Abstract
Artificial intelligence (AI) promises to revolutionize many fields, but its clinical implementation in cardiovascular imaging is still rare despite increasing research. We sought to facilitate discussion across several fields and across the lifecycle of research, development, validation, and implementation to identify challenges and opportunities to further translation of AI in cardiovascular imaging. Furthermore, it seemed apparent that a multi-disciplinary effort across institutions would be essential to overcome these challenges. This paper summarizes the proceedings of the NHLBI-led workshop, creating consensus around needs and opportunities for institutions at several levels to support and advance research in this field and support future translation.
Keywords: artificial intelligence, data science, machine learning, deep learning, AI algorithms, cardiovascular imaging
Introduction
Over the last century, advances in cardiovascular imaging have revolutionized the care of patients with cardiovascular disease (CVD)(1). However, these advances have come at a significant financial and human cost. Healthcare expenditures, as well as the burden on imaging specialists to generate increasingly detailed reports of imaging studies, both continue to rise in parallel with the exponential increase in frequency and variety of cardiovascular imaging tests performed during routine clinical care(2). Thus, the field of cardiovascular imaging is at a breaking point and ripe for a paradigm shift.
Artificial intelligence (AI) is touted as the disruptive innovator needed to solve a wide variety of problems in cardiovascular imaging(3), but whether the field can realize the promise of AI to reduce costs of cardiovascular imaging while advancing human health remains to be seen. In this regard, much can be learned from the tenets of disruptive innovation to prepare the field of cardiovascular imaging to best harness the power of AI to achieve goals(4). Some of these principles include: (1) understanding disruption as a process, rather than a fixed event or solution; (2) acknowledging that true disruption requires scientific, clinical, and business models that are very different from the status quo; and (3) realizing that not all disruptive innovations succeed. From these principles, it is clear that preparing, planning, and collaborating for research, testing, and implementation of any potential disruptive innovation, such as AI in cardiovascular imaging, is key to improving chances for success.
For these reasons, the US National Institutes of Health (NIH) convened a 2-day workshop in June 2022 entitled, “Artificial Intelligence in Cardiovascular Imaging: Translating Science to Patient Care,” which brought together key stakeholders from academia, NIH, the National Science Foundation, industry, the US Food and Drug Administration (FDA), the Centers for Medicare and Medicaid Services (CMS), and cardiovascular imaging societies. The overall goal of the workshop was to discuss challenges and opportunities for AI in cardiovascular imaging, focusing on how these stakeholders can support research and development to move AI from promising proofs of concept to robust, generalizable, equitable, scalable, and implementable AI. Workshop participants included representatives from over 18 research institutions, five medical societies, and four government agencies. The participants were 34 percent female, 50 percent Caucasian, 45 percent Asian, and 5 percent Black, and represented geographic regions across the US, as well as the United Kingdom. In preparation for this workshop, we performed an analysis of NHLBI-funded studies of AI in CV imaging. Although we noted an increase over the last two years, the overall number of funded studies was quite small (Figure 1). Here we describe the deliberations of the working group members, who sought to identify and prioritize the challenges and needs facing researchers in 6 key areas: data, algorithms, infrastructure, regulatory requirements, implementation, and human capital. The resulting proceedings described here provide a blueprint for academia, government institutions, and industry that should accelerate the incorporation of AI in all aspects of cardiovascular imaging, reduce financial and human resources burden, and most importantly, improve the care of patients with CVD.
Figure 1: NHLBI grants supporting AI in cardiovascular imaging over the past ten years.
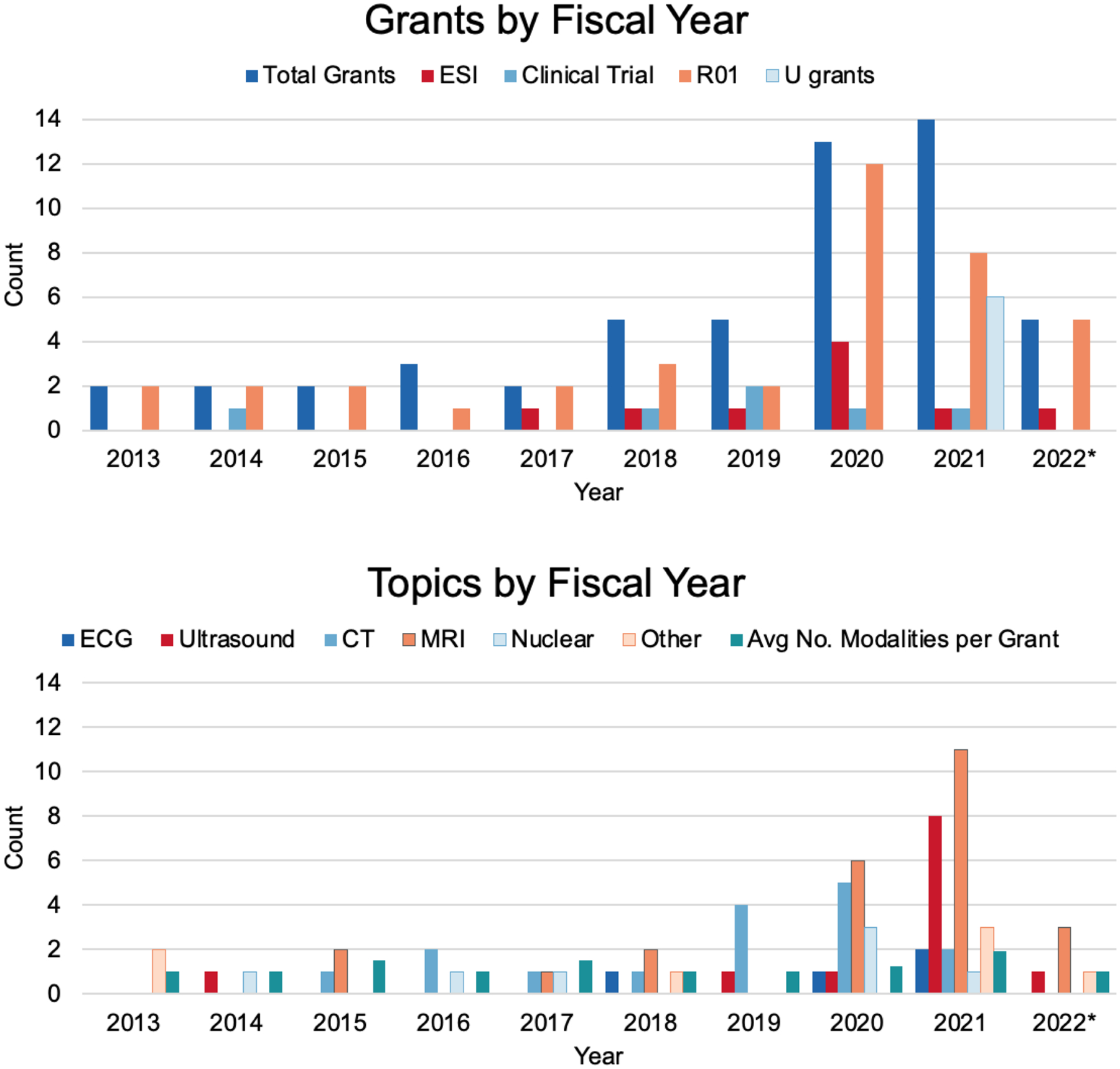
The NIH internal data platform was used to search for grants from fiscal year 2013 to 2022 using free-text search in the title and abstract. The terms used were: (“machine learning” and “imaging”) AND (“deep learning” and “imaging”) AND (“AI” and “imaging”). Fifty-three grants were finalized after removing non-human studies and lung and blood related studies, as well as removing non-target grants by manual curation. The grant information is accessible using the publicly available NIH Research Portfolio Online Reporting Tools (NIH RePORT) system. Early-stage investigator grants, clinical trials, and U-grants have all become more common in recent years. Grants span imaging modalities, but typically focus on only one modality per grant.
1: Data
Data is of fundamental importance to AI development. In cardiovascular imaging, data spans multiple modalities including ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), angiography, and nuclear imaging, each with different underlying physics of acquisition/processing, and involving several vendors. Data spans multiple patient types across diseases and demographics, has high clinical volume, and is large in size. These characteristics present inherent challenges in terms of how to acquire, harmonize, curate, analyze, and store cardiovascular imaging data for AI. The workshop identified several needs and opportunities to advance data-centric research:
How should data quality and diversity be assessed? These challenges exist at a dataset level (e.g., patient representation, missing data), as well as at an image level (e.g., image quality and artifacts).
If multi-center, multi-vendor datasets are superior for training and testing robust AI models, how can we best harmonize and standardize that data? How do we best distribute the responsibility for these tasks among national, institutional, and individual stakeholders?
How can patient privacy best be protected while preserving data access for research?
