Abstract
Purpose
A small percentage of patients with small cell lung cancer (SCLC) experience durable responses to immune checkpoint blockade (ICB). Defining determinants of immune response may nominate strategies to broaden the efficacy of immunotherapy in patients with SCLC. Prior studies have been limited by small numbers and/or concomitant chemotherapy administration.
Methods
CheckMate 032, a multicenter, open-label, phase 1/2 trial evaluating nivolumab alone or with ipilimumab in patients with previously treated advanced or metastatic solid tumors was the largest study of ICB alone in patients with SCLC. We performed comprehensive RNA sequencing of 286 pretreatment SCLC tumor samples from patients enrolled on this study. We evaluated outcome based on defined SCLC subtypes (SCLC-A, -N, -P, and -Y), and explored expression signatures associated with durable benefit, defined as progression-free survival ≥6 months. Potential biomarkers were further explored by immunohistochemistry.
Results
None of the subtypes were associated with progression-free or overall survival. YAP1 gene expression across the dataset was associated with an inflammation signature (R = 0.25, p=0.00014), and SCLC-Y associated with expression of antigen presentation machinery (APM) (p<0.00001). The APM signature (p=0.000032) and presence of ≥ 1% infiltrating CD8+ T cells by immunohistochemistry (HR 0.51; 95% confidence interval 0.27 – 0.95) both correlated with overall survival in patients treated with nivolumab. Pathway enrichment analysis demonstrated association between durable benefit from immunotherapy and antigen processing and presentation.
Conclusions
Tumor antigen processing and presentation is a key correlate of ICB efficacy in patients with SCLC. As antigen presentation machinery is frequently epigenetically suppressed in SCLC, this study defines a targetable mechanism by which we might improve clinical benefit of ICB for patients with SCLC.
Keywords: small cell lung cancer, immune checkpoint blockade, antigen presentation
INTRODUCTION
Small cell lung cancer (SCLC) accounts for 13–15% of all lung cancers, with approximately 250,000 cases diagnosed annually worldwide.1 Two thirds of patients present with metastatic, or extensive-stage, disease at diagnosis. While patients with extensive-stage SCLC typically have robust responses to first line platinum-based chemotherapy, most will experience chemoresistant relapse within the first year. The addition of anti-PD-(L)1 immune checkpoint blockade (ICB) to first line chemotherapy improves both progression-free and overall survival in patients with newly diagnosed extensive-stage SCLC.2-5 While ICB leads to durable benefit in a minority of patients (e.g. increasing 3-year survival from 5.8% to 17.6% with durvalumab), most patients with SCLC derive minimal if any benefit (e.g. the median survival duration improves only from 10.5 to 12.9 months).6 The dichotomy between transformative benefit in a subpopulation and minimal benefit in most patients underscores the need for a better understanding of the underlying biology of SCLC and its interactions with the immune system, to identify predictive biomarkers and differential therapeutic vulnerabilities to inform personalized therapy and ultimately improve patient outcomes.
ICB demonstrates activity in recurrent, metastatic SCLC, even in the absence of chemotherapy. The CheckMate 032 study enrolled patients with recurrent advanced solid tumors including SCLC, and included cohorts treated with nivolumab (anti-PD1), or with nivolumab and ipilimumab (anti-CTLA4).7 The initial phase of this trial demonstrated activity of both regimens in SCLC patients, leading to the launch of a randomized cohort specifically assessing the activity of (1) nivolumab 3mg/kg every 2 weeks, or of (2) nivolumab 1 mg/kg plus ipilimumab 3 mg/kg every 3 weeks for four cycles, followed by nivolumab 3 mg/kg every 2 weeks. Both regimens demonstrated activity, with response rates of 11.6% and 21.9%, respectively.7 As seen with chemoimmunotherapy regimens in the first line setting,2-5 while a minority of patients with recurrent metastatic SCLC respond and derive durable benefit from immunotherapy, most patients do not: median time to progression of SCLC patients in the randomized portion of the CheckMate 032 study was 1.4 – 1.5 months on both arms. Given the exceptionally poor prognosis of recurrent SCLC, identification of potential biomarkers defining patients likely to have responsive disease vs. those likely to experience rapid disease progression on immunotherapy – and who could be quickly directed to other therapeutic options – would be of immediate clinical utility. Defining the mechanisms contributing to durable clinical benefit could inform future therapeutic strategies in patients with SCLC.
