Abstract
The bromodomain acts to recognize acetylated lysine in histones and transcription proteins and plays a fundamental role in chromatin-based cellular processes including gene transcription and chromatin remodeling. Many bromodomain proteins, particularly the bromodomain and extra terminal domain (BET) protein BRD4 have been implicated in cancers and inflammatory disorders and recognized as attractive drug targets. Although clinical studies of many BET bromodomain inhibitors have made substantial progress towards harnessing the therapeutic potential of targeting the bromodomain proteins, the development of this new class of epigenetic drugs is met with challenges, especially on-target dose-limiting toxicity. In this review, we highlight the current development of new-generation small molecule inhibitors for the BET and non-BET bromodomain proteins and discuss the research strategies used to target different bromodomain proteins for a wide array of human diseases including cancers and inflammatory disorders.
Introduction
Lysine acetylation is one of the major post-translational modifications (PTMs) of DNA-packing histones and plays an important role in regulation of gene transcription in chromatin.1 Histone lysine acetylation was originally proposed by Allfrey in 1964 to serve as an ‘on-off switch’ in gene transcription.2 Choudhary and colleagues using high-resolution mass spectrometry identified 3600 lysine acetylation sites in over 1750 proteins including histones and transcription-associated proteins in cells.3 Histone lysine acetylation by histone acetyltransferases (HATs), or deacetylation by histone deacetylases (HDACs) was first reported by Schreiber and Allis labs, respectively (Figure 1A). The discovery of bromodomain (BrD) as the acetyl-lysine (Kac) binding domain by Zhou and colleagues in 1999 (Figure 1B) provided direct experimental evidence that lysine acetylation not only reduces interactions between the histone and DNA by neutralizing the positive charges on histones, but also functions to mediate acetylation-mediated protein-protein interactions in gene transcription in chromatin.4 As the first histone reader, the bromodomain that is found in many chromatins and transcription-associated proteins has since been shown in many studies to play a fundamental role in gene transcription in cellular response to physiological and environmental cues.1 Hence, the bromodomains are well recognized as attractive drug targets for a wide array of human diseases ranging from cancers, autoimmune and inflammatory disorders to cardiovascular diseases. Numerous small molecule inhibitors1,5,6, and in more recent years, heterobifunctional chemical protein degraders7-12, known as proteolysis-targeting chimaeras (PROTACs)13 have been developed for bromodomain proteins. In this review, we describe the recent development of small molecule bromodomain inhibitors and their therapeutic applications.
Figure 1.
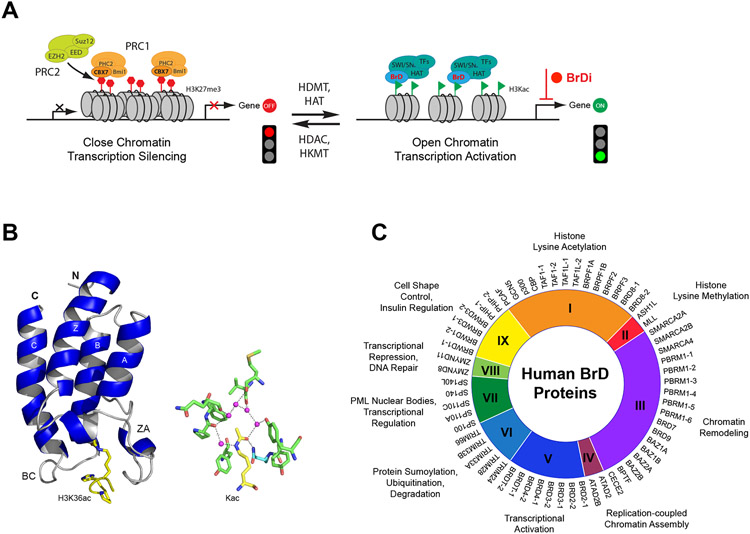
Bromodomain proteins function to regulate gene transcription in chromatin. (A) Scheme depicting the role of histone lysine acetylation (green flag) and methylation (red diamond) in regulation of gene transcription in chromatin. Abbreviations are bromodomain inhibitor (BrDi), histone acetyltransferase (HAT), histone lysine deacetylase (HDAC), histone lysine demethylase (HDMT), histone lysine methyltransferase (HKMT), Polycomb repressive complex (PRC) and transcription factor (TF). (B) Ribbon diagram of the 3D NMR structure of BRD4-BD1 in complex with a H3K36ac peptide (PDB: 2RNX)141. Right panel, side chains of key protein residues of GCN5 BrD involved in acetyl-lysine recognition of H4K16ac are highlighted and color-coded by atom type (PDB: 1E6I)142. The bound water molecules at the base of the acetyl-lysine binding pocket are shown as red spheres. (C) A wheel diagram illustrating classification of human bromodomain proteins based on major known functions.
The Bromodomain as the Histone Acetyl-Lysine Reader in Gene Transcription
The human genome encodes 61 bromodomains in 46 proteins with a diverse set of functions in cellular processes including gene transcription in chromatin (Figure 1C, Figure S1). Note that the bromodomain proteins are grouped based on their functions in cellular processes1 rather than the conventional grouping by sequence similarity of the bromodomains, which is only 25-30%. The latter would yield artificial grouping of the bromodomain proteins with completely different functions. The exitance of evolutionarily conserved bromodomains of about 110 amino acids was initially described by Tamkun et al. in 1992 in Drosophila gene Brahma and female sterile homeostasis proteins.14 The first three-dimensional structure of the bromodomain of human HAT transcriptional coactivator PCAF (p300/CBP-associated protein), determined using nuclear magnetic resonance (NMR) spectroscopy in 1999, revealed a unique structural fold of left-handed four-helix bundle (αZ, αA, αB, and αC) with two inter-helical loops (ZA and BC) that form the acetyl-lysine binding pocket at one end of the helical bundle (Figure 1B).4 The BC-loop contains a well-conserved Asn residue that forms a hydrogen bond between its amide nitrogen and carbonyl oxygen of the acetyl-lysine. The molecular function of the bromodomain as the acetyl-lysine binding domain was uncovered using 2D 1H-15N HSQC (heteronuclear single quantum coherence) NMR spectroscopy, showing that a lysine-acetylated histone H4 peptide caused NMR resonance perturbation of the PCAF bromodomain residues located in the ZA and BC loops in a peptide concentration-dependent and an acetylation sensitive manner.4 Notably, the long ZA-loop has high sequence variations among different BrDs that attribute to structural variability in terms of short helices, turns, and even hairpin insertions in the BrDs.15 The ZA loop is anchored to the αA helix via two conserved backbone hydrogen bonds, forming a cavity, dubbed the ZA channel, an important structural feature that is explored for optimizing the potency and selectivity of BrD inhibitors.
The bromomdomain proteins participate in a diverse set of cellular processes that include histone lysine acetylation and methylation, chromatin assembly and remodeling, transcription activation and repression, protein sumoylation, ubiquitnation and degradation, PML nuclear bodies, DNA repair, cell shape control, and insulin regulation (Figure 1C). Of all bromodomain proteins, the most well characterized is arguably BRD4 of the bromodomain and extra terminal domain (BET) family that consists of BRD2, BRD3, BRD4, and testis-specific BRDT. All four BET proteins, which share similar domain organization, and molecular functions in regulation of gene transcription in chromatin,16 contain two tandem BrDs (BD1 and BD2) followed by an extraterminal (ET) domain, hence the name.4 The BD1s and BD2s of the BET proteins have distinct preference for lysine-acetylated histones and transcription proteins. Specifically, the BD1s prefer binding to dual-acetylated Lys5 and Lys8 of histone H4 (H4K5acK8ac), a well-known transcription activation marker, whereas the BD2s are functionally versatile and can bind to acetylated transcription factors and cyclin T1 of positive transcription elongation factor b (p-TEFb) complex, consisting of cyclin T1 and CDK9. The latter activates p-TEFb from its inactive state in the ribonucleoprotein complex,17,18 which in turn phosphorylates Ser residues of RNA Pol II C-terminal motif (CTM) required for transcriptional elongation and productive gene transcription.
Bromodomain Inhibitors in Clinical Development
Much of our current knowledge of BET proteins as drug targets resulted from the studies of BRD4 using two BET-specific BrD inhibitors, JQ1 (3.1) and I-BET762 (3.2), developed by the researchers at the Structural Genomics Consortium (SGC) and GlaxoSmithKline (GSK), respectively, based on anticancer compounds reported in patents by Mitsubishi Tanabe Pharma. Studies with JQ1 initially focused on rare squamous NUT midline carcinoma (NMC) that is driven by BRD4-NUT fusion protein,19 and expanded to cancers with aberrant c-Myc expression.20 Further, studies by Tarakovsky and colleagues at GSK showed that I-BET762 acts as an immunosuppressant, supporting the notion of therapeutically targeting BRD4 in clinical applications beyond cancer.21
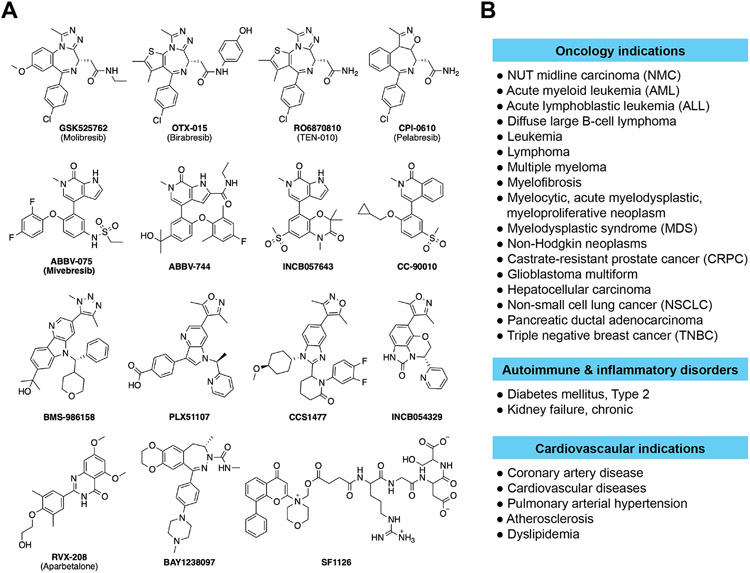
Many BET BrD inhibitors have been evaluated in human clinical studies for the treatment of hematologic malignancies and solid tumors. To date, about 50 clinical trials are in or completed phase 1 trials (Figure 2; Table S1). For instance, the BET inhibitor, OTX015 (MK-8682) has been investigated in phase I trials for patients with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), diffuse large B cell lymphoma (DLBCL), multiple myeloma (MM) (NCT01713582), as well as selected advanced tumors (NCT02259114). Of two GSK BET inhibitors, GSK525762 was evaluated in phase 1/2 studies in combination with fulvestrant in patients with hormone receptor-positive/HER2-negative (HR+/HER2−) advanced/metastatic breast cancer that had prior treatment with at least one line of endocrine therapy (NCT02964507). In open-label, dose escalation study, GSK2820151 was assessed for safety, pharmacokinetics (PK), pharmacodynamics (PD), and clinical activity in subjects 18 years or older with advanced or recurrent solid tumors (NCT02630251). Additionally, BMS-986158, a structurally distinct BET clinical candidate, was administered for a total of 10 doses in phase 1/2 studies for advanced cancers and hematologic indications including myelofibrosis (NCT02419417 and NCT03936465). Given its inhibitory activity of c-MYC expression that blocks tumor growth, ZEN-3694 was tested in phase 2 in patients with metastatic castration-resistant prostate cancer (mCRPC) in combination with Enzalutamide (NCT04986423). RO6870810 was administered subcutaneously for 21 or 14 days of 28- or 21-day cycles, respectively, in patients with NMC, DLBCL or other solid tumors with MYC deregulation (NCT01987362). In addition to monotherapy, Pelabresib (CPI-0610) was studied in combination with JAK inhibitor Ruxolitinib in phases 2/3 trials for myelofibrosis, and other hematologic malignant indications (NCT02158858 and NCT04603495). many other BET inhibitors including ABBV-075, ABBV-744, PLX51107, SF1126, INCB057643, INCB05432, CC-90010, CCS1477, and BAY1238097 have also been evaluated either alone or in combination therapy towards different forms of tumors and myelofibrosis. Finally, Apabetalone, a BET BD2-selective inhibitor of moderate affinity, has been evaluated in phase 2/3 studies in high-risk type-2 diabetes mellitus patients with coronary artery disease increases over the time to major adverse cardiovascular events (NCT02586155, NCT01728467 and NCT01067820).
