of Engineering, JHU) for helpful discussions and resources. We thank Stephen Gould and Shang-Jui Tsai (School of Medicine, JHU) for instrumentation, and Xiaoling Zhang, JHU Ross Research Flow Cytometry Core for cell sorting. This work was supported by NIH grant R35 GM118177 (PND), DARPA HR0011–16-C-0139 (PAI/PND), AFOSR MURI FA95501610052 (PND), and NIH grant S10 OD016374 (S Kuo, JHU Microscope Facility).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luster AD, Alon R, and von Andrian UH (2005). Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol 6, 1182–1190. ni1275 [pii] 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Cassatella MA, Costantini C, and Jaillon S (2011). Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol 11, 519–531. 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 3.Nourshargh S, and Alon R (2014). Leukocyte migration into inflamed tissues. Immunity 41, 694–707. 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Artemenko Y, Axiotakis L Jr., Borleis J, Iglesias PA, and Devreotes PN (2016). Chemical and mechanical stimuli act on common signal transduction and cytoskeletal networks. Proc Natl Acad Sci U S A 113, E7500–E7509. 10.1073/pnas.1608767113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devreotes PN, Bhattacharya S, Edwards M, Iglesias PA, Lampert T, and Miao Y (2017). Excitable Signal Transduction Networks in Directed Cell Migration. Annu Rev Cell Dev Biol 33, 103–125. 10.1146/annurev-cellbio-100616-060739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, and Firtel RA (2007). G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol 178, 185–191. 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai TY, Collins SR, Chan CK, Hadjitheodorou A, Lam PY, Lou SS, Yang HW, Jorgensen J, Ellett F, Irimia D, et al. (2019). Efficient Front-Rear Coupling in Neutrophil Chemotaxis by Dynamic Myosin II Localization. Dev Cell 49, 189–205 e186. 10.1016/j.devcel.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Ku CJ, Zhang ER, Artyukhin AB, Weiner OD, Wu LF, and Altschuler SJ (2013). Identifying network motifs that buffer front-to-back signaling in polarized neutrophils. Cell Rep 3, 1607–1616. 10.1016/j.celrep.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, and Bourne HR (2002). A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol 4, 509–513. 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, and Bourne HR (1999). Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol 1, 75–81. 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Dormann D, Munsterberg AE, and Weijer CJ (2002). Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell 3, 425–437. 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 12.Janssen E, and Geha RS (2019). Primary immunodeficiencies caused by mutations in actin regulatory proteins. Immunol Rev 287, 121–134. 10.1111/imr.12716. [DOI] [PubMed] [Google Scholar]
- 13.SenGupta S, Parent CA, and Bear JE (2021). The principles of directed cell migration. Nat Rev Mol Cell Biol 22, 529–547. 10.1038/s41580-021-00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolaczkowska E, and Kubes P (2013). Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 13, 159–175. 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 15.Kruger P, Saffarzadeh M, Weber AN, Rieber N, Radsak M, von Bernuth H, Benarafa C, Roos D, Skokowa J, and Hartl D (2015). Neutrophils: Between host defence, immune modulation, and tissue injury. PLoS Pathog 11, e1004651. 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artemenko Y, Lampert TJ, and Devreotes PN (2014). Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci 71, 3711–3747. 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagorda A, and Parent CA (2008). Eukaryotic chemotaxis at a glance. J Cell Sci 121, 2621–2624. 10.1242/jcs.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, and Huttenlocher A (2010). Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell 18, 226–236. 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benard V, Bohl BP, and Bokoch GM (1999). Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem 274, 13198–13204. 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, and Bourne HR (2002). Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol 4, 513–518. 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 21.Sun CX, Downey GP, Zhu F, Koh AL, Thang H, and Glogauer M (2004). Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood 104, 3758–3765. 10.1182/blood-2004-03-0781. [DOI] [PubMed] [Google Scholar]
- 22.Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, Welch H, Coadwell J, Turner M, Chilvers ER, et al. (2006). Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nat Cell Biol 8, 1303–1309. 10.1038/ncb1494. [DOI] [PubMed] [Google Scholar]
- 23.Zhao T, Nalbant P, Hoshino M, Dong X, Wu D, and Bokoch GM (2007). Signaling requirements for translocation of P-Rex1, a key Rac2 exchange factor involved in chemoattractant-stimulated human neutrophil function. J Leukoc Biol 81, 1127–1136. 10.1189/jlb.0406251. [DOI] [PubMed] [Google Scholar]
