Abstract
Pathogenic biallelic variants in LSS are associated with three Mendelian rare disease traits including congenital cataract type 44, autosomal recessive hypotrichosis type 14, and alopecia-intellectual disability syndrome type 4 (APMR4). We performed trio research exome sequencing on a family with a four-year-old male with global developmental delay, epilepsy and striking alopecia, and identified novel compound heterozygous LSS splice site (c.14+2T>C) and missense (c.1357 G>A; p.V453I) variant alleles. Rare features associated with APMR4 as cryptorchidism, micropenis, mild cortical brain atrophy and thin corpus callosum were detected. Previously unreported APMR4 findings including cerebellar involvement in the form of unsteady ataxic gait, small vermis with prominent folia, were noted. A review of all reported variants to date in 30 families with LSS-related phenotypes showed an emerging genotype-phenotype correlation. Our report potentially expands LSS-related phenotypic spectrum and highlights the importance of performing brain imaging in LSS-related conditions.
Keywords: LSS-related phenotypes, intellectual disability, hypogenitalism, cerebellar abnormalities, alopecia
Graphical Abstract
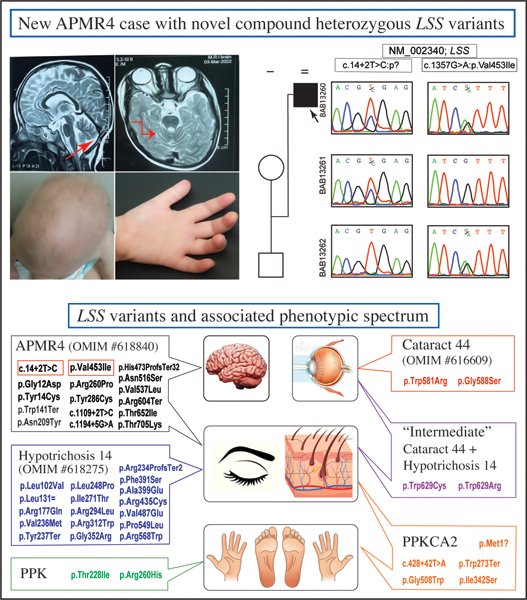
Elbendary et al. report a new Egyptian patient with alopecia-intellectual disability syndrome type 4 (APMR4) and cerebellar involvement due to novel compound heterozygous LSS splice site (c.14+2T>C) and missense (c.1357 G>A; p.V453I) variants. A review of all reported variants to date in 30 families with LSS-related phenotypes showed an emerging genotype-phenotype correlation.
INTRODUCTION
Alopecia-intellectual disability syndrome (APMR) is a rare clinical condition defined by loss of scalp hair, absence of eyebrows, eyelashes, axillary and pubic hair in combination with mild to severe intellectual disability.1 APMR is classified into four subtypes based on the degree of intellectual disability and the associated clinical manifestations.1 Alopecia-intellectual disability syndrome type 4 (APMR4, OMIM #618840) is a very rare, neuro-ectodermal syndrome caused by biallelic variants in LSS located on chromosome 21q22.2
Lanosterol synthase (LSS) is an enzyme involved in the biosynthesis of cholesterol, steroid hormones, and vitamin D.3 It catalyzes the rate-limiting step in the conversion of (S)-2,3-oxidosqualene into lanosterol.4 LSS is expressed in many tissues, including the brain, glomerular podocytes, and skin, potentially explaining multi-system involvement and pleiotropic phenotypic manifestations.5 Biallelic LSS variants are associated with three autosomal recessive rare diseases including congenital cataracts type 44, hypotrichosis simplex type 14, and APMR4 (OMIM #616509, #618275, #618840, respectively). APMR4 is characterized by alopecia universalis, scaly skin, and variable degrees of psychomotor delay.6
A total of 29 families with all three LSS-related disorders have been reported worldwide amongst which are ten families with 15 affected individuals with the APMR4 phenotype.2,5,7–18
We describe an Egyptian patient with biallelic novel variants in LSS and APMR4 to potentially expand the mutational and phenotypic spectrum of the syndrome.
CLINICAL REPORT
The proband is a four-year-old male and the first child of healthy non-consanguineous parents (Figure 1a,b,c). Normal pregnancy and delivery histories and birth growth parameters were recorded. Baldness and absence of eyebrows were noted since early infancy. At the age of 14 months, he presented with global developmental delay, myoclonic epilepsy, and alopecia. On clinical examination, he was able to support the head, recognized the mother and followed objects. His weight was 10kg (−0.5SD), length 76cm (−0.2SD), and head circumference 45cm (−1.2SD). He had alopecia, sparse hair and eyebrows, prominent forehead, wide-spaced eyes, depressed nose, short philtrum and thin upper lip. Neurological evaluation showed generalized hypotonia with normal reflexes. The genitalia showed micropenis (1.5cm) and bilateral cryptorchidism.
