Abstract
TET2 is a member of the ten-eleven translocation (Tet) family of DNA dioxygenases that regulate gene expression by promoting DNA demethylation (enzymatic activity) and partnering with chromatin regulatory complexes (nonenzymatic functions). TET2 is highly expressed in the hematopoietic lineage where its molecular functions are subject of continuous investigations due to the prevalence of TET2 mutations in hematologic malignancies. Previously, we have implicated Tet2 catalytic and noncatalytic functions in regulation of the myeloid and lymphoid lineages, respectively. However, the impact of these functions of Tet2 on hematopoiesis as the bone marrow ages remains unclear. Here, we conducted comparative transplantations and transcriptomic analyses of 3-, 6-, 9-, and 12-month-old Tet2 catalytic mutant (Mut) and knockout (KO) bone marrow. Tet2 Mut bone marrow of all ages exclusively caused hematopoietic disorders of the myeloid lineage. In contrast, young Tet2 KO bone marrow developed both lymphoid and myeloid diseases, while older Tet2 KO bone marrow predominantly elicited myeloid disorders with shorter latency than age-matched Tet2 Mut bone marrow. We identified robust gene dysregulation in Tet2 KO Lin− cells at 6 months that involved lymphoma and myelodysplastic syndrome and/or leukemia-causing genes, many of which were hypermethylated early in life. There was a shift from lymphoid to myeloid gene deregulation in Tet2 KO Lin− cells with age, underpinning the higher incidence of myeloid diseases. These findings expand on the dynamic regulation of bone marrow by Tet2 and show that its catalytic dependent and independent roles have distinct impacts on myeloid and lymphoid lineages with age.
INTRODUCTION
The Ten-eleven translocation (Tet) family of enzymes are DNA dioxygenases that oxide 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine1-3. These modified bases promote both active and passive DNA demethylation4,5. Tet enzymes also regulate gene expression in an enzymatic-independent manner by facilitating the recruitment of several transcription factors and chromatin regulatory complexes to their target sites in the genome4,6,7. For example, we and others have shown that Tet enzymes partner with Sin3a and the polycomb repressive complex (PRC2) to promote H3K27 deacetylation and trimethylation and suppress developmental genes in embryonic stem cells6-9. They also partner with O-linked N-acetylglucosamine transferase (OGT) to promote histone GlcNAcylation and gene activation in various cell types10,11. Therefore, both the catalytic and noncatalytic functions of Tet enzymes are critical for gene regulation and contribute to cellular hemostasis, development, and etiology of human diseases.
TET2 is a key member of this family that is highly expressed in the hematopoietic lineage and is commonly mutated or dysregulated in hematologic disorders and malignancies12-18. TET2 is mutated in myelodysplastic syndrome (MDS), a clonal hematologic disorder characterized by reduced blood cell production and dysplasia. Loss-of-function mutations of TET2 gene have been reported in ~20% of MDS and ~50% of chronic myelomonocytic leukemia (CMML) patients12-18. Loss of Tet2 in mice leads to increased hematopoietic stem cell (HSC) self-renewal and restricted differentiation, and the manifestation of CMML-like disease with multi-organ metastasis18-20. In addition to these critical requirements of Tet2 for the myeloid lineage and suppressing myeloid disorders, Tet2 also influences the lymphoid lineage. TET2 mutations have been reported in ~33% of angioimmunoblastic T-cell lymphomas, ~10% of diffuse large B-cell lymphomas, and 4% of mantle cell lymphoma. Consistently a small subset of Tet2 knockout mice present with abnormal lymphopoiesis21-29. Thus, Tet2 is important for the homeostasis of both myeloid and lymphoid lineages.
Our recent studies have defined distinct catalytic dependent and independent requirements of Tet2 in myeloid and lymphoid lineages, respectively29. Mice lacking Tet2 catalytic activity (Tet2 Mut) and mice knockout for Tet2 (Tet2 KO) developed hematopoietic disorders with similar survival rates, but their disease spectrum was different. Tet2 KO mice developed both myeloid and lymphoid disorders with increased hematopoietic stem and progenitor cell (HSPC) fractions, while Tet2 Mut mice presented with aberrant hematopoiesis restricted to the myeloid lineage. Transplantation of bone marrow from young Tet2 Mut and KO mice recapitulated these results. These findings were accompanied by distinct gene expression changes in the bone marrow Lin− Sca-1+c-Kit+ (LSK) and Lin− compartments of Tet2 Mut and KO mice29. While these findings implicated Tet2 catalytic and noncatalytic functions in the regulation of the myeloid and lymphoid lineages, respectively, their impact on hematopoiesis as the bone marrow ages remains unknown. Here, we have conducted comparative transplantation and transcriptomic analyses of 3-, 6-, 9-, and 12-month-old Tet2 Mut and KO bone marrow. Tet2 Mut bone marrow of all ages caused myeloid disorders only. In contrast, young Tet2 KO bone marrow caused both lymphoid and myeloid phenotypes, while older Tet2 KO bone marrow largely developed myeloid diseases in vivo with shorter latency than the age-matched Tet2 Mut bone marrow. We found a shift from an increased lymphoid to an increased myeloid gene dysregulation in Tet2 KO Lin− cells with age underpinning the higher myeloid and lower lymphoid disease incidence caused by Tet2 KO bone marrow. These findings establish that the catalytic dependent and independent roles of Tet2 have distinct impacts on myeloid and lymphoid lineages with age.
METHODS
Tet2 catalytic mutant and knockout mice
The Tet2 catalytic mutant29 and the Tet2 knockout20 mice were previously characterized in our labs29 and maintained in 129/B6 mixed background. The two mouse strains were first intercrossed and then cohorts of wild-type, Tet2 Mut and Tet2 KO mice were established and housed in an SPF (Specific-pathogen-free) barrier facility for one year. Animals with no overt signs of hematologic disease based on peripheral blood analyses were sacrificed at 3, 6, 9, and 12 months of age (3 to 5 animals/genotype per time point) to harvest bone marrow for transplantation and RNA-seq analyses. All studies were performed in accordance with our IACUC approved protocols overseen by the Institute for Animal Studies at Albert Einstein College of Medicine.