- 24.Stephens L, Milne L, and Hawkins P (2008). Moving towards a better understanding of chemotaxis. Curr Biol 18, R485–494. 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan S, Wang F, Glavas S, Ott A, Hofmann F, Aktories K, Kalman D, and Bourne HR (2003). Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J Cell Biol 160, 375–385. 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, and Kirschner MW (2007). An actin-based wave generator organizes cell motility. PLoS Biol 5, e221. 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graziano BR, Gong D, Anderson KE, Pipathsouk A, Goldberg AR, and Weiner OD (2017). A module for Rac temporal signal integration revealed with optogenetics. J Cell Biol 216, 2515–2531. 10.1083/jcb.201604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graziano BR, Town JP, Sitarska E, Nagy TL, Fosnaric M, Penic S, Iglic A, Kralj-Iglic V, Gov NS, Diz-Munoz A, and Weiner OD (2019). Cell confinement reveals a branched-actin independent circuit for neutrophil polarity. PLoS Biol 17, e3000457. 10.1371/journal.pbio.3000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannigan M, Zhan L, Li Z, Ai Y, Wu D, and Huang CK (2002). Neutrophils lacking phosphoinositide 3-kinase gamma show loss of directionality during N-formyl-Met-Leu-Phe-induced chemotaxis. Proc Natl Acad Sci U S A 99, 3603–3608. 10.1073/pnas.052010699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue T, and Meyer T (2008). Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One 3, e3068. 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Keymeulen A, Wong K, Knight ZA, Govaerts C, Hahn KM, Shokat KM, and Bourne HR (2006). To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J Cell Biol 174, 437–445. 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, and Kirschner MW (2006). Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol 4, e38. 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zigmond SH (1974). Mechanisms of sensing chemical gradients by polymorphonuclear leukocytes. Nature 249, 450–452. 10.1038/249450a0. [DOI] [PubMed] [Google Scholar]
- 34.Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, Cao Q, Sanematsu F, Kanai M, Hasegawa H, et al. (2009). Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science 324, 384–387. 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thevathasan JV, Tan E, Zheng H, Lin YC, Li Y, Inoue T, and Fivaz M (2013). The small GTPase HRas shapes local PI3K signals through positive feedback and regulates persistent membrane extension in migrating fibroblasts. Mol Biol Cell 24, 2228–2237. 10.1091/mbc.E12-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.al-Aoukaty A, Rolstad B, and Maghazachi AA (1999). Recruitment of pleckstrin and phosphoinositide 3-kinase gamma into the cell membranes, and their association with G beta gamma after activation of NK cells with chemokines. J Immunol 162, 3249–3255. [PubMed] [Google Scholar]
- 37.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, and Wymann MP (2000). Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science 287, 1049–1053. 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 38.Knall C, Worthen GS, and Johnson GL (1997). Interleukin 8-stimulated phosphatidylinositol-3-kinase activity regulates the migration of human neutrophils independent of extracellular signal-regulated kinase and p38 mitogen-activated protein kinases. Proc Natl Acad Sci U S A 94, 3052–3057. 10.1073/pnas.94.7.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, and Wu D (2000). Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science 287, 1046–1049. 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 40.Nishio M, Watanabe K, Sasaki J, Taya C, Takasuga S, Iizuka R, Balla T, Yamazaki M, Watanabe H, Itoh R, et al. (2007). Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat Cell Biol 9, 36–44. 10.1038/ncb1515. [DOI] [PubMed] [Google Scholar]
- 41.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, and Williams RL (2000). Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943. 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 42.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, et al. (2000). Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science 287, 1040–1046. 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 43.Vicente-Manzanares M, Rey M, Jones DR, Sancho D, Mellado M, Rodriguez-Frade JM, del Pozo MA, Yanez-Mo M, de Ana AM, Martinez AC, et al. (1999). Involvement of phosphatidylinositol 3-kinase in stromal cell-derived factor-1 alpha-induced lymphocyte polarization and chemotaxis. J Immunol 163, 4001–4012. [PubMed] [Google Scholar]
- 44.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, and Ravichandran KS (2002). Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol 4, 574–582. 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 45.Kunisaki Y, Nishikimi A, Tanaka Y, Takii R, Noda M, Inayoshi A, Watanabe K, Sanematsu F, Sasazuki T, Sasaki T, and Fukui Y (2006). DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J Cell Biol 174, 647–652. 10.1083/jcb.200602142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, Smrcka AV, Wu G, Li L, Liu M, et al. (2003). Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell 114, 215–227. 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 47.Nombela-Arrieta C, Lacalle RA, Montoya MC, Kunisaki Y, Megias D, Marques M, Carrera AC, Manes S, Fukui Y, Martinez AC, and Stein JV (2004). Differential requirements for DOCK2 and phosphoinositide-3-kinase gamma during T and B lymphocyte homing. Immunity 21, 429–441. 10.1016/j.immuni.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, and Kirschner MW (1999). The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231. 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 49.Sanui T, Inayoshi A, Noda M, Iwata E, Stein JV, Sasazuki T, and Fukui Y (2003). DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood 102, 2948–2950. 