Figure 1:
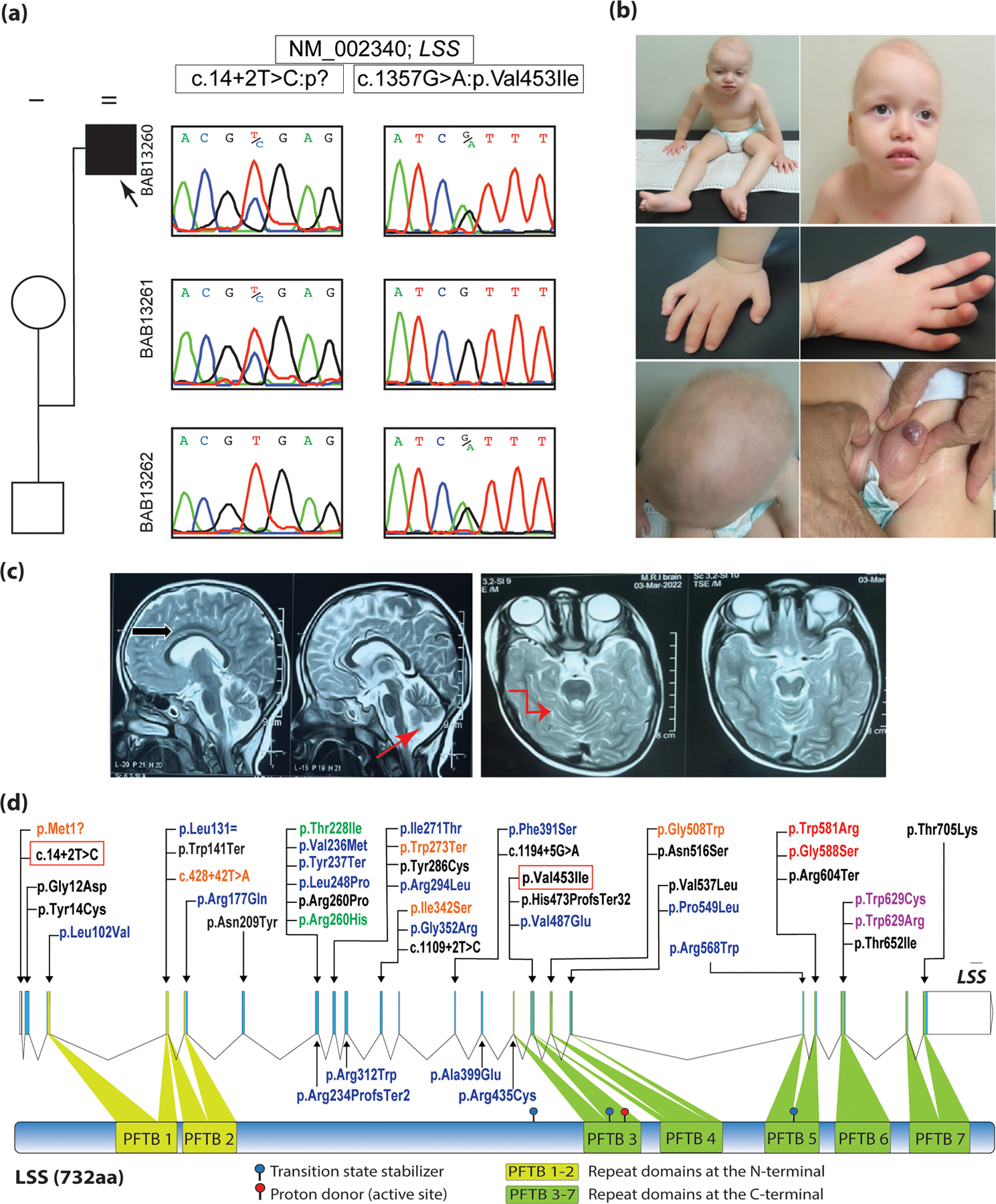
(a) Family pedigree, along with the Sanger validation and segregation next to each corresponding individual.
(b) Facial, hand, and genital images of the proband at the age of 4 years. Note sparse, lanugo-like scalp hair, sparse eyelashes and eyebrows, clinodactyly and micropenis.