Data quality and diversity.
If data quality and diversity are not considered, AI models may reflect and amplify the noise and biases of the underlying datasets. The quality of a cardiovascular imaging dataset includes aspects of the image itself, as well as the timing, location, and methods of acquisition (provenance)(5); purpose of creation; associated metadata; imaging report and other annotations (labels). The methods of annotation, labeler expertise, interobserver variability, and ground truth are all important quality metrics that should accompany datasets and the models that are trained from them. Development of standards for data quality, image acquisition, meta-data completion, image quality analysis, annotation, and reporting are important areas of future research. Ideally, clinical AI models should perform well on datasets of varying quality. Alternatively, AI may be developed to improve image quality (e.g., denoising or super-resolution)(6) for downstream clinical tasks.
Finally, it is important to remember that data quality is a dynamic issue. Developers and users of AI models should be alert for “data shift” that can result from new imaging machines, new technology upgrades, changes to imaging protocols, or a change in utilization.
In some ways, diversity is one aspect of dataset quality. Dataset diversity encompasses patient factors—type of disease pathology, age, sex or gender, race, ethnicity, geography, and socioeconomic status—as well as factors intrinsic to the images themselves, such as scanner manufacturer, artifacts, and modes of acquisition(7).
Lack of patient-level diversity in datasets has the potential to exacerbate health inequalities. For example, in the assessment of chest x-ray datasets, AI models under-diagnosed underserved patients including female, Black, Hispanic, younger, and lower socioeconomic status patients.(8) It is important to engage with communities impacted by health disparities in order to recruit underrepresented minorities for cardiovascular imaging research.
Lack of image-level diversity may also cause healthcare inequalities that are yet unanticipated (e.g., if an AI model performs poorly given a certain imaging artifact or particular brand of scanner).
Diversity is important in both training and testing datasets, and it should be reported along with other quality metrics. Bias mitigation strategies that can be adopted before, during, and after training have been proposed in order to develop fairer algorithms. Identifying appropriate measures of “diversity” for datasets is an important area for future research.
Data curation.
In order to measure quality and track provenance across many images in various datasets at multiple centers, developing and deploying methods to curate and harmonize datasets are key challenges. Meeting this challenge will require innovation in data engineering, management, and deployment, as well as computing infrastructure (Section 3). Specific challenges in curation of cardiovascular imaging at this scale include harmonization across multiple centers, studies, and modalities; dealing with missing data or metadata; ensuring cross-compatibility across vendors and image formats; as well as centralization and deployment of standard image labeling workflows. Optimizing these challenges provides key areas for further research.
Data access, sharing, and safety.
Access to large, high-quality, diverse datasets drives innovation in AI. Data access is also an important democratizer, ensuring that researchers currently underrepresented in the development and use of AI have equal ability to innovate. In addition to curation (above) and infrastructure (Section 3), challenges in data sharing for cardiovascular imaging include data privacy and consent, institutional and national regulatory hurdles, security, and attribution of credit for dataset curation.
Depending on the type of data, different levels of de-identification and security may be required. Concerns about data privacy and consent are well-placed as the technical capabilities around subject identification/re-identification is rapidly changing. For example, facial recognition from scans of the head has been demonstrated(9). Developing “Biosafety levels” around some types of imaging research may help to alleviate these concerns and is an important area for further research. This could include special training, security controls, and monitoring. Different levels of permission and consent may be needed for academic and commercial uses. Synthetic data may also help alleviate some data privacy concerns and warrants further research.
Successful data access and sharing will require both technical and policy support at multiple levels, including governmental, institutional, and individual. One example is the 2023 NIH Data Management and Sharing Policy, which expands its existing data sharing requirements to all extramural research that generates scientific data regardless of funding level(10). Enabling attribution of credit for the hard work of data collection and curation can help promote data sharing. Methods to facilitate this include journals and other resources where datasets can be documented and cited.
2: Algorithms
AI is anticipated to improve cardiovascular imaging through its application to use cases across the entire lifecycle of cardiovascular imaging. AI has been utilized in patient selection and protocoling prior to imaging tests, image acquisition, denoising, registration, and reconstruction during the test acquisition, quantification, interpretation, diagnosis. and reporting, as well as prognostic risk stratification (Figure 2)(11). Early potential was demonstrated using now-standard methods like supervised learning and using networks like convolutional neural networks and U-Nets. This work has sometimes demonstrated impressive results meeting expert human performance; however, AI to date is data-hungry, label-hungry, data-specific, and can also fail in ways that jeopardize user trust. A new generation of AI algorithms must work smarter to meet clinical needs at the level of performance and robustness that clinicians and patients require. Challenges and opportunities in algorithm development include:
How do we design data-efficient, and label-efficient, algorithms to maximize the value of medical imaging data while relieving clinicians of labeling burden?
What are the best methods for leveraging data across cardiovascular imaging modalities, and/or combining cardiovascular imaging with other clinical data modalities?
What are the best methods for evaluation of AI models’ robustness, generalizability, and learned features?
Figure 2: AI can be used to address use cases across the entire cardiovascular imaging workflow.
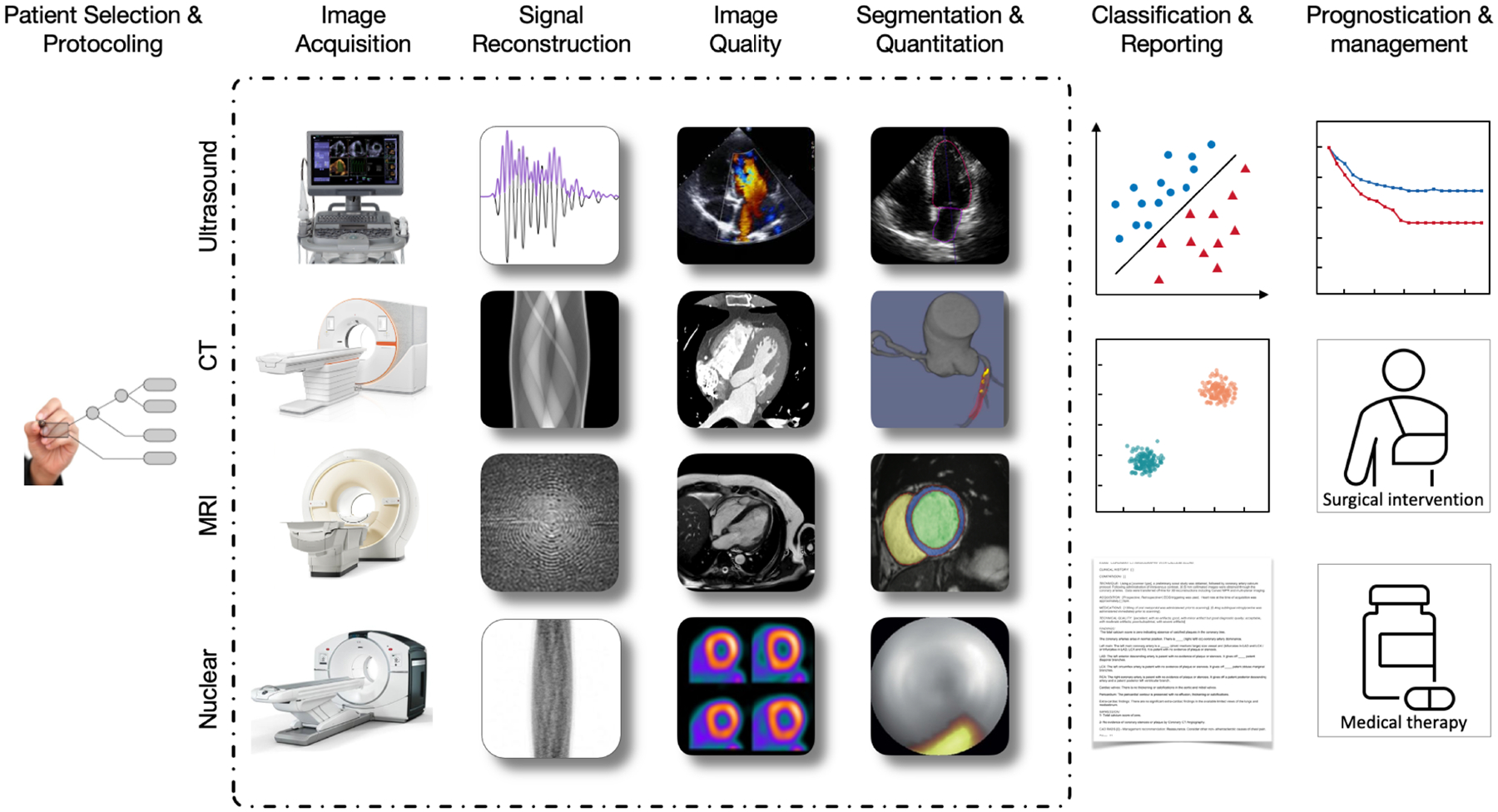
Where AI is typically thought of with respect to downstream applications (e.g., image analysis and prognosis), it is also proving useful upstream, from patient selection and protocoling for imaging studies, to image acquisition at the clinical point of care, signal processing, image denoising, attenuation correction, and reconstruction. These use cases are applicable across cardiovascular imaging modalities. At each step in the clinical workflow, AI can help provide speed and standardization and improve image quality. Finally, improved imaging can impact patient care as it is used to predict outcomes, improve prognostication, and connect to other clinical data modalities (e.g. text reports, lab data).