Analyses of the genomic landscapes of SCLC tumors have defined several recurrent alterations, most notably nearly universal inactivation of the key tumor suppression genes TP53 and RB1, as well as frequent alterations in epigenetic regulators.8, 9 In contrast to lung adenocarcinomas, recurrent genomic alterations in SCLC do not appear to define mutually exclusive subtypes of disease, and typically do not include activating and potentially targetable mutations in mitogenic drivers.10 Epigenetic and transcriptional analyses of both human SCLC tumors and murine models of disease have identified biologically distinct subtypes of SCLC based on differential expression of lineage-defining transcription factors including ASCL1, NEUROD1, and POU2F3.11-16 A fourth subtype with low or absent expression of these three transcription factors has been variously associated with expression of a fourth transcriptional regulator, YAP1, and/or with an immunologically inflamed pattern of gene expression.17-19 An initial consensus among SCLC investigators proposed a nomenclature of SCLC-A, SCLC-N, SCLC-P and SCLC-Y for these subtypes, respectively.16
Transcriptional profiling of SCLC tumors from patients enrolled in the first line IMpower133 study of carboplatin, etoposide and either placebo or atezolizumab defined a trend toward better outcome in patients whose tumors were of the “inflamed” subtype.18 However, as this treatment regimen involved both cytotoxic chemotherapy and immunotherapy, the specific contribution of immunotherapy to outcome could not be definitively determined. Recent work based on limited analysis of human tumors and experimental observations in murine models of SCLC has suggested that epigenetic silencing of critical components of antigen presentation, including but not limited to MHC-I, may abrogate cytotoxic T cell engagement with SCLC tumors.20-23 Key epigenetic factors implicated in silencing MHC-I genes and antigen presentation machinery in SCLC include EZH2 and LSD1.20-23 As potent and specific inhibitors of both EZH2 and LSD1 are in active clinical trials or are approved for use in other disease types, further evidence that lack of antigen presentation by tumors contributes to ICB resistance in patients with SCLC would define immediately testable interventional strategies to overcome resistance. To explore potential predictive biomarkers of clinical benefit from ICB in patients with SCLC, we sought to systematically study baseline tumor samples obtained from patients treated with ICB alone – either nivolumab or nivolumab/ipilimumab – on the CheckMate 032 study.7, 24 In addition to a broad-based exploratory analysis, we specifically sought to identify associations between the transcriptionally defined subtypes of SCLC, expression of key determinants of antigen presentation, and durable clinical benefit from treatment with anti-PD1 or anti-PD1/anti-CTLA-4 blockade.
MATERIALS AND METHODS
Patient Selection
CheckMate 032 (NCT01928394) was a phase 1/2 multicenter, multi-arm open-label trial evaluating safety and efficacy of nivolumab monotherapy or nivolumab combined with ipilimumab in subjects with advanced or metastatic solid tumors. The SCLC cohorts of CheckMate 032 included patients with histologically or cytologically confirmed, limited- or extensive-stage SCLC with progression after one or more platinum-based chemotherapy regimens.24, 25
Tumor biopsy collected prior to study treatment was required for biomarker analyses and TMB has previously been reported on based on WES data.25 Here we report on RNA-seq analyses from data generated on tumor tissue collected from this study. Written consent for these analyses were collected from all patients included in these analyses.
RNA Sequencing
RNA samples were analyzed using the Illumina TruSeq RNA Exome method for library preparation, followed by sequencing on the Illumina NovaSeq platform with a 50bp paired-end strategy with a target read depth of 50M per specimen.
Briefly, first strand cDNA synthesis was primed from total RNA using random primers, followed by the generation of second strand cDNA with dUTP utilized in place of dTTP in the master mix. Double-stranded cDNA subsequently underwent end-repair, A-tailing, and ligation of adapters that included index sequences. The resulting molecules were amplified via PCR, their yield and size distribution determined, and their concentrations normalized in preparation for the enrichment step. Libraries were enriched for the mRNA fraction by positive selection using a cocktail of biotinylated oligos corresponding to coding regions of the genome. Targeted library molecules were then captured via the hybridized biotinylated oligo probe using streptavidin conjugated beads. After two rounds of hybridization/capture reactions, the enriched library molecules were amplified via PCR. Final libraries were assessed using qPCR for quantitation and TapeStation for fragment size assessment. Normalized libraries were pooled and sequenced on the Illumina NovaSeq at a plex-level appropriate to the coverage required.
Immunohistochemistry (IHC) of MHC-I and CD8
MHC-I immunohistochemistry (IHC) was performed at Mosaic Laboratories (Lake Forest, CA) using a monoclonal MHC-I-specific antibody (clone EMR8-5, ab70328, Abcam), that recognizes a shared epitope across in HLA-A, HLA-B and HLA-C. Staining was scored by a board-certified pathologist based on visual review of both frequency and intensity of tumor-cell expression. Scores included an assessment of the total percentage of tumor cells showing expression of MHC-I at any intensity level (MHC-I+ tumor cells/total tumor cells), as well as a “histoscore” (H-score) calculated as: H-score = (1* % of tumor cells with 1+ intensity staining)+( 2* % of tumor cells with 2+ intensity staining)+( 3* % of tumor cells with 3+ intensity staining).
CD8 immunohistochemistry (IHC) was performed by pathology-assisted digital scoring at Mosaic Laboratories (Lake Forest, CA) using a monoclonal CD8-specific antibody (clone C8/144B, cat #M710301–2, Agilent Technologies). CD8 IHC scores were expressed as the percentage of CD8+ immune cells of total cells.
