Abstract
The widespread use of sensitive assays for the detection of viral and cellular RNA sequences has created a need for stable, well-characterized controls and standards. We describe the development of a versatile, novel system for creating RNase-resistant RNA. “Armored RNA” is a complex of MS2 bacteriophage coat protein and RNA produced in Escherichia coli by the induction of an expression plasmid that encodes the coat protein and an RNA standard sequence. The RNA sequences are completely protected from RNase digestion within the bacteriophage-like complexes. As a prototype, a 172-base consensus sequence from a portion of the human immunodeficiency virus type 1 (HIV-1) gag gene was synthesized and cloned into the packaging vector used to produce the bacteriophage-like particles. After production and purification, the resulting HIV-1 Armored RNA particles were shown to be resistant to degradation in human plasma and produced reproducible results in the Amplicor HIV-1 Monitor assay for 180 days when stored at −20°C or for 60 days at 4°C. Additionally, Armored RNA preparations are homogeneous and noninfectious.
In recent years, a variety of techniques for measurement of the absolute concentration of specific RNA sequences have been developed, such as competitive reverse transcription-PCR (RT-PCR), nucleic acid based-sequence amplification, transcription-mediated amplification, and the branched-chain DNA assays (3, 6, 10, 14). These methods are used clinically to measure human immunodeficiency virus (HIV) type 1 (HIV-1) and hepatitis C virus (HCV) concentrations in the plasma of infected patients.
Central to these quantitative assays are reliable RNA preparations which are calibrated to known concentrations. The RNA may serve as (i) a positive “control” to indicate that the assay is performing to its specifications and (ii) a quantitative “standard” by which the samples are measured.
Currently, quantitative RNA standards are produced enzymatically by transcribing a DNA template into RNA by in vitro transcription (7). The positive controls comprised an attenuated or inactivated infectious agent itself or an in vitro-transcribed RNA. A major disadvantage of using a naked RNA is that it is susceptible to degradation by RNases. Because of the prevalence of RNases, the synthesis, purification, and storage of RNA are not trivial. Even if a specific lot of RNA is RNase free, it is susceptible to contamination any time that the storage vessel is opened. For these reasons, there is a need for RNase-resistant RNA controls and standards which are compatible with all of the technologies used to perform viral assays.
RNA coliphages are simple bacteriophages which infect Escherichia coli (for reviews, see references 12 and 16). The genomic RNA packaged within these particles is highly resistant to RNase digestion, and the RNA is easily extracted from the bacteriophage coat protein by conventional methods (1). We reasoned that a recombinant RNA (reRNA) containing the RNA sequence of an infectious agent such as HIV or HCV could be packaged as bacteriophage particles, thereby conferring protection to the reRNA against RNases.
In this article, we describe a method for packaging reRNA into pseudoviral particles. Using “Armored RNA” technology, we have made a positive control compatible with a commercially available HIV-1 diagnostic assay, the Amplicor HIV-1 Monitor assay, and demonstrated that the reRNA in the Armored RNA particles was totally resistant to RNases, even when the particles were stored in human plasma for half of a year. As well, the HIV-1 Armored RNA substituted seamlessly in routine clinical runs for the positive control RNA standard provided with the HIV-1 Monitor kit. A straightforward manufacturing process and reliable performance make this technology ideal for the production of the RNA controls and standards for clinical diagnostics.
MATERIALS AND METHODS
Armored RNA construction.
The details of the synthesis of the packaging vector and the expression and purification of the bacteriophage-like particles have been described previously (5). The AR-QS Armored RNA contains the 142-nucleotide RNA sequence which acts as the internal quantification standard (QS) in the HIV Monitor kit (5).
AR–HIV-B is an HIV-1-positive control standard. Briefly, a consensus 172-bp DNA fragment (Fig. 1) containing a portion of the HIV subtype B (HIV-B) gag, nucleotides 903 to 1074 (9), was designed from the analysis of 32 individual gag sequences contained within the Human Retroviruses and AIDS 1996 nucleotide sequence database (9a). The HIV-B consensus sequence includes the 142-nucleotide gag sequence that serves as the target for the Amplicor HIV-1 Monitor assay with primers SK462 and SK431 (8). De novo construction of the HIV-B consensus gag fragment was performed with polyacrylamide gel electrophoresis-purified oligodeoxynucleotides and by a ligase chain reaction developed for synthetic gene construction (13). The synthetic DNA was amplified by the overlap extension technique to add on an MS2 operator sequence and was then cloned into the packaging vector to produce pAR–HIV-B. This recombinant plasmid was used to synthesize AR–HIV-B.
