Abstract
One of the major goals in the inherited retinal disease (IRD) field is to develop an effective therapy that can be applied to as many patients as possible. Significant progress has already been made toward this end, with gene editing at the forefront. The advancement of gene editing-based tools has been a recent focus of many research groups around the world. Here, we provide an update on the status of CRISPR/Cas-derived gene editors, promising options for delivery of these editing systems to the retina, and animal models that aid in pre-clinical testing of new IRD therapeutics.
1. Introduction
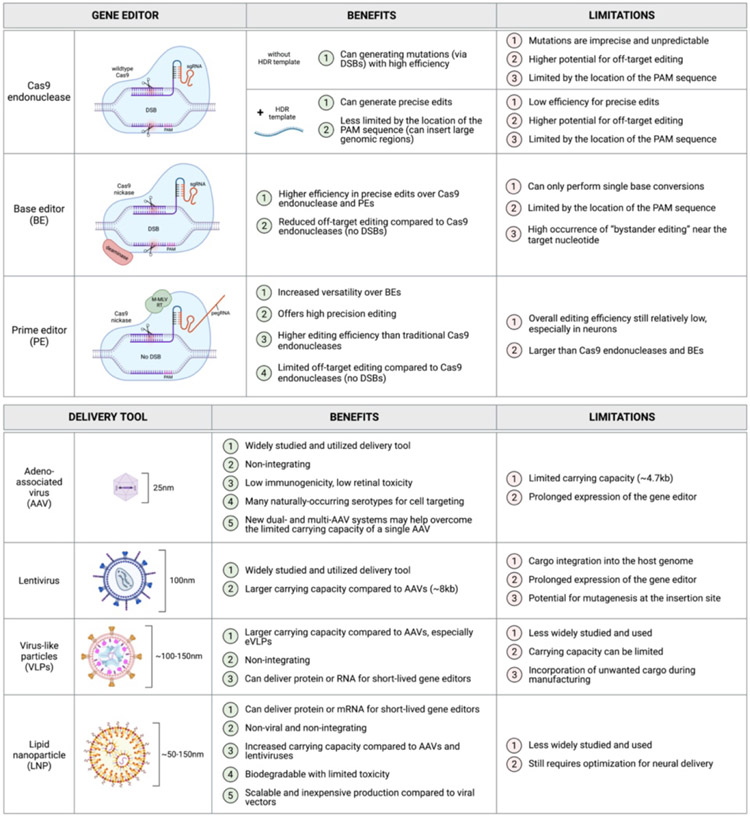
Inherited retinal diseases (IRDs) have an incidence of 1:2000-1:3000 births, making IRDs the leading cause of vision loss in individuals between the ages of 15-45 years old (Bessant et al., 2001, Cremers et al., 2018). Pathogenic variants in at least 250 genes contribute to IRDs, with the majority of those genes being expressed in photoreceptor cells. With the approval of gene augmentation therapy to treat RPE65-associated Leber congenital amaurosis (voretigene neparvovec-rzyl, Luxturna®) (Bennett et al., 2016), and a number of similar therapies in the clinical pipeline, we now have a therapeutic treatment for a handful of IRD patients. While gene augmentation has shown great success as a method of IRD treatment, it is largely limited to autosomal recessive and X-linked IRDs caused by mutations in small genes that can be easily packaged. Therefore, there is a critical need to establish a treatment for the IRDs that fall outside these characteristics, of which there are many. Gene editing has become the standout option for treating virtually any IRD, including those that gene augmentation cannot support. Large insertion or deletion mutations, mutations in large genes, autosomal dominant IRDs, and genes that become silenced upon extended exogenous expression can all benefit from the advancement of gene editing tools. Gene editing-based therapies theoretically offer a permanent, one-time treatment, with the ability to correct any disease-causing mutation in our genome. When developing a gene editing therapeutic, which includes a gene editor system packaged for cell type-optimized delivery, there are a few key factors to consider: safety, delivery and editing efficiency, and broader applicability. Several new technologies have been developed over the past two decades in an attempt to fix IRD-causing mutations, and these include Zinc-finger nucleases (ZFN) (Urnov et al., 2010), transcription activator-like effector nucleases (TALENs) (Joung and Sander, 2013), and most recently, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas endonucleases. In this review, we provide an update on CRISPR-based gene editing tools and promising delivery systems that can be utilized for therapeutic development within the retina (Figure 1 ).
Figure 1.
Benefits and limitations of major gene editor types and delivery methods.
2. Therapeutic CRISPR-based editing approaches
2.1. CRISPR-associated (Cas) endonucleases
Compared to ZFNs and TALENS, the CRISPR/Cas system meets the need for widespread therapeutic editing largely due to its ability to be easily modified to target any gene of interest. The CRISPR/Cas families, which originated as a bacterial and archeal innate immune defense mechanism against secondary bacteriophage infection, are designed to target specific nucleic acid sequences (Sorek et al., 2013). The system consists of CRISPR RNA (crRNA), which is made up of spacer sequences followed by a sequence complementary to the target region, and a tracer-RNA (tracrRNA) molecule, to provide a scaffold for binding by Cas endonucleases (Doudna and Charpentier, 2014). Target recognition requires complementary base pairing between the guide RNA (gRNA) and the genomic region of interest, and this must occur in the presence of a protospacer adjacent motif (PAM). The PAM sequence allows the Cas endonuclease to distinguish between self and non-self sequences when cutting (Marraffini and Sontheimer 2010). Once in position, the Cas protein will cleave both DNA strands at a specific site a few base-pairs from the PAM sequence. This cleavage event produces a double-stranded break (DSB), and once Cas is displaced, cellular DNA repair is triggered.
Six types of CRISPR/Cas systems have been identified and are broadly categorized into two classes based on the structurally and functionally diverse effector complexes used (Makarova et al., 2020). Class I Cas nucleases have multiple smaller Cas effectors, while class II Cas nucleases have a single large effector enzyme. For therapeutic genome editing, Cas9, which is a type II endonuclease within class II, has been most commonly used Cas endonuclease due to its vast characterization and simplicity of the system (Ran et al., 2013). Currently, the most commonly used Cas9 proteins are either derived from Streptococcus pyogenes (SpCas9) (Jinek et al., 2012) or Staphylococcus aureus (SaCas9) (Ran et al., 2015). Since most advancements toward the clinic have been made using SpCas9, we will focus on this subtype, and it will be referred to simply as “Cas9” hereafter. Once in position, the Cas9 protein undergoes a conformational change to cleave both DNA strands using two nuclease domains, HNH and RuvC, each of which is responsible for cleaving one strand of the target dsDNA at a specific site 3-nt from the 5’-NGG-3’ PAM sequence causing a blunt DSB (Jiang et al., 2016). The system is versatile enough that multiple guides can be used to induce breaks in multiple genomic sites simultaneously, allowing for excision of large segments of a gene or even complete chromosomal rearrangements (Zuccaro et al., 2020, Turchiano et al., 2021).
To improve the universality of Cas9, numerous alterations to Cas9 have been generated with regard to PAM recognition. For example, SpCas9-NG and xCas9 use NG as their PAM sequence and show high editing efficiency and low off-target activity (Hu et al., 2018, Nishimasu et al., 2018). Similarly, another variant of Cas9, Cas9SpRY, recognizes an NR (R=A or G) PAM site, which has significantly relaxed PAM requirements for SpCas9 (Walton et al., 2020). Consequently, Cas9SpRY has significantly high off-target activity. Similar efforts are ongoing to engineer other Cas endonuclease to change their PAM recognition sequence to help expand their utility.
2.1.1. DNA repair mechanisms following DSBs
Following endonuclease-induced DSBs, DNA repair mechanisms drive repair to avoid cell death. Two of the most well-studied DNA repair mechanisms include nonhomologous end joining (NHEJ) and homology-directed repair (HDR) (Sung and Klein, 2006, Chang et al., 2017). Of these, NHEJ is most readily used by cells to repair DSBs. During the repair process, NHEJ often introduces random elements into the genome resulting in substitutions, insertions, and/or deletions (indels), which can generate disruptive mutations. This system works well for disrupting a gene or eliminating a segment of DNA, both of which require DSBs in the target genome. As opposed to NHEJ, HDR is a precise repair mechanism that uses a homologous single- or double-stranded DNA template to guide DSB repair. This template can be provided by the host genome during the cell cycle or provided by an exogenous source in the case of targeted genome editing. Importantly, HDR-associated pathways are only active during S and G2-phases of the cell cycle, and are thus typically employed only in dividing cells. When thinking about therapeutic editing for IRD repair, the unpredictable and error-prone nature of the NHEJ repair pathway is not ideal, thus efforts are underway to identify ways to inhibit NHEJ in favor of HDR to drive accurate repair following CRISPR-Cas9 induced DSBs. For example, a small molecule inhibitor, Scr7, targeting DNA ligase IV was used to block NHEJ from occurring both in cells and in mice, resulting in an up to 19-fold increase in HDR (Chu et al., 2015). Small molecules targeting DNA-PKc (NU7441 (Robert et al., 2015)) and KU proteins (STL127705 (Weterings et al., 2016)) have demonstrated a similar promotion of HDR with variable efficiency. Finally, shRNA-based knockdown of Ku proteins or DNA ligase IV resulted in improved HDR repair. Additional small molecules that have been used to boost HDR repair include those targeting CtIP (MLN4924), RAD52 (AICAR), RAD51 (RS-1), and Polθ (ART558 (Schimmel et al., 2023)). Furthermore, a recent publication using a combination of small molecules, so-called “CRISPY mix” (NU7026, Trichostatin A, MLN4924, and NSC 15520) showed a significant additive effect over using one small molecule (Riesenberg and Maricic, 2018).
