Abstract
Background:
Infancy is an important developmental period when the microbiome is shaped. We hypothesized that earlier antiretroviral therapy (ART) initiation would attenuate HIV effects on microbiota in the mouth.
Methods:
Oral swabs were collected from 477 children living with HIV (CLWH) and 123 children without (controls) at two sites in Johannesburg, South Africa. CLWH had started ART <3 years of age; 63% <6 months of age. Most were well-controlled on ART at median age 11 years when the swab was collected. Controls were age-matched and recruited from the same communities. Sequencing of V4 amplicon of 16S rRNA was done. Differences in microbial diversity and relative abundances of taxa were compared between groups.
Results:
CLWH had lower alpha diversity than controls. Genus-level abundances of Granulicatella, Streptococcus and Gemella were greater and Neisseria and Haemophilus less abundant among CLWH than controls. Associations were stronger among boys. Associations were not attenuated with earlier ART initiation. Shifts in genus-level taxa abundances in CLWH relative to controls were most marked in children on lopinavir/ritonavir regimens; with fewer shifts seen if on efavirenz ART regimens.
Conclusions:
A distinct profile of less diverse oral bacterial taxa was observed in school-aged CLWH on ART compared to uninfected controls suggesting modulation of microbiota in the mouth by HIV and/or its treatments. Earlier ART initiation was not associated with microbiota profile. Proximal factors, including current ART regimen, were associated with contemporaneous profile of oral microbiota and may have masked associations with distal factors like age at ART initiation.
Introduction
Advances in technology to characterize the “microbiome,” i.e. the microbial communities that live on and in our bodies, has led to explosive growth in our understanding of how these microorganisms influence health and disease, including HIV.1 Infancy is an important developmental period during which the microbiome is shaped in interaction with breastfeeding and other infant, maternal and environmental factors.2 Sampling from the oral cavity is a convenient and accessible method to describe the microbiota profile. Many studies have investigated associations between oral microbiota and HIV status with most finding shifts in abundances of oral bacterial taxa in adults and children with HIV compared those without.3–11
Perinatally-acquired HIV infection may interfere with healthy microbiome-driven immune development. Early diagnosis and initiation of antiretroviral therapy (ART) as soon as possible has been advised for some time based on the results of the Children with HIV Early antiRetroviral (CHER) trial.12 This trial randomized infants diagnosed in the first few weeks of life to either immediate or delayed ART observing significant reductions in mortality with immediate ART.12 In addition, several groups have shown strong associations between young age at ART initiation and smaller persisting viral reservoirs.13,14
We hypothesized that introducing ART during early life, a critical developmental time window for microbiome development, would attenuate adverse effects of HIV on the microbiome in children living with HIV (CLWH). Here we test this hypothesis examining microbiota in the mouth of CLWH who initiated ART before and after 6 months of age relative to age-matched children who are uninfected.
Methods
Study Population:
A cohort of 553 CLWH was assembled at two sites: (1) Rahima Moosa Mother and Child Hospital (RMMCH), and (2) Perinatal HIV Research Unit (PHRU), Chris Hani Baragwanath Hospital, in Johannesburg, South Africa. Children had started on ART when under three years of age, mostly as part of clinical trials. At RMMCH, children had been participants in two trials evaluating switching from protease-inhibitor-based therapy to Non-Nucleoside analogue Reverse Transcriptase Inhibitor-based therapy; most from the trial that randomized children suppressed on lopinavir-ritonavir at 3–5 years of age to either remain on this regimen or switch to an efavirenz-containing regimen.15,16 At PHRU, children had been participants in CHER12 or had been enrolled in the Wellness services of the unit. All participants were of Black African ancestry consistent with the population who make use of the services at the two sites.
Between February 2013 and August 2014, 553 CLWH were enrolled into an observational cohort study when they were 5–9 years of age. As part of the observational study, children had HIV viral load and CD4+ T-cell counts and percentages monitored every 6 months and information about the child’s current ART regimen was verified and updated. At enrollment into the observational study, pre-ART characteristics, including age at ART initiation, pre-ART viral load and CD4+ T cell counts and percentages, and initial regimen were abstracted. The study was observational and no decisions about clinical management were made as part of the study. In parallel, a cohort of 150 age-matched children without HIV from the same communities were enrolled as controls. Clinical data were collected at each visit including chronic or acute medications (e.g. antibiotics), social and household information, disclosure, weight, height, and mid-upper arm circumference, and oral health. Details are described elsewhere.17–20
Oral sampling:
At the last observational visit, May 2017 to August 2018, when children were a median of 11 years of age (IQR 10–12 years) an oral swab sample was collected from 477 CLWH and 123 controls. The oral swab samples were collected by swabbing multiple locations in the mouth (tongue dorsum, hard palate, buccal mucosa, saliva etc.) to provide a comprehensive snapshot of the microbial communities in the mouth. All samples were immediately placed in coolers on ice and transported to the local laboratory where they were frozen and stored at −80°C within 2 hours of collection until shipment on dry ice to Dr. Grace Aldrovandi’s laboratory the US.
DNA extraction and 16S rRNA sequencing:
DNA from oral samples was extracted using the PowerSoil DNA kit according to the manufacturer’s instruction with the addition of a high temperature heating step and by using Lysing Matrix E tubes (MP Biomedicals, Burlingame, California, USA) which contains a mixture of beads for the bead-beating step since this improve lysis of bacterial and fungal cells. Samples were extracted in batches containing participant cases and controls as well as positive (whole cell bacterial mock community (in-house) and ZymoBionics Microbial Community DNA Standard positive control (Zymo Research, Tustin, CA)) and negative laboratory controls (extraction buffer without sample and PCR no-template-controls) in each batch. Barcoded primer pairs targeting the V4 region of the bacterial 16S rRNA gene were used to amplify extracted DNA and negative controls using the Earth Microbiome Project protocol.21 Amplicons were sequenced on an Illumina MiSeq desktop sequencer using 2×150bp v2 chemistry per manufacturer’s specifications.