Data- and label-efficiency.
Several diagnostic challenges in cardiovascular imaging concern rare conditions where data is scarce. Also, while crowd-sourced labeling has worked well in supervised learning for non-medical imaging tasks, busy clinical experts are often tapped to produce the labor-intensive annotations for cardiovascular imaging. Image segmentation, in particular, is highly useful for several downstream clinical applications, including object detection and quantitative imaging. Creating segmentation labels is also highly time-consuming. Existing clinical annotations, if they are readable by the algorithms, can be used as labels(12), but this approach may propagate human subjectivity into model training and may need data curation.
For these reasons, opportunities exist to further develop both data-efficient AI, requiring the least amount of (labeled or unlabeled) data to reach maximum performance, as well as semi-supervised and self-supervised AI (no labels needed for tasks that typically require them)(13). For example, self-supervised deep learning was recently used to segment all cardiac chambers without any manual labels(14). Tested on over 40 times the amount of data it was trained on, manual labeling for even the relatively small dataset used would have taken a clinician over 2,000 hours. Data-efficient algorithms may include active learning, increasingly lightweight neural networks, and other approaches.
Multi-modal algorithms.
Another opportunity is to further develop AI models that, like good clinicians, can incorporate multiple imaging modalities, and further combine imaging with other clinical data and circulating biomarkers. AI-based combination of quantitative positron emission tomography (PET) and coronary CT angiography atherosclerotic plaque biomarkers with clinical data can improve prediction of future myocardial infarction(15).
Image improvement.
Multi-modal data can even be synthetically generated, leveraging the benefits of multi-modal imaging without the risk and cost of additional patient imaging. For example, deep learning techniques have been utilized to generate pseudo-CT data from single-photon emission computerized tomography (SPECT) emission data to allow attenuation correction on nuclear cardiology scanners which are not equipped with CT.(16) AI-generated CT maps with generative adversarial networks have also found utility in PET to CT image registration for hybrid image registration(17).
Quantitative analysis.
A highly practical application in cardiovascular imaging is assistance in image quantification - a task that is currently subjective and associated with observer variability. Deep learning techniques, such as U-Nets and convolutional Long Short-Term Memory networks, have been successfully applied for this purpose. Fully automated methods have been described for the segmentation of cardiac chambers in ultrasound, CMR, and CT images. AI-enabled quantitative measurement of coronary calcium from chest CT images and of total atherosclerotic plaque from coronary CT Angiography has shown to predict prognostic outcomes(18,19). Use of an AI-based workflow for inline CMR myocardial perfusion analysis enhances objective interpretation of stress perfusion images and provides rapid results on the scanner(20).
Performance evaluation.
AI models typically produce an inference result that must then be put into clinical context; for example, left ventricular segmentation by an inference model must then be utilized for left ventricular size measurement. Like other clinical research, these final clinical results of AI inference can and should be tested using standard biostatistical methods and external validation (Section 5). However, performance evaluation of the AI model itself—what the model is learning—is also a critical challenge. There is opportunity to further develop explainable AI algorithms, ways to measure model robustness and generalizability, and methods to interrogate AI models’ learned features, especially in deep learning.
Current methods to interrogate a neural network’s learned features include saliency maps, gradient-weighted class activation mapping (GradCAM), occlusion/ablation testing, and adversarial testing. These methods can reveal model-learned features for machine learning and deep learning, which can be evaluated as metrics of the AI models. Explainable AI features can even be presented directly to the physician as part of the AI-enabled application(21); and as indicated by preliminary studies, these may lead to improvements and confidence in physician interpretation(22). Finally, transparency is a key aspect for use of AI tools and important for their acceptance by health care providers and patients(23).
3: Computing Infrastructure
Biomedical researchers are in a good position to develop and test AI for cardiovascular imaging due to their domain expertise and skill in experimental design. However, traditionally they may lack access to flexible, high-capacity central processing units (CPU) and graphics processing units (GPU); storage; and mature software for data preparation, analytics, and results communication that are needed to further research.
While many institutions have local computing clusters, cloud providers are an indispensable part of the AI landscape because they have developed standardized, scalable environments with streamlined access to stored data, high-performance compute, and software tools; as well as secure infrastructure to enable federated, multi-center, and/or multi-site collaboration. Examples of major commercial cloud providers include Amazon Web Services (AWS), Microsoft’s Azure, and Google Cloud Platform (GCP), which in turn, leverage GPU hardware from companies, such as NVIDIA. While these core providers offer several software tools for machine learning and computer vision—even some that are specific to medical imaging—they must support and grow for innumerable different users and use cases. Continuing computing challenges include:
How can computing platforms help biomedical researchers stay current with new and ever-evolving computational tools?
How can platforms provide flexibility and interoperability for evolving research needs (new algorithms, new data formats, and cross-platform interoperability for collaboration or data linkage)?
How can platforms and other stakeholders support needs for compute and/or data storage through the uncertainty of grand funding cycles, institutional policy changes, and other research challenges?
Core providers may address some of these challenges, but others will be best addressed by biomedical imaging-specific platforms built on top of core provider services. Table 1 presents several examples.
Table 1.
Selected computing platforms available to researchers.
| Platform | Website | Main Service |
|---|---|---|
| Computing Hardware (Commercial) | https://www.nvidia.com/ | Hardware: GPU, Servers, Desktop |
| NVIDIA | Software development environment/packages; MONAI, FLARE | |
| Software platforms: CLARA | ||
| Core cloud computing providers (Commercial) | ||
| Google Cloud Platform (GCP) | https://cloud.google.com | Traditional and high performance GPU computing, tools, and cloud storage |
| Amazon Web Services (AWS) | https://aws.amazon.com | Traditional and high performance GPU computing, tools, and cloud storage |
| Microsoft Azure | https://azure.microsoft.com | Traditional and high performance GPU computing, tools, and cloud storage |
| Platforms for medical imaging | ||
| Non-commercial | ||
| NIH Medical Imaging and Data Resource Center (MIDRC) | https://www.midrc.org/ | An NIH supported multi-institutional initiative, hosted at the University of Chicago, representing a partnership spearheaded by the medical imaging community and, representatives of the American College of Radiology® (ACR®), the Radiological Society of North America (RSNA), and the American Association of Physicists in Medicine (AAPM) for rapid and flexible collection, artificial intelligence analysis, and dissemination of imaging and associated data with a first common goal to build an AI-ready data commons to fuel COVID-19 machine intelligence research that can expand to other organs, relevant diseases, and datasets. |
| NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative | https://datascience.nih.gov/strides | Broker providing price advantaged access to commercial cloud services for NIH grantees, collaborators, and intramural researchers. |
| American Heart Association Precision Medicine Platform (PMP) | https://precision.heart.org | A data marketplace to find and access datasets and an AWS suppored cloud based virtual environment for analytics, tool development, and discovery. |
| Biodata Catalyst | https://biodatacatalyst.nhlbi.nih.gov/ | Data analysis tools and access to compute platforms internally within NIH and externally through commercial platforms |
| Commercial | ||
| Seven Bridges | https://www.sevenbridges.com | Offers a cloud-based environment for storing and conducting collaborative bioinformatic analyses that is geared toward genomic data and provides tools to optimize processing. |
| Flywheel Biomedical Data Research Platform | https://flywheel.io | The service provides medical image management environment with tools to streamline research data with searchable, web-based data mining and analytics and sharing capabilities. |
| Ambra Health / Intelerad | https://www.intelerad.com/ambra/ | Offers scalable enterprise medical imaging platform and vendor neutral archive that connects provides tools for image management, clinician access, and other researcher needs such as AI driven quality control and anonymization. |
| CLARA | https://developer.nvidia.com | Platform offering AI applications and accelerated frameworks for healthcare developers, researchers, and medical device makers |
| Terra | https://terra.bio | Platform offerings include tools to help connect researchers to various data resources, analytical capabilities, and visualization software |
Tools, outreach, and education.