Bioinformatics Data Analysis and Workflow
RNA sequencing (RNAseq) data was calculated as count per million (CPM) and log transformed and TMM normalized. SCLC subtype definition was calculated as previously described.16 Differential gene and gene signature association between patients with durable benefit from therapy (defined here as progression-free survival of ≥ 6 months) and patients without durable benefit (progression-free survival of < 6 months) was assessed with Hallmark gene sets, KEGG gene sets and immune cell types. Gene signature scores were calculated as the median value of Z-scored expression for the gene set transcripts.
Statistical analysis
Patient characteristics and clinical outcomes were compared between the biomarker-evaluable population and the intent-to-treat population using frequency and descriptive statistics. Hazard ratios (HRs) and confidence intervals (CIs) were estimated using Cox proportional hazards regression models (r package survival v2.44-1.1) to evaluate the association between biomarkers of interest with OS or PFS. Kaplan–Meier plots based on SCLC subtypes were used to illustrate associations with clinical endpoints. All data analyses were performed with R 3.6.1.
Receiver operating curve (ROC) analysis was used to detect the cut-offs with the best predictive value for MHC-I. Cut-off for analyses of predictive value for CD8 was tested at 1% positive cells which was close to median value for the samples analyzed.
RESULTS
Characterizing four distinct transcriptional subtypes of SCLC
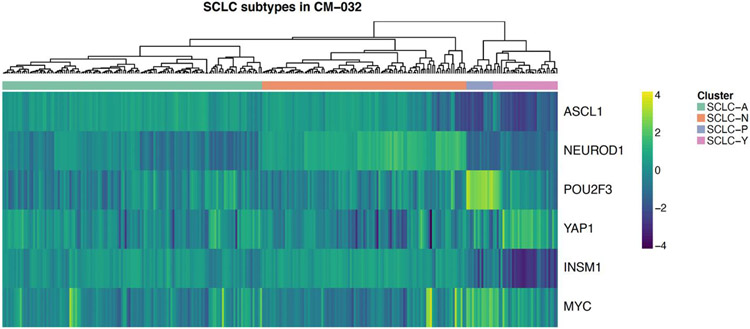
CheckMate 032 enrolled a total of 460 patients with SCLC. Comprehensive bulk RNA sequencing was successfully performed on pre-treatment tumor biopsy samples from a total of 286 of these patients, including 156 treated with single agent nivolumab and 130 treated with combination nivolumab and ipilimumab. Demographic and clinical characteristics of the biomarker cohorts were largely reflective of the larger randomized populations of SCLC patients enrolled to both arms of the study (Table 1). Tumors were assigned to one of four subtypes based on differential expression of genes encoding the transcription factors ASCL1, NEUROD1, POU2F3, and YAP1, using the single highest gene expression to define subtypes SCLC-A, SCLC-N, SCLC-P and SCLC-Y, as previously described 16 (Figure 1). In the nivolumab arm, tumors were distributed among the four different subtypes as follows: SCLC-A (n=49; 32%), SCLC-N (n=47; 30%), SCLC-P (n=21; 14%) and SCLC-Y (n=39; 25%). In the nivolumab/ipilimumab arm, tumors were similarly distributed as follows: SCLC-A (n=40; 31%), SCLC-N (n=47; 36%), SCLC-P (n=14; 11%) and SCLC-Y (n=29; 22%). As in prior analyses of SCLC,16 most cases were of the more highly neuroendocrine subtypes SCLC-A and -N, with neuroendocrine-low SCLC-P and -Y tumors comprising smaller subsets. Consistent with prior reports,26-28 neuroendocrine-high vs. -low SCLC subtypes demonstrated differential expression of NOTCH family members (p=2.7 x 10−6 and 7.6 x 10−14 for NOTCH 1 and 2, respectively), REST (p<2 x 10−16), EZH2 (p<2 x 10−16), and MYC family members (p=7 x 10−12 and 4.1 x 10−5 for MYC and MYCL, respectively) (Supplemental Figure S1).
Table 1:
Baseline demographics and clinical characteristics
| Full study cohort | RNA-Seq cohort | |||
|---|---|---|---|---|
| Nivolumab | Nivo + Ipi | Nivolumab | Nivo + Ipi | |
| N | 245 | 215 | 156 | 130 |
| STAGE | ||||
| Extensive | 186 (76%) | 159 (74%) | 121 (78%) | 100 (77%) |
| Limited | 59 (24%) | 56 (26%) | 35 (22%) | 30 (23%) |
| AGE | ||||
| < 65 | 137 (56%) | 114 (53%) | 86 (55%) | 71 (55%) |
| ≥ 65 | 108 (44%) | 101 (47%) | 70 (45%) | 59 (45%) |
| REGION | ||||
| US | 151 (62%) | 124 (58%) | 102 (65%) | 82 (63%) |
| Rest of World | 94 (38%) | 91 (42%) | 54 (35%) | 48 (37%) |
| FIRST-LINE PLATINUM SENSITIVITY | ||||
| Platinum-resistant | 109 (45%) | 94 (44%) | 77 (49%) | 59 (45%) |
| Platinum-sensitive | 134 (55%) | 115 (54%) | 77 (49%) | 69 (53%) |
| Unknown | 2 (1%) | 6 (3%) | 2 (1%) | 2 (2%) |
| CNS METASTASES | ||||
| No | 224 (91%) | 196 (91%) | 145 (93%) | 114 (88%) |
| Yes | 21 (9%) | 19 (9%) | 11 (7%) | 16 (12%) |
Figure 1. Differential gene expression in tumors of patients with SCLC enrolled on CheckMate 032.