Microbiome data analysis and quality control:
Reads were de-multiplexed using QIIME22 and then processed to generate an amplicon sequence variant (ASV) table using DADA2.23 Taxonomic classification was performed using the RDP naïve Bayes classifier with training set 18.24,25 Contaminant sequences were removed using the ‘decontam’ R package version 1.6.0 with default parameters.
Statistical Analysis:
We compared diversity indices and abundances of different taxa between CLWH and controls. Alpha diversity was measured with Simpson, Shannon, Chao1, and Fisher diversity metrics. Beta diversity was evaluated using Bray-Curtis distance.26 We also compared groups using standard distance-based approaches (e.g. permutational multivariate ANOVA: PERMANOVA27) and visualized these groupings using principal coordinates analysis (PCoA) plots. Standard statistical methods (two-sample t-tests and logistic regression) were utilized to compare microbial diversity and taxa abundances between groups with and without adjustment for age and sex.28 To test an a priori hypothesis that differences in diversity/taxa abundances by HIV status may be different in boys and girls, we tested group differences in indices and taxon abundances stratified by sex, and tested interaction terms between sex and taxon abundances using logistic regression. All p-values were adjusted for multiple testing using the Benjamini-Hochberg (BH) false discovery rate (FDR) method.29 Bonferroni-adjusted p-values were also reported. All statistical analyses were performed using R (version 4.2.0).
Results
Study population
Swab samples were collected from 477 CLWH at the two sites in Johannesburg and from 123 age-matched children without HIV. The mean age was 10.8 years at the time of swab collection. CLWH had been on ART a mean of 10.3 years with 63% having initiated ART under 6 months of age. Most (89%) had initiated ART with a regimen containing ritonavir-boosted lopinavir but at the time of sampling only 61% were on a regimen containing this drug. Most (91%) CLWH had undetectable viral load and had CD4 counts in the normal range (Table 1).
Table 1:
Description of the 600 children with oral swab samples recruited at two sites in Johannesburg, South Africa, 22 May 2017–16 August 2018.
| Children Living with HIV | Children Not infected | |
|---|---|---|
| Total | 477 | 123 |
| [min max] | [7.8 – 14.0] | [7.5 – 14.1] |
| 5 | 4 (<1%) | 0 |
| Female | 260 (55%) | 49 (40%) |
| Other | 11 (2%) | |
| Missing | 2 | |
| Unknown | 3 (2%) | |
| Mean (std) [Min, Max] | 927 (308) [165 to 2358] | |
| >730 | 4 (<1%) | |
| Mean (std) [Min, Max] | 201 (181) [24, 985] | |
| Mean (std) [Min, Max] | 10.3 (1.3) [6.2 to 12.7] | |
| Missing | 66 | |
| Mean (std) [Min, Max] | 26.1 (11.3) [1.4 to 56.1] | |
| Other | 5 (1%) |
