Abstract
Background:
Mast cell activation is critical for the development of allergic diseases. Ligation of Sialic acid-binding immunoglobin-like lectins (Siglecs) such as CD33, Siglec-6, -7 and -8 have been shown to inhibit mast cell activation. Recent studies showed that human mast cells express Siglec-9, an inhibitory receptor also expressed by neutrophils, monocytes, macrophages, and dendritic cells.
Objective:
We aimed to characterize Siglec-9 expression and function in human mast cells in vitro.
Methods:
We assessed the expression of Siglec-9 and Siglec-9 ligands on human mast cell lines and human primary mast cells by qPCR, flow cytometry and confocal microscopy. We used a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) gene editing approach to disrupt the SIGLEC9 gene. We evaluated Siglec-9 inhibitory activity on mast cell function by using native Siglec-9 ligands, glycophorin A (GlycA) and high molecular weight hyaluronic acid (HMW-HA), a monoclonal antibody against Siglec-9, and co-engagement of Siglec-9 with the high affinity receptor for IgE (FcεRI).
Results:
Human mast cells express Siglec-9 and Siglec-9 ligands. SIGLEC9 gene disruption resulted in increased expression of activation markers at baseline and increased responsiveness to IgE-dependent and IgE-independent stimulation. Pre-treatment with GlycA or HMW-HA followed by IgE-dependent or independent stimulation had an inhibitory effect on mast cell degranulation. Co-engagement of Siglec-9 with FcεRI in human mast cells resulted in reduced degranulation, arachidonic acid production, and chemokine release.
Conclusions:
Our study indicates that Siglec-9 and its ligands play an important role in limiting human mast cell activation in vitro.
Clinical implications
Targeting Siglec-9 may limit mast cell contribution to allergic disease by inhibiting mast cell ability to release pro-inflammatory mediators.
Keywords: Mast cells, Siglec-9, sialic acids, CRISPR/Cas9, FcεRI
Capsule summary
Human mast cell expression of functional Siglec-9 limits mast cell activation and mediator release in vitro.
INTRODUCTION
Mast cells are hematopoietic progenitor-derived, granule-containing immune cells that are widely distributed in tissues that interact with the external environment, such as the skin and mucosal tissues1. Mast cells have been proposed to contribute to defense against pathogens, wound healing, and tumor surveillance by responding to a broad array of activating signals, secreting a wide range of inflammatory mediators, and recruiting and activating immune cells2–5. Despite these beneficial roles, numerous preclinical and clinical studies recognize mast cells as key effector cells in urticaria, mastocytosis and allergic disease6–10. These studies have shown that the dysregulated expansion and/or activation of mast cells have detrimental consequences in these disorders. Therefore, there is a need for effective new strategies that inhibit mast cell function and/or deplete active mast cells for the treatment of mast cell driven disorders.
Siglecs are a family of single-pass cell surface receptors characterized by a N-terminal domain that binds sialylated glycans11. Most Siglecs have one or multiple immunoreceptor tyrosine-based inhibitory motifs (ITIM) on the C-terminus that trigger inhibitory signals through the recruitment of tyrosine and inositol phosphatases12. Siglecs are predominantly found on immune cells, with each cell expressing a unique combination of Siglecs that allows them to respond to distinct sialylation patterns13, 14. Prior studies have shown that human mast cells express CD22/Siglec-2, CD33/Siglec-3, Siglec-5, Siglec-6, Siglec-7, Siglec-8, and Siglec-1015. Importantly, it has been shown that Siglecs can reduce the release of mast cell mediators16–19, attenuate mast cell-dependent anaphylaxis18, limit growth of human mast cell lines in a mouse model of mastocytosis20, ameliorate mast cell activation and inflammation in mouse models of non-allergic airway inflammation21, and reduce mast cell numbers in a mouse model of eosinophilic gastrointestinal disease22. Thus, the capability to limit mast cell activation and expansion has made Siglecs an attractive therapeutic target to downregulate mast cell function in allergic and non-allergic diseases.
Siglec-9 is an inhibitory receptor broadly expressed by neutrophils, monocytes, macrophages, dendritic cells, and subsets of B cells, T cells, and natural killer (NK) cells23–26. Studies of Siglec-9 have been mainly focused on its detrimental effects including dampening the innate immune response to certain pathogens27–30 and impairing immune surveillance in certain cancers31–34. However, recent studies support the use of Siglec-9 engagement to inhibit excessive immune cell activation and limit the magnitude of the inflammatory response during arthritis35, colitis36, and severe COVID-1937.
Siglec-9 expression and function in mast cells has been largely unexplored. Publicly available published datasets and online databases indicate that SIGLEC9 mRNA expression levels are low38 or not detected in peripheral blood mononuclear cell derived-cultured mast cells (PBCMCs)39. Similarly, SIGLEC9 mRNA was undetected (Lungmap database) or very low (CZ CellxGene data portal) in human lung mast cells. Based on the CZ CellxGene data portal, human skin mast cells expressed SIGLEC9 mRNA at low levels. However, the FANTOM Consortium database reports that human skin mast cells express higher mRNA levels of SIGLEC9 than SIGLEC7 or SIGLEC840. Recent reports support Siglec-9 protein expression in human skin18 and lung41 mast cells. Here, we confirmed that human mast cell lines and human primary mast cells express Siglec-9. Kinetic assessment of surface expression showed that Siglec-9 expression paralleled the expression of FcεRI during mast cell differentiation. Based on this evidence, we decided to investigate the functional relevance of Siglec-9 expression on human mast cells. Siglec-9 deletion by a CRISPR-Cas9 approach significantly increased the expression of activation markers on mast cells at baseline and mast cell ability to undergo a more robust activation when compared to unedited cells. Importantly, mast cells exhibited a marked reduction in mast cell degranulation when Siglec-9 was engaged with native ligands prior to IgE-dependent and IgE-independent activation. Furthermore, co-aggregating Siglec-9 with FcεRI resulted in decreased degranulation and reduced production of arachidonic acid metabolites and chemokines.
METHODS
Ethics statement
All animal experiments were approved by the Seattle Children’s Research Institutional Animal Care and Use Committee (Protocol #IACUC00020) and performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (8th Edition).
Mice
C57BL/6J mice were purchased from Jackson Laboratories and bred and maintained at the Seattle Children’s Research Institute Vivarium. Mice with Siglec-E-deficiency42 on the C57BL/6 background were kindly provided by Dr. Paul R. Crocker (University of Dundee, Scotland, UK).
Generation of bone marrow derived-cultured mast cells (BMCMCs)
Femoral bone marrow cells from C57BL/6J mice were maintained in vitro for ~6 weeks in Dulbecco’s Modified Eagle Medium (DMEM) containing 10 ng/ml recombinant mouse (rm) interleukin (IL)-3 (Peprotech, Rocky Hill, NJ) until the mast cells represented >95% of the total cells defined as c-Kit and FcεRIα positive cells when analyzed by flow cytometry.
Generation of fetal skin derived-cultured mast cells (FSCMCs)
Fetal skin cells from C57BL/6J mice were obtained as described43 and maintained in vitro for 4–6 weeks in DMEM containing 10 ng/ml rmIL-3 and 10 ng/ml rm stem cell factor (SCF) (Peprotech) until the mast cells represented >95% of the total cells defined as c-Kit and FcεRIα positive cells when assessed by flow cytometry.
Human mast cell lines
The human MCL-derived cell line HMC-1, subclone HMC-1.2 harboring KIT V560G and KIT D816V was kindly provided by Dr. J.H. Butterfield (Mayo Clinic, Rochester, MN)44 and was grown in Iscove’s Modified Dulbecco Media (IMDM) supplemented with 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 10% heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine, and 1% penicillin-streptomycin. LUVA cells were generously provided by Dr. John Steinke (University of Virginia, Charlottesville, VA)45 and maintained in StemPro-34 serum-free medium supplemented with StemPro-34 nutrient supplement (catalog number 10639011, Thermo Fisher Scientific, Waltham, MA), 2 mM L-glutamine, and 1% penicillin/streptomycin. LAD2 cells were kindly provided by Dr. Arnold Kirshenbaum (Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD)46 and maintained in the same media as LUVA cells with the addition of 100 ng/ml recombinant human (rh) SCF.
Human primary mast cells
Human peripheral blood was obtained from anonymous donors to Bloodworks Northwest (Seattle, WA) in accordance with established institutional guidelines, and mast cells were generated as described47. Briefly, CD34+ hematopoietic progenitors were isolated from the peripheral blood mononuclear fraction by immunomagnetic positive selection (catalog number 17856, StemCell Technologies, Vancouver, BC, Canada). CD34+ cells were cultured in StemSpan SFEM II media (StemCell Technologies) supplemented with 10 μg/ml ciprofloxacin (Sigma-Aldrich, St. Louis, MO), 50 ng/ml rhIL-6 (Peprotech), and 3% vol/vol of conditioned medium from Chinese hamster ovary (CHO) cell transfectants secreting murine SCF kindly donated by Dr. Patrice Dubreuil (INSERM, CNRS, Aix-Marseille Universite, Marseille, France) for at least 8 weeks. During the first week of culture, 50 ng/ml rhIL-3 (Peprotech) was added to the culture media. A mature mast cell population containing at least 90% c-Kit and FcεRIα positive cells as assessed by flow cytometry, was observed after 5 weeks of culture. Human skin derived-cultured mast cells (HSCMCs) were generated from mononuclear cells isolated from discarded tissues that were collected at the time of mastectomies or purchased from the National Cancer Institute-supported Cooperative Human Tissue Network kindly provided by Dr. Oskeritzian (University of South Carolina). Mast cell isolation procedure was approved by the Seattle Children’s Research Institute Human Subjects Review Committee and by the University of South Carolina Institutional Review Board. Skin mast cells were harvested and cultured as previously described48. Briefly, the subcutaneous fat was removed from skin specimens and the resultant skin was cut into small pieces and enzymatically digested in Hanks balanced salt solution (HBSS) containing 10% FBS, 10 mM HEPES, 0.035% sodium bicarbonate, 0.5% amphotericin B and 1% penicillin/streptomycin (Thermo Fisher Scientific) and supplemented with 0.7 mg/ml hyaluronidase (Sigma-Aldrich), 592.5 U/ml collagenase type II (Thermo Fisher Scientific), 0.15 mg/ml DNase type I (Sigma-Aldrich), and 1 mM CaCl2 (Sigma-Aldrich). The specimens were digested on a shaker at 37°C for 1 h. The digested mixture was filtered through a 70-μm cell strainer, and the remaining tissue was collected for two additional digestions. Collected cells were washed, spun down, filtered through a 40-μm filter, and washed again. The collected pellets from the three digestions were combined and layered over 75% Percoll in HBSS cushion and centrifuged at 800x g at room temperature for 20 min. Nucleated cells were collected from the buffer/Percoll interface, washed, and plated at a concentration of 5 × 105 cells/ml in serum-free X-Vivo 15 media (catalog number 04–418Q iLonza, Basel, Switzerland) supplemented with 100 ng/ml rhSCF (Peprotech). A mature mast cell population containing at least 90% c-Kit and FcεRIα positive cells as assessed by flow cytometry, was observed after 8 weeks of culture.
Lung mast cells were enriched from healthy tissues provided by the National Disease Research Interchange (NDRI) program as previously described49, 50. Briefly, lung fragments of 0.5–2.0 mm3 were washed twice with DMEM containing 2% FCS (DMEM/FCS) before incubation in the same buffer (1 g tissue/ 4 ml buffer) containing 1.5 mg/ml collagenase type IA, 0.75 mg/ml hyaluronidase, and 0.02 mg/ml DNAse. The specimens were digested on a shaker at 37°C for 1h. The digested mixture was filtered through a 70-μm cell strainer and washed twice with DMEM/FCS. The filtered cell suspension was resuspended in 30% Percoll (one phase) and centrifuged at 780x g for 12 min. Excess Percoll was removed by washing three times in HBSS containing 2% FCS. Siglec-9 expression was assessed by flow cytometry in the enriched lung mast cell population identified as c-Kit and FcεRIα positive cells.
Human neutrophils
Nucleated cells were isolated from the blood of healthy anonymous donors (Bloodworks Northwest) using HetaSep™ (catalog number 07906, StemCell Technologies) following manufacturer instructions. Neutrophils were identified as CD45, CD11b and CD66b positive cells as assessed by flow cytometry.