(c) Brain MRI at 4ys old (T2W sagittal and axial cuts) showing thinning of the corpus collosum (black arrow), and mild superior cerebellar vermian volume loss (red arrows).
(d) Schematic representation of LSS gene (NM_002340) and of LSS protein (NP_002331); arrows point to the locations of the reported variants corresponding to each coding exon. Variants are color coded according to each phenotype sub-category as follows:
Red; Cataract 44, Purple; Cataract 44 + Hypotrichosis 14, Blue; Hypotrichosis 14, Black: Alopecia-intellectual disability syndrome (APMR), Orange; PPKCA2, and Green: PPK.
The current variants (c.14+2T>C) and (c.1357G>A:p.Val453Ile) are highlighted in red rectangles.
Brain MRI identified mildly dilated lateral ventricles, mild cortical volume loss, and cerebellar vermian hypoplasia. Other investigations such as karyotyping, metabolic work up, biotinidase enzyme activity, EMG and NCV were normal. EEG revealed generalized multifocal epileptiform activity. Abdominopelvic ultrasound showed bilateral inguinal testicles.
During his last assessment at the age of 4 years, he was able to sit at 24 months old, and walk with a wide-based gait at 40 months. Delayed cognitive function was still present and he achieved single syllables. Anthropometric measurements were 15kg for weight (−0.8SD), 94cm for height (−2 SD), and 47.5cm (−2SD) for head circumference. Alopecia was evident. Follow-up brain MRI was quite similar to the previous scans. Ophthalmological and hearing evaluations were normal.
The family was enrolled after informed consent under an Institutional Review Board (IRB)- approved research protocol (H-29697) at the Baylor College of Medicine Genomics Research to Elucidate the Genetics of Rare (BCM-GREGoR) and Medical Research Ethics Committee of NRC, Egypt. Trio research exome sequencing was performed. Two novel compound heterozygous LSS variant alleles were identified in the proband: NM_002340:c.14+2T>C and NM_002340:c.1357G>A:p.(Val453Ile), of paternal and maternal origin, respectively. The missense variant c.1357G>A:p.(Val453Ile) was predicted deleterious by multiple prediction models including Polyphen, SIFT and LRT and had a CADD score of 24.5 and a REVEL score of 0.288. The splicing variant was predicted to cause donor loss with a score of 0.57 by SpliceAI. The variants were absent in homozygous state in available control databases (ARIC; GO-ESP; 1000 Genomes Project; and gnomAD v2.1.1) or in the in-house generated BHCMG database (~13,000 exomes) while the missense variant c.1357G>A:p.(Val453Ile) was present in the heterozygous state in a single control subject of South Asian origin in gnomAD v2.1.1 (minor allele frequency of 0.000004).
DISCUSSION
Pathogenic biallelic LSS variants cause three autosomal recessive rare diseases with a broad phenotypic spectrum: congenital cataracts 44 (OMIM #616509), hypotrichosis (HYPT14; OMIM#618275), and a severe neuro-ectodermal syndrome APMR4 (OMIM #618840). Table 1 summarizes variant and trait information for published cases with the three LSS-associated traits.
Table 1:
Review of published cases identified to date
| OMIM phenotype | Family # | No. affected subjects | Clinical features | c.DNA Change (NM_002340.6) |
Amino acids change | Zygosity | Ethnicity | Reference |
|---|---|---|---|---|---|---|---|---|
| Cataract 44 | 1 | 3 | Cataract | c.1762G>A | p.Gly588Ser | Hmz | European | Zhao et al., 2015 |
| 2 | 1 | Cataract | c.1741T>C | p.Trp581Arg | Hmz | N/A | ||
|
“Intermediate”
Cataract 44 + HS 14 |