The 600 study samples and 114 laboratory controls yielded 12,613 unique ASVs. One sample from a CLWH had 27 total reads and was excluded leaving 599 children (476 CLWH and 123 controls) with >3,000 reads per sample and 210 genus-level taxa were represented.
Child HIV status
Overall microbiome compositions were similar between CLWH and controls (Figure 1A), although a statistically significant difference was noted by PERMANOVA (p<0.001). CLWH had less diverse microbiomes than uninfected controls across all alpha diversity indices (Figure 1B).
Figure 1:
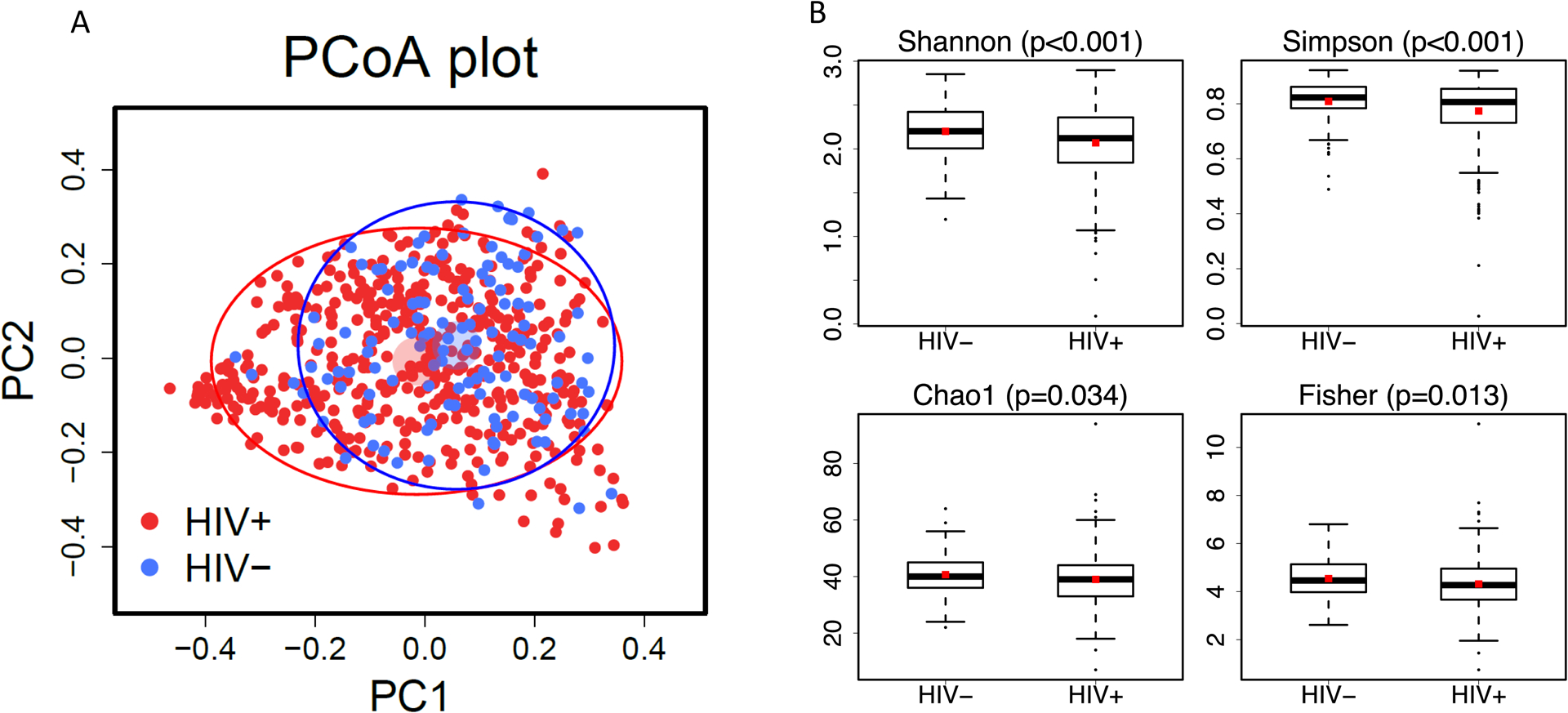
A. Principal coordinates analysis (PCoA) based on Bray-Curtis distances showing 476 children living with HIV as red dots and 123 children who are uninfected as blue dots. The lighter colored circles plot the means of the first two principal coordinates with arbitrary radius. The solid ellipses plot the variation of the principal coordinates with semi-major and semi-minor axes being two standard deviations of the principal coordinates. B. Alpha diversity indices (Shannon, Simpson, Chao1, Fisher) by child HIV status.
At the genus level, Granulicatella, Streptococcus, Gemella and Bulledia were significantly more abundant among CLWH and Neisseria significantly less abundant than in uninfected controls after multiple comparisons adjustment of 210 genus-level taxa (Figure 2A). Only 14 of the 210 genus-level taxa observed had mean abundances ≥1% in at least one of the two groups. Restricting the analysis to these 14, and combining all other taxa into one group, Granulicatella, Streptococcus, Gemella, Rothia, and Schaalia were significantly more abundant and Neisseria and Haemophilus less abundant among CLWH than controls (Figure 2B and C). Inspection of histograms of the abundances of these 15 taxa by HIV status suggested similar distributions across the two groups (Figure S1).
Figure 2:
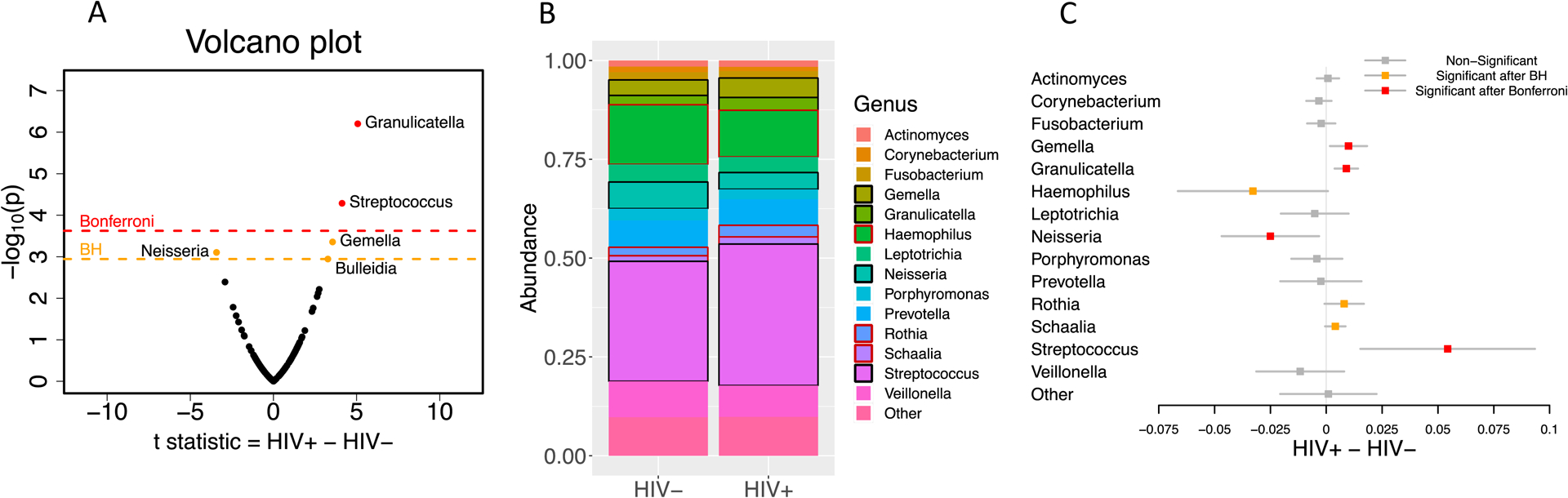
A. Volcano plot using t-statistic testing abundance differences between 476 children living with HIV and 123 children who are uninfected for all 210 genera. B. Taxa plot of abundances of the 14 genus-level taxa (plus a combined group of “other’) represented at ≥1% abundance in at least one of the two groups. Black box means significant after Bonferroni correction and red box means significant after Benjamini-Hochberg adjustment C. Mean differences between the groups in abundances of the 14 genus-level taxa (plus a combined group of “other”) with confidence intervals adjusted by Bonferroni correction.
Within the control group of children who are uninfected, no differences in alpha diversity or taxa abundances were observed between those born to mothers living with HIV (n=42) versus those whose mothers did not have HIV (n=78) (Figure S2). Mother’s HIV status was unknown for three uninfected children.