Gene Editing
To delete Siglec-9 expression, LAD2 cells were transfected with Siglec-9-targeting ribonucleoproteins using Lipofectamine CRISPRMAX reagent (catalog number CMAX00001, Thermo Fisher Scientific). Transfection was performed based on the manufacturer recommendations as follows. Cells were plated at 3 × 105 cells/well of a 24-well dish and transfected with equimolar (12 pmol) Cas9 (catalog number 1081058, Integrated DNA Technologies, Coralville, IA) and sgRNA (5’-GACGAUGCAGAGUUCCGUGA-3’) (catalog number A35533, Thermo Fisher Scientific) complexed with 4 μl Cas9Plus reagent and 1.5 μl CRISPRMAX reagent according to the manufacturer’s mixing instructions. Editing efficiency was evaluated by protein expression measured by flow cytometry.
qPCR
RNA was isolated from primary human mast cells using the RNeasy Plus Micro kit (catalog number 74034, Qiagen, Hilden, Germany). RNA was isolated from mast cell lines using the RNeasy Mini kit (catalog number 74104, Qiagen), converted to first-strand cDNA (SuperScript™ VILO™ cDNA Synthesis Kit, Invitrogen), and cDNA was analyzed for quantitative expression levels with the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific) on a Step One Plus Real-Time PCR System Thermal Cycling Block (Applied Biosystems, Waltham, MA). Results were analyzed using the dCt method normalized to GAPDH Ct. The primers used were as follows: SIGLEC7 forward, 5’-GCCATAAGTTTGCAGCATCTC-3’; SIGLEC7 reverse, 5’- GCCATTGGAAGCTCTATCTGC-3’; SIGLEC8 forward, 5’-GTTGGGGGTGAAGTCAGAAAAG-3’; SIGLEC8 reverse, 5’- GGGTGGGAATCTGGATGAGTT-3’; SIGLEC9 forward, 5’- AATCTGACCTGCTCTGTGCC-3’; SIGLEC9 reverse, 5’- AAGTTCTGAGGCGGGTAGGA-3’; ST3GAL1 forward, 5’- CCGCTGTGGTCATTTAGGAA-3’; ST3GAL1 reverse, 5’- TCCATCTCTGGTCCCCAAAT-3’; ST3GAL3 forward, 5’- CCGCTGTGGTCATTTAGGAA-3’; ST3GAL3 reverse, 5’- GGGGTGAGCTAGAGTGACTA-3’; ST3GAL4 forward, 5’- CTCCCGGGAAGACAGTTTTT-3’; ST3GAL4 reverse, 5’- GTAAGCAGATGGCGTCTTGA-3’; ST3GAL5 forward, 5’- CTCTGTGGCTGCTCTTGTCA-3’; ST3GAL5 reverse TGGTGAGGAGGAGGGAGATG-3’; ST3GAL6 forward, 5’- TTGCGAACAGAGGGTCTTTAG-3’; ST3GAL6 reverse, 5’- AAGACAGCACTCAGGAATATGG-3’; GNE forward, 5’- TGACGGCGTCTGGAACTCTA-3’; GNE reverse, 5’- GTAGCAACACAAACCCGCAG-3’; GAPDH forward, 5’- GAGTCAACGGATTTGGTCGT-3’; GAPDH reverse, 5’- TTGATTTTGGAGGGATCTCG-3’.
Flow cytometry
Single human cell suspensions were stained with a combination of the following antibodies: Alexa Fluor-700-conjugated anti-human Siglec-6 (clone 767329, catalog number FAB2859N-025, R&D Systems, Minneapolis, MN); PE-conjugated anti-human Siglec-7 (clone 194211, catalog number FAB11381P, R&D Systems); BV421-conjugated anti-human Siglec-8 (clone 837535, catalog number 747875, BD Biosciences, Franklin Lakes, NJ); BV421-conjugated anti-human Siglec-9 (clone E10–286, catalog number 743363, BD Biosciences); APC-conjugated anti-human Siglec-9 (clone 191240, catalog number FAB1139A, R&D Systems); BV605-conjugated anti-human c-Kit (clone 104D2, catalog number 562687, BD Biosciences); FITC-conjugated anti-human FcεRIα (clone CRA-1, catalog number 11-5899-42, Invitrogen, Waltham, MA); APC/Cy7-conjugated anti-human lysosomal-associated membrane protein 1 (LAMP-1) (clone H4A3; catalog number 328629, Biolegend); FITC-conjugated anti-human CD45 (clone 2D1, catalog number 11-9459-41, Invitrogen); APC/Cy7-conjugated anti-human CD11b (clone ICRF44, catalog number 301341, Biolegend); BV421-conjugated anti-human CD66b (clone 6/40c, catalog number 392915, Biolegend).
Single mouse cell suspensions were stained with a combination of the following antibodies: anti-APC/Cy7-conjugated anti-mouse c-Kit (clone 2B8, catalog number 105825, Biolegend); PE-conjugated anti-mouse FcεRIα (clone MAR-1, catalog number 134307, Biolegend); BV421-conjugated anti-mouse Siglec-E (clone 750620, catalog number 748154, BD Biosciences). 4,6-Diamidino-2-phenylindole (DAPI) (catalog number D9542, Sigma Aldrich) was used to exclude dead cells.
To determine whether anti-Siglec-9 antibodies can block binding of native Siglec-9 ligands to Siglec-9, LAD2 cells were incubated with 5 μg/ml mouse anti-human Siglec-9 (clone 191240, catalog number MAB1139100, R&D Systems) or mouse IgG2a isotype control antibody (catalog number MAB003, R&D Systems) for 10 min on ice followed by two washes to remove excess antibody. Then, cells were incubated with 50 μg/ml of GlycA (catalog number G5017, Sigma Aldrich) for 30 min on ice. Unbound GlycA was washed off and cells were fixed with 4% paraformaldehyde for 10 min on ice. GlycA bound to mast cells was stained with APC-conjugated anti-human GlycA (clone HIR2, catalog number 306607, Biolegend) and analyzed by flow cytometry.
For Siglec-9 and LAMP-1 staining in PBCMCs and HSCMCs, cells were untreated or stimulated with 100 ng/ml anti-human FcεRIα (clone CRA-1, catalog number 14-5899-82, Invitrogen) for 30 min at 37°C.
In a separate group of experiments, LAD2 cells and HSCMCs were incubated with 10 mU/ml sialidases (catalog number 11080725001, Roche, Indianapolis, IN) for 1 h at 37°C as previously described51 followed by LAMP-1 staining.
Receptor endocytosis was determined by delayed secondary staining as previously described52. Briefly, Siglec-9 was bound with 5 μg/ml unconjugated mouse anti-human Siglec-9 (clone 191240, R&D Systems) or mouse IgG2a isotype control antibody (catalog number MAB003, R&D Systems) for 20 min on ice, two washes to remove unbound antibody, followed by incubation at 37°C for 1, 24, or 48 h. At indicated time points, 1:1000 dilution of an Alexa Fluor-594-conjugated donkey anti-mouse IgG antibody (catalog number A21203, Invitrogen) was added to assess the remaining Siglec-9 on the cell surface by flow cytometry. To assess total Siglec-9 levels, cells were stained with either APC-conjugated anti-human Siglec-9 (clone 191240, catalog number FAB1139A, R&D Systems) or APC-conjugated mouse IgG2a antibody (catalog number IC003A, R&D Systems) for 20 min on ice, unbound antibody was washed off, followed by incubation at 37°C for 1, 24, or 48 h.
The percentage of signal lost was calculated as follows: 100 × (1 – mean intensity fluorescence (MFI) at a specific time/MFI at time 0)
To determine whether native Siglec-9 ligands can cause Siglec-9 to internalize, LAD2 cells were incubated with either 50 μg/mL of GlycA (catalog number G5017, Sigma Aldrich) or 50 μg/ml of HMW-HA (catalog number 53747, Sigma Aldrich) for 30 min on ice. Unbound ligands were washed off and cells were incubated at 37°C for 1 h or 24 h. Cells were fixed with 4% paraformaldehyde for 10 min on ice, stained for Siglec-9 expression using PE-conjugated anti-human Siglec-9 antibody (clone E10–286, catalog number 564251, BD Biosciences), and analyzed by flow cytometry. Cells fixed before the addition of ligands were used as control cells (T= 0).
To determine expression of Siglec-9 ligands, untreated or sialidase-treated mast cells were incubated with 10 μg/ml rhSiglec-9 Fc chimera protein (catalog number 1139-SL-050, R&D Systems) for 30 min on ice followed by incubation with a 1:200 dilution of an Alexa Fluor 488-conjugated anti-human IgG secondary antibody (catalog number 709-546-149, Jackson ImmunoResearch, West Grove, PA) for 30 min on ice. Baseline staining was obtained using PBS followed by the fluorescently labeled secondary antibody.
Mast cell apoptosis was determined in cells cultured with 5 μg/ml mouse anti-human Siglec-9 (clone 191240) or mouse IgG2a isotype control for 20 min on ice followed by washing and incubation at 37°C in warmed media for 1, 24, or 48 h. At indicated time points, cells were collected, stained with propidium iodide (catalog number 421301, Biolegend) and FITC-conjugated Annexin V (catalog number 640905, Biolegend) in Annexin V binding buffer (catalog number 422201, Biolegend), and analyzed by flow cytometry.
Data were acquired on a BD LSR II with the FACSDIVA software and analyzed using FlowJo software (for Windows, version 10, Becton, Dickinson, and Company, Ashland, OR).
Microscopy
For Siglec-9 imaging, cells were plated on chamber slides (catalog number 154526, Thermo Fisher Scientific) coated with 40 μg/ml human plasma fibronectin (catalog number F2006, Sigma Aldrich) for 1 h at 37°C. Cells were fixed with 3% paraformaldehyde for 15 min at room temperature, washed 3 times with PBS, permeabilized with 0.5% Triton X-100 for 10 min, and blocked with 5% normal goat serum for 1 h at room temperature. Cells were then incubated overnight with either mouse anti-human Siglec-9 (clone 191240) or mouse IgG2a isotype control antibody (catalog number MAB003, R&D Systems) at 4°C, washed 3 times with PBS, incubated with 1:200 Alexa Fluor 594-conjugated donkey anti-mouse IgG (catalog number A21203, Invitrogen) for 30 min at room temperature, and washed 3 times with PBS. Samples were mounted using Prolong Diamond DAPI containing mounting media (catalog number P36966, Invitrogen).
For Siglec-9 imaging upon mast cell activation, LAD2 cells were sensitized with 2 μg/ml of human IgE (catalog number ab65866, Abcam) overnight at 37°C. Cells were then washed with PBS and plated on chamber slides (catalog number 154526, Thermo Fisher Scientific) coated with 40 μg/ml human plasma fibronectin (catalog number F2006, Sigma Aldrich) with or without 500 ng/ml anti-human IgE (clone B3102E8, catalog number ab99804, Abcam) at 37°C for 1 h before proceeding with staining as above.
For analysis of endocytosis upon Siglec-9/FcεRI co-crosslinking, cells were fixed, permeabilized, and stained as above, but in suspension rather than adhered to slides and finally resuspended in SlowFade DAPI-containing mounting media (catalog number S36968, Invitrogen) before imaging. Endocytosis analysis was performed with the following additional antibodies: rabbit anti-Rab5a (catalog number 2143S, Cell Signaling Technology, Danvers, MA), rabbit anti-Rab7 (clone D95F2, catalog number 9367S, Cell Signaling Technology), Alexa Fluor 594-conjugated donkey anti-rabbit IgG (catalog number ab150076, Abcam, Cambridge, UK), and Alexa Fluor 488-conjugated donkey anti-mouse IgG (A21202, Invitrogen).
Siglec-9 expression images were acquired on a Leica TCS SP5 confocal inverted-base microscope (Leica Microsystems, IL) with a 63x oil objective and analyzed using ImageJ (NIH).
For Siglec-9 ligand expression images, cells tested for Siglec-9 ligand expression by flow cytometry were attached to a slide by cytospin (500x g, 5 min), fixed using 4% paraformaldehyde for 10 min, washed with PBS, and imaged under oil immersion at 63x using a Leica DM6000 (Leica, Wetzlar, Germany) microscope.
Siglec-9 engagement with native ligands
Mock- and SIGLEC9-edited LAD2 cells sensitized overnight with 2 μg/ml of human IgE (catalog number ab65866, Abcam) and HSCMCs were cultured with either 20–100 μg/ml GlycA (catalog number G5017, Sigma Aldrich) or 20–100 μg/mL HMW-HA (catalog number 53747, Sigma Aldrich) for 20 min. The cells were then activated with the indicated concentrations of stimulants for 30 min at 37°C. LAD2 cell and HSCMC activation was assessed by β-hexosaminidase release and increase in LAMP-1 expression, respectively. Percentage of inhibition of mast cell degranulation was calculated as follows: (% of β-hexosaminidase release or LAMP-1 expression in stimulated cells- β-hexosaminidase release or LAMP-1 expression in untreated cells) - (% of β-hexosaminidase release or LAMP-1 expression in stimulated cells treated with Siglec-9 ligand - β-hexosaminidase release or LAMP-1 expression in untreated cells) / (% of β-hexosaminidase release or LAMP-1 expression in stimulated cells - β-hexosaminidase release or LAMP-1 expression in untreated cells).