Heat-map of gene expression in the 286 SCLC tumors analyzed, arrayed horizontally and sorted by relative expression of ASCL1, NEUROD1, POU2F3, and YAP1, defining subtype assignments to categories SCLC-A, -N, -P, and -Y. Additional genes shown include INSM1, upregulated in the neuroendocrine-high subtypes SCLC-A and -N, and MYC, most highly expressed in SCLC-P.
Effect of SCLC transcriptional subtype on treatment outcomes to immunotherapy
We first sought to explore the correlation between the SCLC transcriptional subtype and progression-free survival (PFS) and overall survival (OS), for both arms of the CheckMate 032 study. No statistically significant correlation with outcome was found among the four identified subtypes comparing each specific subtype against the other three combined, or when comparing the neuroendocrine-high (SCLC-A, SCLC-N) vs. low-neuroendocrine (SCLC-P, SCLC-Y) subtypes (Supplemental Figure S2).
Gene expression analyses and durable response to immunotherapy in SCLC transcriptional subtypes
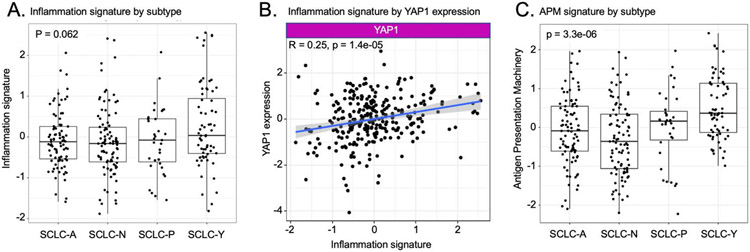
The “SCLC-Y” designation has been questioned by us and others as YAP1 gene expression does not appear to be reflected at the protein level.29 SCLC-Y subtype tumors defined by RNA expression have been reported to be specifically enriched for an immune inflamed pattern of gene expression.19 We evaluated whether this was the case in our dataset of recurrent metastatic SCLC tumors from patients enrolled on CheckMate 032. Analysis of gene expression profiling by RNAseq demonstrated a trend toward higher levels of inflammation gene signature in SCLC-Y tumors at baseline relative to tumors from the other transcriptional subsets, but this did not reach statistical significance (p=0.062; Figure 2A). However, YAP1 gene expression across the dataset did correlate with inflammation signature (R=0.25, p=0.00014), in contrast to ASCL1, NEUROD1, and POU2F3 which did not (Figure 2B and Supplemental Figure S3A).
Figure 2. Analysis of inflammation and antigen presentation signatures by subtype and by differential YAP1 expression.
A. Inflammation gene signature by subtype assignment. Boxes indicate 25th and 75th percentiles, with internal line indicating median. B. Association of inflammation signature and YAP1 expression. Linear correlation line is shown with shaded area reflecting 95% confidence intervals. C. Antigen presentation machinery gene signature by subtype assignment. Boxes indicate 25th and 75th percentiles, with internal line indicating median. P values for A and C are for comparison of subtype SCLC-Y vs. all others (Kruskal-Wallis nonparametric test).
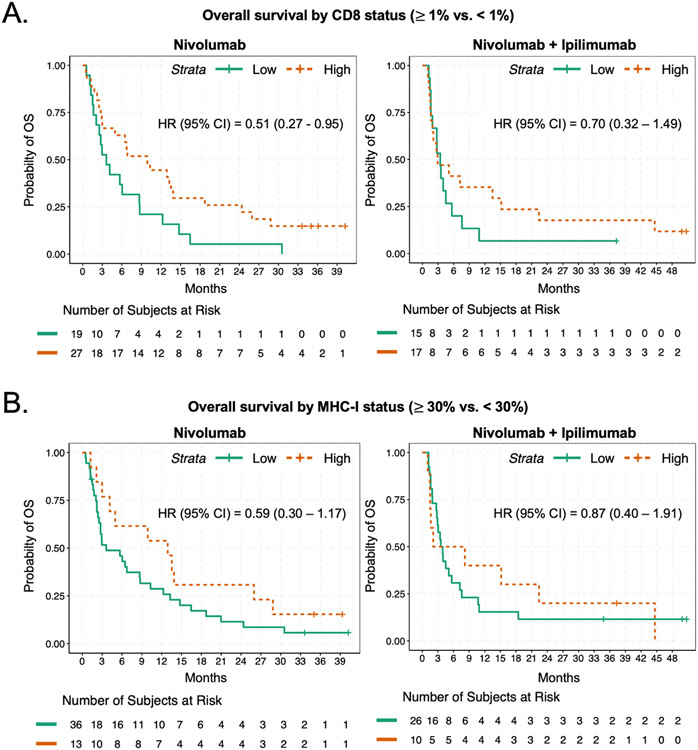
We sought to explore whether CD8+ T cell infiltration as assessed by immunohistochemistry might correlate with the inflammation gene signature, and with patient outcome. Assessing pre-treatment intratumoral CD8+ T cell infiltration as a continuous variable based on percent of tumor cells positive for CD8+ staining by immunohistochemistry, the inflammation gene expression profile across all tumors analyzed correlated with CD8+ T cell infiltration (Supplemental Figure S4A; R=0.64; p=3.1 x 10−10). Dichotomizing CD8 infiltration status as either negative (<1%) or positive (≥ 1% of tumor cells), overall survival was improved in the cohort of patients with CD8 positivity receiving nivolumab relative to patients with low CD8 (HR 0.51, 95% CI 0.27-0.95) with a similar trend observed in patients receiving the combination of nivolumab and ipilimumab (HR 0.7, 95% CI 0.32 – 1.49) (Figure 3A).