Sex differences
Alpha diversity was largely similar between male and female CLWH; whereas among controls, male children generally had higher alpha diversity than female children (Shannon p=0.074, Simpson p=0.221, Chao1 p=0.012, Fisher p=0.011; Figure S3). Likewise, lower alpha diversity associated with HIV status was most marked among male children (Within female: Shannon p=0.110, Within male: Shannon p=0.002; Figure S3).
Taxa abundance differences between CLWH and controls persisted after adjustment for sex and age. Granulicatella, Streptococcus and Gemella remained significantly more abundant and Neisseria and Haemophilus significantly less abundant among CLWH than controls adjusting for sex, age and multiple comparisons.
In sex-stratified analyses, Neisseria abundance was lower and Streptococcus, Granulicatella and Gemella was higher in male CLWH versus male children who are uninfected (significant after multiple comparisons adjustment); whereas among female children only Granulicatella was significantly higher. The interaction term comparing sex differences for Neisseria had the smallest raw p-value of 0.014 but was attenuated to p=0.216 after multiple comparisons adjustment.
Age at ART initiation
Just over a third (172/476 36.1%) of CLWH had started ART <90 days of age, 128 (26.9%) 90–180 days and 176 (37.0%) >180 days of age. Regardless of the timing of ART initiation, alpha diversity was consistently lower in CLWH than in controls i.e. no evidence of attenuation of the HIV association with earlier ART initiation (Figure S4).
Similarly, relative abundances of genus-level taxa in CLWH by the timing of ART initiation revealed no evidence of attenuation of associations with earlier ART initiation. In contrast, CLWH who started ART at young ages (<90 days) had the most significant differences in taxa abundances compared to uninfected children robust to multiple comparisons and adjustment for sex and age. Granulicatella, Streptococcus, Gemella, and Schaalia were significantly more abundant among CLWH who started ART <90 days and Corynebacterium, Haemophilus, Leptotrichia, Neisseria significantly less abundant than in controls after adjustment for sex and age. If ART was started after 180 days of age, only Granulicatella and Streptococcus abundances remained higher and Haemophilus less abundant compared to uninfected children (Figure 3). Of note Pseudomonas had mean abundance >1% among CLWH who started ART <90 days therefore there are 16 combined genus-level taxa for this comparison.
Figure 3:
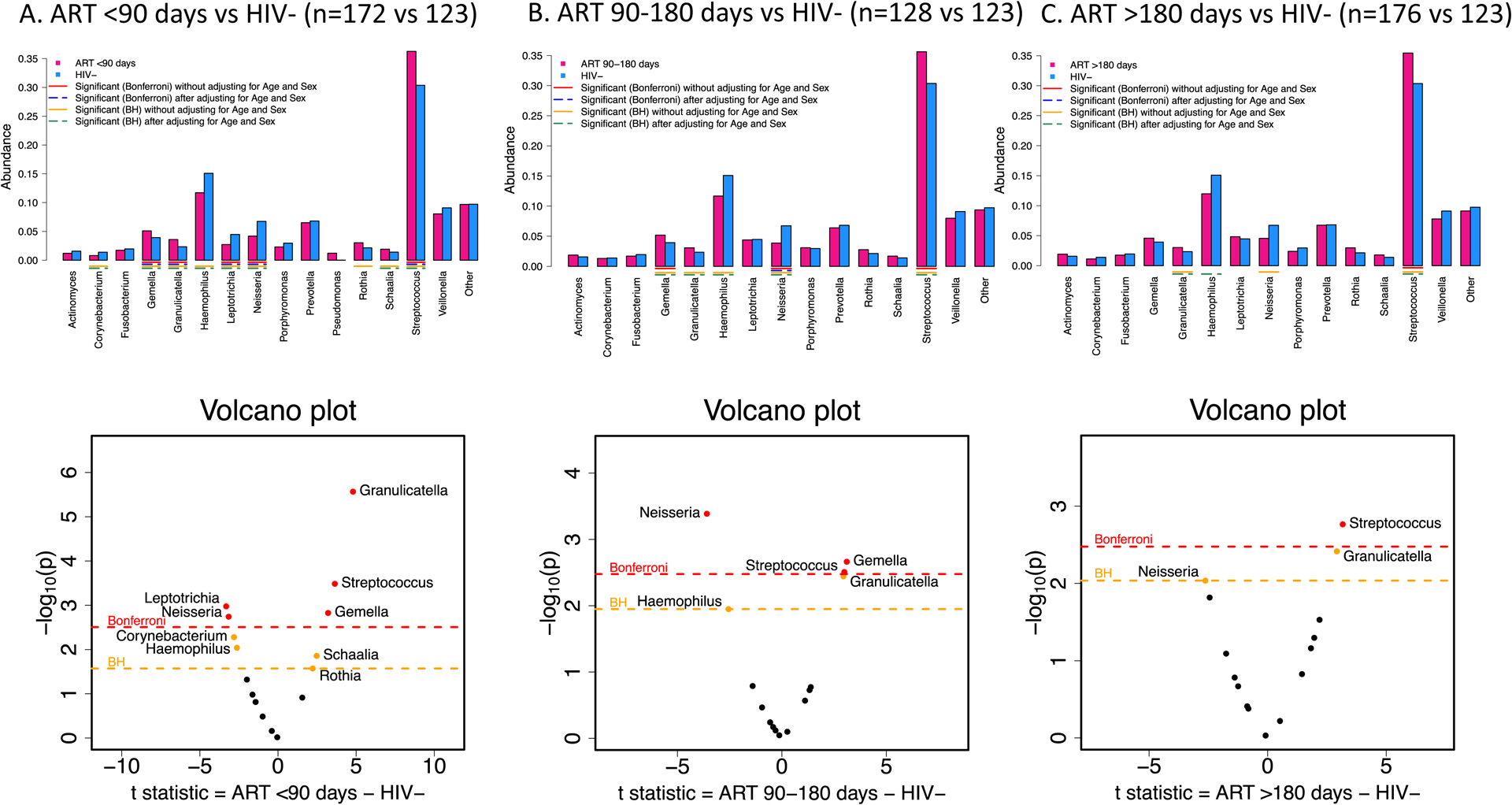
Barplots of mean abundances for each of the 15 Genus-level taxa (16 taxa in panel A with the inclusion of Pseudomonas) and corresponding volcano plots using t-statistic among A. 172 children living with HIV who started ART <90 days of age B. 128 children living with HIV who started ART 90–180 days of age and C. 176 children living with HIV who started ART >180 days of age compared to 123 children who are uninfected with significant differences between the groups before (red line – Bonferroni adjustment, yellow line Benjamini-Hochberg (BH) adjustment) and after (blue dashed line – Bonferroni adjustment, green lines BH adjustment) adjusting for age and sex of the children.