FIG. 1.
Sequence of the HIV RNA packaged within AR–HIV-B. The sequences with which the primers SK462 and SK431 from the HIV-1 Monitor kit hybridize are indicated.
CsCl fractionation.
Approximately 5 to 10 mg of Armored RNA was fractionated for each CsCl gradient. To compare the densities of MS2 and AR–HIV-B, each was loaded in separate gradients. After ultracentrifugation (5), the heat-sealed tube was stabilized in the upright position. An 18-gauge needle was inserted into the top of the tube to equilibrate the pressure in the tube. An 18-gauge needle was slowly inserted into the bottom of the tube, and 0.5-ml fractions were collected.
RT-PCR assay.
To determine viral copy number, Amplicor HIV-1 Monitor assays (Roche Diagnostic Systems, Inc., Branchburg, N.J.) were performed according to the manufacturer’s instructions.
Incubations with purified nucleases.
The RNases were present in the reaction mixtures as a mixture of RNases A and T1 at 0.03 and 1.3 U/μl, respectively, and DNase I (Ambion, Inc., Austin, Tex.) was present in the reaction mixtures at 0.1 U/μl. The reaction mixtures were incubated at 37°C for 60 min. The concentrations of the plasmid DNA (pTRIamp19; Ambion, Inc.), the reRNA isolated from AR–HIV-B, and the intact AR–HIV-B were 0.03, 0.04, and 0.03 mg/ml, respectively. After digestion, the samples were fractionated in a 2% agarose gel, stained with ethidium bromide, and visualized by UV fluorescence.
Stability in plasma and serum.
Purified AR–HIV-B was quantified in duplicate by the HIV-1 Monitor assay. Normal plasma from a single donor containing EDTA as the anticoagulant was clarified by centrifugation at 5,000 × g for 30 min, and sodium azide was added to a concentration of 0.1%. For each study, a single batch of AR–HIV-B spiked into plasma was prepared and aliquoted into single-time-point samples of 0.2 ml, the volume required for the HIV-1 Monitor assay. Samples were incubated at the assigned temperature until they were assayed. For the studies performed at −20°C, the Armored RNA control was assayed in parallel with the HIV-1 Monitor assay high-positive control in regular clinical runs for HIV-1 load comparison. The Armored RNA control and the HIV-1 Monitor assay positive control were assayed two to four times per week. The HIV-1 Monitor assay positive control was used according to the manufacturer’s instructions.
For the study performed at 4°C, AR–HIV-B-spiked plasma samples were removed at each time point and were stored at −80°C, and then all of the samples were assayed in a single run.
Coincubations of Armored RNA and HIV in plasma.
An attenuated HIV-1 strain, HIV-1MC99 (2), and AR-QS were both added to normal human plasma (Roche Diagnostic Systems, Inc.) at approximately 7,500 and 5,000 copies/ml, respectively. Aliquots of 0.2 ml were incubated at 37°C over 30 days. Samples taken at each time point were stored at −80°C and were then processed simultaneously. Samples obtained at each time point were assayed in duplicate and averaged.
Synthesis of bacteriophage lambda Armored RNA particles.
A common 3′ primer was used for the amplification of a series of bacteriophage lambda DNA fragments of increasing lengths. This primer was used in all of the amplification reactions. PCR products, which increased in length, were synthesized with different 5′ primers that hybridized at increasing distances from the 3′ primer. Purified lambda DNA (Ambion, Inc.) was used as the template for PCR. Each of the PCR products was cloned separately into the Armored RNA packaging vector. Purified Armored RNA particles were expressed and isolated as described previously (5).
RNA isolation and Northern blotting.
Packaged RNA from the Armored RNA particles and E. coli RNA were isolated with the RNAqueous RNA isolation kit (Ambion, Inc.). Northern blotting of the purified RNA was performed with the NorthernMAX northern blotting kit (Ambion, Inc.). Oligonucleotide probes used for Northern blotting were 5′ end labeled with 32P by using the KinaseMAX kit (Ambion, Inc.).
RESULTS
General strategy used to produce Armored RNA.