While the above approaches have improved HDR rates, other efforts have also been undertaken to make NHEJ less error-prone. One such method is Homology-Independent Targeted Integration (HITI) (Suzuki et al., 2016). In this approach, a donor sequence is introduced along with Cas9. The donor template here also includes the same targeted sequence for the gRNAs as the host cell but in the reverse direction. Thus, post-Cas9 endonuclease cutting, the template theoretically gets integrated into the cut site. Due to the nature of the design, only correctly-oriented templates get integrated, while reverse-oriented integration would be subjected to recutting by Cas9. This approach was shown to work well in vitro, even in post-mitotic neurons, as well as in vivo in mice and rats with retinitis pigmentosa (Suzuki et al., 2016). This HITI system has also shown recent success in promoting knock-in of large DNA donor minicircle constructs including fluorescence, bioluminescence, and MRI markers into AAVS1 safe harbor locus for long-term tracking of cells (Kelly et al., 2021).
2.2. Base editors (BEs)
To help overcome the innate challenges of the original Cas enzymes, base editors (BEs) were developed as facilitators of precise editing of target genetic loci through single-nucleotide conversions (Komor et al., 2016, Nishida et al., 2016). These conversions are made possible using a catalytically impaired “nicking” Cas nuclease fused to DNA-modifying enzymes that perform the base exchange (deaminases). These exchanges therefore do not require an HDR template or DSBs, eliminating the concern for undesired insertions or deletions in the target DNA.
Existing BE types include 1) cytosine base editors (CBEs), which convert C•G to T•A, 2) the aptly-named C•G to G•C base editors (CGBEs), which convert C•G to G•C and were derived from CBEs, and 3) adenine base editors (ABEs), which convert A•T to G•C. Although BEs provide more efficient editing compared to traditional Cas nucleases, they are limited to single base edits and cannot address larger insertion or deletion mutations, either to remove them or generate them. Furthermore, these editors are more restricted by the location of the PAM sequence, which must lie within about 13-17nt from the desired base edit; this makes BEs more limited in their application. Finally, while the lack of DSBs makes BEs safer than traditional Cas9, there is a reasonable risk of off-target base editing of bystander nucleotides (called, ‘bystander editing’), which often occurs near the target locus if a nucleotide identical to the target nucleotide is present. Initial studies describing BEs reported up to 75% base editing in mammalian cells in vitro, with low indel formation and a high conversion rate within the editing window surrounding the target nucleotide (Komor et al., 2016). Therefore, although limited in its application, base editing is a highly efficient editing strategy to consider for therapeutic development.
2.3. Prime editors (PEs)
Prime editors (PEs) were first developed by Liu and colleagues (Anzalone et al., 2019) to overcome the limitations of the existing Cas9 nucleases and BEs. Like BEs, PEs use a Cas9 nickase to cut a single strand of DNA and avoid DSB generation. The Cas9 nickase is fused to a reverse transcriptase (RT), which allows for the desired edit to be reverse transcribed at the target locus. This eliminates the need for co-delivery of a DNA template, while also getting around the need for engagement of the inefficient HDR repair mechanism. Instead of using the standard single guide RNA (sgRNA) molecule to direct Cas9 to its target within the genome, PE technology utilizes a prime editing guide RNA (pegRNA), which combines the sgRNA with a primer binding site and a template for the RT. The RT template is the portion of the pegRNA that contains the desired edit. With these modifications, prime editing expands the possibilities for gene editing in post-mitotic cells beyond the capabilities of traditional Cas9 and BEs. PEs in human immortalized cell lines have yielded up to 58% editing efficiency, and have been shown to work in post-mitotic cells, including primary neurons, with an average efficiency of 7% (Anzalone et al., 2019, Anzalone et al., 2020). While current versions of PEs (e.g. PE2) still have some off-target editing, off-target editing events are 4.4-fold lower compared with wildtype Cas9 cutting (Anzalone et al., 2019). More recent improvements in the PE system include the development of PE4, PE5, and PEmax systems (Chen et al., 2021). These new PEs introduce transient expression of a mutated DNA mismatch repair (MMR) protein (MLH1dn) which acts as a dominant negative version of MLH1dn to interfere with the suppressive effects of the normal DNA MMR process that exists in cells. The PE4 (PE2+MLH1dn) and PE5 (PE3+MLH1dn) systems improve editing efficiency in iPSCs by an average of 7.7-fold. The ratio of edits to indels was found to increase by 3.4-fold over the PE2 and PE3 alone, respectively. Chen and colleagues further improved editing efficiency with the PEmax system, which introduced additional mutations to Cas9, alterations to the NLS sequence, as well as changes to the peptides that link the Cas9 nickase and the RT. The PEmax changes were applied to all versions of PEs, (PE2, PE3, PE4, PE5), all of which showed an average increase in editing efficiency compared to the corresponding system from which each was derived (Chen and Liu, 2023). Editing can be further optimized using an engineered pegRNA (epegRNA) molecule, which harbors a modification to the original pegRNA to improve its stability (Nelson et al., 2022).
3. Ocular delivery methods
Once the optimal gene editor system has been chosen and optimized for a given study, the appropriate delivery system must then be employed.
3.1. Adeno-associated viruses (AAVs)
One promising approach for the delivery of gene editors to the retina is the use of adeno-associated viruses (AAVs). These naturally-occurring viruses have been applied to the eye in research or clinical settings for nearly three decades, making them one of the more widely studied and utilized delivery tools currently in use. AAVs offer an extremely efficient and biocompatible system for the delivery of non-integrating single-stranded DNA up to ~5kb (~4.7kb of that is available for a transgene cassette). While this cargo size and type may be perfect for exogenous expression of smaller genes, it cannot hold most gene editors, which typically exceed 4.7kb in size. To overcome the limited carrying capacity of a single AAV, dual-AAV and multi-AAV systems have been developed and are currently being studied (Trapani, 2019, Zhi et al., 2022). Another feature of AAVs is that genes delivered using this method exhibit lengthy expression, which is critical for gene augmentation-based therapies. However, for gene editing, it is beneficial to limit the expression time of the gene editor system to reduce the chances of unwanted editing within the target genome. To address this concern, various research groups have presented AAV systems with self-inactivating capabilities for the delivery of CRISPR/Cas9 tools (Ibraheim et al., 2021, A. Li et al., 2019, F. Li et al., 2019); however, these systems exhibit incomplete inactivation and evidence of AAV cargo integration into the target genome, posing an added risk for undesired edits. Finally, at least 12 naturally-occurring AAV serotypes have been discovered, with some variability in tissue tropisms among these serotypes (Li and Samulski, 2020). This variability provides options for optimizing delivery to different cell types in vivo. Cross-packaging can also be used to mix serotypes, further increasing the options for generating efficacious cell type-specific targeting (Hu et al., 2021). AAVs offer a relatively safe (Bucher et al., 2021) and efficient delivery option for gene editors. More extensive reviews covering the current safety profiles, utility, and limitations of AAVs and other viral vectors can be found in (Raguram et al., 2022, Bucher et al., 2021, Schön et al., 2015, Hori et al., 2019).
3.2. Lentiviruses and virus-like particles (VLPs)
Another commonly used virus for delivery of nucleic acids to mitotic and postmitotic cells is the lentivirus, and as such, it provides another option for delivery of gene editing tools (Holmgaard et al., 2019, Raguram et al., 2022b). Lentiviruses offer a larger carrying capacity (~8kb) compared to AAVs, which could allow for packaging of some gene editors into a single lentivirus. In contrast to AAVs, lentiviruses deliver cargo that integrates into the host genome, which could cause undesired prolonged expression of the gene editor. These integration events also have the potential to generate mutations at the insertion site, as they often insert into open chromatin with genes that are actively being expressed. While lentiviruses may be applicable for certain IRD treatments, especially gene augmentation, their limitations with regard to delivery of gene editors may outweigh the benefits when you consider the lentivirus options that are currently available.
Virus-like particles (VLPs) are viral protein assemblies that have the ability to infect target cells, but lack any viral genetic materials. VLPs can carry proteins, such as ribonucleoproteins (RNPs), allowing for short-lived Cas enzymes. These particles avoid the typical risks associated with viral particle delivery, while maintaining tissue- or cell-specific targeting, with the ability to modulate tropism by changing out the envelope glycoproteins used (Mazurov et al., 2023). Banskota and colleagues recently generated engineered VLPs (eVLPs), which have about a 16-fold increase in RNP carrying capacity compared to previous generations of VLPs (Banskota et al., 2022), revealing a promising new option for efficient delivery of large gene editors. The major limitations for VLPs and eVLPs include non-specific co-packaging of proteins or RNA from the host cells in which the VLPs/eVLPs are manufactured, as well as cargo size limitations, especially for VLPs, which become more inefficient to package as the size of the protein cargo increases. Altogether, this recently developed delivery method shows great promise for delivery of therapeutic agents, including gene editors, to treat IRDs.