Flow cytometry analysis
Peripheral blood and bone marrow mononuclear cells (BM-MNCs) were isolated as previously described29-31. For flow cytometric analyses, we used monoclonal antibodies specific for the following: CD135 (A2F10), CD34 (RAM34), Sca-1 (D7), CD3e (145-2C11), CD8 (53-6.72), IgM (II/41), F4/80 (BM8), CD71 (R17217), CD45.2 or Ly5.2 (104), and CD45.1 or Ly5.1 (A20); all were from eBioscience. CD4 (GK1.5), B220 (RA3 6B2), TER-119 (TER-119), CD11b (M1/70), CD19 (1D3), and NK-1.1 (PK136) were from BD Bioscience. CD150 (TC15-12F12.2), CD48 (HM48-1), c-Kit (2B8) and Gr-1 (RB6-8C5) were from BioLegend. A mixture of monoclonal antibodies against CD4, CD8, CD3e, B220, TER-119, CD11b, Gr-1, IgM, CD19, and NK-1.1 was used as a lineage marker (Lineage). In some experiments, Streptavidin-APC-eFluor 780 (47-4317), -PerCP-Cy5.5 (45-4317) or -Pacific Blue (48-4317) was used as a secondary antibody. Samples were analyzed on an LSR II flow cytometer (Becton Dickinson). Data were analyzed using FlowJo 10 (Becton Dickinson). For cell sorting, the BM-MNCs were prepared as described before29-31 and cells were incubated with Anti-Biotin MicroBeads (10 μl beads per BM, Mylteni Biotech) for 10 min at room temperature, then ran through MACS LS column (Mylteni Biotech) for lineage depletion. Cells were sorted directly into StemSPAN™ serum-free expansion medium (SFEM from STEMCELL Technologies) through a BD FACS ARIA II (Becton Dickinson).
Bone marrow transplantation
To analyze hematopoietic regeneration and homeostasis in vivo, 4.0 × 105 BM-MNCs from primary Tet2 Mut, Tet2 KO mice, or littermate controls (Ly5.2) were transplanted into irradiated recipient mice (Ly5.1) with 4.0×105 competitor BMMNCs (Ly5.1) as described before29-31. Repopulation of donor (Ly5.2) myeloid and lymphoid cells was monitored, and in some experiments donor contribution to the immature hematopoietic lineages was also evaluated. When the transplanted mice were moribund or deceased their bone marrow was analyzed.
RNA-seq
Transcriptomic profiling by RNA-seq was performed as described before9,29. Briefly, LSK and Lin− cells were isolated from 3-, 6-, 9- and 12-month-old Tet2 wild-type, Mut, and KO mice (n=3 of each genotype for each timepoint was pooled) and RNA was extracted using Qiagen RNeasy Micro Kit (74004). Libraries were prepared and sequenced by Novogene using their Illumina Novoseq 6000 platform. Trim galore (v0.6.5) was used to trim adaptors. Clean reads were mapped to the mouse genome (mm10) using STAR (v2.7.3a). Mapped reads were assigned to genes and counts were extracted using featureCounts with --largestOverlap parameter. Differential expression analysis was performed on raw counts using edgeR (p-adjusted value <0.05 and fold-change >1.0), following the package manual.
WGBS
Whole-genome bisulfite sequencing (WGBS) was performed as described previously9. LSK and lineage negative cells were isolated from 3-month-old Tet2 WT, Mut, and KO mice (n=3 of each genotype pooled), and DNA was extracted by Quick-DNA miniprep kit (Zymo, D3024) following the manufacturer’s instructions. Bisulfite conversion, library preparation, and sequencing were performed by BGI Genomics (https://en.genomics.cn/). Libraries were subjected to 100 bp pair-end sequencing on a HiSeq 4000 Illumina platform. Adaptors and low-quality reads were filtered out using SOAPnuke (v1.5.5) (https://github.com/BGI-flexlab/SOAPnuke) with the parameters -n 0.001 -l 20 -q 0.4 -A 0.25 -Q 2 -G. Reads were mapped to the mouse genome (mm10) using Bismark (v0.22.3) with default parameters. Reads were de-duplicated using deduplicate_bismark and cytosine methylation status was extracted with bismark_methylation_extractor. MethPipe (v3.4.3) was used to find differentially methylated regions (DMRs) between Tet2 Mut vs. WT, and between Tet2 KO vs. WT in both cell types using standard parameters (regions >5 CpGs, methylation difference >20%, and FDR <0.05). Hypermethylated DMRs were annotated to genomic regions in R with the package ChIPseeker (v1.30.3). For visualization of DNA methylation levels on Integrative Genome Browser (IGV), bedGraph files were converted to bigwig using bedtools (v2.30.0) function bedGraphToBigWig.
Tet2 ChIP-seq analysis
Publicly available Tet2 ChIP-seq data in AML cells was downloaded from GEO (GSE115972) and processed as described before32. Briefly, raw data was trimmed using trim galore (v0.6.5), and clean reads were mapped to the mouse genome (mm10) using Bowtie2 (v 2.3.5.1) with standard parameters. Samtools (v1.9) was used to sort, index, and balance mapped reads. BAM files were removed of duplicates with Picard tools (v2.3.0) and peaks were called using MACS2 (v2.2.7.1). ChIPseeker (v1.30.3) was used to annotate called peaks and extract Tet2- bound genes at different genomic regions. Lists of Tet2-bound genes were intersected with DEGs and hyper-DMR-associated genes in R.
Data Availability:
The RNA-seq and WGBS data are deposited in the Gene Expression Omnibus (GEO) database (Accession: GSE227977).