The RNAs used as controls and standards in clinical assays for the detection of HIV-1 and HCV have an inherent weakness in that they are susceptible to degradation by RNases. Our goal was to produce an RNA preparation that was resistant to RNase digestion, that could be produced in a relatively inexpensive and straightforward manner, that was easily adapted to various RNA sequences, and that would act as a template for reverse transcription. Since the genomic RNA packaged in the E. coli bacteriophage MS2 is resistant to RNase digestion, we hypothesized that non-MS2 RNA sequences could be packaged within a similar structure to confer similar protection from RNases. Bacteriophage MS2 is a simple ribonucleoprotein structure composed of 180 coat protein molecules, one copy of maturase protein, and one copy of the 3.6 kb plus-strand gRNA. The coat protein makes up the bulk of the bacteriophage, assembling into an icosahedral structure of 26 nm in diameter (16).
The initial strategy was to produce viable, recombinant MS2 bacteriophage containing reRNA, but it was rejected for several reasons. First, recombinant coliphages are genetically unstable and quickly delete non-phage RNA sequences. Second, viable reRNA bacteriophage in clinical reference laboratories could proliferate and could cause serious contamination. Finally, the MS2 RNA replicase is a low-fidelity polymerase and would produce point mutations and deletions in an RNA standard.
Since the production of viable, recombinant MS2 bacteriophage was not an option for the packaging of reRNA, the alternative strategy which we adopted was to develop a plasmid-driven packaging system. Several researchers had shown that pseudoviral particles could be synthesized in vivo and in vitro with coat protein alone. In fact, a non-phage RNA sequence could be specifically packaged in E. coli as a pseudoviral particle if the recombinant RNA contained an “operator” sequence (11). The operator is a 19-base sequence bound by coat protein to initiate the assembly of the bacteriophage particle.
In the plasmid packaging system, the DNAs encoding the coat protein, the target RNA sequence, and the MS2 operator sequence were cloned downstream of an inducible lac promoter. This strategy used the high-fidelity E. coli RNA polymerase to transcribe the reRNA. The recombinant packaging vector was transformed into E. coli. Isopropyl-β-d-thiogalactopyranoside was added to induce the transcription of the reRNA and the expression of the pseudoviral particles. As coat protein is translated, it binds to the operator sequence at the 3′ end of the reRNA, initiating the encapsidation of the reRNA to produce pseudoviral particles. Unlike MS2, which is released into the spent medium by lysing E. coli, Armored RNA is localized in the cytoplasmic fraction of E. coli.
Construction of HIV-1 Armored RNA.
To demonstrate the feasibility of the Armored RNA technology, we produced a control compatible for use with the Amplicor HIV-1 Monitor kit. The AR–HIV-B Armored RNA was generated for use as a positive control by packaging an RNA derived from a consensus sequence from the gag region of HIV isolates of clade B. We also produced an Armored RNA version of the QS used in the HIV-1 Monitor kit (AR-QS). In the HIV-1 Monitor assay, the QS RNA is the calibrating RNA which is added to each patient sample and which is used to calculate the patient’s viral concentration.
Homogeneity of Armored RNA.
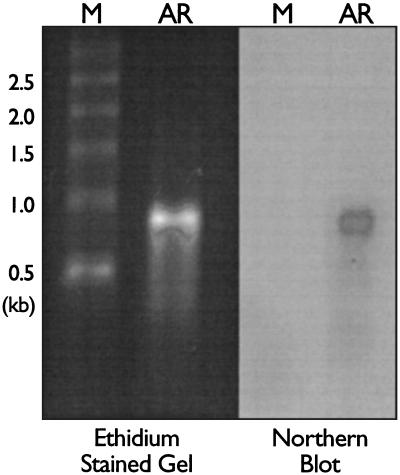
The reRNA was isolated from purified AR–HIV-B. The majority of the reRNA packaged was approximately 900 bases in length, as detected by ethidium bromide staining and Northern blotting (Fig. 2).
FIG. 2.
Characterization of the recombinant RNA packaged in AR–HIV-B. reRNA was isolated from AR–HIV-B, fractionated in a denaturing 1% agarose gel, stained with ethidium bromide, and detected by UV fluorescence. The reRNA was transferred to a membrane and probed with a 32P-labeled oligonucleotide to the 3′ end of the HIV-B sequence. Abbreviations: M, RNA markers; AR, AR–HIV-B reRNA.