3.3. Lipid nanoparticles (LNPs)
Lipid nanoparticles (LNPs) are quickly becoming a promising tool for the safe and efficient delivery of diverse cargos to various tissues, including the eye. LNPs are commonly composed of a cationic or ionizable lipid, structural lipids (such as phospholipids and sterol lipids), and a PEG-conjugated lipid, and are formulated using controlled microfluidic mixing, during which all components are able to self-assemble and encapsulate the desired cargo (Eygeris et al., 2022). With this formulation process, LNPs offer an easily modifiable delivery system, in which lipid composition, size, shape, charge, and other key properties can be adjusted to attain successful delivery to different cell types. Furthermore, LNPs can be manufactured to incorporate cell type-specific targeting peptides, as was done by Herrera-Barrera and colleagues to target photoreceptor cells in vivo in mice and nonhuman primates (Herrera-Barrera et al., 2023).
LNPs have the ability to delivery various types of cargo (DNA, RNA, protein, small molecules, etc.) of theoretically unlimited size. Studies have shown that LNP delivery of mRNA to the retina results in rapid, short-lived expression of the cargo (Patel et al., 2019), which is ideal for limiting unintended editing events that can result from prolonged expression of gene editors. In preliminary studies, we have shown that LNPs can deliver EGFP mRNA to photoreceptor cells in human retinal organoids (data not shown); however, efficiency will need to be improved using further optimized LNP formulations. In contrast, delivery to iPSC-derived RPE cells is highly efficient, likely due to the phagocytic nature of these cells. Altogether, LNPs offer a relatively safe, scalable delivery tool that can be optimized to fit the needs of the therapy type and target tissue, and they are becoming a more active area of research for IRD therapeutic development (Wang et al., 2015, del Pozo-Rodríguez et al., 2013).
4. Advancements in the delivery of gene editors to the retina
Several in vitro and in vivo studies have already been conducted to test the capabilities of gene editor systems delivered to the retina. Rd10 mice (Pde6b mutation), which show features of retinitis pigmentosa, were treated with an AAV8-ABE, introduced via subretinal injection. Sequencing at the genomic DNA and cDNA level was found to average 20.79% and 54.97%, respectively. This study also showed bystander editing reached 8.85% at the target locus. This level of correction in these mice restored Pde6b expression, preserved photoreceptor cells, and rescued photopic ERG amplitude (Su et al., 2023). Another group studied base editing in rd12 mice, which is a model for Rpe65-LCA. 3-week-old rd12 mice were given subretinal injections of lentivirus-packaged ABE to target the Rpe65 mutation in RPE cells. They evaluated Rpe65 editing at three weeks post-treatment and showed up to 40% correction efficiency in cDNA (with average editing = 27±12%) if you exclude corrections with bystander edits. Bystander editing was found to be ~20-30%, which shows a high level of unwanted conversions in the Rpe65 gene, which could have additional unintended consequences, depending on the resulting amino acid changes at those sites. This group also attempted dual-AAV delivery of the ABE and observed 1.6% precision editing efficiency. In a similar study using rd12 mice, sequencing of bulk RPE revealed ~33% correction at 3 months post-treatment with dual AAV-ABE targeting the Rpe65 gene. Bystander editing, however, was present in more than 50% of sequenced cells (Jo et al., 2023). Furthermore, lentiviral delivery of ABE was explored to correct Gnat in cone photoreceptor cells in rd10/Gnat−/− mice, which resulted in rescue of cone function and survival (Choi et al., 2022). Finally, eVLPs were used to deliver ABE RNPs to the RPE cells of rd12 mice, targeting the Rpe65 gene (ABE7.10-NG-eVLPs), and this study was able to achieve 12% correction efficiency, with a lack of bystander editing near the target base (Banskota et al., 2022). While the majority of in vivo BE experiments reveal unwanted bystander editing in retinal cells, efficiency of on-target editing events is rather high, especially when compared to other gene editing modalities. Furthermore, rescue of retinal phenotypes has been observed in mice treated with ABEs targeting two different genes, providing evidence of its utility for therapeutic editing. Although BE is limited to single nucleotide conversions, there are numerous mutations in IRD-causing genes that can be addressed with this method of gene editing, and many of these targets have already been identified (Piotter et al., 2021, Kaukonen et al., 2022, Elsayed et al., 2022, Bellingrath et al., 2022, Lopes da Costa et al., 2023, Fry et al., 2021).
Prime editing is still a relatively new technology; however, a few groups have begun to employ PEs for correction of IRD mutations. Wimmer and colleagues introduced PE2 plasmids into HEK293 cells with an ABCA4 mutation to determine in vitro editing efficiency (Wimmer et al., 2023). Their study, which used a bioluminescence resonance energy transfer (BRAT)-based editing sensor as a readout, found up to 92% correction of the ABCA4 gene. Qin et al. studied PESpRY editing of the Pde6B gene in rd10 mice, using split-AAV-based delivery (Qin et al., 2023). This study yielded an average editing frequency of up to 76% in infected cells and showed recovery of disease phenotypes. Two independent studies in rd12 mice (Jang et al., 2022, She et al., 2023) tested AAV-delivery of PE to assess correction of the Rpe65 mutation. In Jang et al., subretinal injection of AAV2-PE2 showed a delivery efficiency of 23% within the entire RPE layer, and an editing efficiency of 6.4% (4.1-7.4% range). When estimating the editing efficiency just in the cells that received PE2, the average increases to 28%. No off-target editing events were observed in the target region or in common off-target sites. Finally, ERG responses were elevated in treated mice compared to untreated, showing some rescue of the disease (Jang et al., 2022). In She et al., a dual AAV8-split PE3 was delivered subretinally into rd12 mice, which resulted in 11.4±2.3% editing in RPE cells, restoration of RPE65 protein levels, and improved photoreceptor function and survival (She et al., 2023). Based on the data so far, both BEs and PEs should be considered strong candidates for a gene editing-based therapeutic strategy.
5. Preclinical model systems
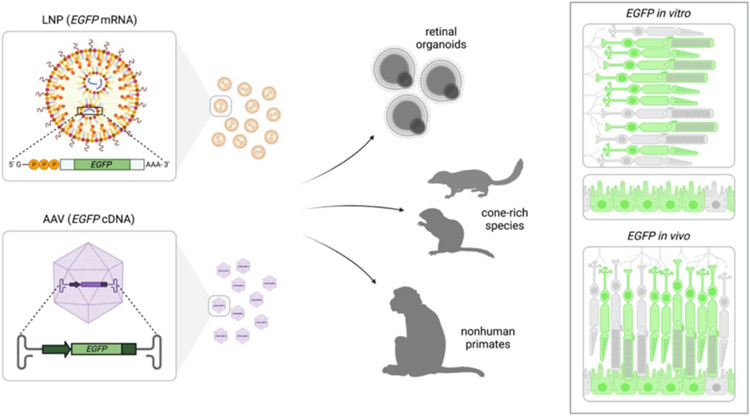
Therapeutic gene editing in patients is already underway; however, the first clinical trial in the eye, focused on the CEP290 gene (EDIT-101), is currently paused due to poor efficacy with a very small responder group (discussed in Section 7). A major challenge for these trials lies with pre-clinical assessment of the clinical product. Rodents, such as mice and rats, are the de facto models for assessment of efficacy and toxicity; however, there are significant differences in retinal structure and function in these model systems when compared to humans. Key differences include a lack of cone-rich regions similar to the macula and/or fovea, differences in the ratio and classifications of cell subtypes (e.g., cones and ganglion cells), and globe size and access to delivery sites. To get around some of these concerns, alternate models are being considered (Figure 2).
Figure 2.
Schematic of lipid nanoparticle (LNP) delivery of EGFP mRNA and AAV delivery of EGFP cDNA to human retinal organoids, cone-rich species, and nonhuman primates. A schematic of EGFP expression is shown in photoreceptor cells and RPE both in vitro and in vivo.