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- 50.Volinsky N, Gantman A, and Yablonski D (2006). A Pak- and Pix-dependent branch of the SDF-1alpha signalling pathway mediates T cell chemotaxis across restrictive barriers. Biochem J 397, 213–222. 10.1042/BJ20051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, and Bourne HR (2000). Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287, 1037–1040. 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yagi M, Kantarci A, Iwata T, Omori K, Ayilavarapu S, Ito K, Hasturk H, and Van Dyke TE (2009). PDK1 regulates chemotaxis in human neutrophils. J Dent Res 88, 1119–1124. 10.1177/0022034509349402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Tang H, Hay N, Xu J, and Ye RD (2010). Akt isoforms differentially regulate neutrophil functions. Blood 115, 4237–4246. 10.1182/blood-2009-11-255323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu G, Bi Y, Wang R, Shen B, Zhang Y, Yang H, Wang X, Liu H, Lu Y, and Han F (2013). Kinase AKT1 negatively controls neutrophil recruitment and function in mice. J Immunol 191, 2680–2690. 10.4049/jimmunol.1300736. [DOI] [PubMed] [Google Scholar]
- 55.Worthen GS, Avdi N, Buhl AM, Suzuki N, and Johnson GL (1994). FMLP activates Ras and Raf in human neutrophils. Potential role in activation of MAP kinase. J Clin Invest 94, 815–823. 10.1172/JCI117401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suire S, Lecureuil C, Anderson KE, Damoulakis G, Niewczas I, Davidson K, Guillou H, Pan D, Jonathan C, Phillip TH, and Stephens L (2012). GPCR activation of Ras and PI3Kc in neutrophils depends on PLCb2/b3 and the RasGEF RasGRP4. EMBO J 31, 3118–3129. 10.1038/emboj.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senoo H, Murata D, Wai M, Arai K, Iwata W, Sesaki H, and Iijima M (2021). KARATE: PKA-induced KRAS4B-RHOA-mTORC2 supercomplex phosphorylates AKT in insulin signaling and glucose homeostasis. Mol Cell 81, 4622–4634 e4628. 10.1016/j.molcel.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Zheng L, Eckerdal J, Dimitrijevic I, and Andersson T (1997). Chemotactic peptide-induced activation of Ras in human neutrophils is associated with inhibition of p120-GAP activity. J Biol Chem 272, 23448–23454. 10.1074/jbc.272.37.23448. [DOI] [PubMed] [Google Scholar]
- 59.Baffi TR, Lorden G, Wozniak JM, Feichtner A, Yeung W, Kornev AP, King CC, Del Rio JC, Limaye AJ, Bogomolovas J, et al. (2021). mTORC2 controls the activity of PKC and Akt by phosphorylating a conserved TOR interaction motif. Sci Signal 14. 10.1126/scisignal.abe4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pylayeva-Gupta Y, Grabocka E, and Bar-Sagi D (2011). RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11, 761–774. 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X, Wen X, Moosa A, Bhimani S, and Jin T (2021). Ras inhibitor CAPRI enables neutrophil-like cells to chemotax through a higher-concentration range of gradients. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2002162118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arai Y, Shibata T, Matsuoka S, Sato MJ, Yanagida T, and Ueda M (2010). Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Natl Acad Sci U S A 107, 12399–12404. 10.1073/pnas.0908278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai H, Das S, Kamimura Y, Long Y, Parent CA, and Devreotes PN (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol 190, 233–245. 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukushima S, Matsuoka S, and Ueda M (2019). Excitable dynamics of Ras triggers spontaneous symmetry breaking of PIP3 signaling in motile cells. J Cell Sci 132. 10.1242/jcs.224121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kae H, Lim CJ, Spiegelman GB, and Weeks G (2004). Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep 5, 602–606. 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kortholt A, Kataria R, Keizer-Gunnink I, Van Egmond WN, Khanna A, and Van Haastert PJ (2011). Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep 12, 1273–1279. 10.1038/embor.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Edwards M, Swaney KF, Singh N, Bhattacharya S, Borleis J, Long Y, Iglesias PA, Chen J, and Devreotes PN (2018). Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration. Proc Natl Acad Sci U S A 115, E9125–E9134. 10.1073/pnas.1809039115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Miao Y, Pal DS, and Devreotes PN (2020). Excitable networks controlling cell migration during development and disease. Semin Cell Dev Biol 100, 133–142. 10.1016/j.semcdb.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pal DS, Li X, Banerjee T, Miao Y, and Devreotes PN (2019). The excitable signal transduction networks: movers and shapers of eukaryotic cell migration. Int J Dev Biol 63, 407–416. 10.1387/ijdb.190265pd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sasaki AT, Chun C, Takeda K, and Firtel RA (2004). Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol 167, 505–518. 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu L, Valkema R, Van Haastert PJ, and Devreotes PN (1995). The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol 129, 1667–1675. 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bolourani P, Spiegelman G, and Weeks G (2010). Determinants of RasC specificity during Dictyostelium aggregation. J Biol Chem 285, 41374–41379. 10.1074/jbc.M110.181115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chubb JR, Wilkins A, Thomas GM, and Insall RH (2000). The Dictyostelium RasS protein is required for macropinocytosis, phagocytosis and the control of cell movement. J Cell Sci 113 (Pt 4), 709–719. 10.1242/jcs.113.4.709. [DOI] [PubMed] [Google Scholar]