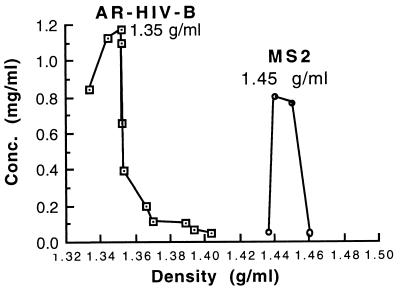
The homogeneity of the AR–HIV-B preparation was demonstrated by taking fractions from a CsCl gradient. The AR–HIV-B banded as a sharp peak at a density of 1.35 g/ml, while native MS2 bacteriophage banded at 1.45 g/ml (Fig. 3). The MS2 particles were denser because they contained three times more RNA and maturase protein.
FIG. 3.
Densities of AR–HIV-B and bacteriophage MS2 particles. MS2 and AR–HIV-B were loaded in separate gradients and centrifuged, and then 0.5-ml fractions were collected and weighed to determine the density of the CsCl. The optical density of each fraction at 260 nm was measured to calculate the Armored RNA and MS2 concentrations.
Durability of Armored RNA.
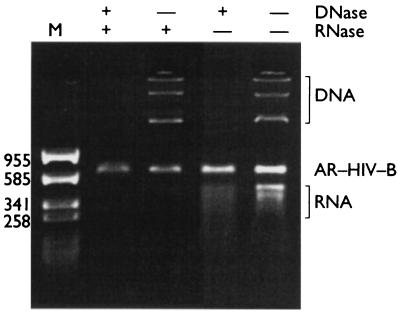
The reRNA packaged within AR–HIV-B was completely resistant to DNase and RNase treatment under conditions in which naked DNA and RNA are both degraded rapidly (Fig. 4). The AR–HIV-B preparation was stable at temperatures of up to 64°C in the presence of 1 mM MgCl2 but was stable only up to 54°C in 1 mM EDTA (data not shown). If the AR–HIV-B particles were heated at 70°C for 5 min, the coat protein was denatured, releasing the packaged reRNA and exposing it to nuclease attack (data not shown).
FIG. 4.
Resistance of purified Armored RNA particles to nucleases. The particles were mixed with plasmid DNA and purified, naked reRNA. The mixture of plasmid DNA, reRNA, and intact Armored RNA was incubated with DNase I and/or the RNases at 37°C for 1 h, fractionated by gel electrophoresis in a 2.0% nondenaturing agarose gel, and detected by ethidium bromide staining and UV fluorescence. The numbers on the left are in base pairs.
Stability at 45°C.
We investigated the stability of the Armored RNA incubated at 45°C for 3 days, which are the standard conditions used to examine shipping compatibility. Preliminary experiments indicated that Armored RNA was not completely stable in 10 mM Tris (pH 7.0)–100 mM NaCl–1 mM MgCl2 (TSM) at low concentrations at room temperature or 45°C. Tenfold dilutions of the AR-HIV preparation were made in TSM, incubated at 45°C for 3 days, and then assayed for reRNA copy number. At concentrations below 0.05 mg/ml, the reRNA copy number of the AR-HIV decreased (data not shown).
We postulated that we could stabilize a specific Armored RNA at a low copy number by formulating it with a “null” Armored RNA (AR-1) at a concentration of 0.05 mg/ml. AR-1 is an Armored RNA in which only MS2 and some of the plasmid RNA sequence is packaged. To demonstrate that AR-1 could stabilize AR-HIV at low concentrations, AR-HIV was diluted to 2.5 × 10−7 mg/ml in a solution of 0.05 mg of AR-1 per ml in TSM and incubated 3 days at 45°C, and the copy number was compared to that of the AR-HIV stored at −20°C. There was no loss in copy number (data not shown). We have observed similar stabilizing effects using L-broth and StabilZyme AP (SurModics, Inc., Eden Prairie, Minn.), whereas StabilGuard (SurModics, Inc.), StabilZyme HRP (SurModics, Inc.), acetylated bovine serum albumin (1 mg/ml), and SeraSub and ProDil (CST Technologies, Inc., Great Neck, N.Y.) did not stabilize the Armored RNA at 45°C (data not shown).
Maximum size of reRNA which can be packaged.