RESULTS
Comparative transplantation of bone marrow from Tet2 Mut and KO mice of various ages
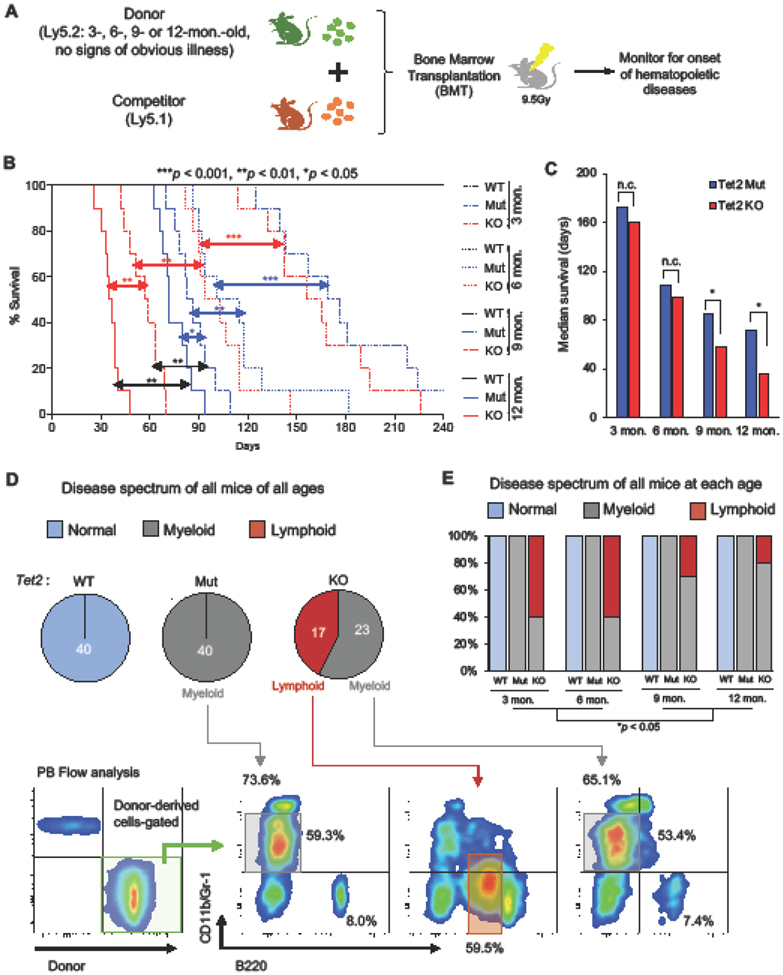
To examine the role of Tet2 in hematopoietic homeostasis and MDS suppression with age, we isolated bone marrow mononuclear cells (BM-MNCs) from 3-, 6-, 9- and 12-month-old Tet2 catalytic mutant (Mut), knockout (KO) and wild-type (WT) mice and subjected each to a competitive bone marrow transplantation (BMT) assay (Fig. 1A). 4 × 105 BM-MNCs (isolated and pooled from three donors without any signs of obvious hematologic disorder based on peripheral blood analysis) were transplanted into lethally irradiated congenic mice in competition with an equal number of BM-MNCs from Ly5.1 mice. In total 10 animals were transplanted for each group and monitored for up to 240 days for the onset of hematopoietic phenotypes. We found that all cohorts of recipient mice transplanted with donor Tet2 Mut or KO BM-MNCs displayed hematological disorders while recipient mice transplanted with WT donor cells remained disease-free during the course of the study. We made three unique observations: First, we found that mice transplanted with older (9- or 12-month) Tet2 Mut or KO BM-MNCs developed hematological disorders much faster than mice transplanted with younger (3- or 6- month) BM-MNCs (Fig. 1B). Second, recipients of younger Tet2 Mut or KO BM-MNCs had similar survival and disease onset, in contrast to recipients of older Tet2 Mut or KO BM-MNCs, where Tet2 Mut donor cells exhibited a survival advantage over the Tet2 KO group (Fig. 1B-C). Third, while recipient mice transplanted with Tet2 KO donor BM-MNCs developed both myeloid and lymphoid malignancies, recipients transplanted with Tet2 Mut donor cells displayed blood phenotypes restricted to the myeloid lineage (Fig. 1D). Strikingly, the disease spectrum initiated by young Tet2 KO donor BM-MNCs differed from that initiated by older Tet2 KO donors. Younger Tet2 KO donor cells led to onset of both myeloid and lymphoid disorders, while older Tet2 KO donor cells led to onset of predominantly myeloid diseases (Fig. 1E). In contrast to 60% recipients of 3- and 6-month-old Tet2 KO donor BM-MNCs developing lymphoid malignancies, 75-80% recipients of 9- or 12-month-old Tet2 KO donor BM-MNCs displayed skewed myeloid/lymphoid ratio with an expansion of the CD11b+Gr-1mid-high fractions (Fig. 1D-E). This shows that with age, Tet2 KO bone marrow becomes more biased toward the onset of myeloid disorders, which is likely initiated between 6 to 9 months of age. This prompted us to characterize in more detail the hematopoietic phenotypes induced by donor cells from 9-month-old mice.
Figure 1. Analysis of the age-related hematological phenotypes induced by Tet2 Mut and KO mouse bone marrow cells.
A. Schematic of transplantation of bone marrow from Tet2 Mut and KO mice at the indicated ages. 4.0 × 105 bone marrow mononuclear cells (BM-MNCs) from 3-, 6-, 9- and 12- month-old Tet2 Mut or KO primary mice with no sign of hematological disorders, or age-matched wild-type mice (Ly5.2) were transplanted into irradiated recipient mice (Ly5.1) with 4.0 × 105 competitor BM-MNCs (Ly5.1).
B. Kaplan-Meier survival curve of transplanted mice (n = 10 per each group). Log-rank test was used to assess statistical significance (p-value).
C. Median survival of transplanted mice. Note: control wild-type cohort survived beyond 240 days.
D. Disease spectrum of the recipient mice from entire cohort (3, 6, 9 and 12 months old, n = 40). Representative flow data and percentages of the indicated fractions of the donor-derived cells in the peripheral blood of recipient mice transplanted with Tet2 Mut and KO donor bone marrow cells which developed hematological disorders are shown in lower panels. Representative gating strategy of donor-derived cells (green) is also shown in bottom left.