Siglec-9 engagement with anti-Siglec-9 antibody
Mast cells were pre-incubated with 2.5–5 μg/ml mouse anti-Siglec-9 (clone 191240) or mouse IgG2a isotype control antibody for 30 min at room temperature prior to activation with the indicated concentrations of stimulants for 30 min at 37°C.
In a separate set of experiments, PBCMCs incubated with anti-Siglec-9 or isotype control were exposed to 5 μg/ml goat anti-mouse IgG (Fc specific) F(ab’)2 fragment antibody (catalog number M0284, Sigma-Aldrich) for 2 min to cross-link Siglec-9 prior to activation.
LAD2 cell activation was assessed by β-hexosaminidase release, and PBCMC and HSCMC activation was assessed by LAMP-1 staining.
Co-crosslinking of FcεRI and Siglec-9
PBCMCs and HSCMCs were incubated with 100 ng/ml anti-human FcεRIα plus 5 μg/ml mouse isotype control antibody (catalog number MAB003, R&D Systems) or anti-Siglec-9 mAb (clone 191240) for 2 min at 4°C. Cells were washed and then incubated with 5 μg/ml goat anti-mouse IgG (Fc specific) F(ab’)2 fragment antibody for 2 min. After an additional PBS wash, cells were resuspended in warm complete medium and incubated for 30 min at 37°C to assess mast cell activation by LAMP-1 staining. Cell supernatants were collected at 20 min and after overnight stimulation to measure the levels of arachidonic acid metabolites and chemokine/cytokines, respectively.
β-hexosaminidase release assay
LAD2 cells were sensitized with 2 μg/ml of human IgE by overnight incubation at 37°C. The cells were then washed with Tyrode’s buffer (10 mM HEPES, pH 7.4, 130 mM NaCl, 5 mM KCl, 1.4 mM CaCl2, 1 mM MgCl2, and 0.1% glucose) and 1 × 105 cells/well were added to a 96-well V-bottom plate. Next, equal volume of a 2x concentration of stimulus (500 ng/ml anti-human IgE (clone B3102E8, catalog number ab99804, Abcam), 10 μM compound 48/80 (c48/80) (catalog number C2313, Sigma Aldrich) or 1 μM A23187 (catalog number C7522, Sigma Aldrich) was added to the appropriate wells, and incubated at 37°C for 1 h. After centrifugation, supernatants were collected, and pellets were lysed with 0.5% Triton X 100. β-hexosaminidase release was quantified by enzyme immunoassay with p-nitrophenyl-N-acetyl-β-d-glucosamine (catalog number N9376, Sigma-Aldrich) substrate, as follows: 10 μl of culture supernatant or lysate was added to the wells of a 96-well flat-bottom plate; 50 μl of 1.3 mg/ml p-nitrophenyl-N-acetyl-β-d-glucosamine solution in 100 mM sodium citrate, pH 4.5, was added, and the plate was incubated at 37°C for 1 h. Next, 150 μl of 200 mM glycine, pH 10.7, was added to stop the reaction, and the optical density (OD405) was determined.
Prostaglandin (PG)D2 and cysteinyl leukotriene (cys-LT) release assays
PGD2 and cys-LT levels were measured in supernatants by ELISA per manufacturer’s instructions (catalog numbers 512031 and 500390 for PGD2 and cys-LT, respectively, Cayman Chemical, Ann Arbor, MI). The assay detection limits were 19.5 pg/ml and 8.6 pg/ml for PGD2 and cys-LT, respectively.
Chemokine release assay
IL-8 and monocyte chemoattractant protein (MCP)-1 levels were measured in cell supernatants using the human ProcartaPlex Multiplex ImmunoAssay (catalog number PPX-07-MXH6CA4, Thermo Fisher Scientific). The assay detection limits were 2.49 pg/ml for IL-8 and 4.16 pg/ml for MCP-1.
Statistical Analyses
Data are presented as means ± SEMs unless otherwise indicated. Statistical significance was determined by Mann Whitney U test, Wilcoxon matched pair signed rank test, or ANOVA and Tukey multiple comparisons test as appropriate, using GraphPad Prism version 9 (San Diego, CA, USA). When appropriate, single outliers were identified and removed from the data using the ROUT outlier test. Statistical differences were considered significant at P <0.05.
RESULTS
Human mast cells express Siglec-9
First, we compared the surface level expression of Siglec-9 across a variety of human mast cell types. We found that human mast cell lines (Fig. E1A), PBCMCs, HSCMCs, and lung mast cells express Siglec-9 (Fig. 1A). Human neutrophils isolated from peripheral blood were used as a positive control for Siglec-9 protein expression (Fig. 1A). When compared to other Siglecs, human primary mast cells expressed Siglec-9 levels comparable to Siglec-7 and Siglec-8 (Fig. 1B). Moreover, Siglec-9 was the only Siglec expressed by all the human mast cell lines tested (Fig. E1B–C). At the mRNA level, human mast cell lines, PBCMCs, and HSCMCs expressed SIGLEC9 mRNA levels comparable to Siglec-7 and Siglec-8 (Fig. E2A–C).
Figure 1. Siglec-9 expression in human primary mast cells.
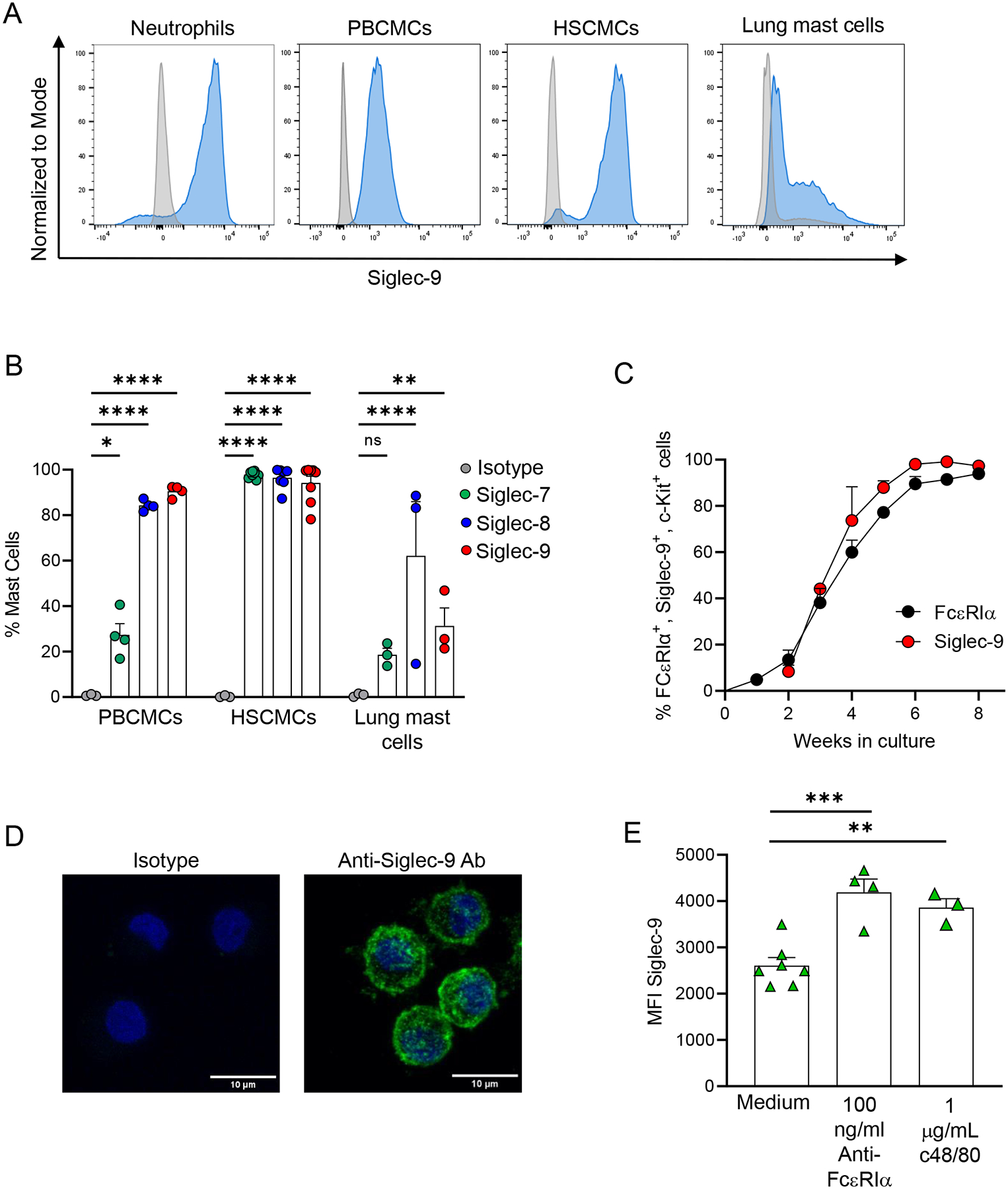
(A-B) Representative flow cytometry analysis of Siglec-9 surface expression in human neutrophils, human peripheral blood mononuclear cell-derived mast cells (PBCMCs), human skin cultured mast cells (HSCMCs), and human lung mast cells (A) and percentage of PBCMCs, HSCMCs and human lung mast cells expressing Siglec-7, Siglec-8, and Siglec-9 (B). (C) Kinetics of Siglec-9 surface expression on CD34+ -derived PBCMCs (n = 8). (D) Representative confocal microscopy images show intracellular and cell surface staining for Siglec-9 in HSCMCs (green fluorescence in right panel). Nuclei were counterstained with DAPI (blue fluorescence). Negative control was performed with isotype control and secondary antibodies (left panel). Scale bar equals 10μm. (E) Mean fluorescence intensity (MFI) of Siglec-9 in HSCMCs stimulated with either an anti-FcεRIα antibody (100 ng/ml), compound 48/80 (c48/80) (1 μg/ml) or maintained in medium alone for 20 min. Flow cytometry data in A and confocal images in D are representative of 2–6 experiments. Data in B, C and E are shown as mean + SEM with circles in B (n = 3–8), triangles in E (n = 3–6) showing values from individual experiments with cells generated from individual donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
As previously reported for Siglec-6 and Siglec-815, our kinetics studies showed that Siglec-9 appeared on the cell surface in parallel with FcεRIα expression (Fig. 1C), indicating that Siglec-9 expression increases with mast cell maturation.
Confocal microscopy for Siglec-9 localization in HSCMCs (Fig. 1D) and LAD2 cells (Fig. E3) showed a punctate distribution of Siglec-9, which has previously been described in other cell types25, 33, 53. Given the punctate intracellular staining of Siglec-9 in HSCMCs, we hypothesized that there is a potential intracellular pool of Siglec-9. To test this, we stimulated HSCMCs in an IgE-dependent and IgE-independent manner and assessed surface expression of Siglec-9. HSCMCs (Fig. 1E) showed a significant increase in Siglec-9 expression after undergoing degranulation. Interestingly, confocal microscopy showed that there was not a significant reduction in punctuate staining for Siglec-9 in LAD2 cells upon IgE-dependent activation (Fig. E3B). These data suggest that a possible translocation of Siglec-9 to the cell surface may not be the only mechanism by which Siglec-9 surface expression increases upon mast cell activation. Together, these data demonstrate that human mast cells express Siglec-9 on their cell surface and intracellularly.
Siglec-9 is an endocytic receptor
Siglec-6 and Siglec-8 are internalized after antibody engagement of the receptor on eosinophils and mast cells14, 16. The endocytic capacity of Siglec-9 has only been described in cancer cells54; where up to 90% of Siglec-9 was internalized after treatment with anti-Siglec-9 antibodies. Therefore, we first tested whether Siglec-9 ligation causes a decrease in Siglec-9 surface expression using the previously described delayed secondary staining method14. After ligation of Siglec-9 with an anti-Siglec-9 monoclonal antibody (clone 191240), there was a gradual decrease in surface Siglec-9 staining on HSCMCs (Figs. 2A and 2B). As shown in Fig. 2C, approximately 30% of the Siglec-9 signal was lost at 24 h after antibody engagement when compared with Siglec-9 signal detected at time 0. Siglec-9 surface expression decreased more rapidly in LAD2 cells (Fig. E4A–B), which exhibited approximately a 30% loss in Siglec-9 surface expression at 1 h after Siglec-9 ligation (Fig. E4C). The MFI slightly increased and the percentage in surface Siglec-9 loss decreased at 24 h after Siglec-9 ligation, suggesting that a small portion of the original Siglec-9 started returning to the LAD2 cell surface (Figs. E4B and E4C). Together, this evidence shows that Siglec-9 surface expression decreases after receptor engagement.