Figure 3. Overall survival estimates by CD8+ T cell infiltration and MHC-I expression.
A. Kaplan-Meier curves of overall survival based on presence or absence of at least 1% of cells within the tumor expressing CD8 by immunohistochemistry, for patients treated with nivolumab (left) or nivolumab and ipilimumab (right). A. Kaplan-Meier curves of overall survival based on presence or absence of at least 30% of cells within the tumor expressing MHC-I by immunohistochemistry, for patients treated with nivolumab (left) or nivolumab and ipilimumab (right).
Tumor recognition by cytolytic CD8+ T cells is entirely dependent on antigen presentation in the context of MHC-I30. Antigen presentation is known to be suppressed in many SCLC, which has been nominated as an explanation for the low response rate of SCLC to ICB despite an exceptionally high tumor mutation burden.31, 32 We therefore specifically interrogated a gene expression signature comprised of genes encoding the antigen presentation machinery (APM) including HLA-A, HLA-B, HLA-C, B2M, TAP1 and TAP2 genes. The APM signature is strongly correlated with inflammation (Supplemental Figure S3B; p=2.2 x 10−16), while being more focused on the specific immunologic deficits that have been noted in SCLC. The APM gene signature was strongly enriched in SCLC-Y (p<10−5) (Figure 2C) and is also correlated with intratumoral percentage of CD8 positivity (Supplemental Figure S4B). High intratumoral expression of MHC-I was also associated with a trend toward better overall survival in both the nivolumab (HR 0.59, 95% CI 0.3 – 1.17) and nivolumab plus ipilimumab (HR 0.87, 95% CI 0.4 – 1.91) treatment cohorts (Figure 3B).
We were interested to explore whether individual components of the APM gene signature were primarily driving the association with tumor inflammation. Remarkably, expression of each one of these genes was strongly associated with every other gene in the signature, and consistently associated with tumor inflammation (Supplemental Figure S5). These data suggest that these factors involved in antigenic peptide transport and comprising different classes and components of MHC-I itself define a tightly co-regulated functional program in SCLC.
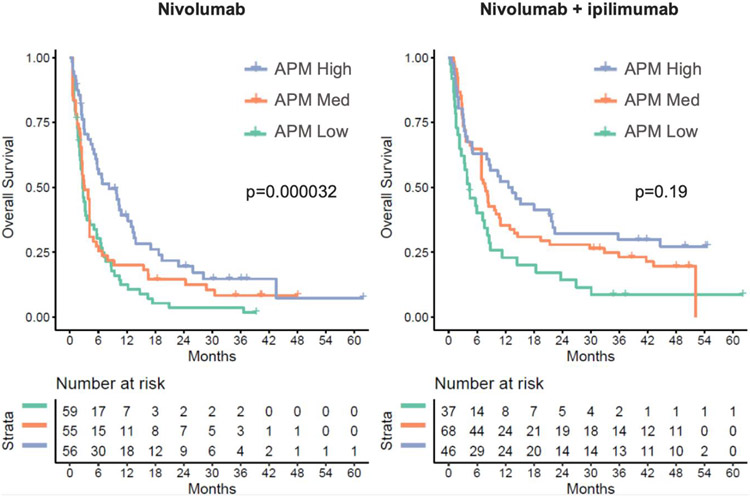
To further explore the APM gene signature as a correlate of outcome to immunotherapy, we assessed patient survival dividing cohorts of SCLC patients treated with either nivolumab or the combination of nivolumab and ipilimumab into tertiles based on APM gene signature. APM expression was significantly correlated with overall survival for patients treated with nivolumab (p=3.2 x 10−4; Cox proportional hazards model), with a trend toward improved survival as well among patients treated with the combination of nivolumab and ipilimumab with higher APM signature expression (p=0.19) (Figure 4).
Figure 4. APM gene signature as a correlate of survival.
Kaplan-Meier curves of overall survival among patients treated with nivolumab only (left) and nivolumab plus ipilimumab (right), by tertiles of antigen presentation machinery (APM) gene expression signature. P values are for cox proportional hazards of OS as a function of APM gene signature.