Current treatment regimen
Almost all children started on a protease-inhibitor-based ART regimen, mostly ritonavir-boosted lopinavir (LPV/R) and those who started on a ritonavir-based regimen were transitioned to LPV/r after 6 months of age. However, by the time of oral sample collection, 176 (37.0%) CLWH were receiving an efavirenz-based regimen and 289 (60.7%) remained on LPV/R-regimen (11 [2.3%] were on other regimens). Oral microbiota of those who remained on LPV/R-based regimens was less diverse compared than controls. CLWH who had switched to an efavirenz-based regimen had alpha diversity to controls (Figure 4C).
Figure 4:
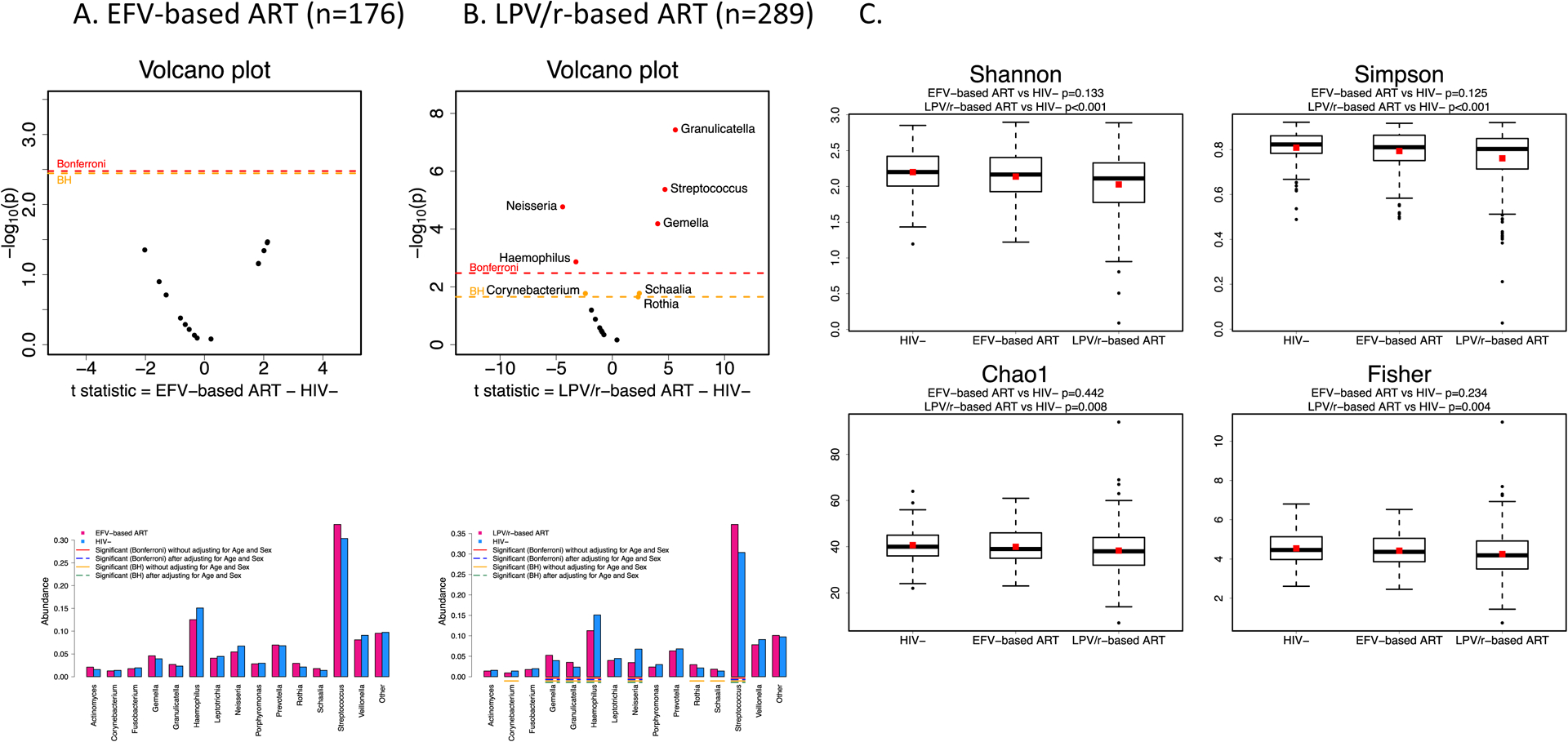
Barplot of mean abundances for each of the 15 Genus-level taxa and corresponding volcano plots using t-statistic comparing A. 176 children living with HIV who are currently on an Efavirenz-based ART regimen and B. 289 children living with HIV who are currently on a ritonvir-boosted lopinavir-based ART regimen compared to 123 children who are uninfected. C. Alpha diversity indices (Shannon, Simpson, Chao1, Fisher) by child HIV status and current treatment regimen.