Figure 2. Siglec-9 is internalized following antibody ligation.
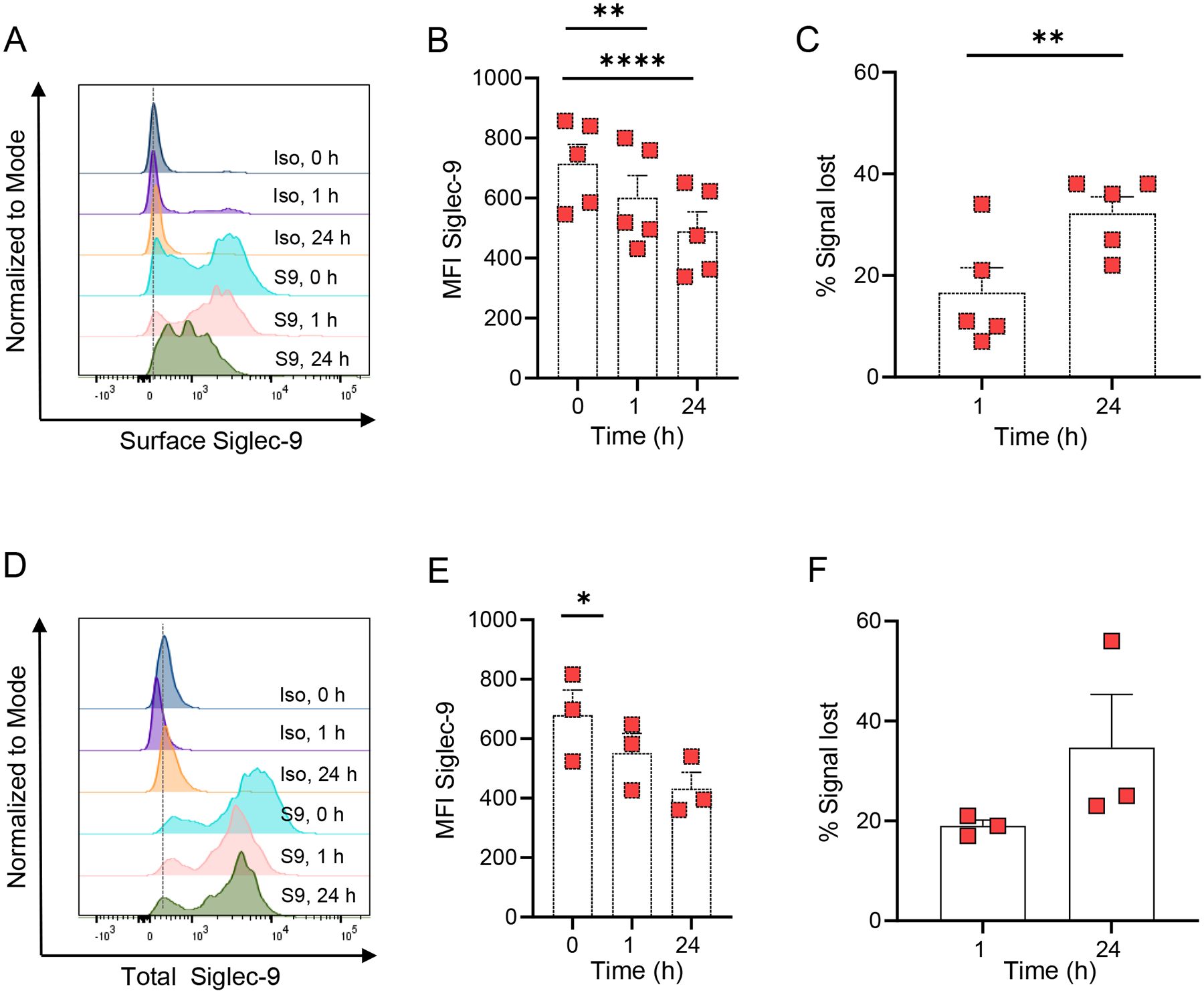
(A and D) Representative flow cytometry analysis of surface (A) and total (D) Siglec-9 expression in HSCMCs treated with 5 μg/ml of either isotype control (Iso) or anti-Siglec-9 antibody (S9) for the indicated time points. Mean fluorescence intensity (B and E) and percentage of signal lost (C and F) at indicated points for surface (B and C) and total (E and F) Siglec-9 expression in HSCMCs treated with 5 μg/ml of either APC-conjugated isotype control or APC-conjugated anti-Siglec-9 antibody (Anti-Sig9) for the indicated time points. The percentage of signal lost was calculated as MFI for Siglec-9 at indicated point minus MFI for Siglec-9 at time 0. Flow cytometry data in A and D are representative of 3–5 independent experiments. Data in B (n = 5), C (n = 5), E (n = 3) and F (n = 3) are shown as mean + SEM with squares showing values from individual experiments with cells generated from individual donors. *P < 0.05, **P < 0.01, ****P < 0.0001.
To further characterize whether the Siglec-9 loss was due to endocytosis, we assessed the expression of surface and internalized Siglec-9 (Total Siglec-9) by using a fluorophore-conjugated anti-Siglec-9 antibody. HSCMCs exhibited a significant loss in total Siglec-9 signal at 1 h after antibody engagement (Fig. 2D). Loss in total Siglec-9 signal was more pronounced in LAD2 cells which exhibited an almost complete loss in total Siglec-9 signal at 24 h after Siglec-9 ligation (Figs. E4D–F). In contrast, LAD2 cells incubated with isotype control did not exhibit any loss in total Siglec-9 expression during the times tested (Fig. E4G). Based on these data, we hypothesized that Siglec-9 was degraded after endocytosis. To address this hypothesis, we examined whether Siglec-9 co-localizes with markers of the endosomal pathway in LAD2 cells after Siglec-9 engagement. Confocal images showed that Siglec-9 co-localized with Rab5, a marker of early endosomes, and with Rab7, a marker of late endosomes, at 1 h and 24 h after Siglec-9 engagement, respectively (Fig. E5). In contrast, Siglec-9 did not co-localize with Rab5 or Rab7 in LAD2 cells treated with isotype control at any timepoint tested (Fig. E5).
Together, these data suggest that Siglec-9 is internalized and degraded upon engagement with anti-Siglec-9 antibodies.
Siglec-9 interactions with sialic acids in cis dampen mast cell susceptibility to activation
Next, we decided to generate a human mast cell line deficient in Siglec-9 to assess the role of Siglec-9 in mast cell function. For this purpose, we deleted Siglec-9 in LAD2 cells using a CRISPR/Cas9 approach. By using a CRISPR/Cas9 approach we were able to generate a large population of LAD2 cells that does not express Siglec-9 (Fig. 3A). In contrast, Siglec-6, Siglec-7, and Siglec-8 expression levels were unaffected by Siglec-9 deletion in LAD2 cells (Fig. 3B). First, we observed that naïve Siglec-9-deleted LAD2 cells exhibited increased cell surface expression levels of LAMP-1 which indicates granule mobilization towards the plasma membrane (Fig. 3C). Moreover, Siglec-9-deleted LAD2 cells were more susceptible to degranulation induced by IgE-dependent (Fig. 3D) and IgE-independent stimuli (Figs. 3E and 3F). These observations support the presence of sialic acid ligands with the ability to trigger an inhibitory signal in mast cells via Siglec-9 engagement. To address this hypothesis, we directly probed mast cells for the presence of Siglec-9 ligands using a recombinant chimera of Siglec-9 (Fc-Siglec-9) as previously described55. HSCMCs showed expression of Siglec-9 ligands by flow cytometry (Figs. 3G and 3H). Imaging of the Siglec-9 ligand expression in HSCMCs showed a punctate pattern of expression, suggesting there is an enrichment of these ligands in certain areas of the cell membrane (Fig. 3I). As shown in Figs. E6A and E6B, LAD2 cells also express significant levels of Siglec-9 ligands when compared with cells treated with secondary antibody alone. The staining for Siglec-9 ligands in LAD2 cells was significantly reduced when cells were treated with sialidases to remove potential Siglec ligands, demonstrating that Fc-Siglec-9 chimeras specifically bind to sialic acid residues on the cell membrane (Figs. E6A and E6B). Notably, Siglec-9-deleted LAD2 cells showed significantly higher staining for Siglec-9 ligands (Figs. E6A and E6B) suggesting that binding of Siglec-9 to sialic acids may interfere with Siglec-9 ligand detection by Fc-Siglec-9 chimeras in unedited LAD2 cells.
Figure 3. Siglec-9 interactions with sialic acids in cis limit mast cell activation.
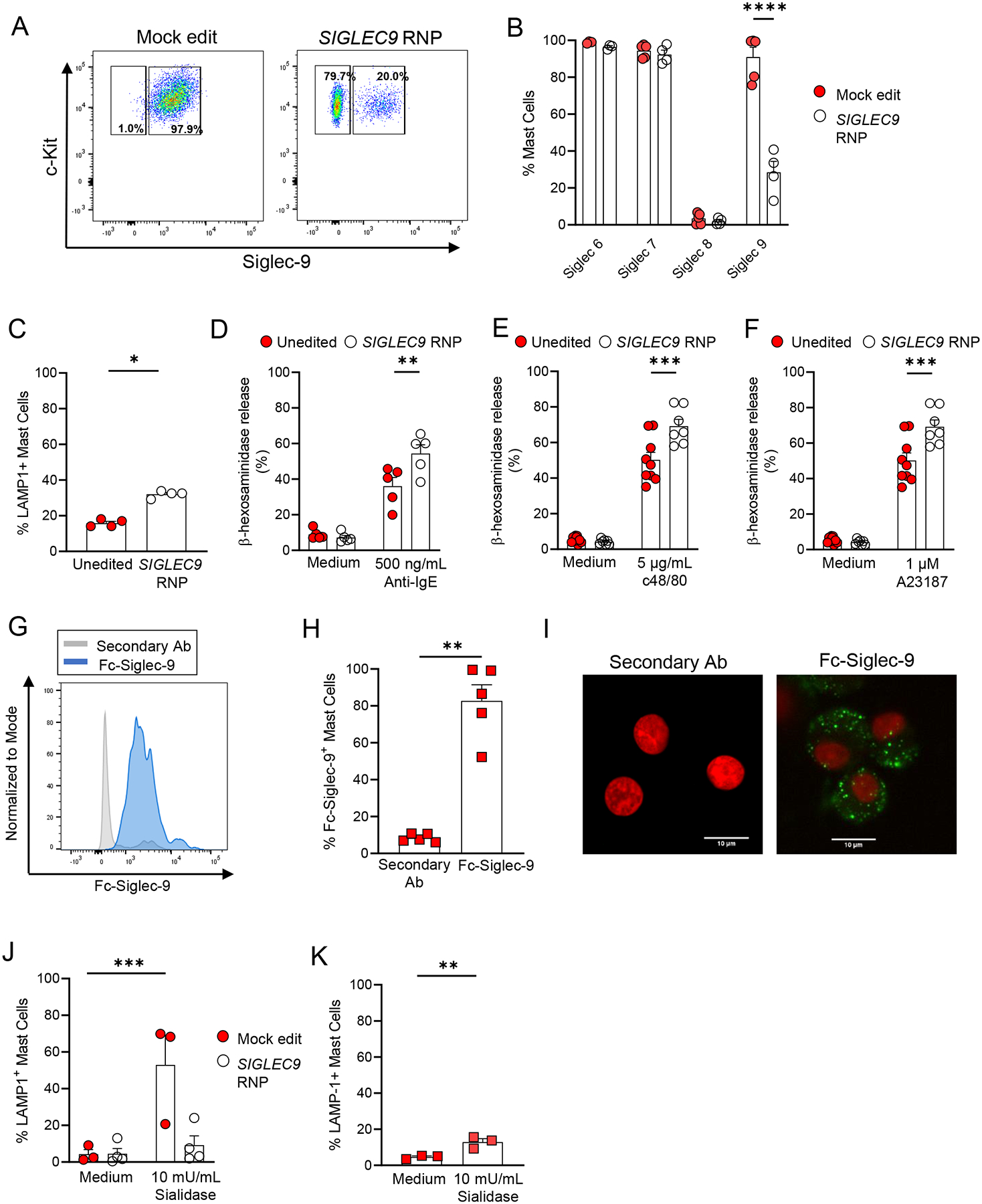
Representative flow cytometry analysis of Siglec-9 expression of mock- and SIGLEC9-edited LAD2 cells. (B) Siglec surface expression in mock- and SIGLEC9-edited deficient LAD2 cells. (C) LAMP-1 surface expression in unedited and SIGLEC9-edited deficient LAD2 cells maintained in medium alone. (D-F) β-hexosaminidase release by unedited and SIGLEC9-edited LAD2 cells upon activation. Unedited and SIGLEC9-edited cells were sensitized with IgE (2 μg/ml) overnight and then were challenged with either anti-human IgE (500 ng/ml) (D), compound 48/80 (c48/80) (5 μg/ml) (E), or calcium ionophore (A23187) (1 μm) (F) for 1 h. (G-I) Representative flow cytometry analysis of Siglec-9 ligand expression (G), percentage (H) of HSCMCs expressing Siglec-9 ligands. (I) Representative fluorescent microscopy images show cell surface binding of Fc chimera Siglec-9 protein in HSCMCs (green in right panel). Nuclei were counterstained with DAPI (red fluorescence). Negative control was performed with secondary antibodies only (left panel). Scale bar equals 10μm. (J-K) LAMP-1 expression in unedited and SIGLEC9-edited LAD2 cells (J), and HSCMCs maintained in medium alone or treated with sialidases (10 mU/ml) for 1 h. Flow cytometry data in A and G, and microscopy images in I are representative of 2–3 independent experiments. Data in B (n = 4), C (n = 4), D (n = 5), E (n = 7), F (n = 5–8), H (n = 5), J (n = 3–4) and K (n = 3) are shown as mean + SEM. Circles in B-F and J show values from individual experiments with LAD2 cells. Squares in H and K show values from individual experiments with cells generated from individual donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P <0.0001.