E. Disease spectrum of each cohort (n = 10 per each group).
In all panels, error bars indicate SD unless otherwise specified. n.c. stands for no significant change. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
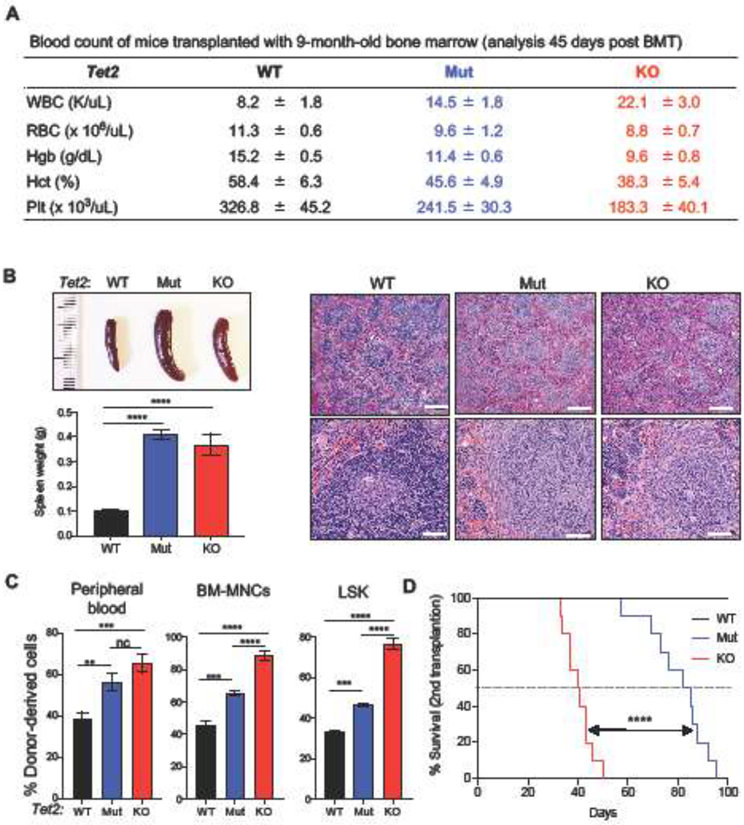
Recipient mice transplanted with 9-month-old Tet2 WT, Mut, and KO BM-MNCs were sacrificed 45 days after 1st transplantation (these animals were part of the cohort in Fig. 1B). Compared to WT controls, both the Tet2 Mut and KO groups had increased white blood cell counts and reductions in hemoglobin, hematocrit, and platelets (Fig. 2A). They also presented with splenomegaly with histopathologic abnormality (Fig. 2B) as well as higher donor chimerism in the peripheral blood and the immature hematopoietic lineages (Fig. 2C). Next, we transplanted 1.0 × 106 BM-MNCs from the primary recipient mice (1st BMT) into irradiated secondary recipient mice (Ly5.1, 2nd BMT). All recipient mice transplanted with either Tet2 KO or Mut donor cells developed lethal hematological disorders within 100 days of transplantation (Fig. 2D). However, mice transplanted with 9-month-old Tet2 Mut donor cells showed a survival advantage over the Tet2 KO group. 50% of Tet2 Mut group died by 83 days in contrast to Tet2 KO group that died by 41 days (Fig. 2D). Together, these analyses suggest that older Tet2 KO bone marrow accelerated disease progression causing more aggressive hematologic defects than Tet2 Mut bone marrow, and that Tet2 KO bone marrow becomes progressively biased towards myeloid defects with age.
Figure 2. Hematopoietic characterization induced by donor bone marrow cells from 9-month-old Tet2 Mut and KO mice.
A. Blood count of recipient mice transplanted with BM-MNCs from 9-month-old Tet2 Mut and KO donor bone marrow cells 45 days after 1st BMT (n = 5 of each genotype).
B. Representative images and weights (left) and H&E staining (right) of spleens of recipient mice transplanted with 9-month-old Tet2 WT, Mut, or KO BM-MNCs 45 days after 1st BMT. Scale bars= 400 μm (in top three panels) and 60 μm (in bottom three panels).
C. Percent donor-derived cells in peripheral blood (left) and bone marrow (BM-MNCs [middle] and LSK [right]) 45 days after 1st BMT.
D. Kaplan-Meier survival curve of recipient mice upon secondary transplantation (2nd BMT, 10 mice per group). Mice were transplanted with donor bone marrow cells from recipients of 1st transplantation. Log-rank test was used to generate p values.
LSK, Lin−Sca-1+c-Kit+. Error bars indicate SD. n.c. stands for no significant change. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Early and robust transcriptomic dysregulation in Tet2 KO Lin− cells in contrast to Tet2 Mut Lin− cells
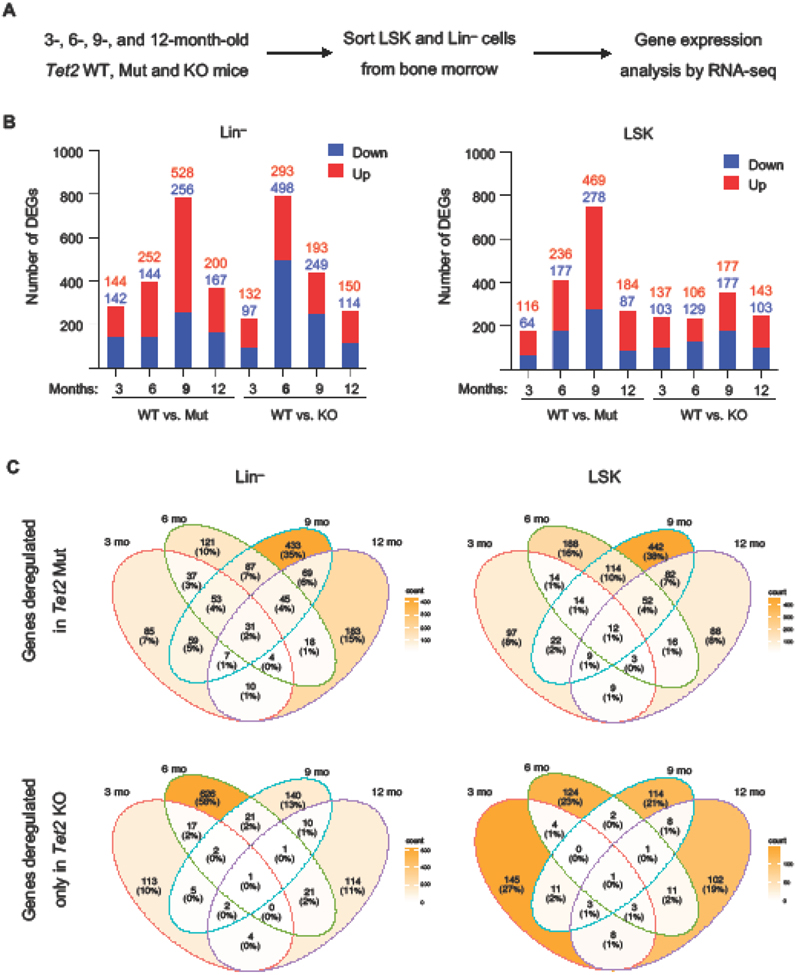
To gain molecular insights into the biology of Tet2 Mut and Tet2 KO bone morrow over time, we sorted progenitor fraction (Lin− cells) from the bone marrow of 3-,6-,9-, and 12-month-old Tet2 Mut, Tet2 KO and WT healthy mice (no signs of lymphoid or myeloid malignancies by peripheral blood analysis) and analyzed their gene expression profiles by RNA-seq (Fig. 3A). To rule out any potential of specific phenotypes enriched in hematopoietic stem and progenitor cells (HSPCs), LSK (Lin− Sca-1+ c-Kit+) cells were isolated from the same animals and subjected to the same unbiased transcriptomic analyses. We found a significant number of both up and down-regulated genes at each time point in Lin− and LSK cells of both genotypes (when compared to their WT counterparts) (Fig. 3B). Overlapping of