Abstract
Bardet-Biedl syndrome (BBS), is an emblematic ciliopathy hallmarked by pleiotropy, phenotype variability, and extensive genetic heterogeneity. BBS is a rare (~1/140,000 to ~1/160,000 in Europe) autosomal recessive pediatric disorder characterized by retinal degeneration, truncal obesity, polydactyly, cognitive impairment, renal dysfunction, and hypogonadism. Twenty-eight genes involved in ciliary structure or function have been implicated in BBS, and explain the molecular basis for ~75–80% of individuals. To investigate the mutational spectrum of BBS in Romania, we ascertained a cohort of twenty-four individuals in twenty-three families. Following informed consent, we performed proband exome sequencing (ES). We detected 17 different putative disease-causing single nucleotide variants or small insertion-deletions and two pathogenic exon disruptive copy number variants in known BBS genes in 17 pedigrees. The most frequently impacted genes were BBS12 (35%), followed by BBS4, BBS7, and BBS10 (9% each) and BBS1, BBS2, and BBS5 (4% each). Homozygous BBS12 p.Arg355* variants were present in seven pedigrees of both Eastern European and Romani origin. Our data show that although the diagnostic rate of BBS in Romania is likely consistent with other worldwide cohorts (74%), we observed a unique distribution of causal BBS genes, including overrepresentation of BBS12 due to a recurrent nonsense variant, that has implications for regional diagnostics.
Keywords: ciliopathy, retinal dystrophy, polydactyly, urogenital malformations, second-site modifiers, pleiotropy
Introduction
Bardet-Biedl syndrome (BBS [OMIM 209900]), is a rare autosomal recessive pediatric disorder caused by dysfunction of primary cilia1; 2. BBS is a multisystem primary ciliopathy characterized by multiple clinical manifestations, most prominently progressive retinal degeneration, postaxial polydactyly, truncal obesity, cognitive impairment, renal dysfunction, and hypogonadism or urogenital malformations. Additional clinical findings of varying frequency that may complicate clinical diagnosis include neurological abnormalities, endocrine and metabolic impairment, cardiovascular defects, brachydactyly/syndactyly, dental anomalies, and gastrointestinal abnormalities1. Clinical diagnostic criteria have been proposed as the presence of either four or three major clinical features in combination with at least two minor or secondary features3. Certain signs are detectable antenatally; these include polydactyly, kidney anomalies or abdominal distension due to genitourinary abnormalities, and thus raise the suspicion of BBS in early childhood4; 5. However, most patients are diagnosed in late childhood or early adulthood, and typically the diagnosis is prompted by the manifestation of retinal dystrophy3; 6. The incidence of BBS varies among different populations and is increased in the regions with a high level of endogamy. For instance, in North America and Europe, its prevalence is estimated at around 1:140,000 to 1:160,000 live births7; 8 while the incidence is elevated in certain isolated populations such as Newfoundland and Kuwait, where the incidence rises to 1:18,000 and 1:13,500, respectively, postulating a founder effect1; 9; 10.
There are six cardinal features of BBS and multiple infrequent clinical symptoms. Progressive retinal degeneration is a highly penetrant feature evident in the first decade 6; 11; 12 with complete loss of visual acuity by second or third decade of life13–15. Obesity usually begins in childhood and becomes obvious during the first 3 years of life1; 16; 17. Polydactyly is observed commonly but not always in affected individuals with BBS; it may occur with syndactyly, brachydactyly, and clinodactyly3; 18; 19. One of the least understood and disputed features of BBS is cognitive impairment; >62% of patients have been reported to have cognitive difficulties although the severity is highly variable3; 15; 20. Functional kidney deficits are variable and often lead to chronic kidney disease (CDK) which is considered a major contributor of morbidity in individuals affected with BBS21. Individuals with BBS also display congenital structural abnormalities22–25. Hypogenitalism and hypogonadism are reported in nearly all males, while hypoplastic labia minora, vaginal atresia, and septate or imperforate vagina are common in females9; 26. Several minor features have also been documented in individuals with BBS, including facial dysmorphism, developmental delay, speech deficit, neurological abnormalities, metabolic and endocrine disturbance, diabetes mellitus, cardiovascular defects and Hirschsprung disease27; 28. Assembly and analysis of consistent and longitudinal clinical data from the Clinical Registry Investigating Bardet-Biedl Syndrome (CRIBBS), have refined further the incidence and variability of clinical phenotypes21; 29; 30.
To date, causal variants in twenty-eight different genes have been linked with BBS (Table S2), all of which are implicated in the structure and/or function of the primary cilium21; 31; 32. Pathogenic variants in primary recessive driver loci explain the molecular basis for 75–80% of cases, suggesting that additional genes remain to be identified4. BBS1 and BBS10 are major contributors, accounting for nearly half of affected individuals33–35, although some regional variation in prevalence exists16. Causal variants identified in BBS12 and ARL6/BBS3 account for ~8% of the clinically diagnosed patients each36–38, and the remainder of genes account for less than 5%39. Copy number variants (CNV) also contribute to the mutational burden of BBS, and exon disruptive CNVs are detectable in up to 18% of clinically assessed cases32; 40; 41. BBS is inherited predominantly in an autosomal recessive manner; however, this classical mode of inheritance has been challenged by extensive molecular and functional investigation reporting second-site variation in other BBS genes which could possibly explain phenotypic variability42–44 45; 46 47–49.
BBS is considered as a model disease to gain insight into the biology of the primary cilium. A subset of disease-associated proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8/TTC8, BBS9, and BBS18/BBIP1) form a multimeric complex known as the BBSome. This multiprotein complex is localized to the base of the cilium and functions as an adaptor for intraflagellar transport (IFT) molecules50; 51. Three chaperonin-like proteins (BBS6/MKKS, BBS10, and BBS12) form a complex with chaperonin containing tailless complex polypeptide 1 or tailless complex polypeptide 1 ring complex (CCT/TRiC) family chaperonins and play an essential role in BBSome assembly52; 53. Other proteins have roles in BBSome localization and activation (ARL6/BBS3)54, entry into cilia (BBS17) or are associated with the BBSome (BBS14)55, whereas the remainder of proteins disrupted in BBS cases are vital to ciliogenesis and ciliary function 56.
Primary cilia function in sensory perception and various signaling pathways including, sonic hedgehog signaling (shh), wingless/integrated (Wnt), notch, salvador warts hippo (SWH), Platelet-derived growth factor receptors (PDGFR), mammalian target of rapamycin (mTOR), and G-protein coupled receptors (GPCR), to regulate developmental processes, tissue plasticity, and organ development57; 58. Dysregulation of these signaling pathways has been associated with ciliary dysfunction59; 60, which results in multiorgan defects. For example, impaired shh signaling may induce digit abnormalities, craniofacial defects, skeletal malformations, and intellectual disability, while dysregulated Wnt signaling is likely a contributor to some renal phenotypes61. BBS belongs to a broader clinical group of disorders termed ciliopathies which share a common organellar etiology; phenotypic overlap with other ciliopathies including McKusick-Kauffman syndrome (MKKS), Joubert syndrome (JBTS), Alstrom syndrome (ALMS), and Meckel Gruber syndrome (MKS) have been discussed elsewhere1; 48; 62.
Here, we report the clinical spectra and genetic analysis of a cohort of 23 families who reside in Romania. Using exome sequencing (ES), we identified 17 different SNVs or small indels and 2 CNVs in 19 families (17 families with a primary recessive locus identified and 2 additional families with heterozygous variants). To analyze CNVs, we characterized breakpoints by long-range PCR and subsequent Sanger sequencing. Additionally, we functionally characterized a rare intronic variant (RefSeq ID: NM_033028.5, BBS4: c.332+8T>C) segregating in trans with a CNV. Finally, our cohort presents a unique distribution of BBS causal genes, due in part to the recurrence of a BBS12 nonsense variant (RefSeq ID: NM_152618.3, c.1063C>T) possibly due to a founder effect.
Materials and Methods
Study Participants, Clinical Evaluation, Ethics Approval, and DNA Extraction
The relevant ethics committees of the University of Medicine and Pharmacy Carol Davila Bucharest and Ann & Robert H. Lurie Children’s Hospital of Chicago approved the study. We recruited twenty-four affected individuals with BBS and their available family members from twenty-three unrelated families. Clinical evaluation was performed after receiving written informed consent from the legal guardian of pediatric participants, and their adult family members. BBS diagnoses were ascertained by a medical geneticist who identified either four or three major features plus two minor or secondary features according to established criteria3. We obtained peripheral blood by venipuncture and extracted genomic DNA from samples using the Purelink® Genomic DNA Extraction kit (Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. Individual DM1569 was reported previously63.