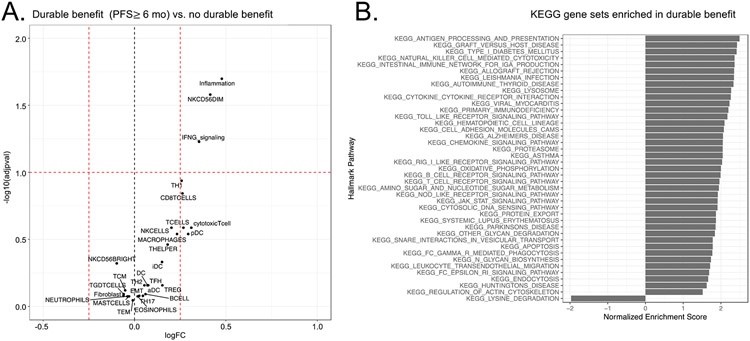
As a benchmark measure of tumor control, we defined durable benefit from ICB as progression-free survival lasting at least 6 months. We explored whether durable benefit to ICB was positively or negatively correlated with expression of either individual genes or defined patterns of gene expression. Tumors from all patients with durable benefit to immunotherapy (both treatment arms; nivolumab alone and nivolumab plus ipilimumab) demonstrated higher expression of multiple inflammatory genes, notably GPR114, GNLY, CXCL9, GZMB, XCL2, and IDO1 (Supplemental Figure S6A). At the pathway level, tumors from patients with durable benefit demonstrated higher levels of inflammation, NK cell activation, and IFN-γ signaling (Figure 5A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis demonstrated multiple immunologically relevant gene sets enriched in the pre-treatment tumors associated with durable benefit, including, most strongly, Antigen Processing and Presentation, as well as multiple pathways associated with T cell proliferation (Figure 5B). Similar T cell proliferative pathways were also identified using the Hallmark gene sets (Supplemental Figure S6B). These gene set and pathway analyses also nominated other immune cell types, including macrophages and natural killer (NK) cells as contributors to anti-tumor immunity in SCLC. Gene expression signatures reflective of both macrophages and NK cells were enriched in the SCLC-Y subtype (p=3.6 x 10−8 and 1.5 x 10−7, respectively; Supplemental Figure S7).
Figure 5. Gene signature and gene set correlates of durable benefit from ICB.
A. Volcano plot of gene signatures associated with durable benefit from ICB, defined as progression-free survival of ≥ 6 months, in patients with SCLC enrolled in CheckMate 032. Positive association with durable benefit on the X axis is to the right of 0.0 (log fold-change); negative association to the left. Y axis represents negative log of adjusted p value, with a significance threshold set at 1.0. B. KEGG gene sets associated with durable benefit. Gene sets positively correlating with durable benefit are indicated by bars to the right of 0; gene sets negatively correlating are to the left. Bar length reflects strength of association.
DISCUSSION
The addition of PD(L)1 blockade with either atezolizumab or durvalumab to platinum-based chemotherapy has redefined the standard of care for first line treatment of patients with metastatic SCLC.2, 3 With the increasing use of combination chemoimmunotherapy for patients with SCLC, the extensive sample collection from patients enrolled to the CheckMate 032 study represented a unique opportunity to assess determinants of response to immunotherapy in patients with SCLC. ICB using either nivolumab or the combination of nivolumab and ipilimumab can induce objective clinical responses in patients with recurrent metastatic SCLC.7 Although the number of patients benefitting from ICB in this context is clearly limited, immune-mediated responses when they do occur can be durable, including some rare patients with SCLC treated with immunotherapy who remain free of clinically evident progressive disease for years. Identifying patients who might have such transformative responses to ICB, and conversely identifying patients who definitively will not, and who could be appropriately directed to other therapeutic options, remains a critical unmet need. This study evaluated correlates of clinical benefit from ICB in a cohort of patients who provided pre-treatment tumor samples for such exploratory analyses, including comprehensive gene expression profiling and immunohistochemistry.
Transcriptional profiling has suggested that SCLC can be subdivided into subtypes of disease based on differential expression of a small number of “master” transcriptional regulators.16 Three of these subtypes, associated with expression of ASCL1, NEUROD1, and POU2F3, have been consistently identified. The remainder of cases, associated in some series with YAP1 gene expression, have been more controversial, and are likely to comprise a more heterogeneous collection of tumors.17 Interestingly, while transcriptional profiling has defined YAP1 gene expression as a putative subtype-defining factor, analysis of SCLC at the protein level has demonstrated low level intratumoral YAP1 expression across subtypes, rarely exceeding an H score of 100 on a standard 300-point scale.17 A comprehensive analysis of RNA vs. protein expression of these factors and additional definitive determinants of subtypes are needed. Here we defined SCLC subtype based simply on the single highest expression of these four genes. The resulting categorization is notably overlapping with, but not identical to, classification based on non-negative matrix factorization of the entire transcriptional dataset defining subtypes SCLC-A, -N, -P, and -I.33
This report represents the largest effort to date to evaluate baseline transcriptional profiles from recurrent SCLC tumors treated with ICB and to assess correlations with clinical outcomes of patients enrolled on a prospective clinical trial. To our knowledge, this is the first report demonstrating that higher abundance of pre-treatment CD8+ T cells is associated with improved survival after ICB. Moreover, we observed that patients with higher levels of MHC-I expression also had greater survival after ICB treatment, which is consistent with retrospective outcomes previously reported from a single center.20 We noted a trend toward higher levels of a global inflammation signature among non-A/N/P SCLC (i.e. SCLC-Y), a more robust linkage was found with a signature specifically focused on determinants of antigen presentation. More importantly, beyond correlation with subtype, we found antigen processing and presentation to be a signal strongly enriched in patients with durable clinical benefit from ICB.