Figure 2.
Bromodomain inhibitors that are being evaluated in human clinical trials. (A) Chemical structures of the BET bromodomain inhibitors being evaluated in human clinical trials. (B) Disease indications ranging from oncology, autoimmune and inflammatory disorders, to cardiovascular indications that are being studied with various BET bromodomain inhibitors in the clinical trials.
While much of these clinical studies of BET inhibitors provide valuable validation of BRD4 as a drug target for cancer and many other indications, the published clinical data revealed on-target dose-limiting toxicity (DLT) for this class of BET inhibitors. The common adverse effects are thrombocytopenia, diarrhea, fatigue, vomiting, anemia, and hyperbilirubinemia. Although much of these adverse effects are reversible, poor and short-term clinical responses limit clinical potential of using pan-BET inhibitors for the clinical treatment of cancer. Notably, the wide scope of these clinical studies has also illustrated a significant room for future development of next-generation target-selective and disease-specific BrD inhibitors to fulfill their clinical benefits.
Small Molecule Inhibitors of Bromodomains
The rapid growing knowledge of the BrD protein functions in many cellular processes associated with gene transcription and their implications in human diseases has greatly fueled the concerted efforts to develop new small molecule inhibitors for BrDs of the BET proteins as well as many other BrDs. Given the extensive coverage of early development of BrD inhibitors in the literature, in this review, we focus on the newly reported BrD inhibitors since 2019 according to the functional classification of target BrD proteins (Figure 1C).
(A). Pan BET Inhibitors
JQ1 (3.1)16 and I-BET762 (3.2) are two of the most recognized BET BrD inhibitors developed based on triazolodiazepine compounds of anticancer activities reported in patents22,23 (Figure 3). Note that the diazepine compounds have long been explored in drug discovery since 1974 and in use in the clinic as anxiolytic and sedative agents.24 This chemical scaffold is considered a privileged drug class that can be further tailored to optimize bioavailability, aqueous solubility, and metabolic stability as therapeutic molecules for BET BrDs.
Figure 3.
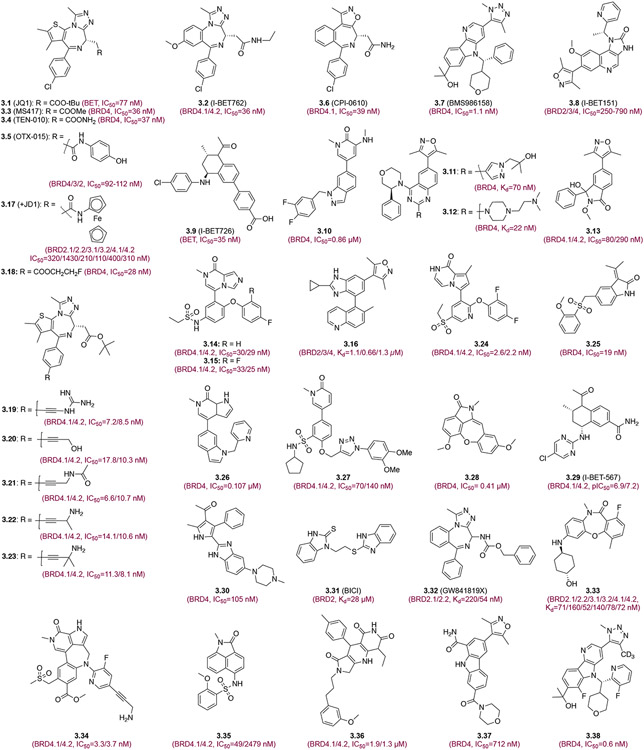
Pan-BET bromodomain inhibitors. Each inhibitor is depicted with its chemical structure and binding affinity to corresponding bromodomains.
JQ1 (3.1) binds in the acetyl-lysine binding pocket of BRD4 BD1 and BD2 with high affinity (Kd=50-100 nM), and suppresses c-MYC expression in many cells.16,20 As shown in the crystal structure, the triazole ring of JQ1 is the key in BRD4 recognition with one triazole atom forming a hydrogen bond to side-chain amide of Asn140, another nitrogen engaged in a water-mediated hydrogen bond to phenyl OH of Tyr97, and the triazole methyl group entering a hydrophobic pocket. Further, the thiophene ring in JQ1 is sandwiched between the “WPF” shelf and Leu92 in ZA loop, and the 4-chloro-phenyl ring in JQ1 occupies the large hydrophobic area between the “WPF” shelf and side chain of Leu146. Although JQ1 exhibits excellent anticancer proliferation activity in preclinical studies, its short in vivo half-life (one hour) limits its clinical applications.25 Because of JQ1’s chemical simplicity and anticancer potency, many diazepine analogs have been developed including I-BET762 (3.2), MS417 (3.3), TEN-010 (3.4), OTX-015 (3.5), and CPI-0610 (3.6), and some are advanced to clinical studies for the treatment of hematological malignancies and solid tumors (Figure 2). Additionally, non-diazepine BET inhibitors have been reported such as BMS-986158 (3.7), I-BET151 (3.8) and I-BET726 (3.9) that have nano-molar potency,26 inhibit cancer cell proliferation in vitro and in mouse xenograft models,27,28 as well as suppress transcriptional expression of proinflammatory cytokines such as IL-6, IL-1β, and INF-γ.29
Many new BET BrD inhibitors of distinct chemical scaffolds have been reported in the recent years (Figure 3). Pyridone derivative 3.10 inhibits BRD4 with decent potency and shows good anti-proliferative activity in MV4-11 cells by reducing c-Myc expression and arresting cells at G0/G1 phase.30 The quinazoline BET inhibitors, 3.11, 3.12 show significant efficacy in Kasumi-1 AML xenograft and in collagen-induced arthritis (CIA) models in mice, suggesting their therapeutic potential for treating cancer and inflammatory diseases.31 Compound 3.13 displays excellent BRD4 inhibition (IC50=80 nM) and anti-proliferation (IC50=365 nM) in human promyelocytic leukemia HL-60 cells.32 Yang et al developed two novel BET inhibitors 3.14 and 3.15 with 7-methylimidazo[1,5-a]pyrazin-8(7H)-one core. Compound 3.14 exhibited excellent BRD4 inhibitory activity (IC50=33 nM) and anti-proliferation potency (IC50=110 nM) in HL-60 cells,33 while 3.15 with modified phenyl substituent exhibited improved PK properties and good anti-tumor efficacy with no significant toxicity in mouse xenograft models of pancreatic cancer.34 Sperandio et al. reported a new BET BrD inhibitor 3.16 that exhibited good cellular potency, high permeability, and sufficient hepatic stability in in vivo efficacy studies.35
Yang et al. reported 3.17 (+)-JD1, a ferrocene analogue of JQ1,36 and radioactive fluorine-18-labeled derivative of JQ1 (3.18)37 that retain JQ1 inhibitory activity on c-Myc expression in cells, and possess excellent brain penetration and reasonable metabolic stability. Further, new JQ1 analogues 3.19-3.23 have been reported with nanomolar activity to BRD4.38 Li et al incorporated 8-methyl-pyrrolo[1,2-a]pyrazin-1(2H)-one to the scaffold of ABBV-075 and this compound 3.24 shows an 500-fold selectivity for the BET BrDs over other BrDs including p300 BrD.39 Orally administered 3.24 achieves a nearly complete inhibition of tumor growth (99.7%) accompanied by good tolerability.39 Indole-2-one derivative (3.25) exhibited remarkable BRD4 inhibitory activities (IC50=19 nM for BD1; 28 nM for BD2) and anti-proliferation potency in HT-29 and HL-60 cells (IC50=4.75 μM and 1.35 μM, respectively). Compound 3.25 has favorable PK properties, and arrests HT-29 cells at G1 phase and reduces c-Myc expression.40 Another class of indanone derivatives such as ZLD2218 (3.26) (indol-6-yl-pyrrolo[2,3-c]pyridin-7-one derivatives) was shown with promising inhibitory activity against BRD4, (IC50=107 nM). Importantly, ZLD2218 alleviated kidney injury and fibrosis in unilateral ureteral obstruction (UUO) mice at doses of 15 and 30 mg/kg/d for 8 consecutive days, with 30 mg/kg/d being comparable to JQ1 at 100 mg/kg/d.41 Phenylisoxazole sulfonamide derivative 3.27 exhibited robust inhibitory potency against BRD4-BD1/BD2 (IC50=70 and 140 nM, respectively). In addition, compound 3.27 significantly suppressed cell proliferation of leukemia cell lines and has better PK profile than JQ1.42 Yoo et al. developed pyrido-benzodiazepinone-based BRD4 inhibitor 3.28 (9-fluorobenzo [f]pyrido [4,3-b][1,4]oxazepin-10-one derivative) that inhibited BRD4 with high affinity and displayed excellent potency (IC50=0.41 μM) in imiquimod (IMQ)-induced psoriasis in mice.43 I-BET-567 (3.29), a BET inhibitor, exhibited high solubility, permeability, low off-target liabilities, and appropriate PK for in vivo studies of cancer and inflammatory disease models in mice, which were all made possible by enhanced LipE.44 The newly synthesized heterocyclic substance 3.30, 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivative, shows promising PK properties and antitumor efficacy in a mouse xenograft model through arresting cancer cells at G0/G1 phase and inducing apoptosis via blocking c-MYC expression.45 Of recently disclosed 3.31-3.38 that show remarkable potency towards BET BrDs, 3.38, a clinical compound derivative (BMS986158), exhibits sub-nanomolar potency, improved PK properties and solubility.46-50 These compounds exhibited satisfactory drug-like profiles including favorable metabolic stability in multiple species, good absorption, low clearance and excellent bioavailability in rats, in keeping with superior antitumor activity in MV4-11 mouse xenograft models.