Next-Generation Sequencing, Variant Filtration, and In Silico Analyses
To identify the causative genes in our cohort, we performed ES on proband genomic DNA according to an established protocol (LC Sciences, LLC). For exome capture, we used the Agilent SureSelect Human All Exon V6 kit (Agilent Technologies, Inc.) and constructed libraries according to the manufacturer’s instructions. Captured libraries were further subjected to paired-end sequencing on an Illumina Novaseq6000 system at Lianchuan Bio, resulting in 150 bp paired-end reads, to a mean depth ranging from 61X-140X across individuals, with 82 % to 96 % of coding regions covered by ≥ 20 reads (Table S1). The detailed methods were adopted for ES as described63. Low-quality reads were removed and reads with minimum coverage of ≥ 10 were considered further for analysis. Varsome clinical (10.1) analysis software and Variant Annotation and Filtering Tool (VarAFT), version 2.17–2 (https://varaft.eu/)64 were used to prioritize SNVs and small indels to retain functional variants with minor allele frequency (MAF) <0.01 in the Genome Aggregation Database (gnomAD v3.1.2) (https://gnomad.broadinstitute.org/), predicted to alter the amino acid sequence and intron-exon junctions in known BBS genes (Table S2). CNVs were identified within the Varsome clinical platform with the ExomeDepth CNV caller (v1.1.11)65. Variant pathogenicity was predicted using the following in silico tools: MutationTaster2021 (https://www.mutationtaster.org/)66, Provean (v1.1) (https://www.jcvi.org/research/provean)67, CADD (v1.6) (https://cadd.gs.washington.edu/)68, and SIFT (v6.2.1) (https://sift.bii.a-star.edu.sg/)69 and categorized according to the American College of Medical Genetics (ACMG) classification system70. Prioritized variants were inspected visually with the Integrated Genomics Viewer (IGV, Broad Institute). Amino acid conservation was visualized by generating multiple sequence alignments using Clustal Omega (EMBL-EBI, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK) (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Long Range PCR for Variant Phasing and CNV Junction Fragment Analyses
To phase rare variants in families for which neither parental genomic DNA was available (DM1574) or to characterize CNV breakpoints (DM1586, DM1576, and DM1566), we performed long-range PCR, cloning and sequencing. The region of interest was amplified using the Phusion High-Fidelity PCR Kit (Thermo Fisher Scientific, F5321L) according to the manufacturer’s instructions. The BBS1, BBS4 and BBS10 amplicons unique to the carrier and not observed in the controls were gel-separated, purified, and subjected to Sanger sequencing.
Sanger Confirmation and Segregation Analysis
For SNV or CNV validation and segregation analysis in genomic DNA from all available family members, we PCR-amplified the targeted regions (primer sequences and PCR conditions are available upon request). PCR products were sequenced bidirectionally using BigDye terminator 3.1 chemistry and an ABI 3730xl DNA analyzer according to the manufacturer’s protocols (ThermoFisher Scientific, Inc.). Sequence chromatograms were analyzed by Sequencher® 5.4.6 (Gene Codes Corporation, Ann Arbor, MI, USA). Variant phasing was possible in 9 families with both parental samples available; confirmed in 1 family by long range PCR (see above); and estimated in the remaining families with 1 parental DNA available (n=5) or neither parental DNA available (n=2).
Establishment and Culture of Lymphoblastoid Cell Lines (LCL)
Whole blood samples were collected in BD Vacutainer® ACD A tubes (BD: 0100195). Peripheral blood mononuclear cells (PBMCs) were separated from whole blood, and then approximately 2.5 X 106 PBMCs were exposed to Epstein Barr virus to establish LCLs as described71. LCLs were cultured in Gibco Roswell Park Memorial Institute (RPMI) supplemented with 10% Heat Inactivated Fetal Bovine Serum and 1% pen-strep (100 IU/ml penicillin and 100 μg/ml streptomycin). The cells were incubated at 37°C in a humidified atmosphere with 5% CO2 using standard cell culture protocols.
RNA Isolation and mRNA Splicing Studies
LCLs were harvested for total RNA isolation using Trizol reagent (ThermoFisher Scientific) and subsequently reverse transcribed to cDNA using the QuantiTect reverse transcription kit (Qiagen) according to the manufacturer’s guidelines. To investigate impaired splicing, we performed PCR on cDNA obtained from a matched control and an affected individual carrying an intronic variant and an exon 3–4 deletion in trans in BBS4. The primer pairs used for PCR annealed to exons 1 and 7, upstream and downstream variant sites, respectively. The amplified product was separated on a 2% agarose gel with 1 kb plus ladder (ThermoFisher Scientific), gel slices were purified separately (QIAquick® Gel Extraction Kit, Qiagen) and confirmed by Sanger sequencing.
Results
Clinical manifestations of individuals with BBS
Twenty-four affected individuals with suspected BBS (23 probands and 1 affected sibling) from twenty-three families who reside in Romania (Table 1) were referred from multiple sites. Their self-reported ethnicity includes Eastern European (n=17), Romani (n=5), and Arab (n=1). Among these, our group previously reported one family as a case report63. We noted a broad age range at the time of clinical ascertainment (2 months to 43 years) with both sexes represented in our cohort (9 males; 15 females). The affected individuals were evaluated by multidisciplinary clinical teams and consented for research.
Table 1.
Summary of main BBS related clinical manifestations and overlapping features
| Family ID | Sex | Age | BMI (kg/m2) | Main clinical features | Minor or secondary clinical features | Others | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RP | PD | Ob | HG | RD | ID | DD | BD | SYN | MY | AST | Str | HAP | SLI | MC | BC | PF | |||||
| DM1564 | F | 10 yrs | 22.1 | + | + | ND | + | ND | ND | ND | − | − | + | − | + | + | − | − | − | − | LD |
| DM1565 | F | 29 yrs | 36.2 | + | + | + | + | + | ND | ND | + | − | + | − | − | − | − | − | + | + | DM, HCL, HH, PCOS SS,HT |
| DM1566 | M | 15 yrs | 32.4 | + | + | + | + | + | mild | + | + | − | + | + | + | − | − | − | + | + | CTEV, ROM, HTG, XN, PP |
| DM1567 | M | 5 yrs | 17.4 | ND | + | ND | + | ND | severe | + | − | − | − | − | − | − | − | − | + | − | Atx, BA, AT, VI, XN, MG, CD, Sn |
| DM1568 | F | 6 yrs | 24.7 | ND | + | + | + | ND | moderate | + | + | + | − | − | + | − | + | − | − | + | S, SVT, HD, BA, AT |
| DM1569 | F | 6 yrs | 31.7 | + | + | + | + | ND | severe | − | + | + | − | − | − | − | − | + | − | − | PMA, BA, XN |
| DM1570 | F | 39 yrs | 52.7 | + | + | + | + | + | moderate | ND | − | + | + | + | + | − | − | − | − | − | HF, HT, DM, NYS, PC |
| DM1571 | F | 42 yrs | 36.4 | + | + | + | + | + | ND | + | − | − | + | − | + | − | − | − | + | − | DBA, ONA, HT, SS, PCOS |
| DM1572 | M | 18 yrs | 35.3 | − | + | + | + | ND | moderate | + | − | + | − | − | − | − | − | − | − | − | HH, ASD, RTG, Sn, BA |
| DM1573–1 | M | 21 yrs | 37 | + | − | + | + | ND | moderate | + | + | + | − | − | − | − | − | + | + | − | T2DM, FL, HT |
| DM1573–2 | F | 16 yrs | 54.7 | + | + | + | ND | ND | severe | + | + | − | − | − | − | − | − | + | − | + | BA, HT, T2DM, HTG, HCM, FL |
| DM1574 | M | 20 yrs | 34.9 | + | + | + | + | + | moderate | + | − | − | + | + | − | − | + | − | − | − | HT, HCL, HTG, FL |
| DM1575 | F | 8 yrs | 30.4 | + | + | + | − | ND | severe | + | − | − | − | − | − | − | + | + | − | + | BA, FL, HM, VSD, Atx |
| DM1576 | M | 16 yrs | 31.9 | + | − | + | + | ND | mild | ND | − | − | − | − | − | + | − | + | − | + | FL, SL |
| DM1582 | M | 33 yrs | 34.7 | + | − | + | + | + | mild | + | − | − | − | − | − | − | − | − | − | − | LD, HCL, FL, CTEV |
| DM1583 | F | 15 yrs | 27 | + | + | + | + | + | mild | + | − | + | + | + | + | + | + | − | + | + | HT, ASD |
| DM1584 | M | 7 yrs | 24.2 | + | + | + | + | ND | mild | − | + | − | − | − | − | − | + | − | − | + | HM, HTG |
| DM1585 | F | 7 yrs | 22.6 | + | + | + | + | ND | severe | + | + | − | − | − | − | − | − | − | + | + | Cat, VI |
| DM1586 | F | 13 yrs | 33.4 | + | + | + | ND | + | moderate | + | − | + | − | − | − | − | − | + | − | + | − |
| DM1587 | F | 2 mo | − | ND | + | + | + | + | ND | − | − | − | − | − | − | + | − | + | + | + | IH |
| DM1588 | F | 13 yrs | 28.1 | + | + | + | + | + | moderate | − | − | − | − | − | − | + | − | − | − | − | AN, VI, PMD |
| DM1589 | M | 3 yrs | 29.5 | ND | + | ND | + | ND | moderate | − | − | − | − | − | − | − | − | − | − | − | VI, PMD |
| DM1590 | F | 5 yrs | 31,2 | ND | + | + | + | + | mild | − | − | − | − | − | − | + | + | + | − | + | CVA, PF, PMA, HTG, HCL |
| DM1591 | F | 2 mo | − | ND | + | ND | + | + | ND | − | − | − | − | − | − | + | − | − | − | + | CVA, UTIs, RRIs |
Phenotypes are indicated as present (+) and absent (−). ND indicates no data available, or subject was not evaluated. ASD, atrial septal defect; AST, astigmatism; AT, autistic trait; Atx, ataxic gait; AN, acantosis nigricans; BA, behavior abnormalities; BC, brachycephaly; BD, brachydactyly; BMI, body mass index; Cat, cataract; CD, clinodactyly; CTEV, congenital talipes equinovarus; CVA, congenital vaginal atresia; DBA, delayed bone age; DD, developmental delay; DM, diabetes mellitus; T2DM, diabetes mellitus type 2; FL, fatty liver; F, female; HAP, high arch palate; HCL, hypercholesterolemia; HD, hypodontia; HF, hepatic fibrosis; HG, hypogonadism; HH, hepatic hemangioma; HM, hepatomegaly; HT, hypothyroidism; HTG, hypertriglyceridemia; ID, intellectual disability; IH, imperforate hymen; HCM, hypertrophic cardiomyopathy; LD, learning difficulties; MC, macrocephaly; M, male; MG, micrognathia; mo, months; MY, myopia; NYS, nystagmus; Ob, obesity; ONA, optic nerve atrophy; PC, presbycusis; PCOS, polycystic ovaries; PD, polydactyly; PF, down slated palpebral fissures; PMA, psychomotor hyperactivity/agitation; PMD, psychomotor delay; PP, puberphonia; RD, renal disorder; ROM, recurrent otitis media; RP, retinitis pigmentosa; RRIs, recurrent respiratory infections; RTG, retrognathia; SL, skin lesions; SLI, speech and language impairment; Sn, synophrys; SS, short stature; Str, strabismus; SYN, syndactyly; UTIs, recurrent urinary tract infections; VI, vision impairment; VSD, ventricular septal defect; XN, night blindness; yrs, years.