- 74.Khosla M, Spiegelman GB, Insall R, and Weeks G (2000). Functional overlap of the dictyostelium RasG, RasD and RasB proteins. J Cell Sci 113 (Pt 8), 1427–1434. 10.1242/jcs.113.8.1427. [DOI] [PubMed] [Google Scholar]
- 75.El-Brolosy MA, and Stainier DYR (2017). Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet 13, e1006780. 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, and Stainier DY (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233. 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 77.Zhang S, Charest PG, and Firtel RA (2008). Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol 18, 1587–1593. 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bell GRR, Rincon E, Akdogan E, and Collins SR (2021). Optogenetic control of receptors reveals distinct roles for actin- and Cdc42-dependent negative signals in chemotactic signal processing. Nat Commun 12, 6148. 10.1038/s41467-021-26371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Beco S, Vaidziulyte K, Manzi J, Dalier F, di Federico F, Cornilleau G, Dahan M, and Coppey M (2018). Optogenetic dissection of Rac1 and Cdc42 gradient shaping. Nat Commun 9, 4816. 10.1038/s41467-018-07286-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hadjitheodorou A, Bell GRR, Ellett F, Shastry S, Irimia D, Collins SR, and Theriot JA (2021). Directional reorientation of migrating neutrophils is limited by suppression of receptor input signaling at the cell rear through myosin II activity. Nat Commun 12, 6619. 10.1038/s41467-021-26622-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karunarathne WK, Giri L, Patel AK, Venkatesh KV, and Gautam N (2013). Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci U S A 110, E1575–1583. 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Neill PR, Castillo-Badillo JA, Meshik X, Kalyanaraman V, Melgarejo K, and Gautam N (2018). Membrane Flow Drives an Adhesion-Independent Amoeboid Cell Migration Mode. Dev Cell 46, 9–22 e24. 10.1016/j.devcel.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Neill PR, and Gautam N (2014). Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell 25, 2305–2314. 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Neill PR, Kalyanaraman V, and Gautam N (2016). Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Mol Biol Cell 27, 1442–1450. 10.1091/mbc.E15-12-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valon L, Etoc F, Remorino A, di Pietro F, Morin X, Dahan M, and Coppey M (2015). Predictive Spatiotemporal Manipulation of Signaling Perturbations Using Optogenetics. Biophys J 109, 1785–1797. 10.1016/j.bpj.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, He L, Wu YI, Hahn KM, and Montell DJ (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 12, 591–597. 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, and Hahn KM (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y, Hyun YM, Lim K, Lee H, Cummings RJ, Gerber SA, Bae S, Cho TY, Lord EM, and Kim M (2014). Optogenetic control of chemokine receptor signal and T-cell migration. Proc Natl Acad Sci U S A 111, 6371–6376. 10.1073/pnas.1319296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arrieumerlou C, and Meyer T (2005). A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell 8, 215–227. 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 90.Gerisch G, and Keller HU (1981). Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J Cell Sci 52, 1–10. 10.1242/jcs.52.1.1. [DOI] [PubMed] [Google Scholar]
- 91.Olguin-Olguin A, Aalto A, Maugis B, Boquet-Pujadas A, Hoffmann D, Ermlich L, Betz T, Gov NS, Reichman-Fried M, and Raz E (2021). Chemokine-biased robust self-organizing polarization of migrating cells in vivo. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2018480118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Surve CR, To JY, Malik S, Kim M, and Smrcka AV (2016). Dynamic regulation of neutrophil polarity and migration by the heterotrimeric G protein subunits Galphai-GTP and Gbetagamma. Sci Signal 9, ra22. 10.1126/scisignal.aad8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang M, Wang M, Shi C, Iglesias PA, Devreotes PN, and Huang CH (2014). Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat Commun 5, 5175. 10.1038/ncomms6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang MJ, Artemenko Y, Cai WJ, Iglesias PA, and Devreotes PN (2014). The directional response of chemotactic cells depends on a balance between cytoskeletal architecture and the external gradient. Cell Rep 9, 1110–1121. 10.1016/j.celrep.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milligan G (2003). Constitutive activity and inverse agonists of G protein-coupled receptors: a current perspective. Mol Pharmacol 64, 1271–1276. 10.1124/mol.64.6.1271. [DOI] [PubMed] [Google Scholar]
- 96.Kirimanjeswara GS, Agosto LM, Kennett MJ, Bjornstad ON, and Harvill ET (2005). Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J Clin Invest 115, 3594–3601. 10.1172/JCI24609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lehmann DM, Seneviratne AM, and Smrcka AV (2008). Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol 73, 410–418. 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shan D, Chen L, Wang D, Tan YC, Gu JL, and Huang XY (2006). The G protein G alpha(13) is required for growth factor-induced cell migration. Dev Cell 10, 707–718. 10.1016/j.devcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 99.Surve CR, Lehmann D, and Smrcka AV (2014). A chemical biology approach demonstrates G protein betagamma subunits are sufficient to mediate directional neutrophil chemotaxis. J Biol Chem 289, 17791–17801. 10.1074/jbc.M114.576827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reuther GW, Lambert QT, Rebhun JF, Caligiuri MA, Quilliam LA, and Der CJ (2002). RasGRP4 is a novel Ras activator isolated from acute myeloid leukemia. J Biol Chem 277, 30508–30514. 10.1074/jbc.M111330200. [DOI] [PubMed] [Google Scholar]