These findings prompted us to explore whether mast cells express enzymes involved in sialic acid biosynthesis. Sialic acid biosynthesis starts with the formation of cytidine monophosphate N-acetylneuraminic acid (CMP-Neu5Ac)56 as an activated sugar donor for the transfer of sialic acids by sialyltransferases (SiaT) to the terminal glycosyl group of glycoproteins and glycolipids as acceptor molecules. As shown in Fig. E7A, we found that human mast cells express mRNA for UDP-GlcNAc 2-epimerase/ManNAc-6 (GNE), the first enzyme in the pathway to generating CMP-Neu5Ac57. Moreover, human mast cells express mRNA for SiaTs that can catalyze the formation of glycosidic linkages found in Siglec-9 ligands including ST3GAL1, ST3GAL3, ST3GAL4, ST3GAL5 and ST3GAL6 (Figs. E7B–F). Siglec-9-deleted LAD2 cells did not exhibit alterations in the expression of mRNA for GNE and SiaTs indicating that Siglec-9 expression does not regulate the expression of enzymes involved in sialic acid biosynthesis (Fig. E7G). Overall, this evidence supports the expression and biosynthesis of Siglec-9 ligands in human mast cells.
To address whether the presence of these sialic acid ligands can trigger an inhibitory signal in mast cells via Siglec-9 engagement, we examined LAMP-1 expression in mast cells devoid of potential Siglec ligands. As shown in Fig. 3J, unedited but not Siglec-9-deleted LAD2 cells exhibited increased LAMP-1 expression after sialidase treatment. Increased LAMP-1 expression was also observed when HSCMCs were treated with sialidases, indicating that sialic acid ligands also limit the extent of human primary mast cell activation (Fig. 3K). Together, these data show that mast cells express Siglec-9 ligands in vitro that can engage Siglec-9 to promote mast cell quiescence.
Siglec-9 ligands reduce mast cell activation
GlycA and HMW-HA have been recognized as ligands for Siglec-9 that can downregulate the activation of immune cells27, 58. Therefore, we assessed the effects of GlycA and HMW-HA on mast cell activation. As shown in Fig. 4A and Figs. E8A–D, optimal concentrations of both ligands inhibited β-hexosaminidase release from LAD2 cells stimulated by IgE-dependent and IgE-independent stimuli. Both GlycA and HMW-HA were also able to inhibit PBCMC degranulation as assessed by a decrease LAMP-1 expression levels upon stimulation by IgE-dependent (Fig. 4B and Figs. E8E and E8F). In contrast, Siglec-9 ligands did not significantly inhibit PBCMC degranulation induced by IgE-independent stimuli (Fig. 4B and Figs. E8G–H).
Figure 4. Siglec-9 ligands inhibit mast cell degranulation.
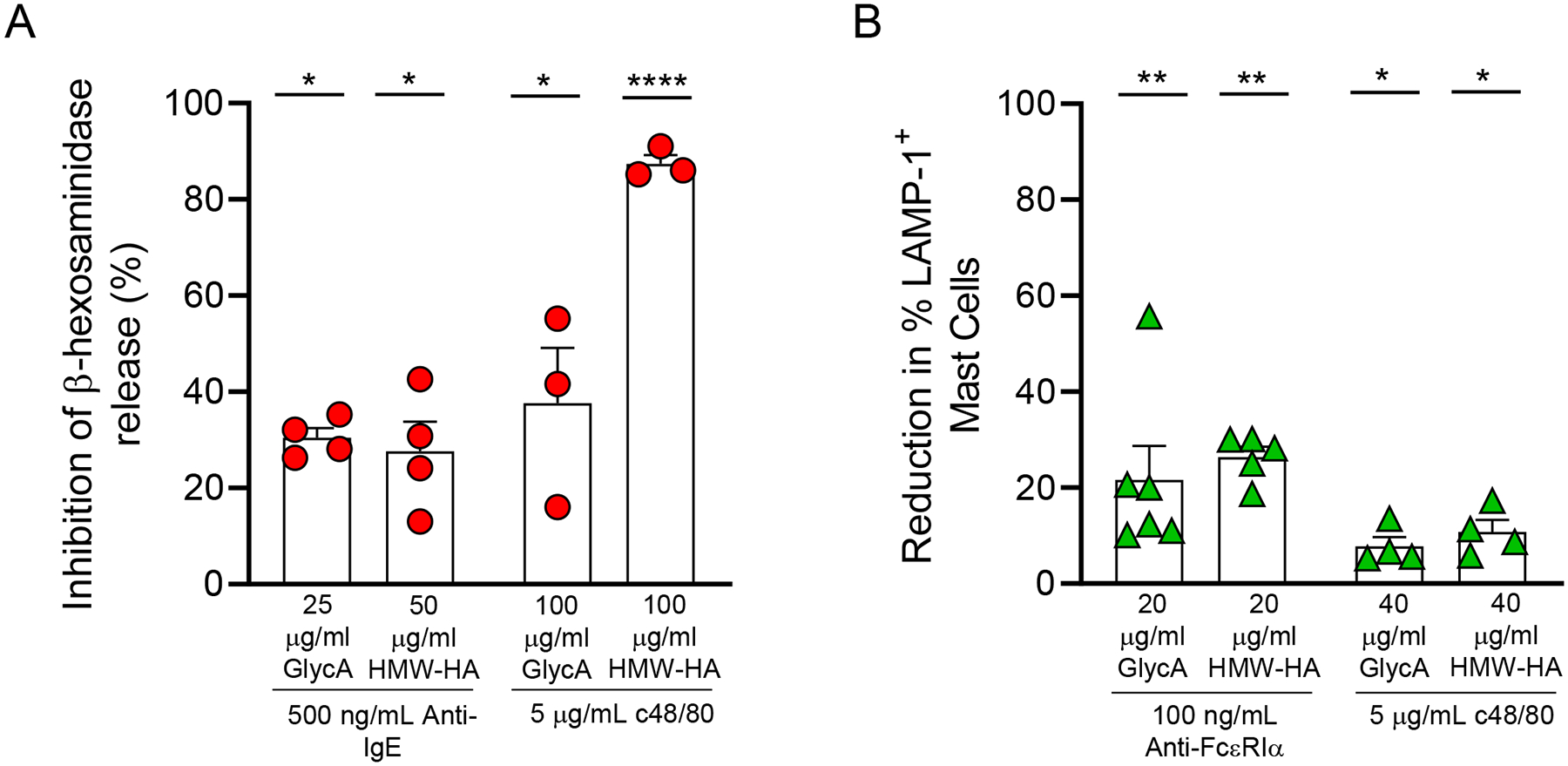
(A-B) Inhibition of β-hexosaminidase release in LAD2 cells (A) and reduction in LAMP-1 expression in PBCMCs (B) treated with either glycophorin A (GlycA) (25–100 μg/ml) or high molecular weight hyaluronic acid (HMW-HA) (20–100 μg/ml) for 20 min before stimulation. For LAD2 cell IgE-dependent activation, cells were sensitized with IgE (2 μg/ml) overnight and then were challenged with anti-human IgE (500 ng/ml) for 1 h. For PBCMC IgE-dependent activation, cells were treated with anti-FcεRIα antibodies (100 ng/ml) for 20 min. For IgE-independent mast cell activation, LAD2 cells and PBCMCs were stimulated with compound 48/80 (c48/80) (5 μg/ml) for 1 h and 20 min, respectively. Data are shown as mean + SEM. Circles in A (n = 3–4) show values from individual experiments with LAD2 cells. Triangles in B (n = 4–6) show values from individual experiments with cells generated from individual donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P <0.0001 vs. cells treated with stimuli alone.
It has been shown that anti- human Siglec-9 antibodies can block the interactions between HMW-HA and Siglec-927. Similarly, Fig. E9A shows a significant reduction in GlycA binding to LAD2 cells pre-treated with anti-human Siglec-9 antibodies. Importantly, Siglec-9 deleted LAD2 cells did not exhibit a reduction in their ability to release β-hexosaminidase when incubated with Siglec-9 ligands (Fig. E9B). This evidence suggests that GlycA and HMW-HA inhibit mast cell activation by specifically engaging Siglec-9.
To assess whether Glyc A and HMW-HA cause Siglec-9 internalization, we assessed Siglec-9 expression in LAD2 cells at 1 h and 24 h after treatment with native ligands. As shown in Fig. E10, LAD2 cells treated with either GlycA or HMW-HA exhibited similar Siglec-9 expression to cells cultured in medium alone, suggesting that GlycA and HMW-HA were unable to induce Siglec-9 internalization.
These results point to naturally occurring Siglec-9 ligands like GlycA and HMW-HA being able to further dampen mast cell activation through their engagement of Siglec-9.
Antibody engagement of Siglec-9 inhibits mast cell activation
Antibodies against Siglecs have been used successfully to engage these inhibitory receptors and modulate immune and inflammatory responses20–22, 59, 60. To determine whether antibody engagement of Siglec-9 reduces mast cell activation, we incubated LAD2 cells with an anti-Siglec-9 monoclonal antibody (clone 191240) for 30 min prior to mast cell stimulation. Siglec-9 antibody engagement slightly inhibited mast cell degranulation induced by IgE-dependent (Fig. E11A) and IgE-independent stimuli (Fig. E11B) in LAD2 cells. In contrast, PBCMCs and HSCMC did not exhibit a reduction in their ability to degranulate when treated with anti-Siglec-9 antibodies (Figs. E11C and E11D). PBCMC degranulation was also not inhibited when anti-Siglec-9 antibodies were crosslinked with a secondary antibody prior to activation to enhance the inhibitory signal (Fig. E11E). As Siglec-9 engagement can induce apoptosis in neutrophils24, we also examined PBCMC survival after Siglec-9 engagement with anti-Siglec-9 antibodies. As shown in Fig. E12, there was not significant decrease in PBCMC viability at any timepoint tested in cells treated with anti-Siglec-9 antibodies or isotype control conditions.