We observed the archetypal BBS features in all affected individuals (Table 1; reported in detail elsewhere)72. Retinal degeneration, male hypogonadism, renal anomalies and learning difficulties are the most predominant features. Retinal dystrophy was noted in 17 of 18 individuals for which data are available, 94%. We documented urogenital anomalies in all males (hypogonadism, 9 of 9; 100%). Among females for which data were available, we observed a high incidence of hypoplastic genitalia (12 of 13, 92%), with concomitant congenital vaginal atresia in a minority (2 of 13, 15%). Among the entire cohort, we observed renal anomalies such as hydronephrosis, polycystic kidney, hypoplastic or atrophic kidney and end stage kidney disease in 12 of 12 (100%) individuals for whom data are available. All individuals with cognitive assessment showed intellectual disability, albeit at varying degrees of severity ranging from mild and moderate to severe (19 of 19, 100%). Additionally, we observed obesity in 20 of 24 cases (83%). However, in the other cases (two girls aged two months, one girl and one boy aged ten years) we cannot preclude the possibility that this feature may manifest later in life. Digit anomalies were present in 21 of 24 (92%) cases; these included postaxial polydactyly, syndactyly and brachydactyly and ranged from a single hand or foot to all four extremities.
In addition to classical BBS clinical manifestations, most cases also exhibited secondary or minor features including psychomotor delay (72%), language and speech delay (64%), other neuropsychiatric abnormalities (28%), cardiovascular involvement (28%), metabolic syndrome (28%) and type 2 diabetes (24%) (Table 1).
Genetic analysis of individuals with BBS
To identify the genetic etiology of BBS in our cohort, we performed ES on all affected individuals. We generated 150 bp paired-end reads on an Illumina platform to acquire average target read depth of 106x (61–140x) with 82–96% of bases covered by >20x (Table S1). Bioinformatic filtering identified 17 different causal variants or small indels and two exon disruptive CNVs in known BBS genes in 17 families. Of these, 12 affected individuals harbor causative homozygous SNVs or indels (52%), 3 have compound heterozygous SNVs or small indels (13%), 1 affected individual carries an SNV in trans with a CNV (4%), and 1 affected individual harbors a homozygous CNV (4%) in BBS genes (Figures 1, 2, 3; Tables 2 and 3, Table S3 63).
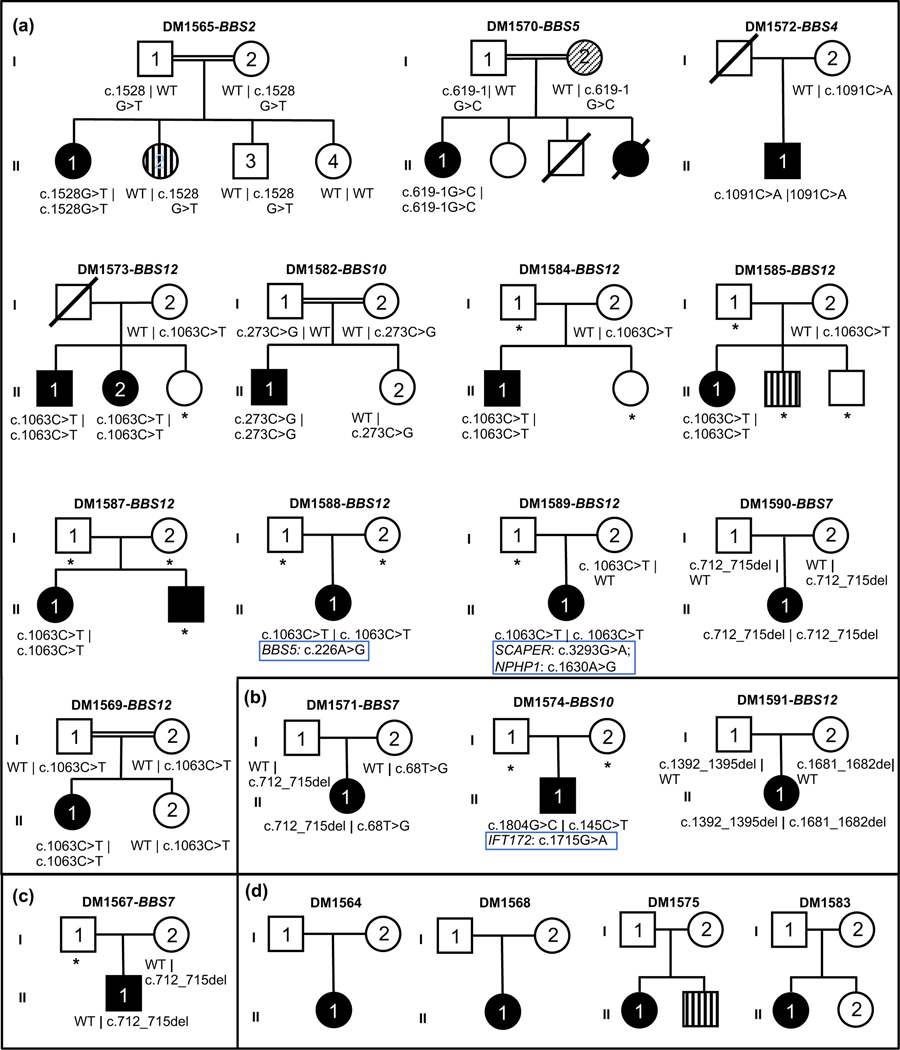
Figure 1. Sixteen pedigrees harboring single nucleotide variants or small indels in BBS genes and four unresolved pedigrees.
Pedigrees and genotyping data of BBS gene variants (SNVs or small indels). Causal gene and family identifier (denoted by DM number) are mentioned on top of each pedigree. Symbols indicate the following: square, male; circle, female; unfilled, unaffected individual; black filled, individual affected with BBS; vertical striped shape, individual affected with neurodevelopmental disorder; diagonal striped shape, individual affected with polycystic kidney disease and end-stage kidney disease; diagonal line, deceased individual; double horizontal lines, consanguinity; WT, wild type; asterisk (*), no DNA available. (a) families carrying homozygous variants; see DM1569 in 63; (b) families with compound heterozygous changes; (c) families with heterozygous variants; (d) unresolved families. Wherever applicable modifiers or secondary loci are listed under the primary causal alleles and highlighted by blue rectangles.
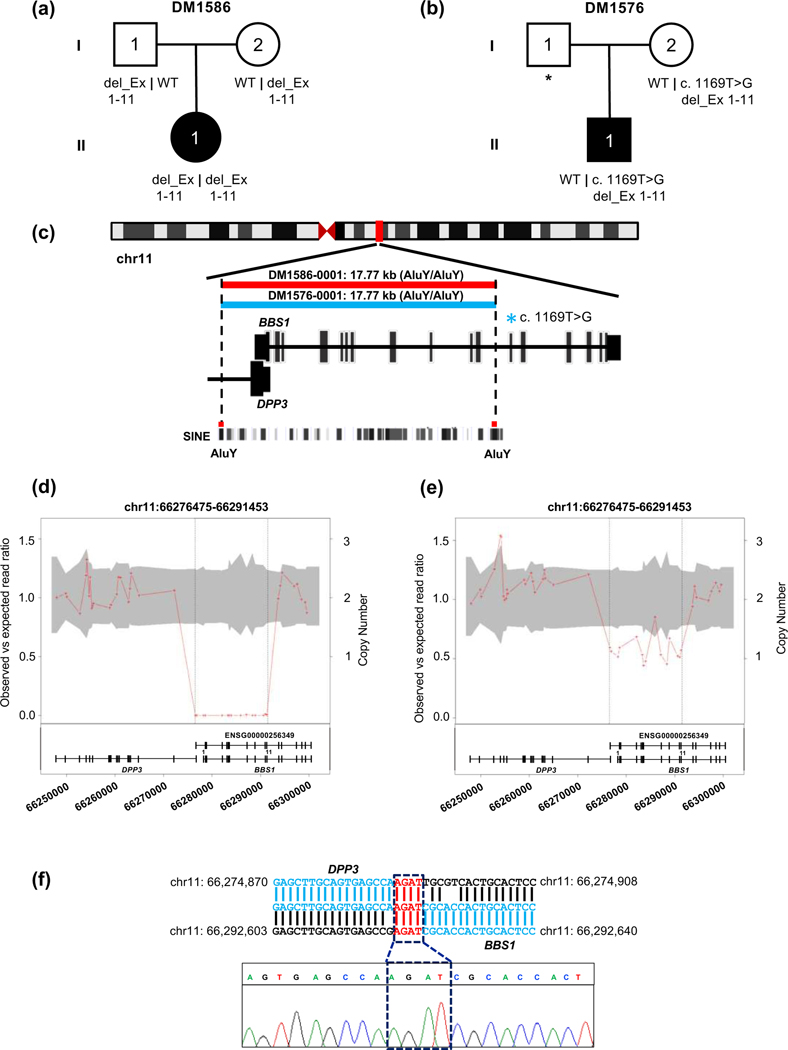
Figure 2. Characterization of a 17.77 kb CNV deletion in BBS1 in family DM1586 and family DM1576.