Rather than have each group of researchers develop their own tools for image preprocessing and labeling, model training, inference and performance analysis, platform tools can help streamline and accelerate research and collaboration. For example, NVIDIA’s Medical Open Network for Artificial Intelligence (MONAI)(24) is an open-source toolkit that can run on several computing platforms. GCP’s Healthcare API(25) offers tools for storage, de-identification, and processing of Digital Imaging and Communications in Medicine (DICOM) files. AWS offers integration with Orthanc(26), an open-source DICOM server system.
While readily available tools and platforms are helpful, researchers may find it challenging to keep pace with fast-evolving, ever-proliferating software. Continued outreach and education from software providers can help improve the learning curve and allow researchers to fully leverage available tools. Outreach should also include institutional leaders, who are often in the position of approving tools or platforms for their own institution.
Flexibility for novel research needs.
Despite available tools, the nature of novel research dictates that at times, researcher needs will not be well-served by existing offerings. For example, early cloud computing environments offered by core providers did not provide the security features required for patient data. Smaller platforms focused on building patient-safe environments on top of core services did not initially allow access to the full range and scope of computing required. Non- DICOM image formats are still not uniformly supported. It is therefore important for platform providers to continuously solicit feedback from researchers and respond to those needs by altering or enhancing platform capabilities. Some researchers may also play a valuable role on platform development teams or as alpha users. Finally, strengthening collaborations between researchers and software providers can help productionize promising tools developed out of research labs.
Interoperability.
While some tools are platform-agnostic, others are proprietary, offered only within specific cloud platforms, or require institutional endorsement that functionally silos their use. Lack of interoperability among platforms can limit exchange, use, and collaboration among researchers from different institutions. Technical barriers to interoperability arise when data and tools do not adhere to FAIR (findability, accessibility, interoperability, reusability)(10) characteristics. Other barriers concern policy, consent, and governance. Meta platforms, such as Seven Bridges (Table 1), can help communicate within and across repositories that are distributed, in-house, or on the cloud, including those that might be in different vendor offerings. They enhance collaboration among researchers through partnerships with biomedical research data platform providers, such as Flywheel (Table 1), which help connect to data across any academic, life sciences research, clinical, or commercial technology organization to provide a common interface for biomedical analysis pipelines.
Support research needs through research uncertainty.
Workforce turnover and funding changes are inherent to biomedical research, and yet, these can jeopardize the integrity and availability of painstakingly developed data, code, and results. Researchers may depend on costly cloud computing, for example, only to face an interruption in grant funding. Several stakeholders have an opportunity to help researchers develop and maintain sustainable data and code. For example, the NIH has funded the development of an infrastructure for open imaging data commons called Medical Imaging and Data Resource Center (MIDRC, Table 1) in order to support tools for machine learning algorithms. It has also developed the BioData Catalyst (Table 1) ecosystem whose mission is to develop and integrate advanced cyberinfrastructure, leading edge tools, and FAIR standards to support the NHLBI research community. Individual institutions have negotiated research discounts with cloud providers; on a national level, the NIH Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) program (Table 1) has also negotiated simplified cloud access and reduced costs for researchers. Additionally, cloud providers have made small amounts of cloud computing credits directly available to researchers through granting programs, and there is an opportunity to expand these offerings.
4: Regulatory Considerations
With biomedical researchers taking an active role in developing AI for cardiovascular imaging, it is natural to want to move innovation to the bedside. However, many researchers may find this their first experience with the regulatory processes required to test and implement new devices in patient care settings. Major challenges include:
Researchers are unfamiliar with the regulatory process, particularly with respect to the rapidly evolving area of AI-enabled device approval
Uncertainty exists about when and how to engage the FDA
In addition to independent AI research and development, there is a need for ongoing regulatory science research toward rigorous performance evaluation of AI devices.
Software as a medical device (SaMD) regulatory process.
An AI/ML-enabled SaMD implements trained algorithmic models intended for well-defined medical purposes (Figure 4). Based on the associated risk, regulatory controls, and the type of review established by the Food and Drug Administration (FDA), medical devices may fall into three classes (Table 2)(27); most AI/ML cardiovascular imaging SaMDs fall within Class II. All three classes require general controls that include adulteration, misbranding, registration, listing, and premarket notification, banned devices, notification and other remedies, records, reports, and unique device identification, and general provisions of the Federal Food, Drug, and Cosmetic Act. Class II devices require special controls that include performance assessment strategies, post-market assessment, patient registries and guidelines in guidance documents. For devices where general and special controls are not sufficient to reasonably assure safety and effectiveness require premarket approval.
Figure 4: Overview of different components of AI/ML-enabled software as a medical device in premarket submissions.
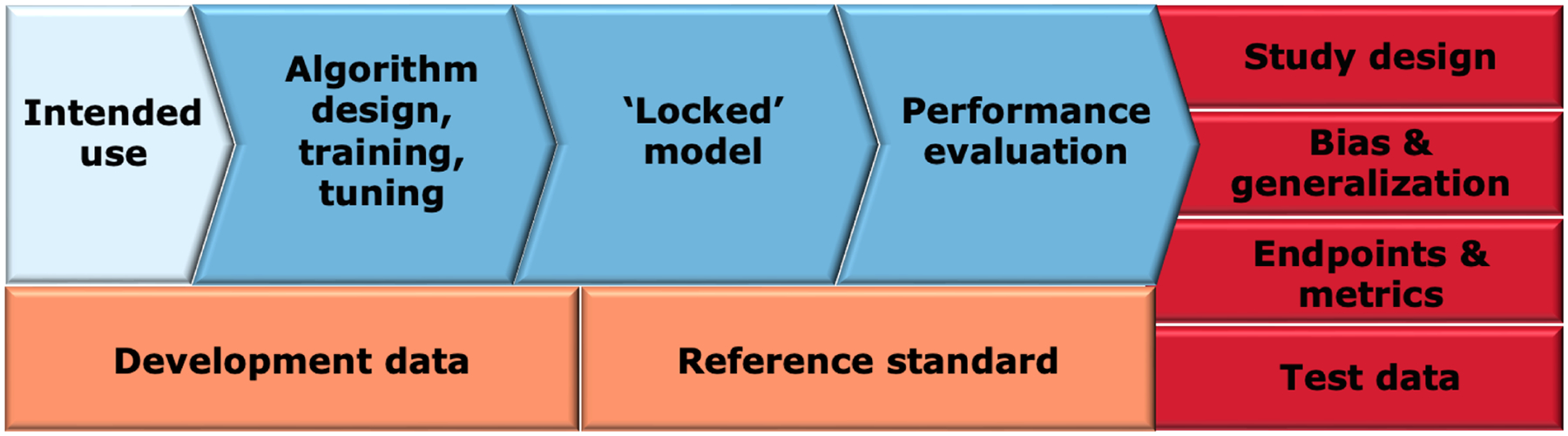
The intended use of the device (light blue) influences the data used for device development and the associated reference standard (orange). The device development process (blue) includes the algorithm development, a resulting locked model, and model evaluation. Clinical performance evaluation (red) consists of several interrelated components such as study design, bias and generalization assessment, selection of appropriate endpoints and metrics and selection of testing data that is representative of the target population.
Table 2.
Device classes and pre-market requirements.
| Device class | Controls | Premarket review process | Highlights |
|---|---|---|---|
| Class I (Low risk) | General controls | Most are exempt | |
| Class II (Moderate/controlled risk) | General controls Special controls | Premarket notification (510(k)) - substantially equivalent to a predicate | Most common way to market |
| De Novo - no predicate | Newer pathway to market | ||
| Class III (High risk) | General controls Premarket approval | Premarket approval (PMA) | High risk devices with most stringent data requirements |
21 CFR Part 860 -- Medical Device Classification Procedures. (27)
Beyond Class, AI/ML SaMDs will fall into different device types based on function and intended use. For example, devices may provide annotations, measure objects of interest in medical images, improve the quality of acquired images, help with the acquisition of images, prioritize patients based on health conditions, alert a clinician to a suspicious region in the image, or provide diagnostic values to aid in medical decisions. Generally, the intended use of the device drives the required type and amount of validation, outcomes data and required details on the clinical reference standard. Similarly, the intended use can drive requirements of how the AI/ML SaMD algorithm must be trained and tested to result in a “locked” model followed by performance evaluation.
With respect to designing studies to evaluate SaMD performance, it is important to incorporate study endpoints and metrics to validate both the standalone performance of the device, as well as its performance within the intended clinical workflow. Careful planning and analysis are needed to ensure that bias is minimized at all stages, including data collection, reference standard determination, algorithm development, metrics, and performance assessment. Through multi-disciplinary experts, FDA makes non-binding recommendations regarding the above considerations, including recommendations on performance assessment of computer-assisted detection devices for radiological imaging and other device data(28–30).
Regulatory challenges for AI/ML SaMDs.