Suppression of antigen processing and presentation may explain in part the lack of ICB efficacy in most patients with SCLC. One might imagine that different SCLC tumors could achieve immune evasion via suppression of antigen presentation by distinct mechanisms of genetic disruption or epigenetic silencing – e.g. reduction or elimination of B2M in some tumors, of the TAP proteins in others, etc. Our data argues against this model and supports an alternative. The striking correlation of gene expression across a diversity of key contributors to MHC-I presentation – HLA-A, HLA-B, HLA-C, B2M, TAP1, and TAP2 – suggests that these genes comprise a co-expression network and implies a shared epigenetic mechanism controlling their transcription.
These data lend urgency to ongoing research into strategies to enhance antigen presentation in SCLC through identification and targeting of the critical epigenetic regulators. Several studies have indeed confirmed that silencing of key determinants of antigen presentation in SCLC is epigenetically controlled.20-23 Epigenetically targeted therapies, notably inhibition of EZH2, the enzymatic subunit of the Polycomb repressive complex 2 (PRC2) which catalyzes trimethylation of histone H3 at lysine 27 leading to gene silencing, have been shown to re-induce MHC-I and lead to enhanced response in preclinical models of high-grade neuroendocrine tumors including SCLC.20, 23, 34 Recent observations by our group and others also point to Lysine-specific demethylase I (LSD1), which removes regulatory methyl groups from histone H3 at lysines at positions 4 and 9, as a key determinant of MHC-I expression and antigen presentation in SCLC.21, 22 The correlative analyses described herein support that pursuing such strategies should be a priority: if antigen processing and presentation is a biomarker of durable response to ICB, re-expressing this set of factors in tumors in which antigen presentation machinery has been epigenetically silenced may substantially increase the population of SCLC patients benefiting from immunotherapy.
This study has several limitations. Most notably, this was an exploratory post hoc analysis, and key results will need to be confirmed in independent data sets. The time of pre-study biospecimen collection relative to start of immunotherapy was variable, some samples being archival diagnostic material that may not fully reflect tumor status at the time of study entry. A prior retrospective study of SCLC and pulmonary carcinoid tumors noted an association between YAP1 and increased IFN-γ, HLA genes, and T cell inflammation gene expression signatures.19 However, only 6 tumors in this cohort of 33 SCLC were classified as SCLC-Y and none of these patients were treated with ICB, precluding any association with immune response. There may be opportunities to more effectively validate the correlations noted here using biospecimens from other completed clinical trials, such as the SCLC cohort of the KEYNOTE-158 study of pembrolizumab.35 We would encourage prospective collection of pre-treatment tumor samples for analyses in ongoing or upcoming clinical studies of ICB in the disease, to help inform therapeutic choices for patients with recurrent and metastatic SCLC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Bristol Myers Squibb through the International Immuno-Oncology Network (II-ON), and by NIH grants R35 CA263816, U24 CA213274 and P30 CA008748 (CMR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Statement of potential conflicts of interest:
CMR has advised on oncology drug development with AbbVie, Amgen, Astra Zeneca, D2G, Daiichi Sankyo, Epizyme, Genentech/Roche, Ipsen, Jazz, Kowa, Lilly, Merck, and Syros, and serves on the scientific advisory boards of Auron, Bridge Medicines, DISCO, Earli, and Harpoon Therapeutics. DB, WJG, and AF are employees and shareholders of Bristol Myers Squibb. Subsequent to their contributions to this work, WVL and MDH became employees and shareholders of Astra Zeneca. MDH reports prior grants from BMS and personal fees from Achilles; Adagene; Adicet; Arcus; AstraZeneca; Blueprint; BMS; DaVolterra; Eli Lilly; Genentech/Roche; Genzyme/Sanofi; Janssen; Immunai; Instil Bio; Mana Therapeutics; Merck; Mirati; Natera; Pact Pharma; Shattuck Labs; and Regeneron; as well as equity options from Factorial, Immunai, Shattuck Labs, Arcus, and Avail Bio. NJC reports institutional research support from AbbVie, Amgen, Harpoon, Merck, and Monte Rosa Therapeutics, and personal fees from G1 Therapeutics, OncLive, Sanofi, and Wolters Kluwer. All other authors state no potential conflicts of interest.