5.1. Nonhuman primates (NHPs)
Nonhuman primates (NHP) appear to be an ideal animal model for testing gene editing-based therapies, due to their shared commonalities with humans in terms of genetics, tissue structure and function, and genetics. Commonly used NHPs in research include cynomolgus (Macaca fascicularis), rhesus (Macaca mulatta), and common marmoset (Callithrix jacchus). With relation to the eye, NHPs share central retinal features including a macula and fovea. A few naturally occurring human retinal disease models have been identified in NHP colonies, including a rhesus colony with mutations in the PDE6C gene leading to achromatopsia (Moshiri et al., 2019). Four related NHPs were detected upon discovery of the photophobic feature that is typically observed in patients with achromatopsia. Sequencing analysis confirmed a homozygous R565Q missense mutation in the catalytic domain of PDE6C, which is a critical cone-specific phototransduction enzyme. The NHPs were confirmed to have preserved rod function, but no cone function, via electroretinograms (ERG). Martha Neuringer’s lab at OHSU recently reported the existence of a rhesus macaque model of Bardet-Biedl syndrome, a multi-system syndromic disorder associated with retinitis pigmentosa (Peterson et al., 2019). This study identified three related NHPs bearing a homozygous mutation (c.160delG) in the BBS7 gene. The animals displayed severe macular degeneration, including loss of photoreceptor layers, RPE degeneration, and vasculature atrophy, all of which are hallmarks of the disorder. The NHPs also had severe functional defects on ERG analysis. Interestingly, CRISPR-based gene editing has been used to generate induced models of retinal degeneration in NHPs. For example, a cynomolgus achromatopsia model was generated by knocking down the CNGB3 gene (Lin et al., 2020). Using a dual-AAV9 system to deliver Cas9 and gRNAs, the animals were injected subretinally at multiple sites to spread the virus over a wider coverage area. Treated animals showed reduced cone activity on a multi-focal ERG at the sites of injection. Similarly, a RHO model was generated by in vivo knockdown using gRNAs targeting the RHO gene, delivered into the eye using AAV (Li et al., 2021). In addition, a human synapsin I (hSyn) promoter was used to drive expression in retinal neurons to prevent bystander effects on glia and RPE. The NHPs showed generalized retinal degeneration with the most prominent loss occurring in the macula, with severe functional deficits. Another recent report described the development of a NHP Usher syndrome model, in which CRISPR/Cas9 was used to disrupt MYO7A in rhesus macaque zygotes. While an overt retinal phenotype was not detected up to 12 months of age, the monkey did show auditory disturbances (Ryu et al., 2022). Currently, the biggest challenge to the use of NHP models is limited availability and high animal costs. Furthermore, higher ethical considerations exist with the use of NHPs, with efforts being made to use them only for disorders where alternate models do not suffice. Finally, the presence of very few naturally occurring mutations in NHPs are a challenge, but an increased effort to identify more naturally occurring mutations should identify more cohorts. Some gene editing-based approaches at the embryo stage are also being employed to generate human disease-associated models for various extra-ocular disorders (Chen et al., 2015, Yang et al., 2019, Tu et al., 2019).
5.2. Cone-rich species
For treatments targeting cone photoreceptors, rodents are particularly impractical due to the diffuse cone distribution. Therefore, species with cone-rich retinae would be ideal for such treatments. Two cone-rich species, the 13-lined ground squirrel (13-LGS; Ictidomys tridecemlineatus) and northern tree shrew (Tupaia belangeri) are extremely promising vision models. Ground squirrels exhibit an ~85% cone population (Kryger et al., 1998, Kandoi et al., 2022). Their lens:globe ratio is much closer to humans than mice or rats, allowing for better in vivo imaging of their photoreceptor mosaic (Gur and Sivak, 1979, Sajdak et al., 2016). The biggest challenge with using this species lies with its hibernation behavior (Remé and Young, 1977); the animals are unavailable for 4-6 months in the Winter and undergo significant cone remodeling during this phase. Tree shrews are another intriguing model, given their close relation to primates (Fan et al., 2013), and the fact that their photoreceptor layer consists predominantly of cones (~95%) (Müller and Peichl, 1989). Although tree shrews are native to South Asia, there are a few inbred colonies in the United States, and they have been used extensively as models for studying myopia (Sherman et al., 1977, McBrien and Norton, 1992) and central visual processing (Fitzpatrick, 1996, Chisum et al., 2003). Like the 13-LGS, tree shrews have large eyes with a more human-like lens:globe ratio, and also exhibit highly visual behaviors (Immel and Fisher, 1985). Both of these models could be well adapted to optimize delivery methods and assess recovery for cone-based therapies.
5.3. Human retinal organoids
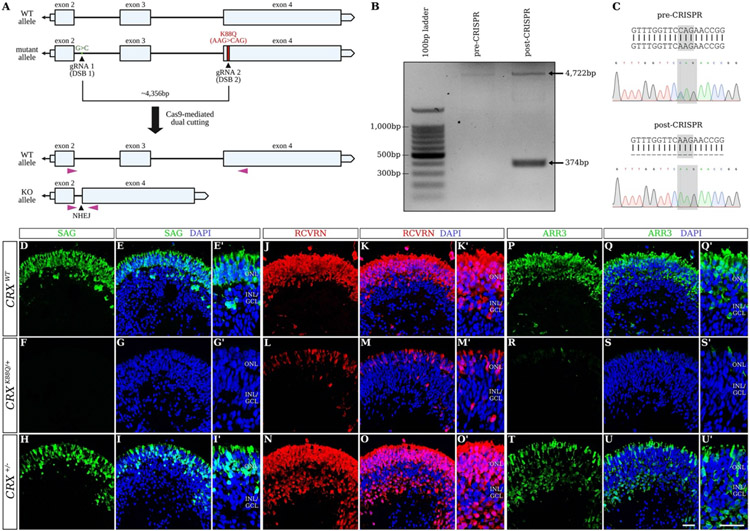
With the discovery of somatic cell reprogramming to generate human induced pluripotent stem cells (iPSCs) (Takahashi et al., 2007, Yu et al., 2007), we now have the opportunity to directly test therapies in patient cells. Furthermore, a number of validated protocols are available to generate retinal organoids (Ohlemacher et al., 2015, Capowski et al., 2019, Chew et al., 2022). These organoids closely mimic the human retinogenesis timeline, structure, and composition (Cowan et al., 2020, Sridhar et al., 2020, Finkbeiner et al., 2022). Furthermore, we and others have demonstrated that organoids generated from patient iPSCs reliably reproduce disease pathogenesis and can be important tools for therapeutic discovery. We recently used LCA7 organoids (harboring CRX mutations) to demonstrate the efficacy of a traditional Cas9-based allele-specific knockdown approach to treat dominant negative disorders (Figure 3 (Chirco et al., 2021)). Similarly, iPSCs from X-linked Retinoschisis (XLRS) patients with RS1 mutations have been used to generate retinal organoids for testing gene editing (Huang et al., 2019). This group demonstrated partial rescue of the disease phenotype using an ABE to repair the C625T mutation in the RS1 gene. The ability to work with human organoids will have distinct advantages for the assessment of on- and off-target editing, as well as methods of delivery to human cells. The key challenge that remains is the slow development of these retinal organoid models, which can take nine months to a year to develop measurable disease phenotypes for early onset disorders, making slow-developing diseases like retinitis pigmentosa even more challenging to model and show therapeutic efficacy.
Figure 3.
Allele-specific editing in hiPSCs harboring a dominant mutation in the CRX gene (from (Chirco et al., 2021)). The CRISPR/Cas9 dual-cutting target sites are mapped onto the mutant allele of the CRX gene (A). PCR was performed using primers shown in (A; purple triangles), revealing an additional 374-bp band representing the edited “knockout (KO) allele” after CRISPR/Cas9 editing (B). Sanger sequencing was also utilized to confirm loss of the K88Q mutation after CRISPR-mediated editing (C). Immunofluorescence staining using antibodies against SAG (green; D-I′), RCVRN (red; J-O′), and ARR3 (green; P-U′) are shown for control (CRXWT; D-E′, J-K′, and P-Q′), CRXK88Q/+ (F-G′, L-M′, and R-S′), and CRX+/− (H-I′, N-O′, and T-U′) retinal organoids at D180 (n=3 organoids per line). Nuclei are counterstained with DAPI (blue). Scale bars (U and U′), 100μm. OPL, outer plexiform layer; INL/GCL, inner nuclear layer/ganglion cell layer.
6. Ethical and regulatory challenges
The least controversial therapeutic application of gene editing is somatic cell editing to treat pre-existing disorders. Most regulatory authorities are primarily concerned with the safety of the product, so the key concern with relation to CRISPR-based editing is off-target editing. This is less of a concern when cells are edited ex vivo prior to re-implantation, as proposed for some hematopoietic populations, since these edited cells can be fully screened for unintended edits. In contrast, direct editing in vivo cannot be further screened in living patients, which is where human organoid models could become useful to better predict off-target effects within the human genome. As opposed to CRISPR-based somatic editing, germline editing is highly controversial and fraught with concerns. Targeting germline cells could have long-term effects on the individual and lead to permanent and unpredictable side effects passed down through future generations (Schleidgen et al., 2020). Another major concern with gene editing in patients involves obtaining proper informed consent from the individual undergoing editing therapy, especially for disorders that may not be life-threatening (Jonlin, 2020). Finally, there are societal concerns around access to such technologies due to high cost of the treatment, meaning that they may only be accessible to the wealthy. A concerted effort is needed to ensure such gene editing therapies will be available to everyone (Muigai, 2022).