(B). BET-BD1 Inhibitors
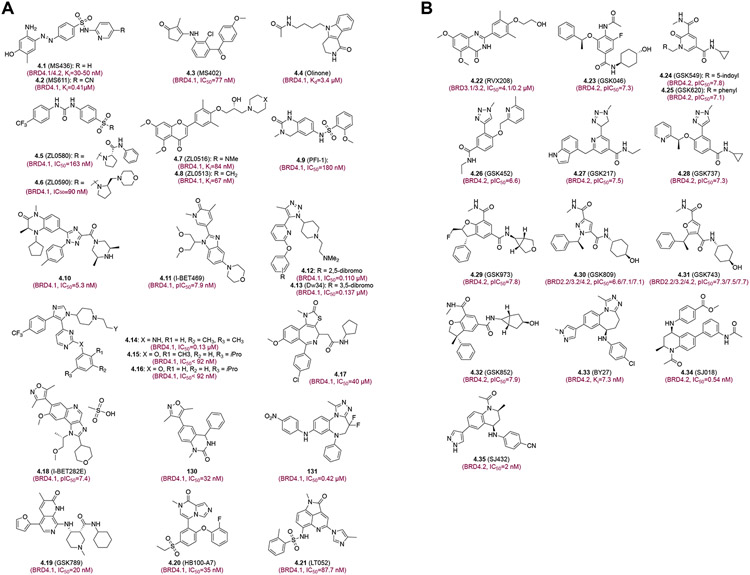
To alleviate the dose-limiting toxicity of pan-BET BrD inhibitors observed in clinical studies, major efforts have been devoted in the field to developing new selective BET BD1 or BD2 inhibitors. Only a few residues are different in the acetyl-lysine binding pocket between the BD1 and BD2. Among them are Gln85 (in ZA channel), Ile146 (gatekeeper residue) and Asp144 (in BC loop) in BRD4 BD1, which correspond to Lys374, Val435 and His433 in BRD4 BD2, respectively. These residues are crucial for the design of BD1 or BD2-selective inhibitors. In addition, the structural water molecules in the acetyl-lysine binding pocket also play a key role in ligand recognition. For BD1-selective inhibitors, the Zhou group made important contributions and discovered different scaffold inhibitors including MS436 (4.1), MS611 (4.2), MS402 (4.3), and Olinone (4.4) (Figure 4A) As the first low-nanomolar BD1 inhibitor, MS436 (110) with the diazobenzene scaffold exhibits 10-fold selectivity towards BRD4-BD1 (Ki=30–50 nM) over BRD4-BD2. The water-mediated hydrogen bond interactions between MS436 with Pro82, Gln85, Lys91, and Asn140 are key determinants to MS436 BD1-selectivity. In murine macrophages, MS436 was shown to effectively inhibit BRD4 transcriptional activity in NF-κB-directed production of proinflammatory cytokine IL-6 and nitric oxide.51 MS402 (4.3), a follow-up inhibitor in this series, displayed nanomolar inhibitory activity against BD1 (Ki=77 nM) with a 9-fold selectivity over BD2 of BET proteins. By blocking BRD4 occupancy at Th17 signature genes loci in chromatin, MS402 was shown to render Th17 gene transcriptional program and cell development, thereby ameliorating T cell-induced colitis in mice.52 Olinone (4.4) is selective for BET-BD1 (Kd=3.4 μM) with >100-fold selectivity over BD2. Olinone promotes the oligodendrocyte progenitor differentiation in primary mouse cells, whereas inhibition of BET BD1 and BD2 by JQ1 hinders oligodendrocyte progenitor differentiation. These studies suggest that selectively modulating individual BDs may enhance cell regenerative activities in aging and neurodegeneration.53
Figure 4.
Selective BET BD1 (A) or BD2 (B) inhibitors. Each inhibitor is depicted with its chemical structure and binding affinity to corresponding bromodomains.
ZL0580 (4.5), a BD1 inhibitor (IC50=163 nM) with 11-fold selectivity over BD2, was shown to cause HIV suppression through inducing repressive chromatin at HIV promoter and inhibiting Tat transactivation. Docking analysis revealed that the phenylurea sulfonamide moiety of ZL0580 binds to BRD4 BD1 by extending to the region between WPF shelf and ZA channel, different from pan-BET inhibitor JQ1.54,55 In addition, ZL0590 (4.6) was identified as a potent, selective BRD4-BD1 inhibitor. The crystal structure revealed that ZL0590 binds to BRD4-BD1 at a non-acetyl-lysine binding site at the αB/αC interface interacting with BRD4 residues such as Glu151unique from other BET BrDs.56 Liu et al developed two BET inhibitors ZL0516 (4.7) and ZL0513 (4.8) with high affinity (IC50=67–84 nM) and good selectivity over other BrDs. Both compounds inhibited Toll-like receptor-induced inflammatory gene transcription and airway inflammation in murine models. Moreover, 4.7 and 4.8 demonstrated excellent DMPK properties including bioavailability >35%, aqueous solubility, metabolic stability, and weak-to-low CYP450 enzymes and hERG inhibitory effects.57 Compound 4.9 (PF-1) is reported as a BRD4 BD1 inhibitor that is structurally orthogonal to triazolodiazepines and isoxazoles and has demonstrated antiproliferative and anti-inflammatory activity.58 The arylquinoxaline derivative 4.10 was shown to have balanced activity, stability, and antitumor efficacy with high selectivity towards BRD4-BD1 over other BD1 and BD2 of the BET proteins.54 Dimethylpyridone benzimidazole derivative I-BET469 (4.11) possesses favorable oral PK properties, displays activity in vivo, and is projected to have a low human efficacious dose.59 Cui et al. described new triazole-based inhibitors 4.12 and 4.13 (DW34) that have excellent affinity to BRD4-BD1 (Kd=12 and 6.4 nM, respectively) and 9–33 fold selective for BRD4-BD1 over other BET BrDs, and can effectively suppress c-MYC expression in MM.1S cells and down-regulate the expression of inflammatory genes such as IL-8 in TNF-α-stimulated human airway epithelial A549 cells.60 Additionally, compounds 4.14-4.16 were reported as potent and selective BRD4-BD1 inhibitors with 500-fold selectivity over BRD2-BD1 and BRD4-BD2.61 While these inhibitors were unable to reduce c-MYC expression at low concentrations in multiple myeloma cells, they down-regulated transcriptional expression of IL-8 and chemokines in inflammation.62 An N-methylthiazolidone heterocyclic compound 4.17 contains a hydrophobic acetyl cyclopentanyl side chain that contributes to high affinity to BRD4-BD1 (IC50 =110 nM) and efficacy in suppressing proliferation of MV-4-11 cells.63 I-BET282E (4.18), a derivative of pan-BET inhibitor I-BET151 (3.8), has excellent selectivity for BRD4-BD1 and in vivo efficacy in a collagen-induced arthritis model.64 GSK reported GSK789 (4.19) that is arguably one of the most selective BET-BD1 inhibitors, with more than 1000-fold selectivity for BD1 over BD2, retains potent antiproliferative, anti-inflammatory, and immunomodulatory activities in vitro, and phenocopies pan-BET inhibitor responses in these readouts.65 HB100-A7 (4.20) was shown to exhibit robust inhibition against BRD4-BD1 (IC50=0.035 μM), and arrest PCa cells (BxPC3) at G0/G1 phase and induce cell apoptosis by regulating the expression of apoptotic genes.66 HB100-A7 has the capacity to inhibit the expression of BRD4 and its downstream effector c-MYCc, a key regulator of cell proliferation, differentiation and apoptosis.66 A pyrroloquinazoline derivative LT052 (4.21) is 100x more selective for BRD4-BD1 over BD2, and was shown to modulate BRD4/NF-kB/NLRP3 inflammatory signaling pathways, and improve gout arthritis symptoms in a rat model.67
(C). BET-BD2 Inhibitors
While RVX208 (4.21) (Figure 4B) is arguably the first BET-BD2 inhibitor that has been evaluated in clinical studies for various inflammatory disorders, it has modest affinity for BRD4 BD2. Several studies reported lead optimization of a fragment-like chemical hit to BET BD2-selective inhibitors that possess good physicochemical properties and 200-to-1000-fold selectivity over BD1s. Preston et al. reported a potent BET-BD2 inhibitor GSK046 (4.23) with pIC50 >7 for BET BD2s and >100-fold selectivity over the BD1s that inhibits MCP-1 cytokine release in a cellular and whole blood context.68 Two BD2-selective inhibitors GSK549 (4.24) and GSK620 (4.25) were reported by Seal et al. to have potent anti-inflammatory activity in human whole blood with excellent in vivo oral PK.69 Aylott et al described the approach to mitigating the genotoxicity risk of GSK046 (4.23) by replacing the acetamide functionality with a heterocyclic ring. They took a structure-based template-hopping and hybridization approach, and developed potent, selective, and bioavailable compounds 4.26 (GSK452), 4.27 (GSK217), and 4.28 (GSK737).70 Compound GSK973 (4.29) has good potency, selectivity and PK property as a BET BD2-selective inhibitor.71 The