Credit Statement:
Charles M Rudin: conceptualization, supervision, writing – original draft preparation, funding acquisition; David Balli: methodology, formal analysis, investigation, writing – review & editing; W. Victoria Lai: investigation, writing – review & editing; Allison L. Richards: methodology, formal analysis, investigation, writing – review & editing; Evelyn Nguyen: investigation, writing – review & editing; Jacklynn V. Egger: investigation, data curation, writing – review & editing; Noura Choudhury: investigation, writing – review & editing; Triparna Sen: investigation, writing – review & editing; Andrew Chow: investigation, writing – review & editing; John T. Poirier: investigation, writing – review & editing; William Geese: resources, supervision, writing – review & editing; Matt D. Hellmann: conceptualization, writing – review & editing, funding acquisition; Ann Forslund: formal analysis, writing – review & editing.
REFERENCES
- 1.Rudin CM, Brambilla E, Faivre-Finn C, et al. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn L, Mansfield AS, Szczesna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220–2229. [DOI] [PubMed] [Google Scholar]
- 3.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929–1939. [DOI] [PubMed] [Google Scholar]
- 4.Leal T, Wang Y, Dowlati A, et al. Randomized phase II clinical trial of cisplatin/carboplatin and etoposide (CE) alone or in combination with nivolumab as frontline therapy for extensive-stage small cell lung cancer (ES-SCLC): ECOG-ACRIN EA5161. Journal of Clinical Oncology 2020;38:9000–9000. [Google Scholar]
- 5.Rudin CM, Awad MM, Navarro A, et al. KEYNOTE-604: Pembrolizumab (pembro) or placebo plus etoposide and platinum (EP) as first-line therapy for extensive-stage (ES) small-cell lung cancer (SCLC). Journal of Clinical Oncology 2020;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paz-Ares L, Chen Y, Reinmuth N, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022;7:100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ready NE, Ott PA, Hellmann MD, et al. Nivolumab Monotherapy and Nivolumab Plus Ipilimumab in Recurrent Small Cell Lung Cancer: Results From the CheckMate 032 Randomized Cohort. J Thorac Oncol 2020;15:426–435. [DOI] [PubMed] [Google Scholar]
- 8.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drapkin BJ, Rudin CM. Advances in Small-Cell Lung Cancer (SCLC) Translational Research. Cold Spring Harb Perspect Med 2021;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YH, Klingbeil O, He XY, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 2018;32:915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McColl K, Wildey G, Sakre N, et al. Reciprocal expression of INSM1 and YAP1 defines subgroups in small cell lung cancer. Oncotarget 2017;8:73745–73756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mollaoglu G, Guthrie MR, Bohm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baine MK, Hsieh MS, Lai WV, et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. J Thorac Oncol 2020;15:1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owonikoko TK, Dwivedi B, Chen Z, et al. YAP1 Expression in SCLC Defines a Distinct Subtype With T-cell-Inflamed Phenotype. J Thorac Oncol 2021;16:464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahadevan NR, Knelson EH, Wolff JO, et al. Intrinsic Immunogenicity of Small Cell Lung Carcinoma Revealed by Its Cellular Plasticity. Cancer Discov 2021;11:1952–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen EM, Taniguchi H, Chan JM, et al. Targeting Lysine-Specific Demethylase 1 Rescues Major Histocompatibility Complex Class I Antigen Presentation and Overcomes Programmed Death-Ligand 1 Blockade Resistance in SCLC. J Thorac Oncol 2022;17:1014–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiatt JB, Sandborg H, Garrison SM, et al. Inhibition of LSD1 with Bomedemstat Sensitizes Small Cell Lung Cancer to Immune Checkpoint Blockade and T-Cell Killing. Clin Cancer Res 2022;28:4551–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burr ML, Sparbier CE, Chan KL, et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 2019;36:385–401 e388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonia SJ, Lopez-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883–895. [DOI] [PubMed] [Google Scholar]
- 25.Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853–861 e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shue YT, Drainas AP, Li NY, et al. A conserved YAP/Notch/REST network controls the neuroendocrine cell fate in the lungs. Nat Commun 2022;13:2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland KD, Ireland AS, Oliver TG. Killing SCLC: insights into how to target a shapeshifting tumor. Genes Dev 2022;36:241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baine MK, Hsieh MS, Lai WV, et al. Small Cell Lung Carcinoma Subtypes Defined by ASCL1, NEUROD1, POU2F3 and YAP1: Comprehensive Immunohistochemical and Histopathologic Characterization. J Thorac Oncol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliveira CC, van Hall T. Alternative Antigen Processing for MHC Class I: Multiple Roads Lead to Rome. Front Immunol 2015;6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle A, Martin WJ, Funa K, et al. Markedly decreased expression of class I histocompatibility antigens, protein, and mRNA in human small-cell lung cancer. J Exp Med 1985;161:1135–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funa K, Gazdar AF, Minna JD, et al. Paucity of beta 2-microglobulin expression on small cell lung cancer, bronchial carcinoids and certain other neuroendocrine tumors. Lab Invest 1986;55:186–193. [PubMed] [Google Scholar]
- 33.Gay CM, Stewart CA, Park EM, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346–360 e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strosberg J, Mizuno N, Doi T, et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Neuroendocrine Tumors: Results From the Phase II KEYNOTE-158 Study. Clin Cancer Res 2020;26:2124–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.