Regulatory science gaps for AI include the lack of consensus methods for enhancing algorithm training for small clinical datasets, the lack of clear definition and understanding of artifacts, limitations, and failure modes for deep-learning-based devices for image denoising and reconstruction, lack of assessment techniques to evaluate trustworthiness of adaptive and autonomous AI/ML devices, and lack of a clear path to updating AI/ML SaMD if the training and/or algorithm that make up the device change.
To address these gaps and prepare for increasing AI/ML-enabled device submissions, the Center for Devices and Radiological Health (CDRH) has begun several initiatives, incorporating feedback from various stakeholders including patients. The digital health Center of Excellence was created to advance digital health technology, including mobile health devices, SaMDs, and wearables used as medical devices. The FDA has created an AI/ML SaMD action plan(28), held a public workshop on transparency of AI/ML-enabled medical devices(29), developed guiding principles for good practice in ML device development, and proposed a regulatory framework for modifications to AI/ML-based SaMDs(30).
Furthermore, the FDA recognizes certain regulatory-grade methods and tools as medical device development tools (MDDTs)(29) in order to improve predictability and efficiency in regulatory review. Medical device sponsors can use MDDTs in the development and evaluation of their devices and be sure they will be accepted by the FDA without the need to reconfirm their suitability.
Interfacing with FDA.
Early and frequent communication between the FDA and developers of cardiovascular imaging SaMDs is vital in bringing high-quality, safe, and effective devices to the market in efficient and scientifically valid ways. The Agency’s Q-Submission program(31) introduced in 2019 provides a mechanism for interactive dialogue with the FDA regarding early stage product development, protocols for clinical and non-clinical testing, and preparation of premarket applications across a wide range of device types and regulatory pathways. Another mechanism to determine device classification of a device is through the 513(g) Request for Classification(32). Through the 513(g), developers can inquire about the appropriate regulatory path for their device which can significantly impact the timeline and resources required to bring the product to market.
Regulatory research in AI.
The regulatory challenges for AI/ML SaMDs mentioned above require both a policy response and ongoing technical research. The Artificial Intelligence and Machine Learning research program in Center for Devices and Radiological Health’s Office of Science and Engineering Laboratories (OSEL) conducts regulatory science research to ensure patient access to safe and effective medical devices using AI/ML. The program is charged with addressing scientific gaps such as methods for AI evaluation, image noise, artifacts, failure modes of AI/ML, trustworthiness, and generalizability.
In addition, OSEL has created a mechanism for developing and sharing regulatory science tools (RSTs)(33) to help assess the safety and effectiveness of emerging technologies. RSTs are computational or physical phantoms, methods, datasets, computational models, and simulation pipelines. RSTs are designed to accelerate the development of technologies and products by providing turn-key solutions for assessment of device performance.
5: Implementation
In addition to regulatory compliance, deployment of AI algorithms in clinical practice must include consideration of effectiveness in real-world settings, workflow integration, trust, and adoption by providers, reimbursement, and continuous monitoring and updating. Challenges in real-world implementation include:
Additional consideration is needed to determine whether and how payer reimbursement, an important incentive to clinical implementation of new devices and services, applies to AI tools for cardiovascular imaging.
Determining how to leverage other incentives to implementation, including outcomes and healthcare utilization data, may help in moving forward clinical translation of AI.
Processes to integrate AI into existing clinical workflows and to perform continuous monitoring and updating require additional development.
Reimbursement.
Reimbursement by the Centers for Medicare and Medicaid Services (CMS) and other payers may be a significant hurdle in the implementation process. From a payer perspective, the use of AI algorithms does not necessarily add specific, measurable benefit beyond current patient care. Electrocardiograms, for example, have long included algorithms for computation of rates and intervals, and diagnoses, but that software is not separated out for payment. Similarly, AI algorithms are generally not considered as a standalone procedure or service but rather as part of additional device or analysis support for providers.
There are, however, some special cases of AI imaging algorithms obtaining CMS approval. Reimbursement requests in CMS are handled through 3 different pathways: Medicare physician fee schedule (MPFS), the Inpatient Prospective Payment System (IPPS), or the Hospital Outpatient Prospective Payment System (HOPPS). Within MPFS, a new taxonomy for classification of an AI-supported service or procedure into one of three categories was recently released. The taxonomy recognizes assistive (machine detects clinically relevant data without analysis or conclusion), augmentative (machine analyzes and/or quantifies data), or autonomous (machine automatically interprets data without physician involvement)(34) algorithms. Furthermore, the New Technology Add-on Payment (NTAP) is part of the IPPS and was recently used for approval of an AI-guided ultrasound platform(35), for coronary artery fractional flow reserve CT analysis(36), and for AI-enabled coronary plaque analysis from CT(37). Of note, temporary add-on payment augmentation is considered to recognize the increased provider cost to furnish an AI-assisted service, rather than recognition of the AI itself as a payable standalone benefit. Additionally, to the extent that AI software takes over physician work, one could argue that physician payment could actually be reduced in the current paradigm of resource-based reimbursement.
In the end, payers, like other stakeholders, will be positively influenced by strong causal evidence of improved health outcomes attributable to AI-augmented management versus current best practices.
Outcomes.
Improved patient outcomes derived from AI tools would incentivize payers, physicians, hospital administrators, data scientists, and patients alike to adopt and implement their use, especially where reimbursement opportunities may be limited. As has been done for drugs and other devices, there is an opportunity to use prospective, multicenter randomized clinical trials (RCTs) to test whether AI tools confer significant benefit and efficacy. For example, a recent trial of AI-enabled left ventricular ejection fraction (LVEF) found that AI-based LVEF was superior to initial sonographer assessment(38). While RCTs are a mainstay of clinical research, post-hoc imaging analyses making use of imaging already in clinical trials and large registries can also show improvement in diagnosis and prognostication for patients compared to standard of care(21,39,40). An equally important aspect requiring evaluation is the potential impact of incidental findings uncovered by AI, although this was not extensively discussed in the workshop.
Healthcare utilization studies may additionally reveal cost and quality incentives for implementing AI. For example, use of a deep neural network-based computational workflow for inline CMR myocardial perfusion analysis not only removes the subjective, qualitative aspect of interpreting stress perfusion images(20), but it also can significantly reduce the time needed for image analysis. Healthcare utilization studies may also reveal cost savings when AI is used to re-purpose imaging acquired for one purpose in order to find additional benefits. For example, deep learning has utilized CT scans performed for non-cardiac purposes to report additional biomarkers of cardiovascular risk such as coronary calcium, epicardial fat, or chamber size(41).
Integration.
Finally, easy integration into existing clinical workflows will incentivize adoption of AI-enabled tools for cardiovascular imaging. Technical challenges to integration include the need for software engineers to clean and package code, the need for user interfaces for the AI tools, and the need for interfaces/integrations with existing vendor-based imaging machines and/or software. The latter may be proprietary and therefore difficult to access without cooperation from vendors. Regulatory challenges to integration include testing, evaluating, and securing approval for modifications to clinical software(28). Finally, integration requires clinicians and hospital administrators to design and test care pathways that incorporate AI tools.
6: Human Capital
When discussing AI, it has become common to pit AI models against humans. However, every aspect of operationalizing AI for cardiovascular imaging previously described needs both AI and humans. Cardiovascular imaging experts are particularly important, as they will ultimately be the ones to use or not use AI to care for patients. Therefore, AI developers must collaborate with cardiovascular imagers in all implementation strategies to ensure the process’s practicality, robustness, and reliability. At the same time, physicians do not typically receive training in data science or AI. Challenges for developing human capital include:
Clinician users are unfamiliar with AI development and validation processes, making it challenging for them to critically evaluate AI and to trust it.
Dissemination of AI and education of the general community is limited to institutional pockets.
Clinical studies and trials occur more commonly at single sites with a narrow focus of disease.
Consensus on scientific rigor and reporting requirements for datasets and algorithms is still diffuse.
Education and training.
AI techniques remain weakly understood outside of the realms of technical experts. For broader adoption, AI requires more education and closer collaboration among computer scientists, researchers, physicians, and technical staff, to identify the most relevant problems to be solved and the best approaches for using data sources to achieve this goal. In addition, given the upsurge of AI techniques in cardiac imaging, we need AI as a topic included in educational curricula offered for professional certifications. Moreover, institutions, administrators, and service-line leaders need to incentivize using AI to overcome health information silos that undermine care coordination and efficiency. Finally, automated tools that allow novices to implement AI models without writing code could help bridge the AI-literacy gap.
The application of AI for medical decision-making is fraught with challenges since treating patients does not imply simple “batch predictions” but more nuanced approaches given the varying needs of image interpretations within a clinical contextual framework. Moreover, applying such predictions for medical decision-making requires application within the framework of a patient-doctor relationship for shared decision-making. Therefore, while adopting and implementing AI, hospital and institutional leaders should invest in suitable human capital, training needs, and team structures and develop the right incentives for adopting AI-integrated workflow solutions.