differentially expressed genes (DEGs) with the publicly available Tet2 ChIP-seq data set in acute myeloid leukemia (AML) cells32 revealed that between 15-25% of DEGs across time points were direct targets of Tet2 (bound by Tet2 at their promoters, gene bodies or distal intergenic regions) (Fig. S1A). The number of DEGs was higher in 6- and 9-month-old Tet2 Mut LSK cells than in the age-matched Tet2 KO LSK cells (Fig. 3B right panel). While in Tet2 Mut Lin− cells the number of DEGs progressively increased from 3- to 6- to 9-months, in Tet2 KO Lin− cells the number of DEGs drastically increased at 6 months (Fig. 3B left panel). Consistently, overlapping DEGs in LSK or Lin− cells of each genotype across time points, revealed that a large number of genes (626) are uniquely deregulated at 6 months of age in Tet2 KO Lin− cells (Fig. 3C, bottom left). In contrast, in Tet2 Mut Lin− cells a large number of genes (433) are uniquely deregulated at 9 months (Fig. 3C, top left). Comparison of the transcriptomes of Tet2 Mut LSK or Lin− cells versus their Tet2 KO counterparts at each time point confirmed that the transcriptome of Tet2 Mut and Tet2 KO Lin− cells were more divergent at 6 months of age (r2=0.87) than at any other time point (Fig. S1B). These findings suggest that Tet2 KO Lin− cells exhibit a very robust transcriptomic dysregulation earlier than Tet2 Mut Lin− cells, which may underlie the reduced survival of mice transplanted with Tet2 KO bone marrow than mice transplanted with Tet2 Mut bone marrow.
Figure 3: Gene expression profiling of Tet2 Mut and KO LSK and Lin− cells harvested at various time points.
A. Schematic of RNA-seq strategy.
B. Quantification of the number of upregulated and downregulated DEGs in Tet2 Mut and KO Lin− and LSK cells vs. WT (log2 fold-change > 1.0, FDR < 0.05) identified by RNA-seq.
C. Venn diagram overlapping DEGs found specifically in Tet2 Mut and KO Lin− cells (left) and LSK cells (right) across different time points (3, 6, 9, and 12 months). Note that different sets of genes are affected at each time point.
Deregulation of lymphoid and MDS/AML genes in 6-month-old Tet2 KO Lin− cells
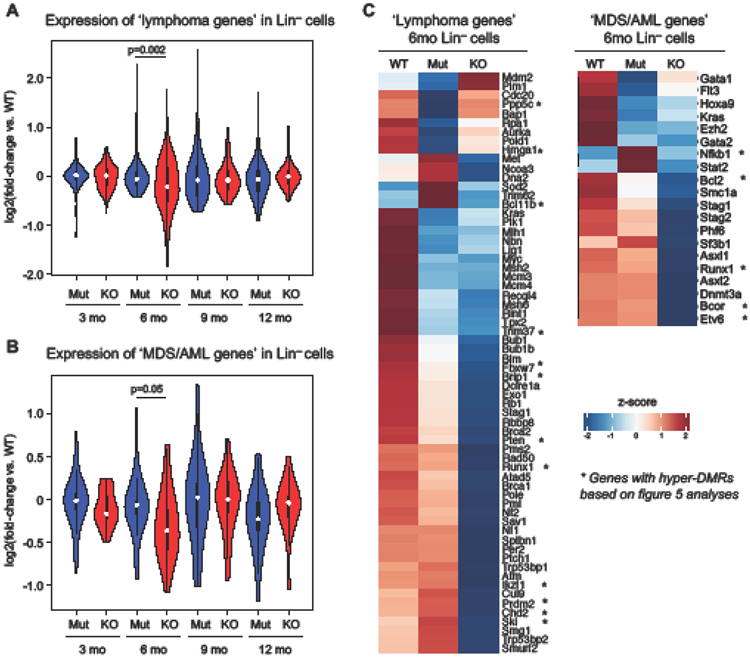
To gain insights into the age-dependent shift from lymphoid to more myeloid disease caused by Tet2 KO bone marrow (Fig. 1E), we examined changes in the expression of genes commonly associated with lymphoid disorders (Increased Lymphoma Incidence Genes GSEA MP:0012431) or myeloid disorders (genes commonly mutated in MDS/AML from the literature33) in Tet2 KO and Tet2 Mut Lin− cells over the four timepoints. While there was an overall trend in less deregulation of lymphoma-associated genes and more deregulation of MDS/AML-associated genes (based on overall fold change) with age in Tet2 KO Lin− cells (compared to Tet2 Mut Lin− cells), the differences were only statistically significant at the pivot timepoint of 6 months. Both categories of genes were significantly more deregulated in 6-month-old Tet2 KO Lin− cells compared to age-matched Tet2 Mut Lin− cells (Fig. 4A-B). A vast majority of these genes were downregulated in Tet2 KO Lin− cells (Fig. 4C). Since this gene expression analysis is from mice that had not developed disease, the data is unlikely skewed by the presence of any particular malignant cell types. Thus, this shift from an increased lymphoid gene deregulation to an increased myeloid gene deregulation in Tet2 KO Lin− cells with age parallels the higher incidence of myeloid disease in mice transplanted with 9- and 12-month-old bone marrow.