7. Status of clinical trials
A number of gene editing-based clinical trials are currently ongoing, and these can be divided into 1) in vivo editing, or 2) ex vivo editing of cells that are then transplanted back into the patient. A number of the trials listed on ClinicalTrials.gov are listed below in Table 1. Of these, there have been two trials with relation to the eye; one for the treatment of refractory keratitis due to HSV-1 infection, and another for the treatment of LCA10. The keratitis trial involved treatment of the cornea with CRISPR and guide mRNA targeting the HSV-1 virus, delivered using lentiviral particles. A total of three patients were treated, and a pre-print of the data shows that the patients were virus-free within 2 months and remained so up to 21 months of follow-up (Wei et al., 2023). The other clinical trial focused on treating LCA10 patients with CEP290 mutations (BRILLIANCE trial by Editas Medicine). AAV5 vector was used to deliver the SaCas9 and CEP290-specific gRNAs to photoreceptor cells by subretinal injection. A GRK1 promoter was used to selectively target the photoreceptors and the gRNAs were targeted against a common variant in the CEP290 gene: IVS26 c.2991+1655 A>G mutation (EDIT-101 (Maeder et al., 2019)). The mutation lies within the intronic region, between exons 26 and 27, and the investigators used two guides to cut out the defect. Preclinical testing was carried out in human retinal explants following punch biopsy to test editing frequency. To test in vivo editing, a human CEP290 IVS26 knock-in mouse model was used, and they observed up to 21% editing that was sustained over 6-9 months. Finally, this study verified editing in NHPs, although the gRNAs needed to be modified due to sequence variation between humans and NHPs (Maeder et al., 2019). Based on the preclinical data, a total of 14 patients were treated in the trial, including 12 adults and two kids. Unfortunately, while the treatment was well tolerated, the trial is currently paused as only three out of 14 subjects met the threshold of clinically meaningful visual improvements. Interestingly, two of the three responders had homozygous mutations, suggesting that correction of one copy is likely not enough for clinical efficacy.
Table 1.
Gene editing-based clinical trials found in ClinicalTrials.gov
| Title | Conditions | Interventions | Trial# |
|---|---|---|---|
| In vivo Eye Diseases | |||
| Safety and Efficacy of CRISPR/Cas9 mRNA Instantaneous Gene Editing Therapy to Treat Refractory Viral Keratitis | Refractory Viral Keratitis, Herpes Simplex Virus Infection | Drug: BD111 Adult single group Dose | NCT04560790 |
| Single Ascending Dose Study in Participants With LCA10 | Leber Congenital Amaurosis 10, Inherited Retinal Dystrophies | Drug: EDIT-101 | NCT03872479 |
| In Vivo Non-Eye Disorders | |||
| Study to Evaluate Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of NTLA-2001 in Patients With Hereditary Transthyretin Amyloidosis With Polyneuropathy (ATTRv-PN) and Patients With Transthyretin Amyloidosis-Related Cardiomyopathy (ATTR-CM) | Transthyretin-Related (ATTR) Familial Amyloid Polyneuropathy | Biological: NTLA-2001 | NCT04601051 |
| A Safety and Efficacy Study of TALEN and CRISPR/Cas9 in the Treatment of HPV-related Cervical Intraepithelial Neoplasia | Human Papillomavirus-Related Neoplasm | Biological: CRISPR/Cas9 or TALEN | NCT03057912 |
| Study of EBT-101 in Aviremic HIV-1 Infected Adults on Stable ART | HIV-1-infection | Biological: EBT-101 | NCT05144386 |
| NTLA-2002 in Adults With Hereditary Angioedema | Hereditary Angioedema | Biological NTLA-2002 | NCT05120830 |
| Treatment of a Single Patient With CRD-TMH-001 | Duchenne Muscular Dystrophy | Drug: CRD-TMH-001 | NCT05514249 |
| Ex vivo edited non-blood cells | |||
| An Open-Label, FIH Study Evaluating the Safety, Tolerability, and Efficacy of VCTX211 Combination Product in Subjects With T1D | Diabetes Mellitus, Type 1 | Combination Product: VCTX211 | NCT05565248 |
| An Open-Label, FIH Study Evaluating the Safety and Tolerability of VCTX210A Combination Product in Subjects With T1D | Diabetes Mellitus, Type 1 | Combination Product: VCTX210A unit | NCT05210530 |
| Ex vivo edited hematopoietic cells | |||
| Study of CRISPR-Cas9 Mediated PD-1 and TCR Gene-knocked Out Mesothelin-directed CAR-T Cells in Patients With Mesothelin Positive Multiple Solid Tumors. | Solid Tumor, Adult | Biological: anti-mesothelin CAR-T cells | NCT03545815 |
| EDIT-301 for Autologous Hematopoietic Stem Cell Transplant (HSCT) in Participants With Transfusion-Dependent Beta Thalassemia (TDT) | Transfusion Dependent Beta Thalassemia | Genetic: EDIT-301 | NCT05444894 |
| Transplantation of Clustered Regularly Interspaced Short Palindromic Repeats Modified Hematopoietic Progenitor Stem Cells (CRISPR_SCD001) in Patients With Severe Sickle Cell Disease | Sickle Cell Disease | Drug: CRISPR_SCD001 | NCT04774536 |
| CISH Inactivated TILs in the Treatment of NSCLC | Metastatic Non Small Cell Lung Cancer | Other: CISH Inactivated T cells | NCT05566223 |
| A Safety and Efficacy Study Evaluating CTX120 in Subjects With Relapsed or Refractory Multiple Myeloma | Multiple Myeloma | Biological: CTX120 | NCT04244656 |
| Evaluation of Efficacy and Safety of a Single Dose of CTX001 in Participants With Transfusion-Dependent Î2-Thalassemia and Severe Sickle Cell Disease | Beta-Thalassemia, Sickle Cell Disease | Biological: CTX001 | NCT05477563/NCT03655678 |
| A Safety and Efficacy Study Evaluating CTX130 in Subjects With Relapsed or Refractory Renal Cell Carcinoma (COBALT-RCC) | Renal Cell Carcinoma | Biological: CTX130 | NCT04438083 |
| A Safety and Efficacy Study Evaluating CTX112 in Subjects With Relapsed or Refractory B-Cell Malignancies | B-cell Lymphoma | Biological: CTX112 | NCT05643742 |
| A Safety and Efficacy Study Evaluating CTX130 in Subjects With Relapsed or Refractory T or B Cell Malignancies (COBALT-LYM) | T Cell Lymphoma | Biological: CTX130 | NCT04502446 |
| A Safety and Efficacy Study Evaluating CTX110 in Subjects With Relapsed or Refractory B-Cell Malignancies (CARBON) | B-cell Malignancy, Non-Hodgkin Lymphoma | Biological: CTX110 | NCT04035434 |
| CRISPR-Edited Allogeneic Anti-CD19 CAR-T Cell Therapy for Relapsed/Refractory B Cell Non-Hodgkin Lymphoma | B Cell Non-Hodgkin's Lymphoma | Genetic: CB-010 | NCT04637763 |
| iHSCs With the Gene Correction of HBB Intervent Subjects With Î2-thalassemia Mutations | Thalassemia | Biological: iHSCs treatment group | NCT03728322 |
| CRISPR-Edited Allogeneic Anti-BCMA CAR-T Cell Therapy in Patients With Relapsed/Refractory Multiple Myeloma | Relapsed/Refractory Multiple Myeloma | Biological: CB-011 | NCT05722418 |
| A Safety and Efficacy Study Evaluating ET-01 in Subjects With Transfusion Dependent Î2-Thalassaemia | Transfusion Dependent Beta-Thalassemia | Biological: ET-01 | NCT04925206 |
| Study of PD-1 Gene-knocked Out Mesothelin-directed CAR-T Cells With the Conditioning of PC in Mesothelin Positive Multiple Solid Tumors | Mesothelin+ Solid Tumor | Biological: Mesothelin-directed CAR-T cells | NCT03747965 |
| A Study Evaluating UCART019 in Patients With Relapsed or Refractory CD19+ Leukemia and Lymphoma | B Cell Leukemia/Lymphoma | Biological: UCART019 | NCT03166878 |
| PD-1 Knockout Engineered T Cells for Advanced Esophageal Cancer | Esophageal Cancer | Other: PD-1 KO T Cells | NCT03081715 |
| Safety and Efficacy Evaluation of BRL-101 in Subjects With Transfusion-Dependent Î2-Thalassemia | Beta-Thalassemia | Drug: BRL-101 | NCT05577312 |
| A Feasibility and Safety Study of Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cell Immunotherapy for Relapsed or Refractory Leukemia and Lymphoma | B Cell Leukemia/ Lymphoma | Biological: CD19 and CD20 or CD22 CAR-T Cells | NCT03398967 |
| Genetic Ablation of CD33 in HSC to Broaden the Therapeutic Index of CD33-directed Immunotherapy in Patients With AML | Refractory Acute Myeloid Leukemia (AML) | Biological: CD34+ HSC with CD33 deletion | NCT05662904 |
| Gene Correction in Autologous CD34+ Hematopoietic Stem Cells (HbS to HbA) to Treat Severe Sickle Cell Disease | Sickle Cell Disease | Genetic: GPH101 Drug Product | NCT04819841 |
| Study Investigating NTLA-5001 in Subjects with Acute Myeloid Leukemia | Acute Myeloid Leukemia | Genetic: NTLA-5001 | NCT05066165 |
| TRAC Locus-inserted CD19-targeting STAR-T Cell Therapy in r/r B-NHL | B Cell Non-Hodgkin Lymphoma | Biological: Autologous CD19-STAR-T cell | NCT05631912 |
| TGFÎ2R-KO CAR-EGFR T Cells in Previously Treated Advanced EGFR-positive Solid Tumors | EGFR+ Solid Tumors | Biological: TGFÎ2R-KO CAR-EGFR T Cells | NCT04976218 |
| PD-1 Knockout Engineered T Cells for Metastatic Non-small Cell Lung Cancer | Metastatic Non-small Cell Lung Cancer | Other: PD-1 Knockout T Cells | NCT02793856 |
| Safety and Efficacy of CT125A Cells for Treatment of Relapsed/Refractory CD5+ Hematopoietic Malignancies | CD5+ Relapsed/Refractory Hematopoietic Malignancies | Biological: CT125A cells | NCT04767308 |
| Study of Base Edited CAR7 T Cells to Treat T Cell Malignancies (TvT CAR7) | Refractory T-cell Acute Lymphoid Leukaemia | Biological: BE CAR7 T cells | NCT05397184 |
| A Long-term Follow-up Study of Patients Who Received VOR33 | Leukemia, Myeloid, Acute | Genetic: VOR33 | NCT05309733 |
| Safety of Transplantation of CRISPR CCR5 Modified CD34+ Cells in HIV-infected Subjects With Hematological Malignances | HIV-1-infection | Genetic: CCR5 gene modification | NCT03164135 |
| CRISPR (HPK1) Edited CD19-specific CAR-T Cells (XYF19 CAR-T Cells) for CD19+ Leukemia or Lymphoma. | Refractory CD19+ Leukemia or Lymphoma | Genetic: XYF19 CAR-T cell | NCT04037566 |
8. Conclusion
One of the ultimate goals in the IRD field is to develop a universal or easily modifiable, scalable therapy that can be applied to as many patients as possible. Substantial progress has already been made toward this end, with gene editing at the forefront. Gene editing seems like the ideal therapy to mitigate IRD progression, as it provides a permanent fix to disease-causing mutations. Here, we discuss the most promising tools currently available, including gene editors, delivery systems, and model systems for pre-clinical testing. Much work is still required to attain a safe, efficient, and broadly applicable gene editing-based therapeutic; however, significant progress has been made in this active area of research. With the ongoing efforts of many research groups, we can hopefully achieve our collective goal in the near future, and help to improve vision for countless individuals.