Real-life Translation.
Improved education about AI can enable clinical researchers to study its translational impact. One obstacle to incorporating AI-based imaging tools is the failure of models to generalize when deployed across institutions with heterogeneous populations which differ in disease prevalence, racial and gender diversity, comorbidities, and other non-biological factors like referral patterns, equipment, imaging protocols, and archival. Indeed, only a small fraction of published studies has reported robust sets of prospective real-life external validation. For example, despite independent validation, a deep-learning diabetic-retinopathy screening imaging tool developed by Google Health faced challenges when deployed in a clinic workflow(42). As a result, there is growing emphasis on continuous evaluations in real-world settings and pragmatic clinical trials. The recently published extensions of the CONSORT and Standard Protocol Items: Recommendations for Interventional Trials (SPIRIT) statements for RCTs of AI-based interventions (namely Consolidated Standards of Reporting Trials (CONSORT)-AI and SPIRIT-AI) are beginning to provide such a framework(43,44).
The role of professional medical societies in translation of AI to the bedside.
Societies can help develop guidelines and standards that can facilitate interoperability of data and algorithms among developers, industry, and healthcare institutions. They can also aid research, benchmarking, and quality improvement by developing national imaging registries that are easily available to developers and researchers(45). For example, the development of the UK Biobank(46) imaging data has attracted investigators to embrace new ways to diagnose, treat, and potentially predict the onset of cardiovascular diseases. Societies may build interest and collaboration around cardiovascular imaging use cases by hosting hackathons or data science competitions. They may also sponsor physicians to pursue training in AI, through educational content at conferences as well as dedicated training. Finally, societies can serve as alpha- and beta-users of AI, offering feedback on AI-human interactions, user interface, impact evaluation compared to the standard of care, and other implementation issues.
Dissemination of AI through journals and publications.
Once considered a niche for mainstream high-impact journals, AI-related research has been increasingly presented in cardiac imaging journals. By thoughtfully curating specific topics that engage AI algorithms’ downstream impact, journal editors can contribute significantly to responsible AI research. Moreover, improving the ability of non-technical experts and reviewers to assess the potential impact of AI research is critical for timely, constructive peer review and acceptance. Guidelines for scientific rigor and reporting in AI, as well as general educational opportunities, will create a deeper bench of qualified reviewers for cardiovascular imaging AI papers. The development of such steps enables a significant proportion of non-technical clinical reviewers to assess complex AI-related topics while focusing primarily on clinical translation of the AI approaches. Finally, expanding the use of preprint servers in partnership with peer-reviewed journals can improve timely dissemination of novel research.
Conclusion
There are several multi-disciplinary research opportunities and additional developmental work that are all important to translate AI of cardiovascular imaging forward into clinical practice. Some of these opportunities that were identified in the workshop include:
Developing methods to assess data quality and diversity, data harmonization, and data security, all while also promoting a diverse AI workforce
Expanding methods for data- and label-efficient algorithms, multi-modality algorithms, and methods for evaluation of AI models’ robustness, generalizability, and learned features
Developing methods for interoperable and sustainable code across computing platforms that are easy for biomedical researchers to learn and use
In regulatory science, addressing the lack of consensus methods for enhancing algorithm training for small clinical datasets, a better understanding of failure modes for AI devices, developing assessment methods to evaluate adaptive and autonomous devices, and forging a clear path to updating AI-based SaMD as technology rapidly evolves
Formulating clinical trials, healthcare utilization studies, and other implementation studies to demonstrate AI’s ability to impact clinical outcomes and thus help facilitate payer reception of AI-based tools
Educating clinicians in AI to improve trust in the methods and thus promote evaluation of AI for clinical cardiology by leading clinical trials and contributing to quality improvement, and engaging societies in the education process, in supporting AI-friendly datasets and registries that are readily accessible to users, and in helping to create guidelines
Clinical cardiology is well-positioned for implementation of artificial intelligence that is expected to bring improvement across cardiovascular imaging workflow, diagnosis, management, and prognosis. Much work remains to be done in all areas of artificial intelligence in cardiovascular imaging, but with clear challenges and opportunities set out by a multi-disciplinary, cohesive community, progress can and will be achieved.
Supplementary Material
Figure 3: The computing infrastructure landscape for AI is complex, including several types of stakeholders and several computing resources and tools.
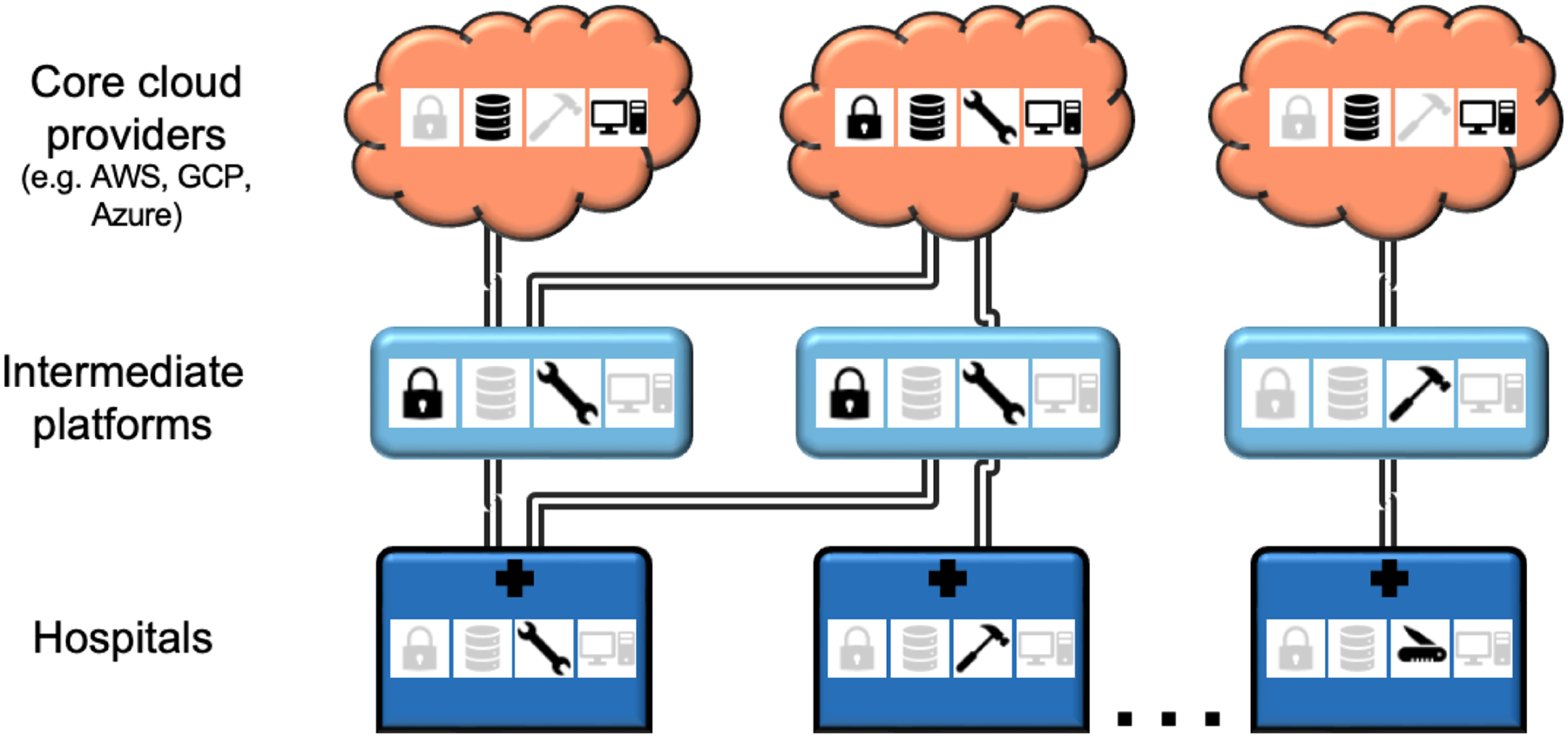
While a crowded playing field has its advantages, the complexity could also make it more difficult for medical centers to participate in AI research and to collaborate with each other. Stakeholders include medical centers (dark blue), large cloud providers (orange), as well as intermediate platforms (light blue) that offer certain computing features and tools but ultimately rely on large cloud providers for core services. Given the complexity of the computing landscape, the computing option(s) chosen by an institution affect availability of certain tools and/or datasets and can affect ease of collaboration with other institutions. AWS, Amazon Web Services. GCP, Google Cloud Platform.
Central Illustration: Translating AI into patient care for cardiovascular imaging across modalities requires several components and has potential for improving care.