Figure 4: Lymphoma and leukemia genes are downregulated in Tet2 KO Lin− cells.
A. Expression levels of lymphoma signature genes (from GSEA) in Tet2 Mut and KO Lin− cells across time points. Fold-change vs. WT is shown. Welch two sample t-test.
B. Expression levels of selected MDS/AML signature genes (reported as commonly mutated in the literature) in Tet2 Mut and KO Lin− cells across time points. Fold-change vs. WT is shown. Welch two sample t-test.
C. Heatmap showing lymphoma and MDS/AML signature genes downregulated in Tet2 KO Lin− cells at 6 months. Relative expression levels from normalized counts are shown. * indicated genes containing a hyper-DMR (per the analyses in figure 5).
Deregulated genes in Tet2 KO Lin− and LSK cells are associated with differentially hyper-methylated regions (hyper-DMRs) from early in life
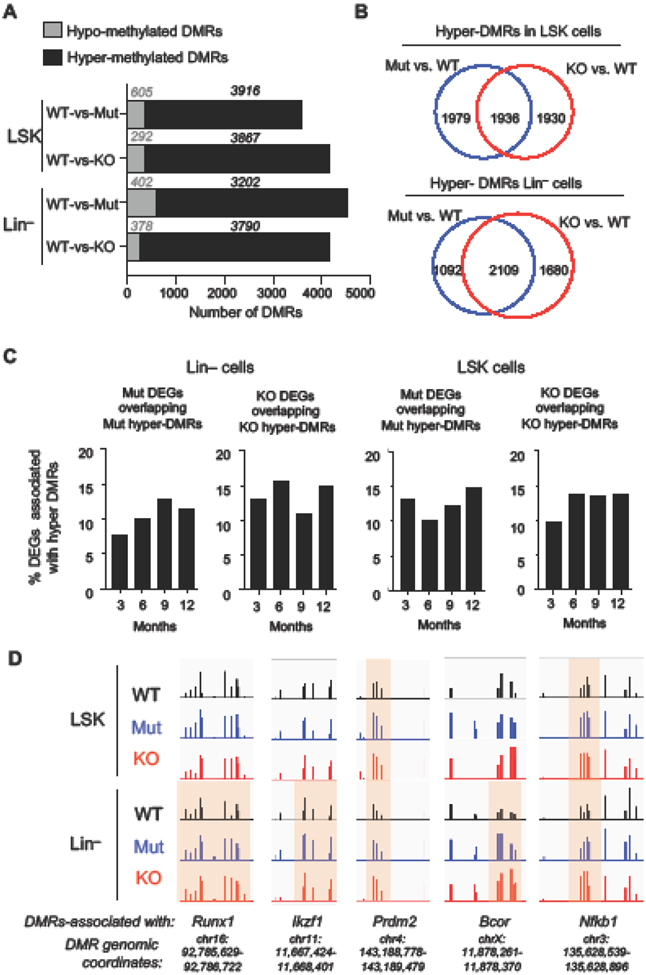
To examine how transcriptomic changes are influenced by early changes in DNA methylation, we mapped the genomic distribution of 5-methylcytosine (5mC) in Tet2 KO and Tet2 Mut bone marrow cells isolated from 3-month-old healthy mice by whole genome bisulfite sequencing (WGBS). We identified a comparable number of differentially methylated regions (DMRs, methylation difference vs. WT >20%, FDR <0.05) in progenitor cells (Lin− cells) and HSPCs (LSK) from Tet2 KO and Tet2 Mut mice, with majority of the DMRs being hypermethylated consistent with a role for Tet2 in DNA demethylation (Fig. 5A). A large number of DMRs were at distal intergenic regions and gene bodies followed by promoters (Fig. S2A-B). In LSK cells, nearly half of the hyper-DMRs (~1900) were common between Tet2 KO and Tet2 Mut cells, while the other half were unique to each genotype (Fig. 5B top). In Lin− cells, ~2100 hyper-DMRs were common between Tet2 KO and Tet2 Mut cells, ~1100 were unique to Tet2 Mut, and ~1700 were unique to Tet2 KO (Fig. 5B bottom). The overlap of hypo-DMRs was not robust (Fig. S2C). Integration of DNA methylation and gene expression data revealed that ~15% of DEGs were associated with hyper-DMRs (Fig. 5C). We found that many lymphoma and MDS/AML genes such as Runx1, Prdm2, Ikzf1, Bcor and Nfkb2 contained hypermethylated DMRs (Fig. 5D). Of the 17 genes associated with hypermethylated DMRs (denoted with asterisk in Fig 4C), 13 were downregulated in Tet2 KO Lin− cells suggesting a positive correlation between DNA hyper-methylation and gene downregulation. The other 4 genes were unaffected in Tet2 KO but dysregulated in Tet2 Mut implying that hypermethylation of the specific DMRs at these genes did not affect their expression, and that other direct or indirect mechanisms are involved in their deregulation. Overall our findings reveal a correlation between gene deregulation and hyper-DMRs established early in life in LSK and Lin− cells due to Tet2 deficiency.
Figure 5: DNA methylation profiling of Tet2 Mut and KO LSK and Lin− cells at 3 months of age.
A. Number of hypermethylated and hypomethylated differentially methylated regions (DMRs) found in Tet2 Mut and KO LSK and Lin− cells by whole genome bisulfite sequencing (WGBS).
B. Overlap of differentially hyper-methylated regions (hyper-DMRs) between Tet2 Mut and KO LSK and Lin– cells.
C. Overlap of DEGs in Tet2 Mut and KO LSK and Lin− cells with hypermethylated DMRs across time points. %DEGs that overlapped with hyper-DMR-associated genes are shown.