- 101.Yang Y, Li L, Wong GW, Krilis SA, Madhusudhan MS, Sali A, and Stevens RL (2002). RasGRP4, a new mast cell-restricted Ras guanine nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Identification of defective variants of this signaling protein in asthma, mastocytosis, and mast cell leukemia patients and demonstration of the importance of RasGRP4 in mast cell development and function. J Biol Chem 277, 25756–25774. 10.1074/jbc.M202575200. [DOI] [PubMed] [Google Scholar]
- 102.Rottner K, and Schaks M (2019). Assembling actin filaments for protrusion. Curr Opin Cell Biol 56, 53–63. 10.1016/j.ceb.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 103.Hetrick B, Han MS, Helgeson LA, and Nolen BJ (2013). Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol 20, 701–712. 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saito S, Cao DY, Victor AR, Peng Z, Wu HY, and Okwan-Duodu D (2021). RASAL3 Is a Putative RasGAP Modulating Inflammatory Response by Neutrophils. Front Immunol 12, 744300. 10.3389/fimmu.2021.744300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boneschansker L, Jorgensen J, Ellett F, Briscoe DM, and Irimia D (2018). Convergent and Divergent Migratory Patterns of Human Neutrophils inside Microfluidic Mazes. Sci Rep 8, 1887. 10.1038/s41598-018-20060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Campbell JJ, Foxman EF, and Butcher EC (1997). Chemoattractant receptor cross talk as a regulatory mechanism in leukocyte adhesion and migration. Eur J Immunol 27, 2571–2578. 10.1002/eji.1830271016. [DOI] [PubMed] [Google Scholar]
- 107.Foxman EF, Campbell JJ, and Butcher EC (1997). Multistep navigation and the combinatorial control of leukocyte chemotaxis. J Cell Biol 139, 1349–1360. 10.1083/jcb.139.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heit B, Tavener S, Raharjo E, and Kubes P (2002). An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J Cell Biol 159, 91–102. 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Azzi J, Moore RF, Elyaman W, Mounayar M, El Haddad N, Yang S, Jurewicz M, Takakura A, Petrelli A, Fiorina P, et al. (2012). The novel therapeutic effect of phosphoinositide 3-kinase-gamma inhibitor AS605240 in autoimmune diabetes. Diabetes 61, 1509–1518. 10.2337/db11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Francon B, et al. (2005). Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med 11, 936–943. 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 111.Vlahos CJ, Matter WF, Hui KY, and Brown RF (1994). A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 269, 5241–5248. [PubMed] [Google Scholar]
- 112.Hoang B, Frost P, Shi Y, Belanger E, Benavides A, Pezeshkpour G, Cappia S, Guglielmelli T, Gera J, and Lichtenstein A (2010). Targeting TORC2 in multiple myeloma with a new mTOR kinase inhibitor. Blood 116, 4560–4568. 10.1182/blood-2010-05-285726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aoki M, Batista O, Bellacosa A, Tsichlis P, and Vogt PK (1998). The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci U S A 95, 14950–14955. 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meili R, Ellsworth C, and Firtel RA (2000). A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr Biol 10, 708–717. 10.1016/s0960-9822(00)00536-4. [DOI] [PubMed] [Google Scholar]
- 115.Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, and Firtel RA (1999). Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J 18, 2092–2105. 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hoxhaj G, and Manning BD (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 20, 74–88. 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Heit B, Robbins SM, Downey CM, Guan Z, Colarusso P, Miller BJ, Jirik FR, and Kubes P (2008). PTEN functions to ‘prioritize’ chemotactic cues and prevent ‘distraction’ in migrating neutrophils. Nat Immunol 9, 743–752. 10.1038/ni.1623. [DOI] [PubMed] [Google Scholar]
- 118.Lundgren SM, Rocha-Gregg BL, Akdoǧan E, Mysore MN, Hayes SL, and Collins SR (2022). Signaling dynamics distinguish high and low priority neutrophil attractant receptors. bioRxiv, 2022.2005.2026.493652. 10.1101/2022.05.26.493652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang F (2009). The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol 1, a002980. 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, and Weiner OD (2012). Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell 148, 175–188. 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Diz-Munoz A, Fletcher DA, and Weiner OD (2013). Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol 23, 47–53. 10.1016/j.tcb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Diz-Munoz A, Thurley K, Chintamen S, Altschuler SJ, Wu LF, Fletcher DA, and Weiner OD (2016). Membrane Tension Acts Through PLD2 and mTORC2 to Limit Actin Network Assembly During Neutrophil Migration. PLoS Biol 14, e1002474. 10.1371/journal.pbio.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, Crabbe T, Finan P, Jones G, Jackson S, et al. (2007). PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol 9, 86–91. 