(a-b) Pedigrees and segregation of BBS1 exon disruptive deletion. (c) Schematic representation of human chromosome 11 and location of BBS1 CNV deletion is indicated with vertical red bar; enlarged view shows schematic of BBS1 transcript and location of AluY-AluY repeats elements. Short interspersed nuclear elements (SINE) (d-e) CNV plot showing homozygous and heterozygous BBS1 deletion, the gray area marks 95% confidence interval and the vertical black dotted lines indicate the location of the CNV; bottom, schematic of BBS1 locus: vertical bars, exons; horizontal line, intronic region; coordinates on chromosome 11 (hg19) are shown. (f) BBS1 breakpoint junction and sequence chromatograms amplified from genomic DNA of DM1586-0001 (II-1); a 4 bp microhomology region is present at the junction of DPP3 and BBS1, highlighted in red.
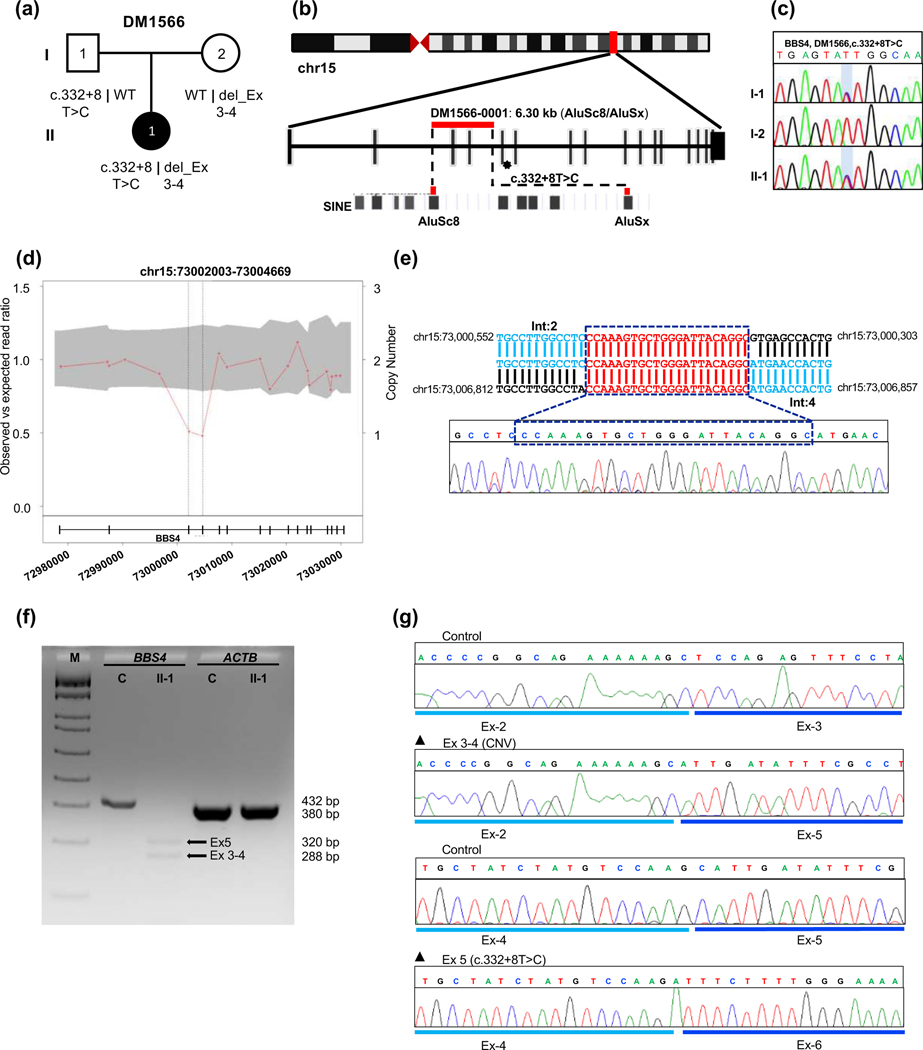
Figure 3. Genetic analysis of DM1566 harboring a biallelic two-exon deletion in trans with a pathogenic splicing variant in BBS4.
(a) Pedigree and segregation analysis of BBS4 variants in DM1566. (b) Schematic representation of human chromosome 15 showing the location of BBS4 with vertical red bar; enlarged view of CNV and SNV bearing region of BBS4 correspond to red horizontal bar and asterisk respectively; AluSc8 and AluSx are indicated at bottom. (c) Sequence chromatogram of paternally-inherited c.332+8T>C change, the variant position is shaded with a light blue vertical bar. (d) CNV plot showing a pathogenic heterozygous two-exon deletions in BBS4, the gray area marks a 95% confidence interval. Black vertical dotted lines indicate the position of the CNV. Bottom, schematic of the BBS4 locus; vertical bars, exons; horizontal line, intronic region; numbers indicate genomic coordinates on chromosome 15 (hg19). (e) BBS4 breakpoint junction sequence in genomic DNA; reference location highlighted in blue and a 22 bp microhomology region present at the junction is shown in red; Int, intron. (f) RT-PCR results show impaired splicing in proband cDNA; 2% agarose gel showing BBS4 amplification products in unaffected control (C, 432 bp expected wild type product) and affected individual II-1 (320 bp band showing exon-5 skipping due to splicing variant and 288 bp band indicating exon 3-4 deletion due to CNV); ACTB (380 bp) was amplified to ensure RNA integrity for both control and proband; M, DNA marker; Ex, exon. (g) Sequence chromatograms of RT-PCR products from unaffected control (top) and DM1566 proband show aberrant mRNA splicing outcomes from maternally inherited exon 3-4 deletion (middle) and paternally inherited exon 5 skipping (bottom).
Table 2.
SNVs and small indels identified in known BBS genes in 18 affected individuals with BBS (primary causal alleles or secondary alleles)
| Family ID | Ethnic Origin | Cons. | BBS gene | Transcript ID | Nucleotide change | Amino acid change | ACMG Class | dbSNP ID | Ref | gnomAD* | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele counts (Hom/Het/Wt) | Frequency | ||||||||||
| DM1565 | Arab | YES | BBS2 | NM_031885.5 | c.1528G>T (H) | p.Val510Phe | LP | − | 85 | − | − |
| DM1566 | E. Eur | NO | BBS4 | NM_033028.5 | c.332+8T>C (h) | p.Ala74Aspfs*7 | P | rs1456405256 | − | 0/01/250944 | 3.98e-6 |
| DM1567 | E. Eur | NO | BBS7 | NM_176824.3 | c.712_715del (h) | p.Arg238Glufs*59 | P | rs760165634 | 86 | 0/13/250940 | 5.18e-5 |
| DM1569 | Rom | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| DM1570 | E. Eur | NO | BBS5 | NM_152384.3 | c.619–1G>C (H) | − | P | rs753234582 | 78 | 0/04/250308 | 1.60e-5 |
| DM1571 | E. Eur | NO |
BBS7
BBS7 |
NM_176824.3 | c.68T>G (h) c.712_715del (h) |
p.Leu23Arg p.Arg238Glufs*59 |
LP P |
rs1727380420 rs760165634 |
**
86 |
−0/13/250940 | −5.18e-5 |
| DM1572 | E. Eur | NO | BBS4 | NM_033028.5 | c.1091C>A (H) | p.Ala364Glu | LP | rs28938468 | 87 | − | − |
| DM1573 | Rom | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| DM1574 | E. Eur | NO |
BBS10
BBS10 |
NM_024685.4 | c.145C>T (h) c.1804G>C (h) |
p.Arg49Trp p.Val602Leu |
LP LP |
rs768933093 rs778431173 |
88
88 |
0/15/279274 0/01/248576 |
5.37e-5 4.02e-6 |
| IFT172 | NM_015662.3 | c.1715G>A (h) | p.Arg572Gln | VUS | rs764302265 | − | 0/13/282810 | 4.60e-5 | |||
| DM1576 | E. Eur | NO | BBS1 | NM_024649.5 | c.1169T>G (h) | p.Met390Arg | P | rs113624356 | 79 | 0/444/282790 | 1.57e-3 |
| DM1582 | E. Eur | YES | BBS10 | NM_024685.4 | c.273C>G (H) | p.Cys91Trp | LP | rs148374859 | 89 | 0/07/248572 | 2.82e-5 |
| DM1584 | E. Eur | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| DM1585 | E. Eur | YES | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| DM1587 | Rom | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| DM1588 | Rom | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| BBS5 | NM_152384.3 | c.226A>G (h) | p.Ile76Val | VUS | rs763696357 | − | 0/05/250968 | 1.99e-5 | |||
| DM1589 | E. Eur | NO | BBS12 | NM_152618.3 | c.1063C>T (H) | p.Arg355* | P | rs121918327 | 37 | 0/07/251322 | 2.79e-5 |
| SCAPER | NM_020843.4 | c.3293G>A (h) | p.Arg1098Gln | VUS | rs771454836 | − | 0/05/248920 | 2.01e-5 | |||
| NPHP1 | NM_000272.5 | c.1630A>G (h) | p.Met544Val | VUS | rs373953762 | − | 0/01/251420 | 3.98e-6 | |||
| DM1590 | E. Eur | NO | BBS7 | NM_176824.3 | c.712_715del (H) | p.Arg238Glufs*59 | P | rs760165634 | 86 | 0/13/250940 | 5.18e-5 |
| DM1591 | E. Eur | NO |
BBS12
BBS12 |
NM_152618.3 | c.1392_1395del (h) c.1682_1682del (h) |
p.Cys464Trpfs*7 p.Glu561Lysfs*10 |
P P |
− − |
− − |
− − |
− − |
The Genome Aggregation Database v2.1.1 (https://gnomad.broadinstitute.org/); all populations;
ClinVar accession VCV000940124.1. Abbreviations: Hom, homozygous; Het, heterozygous; WT, wild type reference. Abbreviations: Cons., consanguinity; E. Eur, European; H, homozygous; h, heterozygous; LP, likely pathogenic; P, pathogenic; Rom, Romani, VUS, variant of uncertain significance. Modifiers or secondary alleles are listed under primary causal loci wherever applicable.