related pyrazole derivative GSK809 (4.30) and furan derivative GSK743 (4.31) exhibited further improved potency and selectivity owing to an added benzylic methyl group that makes favorable hydrophobic interactions with the ZA channel of the BD2, which was not accessible by the pyridone GSK973 (4.29).71 4.31 (GSK743) was noted with excellent selectivity, no obvious off-target liabilities, and good synthetic tractability.72 Insertion of a quaternary center into the 2,3-dihydrobenzofuran core blocked a key site of metabolism and improved the solubility. The lead bezofuran derivative 4.32 (GSK852) is 1000-fold selective and highly soluble with good in vivo PK in rats and dogs.73 Notably, Chen et al reported an exo-cyclic aromatic amine compound BY27 (4.33) that has 11-fold selectivity for BET BD2s over BD1s, as shown by Ki of 80 nM and 7.3 nM for BRD4-BD1 and BRD4-BD2, respectively.74 BY27 has unique structural feature and was shown to exert good tumor growth inhibition (67%) and less toxicity than pan-BET inhibitors in mice, which makes this compound a good starting point for the design and synthesis of next-generation BET BD2-selective inhibitors.74 Finally, tetrahydroquinoline derivatives 4.34 and 4.35 were reported to have nano-molar potency and 50-fold selectivity for BET BD2s, and excellent inhibitory activity in blocking c-MYC expression and proliferation of a set of pediatric cancer cell lines while being only mildly toxic to non-tumorigenic cells.75
(D). Bivalent BrD inhibitors
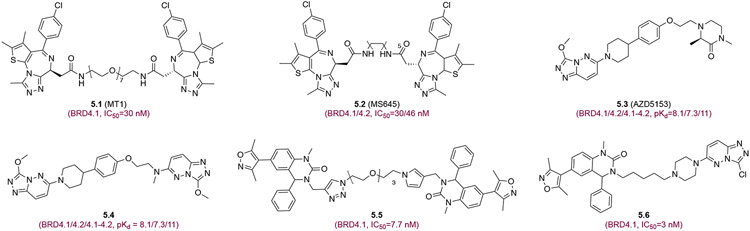
Arnold et al. described in a patent new BET BrD inhibitors that are capable of a bivalent binding mode to the tandem BET BD1-BD2.76 Minoru et al. reported that a thienodiazepine-based (JQ1) bivalent BET inhibitor MT1 (5.1) (Figure 5) is >100-fold more potent in cellular assays than the monovalent JQ1 and delayed leukemia progression in mice.77 Another thienodiazepine bivalent BrD inhibitor, MS645 (5.2), exerts spatially constrained tandem BD1-BD2 inhibition and affords sustained repression of BRD4 transcriptional activity in blocking proliferation of solid-tumor cells including a panel of triple-negative breast cancer (TNBC) cells by down-regulating the expression of pro-inflammatory cytokines and genes for cell-cycle control and DNA damage repair that are largely unaffected by monovalent JQ1.78 Rhyasen and colleagues characterized in-cell bivalent binding mode of AZD5153 (5.3) (triazolopyridazine) and demonstrated its inhibition of transcriptional programs of c-MYC, E2F, and mTOR required for AML, MM, and DLBCL malignant cell proliferation.79,80 In addition, di-traizolepyridazine derivative (5.4) showed high cellular potency for BRD4 binding and disruption of BRD4/mediator complex (EC50=100 pM).81 Yang et al. reported a dimeric 3,5-dimethylisoxazole bivalent BET inhibitor, 5.5 that inhibits c-MYC expression , induces HEXIMI production, and blocks proliferation of colorectal cancer HCT116 cells (IC50=162 nM) with 20-fold antiproliferative activity higher than monovalent JQ1.82 Finally, Fang developed a dimethylisoxazole-triazolopyridazine hybrid bivalent BET inhibitor 5.6 that possesses excellent inhibitory activity towards BRD4 (IC50=3 nM).83
Figure 5.
Bivalent BET bromodomain inhibitors. Each inhibitor is depicted with its chemical structure and binding affinity to corresponding bromodomains.
(E). Dual Target BrD Inhibitors
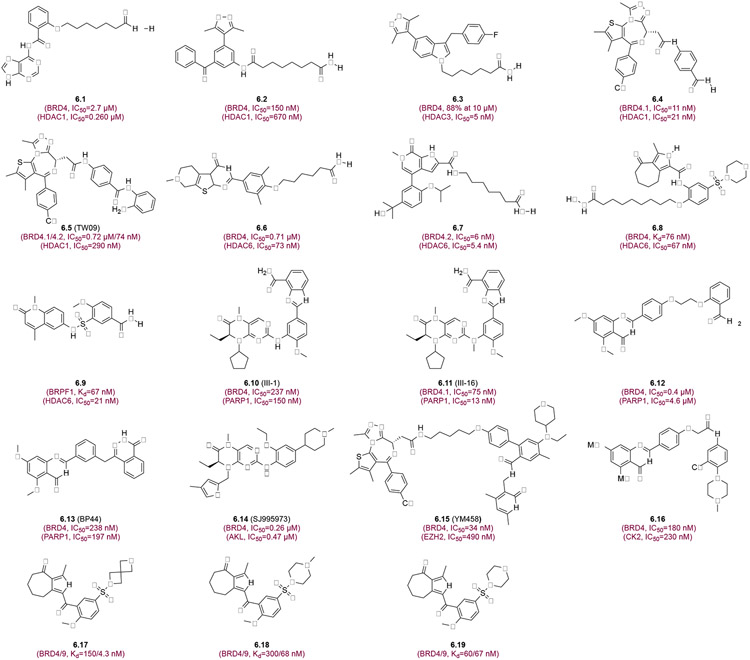
Another promising strategy for developing novel therapeutics reported is to create molecules that bind both a BrD and another protein target such as HDAC or kinase.84 Schönbrunn and colleagues reported in 2013 that CDK inhibitor Dinaciclib interacts with the Kac-binding pocket of BRDT, providing the first evidence that kinase inhibitors can be used to rationally design next-generation BrD/kinase inhibitors.85 Many dual BET/kinase inhibitors have been developed on the premise that poly-pharmacology inhibitors for multiple targets would have a more robust action.84 Because of HDACs’ role in regulation of gene transcription, it is logical to target BET proteins and HDACs for more efficient repression of oncogenic gene transcription and tumor cell proliferation.86 Purine-hydroxamide derivative (6.1) (Figure 6) that target BRD4 and HDAC1 was reported by Amemiya et al and shown to robustly inhibit HL-60 cell growth and induce differentiation.87 An arylisoxazole-based dual inhibitor 6.2 exhibited efficient activities towards BRD4 and HDAC1 and showed potent anti-proliferative activities against human leukemia cell line K562 and MV4-11.88 Further, 6.3, an indole substituent dual inhibitor for BRD4 and HDAC3 demonstrated impressive binding affinity against BRD4 (88% inhibition at 10 μM) and HDAC3 (IC50= 5 nM).89 Recently, Sheng et al. reported a JQ1-based hydroxamic acid compound 6.4 as a dual BRD4/HDAC1 inhibitor and demonstrated remarkable antitumor activity BRD4-BD1 (IC50=11 nM) and HDAC1 (IC50=21 nM) against Capan-1 cell lines. Importantly, compound 6.4 was shown to possess excellent in vivo antitumor efficacy in Capan-1 xenograft models (TGI=87.7%, 20 mg/kg, ip, bid), which was much more potent than JQ1 and Vorinostat used alone or in combination.90 Further, a JQ1/MS275-based dual inhibitor ortho-aminoanilide Tw09 (6.5) was reported to induce cell death and mitochondrial apoptosis in rhabdomyosarcoma (RMS) and PDAC cells.91,92 Pan et al. developed thieno[2,3-d]pyrimidine-based hydroxamic acid derivative 6.6 as new BRD4/HDAC dual inhibitor by combining RVX-208 and Vorinostat and showed that 6.6 exhibited nanomolar potency toward colorectal cancer, resulting in reduced c-MYC expression and increased histone H3 acetylation.93 Further, Chen et al. developed a novel HDAC6/BRD4 dual inhibitor (6.7), based on the selective BD2 inhibitor ABBV-744, which exhibited synergistic effects on inducing G0/G1 arrest and apoptosis in AML MV-4-11 cells.94 Another XD14/Varinoistat hydride 6.8 from known pharmacophores of XD14 (BET inhibitor) and Varinostat (HDAC inhibitor), was show to inhibit BET and HDAC proteins with potent and balanced inhibition and induce significantly apoptosis.95 Another BRPF1/HDAC6/8 dual inhibitor 6.9 was developed by Ghazy et al., which exhibited nanomolar potency towards both target proteins, but a weak effect on AML cells due to poor permeability.96 Because BRD4 plays a critical role in DNA repair, inhibition of BRD4 shows synthetic lethality with poly-(ADP-ribose) polymerase-1 (PARP1) inhibitors due to homologous recombination (HR) DNA repair deficiency, providing a strategy for cancer treatment. Wang et al. reported selective dual PARP/BRD4 inhibitors III-1 (6.10) and III-16 (6.11), and the latter showed favorable synergistic antitumor efficacy in pancreatic cancer cells and xenografts in mice by arresting cell cycle progression, inhibiting DNA damage repair, and promoting autophagy-associated cell death.97 dual BRD4/PARP1 inhibitors 6.12 and 6.13 (BP44) were designed with the conception of synthetic lethality,98 and 6.13 was shown to inhibit homologous recombination in TNBC cells and trigger synthetic lethality, leading to cell cycle arrest and cancer cell apoptosis.99
Figure 6.
Dual-target bromodomain inhibitors. Each inhibitor is depicted with its chemical structure and binding affinity to corresponding bromodomains and other protein targets.