To define the size limits for reRNA packaging, we created constructs designed to package bacteriophage lambda RNA sequences of 0.5, 1, 1.5, 2, 3, and 4 kb. These particles were expressed and purified, and the RNA was isolated from each of these constructs. Only the construct encoding the 0.5-kb bacteriophage lambda RNA contained a reRNA of the expected size, as determined by ethidium bromide staining. The other constructs contained RNA which was heterogeneous in length (data not shown). Northern blotting of the purified recombinant RNA with probes directed to the 3′ terminus of the bacteriophage lambda sequence revealed that packaging of 500 bases of RNA was very efficient but that packaging of the 1- and 1.5-kb amounts of RNA was inefficient. As the size of the reRNA was increased, greater amounts of host (E. coli) RNA was packaged in preference to the amount of reRNA that was packaged. Although the 1.0- and 1.5-kb amounts of bacteriophage lambda RNA were detectable by Northern blotting, they were not detectable as discrete RNA species by ethidium bromide staining and UV fluorescence.
Stability of Armored RNA in plasma.
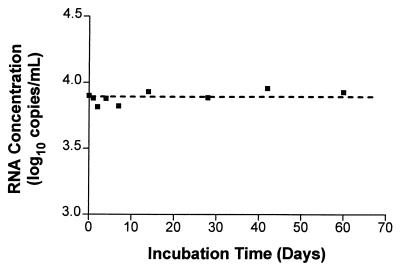
AR-QS was diluted in human serum or in plasma spiked with acid citrate dextrose, sodium citrate, or EDTA to inhibit coagulation, incubated for 1 h at 21°C, and then processed by the HIV-1 Monitor assay. No loss of signal was observed in any of these samples, indicating that AR-QS was stable in any of these blood products (data not shown). AR–HIV-B in EDTA-anticoagulated plasma was stable after five freeze-thaw cycles (data not shown). Incubation of AR–HIV-B at 4°C for 60 days in EDTA-anticoagulated human plasma did not compromise the original signal (Fig. 5).
FIG. 5.
Stability study of AR–HIV-B spiked into EDTA-anticoagulated human plasma at 4°C. AR–HIV-B was added to clarified plasma to a final concentration of ∼7,500 copies/ml. Samples were incubated at 4°C for 0, 1, 2, 4, 7, 14, 28, 42, and 60 days. Samples from each time point were assayed in duplicate, and the copy number determinations were averaged. The mean for all of the samples was 7,780 copies per ml (3.8 log10; range, 6,530 to 9,020 copies per ml [range, 3.81 to 3.96 log10]), and the coefficient of variation was 10.7%. The dashed line represents the mean.
Armored RNA as a positive control in a clinical assay.
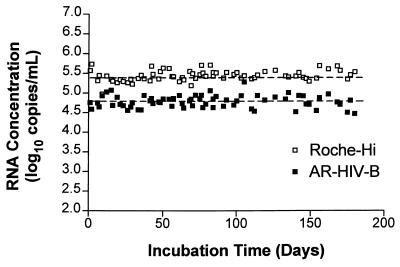
AR–HIV-B was diluted in EDTA-anticoagulated human plasma at 65,000 copies/ml and was stored at −20°C in aliquots of 0.2 ml. To assess the performance of an Armored RNA control in a clinical setting, AR–HIV-B was used as the positive control in alternate runs of HIV-1 Monitor assay with clinical samples in place of the HIV-1 Monitor assay positive control (a naked RNA) provided with the HIV-1 Monitor kit. The Armored RNA positive control performed reliably over 180 days, with results comparable to those obtained with the high-positive control provided with the kit (Fig. 6).
FIG. 6.
Comparison of Armored RNA positive control and the HIV-1 Monitor assay high-positive control (Roche-Hi) used in a clinical setting over 180 days. AR–HIV-B was added to clarified EDTA-anticoagulated plasma to a final concentration of ∼65,000 copies/ml, aliquoted into 0.2-ml samples, and stored at −20°C until it was used in the HIV-1 Monitor assay to determine the RNA copy number. The Armored RNA positive control and the HIV-1 Monitor assay high-positive control were used in clinical runs two to four times per week for 180 days. For the Armored RNA standard, the mean was 64,598 copies/ml, the range was 31,760 to 191,716 copies/ml (4.50 to 5.28 log10), and the coefficient of variation was 40%. For the HIV-1 Monitor assay high-positive control, the mean was 290,537 copies/ml; the range was 94,345 to 544,737 copies/ml (4.97 to 5.74 log10), and the coefficient of variation was 32%. The dashed lines represent the means for the two different positive controls.