Table 3.
Breakpoint features of CNVs identified in 3-affected individuals
| Family ID | Origin | Cons. | Locus | hg19 CNV Coordinates | Result | Size (bp) | Rearrangement type | Ref | Breakpoint features | % Identity | ACMG Class |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DM1586 | E. Eur | NO | BBS1 | chr11:66,274,870- 66,292,647 |
Hom del: | 17,777 | Simple non-recurrent | 32 | AluY-AluY | 92% | P |
| Exon1_11(H) | |||||||||||
| DM1576 | E. Eur | NO | BBS1 | chr11:66,274,870- 66,292,647 |
Het del: | 17,777 | Simple non-recurrent | 32 | AluY-AluY | 92% | P |
| Exon1_11(h) | |||||||||||
| DM1566 | E. Eur | NO | BBS4 | chr15:73,000,552- 73,006,857 |
Het del: | 6,305 | Simple non-recurrent | 32 | AluSc8-AluSx | 83% | P |
| Exon3_4(h) |
Abbreviations are as follows: CNV, copy number variation; Cons., consanguinity; del, deletion; H, homozygous; h, heterozygous; E. Eur, Eastern European
Biallelic SNVs or small indels in known BBS genes are predominant in our cohort
A majority of our cohort carry biallelic pathogenic or likely pathogenic SNVs or small indels in established BBS genes. Eleven variants have been reported previously in affected individuals, have high amino acid conservation, and segregated with disease in available family members (Table 2, Table S3, Figure 1, Figures S1 and S2). The majority of previously reported variants were non-recurrent in our cohort, however, we observed a BBS12 p.Arg355* allele in 7 of 23 probands. Additionally, we identified two hitherto unreported variants in cases: BBS12 p.Cys464Trpfs*7 and p.Glu561Lysfs*10 leading to a frameshift and predicted premature termination (Table 2, Table S3, Figure 1, Figure S1).
Identification of causal genes disrupted by CNV deletions in known BBS loci
Rare CNV deletions and duplications have been identified in a broad spectrum of human diseases including autism, intellectual disability, and BBS32; 40; 73; 74. We found 2 of 17 individuals who carry previously reported recurrent exon disruptive deletions in BBS1 and BBS432; 40, respectively, that are inherited under a recessive paradigm.
In family DM1586, we used exome read depth to detect a homozygous 17.7 kb CNV in BBS1 that deletes exons 1–11 (Figure 2, Table 3). To refine the CNV breakpoint and map to the precise genomic location, we performed long-range PCR and subsequent Sanger sequencing. The CNV junction was located within substrate pairs of Alu elements from the same family with 92% sequence identity, suggesting that the deletion was mediated by Alu-Alu recombination. This recombination forms an Alu hybrid which is the most prominent mechanism underlying the formation of a pathogenic CNV75–77. This deletion variant was confirmed heterozygous in both carrier parents, has been reported previously in trans with an p.Glu549* SNV in an affected individual with BBS and was deemed as pathogenic32.
In family DM1566, the affected individual harbored a two-exon deletion in trans with an intronic SNV in BBS4. This phenomenon was reported previously to contribute to pathology in BBS1, BBS7, and IFT7432. We first characterized the maternally inherited CNV spanning exons 3 and 4. The breakpoint sequencing and analysis of junction fragments showed that the deletion was located within distinct substrate pairs (AluSc8-AluSx) with 83% sequence identity; this CNV was reported previously in homozygosity in a BBS case32. Next, we evaluated the impact of the paternally inherited heterozygous intronic variant (c.332+8T>C) on mRNA splicing. This change was found once in 250,944 alleles in gnomAD (accessed November 2022). To test whether the variant affects splicing, we established an LCL from whole blood of the affected individual and extracted total RNA. Sequencing of TOPO-cloned RT-PCR product spanning exons 1–7 showed impaired mRNA splicing that results in exon 5 exclusion, resulting in a frameshift p.Arg74Aspfs*7 deletion and putative premature protein termination (Table 2, Figure 3f-g). Sequencing of cloned RT-PCR products also revealed that the maternally inherited CNV results in aberrant mRNA splicing of exons 2 and 5, resulting in an in-frame deletion of 48 amino acids. Together, the segregation data, previous report in BBS cohorts, and our RT-PCR data suggest that these variants are pathogenic.
Identification of secondary contributing variants in BBS genes
We have shown previously that the presence of BBS gene mutational burden is significantly enriched in BBS individuals compared to matched unaffected controls47. Among the pedigrees for which we could identify the primary causal locus, three families harbored additional rare heterozygous changes in BBS loci. In family DM1574, with a primary causal BBS10 locus (with previously reported alleles p.Arg49Trp37 and Val620Leu78), we identified a rare and phylogenetically conserved p.Arg572Gln variant in IFT172 (Table 2, Table S3; Figures 1, Figure S1 and S2). In family DM1588, with a primary causal BBS12 locus (homozygous p.Arg355*), we detected a heterozygous p.IIe76Val change in a conserved residue of BBS5 (Table 2, Table S3; Figures 1, Figure S1 and S2). In family DM1589, also with the same primary causal BBS12 locus, we identified two rare heterozygous second-site missense variants: NPHP1, p.Met544Val and SCAPER, p.Arg1098Gln (Table 2, Table S3; Figures 1, Figure S1 and S2).
A subset of BBS cases has unresolved molecular etiology
We did not identify a causal locus for a modest fraction of our cohort (n=6). In two families, we identified heterozygous rare variants in an established BBS gene, each of which were inherited from a single parent. In DM1576, we identified a maternally-inherited BBS1 allele with two pathogenic changes in cis: the recurrent 17.7 kb exon 1–11 disruptive deletion32 (also identified in DM1586) and the common p.Met390Arg79 variant (Tables 2 and 3, Table S3; Figures 1 and 2, Figure S1). Additionally, family DM1567 harbors a paternally-inherited putative truncating variant in BBS7 (p.Arg238Glufs*59). (Table 2; Figure 1, Figure S1). The remaining four families were bereft of rare SNVs, small indels or CNVs in known BBS genes (Figure 1d). Furthermore, unbiased filtering of the exome for any rare, functional biallelic variants yielded no likely causal candidate genes. These families may harbor deep intronic or large structural variants that are intractable to ES.
Discussion
Here, we report the molecular analysis of 24 individuals in 23 families who reside in Romania and fulfill clinical diagnostic criteria for BBS. ES of affected individuals, bioinformatic filtering, and segregation analysis enabled the detection of biallelic likely pathogenic or pathogenic SNVs or CNVs that could potentially inform disease causality in 17 of 23 families. We achieved an overall diagnostic rate of 74%, which is consistent with previous genetic studies on BBS cohorts 70–80%16; 34; 80; 81. For one family, we leveraged mRNA profiling from primary LCLs to simultaneously characterize the functional effects of an intronic SNV and an exon disruptive CNV, thus enabling more accurate assessment of variant pathogenicity.