TG101209 and BI-2536, originally reported as kinase inhibitors, were found to have strong inhibitory activity against BRD4 through binding to the Kac binding pocket, providing a rationale for the development of dual-target inhibitors.100,101 Concomitant inhibition of anaplastic lymphoma kinase (ALK) and BRD4 is a potential therapeutic strategy for targeting two key oncogenic drivers that co-segregate in a significant proportion of high-risk neuroblastoma patients with mutation of ALK and amplification of MYCN. Exploring a structure-based design, Watts et al. reported their development of compound (R)-2-((2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl)amino)-7-ethyl-5-methyl-8-((4-methylthiophen-2-yl)methyl)-7,8-dihydropteridin-6(5H)-one (6.14) as an ALK/BRD4 dual inhibitor, which possess balanced inhibitory activities for ALK (IC50=17 nM) and BRD4 (Kd=44 nM).102 EZH2 of Polycomb repression complex 2 (PRC2) is histone lysine transferase responsible for histone H3 Lys27 trimethylation (H3K27me3) and PRC2-medidated gene transcriptional silencing. Guo et al. discovered YM458 (6.15), a potent EZH2/BRD4 inhibitor, which exhibited a remarkable antiproliferative capacity against 11 solid cancer cell lines tested.103 YM458 through its inhibition of EZH2/BRD4 reduced the expression of anti-apoptotic proteins Bcl-2 and Bcl-xl, reversed the expression of H3K27ac, and increased the expression of tumor suppressor p21.103 Inhibiting the phosphorylation of BRD4 by casein kinase 2 (CK2) is another potential strategy to overcome drug resistance in cancer therapy. Zhang et al developed a BRD4/CK2 dual inhibitor 6.16, which possesses potent and balanced activities against BRD4 (IC50=180 nM) and CK2 (IC50= 230 nM), and was shown to inhibit proliferation and induce autophagy-associated cell death of MDA-MB-231 and MDA-MB-468 cells.104 Finally, Hungle et al. reported BET/BRD7/9 dual inhibitors of spiro, piperidine, and morpholine derivatives 6.17, 6.18, and 6.19 that exhibited nanomolar affinity to BRD4-BD1 and BRD9 and synergistic activities in inhibiting breast cancer and melanoma cell proliferation.105
(F). Non-BET BrD Inhibitors
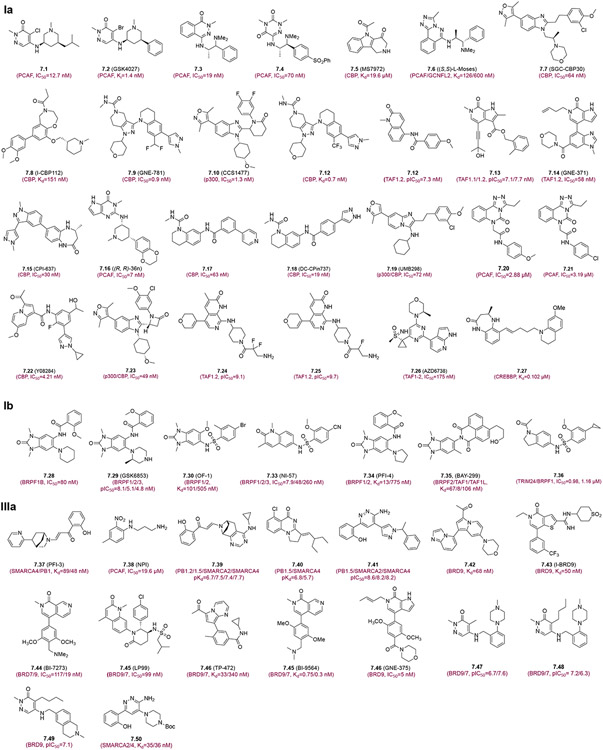
Non-BET BrD proteins play an important role in a variety of cellular processes in biology (see Figure 1C) and have been implicated in human diseases including cancer, inflammation, and cardiovascular disease. The HAT co-activators CBP/p300 (Figure 7, Group Ia) are likely the most investigated non-BET BrD targets for drug discovery.106 The non-BET BrD inhibitors reported prior to 2019 including 7.1-7.14 and their biological activity have been reviewed in detail previously.1 CPl-637 (7.15) is an excellent potent and selective CBP/p300 BrD inhibitor (IC50 of 30 nM for CBP, 51 nM for p300; >700-fold selectivity over other BrDs), and was shown to suppress MYC expression with EC50 of 0.6 μM, which is more effective than CBP30 (7.7) (>20 μM) and I-CBP112 (7.8) (2.7 μM).107 Huang et a/. developed the pyrrolpyrimidinone analog (R, R)-36n (7.16), which is most active against PCAF BrD (IC50=7 nM) with some activity for GCN5 and BPTF (FALZ) BrDs, but shows minimal or no activity against the other 29 BrDs and 422 kinases tested.108 Using structure-based drug design, Luo et al. developed tetrahydroquinolin derivative DC-CPin-711 (7.17)109 and DC-CPin734 (7.18) as a potent and selective CBP BrD inhibitors (IC50= 63.3 nM, 19.5 nM, and >150 and >400-fold selectivity over other BrDs, respectively) that effectively inhibit c-MYC expression.110 Muthengi et al. reported a dimethyl isoxazole-attached imidazopyridine UMB298 (7.19) as a potent CBP/p300 BrD inhibitor (IC50=72 nM).111 Triazoloquinazoline derivatives (7.20 and 7.21) were reported as potent PCAF inhibitors with significant cytotoxicity in that they can arrest cell cycle at G2/M phase and induce apoptotic cell death of PC3 PCa cells.112 Xiang and colleagues developed a 1-(indolizin-3-yl)ethan-1-one compound (7.22) as selective CBP BrD inhibitor that exhibits good liver microsomal stability and PK properties, and inhibits PCa cell growth and colony formation.113 7.23, a novel CBP30-based compound, was developed using bioisosterism and conformational restriction strategy and demonstrated more potent inhibitory activity (IC50=49 nM) against p300 BrD and anti-proliferative activity in a panel of cancer cell lines than CBP-30 (7.9) through inhibition of c-MYC expression and inducing G1/Go phase arrest and apoptosis in OPM-2 cells.114 Compounds 7.24 and 7.25, reported as TAF1(2) binding pendant amines, exhibit excellent potency (pKi=9.1 and 9.7) and >1000-fold selectivity over BRD4, BAZ2B, CECR2, and BRPF1 BrDs.115 The ATR kinase inhibitor 7.26 (AZD6738, Ceralasertib) displays significantly higher binding affinity for TAF1-2 BrDs with IC50=175 nM.116 Brand et al. reported a high-affinity inhibitor 7.27 for CBP/p300 BrDs that has a selectivity of >100-fold over the BET BrDs.117 This compound was shown to significantly inhibit CBP/P300 BrDs in HCT116 colon cancer cells, reducing c-MYC expression and acetylation levels at H3K18 and H3K27.117
Figure 7.
Non-BET bromodomain inhibitors. The bromodomain inhibitors are grouped based on the classification of human bromodomain proteins as in Figure 1. Each inhibitor is depicted with its chemical structure and binding affinity to corresponding bromodomains.
Several benzimidazolone-based compounds, developed from hits of high-throughput chemical screening, were reported as potent and selective BrD inhibitors of BRPF1B (Figure 7, Group lb). These include compound 7.28 (IC50=80 nM)118, GSK6853 (7.29) (pIC50=8.1nM)119, OF-1 (7.30) (Kd=101 nM)120 Nl-42 (7.31) (IC50=7.9 nM)121 and PFl-4 (7.32) (Kd=13 nM)120. The most potent compound BAY-299 (7.33) is a dual inhibitor with excellent selectivity towards BRPF2, TAF1-BD2 and TAF1L-BD2 (IC50=67 nM, 8 nM and 106 nM, respectively).122,123 Xinag and colleagues reported a potent TRIM24/BRPF1 dual BrD inhibitor 7.34 (1-(indolin-1-yl)ethan-1-ones) (IC50=0.98 and 1.16 μM, respectively), and demonstrated that this compound inhibits PCa tumor growth (50 mg/kg/day, TGI=53%) in mouse xenograft model without exhibiting significant toxicity.124
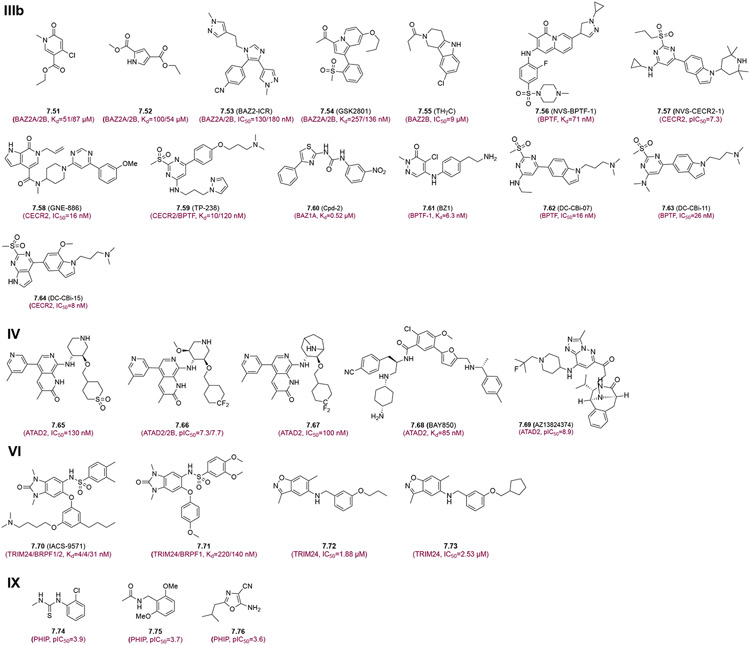
The BAF and PBAF chromatin remodeling complexes, whic are two of the most frequently mutated complexes in cancers, contain BRD9 and BRD7 (Figure 7, Group Illa), making them potentially attractive targets for oncology. BRD7/9 BrD inhibitors including 7.35-7.46 and their characterization were covered in our recent review.1 Recently, Clegg et al. reported pyridazinone-based BRD7/9 inhibitors 7.47, 7.48 and 7.49 that show superior selectivity and optimized solubility and cell-penetration.125 Inhibitor 7.50 was reported as a highly selective inhibitor within a panel of 25 BRDs and exhibits strong affinity against SMARCA2/4 and PB1(5) BrDs with Kd=35, 36 and 13 nM, respectively.126 Intriguingly, compound 7.50 blocked adipogenesis of 3T3-L1 murine fibroblasts by reducing the mRNA levels of key adipocyte differentiation markers such as PPARy, C/EBPa, and FABP4.126 Group IIIb inhibitors 7.51-7.59 were also reviewed previously (Figure 7. Group lllb)1. Yang and colleagues developed first selective inhibitor Cpd-2 (7.60) for BAZ1A that has Kd=0.52 μM, and exhibits good anti-proliferation activity against four cancer cell lines THP-1, ZR-75-30, BT474, and H1975 that have high expression levels of BAZ1A (IC50=5.08 μM, 4.29 μM, 10.65 μM, and 7.70 μM, respectively).127 Zahid et al. developed new pyridazinone-based BPTF inhibitor BZ1 (7.61) that has high affinity for BPTF (Kd= 6.3 nM) and >350-fold selectivity toward BRD7/9, and was shown with on-target BPTF activity in breast cancer cell lines.128 Based on the known BPTF inhibitor TP-238 (7.59), Lu et al identified the privileged fragments DC-BPi-07 (7.62) and DC-BPi-11 (7.63) that have high affinity for BPTF with 100-fold selectivity over other BrDs.129 These indole based potent inhibitors (7.62 and 7.63) were show to reduce proliferation, block cell cycle, and induce apoptosis, consistent with the parent compound TP-238 (7.59).129 Further, DC-CBi-22 (7.64) was reported with connecting with pyrrolo[2,3-d] pyrimidine with indole that displays high potency for CECR2 BrD (IC50=8.0 ± 1.4 nM) as well as an excellent 24.9-fold selectivity for CECR2 BrD over BPTF BrD.130
ATAD2 (Figure 7, Group IV) is an important cofactor for transcription factors such as E2F, MYC, estrogen receptors, and androgen receptors, and reportedly contributes to the progression of aggressive cancers such as TNBC, PCa, lung cancer, cervical cancer, endometrial carcinoma, and hepatocellular carcinoma.131,132 The first reported ATAD2 BrD inhibitor 7.65 by Bamborough et al. has nanomolar affinity, but low hydrophilicity and cell permeability limiting its use for cellular study.131 The authors developed next-generation potent and selectivity ATAD2 BrD inhibitor 7.66 using CF2 as a sulfone bio-isostere to explore the uniqueness of fluorine. This modification preserved the sulfone group’s favorable ATAD2 interactions and selectivity over BET BrDs while providing a dramatic improvement in logD and passive permeability.133 GSK8814 (7.67), another ATAD2 selective inhibitor from the same group, was synthesized by converting the piperidine ring to tropane. This inhibitor retains excellent ATAD2 potency and selectivity over BET BrDs.134,135 BAY850 (7.68), an interesting BrD inhibitor, exhibits potent activity only for ATAD2 but not to ATAD2B or other BrDs tested. It is an isoform-specific inhibitor as it specifically induces ATAD2 BrD dimerization and prevents interactions with lysine-acetylated histones in chromatin.136 AZ13824374 (7.69) is a recently reported potent and selective ATAD2 inhibitor with cellular target engagement and antiproliferative activity in a panel of breast cancer cells. With high permeability, compound 7.69 was found to be more than 100-fold selective against all bromodomain targets.137