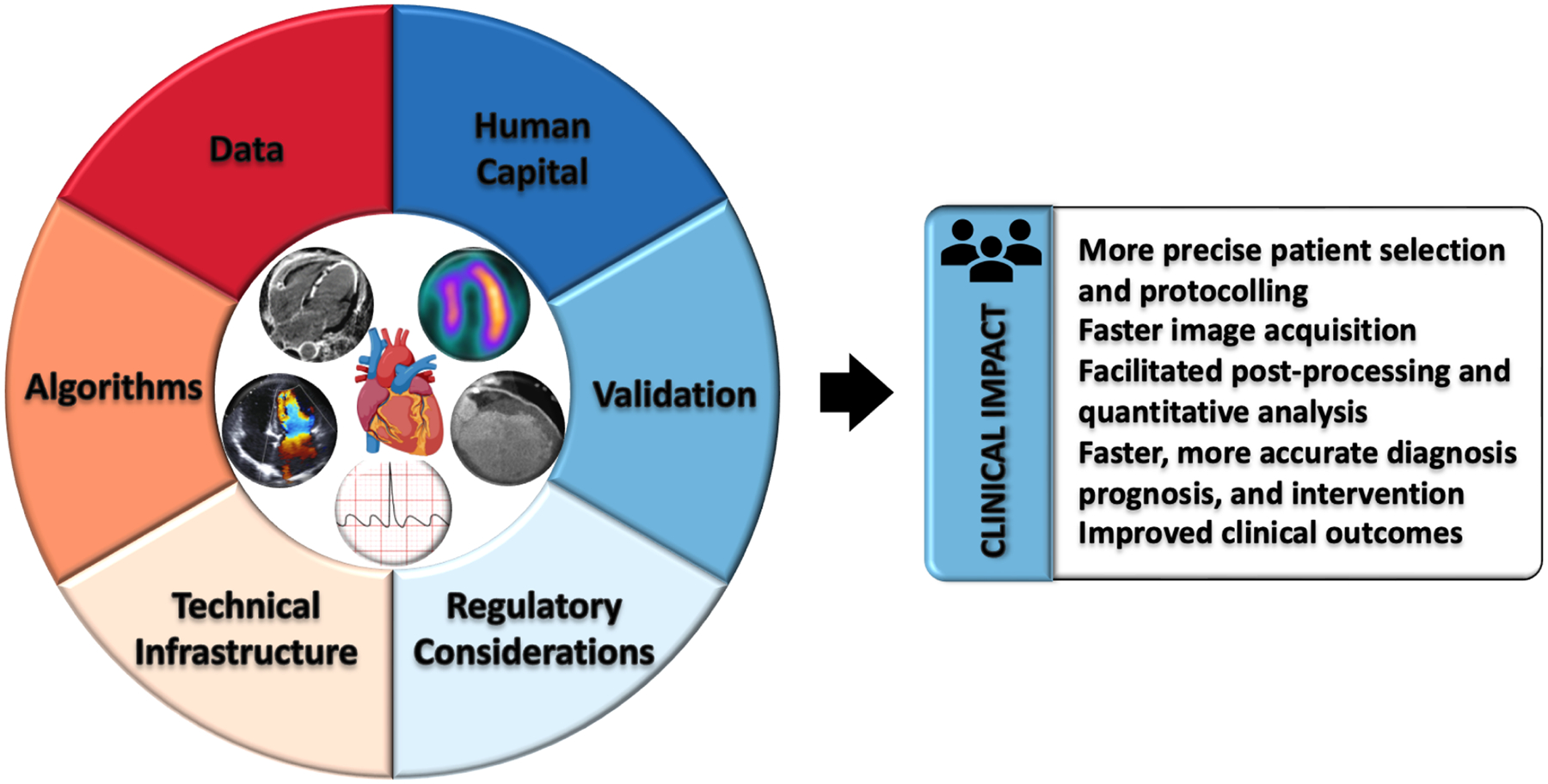
Data management, algorithm innovation, technical infrastructure, regulatory policies, and human capital must all continue to be developed in concert in order to enable clinical validation and testing, to ultimately achieve clinical impact for patient care.
Highlights.
Despite increasing research, clinical use of artificial intelligence (AI) for cardiovascular imaging is still rare
We identified key stakeholder groups to build consensus around challenges and priorities in supporting this research through to implementation
Major needs to advance the field include methods and policies supporting analysis of data quality, content, and diversity; scalable, accessible, and flexible computing platforms; and clinical testing for AI algorithms
Disclosures and Funding Support
The National Heart, Lung, and Blood Institute (NHLBI) Workshop on Artificial Intelligence in Cardiovascular Imaging: Translating Science to Patient Care, held on June 27th and 28th, 2022, was supported by the Division of Cardiovascular Sciences, NHLBI.
Dr. Antani is supported by the Intramural Research Program of the National Library of Medicine (NLM) and National Institutes of Health.
Dr. Arnaout is supported by the National Institutes of Health, the Department of Defense, and the Gordon and Betty Moore Foundation.
Dr. Dey receives software royalties from Cedars Sinai. She receives funding support from the National Institutes of Health/National Heart, Lung, and Blood Institute grants 1R01HL148787-01A1and 1R01HL151266.
Dr. Leiner serves on the Advisory Board for Cart-Tech B.V. and AI4Med, is a Clinical Advisor for Quantib B.V., and a Consultant for Guerbet. He receives funding support from the Netherlands Heart Foundation.
Dr. Sengupta serves on the Advisory Boards of Echo IQ and RCE Technologies, and he receives funding support from NSF Award: 2125872 and NRT-HDR: Bridges in Digital Health.
Dr. Shah receives consulting fees from AstraZeneca, Amgen, Aria CV, Axon Therapies, Bayer, Boehringer-Ingelheim, Boston Scientific, Bristol Myers Squib, Cytokinetics, Edwards Lifesciences, Eidos, Gordian, Intellia, Ionis, Merck, Novartis, Novo Nordisk, Pfizer, Prothena, Regeneron, Rivus, Sardocor, Shifamed, Tenax, Tenaya, and United Therapeutics, and he receives funding support from the National Institutes of Health (U54 HL160273, R01 HL140731, R01 HL149423), Corvia, and Pfizer.
Dr. Slomka receives software royalties from Cedars Sinai, and he receives funding support from the National Institutes of Health/National Heart, Lung, and Blood Institute grant 1R35HL161195-01.
Dr. Williams is a Speaker at lectures sponsored by Canon Medical Systems and the Siemens Healthineers. She receives funding support from the British Heart Foundation (FS/ICRF/20/26002).
The remaining authors have no disclosures to report.
Abbreviations:
- AI
artificial intelligence
- AWS
Amazon Web Services
- CPU
central processing unit
- DICOM
Digital Imaging and Communications in Medicine
- FAIR
findability, accessibility, interoperability, reusability
- GPU
graphics processing unit
- ML
machine learning
- MDDTs
medical device development tools
- MIDRC
Medical Imaging and Data Resource Center
- SaMD
software as a medical device
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer: The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Heart, Lung, and Blood Institute, National Institutes of Health, or the United States Department of Health and Human Services.