Acknowledgments
Figures 1 and 2 were created using BioRender.com.
Funding
This work was supported in part by NIH research grants: K99EY033833 (NEI) to K.R.C., and R01EY032197 (NEI) to D.A.L. Support was also provided by a NEI P30 Vision Core grant to the Casey Eye Institute at OHSU, an unrestricted grant to the Casey Eye Institute at OHSU from Research to Prevent Blindness, a NEI P30 Vision Core grant to the UCSF Department of Ophthalmology, and an unrestricted grant to the UCSF Department of Ophthalmology from Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CRediT authorship contribution statement
K.R.C.: Conceptualization and writing (original draft, review, and editing). C.M.: Writing (original draft). D.A.L.: Conceptualization and writing (original draft, review, and editing).
Declaration of Competing Interest
All authors have declared that no conflict of interest exists.
References
- Anzalone AV, Koblan LW, Liu DR, 2020. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844. 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR, 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banskota S, Raguram A, Suh S, Du SW, Davis JR, Choi EH, Wang X, Nielsen SC, Newby GA, Randolph PB, Osborn MJ, Musunuru K, Palczewski K, Liu DR, 2022. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 185, 250–265.e16. 10.1016/j.cell.2021.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingrath J-S, McClements ME, Shanks M, Clouston P, Fischer MD, MacLaren RE, 2022. Envisioning the development of a CRISPR-Cas mediated base editing strategy for a patient with a novel pathogenic CRB1 single nucleotide variant. Ophthalmic Genet 43, 661–670. 10.1080/13816810.2022.2073599 [DOI] [PubMed] [Google Scholar]
- Bennett J, Wellman J, Marshall KA, McCague S, Ashtari M, DiStefano-Pappas J, Elci OU, Chung DC, Sun J, Wright JF, Cross DR, Aravand P, Cyckowski LL, Bennicelli JL, Mingozzi F, Auricchio A, Pierce EA, Ruggiero J, Leroy BP, Simonelli F, High KA, Maguire AM, 2016. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet 388, 661–672. 10.1016/S0140-6736(16)30371-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessant DA, Ali RR, Bhattacharya SS, 2001. Molecular genetics and prospects for therapy of the inherited retinal dystrophies. Curr Opin Genet Dev 11, 307–316. 10.1016/s0959-437x(00)00195-7 [DOI] [PubMed] [Google Scholar]
- Bucher K, Rodríguez-Bocanegra E, Dauletbekov D, Fischer MD, 2021. Immune responses to retinal gene therapy using adeno-associated viral vectors - Implications for treatment success and safety. Prog Retin Eye Res 83, 100915. 10.1016/j.preteyeres.2020.100915 [DOI] [PubMed] [Google Scholar]
- Capowski EE, Samimi K, Mayerl SJ, Phillips MJ, Pinilla I, Howden SE, Saha J, Jansen AD, Edwards KL, Jager LD, Barlow K, Valiauga R, Erlichman Z, Hagstrom A, Sinha D, Sluch VM, Chamling X, Zack DJ, Skala MC, Gamm DM, 2019. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 146. 10.1242/dev.171686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR, 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18, 495–506. 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen P-F, Chen C, Nelson JW, Newby GA, Sahin M, Osborn MJ, Weissman JS, Adamson B, Liu DR, 2021. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29. 10.1016/j.cell.2021.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Liu DR, 2023. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet 24, 161–177. 10.1038/s41576-022-00541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Kang Y, Yang W, Niu Y, Guo X, Tu Z, Si C, Wang H, Xing R, Pu X, Yang S-H, Li S, Ji W, Li X-J, 2015. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet 24, 3764–3774. 10.1093/hmg/ddv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SH, Martinez C, Chirco KR, Kandoi S, Lamba DA, 2022. Timed Notch Inhibition Drives Photoreceptor Fate Specification in Human Retinal Organoids. Invest Ophthalmol Vis Sci 63, 12. 10.1167/iovs.63.10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirco KR, Chew S, Moore AT, Duncan JL, Lamba DA, 2021. Allele-specific gene editing to rescue dominant CRX-associated LCA7 phenotypes in a retinal organoid model. Stem Cell Reports 16, 2690–2702. 10.1016/j.stemcr.2021.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisum HJ, Mooser F, Fitzpatrick D, 2003. Emergent properties of layer 2/3 neurons reflect the collinear arrangement of horizontal connections in tree shrew visual cortex. J Neurosci 23, 2947–2960. 10.1523/JNEUROSCI.23-07-02947.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EH, Suh S, Foik AT, Leinonen H, Newby GA, Gao XD, Banskota S, Hoang T, Du SW, Dong Z, Raguram A, Kohli S, Blackshaw S, Lyon DC, Liu DR, Palczewski K, 2022. In vivo base editing rescues cone photoreceptors in a mouse model of early-onset inherited retinal degeneration. Nat Commun 13, 1830. 10.1038/s41467-022-29490-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R, 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33, 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Cowan CS, Renner M, De Gennaro M, Gross-Scherf B, Goldblum D, Hou Y, Munz M, Rodrigues TM, Krol J, Szikra T, Cuttat R, Waldt A, Papasaikas P, Diggelmann R, Patino-Alvarez CP, Galliker P, Spirig SE, Pavlinic D, Gerber-Hollbach N, Schuierer S, Srdanovic A, Balogh M, Panero R, Kusnyerik A, Szabo A, Stadler MB, Orgül S, Picelli S, Hasler PW, Hierlemann A, Scholl HPN, Roma G, Nigsch F, Roska B, 2020. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 182, 1623–1640.e34. 10.1016/j.cell.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers FPM, Boon CJF, Bujakowska K, Zeitz C, 2018. Special Issue Introduction: Inherited Retinal Disease: Novel Candidate Genes, Genotype-Phenotype Correlations, and Inheritance Models. Genes (Basel) 9, 215. 10.3390/genes9040215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo-Rodríguez A, Delgado D, Gascón AR, Solinís MÁ, 2013. Lipid nanoparticles as drug/gene delivery systems to the retina. J Ocul Pharmacol Ther 29, 173–188. 10.1089/jop.2012.0128 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E, 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Elsayed MEAA, Kaukonen M, Kiraly P, Kapetanovic JC, MacLaren RE, 2022. Potential CRISPR Base Editing Therapeutic Options in a Sorsby Fundus Dystrophy Patient. Genes (Basel) 13, 2103. 10.3390/genes13112103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eygeris Y, Gupta M, Kim J, Sahay G, 2022. Chemistry of Lipid Nanoparticles for RNA Delivery. Acc Chem Res 55, 2–12. 10.1021/acs.accounts.1c00544 [DOI] [PubMed] [Google Scholar]
- Fan Y, Huang Z-Y, Cao C-C, Chen C-S, Chen Y-X, Fan D-D, He J, Hou H-L, Hu L, Hu X-T, Jiang X-T, Lai R, Lang Y-S, Liang B, Liao S-G, Mu D, Ma Y-Y, Niu Y-Y, Sun X-Q, Xia J-Q, Xiao J, Xiong Z-Q, Xu L, Yang L, Zhang Y, Zhao W, Zhao X-D, Zheng Y-T, Zhou J-M, Zhu Y-B, Zhang G-J, Wang J, Yao Y-G, 2013. Genome of the Chinese tree shrew. Nat Commun 4, 1426. 10.1038/ncomms2416 [DOI] [PubMed] [Google Scholar]
- Finkbeiner C, Ortuño-Lizarán I, Sridhar A, Hooper M, Petter S, Reh TA, 2022. Single-cell ATAC-seq of fetal human retina and stem-cell-derived retinal organoids shows changing chromatin landscapes during cell fate acquisition. Cell Rep 38, 110294. 10.1016/j.celrep.2021.110294 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D, 1996. The functional organization of local circuits in visual cortex: insights from the study of tree shrew striate cortex. Cereb Cortex 6, 329–341. 10.1093/cercor/6.3.329 [DOI] [PubMed] [Google Scholar]
- Fry LE, McClements ME, MacLaren RE, 2021. Analysis of Pathogenic Variants Correctable With CRISPR Base Editing Among Patients With Recessive Inherited Retinal Degeneration. JAMA Ophthalmol 139, 319–328. 10.1001/jamaophthalmol.2020.6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur M, Sivak JG, 1979. Refractive state of the eye of a small diurnal mammal: the ground squirrel. Am J Optom Physiol Opt 56, 689–695. 10.1097/00006324-197911000-00004 [DOI] [PubMed] [Google Scholar]