Molecular diagnosis of BBS has been notoriously challenging for three reasons: (1) there are >20 causal genes; (2) a majority of variants are private, and there are few recurrent variants that can be utilized for targeted screening; and (3) a notable portion of contributory BBS alleles are CNVs ranging from small indels to large deletion/complex intragenic duplication events32. To potentially circumvent limitations associated with targeted screening, we performed ES, and consistent with previous reports, we identified an allelic series of causal variants that was non-recurrent within our cohort; this includes 13 SNVs or indels. The contribution of CNVs to causality and overall mutational burden has been often under-recognized, and previous studies found that 18% of individuals affected with BBS harbor at least one exon disruptive CNV32. Similarly, we detected CNVs in 13% of individuals in our cohort confirming the importance of systematically querying for structural variants.
Notably, we observed three changes in more than one family within our cohort: BBS1 exon 1–11 CNV deletion; BBS7 p.Arg238Glnfs*59; and BBS12 p.Arg355*. The latter SNV was detected in a surprising fraction of families (7 of 23), with no correlation to self-reported ethnicity (4 Romani and 3 Eastern European). Accordingly, p.Arg355* has been reported in BBS cases of Romani origin37 but is present in gnomAD in both Latino/Admixed American and non-Finnish European populations. Additionally, while all p.Arg355* alleles were found in homozygosity, only 1 of 7 pedigrees self-reported as consanguineous (DM1585). However, given the lack of parental DNAs for a subset of these families, we cannot exclude the possibility of uniparental disomy. Further studies will be required to determine whether p.Arg355* is a founder allele or mutational hotspot. Further, we note with interest that among p.Arg355* homozygotes, 3 of 7 individuals with recorded cognitive testing have severe intellectual disability, however our cohort is too small to determine whether there is a significant genotype-phenotype correlation.
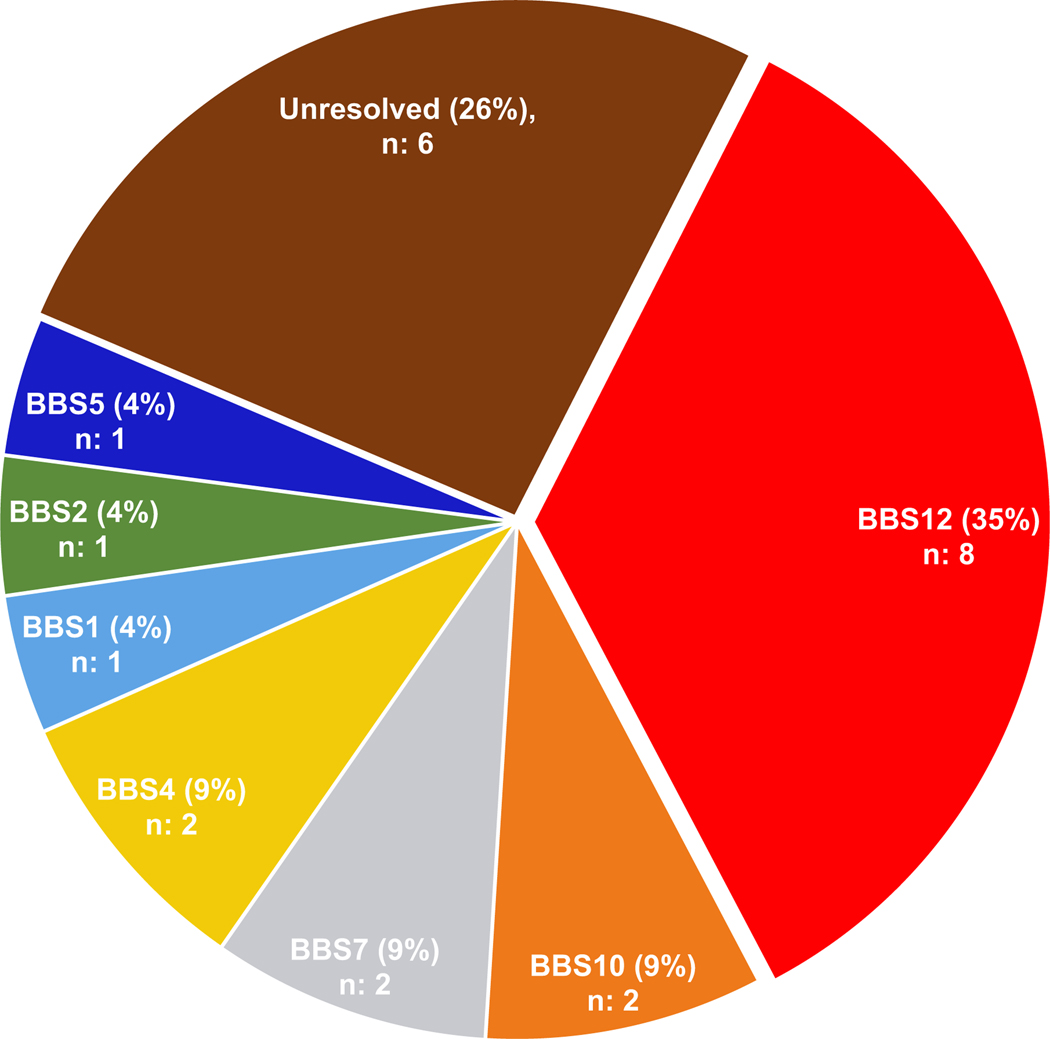
In our cohort, the frequently impacted genes were BBS12 (35%), followed by BBS4, BBS7, and BBS10 (9%; 2 families each), whereas BBS1, BBS2, and BBS5 were detected in (4%; 1 family each) of the cases (Figure 4). Although our cohort size is small, the relative causal gene contribution differs from what has been reported from BBS population studies comprised of individuals of northern European descent. BBS1 and BBS10 are reported to be predominant drivers of BBS, largely due to the recurrent p.Met390Arg and p.Cys91Leufs*5 variants, respectively33; 36; 82; 83. However, there were a paucity of these two common changes in our Romanian cohort (DM1576, BBS1 p.Met390Arg in cis with exon 1–11 CNV deletion; BBS10 p.Cys91Leufs*5 was not detected). Instead, BBS12 p.Arg355* was overabundant in our cohort and contributed to BBS12 emerging as the most common causal gene. Still, BBS12 is among the major contributors to BBS accounting for 8–11% in most reported cohorts36; 37, including a recent study of 99 affected individuals for which BBS12 was causal in 14% of cases43.
Figure 4: Distribution of BBS gene contribution to the molecular etiology of our cohort.
Pie chart shows the genetic architecture of primary causal BBS genes in a cohort of 23 unrelated families.
A minority of affected individuals in our cohort harbor secondary variants outside of their primary causal BBS locus (3 of 17, Table 2), but these cases have no apparent distinguishing clinical features or increased severity. Although the rarity of BBS and underlying genetic heterogeneity limit conclusive statements about commonly observed gene pairings, some genes appear to be more frequently implicated in oligogenic phenomena. In a recent study, BBS1, BBS4, BBS2, CFAP418/BBS21, and BBS12 were reported as common driver genes involved in oligogenic phenomena43. Accordingly, we detected a third allele in two families with primary causal variants in BBS12. Further, we have speculated previously that gene pairings involving proteins known to function in different molecular complexes drive more potent phenotypes (e.g. chaperonin-BBSome or chaperonin-IFT pairings) than inter-module pairings (e.g. BBSome-BBSome or chaperonin-chaperonin)47. Consistent with this notion, all three families with secondary variants involve gene combinations encoding different complexes. However, we are cautious about drawing formal conclusions given the small sample size of our cohort.
Finally, a small fraction of families in our cohort remains molecularly undiagnosed (6 of 23; 26%). We detected heterozygous rare variants in two families, but further investigation will be required to determine whether they are primary causal alleles with the second variant undetected with our current ES methodology, or they are secondary contributor variants. The remaining families harbored neither rare heterozygous variants in known BBS genes, nor rare variants in hitherto unreported ciliopathy genes elsewhere in the exome. We speculate that they might harbor regulatory variants in non-coding regions or large deletions that are intractable to ES. The eventual transition to the whole genome sequencing combined with RNA sequencing will likely overcome this challenge84.
Supplementary Material
Acknowledgments
We are grateful to the families reported in this study for their participation and support of our research. This work was supported by grants from the US National Institute of Health (R01HD042601 and R01DK072301) to EED. SK was funded by an International Research Support Initiative Program (IRSIP) fellowship from the Higher Education Commission of Pakistan. EED is the Ann Marie and Francis Klocke, MD Research Scholar. We Thank Dr. Tahir Khan and Dr. Kamal Khan for critical reading and reviewing of the manuscript.
Footnotes
Ethics Approval/Patient Consent
This study was approved by Institutional Board of the University and Pharmacy Carol Davila Bucharest (approval no. 29700, T.42; Oct 01, 2015) and Ann & Robert H. Lurie Children’s Hospital of Chicago (approval no. IRB 2019–3057, August 5, 2019). The families were enrolled in this study under informed consent and all the experiments conformed with the guidelines of the Declaration of Helsinki.
Conflict of Interest
The authors have no competing interests to declare.