Stability of Armored RNA compared to HIV in plasma.
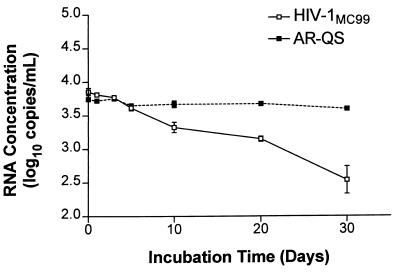
AR-QS and cultured HIV-1MC99 were coincubated in normal human plasma at 37°C for 30 days. Samples were taken in duplicate at seven different time points. AR-QS contains the same RNA sequence as the naked QS RNA standard in the HIV-1 Monitor kit. The HIV and QS sequences are amplified by the same primer set, but they can be distinguished by different internal capture sequences. Over the 30-day period, the HIV-1MC99 copy number declined by ∼80% compared to the original input. The AR-QS was stable over the time course (Fig. 7). The mean for all the AR-QS samples was 4,553 copies/ml (3.66 log10), and the coefficient of variation was 9.8%.
FIG. 7.
Stability of Armored RNA and HIV coincubated in normal human plasma. AR-QS and HIV-1MC99 were coincubated in normal human plasma over 30 days at 37°C. The concentrations of the QS and HIV RNA sequences were determined by the HIV-1 Monitor assay.
DISCUSSION
The use of nucleic acid-based assays for the diagnosis and monitoring of HCV and HIV loads is a relatively new technology. Most of these assays depend on the use of RNA synthesized by in vitro transcription for the positive control and internal or external standards. It is essential, after calibrating the RNA standard, that it be possible to place the RNA in long-term storage without degradation. Several factors can lead to the early demise of an RNA molecule. High pH, high temperatures, and divalent cations such as magnesium and manganese will promote the hydrolysis of RNA. As well, RNases are ubiquitous and RNA is highly susceptible to even minor contamination with RNase. Thus, development of an environment for the synthesis and long-term maintenance of full-length RNA is not a trivial process.
Armored RNA technology was developed to overcome the weaknesses associated with the manufacturing and use of naked RNA as a standard or control in clinical diagnostic assays. With this technology, RNA strands are synthesized in E. coli and assembled into pseudoviral particles, thereby protecting the packaged RNA from RNase attack. Thus, the production of Armored RNA is not dependent on an RNase-free environment. In fact, the protocol for purifying the particles from E. coli involves incubation of the preparation with a high concentration of micrococcal nuclease to digest contaminating host RNA and DNA. Thus, the production procedure is much more forgiving than is the synthesis of RNA by in vitro transcription.
A single lot of Armored RNA produced from 1 liter of E. coli cells can generate up to 1015 particles. These large lot sizes and the stability of the material allow cost-effective production.
An additional benefit of using Armored RNA rather than naked RNA as the positive control or the calibrator is the improved reliability of diagnostic assays. A naked RNA can be inadvertently contaminated with RNase during a clinical run, causing the failure of an entire run. Such failures are time-consuming and expensive. In addition, partial degradation of the calibrator may not be detected and may lead to erroneous results.
As an alternative to naked RNA, intact HIV and HCV are also used as standards or positive controls. The use of Armored RNA has many advantages over the use of intact virus as a positive control. It is noninfectious, decreasing the chance that a laboratory worker could be infected during either its production or its use in an assay. Shipping of Armored RNA requires less expense and less preparation than shipping of an infectious HIV standard or control. It is also more stable than HIV in plasma, and therefore, it can be shipped at ambient temperatures, decreasing the cost compared to those associated with dry-ice shipments. The manufacture of Armored RNA is easier, faster, and less hazardous than that of HIV. In addition, HIV has a high mutation rate, and therefore, it is impossible to know the precise sequence of such a standard or control, whereas the RNA in an Armored RNA preparation is homogeneous in its sequence.
Currently, there is little automation in HIV load assays. However, many companies are developing highly automated assays. Armored RNA materials will be ideal onboard reagents which are stable at room temperature for extended time periods. Armored RNA internal standards and positive controls could both be used in an automated assay without concern that they might degrade.