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- 124.He Y, Li D, Cook SL, Yoon MS, Kapoor A, Rao CV, Kenis PJ, Chen J, and Wang F (2013). Mammalian target of rapamycin and Rictor control neutrophil chemotaxis by regulating Rac/Cdc42 activity and the actin cytoskeleton. Mol Biol Cell 24, 3369–3380. 10.1091/mbc.E13-07-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu L, Das S, Losert W, and Parent CA (2010). mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 19, 845–857. 10.1016/j.devcel.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Funamoto S, Meili R, Lee S, Parry L, and Firtel RA (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611–623. 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 127.Iijima M, and Devreotes P (2002). Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610. 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 128.Wessels D, Lusche DF, Kuhl S, Heid P, and Soll DR (2007). PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J Cell Sci 120, 2517–2531. 10.1242/jcs.010876. [DOI] [PubMed] [Google Scholar]
- 129.Veltman DM, Keizer-Gunnik I, and Van Haastert PJ (2008). Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J Cell Biol 180, 747–753. 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Banerjee T, Biswas D, Pal DS, Miao Y, Iglesias PA, and Devreotes PN (2022). Spatiotemporal dynamics of membrane surface charge regulates cell polarity and migration. Nat Cell Biol 24, 1499–1515. 10.1038/s41556-022-00997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lien EC, Dibble CC, and Toker A (2017). PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol 45, 62–71. 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kamimura Y, and Devreotes PN (2010). Phosphoinositide-dependent protein kinase (PDK) activity regulates phosphatidylinositol 3,4,5-trisphosphate-dependent and -independent protein kinase B activation and chemotaxis. J Biol Chem 285, 7938–7946. 10.1074/jbc.M109.089235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. (2001). Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15, 2203–2208. 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, et al. (2003). Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clin Invest 112, 197–208. 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ackah E, Yu J, Zoellner S, Iwakiri Y, Skurk C, Shibata R, Ouchi N, Easton RM, Galasso G, Birnbaum MJ, et al. (2005). Akt1/protein kinase Balpha is critical for ischemic and VEGF-mediated angiogenesis. J Clin Invest 115, 2119–2127. 10.1172/JCI24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, and Slamon DJ (2003). Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res 63, 196–206. [PubMed] [Google Scholar]
- 137.Higuchi M, Masuyama N, Fukui Y, Suzuki A, and Gotoh Y (2001). Akt mediates Rac/Cdc42-regulated cell motility in growth factor-stimulated cells and in invasive PTEN knockout cells. Curr Biol 11, 1958–1962. 10.1016/s0960-9822(01)00599-1. [DOI] [PubMed] [Google Scholar]
- 138.Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, and Brugge JS (2005). Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol 171, 1023–1034. 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, and Toker A (2005). Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell 20, 539–550. 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 140.Miao Y, Bhattacharya S, Banerjee T, Abubaker-Sharif B, Long Y, Inoue T, Iglesias PA, and Devreotes PN (2019). Wave patterns organize cellular protrusions and control cortical dynamics. Mol Syst Biol 15, e8585. 10.15252/msb.20188585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.de Oliveira S, Rosowski EE, and Huttenlocher A (2016). Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol 16, 378–391. 10.1038/nri.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Millius A, and Weiner OD (2010). Manipulation of neutrophil-like HL-60 cells for the study of directed cell migration. Methods Mol Biol 591, 147–158. 10.1007/978-1-60761-404-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rincon E, Rocha-Gregg BL, and Collins SR (2018). A map of gene expression in neutrophil-like cell lines. BMC Genomics 19, 573. 10.1186/s12864-018-4957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meshik X, O’Neill PR, and Gautam N (2018). Optogenetic Control of Cell Migration. Methods Mol Biol 1749, 313–324. 10.1007/978-1-4939-7701-7_22. [DOI] [PubMed] [Google Scholar]
- 145.Yusa K, Rad R, Takeda J, and Bradley A (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods 6, 363–369. 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhan H, Bhattacharya S, Cai H, Iglesias PA, Huang CH, and Devreotes PN (2020). An Excitable Ras/PI3K/ERK Signaling Network Controls Migration and Oncogenic Transformation in Epithelial Cells. Dev Cell 54, 608–623 e605. 10.1016/j.devcel.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shirai YM, Tsunoyama TA, Hiramoto-Yamaki N, Hirosawa KM, Shibata ACE, Kondo K, Tsurumune A, Ishidate F, Kusumi A, and Fujiwara TK (2017). Cortical actin nodes: Their dynamics and recruitment of podosomal proteins as revealed by super-resolution and single-molecule microscopy. PLoS One 12, e0188778. 10.1371/journal.pone.0188778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li X, Pal DS, Biswas D, Iglesias PA, and Devreotes PN (2021). Reverse fountain flow of phosphatidylinositol-3,4-bisphosphate polarizes migrating cells. EMBO J 40, e105094. 