Due to the lack of anti-Siglec-9 inhibitory effect on human primary mast cell activation, we assessed whether co-engaging Siglec-9 with FcεRI could enhance Siglec-9 inhibitory function as shown for Siglec -3, -6, -7 and -8 on mast cells16–18, 61. For this purpose, we co-aggregated Siglec-9 and FcεRI on PBCMCs and HSCMC using a secondary cross-linking antibody that recognizes both anti-Siglec-9 and anti-FcεRIα antibodies. We observed a significant reduction in PBCMC (Fig. 5A) and HSCMC (Fig. 5B) activation after Siglec-9 co-engagement that was even more pronounced in HSCMCs than in PBCMCs (Fig. 5C). Cross-linking of Siglec-9 with FcεRIα also resulted in reduced production of the arachidonic acid metabolites cys-leukotrienes (cys-LT) (Fig. 5D and Figs. E13A and E13D) and prostaglandin (PG)D2 (Fig. 5E and Figs. E13B and E13E) in both PBCMC and HSCMC. Finally, we tested the effect of Siglec-9-FcεRI co-engagement on mast cells’ ability to generate chemokines upon IgE-dependent activation. As shown in Fig. 5F and Figs. E13C and E13F, both PBCMC and HSCMC released significant reduced amounts of IL-8 when Siglec-9 was cross-engaged with FcεRIα. Moreover, HSCMCs but not PBCMCs were inhibited in their ability to release MCP-1 upon Siglec-9 and FcεRI co-engagement (46.5% ± 12.5% inhibition, P < 0.005 vs. cells treated with isotype control, n = 6 experiments with cells obtained from individual donors). Overall, these data suggest that Siglec-9 proximity to FcεRI increases the effectiveness of the Siglec-9 mediated inhibitory signal.
Figure 5. Co-engagement of FcεRI and Siglec-9 inhibits mast cell degranulation, production of arachidonic acid metabolites, and IL-8 release.
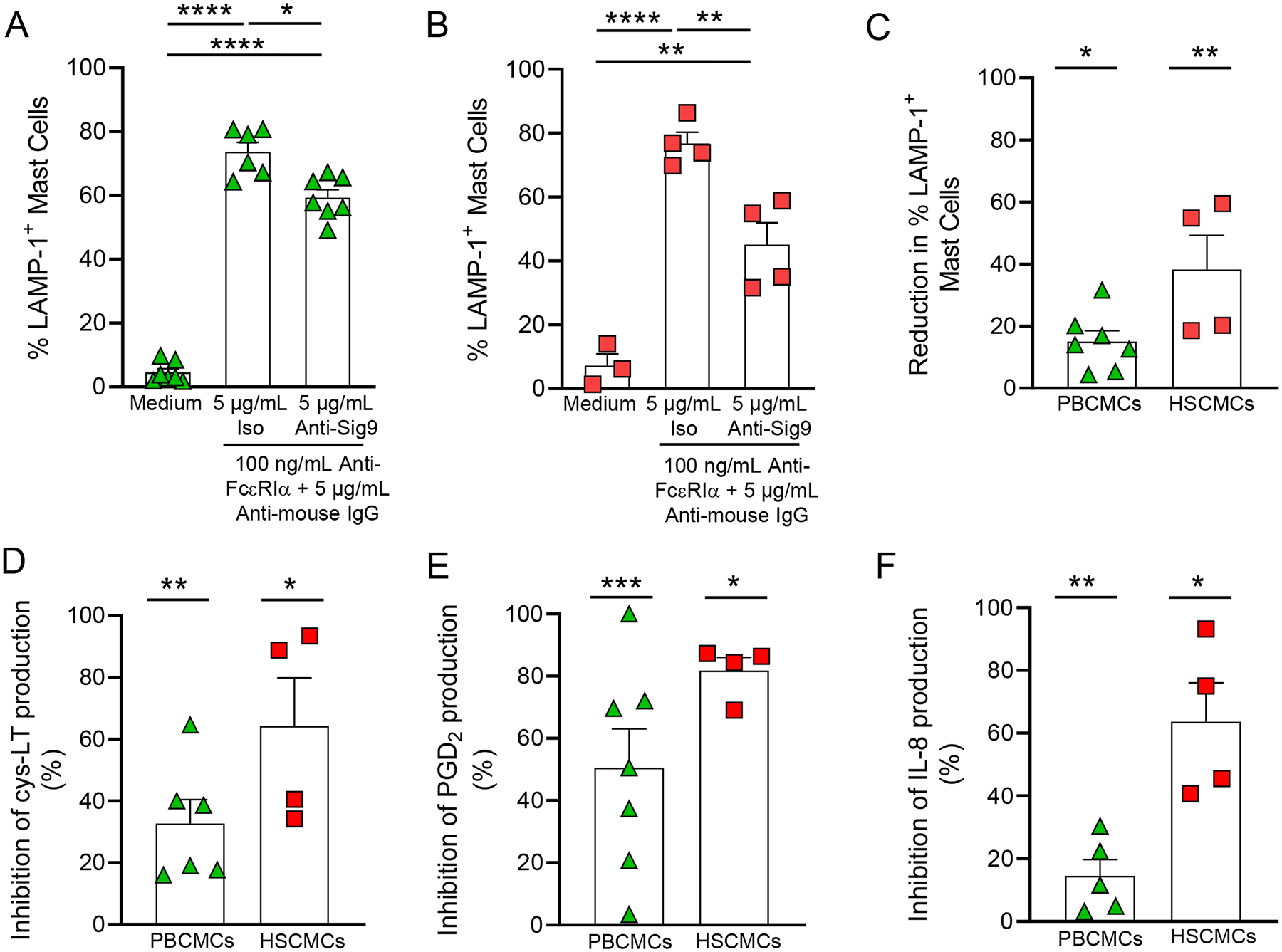
(A-F) LAMP-1 expression in PBCMCs (A) and HSCMCs (B), and reduction in LAMP-1 expression (C), cys-LT (D), PGD2 (E) and IL-8 production in PBCMCs and HSCMCs maintained in medium alone or incubated with either isotype control or mouse anti-Siglec-9 (5 μg/ml) and stimulated with anti-human FcεRIα (100 ng/ml) and a goat anti-mouse IgG (Fc specific) F(ab’)2 fragment antibody (5 μg/ml) to cross-link FcεRIα and Siglec-9. Data are shown as mean + SEM. Triangles in A (n = 6–7), C (n = 7), D (n = 4), E (n = 7) and F (n = 5), and squares in B (n = 3–4) C (n = 4), D (n = 4), E (n = 4) and F (n = 4), show values from individual experiments with cells generated from individual donors. In A and B: *P < 0.05, **P < 0.01, ****P <0.0001. In C-F: *P < 0.05, **P < 0.01, ***P <0.001 vs. cells treated with isotype control.
DISCUSSION
In this study, we report for the first time that human mast cells express functional Siglec-9 in vitro. Specifically, we show that Siglec-9 can inhibit human mast cell degranulation upon engagement with Siglec-9 ligands, GlycA and HA-HMW. Moreover, we observed that co-engagement of Siglec-9 with FcεRI can reduce human mast cell degranulation and limit their ability to produce arachidonic acid metabolites, MCP-1, and IL-8.
Prior studies reported that freshly isolated human skin18 and lung41 mast cells express surface Siglec-9. In the current study, we confirmed that Siglec-9 is expressed in freshly isolated human lung mast cells, HSCMCs, and PBCMCs (Fig. 1A). The levels of Siglec-9 mRNA and protein expression were comparable to those found for Siglec-7 and Siglec-8 in the human mast cells tested (Fig. E2). Previous studies showed that cord blood cultured mast cells and PBCMC do not express mRNA for Siglec-915. We cannot fully explain the discrepancy between these data and the findings from our groups and others. However, we observed that the kinetics of expression of Siglec-9 parallels the expression of FcεRI in CD34+ cells as they differentiate into mast cells in vitro (Fig. 1C). This observation suggests that variations in the culture conditions for CD34+-derived cultured mast cells may affect the levels of Siglec-9 expression levels detected by different labs. The correlation between Siglec-9 expression and cell maturation is not unique to mast cells as it has been noted previously in other immune cells like NK cells31. Importantly, Siglec-6 and Siglec-8 expression also matches an increase in CD51 and FcεRIα expression and histamine content in PBCMCs15 suggesting that mast cells upregulate the expression of a set of inhibitory receptors capable to engage an array of potential Siglec ligands during maturation to maintain mast cell quiescence. We addressed this hypothesis in part by showing that Siglec-9 deletion by a CRISPR-editing approach led to an increase in LAMP-1 expression in LAD2 cells at steady state (Fig. 3C). Moreover, Siglec-9-deleted LAD2 cells were more susceptible to IgE-dependent and IgE-independent stimuli (Figs. 3D–F). To our knowledge, there are no reports on how Siglec-9 deficiency can alter the function of human immune cells. However, studies using mice deficient in Siglec-E, the murine homologue of Siglec-9, suggest that Siglec-E-deficiency can impact the activation status of immune cells as observed for Siglec-9-deficient mast cells. For example, it has been shown that microglia from Siglec-E-deficient mice displays aggravated pro-inflammatory characteristics when exposed to neural debris damage-associated molecular patterns (DAMPs)62 or lipopolysaccharide (LPS)63. Similarly, Siglec-E-deficient neutrophils exhibited increased ability to migrate to the lung in an acute lung airway inflammation model induced by aerosolized LPS42. We could not examine whether Siglec-E-deficiency can impact mast cell function as we found that bone marrow-derived cultured mast cells (BMCMCs), peritoneal mast cells, and fetal skin-derived cultured mast cells showed very low surface expression of Siglec-E (Fig. E14A). These observations are in agreement with the RNAseq database from The Immunological Genome Project (ImmGen, http://rstats.immgen.org/Skyline/skyline.html) showing that mast cells from the peritoneum, esophagus, trachea, tongue, or skin express very low gene expression levels of Siglec-E (Fig. E14B) when compared with blood neutrophils.
Siglec ligands can act in trans, on adjacent cells, and in cis, where they cluster Siglecs on the same cell’s membrane and maintain a basal level of inhibitory signaling that increases the threshold for immune cell activation. Depletion of Siglec ligands in cis by sialidases or oxidative cleavage has been linked to increased activity of B cells64, macrophages65, microglia66, and monocyte-derived dendritic cells67, and prolonged inhibition of sialic acid biosynthesis renders phagocytes more prone to activation68. The increased expression of activation markers in Siglec-9-deleted LAD2 cells points to a possible role for Siglec-9 interactions with sialic acid in cis in mast cell homeostasis. Previously, the ligands for Siglec-9 had only been found on endothelial cells69, red blood cells58, the upper airway70, and the human aorta71. In this report, we demonstrate for the first time that human mast cells express ligands for Siglec-9 (Figs. 3G & E6). We also report that human mast cells highly express mRNA for enzymes involved in sialic acid biosynthesis and sialylation (Fig. E7). Importantly, sialic acid removal in LAD2 cells, and to a lesser degree in HSCMCs, led to an increase in the expression of mast cell activation markers as we observed in Siglec-9-deleted mast cells (Fig. 3J–K) supporting our hypothesis that Siglec-9 interactions with sialic acids in cis contribute to mast cell quiescence.
Despite the progress made to identify Siglec ligands, their carrier proteins and their tissue expression patterns, the identities and physiological roles of endogenous Siglec ligands relevant to mast cell biology are unknown. Using synthetic glycan arrays, it has been shown that Siglec-9 bind preferentially to sulfated sialylated trisaccharide structure (NeuAc α2,3 Gal β1,4 6-SO3-GlcNAc). The search for endogenous ligands with the sulfated sialylated trisaccharide structures required for binding to Siglec-9 has been most successful for this receptor. In human upper airway tissues, Siglec-9 can bind to the glycans of the mucin MUC5B70. Siglec-9 can also bind the erythrocyte sialoglycoprotein, GlycA in a sialic acid dependent manner58. Importantly, Siglec-9-GlycA interactions have been shown to have immunosuppressive effects on neutrophils including decreased degranulation, reactive oxygen species (ROS) and neutrophil extracellular trap (NET) production, chemotaxis, and bacterial killing58. Siglec-9 mediated inhibition of neutrophils was also observed after treatment with the glycosaminoglycan, HMW-HA27. In contrast to GlycA, HMW-HA binding to Siglec-9 has been shown to be sialic acid-independent27. Based on this evidence, we investigated GlycA and HMW-HA effects on mast cell function. We observed that mast cell treatment with either GlycA or HMW-HA, was able to decrease mast cell activation by IgE-dependent or IgE-independent stimuli (Fig. 4A). Although mast cells express CD44, the primary receptor of HMW-HA72, the inhibitory effect of HMW-HA was specific to Siglec-9 in our system since LAD2 cells deficient in Siglec-9 showed no decrease in their ability to degranulate when activated in the presence of HMW-HA. Overall, these in vitro data demonstrate that Siglec-9 and its ligands, whether acting in trans or cis, play an important role in maintaining mast cell quiescence and limiting their activation.