TRIM (TRIM24, TRIM28, and TRIM33) and BRPF1 proteins have been reported to exert multiple functions in gene transcription in chromatin, including their ability to act as E3 ligases for ubiquitin and small-ubiquitin-like modifier (SUMO), making these proteins interesting targets for modulating epigenetic control of gene expression via bromodomain inhibition (Figure 7, Group VI). 7.70 (IACS-9571) and 7.71, potent and selective inhibitors with low nanomolar affinities for TRIM24/BRPF1, were developed using a novel binding mode. These high-quality chemical probes (7.70 and 7.71) for the evaluation of TRIM24 and/or BRPF1 bromodomain function in vitro and in vivo due to its excellent cellular potency (EC50=50 nM) and favorable pharmacokinetic properties (F=29%).138 Recently, two potent and selective compounds (7.72 and 7.73) have been developed as TRIM24 BrD inhibitors with IC50=1.88 μM and 2.53 μM, respectively. These two isoxazole derivatives (7.72 and 7.73) were shown to inhibit cell proliferation of lung cancer cells and PCa cells. These compounds can serve as lead compounds for further optimization to develop more potent TRIM24 BrD inhibitors as potential new therapy for intractable CRPC.139
There are several atypical BrDs found in the BrD family proteins, which lack the conserved Asn residue that is required for acetyl-lysine recognition by forming a hydrogen bond between its side chain amide and carbonyl oxygen of the acetyl group (see Figure 1B). Thus far, the biological function of these atypical BrDs remains elusive. PHIP, which has been implicated in disease progression of diabetes and cancers,140 contains two tandem BrDs, of which BD2 is an atypical BrD (see Figure S1). Cox and colleagues reported three different structural compounds such as thiourea derivative (7.74), arylamide (7.75) and oxazole derivative (7.76) (Figure 7, Group IX), which are highly selective towards PHIP-BD2 with sub micromolar activity.140 Three compounds can be developed into viable chemical probes for PHIP-BD2.
Conclusion and Future Perspectives
The bromodomains, as the acetyl-lysine histone readers, play a fundamental role in directing gene transcription in chromatin through acetylation-mediated protein-protein interactions in numerous cellular processes including but not limited to histone lysine acetylation, chromatin remodeling and transcription factor recruitment. Many bromodomain proteins, particularly BET protein BRD4 have been implicated in human cancers and inflammatory disorders, thus emerging as attractive new epigenetic drug targets. Although many BET BrD inhibitors have entered clinical trials and have had a substantial impact in the development of this new class of epigenetic drugs, their development is met with challenges, especially on-target dose-limiting toxicity that include fatigue, hypertension, thrombocytopenia, and gastrointestinal bleeding. While these adverse effects by pan-BET BrD inhibitors are reversible, they limit the therapeutic potential for this class of BET BrD inhibitors, particularly as drugs for potential chronic treatment of human disorders.
These challenges have motivated scientists to overcome the issues associated with BET BrD drugs with different tactics including intermittent drug dosing, developing disease specific BrD inhibitors, and combination therapy. Notably, recent studies have shown that BET-BD1 or BET-BD2 selective inhibitors are efficacious with indications including certain cancers as well as inflammation, metabolic diseases, and fibrosis with eliciting much less adverse toxicity than pan-BET BrD inhibitors. Thus, the potential advantage of BET-BD1 or -BD2 selective inhibitors is that while retaining antitumor activity, they have reduced toxicity and better tolerability, making them better drug candidates at least for certain diseases. Compared to BET BrD-selective inhibitors, the more circumscribed activity of CBP/p300 BrD-selective inhibitors may benefit the clinical development with properly selected patients. Additionally, dual-target inhibitors against BrDs and other well-recognized protein targets including HDACs and kinases, represent another promising therapeutic strategy. However, actual development of such drugs with poly-pharmacology activity remains a challenge as the two protein targets associated with certain diseases are structurally different. Nevertheless, when designed appropriately such dual-target inhibitors likely reduce the common liabilities that are associated with combination therapy including incomparability of PK properties, off-target toxicities, drug–drug interactions, and additive effects.
The recent preclinical studies indicate that the new generation selective BET BrD inhibitors as well as dual-target BrD inhibitors may potentially overcome the on-target dose-limiting toxicity observed commonly with the first-generation pan-BET BrD inhibitors, the final verdict awaits new clinical studies. Importantly, the rapidly growing number of non-BET BrD inhibitors developed in the recent years will greatly expand our knowledge about the vast biological functions of different BrD proteins in biology and diseases. It is also expected that many of these new BrD inhibitors being developed with novel chemical scaffold and excellent drug-like properties will catalyze the discovery and validation of more BrD proteins as new therapeutic targets for the treatment of a wide variety of human diseases including cancers and inflammatory diseases.
Supplementary Material
Acknowledgements
We would like to thank the current and past members of the Zhou Group for helpful discussion. This work was supported in part by the research grants from the National Institutes of Health (to M.-M.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
M.-M.Z. is a scientific founder, director and shareholder of Parkside Scientific Inc. The other author does not declare any competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ming-Ming Zhou reports a relationship with Parkside Scientific Inc. that includes: equity or stocks.
References:
- 1. Zaware N & Zhou MM Bromodomain biology and drug discovery. Nat Struct Mol Biol 26, 870–879 (2019). ** A recent review on the bromodomain biology and small molecule inhibitors.
- 2.Allfrey VG, Faulkner R & Mirsky AE ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proc Natl Acad Sci U S A 51, 786–794 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary C et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009). [DOI] [PubMed] [Google Scholar]
- 4. Dhalluin C et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999). ** First three-dimensional structure of the BrD and discovery of the BrD as the lysine-acetylated histone reader.
- 5.Xiong Y, Zhang M & Li Y Recent Advances in the Development of CBP/p300 Bromodomain Inhibitors. Curr Med Chem 27, 5583–5598 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Tang P, Zhang J, Liu J, Chiang CM & Ouyang L Targeting Bromodomain and Extraterminal Proteins for Drug Discovery: From Current Progress to Technological Development. J Med Chem 64, 2419–2435 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raina K et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 113, 7124–7129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zengerle M, Chan KH & Ciulli A Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem Biol 10, 1770–1777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J et al. Harnessing the E3 Ligase KEAP1 for Targeted Protein Degradation. J Am Chem Soc 143, 15073–15083 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai AC & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamkun JW et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561–572 (1992). [DOI] [PubMed] [Google Scholar]
- 15. Filippakopoulos P et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012). * This study provided a set of 3D structures of human bromodomains in complex with lysine-acetylated histone peptides.
- 16. Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). * Demonstration of the BET family protein BRD4 as a new anticancer drug target.
- 17.Yang Z, He N & Zhou Q Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28, 967–976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schröder S et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem 287, 1090–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisgrove DA, Mahmoudi T, Henklein P & Verdin E Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A 104, 13690–13695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). * This study showed that c-MYC is a direct target gene of BRD4.
- 21.Prinjha R & Tarakhovsky A Chromatin targeting drugs in cancer and immunity. Genes Dev 27, 1731–1738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi K et al. Thienotriazolodiazepine compound and medicinal use thereof. PCT/JP2006/310709 (2006). [Google Scholar]
- 23.Miyoshi S, Ooike S, Iwata K,Hikawa H & Sugahara K . Antitumor agents. US20100286127 A1. (2008). [Google Scholar]
- 24.Smith SG, Sanchez R & Zhou MM Privileged diazepine compounds and their emergence as bromodomain inhibitors. Chem Biol 21, 573–583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwood WW, Zejnullahu K, Bradner JE & Varmus H Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A 109, 19408–19413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson MA et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G, Smith SG & Zhou MM Discovery of Chemical Inhibitors of Human Bromodomains. Chem Rev 115, 11625–11668 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Shi J et al. Disrupting the Interaction of BRD4 with Diacetylated Twist Suppresses Tumorigenesis in Basal-like Breast Cancer. Cancer Cell 25, 210–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicodeme E et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010). * Demonstration of the BET family protein BRD4 as a new anti-inflammation drug target.