References:
- 1.de Roos A, Higgins CB. Cardiac radiology: centenary review. Radiology 2014;273(2 Suppl):S142–159. Doi: 10.1148/radiol.14140432. [DOI] [PubMed] [Google Scholar]
- 2.Birger M, Kaldjian AS., Roth GA., Moran AE., Dieleman JL., Bellows BK. Spending on Cardiovascular Disease and Cardiovascular Risk Factors in the United States: 1996 to 2016. Circulation 2021;144(4):271–82. Doi: 10.1161/CIRCULATIONAHA.120.053216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen SE., Abdulkareem M, Leiner T Artificial Intelligence Will Transform Cardiac Imaging-Opportunities and Challenges. Front Cardiovasc Med 2019;6:133. Doi: 10.3389/fcvm.2019.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen CM., Raynor ME., McDonald R. What Is Disruptive Innovation? Harvard Business Review 2015. [Google Scholar]
- 5.Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 2021;3(3):199–217. Doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 6.Gupta R, Sharma A, Kumar A Super-Resolution using GANs for Medical Imaging. Procedia Computer Science 2020;173:28–35. Doi: 10.1016/j.procs.2020.06.005. [DOI] [Google Scholar]
- 7.Chinn E, Arora R, Arnaout R, Arnaout R ENRICH: Exploiting Image Similarity to Maximize Efficient Machine Learning in Medical Imaging 2021:2021.05.22.21257645. Doi: 10.1101/2021.05.22.21257645. [DOI] [Google Scholar]
- 8.Seyyed-Kalantari L, Zhang H, McDermott MBA., Chen IY., Ghassemi M Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in underserved patient populations. Nat Med 2021;27(12):2176–82. Doi: 10.1038/s41591-021-01595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazura JC., Juluru K, Chen JJ., Morgan TA., John M, Siegel EL. Facial Recognition Software Success Rates for the Identification of 3D Surface Reconstructed Facial Images: Implications for Patient Privacy and Security. J Digit Imaging 2012;25(3):347–51. Doi: 10.1007/s10278-011-9429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson MD., Dumontier M, Aalbersberg IjJ., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3(1):160018. Doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramon AJ., Yang Y, Pretorius PH., Johnson KL., King MA., Wernick MN. Improving Diagnostic Accuracy in Low-Dose SPECT Myocardial Perfusion Imaging With Convolutional Denoising Networks. IEEE Trans Med Imaging 2020;39(9):2893–903. Doi: 10.1109/TMI.2020.2979940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang D, He B, Ghorbani A, et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 2020;580(7802):252–6. Doi: 10.1038/s41586-020-2145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endo M, Krishnan R, Krishna V, Ng AY., Rajpurkar P Retrieval-Based Chest X-Ray Report Generation Using a Pre-trained Contrastive Language-Image Model. Proceedings of Machine Learning for Health. PMLR; 2021. p. 209–19. [Google Scholar]
- 14.Ferreira DL., Salaymang Z, Arnaout R Label-free segmentation from cardiac ultrasound using self-supervised learning 2022. [Google Scholar]
- 15.Kwiecinski J, Tzolos E, Meah MN., et al. Machine Learning with 18F-Sodium Fluoride PET and Quantitative Plaque Analysis on CT Angiography for the Future Risk of Myocardial Infarction. J Nucl Med 2022;63(1):158–65. Doi: 10.2967/jnumed.121.262283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanbhag AD., Miller RJH., Pieszko K, et al. Deep Learning-based Attenuation Correction Improves Diagnostic Accuracy of Cardiac SPECT. J Nucl Med 2022:jnumed.122.264429. Doi: 10.2967/jnumed.122.264429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Kwiecinski J, Cadet S, et al. Automated nonlinear registration of coronary PET to CT angiography using pseudo-CT generated from PET with generative adversarial networks. J Nucl Cardiol 2022. Doi: 10.1007/s12350-022-03010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieszko K, Shanbhag A, Killekar A, et al. Deep Learning of Coronary Calcium Scores From PET/CT Attenuation Maps Accurately Predicts Adverse Cardiovascular Events. JACC: Cardiovascular Imaging 2022. Doi: 10.1016/j.jcmg.2022.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Lin A, Manral N, McElhinney P, et al. Deep learning-enabled coronary CT angiography for plaque and stenosis quantification and cardiac risk prediction: an international multicentre study. The Lancet Digital Health 2022;4(4):e256–65. Doi: 10.1016/S2589-7500(22)00022-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue H, Davies RH., Brown LAE., et al. Automated Inline Analysis of Myocardial Perfusion MRI with Deep Learning. Radiol Artif Intell 2020;2(6):e200009. Doi: 10.1148/ryai.2020200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Kwiecinski J, Miller RJH., et al. Deep Learning for Explainable Estimation of Mortality Risk From Myocardial Positron Emission Tomography Images. Circ Cardiovasc Imaging 2022;15(9):e014526. Doi: 10.1161/CIRCIMAGING.122.014526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rjh M, K K, A S, et al. Explainable Deep Learning Improves Physician Interpretation of Myocardial Perfusion Imaging. Journal of Nuclear Medicine : Official Publication, Society of Nuclear Medicine 2022. Doi: 10.2967/jnumed.121.263686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiseleva A, Kotzinos D, De Hert P Transparency of AI in Healthcare as a Multilayered System of Accountabilities: Between Legal Requirements and Technical Limitations. Front Artif Intell 2022;5:879603. Doi: 10.3389/frai.2022.879603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MONAI - Home. Available at: https://monai.io/. Accessed October 18, 2022.
- 25.Cloud Healthcare API. Google Cloud. Available at: https://cloud.google.com/healthcare-api. Accessed October 11, 2022.
- 26.Orthanc - DICOM Server. Available at: https://www.orthanc-server.com/. Accessed October 11, 2022.
- 27.21 CFR Part 860 -- Medical Device Classification Procedures. Available at: https://www.ecfr.gov/current/title-21/chapter-I/subchapter-H/part-860. Accessed March 12, 2023.
- 28.U.S. Food and Drug Administration. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD): Discussion Paper and Request for Feedback. April 2019. Accessed March 13, 2023. https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf.
- 29.U.S. Food and Drug Administration, Health Canada, and United Kingdom Medicines and Healthcare products Regulatory Agency. Good Machine Learning Practice for Medical Device Development: Guiding Principles. October 2021. Accessed March 13, 2023. https://www.fda.gov/media/153486/download.
- 30.Sengupta PP., Shrestha S, Berthon B, et al. Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME): A Checklist: Reviewed by the American College of Cardiology Healthcare Innovation Council. JACC Cardiovasc Imaging 2020;13(9):2017–35. Doi: 10.1016/j.jcmg.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.U.S. Food and Drug Administration. Requests for Feedback and Meetings for Medical Device Submissions: The Q-Submission Program, Guidance for Industry and Food and Drug Administration Staff. January 2021. Accessed March 13, 2023. https://www.fda.gov/media/114034/download.
- 32.U.S. Food and Drug Administration. FDA and Industry Procedures for Section 513(g) Requests for Information under the Federal Food, Drug, and Cosmetic Act, Guidance for Industry and Food and Drug Administration Staff. December 2019. Accessed March 13, 2023. https://www.fda.gov/media/78456/download.
- 33.U.S. Food and Drug Administration. Catalog of Regulatory Science Tools to Help Assess New Medical Devices. Accessed March 13, 2023. https://www.fda.gov/medical-devices/science-and-research-medical-devices/catalog-regulatory-science-tools-help-assess-new-medical-devices.
- 34.CPT Appendix S: AI taxonomy for medical services & procedures | American Medical Association. Available at: https://www.ama-assn.org/practice-management/cpt/cpt-appendix-s-ai-taxonomy-medical-services-procedures. Accessed October 11, 2022.
- 35.MEARIS™. Available at: https://mearis.cms.gov/public/resources?app=ntap. Accessed October 18, 2022.
- 36.LCD - Non-Invasive Fractional Flow Reserve (FFR) for Stable Ischemic Heart Disease (L39075). Available at: https://www.cms.gov/medicare-coverage-database/view/lcd.aspx?lcdid=39075&ver=7. Accessed October 11, 2022.
- 37.HHS.gov Guidance Document October-2022 Update Hospital Outpatient Prospective Payment System (OPPS). Available at: https://hhsgovfedramp.gov1.qualtrics.com/jfe/form/SV_3Jyxvg4zv8sPGrH?Q_CHL=si&Q_CanScreenCapture=1. Accessed October 18, 2022.
- 38.Ouyang D Blinded Randomized Controlled Trial of Artificial Intelligence Guided Assessment of Cardiac Function. clinicaltrials.gov; 2022.
- 39.Lin A, Wong ND., Razipour A, et al. Metabolic syndrome, fatty liver, and artificial intelligence-based epicardial adipose tissue measures predict long-term risk of cardiac events: a prospective study. Cardiovasc Diabetol 2021;20(1):27. Doi: 10.1186/s12933-021-01220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knott KD., Seraphim A, Augusto JB., et al. The Prognostic Significance of Quantitative Myocardial Perfusion. Circulation 2020;141(16):1282–91. Doi: 10.1161/CIRCULATIONAHA.119.044666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greco F, Salgado R, Van Hecke W, Del Buono R, Parizel PM., Mallio CA. Epicardial and pericardial fat analysis on CT images and artificial intelligence: a literature review. Quant Imaging Med Surg 2022;12(3):2075–89. Doi: 10.21037/qims-21-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beede E, Baylor E, Hersch F, et al. A Human-Centered Evaluation of a Deep Learning System Deployed in Clinics for the Detection of Diabetic Retinopathy. Proceedings of the 2020 CHI Conference on Human Factors in Computing Systems. New York, NY, USA: Association for Computing Machinery; 2020. p. 1–12. [Google Scholar]
- 43.Liu X, Rivera SC., Moher D, Calvert MJ., Denniston AK. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI Extension. BMJ 2020;370:m3164. Doi: 10.1136/bmj.m3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera SC., Liu X, Chan A-W., Denniston AK., Calvert MJ. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI Extension. BMJ 2020;370:m3210. Doi: 10.1136/bmj.m3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ACC/AHA/STS Statement on the Future of Registries and the Performance Measurement Enterprise | Circulation: Cardiovascular Quality and Outcomes. Available at: https://www.ahajournals.org/doi/full/10.1161/HCQ.0000000000000013. Accessed October 11, 2022. [DOI] [PubMed]
- 46.UK Biobank - UK Biobank. Available at: https://www.ukbiobank.ac.uk/. Accessed October 11, 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