D. DNA methylation tracks at selected DMRs associated with deregulated lymphoma or MDS/AML-causing genes in LSK and Lin− cells.
DISCUSSION
Previously, we have shown that Tet2 catalytic and noncatalytic functions are important for the regulation of the myeloid and lymphoid lineages, respectively29, but how they influence the bone marrow with age is not defined. In this study, we provide five lines of evidence supporting an age-dependent involvement of Tet2 functions in the bone marrow. (1) Loss of Tet2 or deficiency of its catalytic activity alone causes progressively more aggressive malignancies with age. (2) Older Tet2 KO bone marrow promotes more lethal malignancies than age-matched Tet2 catalytic deficient bone marrow. (3) With age, Tet2 KO bone marrow exhibits a shift from equally promoting lymphoid and myeloid malignancies to predominantly causing myeloid malignancies. (4) Tet2 loss leads to an early and robust gene dysregulation in Lin− cells which involves lymphoma and MDS/AML-causing genes. (5) There is an overall shift from an increased lymphoid to an increased myeloid gene deregulation in Tet2 KO Lin− cells with age underpinning the higher myeloid and lower lymphoid disease development. These findings suggest that Tet2 catalytic roles are the main regulators of the myeloid lineage and critical for suppressing myeloid defects in the bone marrow of all ages, while noncatalytic roles of Tet2 regulate the lymphoid lineage in young bone marrow and contribute to preventing aggressive myeloid disease in aged bone marrow.
Our bone marrow transplantation studies allow for examining the cell-autonomous requirements of Tet2 in the bone marrow of different ages in a WT niche. This better recapitulates the disease onset and progression observed in patients. We find an age-dependent shift in the spectrum of disease caused by Tet2 KO donor bone marrow. Younger Tet2 KO bone marrow induces both lymphoid and myeloid defects in contrast to older bone marrow, which mainly promotes myeloid disease. This different spectrum of disease caused by transplantation of Tet2 Mut and KO bone marrow is consistent with our prior studies of germline Tet2 Mut or KO mice which lacked Tet2 or its catalytic activity in all tissues29. However, the onset of disease in transplanted mice is faster than the onset of disease in germline Tet2 Mut or KO mice, where only 50% of mice presented with both myeloid and lymphoid malignancies by two years (and mice of neither genotype succumbed to malignancies at 6 months). This suggests that Tet2 Mut and KO bone marrow may promote disease faster in a WT niche and/or stress hematopoiesis (i.e. bone marrow regeneration after transplantation) facilitates hematopoietic phenotypes induced by Tet2 Mut and KO donor cells. Moreover, it is known that with age there is an expansion of the myeloid compartment of the bone marrow34. Therefore, the increased myeloid disorders caused by older transplanted bone marrow could also be due to age-dependent myeloid-biased hematopoiesis. A separate transplantation of lymphoid and myeloid progenitor cells isolated from young and old Tet2 KO or Mut mice may provide more direct insights into this lineage bias. We note that both the Tet2 KO and Mut mice were maintained in a mixed 129/B6 background. Moreover, the two strains were intercrossed first and then bred to generate Tet2 Mut and KO mice for the study. Therefore, the differences in disease spectrum between the two strains is not due to any variations in their genetic backgrounds. The onset of myeloid and lymphoid disease in our current and prior Tet2 KO mouse studies is also consistent with the prevalence of human TET2 mutations in myeloid12-18 and a subset of lymphoid21,23 malignancies.
The potential of old Tet2 KO bone marrow to form more aggressive malignancies than its catalytic mutant counterparts suggests that the noncatalytic functions of Tet2 likely underpins the shorter latency of myeloid malignancies. This is a previously unknown noncatalytic requirement of Tet2 in the aging of the myeloid lineage. This finding expands on our previous work where in young mice we had distinctly implicated catalytic and noncatalytic functions of Tet2 to myeloid and lymphoid lineages, respectively. Better understanding of these noncatalytic functions will require further molecular and biochemical studies in the myeloid compartment of the bone marrow, including finding the binding partners and genomic distribution of Tet2 with respect to epigenetic regulators and transcription factors. For example, OGT has been identified as a key partner of Tet2 in the bone marrow where it interacts with SET1/COMPASS to promote H3K4 trimethylation and gene activation. This noncatalytic mechanism is shown to repress genes in Tet2 KO bone marrow11. We also note that some non-coding RNAs that are commonly deregulated in blood disorders and leukemia35,36 may be influenced by the noncatalytic functions of Tet2. Future analysis of the noncoding transcriptome of Tet2 KO and Mut LSK and Lin− cells may reveal some of them. This will present yet another angle through which Tet2 noncatalytic roles could regulate the bone marrow.
Given that our molecular analyses were performed using mice that had not developed disease, our data is unlikely skewed by disproportionate presence of any particular malignant cell types. Therefore, the robust transcriptomic changes at 6 months of age in Tet2 KO Lin− cells involving lymphoma and MDS/AML-causing genes, and the shift to more myeloid gene dysregulation with age, provides molecular insights into the transcriptional changes preceding disease onset. We also found that DMRs hypermethylated early in life at 3 months of age are associated with DEGs at later ages. Future studies probing DNA methylation changes across all time points will illuminate if methylation progressively increases and how that impacts gene dysregulation. While this study systematically characterizes the disease spectrum and gene expression changes of Tet2 Mut and KO bone marrow at different ages, a key limitation of this approach is that it does not take into account the transcriptional and epigenetic heterogeneity within the bone marrow. Future work using single-cell RNA-seq and WGBS approaches to analyze the expression and methylation landscapes of each bone marrow cell type and their niche with age can provide more in-depth insights into the dual functions of Tet2 in the bone marrow for proper hematopoiesis.
In summary, this work expands on our prior findings29 by showing that the noncatalytic roles of Tet2 have an age-dependent impact on the myeloid and lymphoid compartments. While it regulate the lymphoid lineage in young bone marrow, it contributes to preventing aggressive myeloid disease in aged bone marrow. Our findings have implications for patients with hematologic disorders and malignancies where Tet2 is downregulated or mutated (truncated/unstable proteins leading to loss of Tet2) causing reduced Tet2 protein levels (hence loss of both its catalytic and noncatalytic functions). Future studies aimed at rescuing hematologic phenotypes of Tet2-deficient patient bone marrow cell lines with catalytically dead Tet2 can functionally test and establish the contribution of Tet2 catalytic and noncatalytic roles in disease etiology.