With the Armored RNA packaging system, there exists the flexibility of introducing a variety of different RNA sequences. Thus, standards for HCV, equine encephalitis virus, enterovirus, and other pathogenic RNA viruses can be engineered. For example, we have already produced and tested an HCV Armored RNA control (4, 15) compatible with both the HCV Monitor assay (Roche Diagnostic Systems, Inc.) and the HCV Quantiplex assay (Chiron Corp., Emeryville, Calif.), thereby producing a “universal” HCV standard for use in direct comparisons of assays.
The efficiency of packaging decreased quickly as the size of the RNA increased beyond 500 bases. Although most nucleic acid-based assays do not target RNA sequences longer than 500 bases, there are applications in which it would be useful to be able to package several thousand bases. For example, the HIV Quantiplex assay (Chiron Corp.) uses a standard which is about 3 kb in length, and therefore, it is not possible to produce a single Armored RNA standard for this assay. However, it may be possible to pool several different Armored RNA standards which collectively encode the entire control sequence. Also, if RNA sequences of several kilobases could be packaged, then a single Armored RNA standard could meet the needs of a variety of different viral assays designed to detect different regions of a viral genome. With such a standard, different research groups and clinical laboratories could make direct comparisons of their quantitative data.
Armored RNA standards can be used for applications other than infectious disease detection. Cytokine Armored RNA standards have been prepared for competitive RT-PCR. The QuantiKit assay (Ambion, Inc.) contains Armored RNA standards for determination of the concentration of cytokine mRNA. Since the reRNA in Armored RNA can be released from its packaging by heating at 70°C for 5 min, an Armored RNA standard can be added directly to a total RNA sample and the mixture can be heated to release the reRNA (data not shown). The heated sample may then be used directly in an RT reaction followed by PCR.
The production, maintenance, and use of intact RNA as standards and controls are not trivial processes. The use of Armored RNA technology offers a simple and reliable alternative to the use of naked RNA for viral assays which must contain dependable RNA standards and controls.
ACKNOWLEDGMENT
This research was supported in part by the National Institutes of Health (grant 1 R43 AI40529-01A1 from the National Institute of Allergy and Infectious Diseases).
REFERENCES
- 1.Argetsinger J E, Gussin G. Intact ribonucleic acid from defective particles of bacteriophage R17. J Mol Biol. 1966;21:421–434. doi: 10.1016/0022-2836(66)90016-7. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Boyle T, Malim M, Cullen B, Lyerly H. Derivation of a biologically contained replication system for human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:7678–7682. doi: 10.1073/pnas.89.16.7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins M L, Irvine I, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L-P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuBois D B, WalkerPeach C, Pasloske B L, Winkler M. Universal ribonuclease resistant RNA standards (Armored RNA) for RT-PCR and bDNA-based hepatitis virus RNA assays. Clin Chem. 1997;43:2218. . (Abstract 28.) [PubMed] [Google Scholar]
- 5.DuBois, D. B., M. M. Winkler, and B. L. Pasloske. October 1997. U.S. patent 5,677,124.
- 6.Kacian, D. L., and T. J. Fultz. March 1995. U.S. patent 5,399,491.
- 7.Kreig P A, Melton D A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 8.Kwok S, Sninsky J J. PCR detection of human immunodeficiency virus type 1 proviral DNA sequence. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 309–315. [Google Scholar]
- 9.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1: in vivo evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Los Alamos National Laboratory. Human retroviruses and AIDS 1996. Los Alamos, N.M: Los Alamos National Laboratory; 1996. [Google Scholar]
- 10.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickett G G, Peabody D S. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res. 1993;21:4621–4626. doi: 10.1093/nar/21.19.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stockley P G, Stonehouse N J, Valegård K. Molecular mechanism of RNA phage morphogenesis. Int J Biochem. 1994;26:1249–1260. doi: 10.1016/0020-711x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 13.Sutton D W, Havstad P K, Kemp J D. Synthetic cryIIIA gene from Bacillus thuringiensis improved for high expression in plants. Transgen Res. 1992;1:228–236. doi: 10.1007/BF02524753. [DOI] [PubMed] [Google Scholar]
- 14.van Gemen B, van Beuningen R, Nabbe A, van Strijp D, Jurriaans S, Lens P, Kievits T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–168. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 15.WalkerPeach, C., M. Winkler, D. DuBois, and B. L. Pasloske. Unpublished data.
- 16.Witherell G W, Gott J M, Uhlenbeck O C. Specific interaction between RNA phage coat proteins and RNA. Proc Nucleic Acids Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]