Data Availability
Variants identified have been deposited in ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/
References
- 1.Forsythe E, and Beales PL. (2013). Bardet-Biedl syndrome. Eur J Hum Genet 21, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrandt F, Benzing T, and Katsanis N. (2011). Ciliopathies. N Engl J Med 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beales PL, Elcioglu N, Woolf AS, Parker D, and Flinter FA. (1999). New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 36, 437–446. [PMC free article] [PubMed] [Google Scholar]
- 4.Forsythe E, Kenny J, Bacchelli C, and Beales PL. (2018). Managing Bardet-Biedl Syndrome-Now and in the Future. Front Pediatr 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mary L, Chennen K, Stoetzel C, Antin M, Leuvrey A, Nourisson E, Alanio-Detton E, Antal MC, Attie-Bitach T, Bouvagnet P, et al. (2019). Bardet-Biedl syndrome: Antenatal presentation of forty-five fetuses with biallelic pathogenic variants in known Bardet-Biedl syndrome genes. Clin Genet 95, 384–397. [DOI] [PubMed] [Google Scholar]
- 6.Weihbrecht K, Goar WA, Pak T, Garrison JE, DeLuca AP, Stone EM, Scheetz TE, and Sheffield VC. (2017). Keeping an Eye on Bardet-Biedl Syndrome: A Comprehensive Review of the Role of Bardet-Biedl Syndrome Genes in the Eye. Med Res Arch 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beales PL, Warner AM, Hitman GA, Thakker R, and Flinter FA. (1997). Bardet-Biedl syndrome: a molecular and phenotypic study of 18 families. J Med Genet 34, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croft JB, Morrell D, Chase CL, and Swift M. (1995). Obesity in heterozygous carriers of the gene for the Bardet-Biedl syndrome. Am J Med Genet 55, 12–15. [DOI] [PubMed] [Google Scholar]
- 9.Moore SJ, Green JS, Fan Y, Bhogal AK, Dicks E, Fernandez BA, Stefanelli M, Murphy C, Cramer BC, Dean JC, et al. (2005). Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am J Med Genet A 132A, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farag TI, and Teebi AS. (1989). High incidence of Bardet Biedl syndrome among the Bedouin. Clin Genet 36, 463–464. [DOI] [PubMed] [Google Scholar]
- 11.Meng X, Long Y, Ren J, Wang G, Yin X, and Li S. (2021). Ocular Characteristics of Patients With Bardet-Biedl Syndrome Caused by Pathogenic BBS Gene Variation in a Chinese Cohort. Front Cell Dev Biol 9, 635216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone EM, Andorf JL, Whitmore SS, DeLuca AP, Giacalone JC, Streb LM, Braun TA, Mullins RF, Scheetz TE, Sheffield VC, et al. (2017). Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmology 124, 1314–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berezovsky A, Rocha DM, Sacai PY, Watanabe SS, Cavascan NN, and Salomao SR. (2012). Visual acuity and retinal function in patients with Bardet-Biedl syndrome. Clinics (Sao Paulo) 67, 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams NA, Awadein A, and Toma HS. (2007). The retinal ciliopathies. Ophthalmic Genet 28, 113–125. [DOI] [PubMed] [Google Scholar]
- 15.Mockel A, Perdomo Y, Stutzmann F, Letsch J, Marion V, and Dollfus H. (2011). Retinal dystrophy in Bardet-Biedl syndrome and related syndromic ciliopathies. Prog Retin Eye Res 30, 258–274. [DOI] [PubMed] [Google Scholar]
- 16.Hjortshoj TD, Gronskov K, Philp AR, Nishimura DY, Riise R, Sheffield VC, Rosenberg T, and Brondum-Nielsen K. (2010). Bardet-Biedl syndrome in Denmark--report of 13 novel sequence variations in six genes. Hum Mutat 31, 429–436. [DOI] [PubMed] [Google Scholar]
- 17.Tobin JL, and Beales PL. (2007). Bardet-Biedl syndrome: beyond the cilium. Pediatr Nephrol 22, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niederlova V, Modrak M, Tsyklauri O, Huranova M, and Stepanek O. (2019). Meta-analysis of genotype-phenotype associations in Bardet-Biedl syndrome uncovers differences among causative genes. Hum Mutat 40, 2068–2087. [DOI] [PubMed] [Google Scholar]
- 19.Katsanis N, Lupski JR, and Beales PL. (2001). Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet 10, 2293–2299. [DOI] [PubMed] [Google Scholar]
- 20.Kerr EN, Bhan A, and Heon E. (2016). Exploration of the cognitive, adaptive and behavioral functioning of patients affected with Bardet-Biedl syndrome. Clin Genet 89, 426–433. [DOI] [PubMed] [Google Scholar]
- 21.Meyer JR, Krentz AD, Berg RL, Richardson JG, Pomeroy J, Hebbring SJ, and Haws RM. (2022). Kidney failure in Bardet-Biedl syndrome. Clin Genet 101, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putoux A, Attie-Bitach T, Martinovic J, and Gubler MC. (2012). Phenotypic variability of Bardet-Biedl syndrome: focusing on the kidney. Pediatr Nephrol 27, 7–15. [DOI] [PubMed] [Google Scholar]
- 23.Forsythe E, Sparks K, Best S, Borrows S, Hoskins B, Sabir A, Barrett T, Williams D, Mohammed S, Goldsmith D, et al. (2017). Risk Factors for Severe Renal Disease in Bardet-Biedl Syndrome. J Am Soc Nephrol 28, 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchese E, Ruoppolo M, Perna A, Capasso G, and Zacchia M. (2020). Exploring Key Challenges of Understanding the Pathogenesis of Kidney Disease in Bardet-Biedl Syndrome. Kidney Int Rep 5, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou CC, Wu KL, Chi Y, Hu WP, Yu SJ, Lee GH, Lin CL, and Chou PT. (2011). Ruthenium(II) sensitizers with heteroleptic tridentate chelates for dye-sensitized solar cells. Angew Chem Int Ed Engl 50, 2054–2058. [DOI] [PubMed] [Google Scholar]
- 26.Tsang SH, Aycinena ARP, and Sharma T. (2018). Ciliopathy: Bardet-Biedl Syndrome. Adv Exp Med Biol 1085, 171–174. [DOI] [PubMed] [Google Scholar]
- 27.Olson AJ, Krentz AD, Finta KM, Okorie UC, and Haws RM. (2019). Thoraco-Abdominal Abnormalities in Bardet-Biedl Syndrome: Situs Inversus and Heterotaxy. J Pediatr 204, 31–37. [DOI] [PubMed] [Google Scholar]
- 28.Branfield Day L, Quammie C, Heon E, Bhan A, Batmanabane V, Dai T, and Kamath BM. (2016). Liver anomalies as a phenotype parameter of Bardet-Biedl syndrome. Clin Genet 89, 507–509. [DOI] [PubMed] [Google Scholar]
- 29.Pomeroy J, VanWormer JJ, Meilahn JR, Maki T, Murali HR, and Haws RM. (2021). Sleep and physical activity patterns in adults and children with Bardet-Biedl syndrome. Orphanet J Rare Dis 16, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pomeroy J, Krentz AD, Richardson JG, Berg RL, VanWormer JJ, and Haws RM. (2021). Bardet-Biedl syndrome: Weight patterns and genetics in a rare obesity syndrome. Pediatr Obes 16, e12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Florea L, Caba L, and Gorduza EV. (2021). Bardet-Biedl Syndrome-Multiple Kaleidoscope Images: Insight into Mechanisms of Genotype-Phenotype Correlations. Genes (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindstrand A, Frangakis S, Carvalho CM, Richardson EB, McFadden KA, Willer JR, Pehlivan D, Liu P, Pediaditakis IL, Sabo A, et al. (2016). Copy-Number Variation Contributes to the Mutational Load of Bardet-Biedl Syndrome. Am J Hum Genet 99, 318–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, Salem N, Chouery E, Corbani S, et al. (2006). BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet 38, 521–524. [DOI] [PubMed] [Google Scholar]
- 34.Muller J, Stoetzel C, Vincent MC, Leitch CC, Laurier V, Danse JM, Helle S, Marion V, Bennouna-Greene V, Vicaire S, et al. (2010). Identification of 28 novel mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Hum Genet 127, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esposito G, Testa F, Zacchia M, Crispo AA, Di Iorio V, Capolongo G, Rinaldi L, D’Antonio M, Fioretti T, Iadicicco P, et al. (2017). Genetic characterization of Italian patients with Bardet-Biedl syndrome and correlation to ocular, renal and audio-vestibular phenotype: identification of eleven novel pathogenic sequence variants. BMC Med Genet 18, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan SA, Muhammad N, Khan MA, Kamal A, Rehman ZU, and Khan S. (2016). Genetics of human Bardet-Biedl syndrome, an updates. Clin Genet 90, 3–15. [DOI] [PubMed] [Google Scholar]
- 37.Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, Vicaire S, Jacquelin C, Plewniak F, Leitch CC, Sarda P, et al. (2007). Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet 80, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta N, D’Acierno M, Zona E, Capasso G, and Zacchia M. (2022). Bardet-Biedl syndrome: The pleiotropic role of the chaperonin-like BBS6, 10, and 12 proteins. Am J Med Genet C Semin Med Genet 190, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelman CD, Brady HL, Baron AE, Norris JM, and Framingham Heart S. (2003). Comparison between two analytic strategies to detect linkage to obesity with genetically determined age of onset: the Framingham Heart Study. BMC Genet 4 Suppl 1, S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindstrand A, Davis EE, Carvalho CM, Pehlivan D, Willer JR, Tsai IC, Ramanathan S, Zuppan C, Sabo A, Muzny D, et al. (2014). Recurrent CNVs and SNVs at the NPHP1 locus contribute pathogenic alleles to Bardet-Biedl syndrome. Am J Hum Genet 94, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redin C, Le Gras S, Mhamdi O, Geoffroy V, Stoetzel C, Vincent MC, Chiurazzi P, Lacombe D, Ouertani I, Petit F, et al. (2012). Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom syndromes. J Med Genet 49, 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, and Lupski JR. (2001). Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293, 2256–2259. [DOI] [PubMed] [Google Scholar]