Supplementary Material
Highlights.
Young and old Tet2 catalytic mutant bone marrow promote mainly myeloid defects
Young Tet2 KO bone marrow causes lymphoid and myeloid defects while old causes only myeloid defects
Old Tet2 KO bone marrow promotes more lethal malignancy than old Tet2 catalytic mutant bone marrow
Tet2 loss leads to an early and robust gene dysregulation in hematopoietic progenitors
Lymphoma and MDS/AML-causing genes are deregulated and hypermethylated at early age
Acknowledgments
We are grateful to Fei Xu and Masako Suzuki for their advice on bioinformatics pipelines and analyses. We thank the flow cytometry (NIH P30CA013330) and histopathology cores for their support in hematopoietic characterization of mice. We also thank Lidiane Torres for critical reading of the manuscript and members of the Dawlaty and Ito labs for helpful suggestions and discussions.
Funding:
This work was supported by NIH/NHLBI (R01HL148852, to Ke.I. and M.M.D as multi-PIs). M.M.D was also supported by NIH R01GM122839, Hirschle Trust Funds, and funds from Albert Einstein College of Medicine. Ke.I was also supported by NIH/NIDDK (R01DK98263, R01DK115577, and R01HL069438), and Albert Einstein College of Medicine, and is a Research Scholar of the Leukemia and Lymphoma Society (#1360-19). J.C.F. was supported by NIH F31 predoctoral fellowship award F31GM140554.
Footnotes
Disclosure of conflict of interests
The authors declare no competing financial or other conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tahiliani M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935, doi: 10.1126/science.1170116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S. et al. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 333, 1300–1303, doi: 10.1126/science.1210597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He YF et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307, doi: 10.1126/science.1210944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastor WA, Aravind L & Rao A TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14, 341–356, doi: 10.1038/nrm3589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu SC & Zhang Y Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11, 607–620, doi: 10.1038/nrm2950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348, doi: 10.1038/nature10066 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393, doi: 10.1038/nature09934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chrysanthou S, Flores JC & Dawlaty MM Tet1 Suppresses p21 to Ensure Proper Cell Cycle Progression in Embryonic Stem Cells. Cells 11, doi: 10.3390/cells11081366 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chrysanthou S. et al. The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res, doi: 10.1093/nar/gkac089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Chen Y, Bian C, Fujiki R & Yu X TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564, doi: 10.1038/nature11742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deplus R. et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32, 645–655, doi: 10.1038/emboj.2012.357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delhommeau F. et al. Mutation in TET2 in myeloid cancers. N Engl J Med 360, 2289–2301, doi: 10.1056/NEJMoa0810069 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Tefferi A. et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia 23, 1343–1345, doi: 10.1038/leu.2009.59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tefferi A. et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23, 905–911, doi: 10.1038/leu.2009.47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O. et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114, 144–147, doi: 10.1182/blood-2009-03-210039 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langemeijer SM et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41, 838–842, doi: 10.1038/ng.391 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Kosmider O. et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica 94, 1676–1681, doi: 10.3324/haematol.2009.011205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song SJ et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13, 87–101, doi: 10.1016/j.stem.2013.06.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran-Crusio K. et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24, doi: 10.1016/j.ccr.2011.06.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z. et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518, doi: 10.1182/blood-2010-12-325241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asmar F. et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica 98, 1912–1920, doi: 10.3324/haematol.2013.088740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez PM et al. TET2 Deficiency Causes Germinal Center Hyperplasia, Impairs Plasma Cell Differentiation, and Promotes B-cell Lymphomagenesis. Cancer Discov 8, 1632–1653, doi: 10.1158/2159-8290.CD-18-0657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemonnier F. et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120, 1466–1469, doi: 10.1182/blood-2012-02-408542 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Mouly E. et al. B-cell tumor development in Tet2-deficient mice. Blood Adv 2, 703–714, doi: 10.1182/bloodadvances.2017014118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan F. et al. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat Commun 8, 15102, doi: 10.1038/ncomms15102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quivoron C. et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20, 25–38, doi: 10.1016/j.ccr.2011.06.003 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Reddy A. et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171, 481–494 e415, doi: 10.1016/j.cell.2017.09.027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leca J. et al. IDH2 and TET2 mutations synergize to modulate T Follicular Helper cell functional interaction with the AITL microenvironment. Cancer Cell 41, 323–339 e310, doi: 10.1016/j.ccell.2023.01.003 (2023). [DOI] [PubMed] [Google Scholar]
- 29.Ito K. et al. Non-catalytic Roles of Tet2 Are Essential to Regulate Hematopoietic Stem and Progenitor Cell Homeostasis. Cell Rep 28, 2480–2490 e2484, doi: 10.1016/j.celrep.2019.07.094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito K. et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 354, 1156–1160, doi: 10.1126/science.aaf5530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morganti C. et al. NPM1 ablation induces HSC aging and inflammation to develop myelodysplastic syndrome exacerbated by p53 loss. EMBO Rep 23, e54262, doi: 10.15252/embr.202154262 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen KD et al. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res 29, 564–575, doi: 10.1101/gr.239277.118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M. et al. Gene mutation spectrum of patients with myelodysplastic syndrome and progression to acute myeloid leukemia. Int J Hematol Oncol 10, IJH34, doi: 10.2217/ijh-2021-0002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beerman I. et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A 107, 5465–5470, doi: 10.1073/pnas.1000834107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Chen K, Guo G & Chen JL Noncoding RNAs and their functional involvement in regulation of chronic myeloid leukemia. Brief Funct Genomics 15, 239–248, doi: 10.1093/bfgp/elv059 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Roden C & Lu J MicroRNAs in Control of Stem Cells in Normal and Malignant Hematopoiesis. Curr Stem Cell Rep 2, 183–196, doi: 10.1007/s40778-016-0057-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and WGBS data are deposited in the Gene Expression Omnibus (GEO) database (Accession: GSE227977).