CLWH who remained on LPV/R-based ART had significant shifts in relative abundances of genus-level taxa compared to uninfected controls. Granulicatella, Streptococcus, and Gemella were significantly more abundant among CLWH on LPV/R-based ART and Haemophilus and Neisseria were significantly less abundant than in controls. No differences in genus-level taxa abundances were observed between CLWH on efavirenz-based regimens and uninfected controls (Figure 4A/B).
Markers of HIV disease severity
Oral bacterial diversity was consistently lower among CLWH compared to controls regardless of pre-ART viral load or pre-ART CD4+ T-cell percent. Abundances of genus-level taxa in CLWH compared to controls were also reasonably consistent regardless of pre-ART viral load with a few exceptions. Granulicatella, Gemella, and Streptococcus were significantly more abundant among CLWH and Neisseria significantly less abundant than in controls irrespective of pre-ART viral load. However, greater abundances of Rothia and Schaalia were only observed in CLWH who had pre-ART viral load <750,000 copies/ml; and a lower abundance of Haemophilus was only observed in CLWH with pre-ART viral load ≥750,000 copies/ml (Figure S5A).
CLWH who had pre-ART CD4+ T-cell percentages closer to the normal range >25% had more marked shifts away from uninfected children than those who had had lower CD4+ T-cell percentages pre-ART. Granulicatella, Streptococcus, Gemella, and Rothia were significantly more abundant and Neisseria and Haemophilus less abundant among CLWH with higher pre-ART CD4+ T-cell percentages; genus-level abundances were largely similar to controls among CLWH with pre-ART CD4+ T-cell percentages <15 with the exception of lower abundance of Veillonella (Figure S5B).
Most (91%) CLWH had viral load <50 copies/ml) at the time of sampling. There were no differences in alpha diversity between those suppressed or not suppressed. Genus-level taxa abundances were largely similar between suppressed and unsuppressed, the only exception being a greater abundance of Corynebacterium in the suppressed.
Sixty percent of CLWH had consistently tested <400 copies/ml across all measurements over the prior 5 years. In later years, assays with sensitivity to <50 copies/ml were used, in earlier years, assays with sensitivity <400 copies/ml were used which is the reason for using the higher threshold. 11% of children had had one or more value between 400 and 1000 copies/ml over this time frame; and 29% had had one or more value >1000 copies/ml. Regardless of past viral control history, CLWH had lower alpha diversity than controls. Abundances of genus-level taxa in CLWH compared to controls were also reasonably consistent regardless of prior viral control history with most marked differences observed in the always suppressed group (Figure S6).
As a confirmatory analysis, we divided the CLWH into 2 groups based on their microbiota Bray-Curtis distances on the Principal Coordinates Analysis (PCoA) with Group 1 (n=64) being those with taxa most distinct from controls and Group 2 being the remainder (n=412). The only covariate that differed was current ART regimen with children taking LPV/R-based regimens over-represented in Group 1. Age, sex, age at ART start, and other markers of disease severity were distributed similarly in Group 1 versus 2 (Figure S7).
Discussion
We observed lower alpha diversity and shifts in the abundances of certain taxa in CLWH well-controlled on ART at a median of 11 years of age relative to uninfected children of the same age from the same communities. Granulicatella, Streptococcus, and Gemella were significantly more abundant and Neisseria and Haemophilus significantly less abundant in the mouth among CLWH than in children who are uninfected. These differences were robust to adjustment for multiple comparisons and remained after adjusting for age and sex. Moreover, the shifts in genus-level taxa abundances relative to uninfected controls were most marked in children on ART regimens containing lopinavir/ritonavir; with few shifts seen if on ART regimens containing efavirenz. There was no evidence of attenuation of the HIV association with younger age at ART initiation or with markers of less severe disease pre-ART.
Multiple studies have investigated associations between oral microbiota and HIV status.10 Oral pathologies are more common in persons with HIV, even those on ART, and oral sampling is a convenient and non-invasive method of assembling informative samples for microbiota characterization making this a feasible line of investigation. Most studies have been in adults and have generally found shifts in abundances of oral bacterial taxa in persons with HIV compared those without.3–9 While differences in approach make direct comparison across studies difficult, some consistencies with our findings are observed. For example, alpha diversity is generally lower in persons with HIV and increases in Streptococcus abundances and deceases in Neisseria abundances in persons with HIV relative to persons without HIV have been reported.5,8,9
Three studies of associations between oral microbiota and HIV in children could be located.11,30,31 One study was small and lacked sufficient numbers of controls for meaningful conclusions.30 A second did not find differences between older children and youth living with HIV compared to a control group of youth who had been born to mothers living with HIV but who had not acquired infection themselves.31 The third study found quite marked differences between infants with HIV and infants who are uninfected born to both women with and without HIV.11 This study had the strength of investigating the profile of oral microbiota at very young ages and found that both HIV infection, and particularly immune compromise, as well as HIV exposure without infection influenced the developing infant microbiome.11
Dental disease and oral microbiota have a bi-directional relationship.32 CLWH have been shown to have an increased prevalence of periodontal diseases33 and dental caries34 compared to children who are uninfected, which may explain our findings. However, a recent study which controlled for other factors including gingivitis, periodontal disease, caries and smoking reported that persons with HIV had persisting shifts in oral microbiota composition compared to persons without HIV even after adjustment for these factors.7 We selected controls matching by age and community to control for environmental factors that may confound associations with HIV status. Future longitudinal comprehensive dental clinic exams in conjunction with oral microbiota data are needed to determine the casual relationship between the two. Despite this, there remains the inevitable limitation in this type of study that factors unrelated to HIV or oral disease may, in fact, be the real drivers of the observed shifts in microbiota.