Siglecs are endocytic receptors that either constitutively cycle between the cell surface and intracellular endosomes, or can undergo endocytosis upon ligation by antibody or multivalent ligands11. Compared to the endocytic capacity of Siglec-6 and Siglec-8 on human skin mast cells16, the internalization kinetics of Siglec-9 closely resembled Siglec-6, with most of the receptor remaining on the cell surface at 24 h after Siglec-9 ligation. Contrastingly, 50% of Siglec-8 is endocytosed at 2 h post engagement and almost none of the receptor could be detected on mast cell surface at 24 h post engagement16. In contrast, there was no detectable Siglec-9 internalization after engagement of Siglec-9 with GlycA or HMW-HA (Fig. E10). Additional studies are required to understand the mechanisms involved in the regulation of endocytosis and expression of different Siglecs on mast cell surface upon engagement. These studies have the potential to further advance the use of Siglecs as antigen delivery therapeutics to target mast cells based on the internalization kinetics of different Siglecs73.
The use of antibodies against Siglecs20–22, 59, 60 and glycomimetic Siglec ligands on multivalent scaffolds, like nanoparticles, liposomes, or polymers that aggregate Siglec receptors16, 37, 73 has demonstrated that Siglec engagement can be leveraged to modulate immune and inflammatory responses. Here, we report that Siglec-9 co-engagement with FcεRI can reduce human mast cell degranulation, arachidonic acid production, and chemokine release (Fig. 5). Similarly, several studies have demonstrated that clustering of CD3318, Siglec-616, Siglec-717, or Siglec-819 with FcεRI is necessary for optimal inhibition of mast cells responses in vitro. Prior studies have shown that several CD33-related Siglecs, namely CD33, Siglec-6, Siglec-7, and Siglec-8, can prevent the release of mast cell mediators, mast cell-dependent anaphylaxis or inflammation in mouse models of disease74. Based on these findings, studies are on their way in our lab to address the impact of Siglec-9 inhibitory effects on mast cell function in vivo.
The possible addition of Siglec-9 to the repertoire of inhibitory receptors that can modulate mast cell function is significant for two reasons. First, mast cells may exhibit differential responsiveness to Siglec modulation depending on the tissue where they reside or the inflammatory microenvironment that may change over time. Accordingly, a broad repertoire of functional Siglecs may be needed for mast cell targeting in different conditions. The rationale for this is that inflammatory environmental changes are known to affect the expression of Siglecs and Siglec ligands. For example, it has been shown that patients with COPD75, chronic rhinosinusitis70, and rheumatoid arthritis76 exhibited an up-regulation in the expression of Siglec-9 and Siglec-9 ligands. In relation to mast cells, our study shows that Siglec-9 inhibitory effects were more pronounced in HSCMC than in PBCMCs which may correlate to the expression levels of Siglec-9 and its ligands in these cells (Fig. 5). Second, certain Siglecs may play an important role in cell homeostasis that may preclude the use of a Siglec-targeted therapy in certain conditions. For example, Siglec-6 is specifically expressed on mast cells when compared with other immune cells16, but it is also expressed on trophoblast cells of the placenta77. Importantly, placental Siglec-6 expression correlates with preterm preeclampsia78 suggesting that Siglec-6 may contribute or represent a response to preeclampsia pathogenesis. This study highlights the need for additional studies to better understand the role of Siglecs in non-immune cells whose function may be negatively impacted by anti-Siglec therapies potentially causing undesired effects.
It is important to point out that downmodulation of mast cell function via Siglec-9 targeting has the potential to negate the beneficial effects of these cells. Strong evidence of immune subversion by bacterial exploitation of Siglec-9 function has been reported27–29. With relevance to mast cells, it has been shown that the sialylated capsule of Group B Streptococcus is recognized by Siglec-9 resulting in dampening of the innate immune response and increased GBS colonization28, 79. We have shown that mast cells protect against GBS in models of systemic infection and pre-term birth80–82. Accordingly, studies are underway to determine how interactions between GBS and Siglec-9 may affect mast cell ability to protect against this bacterium. In the context of cancer, the expression levels of Siglec-7 and Siglec-9 ligands are greatly increased on a variety of cancer cells, which decreases their susceptibility to killing by NK cells31, 83. However, the impact of a Siglec-9 targeted therapy on mast cell contribution to cancer is uncertain as it may largely depend on unknown conditions that lead to a pro-tumoral or anti-tumoral mast cell phenotype84.
In summary, our study provides evidence that human mast cells express Siglec-9 with inhibitory capabilities that may impact mast cell function in normal conditions and disease. Overall, we think that further understanding of Siglec-9 and other Siglecs’ contribution to mast function in different compartments, in normal conditions and disease contexts may be needed to tailor potential Siglec targeted therapies for treatment of mast cell associated disorders.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. T. Eoin West and Shelton Wright (University of Washington School of Medicine, Seattle, WA) for providing lung tissues. We also acknowledge the use of lung tissues procured by the National Disease Research Interchange (NDRI) program with support from NIH grant U42OD11158.
Funding:
This work was supported by seed funds provided by the Seattle Children’s Research Institute and in part by the National Institutes of Health/National Institute of General Medical Sciences (NIGMS) Pilot Project P20GM103641 to C.A.O.
Abbreviations used
- BMCMCs
bone marrow-derived cultured mast cells
- Cas9
CRISPR associated protein 9
- CD
cluster of differentiation
- CHO
Chinese hamster ovary
- CMP-Neu5Ac
cytidine monophosphate N-acetylneuraminic acid
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cys-LT
cysteinyl leukotriene
- DAMPS
damage-associated molecular patterns
- DAPI
diamidino-2-phenylindole (DAPI)
- DMEM
Dulbecco’s Modified Eagle Medium
- FBS
fetal bovine serum
- FcεRIα
alpha subunit of the high affinity receptor for IgE
- FcεRI
high affinity receptor for IgE
- GlycA
glycophorin A
- GNE
UDP-GlcNAc 2-epimerase/ManNAc-6
- HBSS
Hanks balanced salt solution
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HMW-HA
high molecular weight hyaluronic acid
- HSCMCs
human skin derived-cultured mast cells
- Ig
immunoglobulin
- IL
interleukin
- IMDM
Iscove’s Modified Dulbecco Media
- ITIM
immunoreceptor tyrosine-based inhibitory motifs
- LAMP-1
lysosomal-associated membrane protein -1
- LPS
lipopolysaccharide
- MCP-1
monocyte chemoattractant protein-1
- MFI
mean intensity fluorescence
- NETs
neutrophil extracellular traps
- NK
natural killer cells
- PBCMCs
human peripheral blood mononuclear cell-derived cultured mast cells
- PG
prostaglandin
- Rh
recombinant human
- Rm
recombinant mouse
- ROS
reactive oxygen species
- SiaT
sialyltransferases
- Siglec
sialic acid-binding immunoglobin-like lectins
- SCF
stem cell factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Conflict of interest: The authors declare that they have no conflict of interests.
REFERENCES
- 1.Piliponsky AM, Romani L. The contribution of mast cells to bacterial and fungal infection immunity. Immunol Rev 2018; 282:188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gri G, Frossi B, D’Inca F, Danelli L, Betto E, Mion F, et al. Mast cell: an emerging partner in immune interaction. Front Immunol 2012; 3:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varricchi G, Rossi FW, Galdiero MR, Granata F, Criscuolo G, Spadaro G, et al. Physiological Roles of Mast Cells: Collegium Internationale Allergologicum Update 2019. Int Arch Allergy Immunol 2019; 179:247–61. [DOI] [PubMed] [Google Scholar]
- 4.Henz BM, Maurer M, Lippert U, Worm M, Babina M. Mast cells as initiators of immunity and host defense. Exp Dermatol 2001; 10:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Dahlin JS, Maurer M, Metcalfe DD, Pejler G, Sagi-Eisenberg R, Nilsson G. The ingenious mast cell: Contemporary insights into mast cell behavior and function. Allergy 2022; 77:83–99. [DOI] [PubMed] [Google Scholar]
- 6.Metcalfe DD. Mast cells and mastocytosis. Blood 2008; 112:946–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med 2012; 18:693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishizaka T, Conrad DH. Binding characteristics of human IgE receptors and initial triggering events in human mast cells for histamine release. Monogr Allergy 1983; 18:14–24. [PubMed] [Google Scholar]
- 9.Nadler MJ, Matthews SA, Turner H, Kinet JP. Signal transduction by the high-affinity immunoglobulin E receptor Fc epsilon RI: coupling form to function. Adv Immunol 2000; 76:325–55. [DOI] [PubMed] [Google Scholar]
- 10.Theoharides TC, Valent P, Akin C. Mast Cells, Mastocytosis, and Related Disorders. N Engl J Med 2015; 373:163–72. [DOI] [PubMed] [Google Scholar]
- 11.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol 2014; 14:653–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science 2000; 290:84–9. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Gil A, Schnaar RL. Siglec Ligands. Cells 2021; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Sullivan JA, Chang AT, Youngblood BA, Bochner BS. Eosinophil and mast cell Siglecs: From biology to drug target. J Leukoc Biol 2020; 108:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoi H, Myers A, Matsumoto K, Crocker PR, Saito H, Bochner BS. Alteration and acquisition of Siglecs during in vitro maturation of CD34+ progenitors into human mast cells. Allergy 2006; 61:769–76. [DOI] [PubMed] [Google Scholar]
- 16.Robida PA, Rische CH, Morgenstern NB, Janarthanam R, Cao Y, Krier-Burris RA, et al. Functional and Phenotypic Characterization of Siglec-6 on Human Mast Cells. Cells 2022; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizrahi S, Gibbs BF, Karra L, Ben-Zimra M, Levi-Schaffer F. Siglec-7 is an inhibitory receptor on human mast cells and basophils. J Allergy Clin Immunol 2014; 134:230–3. [DOI] [PubMed] [Google Scholar]
- 18.Duan S, Koziol-White CJ, Jester WF Jr., Smith SA, Nycholat CM, Macauley MS, et al. CD33 recruitment inhibits IgE-mediated anaphylaxis and desensitizes mast cells to allergen. J Clin Invest 2019; 129:1387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoi H, Choi OH, Hubbard W, Lee HS, Canning BJ, Lee HH, et al. Inhibition of FcepsilonRI-dependent mediator release and calcium flux from human mast cells by sialic acid-binding immunoglobulin-like lectin 8 engagement. J Allergy Clin Immunol 2008; 121:499–505 e1. [DOI] [PubMed] [Google Scholar]
- 20.Landolina N, Zaffran I, Smiljkovic D, Serrano-Candelas E, Schmiedel D, Friedman S, et al. Activation of Siglec-7 results in inhibition of in vitro and in vivo growth of human mast cell leukemia cells. Pharmacol Res 2020; 158:104682. [DOI] [PubMed] [Google Scholar]
- 21.Schanin J, Gebremeskel S, Korver W, Falahati R, Butuci M, Haw TJ, et al. A monoclonal antibody to Siglec-8 suppresses non-allergic airway inflammation and inhibits IgE-independent mast cell activation. Mucosal Immunol 2021; 14:366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youngblood BA, Brock EC, Leung J, Falahati R, Bochner BS, Rasmussen HS, et al. Siglec-8 antibody reduces eosinophils and mast cells in a transgenic mouse model of eosinophilic gastroenteritis. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higuchi H, Shoji T, Iijima S, Nishijima K. Constitutively expressed Siglec-9 inhibits LPS-induced CCR7, but enhances IL-4-induced CD200R expression in human macrophages. Biosci Biotechnol Biochem 2016; 80:1141–8. [DOI] [PubMed] [Google Scholar]
- 24.von Gunten S, Yousefi S, Seitz M, Jakob SM, Schaffner T, Seger R, et al. Siglec-9 transduces apoptotic and nonapoptotic death signals into neutrophils depending on the proinflammatory cytokine environment. Blood 2005; 106:1423–31. [DOI] [PubMed] [Google Scholar]
- 25.Avril T, Floyd H, Lopez F, Vivier E, Crocker PR. The membrane-proximal immunoreceptor tyrosine-based inhibitory motif is critical for the inhibitory signaling mediated by Siglecs-7 and -9, CD33-related Siglecs expressed on human monocytes and NK cells. J Immunol 2004; 173:6841–9. [DOI] [PubMed] [Google Scholar]
- 26.Zhang JQ, Nicoll G, Jones C, Crocker PR. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem 2000; 275:22121–6. [DOI] [PubMed] [Google Scholar]