10.15252/embj.2020105094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, and Naldini L (1998). A third-generation lentivirus vector with a conditional packaging system. J Virol 72, 8463–8471. 10.1128/JVI.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Banerjee T, Matsuoka S, Biswas D, Miao Y, Pal DS, Kamimura Y, Ueda M, Devreotes PN, and Iglesias PA (2023). A dynamic partitioning mechanism polarizes membrane protein distribution. bioRxiv, 2023.2001.2003.522496. 10.1101/2023.01.03.522496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, and De Camilli P (2012). Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A 109, E2316–2323. 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hsu AY, Liu S, Syahirah R, Brasseale KA, Wan J, and Deng Q (2019). Inducible overexpression of zebrafish microRNA-722 suppresses chemotaxis of human neutrophil like cells. Mol Immunol 112, 206–214. 10.1016/j.molimm.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ocana-Morgner C, Reichardt P, Chopin M, Braungart S, Wahren C, Gunzer M, and Jessberger R (2011). Sphingosine 1-phosphate-induced motility and endocytosis of dendritic cells is regulated by SWAP-70 through RhoA. J Immunol 186, 5345–5355. 10.4049/jimmunol.1003461. [DOI] [PubMed] [Google Scholar]
- 154.Chronopoulos A, Thorpe SD, Cortes E, Lachowski D, Rice AJ, Mykuliak VV, Rog T, Lee DA, Hytonen VP, and Del Rio Hernandez AE (2020). Syndecan-4 tunes cell mechanics by activating the kindlin-integrin-RhoA pathway. Nat Mater 19, 669–678. 10.1038/s41563-019-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Global Ras activation promotes F-actin-driven lamellipodia formation, cell spreading, polarity, and random motility. Related to Figures 1 and 2. (A) Global recruitment of HRas G12V increases random motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-HRas G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘08:10’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Global recruitment of KRas4B G12V promotes random motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-KRas4B G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:01’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Recruitment of KRas4B G12V recruitment in presence of G-protein inhibition. Time-lapse confocal microscopy of pertussis toxin- and gallein-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-KRas4B G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with pertussis toxin (PTX, Gαi inhibitor) and gallein (Gβγ inhibitor) for 20 hrs and 10 mins, respectively, before imaging, and kept in presence of both inhibitors throughout imaging. Pink arrows in both panels denote representative cell. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘03:23’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (D) Global recruitment of RasGRP4 improves spread and motility. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:47’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (E) Global recruitment of empty CTRL does not affect migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-CTRL(control, red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:08’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Video S2. Local Ras activation or inhibition rearranges pre-existing polarity. Related to Figure 3. (A) Local recruitment of RasGRP4 at the cell rear reverses polarity and makes new protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of RasGRP4 recruitment is marked by solid yellow box in the blue panel. Top right corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local recruitment of HRas G12V at the back reverses polarity and creates new protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-HRas G12V ΔCAAX (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. HRas G12V ΔCAAX was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of HRas G12V ΔCAAX recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Local recruitment of RASAL3 at the cell front abrogated existing protrusions. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited exclusively to the front of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar: 5 μm. (D) RASAL3 recruitment inhibits protrusions in activated macrophages. Time-lapse confocal microscopy of FKP-(D-Cha)-Cha-r-treated RAW264.7 macrophage expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeAct-Halo (cyan; right panel). LifeAct-Halo was visualized by standard Janelia Fluor 646 HaloTag labeling protocol. The membrane anchor, untagged CIBN-CAAX, was expressed. Imaging was started within a minute of adding the chemoattractant, FKP-(D-Cha)-Cha-r, to macrophages. RASAL3 was recruited to protrusions by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (E) RASAL3 recruitment silences protrusions in neutrophils. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 is recruited to protrusions by transiently applying 488 nm laser around the cell perimeter, when the solid yellow box appears in the red panel. Disappearance of protrusions at the site of RASAL3 recruitment is marked by solid yellow box in the blue panel. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Top left corner shows time in min:sec format. Scale bar: 5 μm. (F) Local recruitment of CTRL to the rear does not reverse polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-CTRL (control, red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. CTRL was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. No formation of new protrusions at the site of CTRL recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 7 frames per second.