- 30.Rong J et al. Design, synthesis and biological evaluation of 3,5-dimethylisoxazole and pyridone derivatives as BRD4 inhibitors. Bioorg Med Chem Lett 29, 126577 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Yang SM et al. Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases. Bioorg Med Chem Lett 29, 1220–1226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P et al. 3-Hydroxyisoindolin-1-one derivates: Synthesis by palladium-catalyzed CH activation as BRD4 inhibitors against human acute myeloid leukemia (AML) cells. Bioorg Chem 86, 119–125 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Yang Y et al. Exploiting the 7-methylimidazo[1,5-a]pyrazin-8(7H)-one scaffold for the development of novel chemical inhibitors for Bromodomain and Extraterminal Domain (BET) family. Bioorg Chem 90, 103044 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Yang Y et al. Pharmacokinetics-Driven Optimization of 7-Methylimidazo[1,5-a]pyrazin-8(7H)-one as Novel BRD4 Inhibitors. Acs Med Chem Lett 10, 1680–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio D et al. Structure-guided discovery of a novel, potent, and orally bioavailable 3,5-dimethylisoxazole aryl-benzimidazole BET bromodomain inhibitor. Bioorg Med Chem 27, 457–469 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Hassell-Hart S et al. Synthesis and Biological Investigation of (+)-JD1, an Organometallic BET Bromodomain Inhibitor. Organometallics 39, 408–416 (2020). [Google Scholar]
- 37.Bai P et al. Design, Synthesis, and Evaluation of Thienodiazepine Derivatives as Positron Emission Tomography Imaging Probes for Bromodomain and Extra-Terminal Domain Family Proteins. J Med Chem 64, 14745–14756 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Sabnis RW Novel Bromodomain BRD4 Inhibitors for Treating Cancer. Acs Med Chem Lett 12, 951–952 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z et al. Discovery of 8-Methyl-pyrrolo[1,2-a]pyrazin-1(2H)-one Derivatives as Highly Potent and Selective Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitors. J Med Chem 63, 3956–3975 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Xu Y et al. Design, synthesis and biological evaluation of indole-2-one derivatives as potent BRD4 inhibitors. Eur J Med Chem 208, 112780 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Tao S et al. Discovery of indol-6-yl-pyrrolo[2,3-c]pyridin-7-one derivatives as bromodomain-containing protein 4 (BRD4) inhibitors for the treatment of kidney fibrosis. Eur J Med Chem 231, 114153 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Feng Z et al. Design, synthesis, and biological activity evaluation of a series of novel sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia. Bioorg Chem 111, 104849 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Yoo M et al. Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research. Molecules 26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphreys PG et al. Design, Synthesis, and Characterization of I-BET567, a Pan-Bromodomain and Extra Terminal (BET) Bromodomain Oral Candidate. J Med Chem 65, 2262–2287 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Kong B et al. Discovery of 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivatives as novel and potent bromodomain and extra-terminal (BET) inhibitors with anticancer efficacy. Eur J Med Chem 227, 113953 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Hill MD et al. Development of BET inhibitors as potential treatments for cancer: A search for structural diversity. Bioorg Med Chem Lett 44, 128108 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Sato M, Kondo T, Kohno Y & Seto S Discovery of benzo[f]pyrido[4,3-b][1,4]oxazepin-10-one derivatives as orally available bromodomain and extra-terminal domain (BET) inhibitors with efficacy in an in vivo psoriatic animal model. Bioorg Med Chem 34, 116015 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Jiang F et al. Discovery of novel small molecule induced selective degradation of the bromodomain and extra-terminal (BET) bromodomain protein BRD4 and BRD2 with cellular potencies. Bioorg Med Chem 28, 115181 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Gavai AV et al. Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design. J Med Chem 64, 14247–14265 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Ito T et al. Real-time imaging of histone H4K12-specific acetylation determines the modes of action of histone deacetylase and bromodomain inhibitors. Chem Biol 18, 495–507 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Zhang G et al. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem 56, 9251–9264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cheung K et al. BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc Natl Acad Sci U S A 114, 2952–2957 (2017). ** This study demonstrated that BRD4 BD1 inhibitor MS402 can selectively block Th17 cell differentiation, but not affecting other Th subtypes.
- 53. Gacias M et al. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol 21, 841–854 (2014). * This study reported that BRD4 BD1 inhinitor Olinone can promote oligodendrocyte lineage differentiation.
- 54.Hu J et al. Structure-Based Discovery and Development of a Series of Potent and Selective Bromodomain and Extra-Terminal Protein Inhibitors. J Med Chem 62, 8642–8663 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Niu Q et al. Structure-guided drug design identifies a BRD4-selective small molecule that suppresses HIV. J Clin Invest 129, 3361–3373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Z et al. Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site. J Med Chem 65, 2388–2408 (2022). * This study reported a novel class of BRD4 BD1 selective inhibitors.
- 57.Liu Z et al. Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation. J Med Chem 63, 5242–5256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fish PV et al. Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit. J Med Chem 55, 9831–9837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wellaway CR et al. Discovery of a Bromodomain and Extraterminal Inhibitor with a Low Predicted Human Dose through Synergistic Use of Encoded Library Technology and Fragment Screening. J Med Chem 63, 714–746 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Cui H et al. 4-Methyl-1,2,3-Triazoles as N-Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity. J Med Chem 64, 10497–10511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui H et al. Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*. Angew Chem Int Ed Engl 60, 1220–1226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui H et al. A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes. J Med Chem 65, 2342–2360 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q et al. Design, synthesis and biological evaluation of novel 6-phenyl-1,3a,4,10b-tetrahydro-2H-benzo[c]thiazolo[4,5-e]azepin-2-one derivatives as potential BRD4 inhibitors. Bioorg Med Chem 28, 115601 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Jones KL et al. Discovery of a Novel Bromodomain and Extra Terminal Domain (BET) Protein Inhibitor, I-BET282E, Suitable for Clinical Progression. J Med Chem 64, 12200–12227 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Watson RJ et al. GSK789: A Selective Inhibitor of the First Bromodomains (BD1) of the Bromo and Extra Terminal Domain (BET) Proteins. J Med Chem 63, 9045–9069 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Yang Y et al. Design, synthesis and biological evaluation of imidazolopyridone derivatives as novel BRD4 inhibitors. Bioorg Med Chem 29, 115857 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Jiang F et al. Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis. J Med Chem 62, 11080–11107 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Preston A et al. Design and Synthesis of a Highly Selective and In Vivo-Capable Inhibitor of the Second Bromodomain of the Bromodomain and Extra Terminal Domain Family of Proteins. J Med Chem 63, 9070–9092 (2020). [DOI] [PubMed] [Google Scholar]
- 69. Seal JT et al. The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor. J Med Chem 63, 9093–9126 (2020). * This study reported a new class of BRD4 BD2 selective inhibitors.
- 70.Aylott HE et al. Template-Hopping Approach Leads to Potent, Selective, and Highly Soluble Bromo and Extraterminal Domain (BET) Second Bromodomain (BD2) Inhibitors. J Med Chem 64, 3249–3281 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Preston A et al. GSK973 Is an Inhibitor of the Second Bromodomains (BD2s) of the Bromodomain and Extra-Terminal (BET) Family. Acs Med Chem Lett 11, 1581–1587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seal JT et al. Fragment-based Scaffold Hopping: Identification of Potent, Selective, and Highly Soluble Bromo and Extra Terminal Domain (BET) Second Bromodomain (BD2) Inhibitors. J Med Chem 64, 10772–10805 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Lucas SCC et al. Optimization of a Series of 2,3-Dihydrobenzofurans as Highly Potent, Second Bromodomain (BD2)-Selective, Bromo and Extra-Terminal Domain (BET) Inhibitors. J Med Chem 64, 10711–10741 (2021). [DOI] [PubMed] [Google Scholar]
- 74.Chen D et al. Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins. Eur J Med Chem 182, 111633 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Slavish PJ et al. Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer. Cancer Res 80, 3507–3518 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnold LD, Foreman KW, Jin M, Wanner J & Werner D Bromodomain ligands capable of dimerizing in an aqueous solution, and methods of using same. US patent 20140243286 (2014). (2014).
- 77.Tanaka M et al. Design and characterization of bivalent BET inhibitors. Nat Chem Biol 12, 1089–1096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ren C et al. Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth. Proc Natl Acad Sci U S A 115, 7949–7954 (2018). * This study reported a novel bivalent BRD4 BrD inhibitor highly effective in blocking cell proliferation of solid tumors including TNBC.
- 79.Bradbury RH et al. Optimization of a Series of Bivalent Triazolopyridazine Based Bromodomain and Extraterminal Inhibitors: The Discovery of (3R)-4-[2-[4-[1-(3-Methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phen oxy]ethyl]-1,3-dimethyl-piperazin-2-one (AZD5153). J Med Chem 59, 7801–7817 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Rhyasen GW et al. AZD5153: A Novel Bivalent BET Bromodomain Inhibitor Highly Active against Hematologic Malignancies. Mol Cancer Ther 15, 2563–2574 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Waring MJ et al. Potent and selective bivalent inhibitors of BET bromodomains. Nat Chem Biol 12, 1097–1104 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Yang Y, Fang L, Chen P, Zhang H & Zhou J Identification of 3,5-Dimethylisoxazole Derivatives as BRD4 Inhibitors for the Treatment of Colorectal Cancer. Acs Med Chem Lett 11, 2174–2181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang L et al. Discovery of 3,5-dimethylisoxazole derivatives as novel, potent inhibitors for bromodomain and extraterminal domain (BET) family. Bioorg Med Chem 39, 116133 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Carlino L & Rastelli G Dual Kinase-Bromodomain Inhibitors in Anticancer Drug Discovery: A Structural and Pharmacological Perspective. J Med Chem 59, 9305–9320 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Martin MP, Olesen SH, Georg GI & Schönbrunn E Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol 8, 2360–2365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan Z et al. Discovery of Thieno[2,3-d]pyrimidine-Based Hydroxamic Acid Derivatives as Bromodomain-Containing Protein 4/Histone Deacetylase Dual Inhibitors Induce Autophagic Cell Death in Colorectal Carcinoma Cells. Journal of Medicinal Chemistry 63, 3678–3700 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Amemiya S, Yamaguchi T, Hashimoto Y & Noguchi-Yachide T Synthesis and evaluation of novel dual BRD4/HDAC inhibitors. Bioorg Med Chem 25, 3677–3684 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Zhang Z et al. Targeting epigenetic reader and eraser: Rational design, synthesis and in vitro evaluation of dimethylisoxazoles derivatives as BRD4/HDAC dual inhibitors. Bioorg Med Chem Lett 26, 2931–2935 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Cheng G et al. Design, synthesis and biological evaluation of novel indole derivatives as potential HDAC/BRD4 dual inhibitors and anti-leukemia agents. Bioorg Chem 84, 410–417 (2019). [DOI] [PubMed] [Google Scholar]
- 90. He S et al. Potent Dual BET/HDAC Inhibitors for Efficient Treatment of Pancreatic Cancer. Angew Chem Int Ed Engl 59, 3028–3032 (2020). * This study reported a new class of BET/HDAC dual inhibitors.