- Herrera-Barrera M, Ryals RC, Gautam M, Jozic A, Landry M, Korzun T, Gupta M, Acosta C, Stoddard J, Reynaga R, Tschetter W, Jacomino N, Taratula O, Sun C, Lauer AK, Neuringer M, Sahay G, 2023. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci Adv 9, eadd4623. 10.1126/sciadv.add4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgaard A, Alsing S, Askou AL, Corydon TJ, 2019. CRISPR Gene Therapy of the Eye: Targeted Knockout of Vegfa in Mouse Retina by Lentiviral Delivery. Methods Mol Biol 1961, 307–328. 10.1007/978-1-4939-9170-9_19 [DOI] [PubMed] [Google Scholar]
- Hori T, Fukutome M, Koike C, 2019. Adeno Associated Virus (AAV) as a Tool for Clinical and Experimental Delivery of Target Genes into the Mammalian Retina. Biol Pharm Bull 42, 343–347. 10.1248/bpb.b18-00913 [DOI] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR, 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ML, Edwards TL, O’Hare F, Hickey DG, Wang J-H, Liu Z, Ayton LN, 2021. Gene therapy for inherited retinal diseases: progress and possibilities. Clin Exp Optom 104, 444–454. 10.1080/08164622.2021.1880863 [DOI] [PubMed] [Google Scholar]
- Huang K-C, Wang M-L, Chen S-J, Kuo J-C, Wang W-J, Nhi Nguyen PN, Wahlin KJ, Lu J-F, Tran AA, Shi M, Chien Y, Yarmishyn AA, Tsai P-H, Yang T-C, Jane W-N, Chang C-C, Peng C-H, Schlaeger TM, Chiou S-H, 2019. Morphological and Molecular Defects in Human Three-Dimensional Retinal Organoid Model of X-Linked Juvenile Retinoschisis. Stem Cell Reports 13, 906–923. 10.1016/j.stemcr.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheim R, Tai PWL, Mir A, Javeed N, Wang J, Rodríguez TC, Namkung S, Nelson S, Khokhar ES, Mintzer E, Maitland S, Chen Z, Cao Y, Tsagkaraki E, Wolfe SA, Wang D, Pai AA, Xue W, Gao G, Sontheimer EJ, 2021. Self-inactivating, all-in-one AAV vectors for precision Cas9 genome editing via homology-directed repair in vivo. Nat Commun 12, 6267. 10.1038/s41467-021-26518-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel JH, Fisher SK, 1985. Cone photoreceptor shedding in the tree shrew (Tupaia belangerii). Cell Tissue Res 239, 667–675. 10.1007/BF00219247 [DOI] [PubMed] [Google Scholar]
- Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G, Gopalappa R, Kim D, Cho S-R, Kim JH, Kim HH, 2022. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng 6, 181–194. 10.1038/s41551-021-00788-9 [DOI] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA, 2016. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871. 10.1126/science.aad8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo DH, Jang H-K, Cho CS, Han JH, Ryu G, Jung Y, Bae S, Kim JH, 2023. Visual function restoration in a mouse model of Leber congenital amaurosis via therapeutic base editing. Mol Ther Nucleic Acids 31, 16–27. 10.1016/j.omtn.2022.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonlin EC, 2020. Informed Consent for Human Embryo Genome Editing. Stem Cell Reports 14, 530–537. 10.1016/j.stemcr.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD, 2013. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14, 49–55. 10.1038/nrm3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoi S, Martinez C, Merriman DK, Lamba DA, 2022. Characterization of Retinal Development in 13-Lined Ground Squirrels. Transl Vis Sci Technol 11, 17. 10.1167/tvst.11.11.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukonen M, McClements ME, MacLaren RE, 2022. CRISPR DNA Base Editing Strategies for Treating Retinitis Pigmentosa Caused by Mutations in Rhodopsin. Genes (Basel) 13, 1327. 10.3390/genes13081327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JJ, Saee-Marand M, Nyström NN, Evans MM, Chen Y, Martinez FM, Hamilton AM, Ronald JA, 2021. Safe harbor-targeted CRISPR-Cas9 homology-independent targeted integration for multimodality reporter gene-based cell tracking. Sci Adv 7, eabc3791. 10.1126/sciadv.abc3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryger Z, Galli-Resta L, Jacobs GH, Reese BE, 1998. The topography of rod and cone photoreceptors in the retina of the ground squirrel. Vis Neurosci 15, 685–691. 10.1017/s0952523898154081 [DOI] [PubMed] [Google Scholar]
- Li A, Lee CM, Hurley AE, Jarrett KE, De Giorgi M, Lu W, Balderrama KS, Doerfler AM, Deshmukh H, Ray A, Bao G, Lagor WR, 2019. A Self-Deleting AAV-CRISPR System for In Vivo Genome Editing. Mol Ther Methods Clin Dev 12, 111–122. 10.1016/j.omtm.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Samulski RJ, 2020. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 21, 255–272. 10.1038/s41576-019-0205-4 [DOI] [PubMed] [Google Scholar]
- Li F, Hung SSC, Mohd Khalid MKN, Wang J-H, Chrysostomou V, Wong VHY, Singh V, Wing K, Tu L, Bender JA, Pébay A, King AE, Cook AL, Wong RCB, Bui BV, Hewitt AW, Liu G-S, 2019. Utility of Self-Destructing CRISPR/Cas Constructs for Targeted Gene Editing in the Retina. Hum Gene Ther 30, 1349–1360. 10.1089/hum.2019.021 [DOI] [PubMed] [Google Scholar]
- Li S, Hu Y, Li Y, Hu M, Wang W, Ma Y, Cai Y, Wei M, Yao Y, Wang Y, Dong K, Gu Y, Zhao H, Bao J, Qiu Z, Zhang M, Hu X, Xue T, 2021. Generation of nonhuman primate retinitis pigmentosa model by in situ knockout of RHO in rhesus macaque retina. Sci Bull (Beijing) 66, 374–385. 10.1016/j.scib.2020.09.008 [DOI] [PubMed] [Google Scholar]
- Lin Q, Lv J-N, Wu K-C, Zhang C-J, Liu Q, Jin Z-B, 2020. Generation of Nonhuman Primate Model of Cone Dysfunction through In Situ AAV-Mediated CNGB3 Ablation. Mol Ther Methods Clin Dev 18, 869–879. 10.1016/j.omtm.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Costa B, Kolesnikova M, Levi SR, Cabral T, Tsang SH, Maumenee IH, Quinn PMJ, 2023. Clinical and Therapeutic Evaluation of the Ten Most Prevalent CRB1 Mutations. Biomedicines 11, 385. 10.3390/biomedicines11020385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, Dass A, Dhanapal V, Fennell TJ, Friedland AE, Giannoukos G, Gloskowski SW, Glucksmann A, Gotta GM, Jayaram H, Haskett SJ, Hopkins B, Horng JE, Joshi S, Marco E, Mepani R, Reyon D, Ta T, Tabbaa DG, Samuelsson SJ, Shen S, Skor MN, Stetkiewicz P, Wang T, Yudkoff C, Myer VE, Albright CF, Jiang H, 2019. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med 25, 229–233. 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV, 2020. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83. 10.1038/s41579-019-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurov D, Ramadan L, Kruglova N, 2023. Packaging and Uncoating of CRISPR/Cas Ribonucleoproteins for Efficient Gene Editing with Viral and Non-Viral Extracellular Nanoparticles. Viruses 15, 690. 10.3390/v15030690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrien NA, Norton TT, 1992. The development of experimental myopia and ocular component dimensions in monocularly lid-sutured tree shrews (Tupaia belangeri). Vision Res 32, 843–852. 10.1016/0042-6989(92)90027-g [DOI] [PubMed] [Google Scholar]
- Moshiri A, Chen R, Kim S, Harris RA, Li Y, Raveendran M, Davis S, Liang Q, Pomerantz O, Wang J, Garzel L, Cameron A, Yiu G, Stout JT, Huang Y, Murphy CJ, Roberts J, Gopalakrishna KN, Boyd K, Artemyev NO, Rogers J, Thomasy SM, 2019. A nonhuman primate model of inherited retinal disease. J Clin Invest 129, 863–874. 10.1172/JCI123980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muigai AWT, 2022. Expanding global access to genetic therapies. Nat Biotechnol 40, 20–21. 10.1038/s41587-021-01191-0 [DOI] [PubMed] [Google Scholar]
- Müller B, Peichl L, 1989. Topography of cones and rods in the tree shrew retina. J Comp Neurol 282, 581–594. 10.1002/cne.902820409 [DOI] [PubMed] [Google Scholar]
- Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An M, Newby GA, Chen JC, Hsu A, Liu DR, 2022. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 40, 402–410. 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A, 2016. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729. 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O, 2018. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262. 10.1126/science.aas9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemacher SK, Iglesias CL, Sridhar A, Gamm DM, Meyer JS, 2015. Generation of highly enriched populations of optic vesicle-like retinal cells from human pluripotent stem cells. Curr Protoc Stem Cell Biol 32, 1H.8.1–1H.8.20. 10.1002/9780470151808.sc01h08s32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Ryals RC, Weller KK, Pennesi ME, Sahay G, 2019. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J Control Release 303, 91–100. 10.1016/j.jconrel.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SM, McGill TJ, Puthussery T, Stoddard J, Renner L, Lewis AD, Colgin LMA, Gayet J, Wang X, Prongay K, Cullin C, Dozier BL, Ferguson B, Neuringer M, 2019. Bardet-Biedl Syndrome in rhesus macaques: A nonhuman primate model of retinitis pigmentosa. Exp Eye Res 189, 107825. 10.1016/j.exer.2019.107825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotter E, McClements ME, MacLaren RE, 2021. The Scope of Pathogenic ABCA4 Mutations Targetable by CRISPR DNA Base Editing Systems-A Systematic Review. Front Genet 12, 814131. 10.3389/fgene.2021.814131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Zhang W, Zhang S, Feng Y, Xu W, Qi J, Zhang Q, Xu C, Liu S, Zhang J, Lei Y, Liu W, Feng S, Wang J, Fu X, Xu Z, Li P, Yao K, 2023. Vision rescue via unconstrained in vivo prime editing in degenerating neural retinas. J Exp Med 220, e20220776. 10.1084/jem.20220776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguram A, Banskota S, Liu DR, 2022a. Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827. 10.1016/j.cell.2022.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguram A, Banskota S, Liu DR, 2022b. Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827. 10.1016/j.cell.2022.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remé CE, Young RW, 1977. The effects of hibernation on cone visual cells in the ground squirrel. Invest Ophthalmol Vis Sci 16, 815–840. [PubMed] [Google Scholar]
- Riesenberg S, Maricic T, 2018. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun 9, 2164. 10.1038/s41467-018-04609-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J, 2015. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7, 93. 10.1186/s13073-015-0215-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Statz JP, Chan W, Burch FC, Brigande JV, Kempton B, Porsov EV, Renner L, McGill T, Burwitz BJ, Hanna CB, Neuringer M, Hennebold JD, 2022. CRISPR/Cas9 editing of the MYO7A gene in rhesus macaque embryos to generate a primate model of Usher syndrome type 1B. Sci Rep 12, 10036. 10.1038/s41598-022-13689-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajdak B, Sulai YN, Langlo CS, Luna G, Fisher SK, Merriman DK, Dubra A, 2016. Noninvasive imaging of the thirteen-lined ground squirrel photoreceptor mosaic. Vis Neurosci 33, e003. 10.1017/S0952523815000346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Muñoz-Subirana N, Kool H, van Schendel R, van der Vlies S, Kamp JA, de Vrij F, Kushner SA, Smith GCM, Boulton SJ, Tijsterman M, 2023. Modulating mutational outcomes and improving precise gene editing at CRISPR-Cas9-induced breaks by chemical inhibition of end-joining pathways. Cell Rep 42, 112019. 10.1016/j.celrep.2023.112019 [DOI] [PubMed] [Google Scholar]
- Schleidgen S, Dederer H-G, Sgodda S, Cravcisin S, Lüneburg L, Cantz T, Heinemann T, 2020. Human germline editing in the era of CRISPR-Cas: risk and uncertainty, inter-generational responsibility, therapeutic legitimacy. BMC Med Ethics 21, 87. 10.1186/s12910-020-00487-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön C, Biel M, Michalakis S, 2015. Retinal gene delivery by adeno-associated virus (AAV) vectors: Strategies and applications. Eur J Pharm Biopharm 95, 343–352. 10.1016/j.ejpb.2015.01.009 [DOI] [PubMed] [Google Scholar]
- She K, Liu Y, Zhao Q, Jin X, Yang Yiliu, Su J, Li R, Song L, Xiao J, Yao S, Lu F, Wei Y, Yang Yang, 2023. Dual-AAV split prime editor corrects the mutation and phenotype in mice with inherited retinal degeneration. Signal Transduct Target Ther 8, 57. 10.1038/s41392-022-01234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman SM, Norton TT, Casagrande VA, 1977. Myopia in the lid-sutured tree shrew (Tupaia glis). Brain Res 124, 154–157. 10.1016/0006-8993(77)90872-1 [DOI] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B, 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82, 237–266. 10.1146/annurev-biochem-072911-172315 [DOI] [PubMed] [Google Scholar]
- Sridhar A, Hoshino A, Finkbeiner CR, Chitsazan A, Dai L, Haugan AK, Eschenbacher KM, Jackson DL, Trapnell C, Bermingham-McDonogh O, Glass I, Reh TA, 2020. Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep 30, 1644–1659.e4. 10.1016/j.celrep.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, She K, Song L, Jin X, Li R, Zhao Q, Xiao J, Chen D, Cheng H, Lu F, Wei Y, Yang Y, 2023. In vivo base editing rescues photoreceptors in a mouse model of retinitis pigmentosa. Mol Ther Nucleic Acids 31, 596–609. 10.1016/j.omtn.2023.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Klein H, 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7, 739–750. 10.1038/nrm2008 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nuñez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu G-H, Magistretti P, Zhang Kun, Callaway EM, Zhang Kang, Belmonte JCI, 2016. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Trapani I, 2019. Adeno-Associated Viral Vectors as a Tool for Large Gene Delivery to the Retina. Genes (Basel) 10, 287. 10.3390/genes10040287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Zhao H, Li B, Yan S, Wang L, Tang Y, Li Z, Bai D, Li C, Lin Y, Li Y, Liu J, Xu H, Guo X, Jiang Y-H, Zhang YQ, Li X-J, 2019. CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum Mol Genet 28, 561–571. 10.1093/hmg/ddy367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchiano G, Andrieux G, Klermund J, Blattner G, Pennucci V, El Gaz M, Monaco G, Poddar S, Mussolino C, Cornu TI, Boerries M, Cathomen T, 2021. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell 28, 1136–1147.e5. 10.1016/j.stem.2021.02.002 [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11, 636–646. 10.1038/nrg2842 [DOI] [PubMed] [Google Scholar]
- Walton RT, Christie KA, Whittaker MN, Kleinstiver BP, 2020. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rajala A, Rajala RVS, 2015. Lipid Nanoparticles for Ocular Gene Delivery. J Funct Biomater 6, 379–394. 10.3390/jfb6020379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A, Yin D, Zhai Z, Ling S, Le H, Tian L, Xu J, Paludan SR, Cai Y, Hong J, 2023. In Vivo CRISPR Gene Editing in Patients with Herpes Stromal Keratitis (preprint). Genetic and Genomic Medicine. 10.1101/2023.02.21.23285822 [DOI] [PubMed] [Google Scholar]
- Weterings E, Gallegos AC, Dominick LN, Cooke LS, Bartels TN, Vagner J, Matsunaga TO, Mahadevan D, 2016. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair (Amst) 43, 98–106. 10.1016/j.dnarep.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Wimmer T, Sawinski H, Urban AM, Motlik J, Stieger K, 2023. Rapid and Reliable Quantification of Prime Editing Targeting Within the Porcine ABCA4 Gene Using a Bioluminescence Resonance Energy Transfer-Based Sensor. Nucleic Acid Ther. 10.1089/nat.2022.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Liu Y, Tu Z, Xiao C, Yan S, Ma X, Guo X, Chen X, Yin P, Yang Z, Yang S, Jiang T, Li S, Qin C, Li X-J, 2019. CRISPR/Cas9-mediated PINK1 deletion leads to neurodegeneration in rhesus monkeys. Cell Res 29, 334–336. 10.1038/s41422-019-0142-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA, 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zhi S, Chen Y, Wu G, Wen J, Wu J, Liu Q, Li Y, Kang R, Hu S, Wang J, Liang P, Huang J, 2022. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol Ther 30, 283–294. 10.1016/j.ymthe.2021.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccaro MV, Xu J, Mitchell C, Marin D, Zimmerman R, Rana B, Weinstein E, King RT, Palmerola KL, Smith ME, Tsang SH, Goland R, Jasin M, Lobo R, Treff N, Egli D, 2020. Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell 183, 1650–1664.e15. 10.1016/j.cell.2020.10.025 [DOI] [PubMed] [Google Scholar]