Video S3. Local Ras activation directs migration in absence or presence of chemoattractant gradients, primarily through mTorC2 complex. Related to Figures 4 and 5. (A) Local RasGRP4 recruitment steers persistent neutrophil migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 and LifeActmiRFP703 (cyan). The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited locally by continuously applying 488 nm laser near the cell, as denoted by the solid yellow box in the cyan panel. The box was repositioned as the neutrophil migrated, to maintain an approximately constant distance between the box and cell boundary. Top left corner shows time in min:sec format. Neutrophil was not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local RasGRP4 recruitment directs migration in macrophages. Time-lapse confocal microscopy of quiescent RAW264.7 macrophage expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel). The right panel is DIC. The membrane anchor, untagged CIBN-CAAX, was expressed. RasGRP4 was recruited to one side of the cell by consistently applying 488 nm laser near it, as denoted by the solid yellow box in the red panel. Appearance of protrusions at the RasGRP4 recruitment site, and subsequent migration, is marked by solid yellow box in the DIC channel. Top right corner shows time in min:sec format. Macrophages were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Local RASAL3 recruitment stops neutrophil chemotaxis in fMLP gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the N-formyl-met-leu-phe (fMLP) source in the right chamber. This is denoted as ‘−/+fMLP’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (D) Local RASAL3 recruitment stops neutrophil chemotaxis in C5a agonist gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the C5a receptor agonist (ChaCha peptide) source in the right chamber. This is denoted as ‘−/+ChaCha’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (E) Local RASAL3 recruitment stops neutrophil chemotaxis in LTB4 gradient. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RASAL3 and LifeActmiRFP703 (cyan) migrating towards the Leukotriene B4 (LTB4) source in the right chamber. This is denoted as ‘−/+LTB4’ on top of the video. The membrane anchor, untagged CIBN-CAAX, was expressed. RASAL3 was recruited to the front of the chemotaxing cell by continuously applying 488 nm laser near it, as denoted by solid yellow box in the cyan panel. Top left corner shows time in min:sec format. Scale bar represents 5 μm. (F) Global RasGRP4 recruitment bypasses PI3Kγ inhibition in neutrophils. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging as denoted by ‘+AS605240’ on top of the video. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘03:09’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (G) Global RasGRP4 recruitment bypasses pan-PI3K inhibition in neutrophils. Time-lapse confocal microscopy of LY294002-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Cells were pre-treated with LY294002 (pan-PI3K inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:54’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (H) Global RasGRP4 recruitment cannot overcome mTorC2 inhibition. Time-lapse confocal microscopy of PP242-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-RasGRP4 (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with PP242 (mTor inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘06:11’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Video S4. Local Akt activation directly promotes F-actin rich protrusion formation and polarity rearrangement. Related to Figure 6. (A) Local recruitment of Akt1 to the cell rear generates protrusions and reverses pre-existing polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. AKT1 WT was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. Nascent protrusions originating from site of AKT1 WT recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Local recruitment of inactive Akt1T308A to the cell rear does not reverse pre-existing polarity. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 T308A (control, red; left panel) and LifeActmiRFP703 (cyan; right panel). The membrane anchor, untagged CIBN-CAAX, was expressed. AKT1 T308A was recruited exclusively to the back of the cell by transiently applying 488 nm laser near it, when the solid yellow box appears in the red panel. No formation of new protrusions at the site of AKT1 T308A recruitment is marked by solid yellow box in the blue panel. Top left corner shows time in min:sec format. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 7 frames per second.
Video S5. Global Akt activation promotes random migration in an evolutionary conserved manner even when PI3K signaling is strongly suppressed. Related to Figure 6. (A) Global recruitment of Akt2 polarizes neutrophil migration. Time-lapse confocal microscopy of differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT2 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:26’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (B) Global PKBA recruitment polarizes Dictyostelium migration. Time-lapse confocal microscopy of vegetative Dictyostelium expressing tgRFPt-SSPB R73Q-PKBA (red; left panel), before or after 488 nm laser was switched on globally. Right panel is DIC. The membrane anchor, Venus-iLID-CAAX, was expressed but not shown here. Pink and green arrows in both panels denote cells of interest. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:05’ once ‘488 nm ON’ appears at the top of the video. Dictyostelium cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (C) Global CTRL recruitment does not polarize Dictyostelium. Time-lapse confocal microscopy of vegetative Dictyostelium expressing tgRFPt-SSPB R73Q-CTRL (control, red; left panel), before or after 488 nm laser was switched on globally. Right panel is DIC. The membrane anchor, Venus-iLID-CAAX, was expressed but not shown here. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:10’ once ‘488 nm ON’ appears at the top of the video. Dictyostelium cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (D) Global Akt1 recruitment overcomes PI3Kγ inhibitor. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Pink arrows in both panels denote cell of interest. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘08:24’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (E) Global Akt1 recruitment overcomes pan-PI3K inhibitor. Time-lapse confocal microscopy of LY294002-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 WT (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Cells were pre-treated with LY294002 (pan-PI3K inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘04:26’ once ‘488 nm ON’ appears at the top of the video. Cells were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. (F) Inactive Akt1T308A recruitment cannot overcome PI3Kγ inhibitor. Time-lapse confocal microscopy of AS605240-treated differentiated HL-60 neutrophil expressing CRY2PHR-mcherry2-AKT1 T308A (red; left panel) and LifeActmiRFP703 (cyan; right panel), before or after 488 nm laser was switched on globally. The membrane anchor, untagged CIBN-CAAX, was expressed. Neutrophils were pre-treated with AS605240 (PI3Kγ inhibitor) for 10 mins before imaging, and kept in presence of the inhibitor throughout imaging. Top left corner shows time in min:sec format. To initiate recruitment (red; left panel), the laser was switched on at ‘05:50’ once ‘488 nm ON’ appears at the top of the video. Neutrophils were not exposed to any chemoattractant during the course of this experiment. Scale bar represents 5 μm. Frame rate: 20 frames per second.
Data Availability Statement
All data are provided in the main or supplementary text. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.