- 27.Secundino I, Lizcano A, Roupe KM, Wang X, Cole JN, Olson J, et al. Host and pathogen hyaluronan signal through human siglec-9 to suppress neutrophil activation. J Mol Med (Berl) 2016; 94:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 2009; 113:3333–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khatua B, Bhattacharya K, Mandal C. Sialoglycoproteins adsorbed by Pseudomonas aeruginosa facilitate their survival by impeding neutrophil extracellular trap through siglec-9. J Leukoc Biol 2012; 91:641–55. [DOI] [PubMed] [Google Scholar]
- 30.Saha S, Coady A, Sasmal A, Kawanishi K, Choudhury B, Yu H, et al. Exploring the Impact of Ketodeoxynonulosonic Acid in Host-Pathogen Interactions Using Uptake and Surface Display by Nontypeable Haemophilus influenzae. mBio 2021; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jandus C, Boligan KF, Chijioke O, Liu H, Dahlhaus M, Demoulins T, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest 2014; 124:1810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez E, Boelaars K, Brown K, Eveline Li RJ, Kruijssen L, Bruijns SCM, et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat Commun 2021; 12:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laubli H, Pearce OM, Schwarz F, Siddiqui SS, Deng L, Stanczak MA, et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci U S A 2014; 111:14211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas Q, Boligan KF, Jandus C, Schneider C, Simillion C, Stanczak MA, et al. Siglec-9 Regulates an Effector Memory CD8(+) T-cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol Res 2019; 7:707–18. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto T, Takahashi N, Kojima T, Yoshioka Y, Ishikawa J, Furukawa K, et al. Soluble Siglec-9 suppresses arthritis in a collagen-induced arthritis mouse model and inhibits M1 activation of RAW264.7 macrophages. Arthritis Res Ther 2016; 18:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang EA, Soh H, Park S, Lee HJ, Im JP, Kim JS. Soluble Siglec-9 alleviates intestinal inflammation through inhibition of the NF-kappaB pathway. Int Immunopharmacol 2020; 86:106695. [DOI] [PubMed] [Google Scholar]
- 37.Delaveris CS, Wilk AJ, Riley NM, Stark JC, Yang SS, Rogers AJ, et al. Synthetic Siglec-9 Agonists Inhibit Neutrophil Activation Associated with COVID-19. ACS Cent Sci 2021; 7:650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cildir G, Toubia J, Yip KH, Zhou M, Pant H, Hissaria P, et al. Genome-wide Analyses of Chromatin State in Human Mast Cells Reveal Molecular Drivers and Mediators of Allergic and Inflammatory Diseases. Immunity 2019; 51:949–65 e6. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Baruch Morgenstern N, Ballaban AY, Wen T, Shoda T, Caldwell JM, Kliewer K, et al. Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission. J Allergy Clin Immunol 2022; 149:2062–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motakis E, Guhl S, Ishizu Y, Itoh M, Kawaji H, de Hoon M, et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood 2014; 123:e58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronnberg E, Boey DZH, Ravindran A, Safholm J, Orre AC, Al-Ameri M, et al. Immunoprofiling Reveals Novel Mast Cell Receptors and the Continuous Nature of Human Lung Mast Cell Heterogeneity. Front Immunol 2021; 12:804812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMillan SJ, Sharma RS, McKenzie EJ, Richards HE, Zhang J, Prescott A, et al. Siglec-E is a negative regulator of acute pulmonary neutrophil inflammation and suppresses CD11b beta2-integrin-dependent signaling. Blood 2013; 121:2084–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada N, Matsushima H, Tagaya Y, Shimada S, Katz SI. Generation of a large number of connective tissue type mast cells by culture of murine fetal skin cells. J Invest Dermatol 2003; 121:1425–32. [DOI] [PubMed] [Google Scholar]
- 44.Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res 1988; 12:345–55. [DOI] [PubMed] [Google Scholar]
- 45.Laidlaw TM, Steinke JW, Tinana AM, Feng C, Xing W, Lam BK, et al. Characterization of a novel human mast cell line that responds to stem cell factor and expresses functional FcepsilonRI. J Allergy Clin Immunol 2011; 127:815–22 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13). Blood 1999; 94:2333–42. [PubMed] [Google Scholar]
- 47.Folkerts J, Gaudenzio N, Maurer M, Hendriks RW, Stadhouders R, Tam SY, et al. Rapid identification of human mast cell degranulation regulators using functional genomics coupled to high-resolution confocal microscopy. Nat Protoc 2020; 15:1285–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caslin HL, Taruselli MT, Paranjape A, Kiwanuka K, Haque T, Chumanevich AP, et al. The Use of Human and Mouse Mast Cell and Basophil Cultures to Assess Type 2 Inflammation. Methods Mol Biol 2018; 1799:81–92. [DOI] [PubMed] [Google Scholar]
- 49.Okayama Y, Hunt TC, Kassel O, Ashman LK, Church MK. Assessment of the anti-c-kit monoclonal antibody YB5.B8 in affinity magnetic enrichment of human lung mast cells. J Immunol Methods 1994; 169:153–61. [DOI] [PubMed] [Google Scholar]
- 50.Ravindran A, Ronnberg E, Dahlin JS, Mazzurana L, Safholm J, Orre AC, et al. An Optimized Protocol for the Isolation and Functional Analysis of Human Lung Mast Cells. Front Immunol 2018; 9:2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu CK, Wei G, Atwood WJ. Infection of glial cells by the human polyomavirus JC is mediated by an N-linked glycoprotein containing terminal alpha(2–6)-linked sialic acids. J Virol 1998; 72:4643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Sullivan JA, Carroll DJ, Cao Y, Salicru AN, Bochner BS. Leveraging Siglec-8 endocytic mechanisms to kill human eosinophils and malignant mast cells. J Allergy Clin Immunol 2018; 141:1774–85 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikehara Y, Ikehara SK, Paulson JC. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem 2004; 279:43117–25. [DOI] [PubMed] [Google Scholar]
- 54.Biedermann B, Gil D, Bowen DT, Crocker PR. Analysis of the CD33-related siglec family reveals that Siglec-9 is an endocytic receptor expressed on subsets of acute myeloid leukemia cells and absent from normal hematopoietic progenitors. Leuk Res 2007; 31:211–20. [DOI] [PubMed] [Google Scholar]
- 55.Ibarlucea-Benitez I, Weitzenfeld P, Smith P, Ravetch JV. Siglecs-7/9 function as inhibitory immune checkpoints in vivo and can be targeted to enhance therapeutic antitumor immunity. Proc Natl Acad Sci U S A 2021; 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hugonnet M, Singh P, Haas Q, von Gunten S. The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front Immunol 2021; 12:799861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stasche R, Hinderlich S, Weise C, Effertz K, Lucka L, Moormann P, et al. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Molecular cloning and functional expression of UDP-N-acetyl-glucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem 1997; 272:24319–24. [DOI] [PubMed] [Google Scholar]
- 58.Lizcano A, Secundino I, Dohrmann S, Corriden R, Rohena C, Diaz S, et al. Erythrocyte sialoglycoproteins engage Siglec-9 on neutrophils to suppress activation. Blood 2017; 129:3100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerr SC, Gonzalez JR, Schanin J, Peters MC, Lambrecht BN, Brock EC, et al. An anti-siglec-8 antibody depletes sputum eosinophils from asthmatic subjects and inhibits lung mast cells. Clin Exp Allergy 2020; 50:904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebremeskel S, Schanin J, Coyle KM, Butuci M, Luu T, Brock EC, et al. Mast Cell and Eosinophil Activation Are Associated With COVID-19 and TLR-Mediated Viral Inflammation: Implications for an Anti-Siglec-8 Antibody. Front Immunol 2021; 12:650331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan S, Arlian BM, Nycholat CM, Wei Y, Tateno H, Smith SA, et al. Nanoparticles Displaying Allergen and Siglec-8 Ligands Suppress IgE-FcepsilonRI-Mediated Anaphylaxis and Desensitize Mast Cells to Subsequent Antigen Challenge. J Immunol 2021; 206:2290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Claude J, Linnartz-Gerlach B, Kudin AP, Kunz WS, Neumann H. Microglial CD33-related Siglec-E inhibits neurotoxicity by preventing the phagocytosis-associated oxidative burst. J Neurosci 2013; 33:18270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Chen Y, Sluter MN, Hou R, Hao J, Wu Y, et al. Ablation of Siglec-E augments brain inflammation and ischemic injury. J Neuroinflammation 2022; 19:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, Kiessling LL. Sialylated multivalent antigens engage CD22 in trans and inhibit B cell activation. Proc Natl Acad Sci U S A 2009; 106:2500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haney MS, Bohlen CJ, Morgens DW, Ousey JA, Barkal AA, Tsui CK, et al. Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat Genet 2018; 50:1716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pluvinage JV, Haney MS, Smith BAH, Sun J, Iram T, Bonanno L, et al. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 2019; 568:187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva M, Silva Z, Marques G, Ferro T, Goncalves M, Monteiro M, et al. Sialic acid removal from dendritic cells improves antigen cross-presentation and boosts anti-tumor immune responses. Oncotarget 2016; 7:41053–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bull C, Collado-Camps E, Kers-Rebel ED, Heise T, Sondergaard JN, den Brok MH, et al. Metabolic sialic acid blockade lowers the activation threshold of moDCs for TLR stimulation. Immunol Cell Biol 2017; 95:408–15. [DOI] [PubMed] [Google Scholar]
- 69.Aalto K, Autio A, Kiss EA, Elima K, Nymalm Y, Veres TZ, et al. Siglec-9 is a novel leukocyte ligand for vascular adhesion protein-1 and can be used in PET imaging of inflammation and cancer. Blood 2011; 118:3725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia Y, Yu H, Fernandes SM, Wei Y, Gonzalez-Gil A, Motari MG, et al. Expression of ligands for Siglec-8 and Siglec-9 in human airways and airway cells. J Allergy Clin Immunol 2015; 135:799–810 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Zheng Y, Li J, Nie L, Hu Y, Wang F, et al. Immunoregulatory Siglec ligands are abundant in human and mouse aorta and are up-regulated by high glucose. Life Sci 2019; 216:189–99. [DOI] [PubMed] [Google Scholar]
- 72.Fukui M, Whittlesey K, Metcalfe DD, Dastych J. Human mast cells express the hyaluronic-acid-binding isoform of CD44 and adhere to hyaluronic acid. Clin Immunol 2000; 94:173–8. [DOI] [PubMed] [Google Scholar]
- 73.Chen WC, Kawasaki N, Nycholat CM, Han S, Pilotte J, Crocker PR, et al. Antigen delivery to macrophages using liposomal nanoparticles targeting sialoadhesin/CD169. PLoS One 2012; 7:e39039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bochner BS, O’Sullivan JA, Chang AT, Youngblood BA. Siglecs in allergy and asthma. Mol Aspects Med 2022:101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng Z, Li M, Wang M, Wu X, Li Q, Ning Q, et al. Increased expression of Siglec-9 in chronic obstructive pulmonary disease. Sci Rep 2017; 7:10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Liu D, Ning Y, Liu J, Wang X, Tu R, et al. Siglec-9 is upregulated in rheumatoid arthritis and suppresses collagen-induced arthritis through reciprocal regulation of Th17-/Treg-cell differentiation. Scand J Immunol 2017; 85:433–40. [DOI] [PubMed] [Google Scholar]
- 77.Brinkman-Van der Linden EC, Hurtado-Ziola N, Hayakawa T, Wiggleton L, Benirschke K, Varki A, et al. Human-specific expression of Siglec-6 in the placenta. Glycobiology 2007; 17:922–31. [DOI] [PubMed] [Google Scholar]
- 78.Rumer KK, Uyenishi J, Hoffman MC, Fisher BM, Winn VD. Siglec-6 expression is increased in placentas from pregnancies complicated by preterm preeclampsia. Reprod Sci 2013; 20:646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang YC, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, et al. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog 2014; 10:e1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gendrin C, Vornhagen J, Ngo L, Whidbey C, Boldenow E, Santana-Ufret V, et al. Mast cell degranulation by a hemolytic lipid toxin decreases GBS colonization and infection. Sci Adv 2015; 1:e1400225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gendrin C, Shubin NJ, Boldenow E, Merillat S, Clauson M, Power D, et al. Mast cell chymase decreases the severity of group B Streptococcus infections. J Allergy Clin Immunol 2018; 142:120–9 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piliponsky AM, Sharma K, Quach P, Brokaw A, Nguyen S, Orvis A, et al. Mast cell-derived factor XIIIA contributes to sexual dimorphic defense against group B streptococcal infections. J Clin Invest 2022; 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol 2014; 10:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Segura-Villalobos D, Ramirez-Moreno IG, Martinez-Aguilar M, Ibarra-Sanchez A, Munoz-Bello JO, Anaya-Rubio I, et al. Mast Cell-Tumor Interactions: Molecular Mechanisms of Recruitment, Intratumoral Communication and Potential Therapeutic Targets for Tumor Growth. Cells 2022; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