- 91.Laszig S, Boedicker C, Weiser T, Knapp S & Fulda S The novel dual BET/HDAC inhibitor TW09 mediates cell death by mitochondrial apoptosis in rhabdomyosarcoma cells. Cancer Lett 486, 46–57 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Zhang X et al. Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma. Int J Cancer 147, 2847–2861 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Pan Z et al. Discovery of Thieno[2,3-d]pyrimidine-Based Hydroxamic Acid Derivatives as Bromodomain-Containing Protein 4/Histone Deacetylase Dual Inhibitors Induce Autophagic Cell Death in Colorectal Carcinoma Cells. J Med Chem 63, 3678–3700 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Chen J et al. Discovery of selective HDAC/BRD4 dual inhibitors as epigenetic probes. Eur J Med Chem 209, 112868 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Schaker-Hubner L et al. 4-Acyl Pyrrole Capped HDAC Inhibitors: A New Scaffold for Hybrid Inhibitors of BET Proteins and Histone Deacetylases as Antileukemia Drug Leads. J Med Chem 64, 14620–14646 (2021). [DOI] [PubMed] [Google Scholar]
- 96.Ghazy E et al. Design, synthesis, and biological evaluation of dual targeting inhibitors of histone deacetylase 6/8 and bromodomain BRPF1. Eur J Med Chem 200, 112338 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Wang SP et al. Discovery of Potent and Novel Dual PARP/BRD4 Inhibitors for Efficient Treatment of Pancreatic Cancer. J Med Chem 64, 17413–17435 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Chang X et al. Design, synthesis, and biological evaluation of quinazolin-4(3H)-one derivatives co-targeting poly(ADP-ribose) polymerase-1 and bromodomain containing protein 4 for breast cancer therapy. Acta Pharm Sin B 11, 156–180 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J et al. Discovery of 4-Hydroxyquinazoline Derivatives as Small Molecular BET/PARP1 Inhibitors That Induce Defective Homologous Recombination and Lead to Synthetic Lethality for Triple-Negative Breast Cancer Therapy. J Med Chem 65, 6803–6825 (2022). [DOI] [PubMed] [Google Scholar]
- 100.Ember SW et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem Biol 9, 1160–1171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ciceri P et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol 10, 305–312 (2014). * This study reported a new class of BRD4/kinase dual inhibitors.
- 102.Watts E et al. Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity. J Med Chem 62, 2618–2637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo Z et al. Design and Synthesis of Dual EZH2/BRD4 Inhibitors to Target Solid Tumors. J Med Chem 65, 6573–6592 (2022). [DOI] [PubMed] [Google Scholar]
- 104.Zhang J et al. Discovery of Novel Dual-Target Inhibitor of Bromodomain-Containing Protein 4/Casein Kinase 2 Inducing Apoptosis and Autophagy-Associated Cell Death for Triple-Negative Breast Cancer Therapy. J Med Chem 64, 18025–18053 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hugle M et al. 4-Acyl Pyrroles as Dual BET-BRD7/9 Bromodomain Inhibitors Address BETi Insensitive Human Cancer Cell Lines. J Med Chem 63, 15603–15620 (2020). [DOI] [PubMed] [Google Scholar]
- 106.Theodoulou NH, Tomkinson NC, Prinjha RK & Humphreys PG Progress in the Development of non-BET Bromodomain Chemical Probes. ChemMedChem 11, 477–487 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Taylor AM et al. Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637). Acs Med Chem Lett 7, 531–536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang L et al. Discovery of Pyrrolo[3,2-d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain. J Med Chem 62, 4526–4542 (2019). [DOI] [PubMed] [Google Scholar]
- 109.Chen Y et al. Design, synthesis, and biological evaluation of tetrahydroquinolin derivatives as potent inhibitors of CBP bromodomain. Bioorg Chem 101, 103991 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Bi X et al. Structure-based drug optimization and biological evaluation of tetrahydroquinolin derivatives as selective and potent CBP bromodomain inhibitors. Bioorg Med Chem Lett 30, 127480 (2020). [DOI] [PubMed] [Google Scholar]
- 111.Muthengi A et al. Development of Dimethylisoxazole-Attached Imidazo[1,2-a]pyridines as Potent and Selective CBP/P300 Inhibitors. J Med Chem 64, 5787–5801 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.El-Shershaby MH et al. From triazolophthalazines to triazoloquinazolines: A bioisosterism-guided approach toward the identification of novel PCAF inhibitors with potential anticancer activity. Bioorg Med Chem 42, 116266 (2021). [DOI] [PubMed] [Google Scholar]
- 113.Xiang Q et al. Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer. J Med Chem 65, 785–810 (2022). [DOI] [PubMed] [Google Scholar]
- 114.Chen Z et al. Discovery of novel benzimidazole derivatives as potent p300 bromodomain inhibitors with anti-proliferative activity in multiple cancer cells. Bioorg Med Chem 66, 116784 (2022). [DOI] [PubMed] [Google Scholar]
- 115.Clegg MA et al. Optimization of Naphthyridones into Selective TATA-Binding Protein Associated Factor 1 (TAF1) Bromodomain Inhibitors. Acs Med Chem Lett 12, 1308–1317 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karim RM et al. Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain. J Med Chem 65, 4182–4200 (2022). [DOI] [PubMed] [Google Scholar]
- 117.Brand M et al. Controlling Intramolecular Interactions in the Design of Selective, High-Affinity Ligands for the CREBBP Bromodomain. J Med Chem 64, 10102–10123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Demont EH et al. 1,3-Dimethyl Benzimidazolones Are Potent, Selective Inhibitors of the BRPF1 Bromodomain. Acs Med Chem Lett 5, 1190–1195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bamborough P et al. GSK6853, a Chemical Probe for Inhibition of the BRPF1 Bromodomain. Acs Med Chem Lett 7, 552–557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meier JC et al. Selective Targeting of Bromodomains of the Bromodomain-PHD Fingers Family Impairs Osteoclast Differentiation. ACS Chem Biol 12, 2619–2630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Igoe N et al. Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies. J Med Chem 60, 668–680 (2017). [DOI] [PubMed] [Google Scholar]
- 122.Zhou L, Yao Q, Ma L, Li H & Chen J TAF1 inhibitor Bay-299 induces cell death in acute myeloid leukemia. Transl Cancer Res 10, 5307–5318 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bouché L et al. Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains. J Med Chem 60, 4002–4022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiang Q et al. Discovery, optimization and evaluation of 1-(indolin-1-yl)ethan-1-ones as novel selective TRIM24/BRPF1 bromodomain inhibitors. Eur J Med Chem 236, 114311 (2022). [DOI] [PubMed] [Google Scholar]
- 125. Clegg MA et al. Application of Atypical Acetyl-lysine Methyl Mimetics in the Development of Selective Inhibitors of the Bromodomain-Containing Protein 7 (BRD7)/Bromodomain-Containing Protein 9 (BRD9) Bromodomains. J Med Chem 63, 5816–5840 (2020). * This study reported a new group of BRD7/BRD9 inhibitors.
- 126.Wanior M et al. Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis. J Med Chem 63, 14680–14699 (2020). [DOI] [PubMed] [Google Scholar]
- 127.Yang Z, Zhou Y & Zhong L Discovery of BAZ1A bromodomain inhibitors with the aid of virtual screening and activity evaluation. Bioorg Med Chem Lett 33, 127745 (2021). [DOI] [PubMed] [Google Scholar]
- 128.Zahid H et al. New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition. J Med Chem 64, 13902–13917 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu T et al. Discovery of High-Affinity Inhibitors of the BPTF Bromodomain. J Med Chem 64, 12075–12088 (2021). [DOI] [PubMed] [Google Scholar]
- 130.Lu H et al. Discovery of a highly potent CECR2 bromodomain inhibitor with 7H-pyrrolo[2,3-d] pyrimidine scaffold. Bioorg Chem 123, 105768 (2022). [DOI] [PubMed] [Google Scholar]
- 131.Bamborough P et al. Structure-Based Optimization of Naphthyridones into Potent ATAD2 Bromodomain Inhibitors. J Med Chem 58, 6151–6178 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Nayak A, Dutta M & Roychowdhury A Emerging oncogene ATAD2: Signaling cascades and therapeutic initiatives. Life Sci 276, 119322 (2021). [DOI] [PubMed] [Google Scholar]
- 133.Bamborough P et al. A Chemical Probe for the ATAD2 Bromodomain. Angew Chem Int Ed Engl 55, 11382–11386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yao D, Zhang J, Wang J, Pan D & He Z Discovery of novel ATAD2 bromodomain inhibitors that trigger apoptosis and autophagy in breast cells by structure-based virtual screening. J Enzyme Inhib Med Chem 35, 713–725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bamborough P et al. Aiming to Miss a Moving Target: Bromo and Extra Terminal Domain (BET) Selectivity in Constrained ATAD2 Inhibitors. J Med Chem 61, 8321–8336 (2018). [DOI] [PubMed] [Google Scholar]
- 136.Fernández-Montalván AE et al. Isoform-Selective ATAD2 Chemical Probe with Novel Chemical Structure and Unusual Mode of Action. ACS Chem Biol 12, 2730–2736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Winter-Holt JJ et al. Discovery of a Potent and Selective ATAD2 Bromodomain Inhibitor with Antiproliferative Activity in Breast Cancer Models. J Med Chem 65, 3306–3331 (2022). [DOI] [PubMed] [Google Scholar]
- 138.Palmer WS et al. Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor. J Med Chem 59, 1440–1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu Q et al. Discovery and optimization of novel N-benzyl-3,6-dimethylbenzo[d]isoxazol-5-amine derivatives as potent and selective TRIM24 bromodomain inhibitors with potential anti-cancer activities. Bioorg Chem 94, 103424 (2020). [DOI] [PubMed] [Google Scholar]
- 140.Cox OB et al. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem Sci 7, 2322–2330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N & Zhou MM Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16, 643–652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Owen D et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. Embo J 19, 6141–6149 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.