CLWH had initiated ART under 3 years of age and were mostly well-controlled on ART. Like other studies in this field, the separate effects of HIV infection from its treatments are almost impossible to untangle. Several strands of evidence, however, suggest that antiretroviral drugs themselves have an independent contribution.3,6,7,9 Our data showed that children on regimens containing lopinavir/ritonavir had the most marked shifts in taxa abundances compared to uninfected controls. Almost half of the participants in our study had been part of a clinical trial that randomized children who had been started on the recommended lopinavir/ritonavir-based regimen to either stay on this regimen or switch to an efavirenz-based regimen.15 We have previously reported better metabolic and bone outcomes associated with the switch to efavirenz in this cohort.35–39 Given accumulating data on the relevance of oral microbiota for metabolic disease,40 the role of oral microbiota in accounting for some of the metabolic complications associated with HIV and with some antiretroviral drugs is likely to be a fruitful line of investigation.
Our epidemiologic study was designed to provide a cross-sectional description of the abundances of different oral bacterial taxa in the mouth and was not designed to identify pathways by which they influence HIV pathogenesis. Nevertheless, two of the oral taxa, Streptococcus and Granulicatella, which we found to be more abundant in CLWH are known to degrade Hyaluronan (hyaluronic acid) an enzyme which may help reduce the virulence of infections.41 Abundance of Granulicatella in the mouth has also been linked to salivary levels of the proinflammatory cytokine IL-8.42 Neisseria, which we found to be less common in CLWH, has been associated with suppressed salivary levels of IL-6.42 Lower abundances of Neisseria in oral samples has been associated with more severe outcomes in influenza and COVID-19.43,44
We observed more marked shifts in the taxa abundances associated with HIV among male children although direct tests of effect modification were only significant for Neisseria. In the pediatric nutrition field, other studies have suggested sex-dimorphic microbiome associations with stronger associations between growth and perinatal factors like antibiotic use and pregnancy exposures among boys.45,46 Sex differences in response to ART in children are complex.47 In contrast to the suggestion in this study, our previous work in the same cohort found a stronger associations between metabolic complications and lopinavir in girls.47 The current study was not powered to adequately investigate these associations but it remains intriguing.
In contradiction to our original hypothesis, we did not observe the predicted attenuation in any HIV-related effects among children who initiated ART at younger ages. We also did not observe attenuation with any marker of less severe disease at the start of treatment. Proximal factors, including current regimen, were, however, associated with current profile of oral microbiota. Many years of ART may overshadow associations with more distal factors like age at ART initiation. While there may be shifts in microbiota that occur with early treatment, at the older age that we studied here, these shifts are no longer detectable. Prospective studies at younger ages may be more informative.
In conclusion, a distinct profile of less diverse oral bacterial taxa was observed in school-age CLWH on ART versus age-matched children who are uninfected suggesting persisting interference of HIV and/or its treatments on microbiota in the mouth. However, attenuation of this association by earlier ART initiation was not detectable at this age.
Supplementary Material
Acknowledgements:
The study was supported in part by grants from the National Institute of Dental and Craniofacial Research (DE028135) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (HD073952 and HD073977). CTT is supported by the South African Research Chairs Initiative of the Department of Science and Innovation and National Research Foundation of South Africa (84177).
References
- 1.Koay WLA, Siems LV, Persaud D. The microbiome and HIV persistence: implications for viral remission and cure. Current opinion in HIV and AIDS 2018; 13(1): 61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaufin T, Tobin NH, Aldrovandi GM. The importance of the microbiome in pediatrics and pediatric infectious diseases. Current opinion in pediatrics 2018; 30(1): 117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navazesh M, Mulligan R, Pogoda J, et al. The effect of HAART on salivary microbiota in the Women’s Interagency HIV Study (WIHS). Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics 2005; 100(6): 701–8. [DOI] [PubMed] [Google Scholar]
- 4.Beck JM, Schloss PD, Venkataraman A, et al. Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. American journal of respiratory and critical care medicine 2015; 192(11): 1335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee PK, Chandra J, Retuerto M, Tatsuoka C, Ghannoum MA, McComsey GA. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PloS one 2018; 13(7): e0200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewy T, Hong BY, Weiser B, et al. Oral Microbiome in HIV-Infected Women: Shifts in the Abundance of Pathogenic and Beneficial Bacteria Are Associated with Aging, HIV Load, CD4 Count, and Antiretroviral Therapy. AIDS research and human retroviruses 2019; 35(3): 276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffen AL, Thompson ZA, Beall CJ, et al. Significant effect of HIV/HAART on oral microbiota using multivariate analysis. Scientific reports 2019; 9(1): 19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Dunlap DG, Qin S, et al. Alterations in Oral Microbiota in HIV Are Related to Decreased Pulmonary Function. American journal of respiratory and critical care medicine 2020; 201(4): 445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annavajhala MK, Khan SD, Sullivan SB, et al. Oral and Gut Microbial Diversity and Immune Regulation in Patients with HIV on Antiretroviral Therapy. mSphere 2020; 5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coker MO, Cairo C, Garzino-Demo A. HIV-Associated Interactions Between Oral Microbiota and Mucosal Immune Cells: Knowledge Gaps and Future Directions. Frontiers in immunology 2021; 12: 676669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coker MO, Mongodin EF, El-Kamary SS, et al. Immune status, and not HIV infection or exposure, drives the development of the oral microbiota. Scientific reports 2020; 10(1): 10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Violari A, Cotton MF, Gibb DM, et al. Early antiretroviral therapy and mortality among HIV-infected infants. The New England journal of medicine 2008; 359(21): 2233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn L, Paximadis M, Da Costa Dias B, et al. Age at antiretroviral therapy initiation and cell-associated HIV-1 DNA levels in HIV-1-infected children. PloS one 2018; 13(4): e0195514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uprety P, Chadwick EG, Rainwater-Lovett K, et al. Cell-Associated HIV-1 DNA and RNA Decay Dynamics During Early Combination Antiretroviral Therapy in HIV-1-Infected Infants. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2015; 61(12): 1862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murnane PM, Strehlau R, Shiau S, et al. Switching to Efavirenz Versus Remaining on Ritonavir-boosted Lopinavir in Human Immunodeficiency Virus-infected Children Exposed to Nevirapine: Long-term Outcomes of a Randomized Trial. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 2017; 65(3): 477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn L, Coovadia A, Strehlau R, et al. Switching children previously exposed to nevirapine to nevirapine-based treatment after initial suppression with a protease-inhibitor-based regimen: long-term follow-up of a randomised, open-label trial. The Lancet Infectious diseases 2012; 12(7): 521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murnane PM, Sigamoney SL, Pinillos F, et al. Extent of disclosure: what perinatally HIV-infected children have been told about their own HIV status. AIDS care 2017; 29(3): 378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M, Shiau S, Strehlau R, et al. Disclosure to South African children about their own HIV status over time. AIDS care 2021: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramteke SM, Shiau S, Foca M, et al. Patterns of Growth, Body Composition, and Lipid Profiles in a South African Cohort of Human Immunodeficiency Virus-Infected and Uninfected Children: A Cross-Sectional Study. Journal of the Pediatric Infectious Diseases Society 2018; 7(2): 143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel F, Thurman C, Liberty A, et al. Negative Diagnostic PCR Tests in School-Aged, HIV-Infected Children on Antiretroviral Therapy Since Early Life in Johannesburg, South Africa. Journal of acquired immune deficiency syndromes (1999) 2020; 83(4): 381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, Knight R. DNA extraction for streamlined metagenomics of diverse environmental samples. BioTechniques 2017; 62(6): 290–3. [DOI] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods 2010; 7(5): 335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods 2016; 13(7): 581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 2007; 73(16): 5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan B RDP taxonomic training data formatted for DADA2 (RDP trainset 18/release 11.5) https://zenodo.org/record/4310151#.Yz3Y23bMKUk. 2020. [Google Scholar]
- 26.Bray J, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs 1957; 27(4): 326–49. [Google Scholar]
- 27.Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley statsref: statistics reference online 2017; 10.1002/9781118445112.stat07841: 1–15. [DOI] [Google Scholar]
- 28.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal 2012; 6(3): 610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995; 57(1): 289–300. [Google Scholar]
- 30.Goldberg BE, Mongodin EF, Jones CE, et al. The Oral Bacterial Communities of Children with Well-Controlled HIV Infection and without HIV Infection. PloS one 2015; 10(7): e0131615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr JR, Huang Y, Lee KH, et al. Oral microbiota in youth with perinatally acquired HIV infection. Microbiome 2018; 6(1): 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varoni EM, Rimondini L. Oral Microbiome, Oral Health and Systemic Health: A Multidirectional Link. Biomedicines 2022; 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam PPY, Zhou N, Wong HM, Yiu CKY. Oral Health Status of Children and Adolescents Living with HIV Undergoing Antiretroviral Therapy: A Systematic Review and Meta-Analysis. Int J Environ Res Public Health 2022; 19(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhigbe P, Chukwumah NM, Folayan MO, et al. Age-specific associations with dental caries in HIV-infected, exposed but uninfected and HIV-unexposed uninfected children in Nigeria. BMC Oral Health 2022; 22(1): 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arpadi S, Shiau S, Strehlau R, et al. Metabolic abnormalities and body composition of HIV-infected children on Lopinavir or Nevirapine-based antiretroviral therapy. Archives of disease in childhood 2013; 98(4): 258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arpadi SM, Shiau S, Strehlau R, et al. Efavirenz is associated with higher bone mass in South African children with HIV. AIDS (London, England) 2016; 30(16): 2459–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiau S, Yin MT, Strehlau R, et al. Bone turnover markers in children living with HIV remaining on ritonavir-boosted lopinavir or switching to efavirenz. Bone 2020; 138: 115500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strehlau R, Coovadia A, Abrams EJ, et al. Lipid profiles in young HIV-infected children initiating and changing antiretroviral therapy. Journal of acquired immune deficiency syndromes (1999) 2012; 60(4): 369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su J, Shiau S, Arpadi SM, et al. Switch to Efavirenz Attenuates Lipoatrophy in Girls With Perinatal HIV. Journal of pediatric gastroenterology and nutrition 2021; 72(1): e15–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minty M, Canceil T, Serino M, Burcelin R, Tercé F, Blasco-Baque V. Oral microbiota-induced periodontitis: a new risk factor of metabolic diseases. Reviews in endocrine & metabolic disorders 2019; 20(4): 449–59. [DOI] [PubMed] [Google Scholar]
- 41.Yabuuchi S, Oiki S, Minami S, Takase R, Watanabe D, Hashimoto W. Enhanced propagation of Granulicatella adiacens from human oral microbiota by hyaluronan. Scientific reports 2022; 12(1): 10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar A, Kuehl MN, Alman AC, Burkhardt BR. Linking the oral microbiome and salivary cytokine abundance to circadian oscillations. Scientific reports 2021; 11(1): 2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye C, You M, Huang P, et al. Clinical study showing a lower abundance of Neisseria in the oral microbiome aligns with low birth weight pregnancy outcomes. Clinical oral investigations 2022; 26(3): 2465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demirci M Could Neisseria in oral microbiota modulate the inflammatory response of COVID-19? Oral diseases 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozyrskyj AL, Kalu R, Koleva PT, Bridgman SL. Fetal programming of overweight through the microbiome: boys are disproportionately affected. Journal of developmental origins of health and disease 2016; 7(1): 25–34. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Kim M, Paye S, Benayoun BA. Sex as a Biological Variable in Nutrition Research: From Human Studies to Animal Models. Annual review of nutrition 2022; 42: 227–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiau S, Kuhn L, Strehlau R, et al. Sex differences in responses to antiretroviral treatment in South African HIV-infected children on ritonavir-boosted lopinavir- and nevirapine-based treatment. BMC pediatrics 2014; 14: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