- 43.Perea-Romero I, Solarat C, Blanco-Kelly F, Sanchez-Navarro I, Bea-Mascato B, Martin-Salazar E, Lorda-Sanchez I, Swafiri ST, Avila-Fernandez A, Martin-Merida I, et al. (2022). Allelic overload and its clinical modifier effect in Bardet-Biedl syndrome. NPJ Genom Med 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allison SJ. (2021). Secondary-variant genetic burden in Bardet-Biedl syndrome. Nat Rev Nephrol 17, 14. [DOI] [PubMed] [Google Scholar]
- 45.Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, Beales PL, Dietz HC, Fisher S, and Katsanis N. (2006). Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature 439, 326–330. [DOI] [PubMed] [Google Scholar]
- 46.Cardenas-Rodriguez M, Irigoin F, Osborn DP, Gascue C, Katsanis N, Beales PL, and Badano JL. (2013). The Bardet-Biedl syndrome-related protein CCDC28B modulates mTORC2 function and interacts with SIN1 to control cilia length independently of the mTOR complex. Hum Mol Genet 22, 4031–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kousi M, Soylemez O, Ozanturk A, Mourtzi N, Akle S, Jungreis I, Muller J, Cassa CA, Brand H, Mokry JA, et al. (2020). Evidence for secondary-variant genetic burden and non-random distribution across biological modules in a recessive ciliopathy. Nat Genet 52, 1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaghloul NA, and Katsanis N. (2009). Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest 119, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, et al. (2011). TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 51.Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, and Nachury MV. (2008). A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell 15, 854–865. [DOI] [PubMed] [Google Scholar]
- 52.Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, Slusarski DC, and Sheffield VC. (2010). BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A 107, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez-Satta M, Castro-Sanchez S, and Valverde D. (2017). Bardet-Biedl Syndrome as a Chaperonopathy: Dissecting the Major Role of Chaperonin-Like BBS Proteins (BBS6-BBS10-BBS12). Front Mol Biosci 4, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, and Nachury MV. (2010). The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbelanne M, Hossain D, Chan DP, Peranen J, and Tsang WY. (2015). Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum Mol Genet 24, 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shamseldin HE, Shaheen R, Ewida N, Bubshait DK, Alkuraya H, Almardawi E, Howaidi A, Sabr Y, Abdalla EM, Alfaifi AY, et al. (2020). The morbid genome of ciliopathies: an update. Genet Med 22, 1051–1060. [DOI] [PubMed] [Google Scholar]
- 57.Satir P, and Christensen ST. (2007). Overview of structure and function of mammalian cilia. Annu Rev Physiol 69, 377–400. [DOI] [PubMed] [Google Scholar]
- 58.Wheway G, Nazlamova L, and Hancock JT. (2018). Signaling through the Primary Cilium. Front Cell Dev Biol 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerdes JM, Liu Y, Zaghloul NA, Leitch CC, Lawson SS, Kato M, Beachy PA, Beales PL, DeMartino GN, Fisher S, et al. (2007). Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet 39, 1350–1360. [DOI] [PubMed] [Google Scholar]
- 60.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, Chuang PT, and Reiter JF. (2008). Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol 10, 70–76. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura T. (2009). [Expectation for alleviation of joint destruction in rheumatoid arthritis by new molecular targeting pharmaceutics including anti-rankle antibody]. Clin Calcium 19, 381–386. [PubMed] [Google Scholar]
- 62.Badano JL, Mitsuma N, Beales PL, and Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 7, 125–148. [DOI] [PubMed] [Google Scholar]
- 63.Focsa IO, Budisteanu M, Burloiu C, Khan S, Sadeghpour A, Bohiltea LC, Davis EE, and Balgradean M. (2021). A case of Bardet-Biedl syndrome caused by a recurrent variant in BBS12: A case report. Biomed Rep 15, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desvignes JP, Bartoli M, Delague V, Krahn M, Miltgen M, Beroud C, and Salgado D. (2018). VarAFT: a variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res 46, W545–W553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, et al. (2012). A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinhaus R, Proft S, Schuelke M, Cooper DN, Schwarz JM, and Seelow D. (2021). MutationTaster2021. Nucleic Acids Res 49, W446–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi Y, and Chan AP. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, and Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaser R, Adusumalli S, Leng SN, Sikic M, and Ng PC. (2016). SIFT missense predictions for genomes. Nat Protoc 11, 1–9. [DOI] [PubMed] [Google Scholar]
- 70.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis EE, Savage JH, Willer JR, Jiang YH, Angrist M, Androutsopoulos A, and Katsanis N. (2014). Whole exome sequencing and functional studies identify an intronic mutation in TRAPPC2 that causes SEDT. Clin Genet 85, 359–364. [DOI] [PubMed] [Google Scholar]
- 72.Focsa IO, Budisteanu M, Stoica C, Nedelea F, Jurca C, Caba L, Butnariu L, Panzaru M, Rusu C, and Balgradean M. (2022). Clinical Aspects of a Rare Disease: Bardet Biedl Syndrome. Modern Medicine 29, 37–42. [Google Scholar]
- 73.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, et al. (2011). An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med 13, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins RL, Glessner JT, Porcu E, Lepamets M, Brandon R, Lauricella C, Han L, Morley T, Niestroj LM, Ulirsch J, et al. (2022). A cross-disorder dosage sensitivity map of the human genome. Cell 185, 3041–3055 e3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, et al. (2015). Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet 24, 4061–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, Beck CR, Shaw CJ, Stankiewicz P, Moretti P, et al. (2014). The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet 95, 143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verdin H, D’Haene B, Beysen D, Novikova Y, Menten B, Sante T, Lapunzina P, Nevado J, Carvalho CM, Lupski JR, et al. (2013). Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet 9, e1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feuillan PP, Ng D, Han JC, Sapp JC, Wetsch K, Spaulding E, Zheng YC, Caruso RC, Brooks BP, Johnston JJ, et al. (2011). Patients with Bardet-Biedl syndrome have hyperleptinemia suggestive of leptin resistance. J Clin Endocrinol Metab 96, E528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, et al. (2002). Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31, 435–438. [DOI] [PubMed] [Google Scholar]
- 80.Harville HM, Held S, Diaz-Font A, Davis EE, Diplas BH, Lewis RA, Borochowitz ZU, Zhou W, Chaki M, MacDonald J, et al. (2010). Identification of 11 novel mutations in eight BBS genes by high-resolution homozygosity mapping. J Med Genet 47, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janssen S, Ramaswami G, Davis EE, Hurd T, Airik R, Kasanuki JM, Van Der Kraak L, Allen SJ, Beales PL, Katsanis N, et al. (2011). Mutation analysis in Bardet-Biedl syndrome by DNA pooling and massively parallel resequencing in 105 individuals. Hum Genet 129, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mykytyn K, Nishimura DY, Searby CC, Beck G, Bugge K, Haines HL, Cornier AS, Cox GF, Fulton AB, Carmi R, et al. (2003). Evaluation of complex inheritance involving the most common Bardet-Biedl syndrome locus (BBS1). Am J Hum Genet 72, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delvallee C, Nicaise S, Antin M, Leuvrey AS, Nourisson E, Leitch CC, Kellaris G, Stoetzel C, Geoffroy V, Scheidecker S, et al. (2021). A BBS1 SVA F retrotransposon insertion is a frequent cause of Bardet-Biedl syndrome. Clin Genet 99, 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cummings BB, Karczewski KJ, Kosmicki JA, Seaby EG, Watts NA, Singer-Berk M, Mudge JM, Karjalainen J, Satterstrom FK, O’Donnell-Luria AH, et al. (2020). Transcript expression-aware annotation improves rare variant interpretation. Nature 581, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Knopp C, Rudnik-Schoneborn S, Eggermann T, Bergmann C, Begemann M, Schoner K, Zerres K, and Ortiz Bruchle N. (2015). Syndromic ciliopathies: From single gene to multi gene analysis by SNP arrays and next generation sequencing. Mol Cell Probes 29, 299–307. [DOI] [PubMed] [Google Scholar]
- 86.Bin J, Madhavan J, Ferrini W, Mok CA, Billingsley G, and Heon E. (2009). BBS7 and TTC8 (BBS8) mutations play a minor role in the mutational load of Bardet-Biedl syndrome in a multiethnic population. Hum Mutat 30, E737–746. [DOI] [PubMed] [Google Scholar]
- 87.Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, Scambler PJ, Beales PL, and Lupski JR. (2002). BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet 71, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Daniels AB, Sandberg MA, Chen J, Weigel-DiFranco C, Fielding Hejtmancic J, and Berson EL. (2012). Genotype-phenotype correlations in Bardet-Biedl syndrome. Arch Ophthalmol 130, 901–907. [DOI] [PubMed] [Google Scholar]
- 89.Gerth C, Zawadzki RJ, Werner JS, and Heon E. (2008). Retinal morphology in patients with BBS1 and BBS10 related Bardet-Biedl Syndrome evaluated by Fourier-domain optical coherence tomography. Vision Res 48, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variants identified have been deposited in ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/