3 | 1 | Cataract, hypotrichosis | c.1025T>G;c.1887G>T | p.Ile342Ser;p.Trp629Cys | cHet | Chinese | Chen et al., 2017 |
| HS 14 | 4 | 2 | HS | c.1172T>C | p.Phe391Ser | Hmz | Arab | Romano et al. 2018 |
| 5 | 4 | HS | c.304C>G;c.743T>C c.743T>C |
p.Leu102Val;p.Leu248Pro p.Leu248Pro |
cHet Hmz |
Afghani | ||
| 6 | 3 | HS | c.1054G>A;c.1885T>A | p.Gly352Arg;p.Trp629Arg | cHet | Chinese | Li et al. 2019 | |
| 7 | 2 | HS ,midline anomalies, ACC, cleft palate | c. 530G>A; c.701_716del |
p.Arg177Gln;p.Arg234ProfsTer2 | cHet | Japanese | Wada et al., 2020 | |
| 8 | 2 | HS | c.934C>T;c.881G>T | p.Arg312Trp;p.Arg294Leu | cHet | Georgian | Cesarato et al.,2021 | |
| 9 | 1 | HS | c.1702C>T | p.Arg568Trp | Hmz | Syrian | ||
| 10 | 1 | HS | c.393G>A | p.Leu131= | Hmz | Afghani | ||
| 11 | 1 | HS | c.530G>A;c.1460T>A | p.Arg177Gln;p.Val487Glu | cHet | Japanese | Murata et al., 2021 | |
| 12 | 1 | HS | c.711C>G;c.1646C>T | p.Tyr237Ter;p.Pro549Leu | cHet | Japanese | ||
| 13 | 2 | HS | c.706G>A;c.1196C>A | p.Val236Met;p.Ala399Glu | cHet | Chinese | Hua et al., 2021 | |
| 14 | 1 | HS | c.1303C>T;c.1887G>T | p.Arg435Cys;p.Trp629Cys | cHet | Chinese | ||
| 15 | 2 | HS | c.812T>C | p.Ile271Thr | Hmz | Chinese | Zhao et al., 2023 | |
| APMR4 | 16 | 2 | HS + ID | c.625A>T;c.423G>A | p.Asn209Tyr;p.Trp141Ter | cHet | Swiss | Romano et al., 2018 |
| 17 | 2 | Alopecia with ID | c.1547A>G;c.2114C>A | p.Asn516Ser;p.Thr705Lys | cHet | N/A | Besnard et al.,2019 | |
| 18 | 2 | Alopecia with ID | c.779G>C;c.1194+5G>A | p.Arg260Pro;p.? | cHet | Turkish | ||
| 19 | 2 | Alopecia with ID | c.1109+2T>C | p.? | Hmz | N/A | ||
| 20 | 2 | Alopecia with ID | c.857A>G;c.1810C>T | p.Tyr286Cys;p.Arg604Ter | cHet | N/A | ||
| 21 | 1 | Alopecia with ID | c.41A>G;c.1417 dup | p.Tyr14Cys; p.His473ProfsTer32 |
cHet | N/A | ||
| 22 | 1 | Alopecia with ID | c.1955C>T;? | p.Thr652Ile;? | cHet | N/A | ||
| 23 | 1 | Alopecia with ID | c.35G>A | p.Gly12Asp | Hmz | Qatari | ||
| 24 | 1 | Alopecia with ID | c.530G>A | p.Arg177Gln | Hmz | Iraqi | Cesarato et al., 2021 | |
| 25 | 1 | Alopecia with ID GR and teeth mineralization defect, thin CC |
c.1609G>T | p.Val537Leu | Hmz | Egyptian | Elaraby et al., 2022 | |
| 26 | 1 | Alopecia with ID GR, ACC ,hypogenitalism |
c.14+2T>C;c.1357G>A | p?;p.Val453Ile | cHet | Egyptian | This Report 2022 | |
| PPKCA2 | 27 | 1 | Cataract, hypotrichosis , PPK | c.818G>A;c.1025T>G | p.Trp273Ter;p.Ile342Ser | cHet | Chinese | Ho et al., 2022 |
| 28 | 1 | PPK with Alopecia, Cataract, pseudoainhum, ACC | c.3G>A; c.1025T>G |
p.Met1?;p.Ile342Ser | cHet | Chinese | Yang et al., 2022 | |
| 29 | 1 | PPK with Alopecia, Cataract, pseudoainhum, ACC | c.1522G>T;c.428+42T>A | p.Gly508Trp;p.? | cHet | Chinese | ||
| PPK | 30 | 1 | PPK | c.683C > T; c.779G > A | p.Thr228Ile;p.Arg260His | cHet | Chinese | Zhou et al., 2023 |
APMR4; Alopecia-intellectual disability syndrome type 4, ACC; agenesis of corpus callosum, CC; corpus callosum. cHet; compound heterozygous, GR; growth retardation, Hmz; homozygous, ID; intellectual disability, HS; hypotrichosis simplex, PPK; palmoplantar keratoderma, PPKCA2; palmoplantar keratoderma-congenital alopecia syndrome type 2.
LSS pathogenic variants were first associated with a Mendelian trait to cause congenital cataract in rats and humans.7,16,19 In 2015, Zhao et al. reported two families of European origin with congenital cataract due to biallelic LSS variants.7 This was followed by a report of a Chinese male with an “intermediate phenotype” of co-existing cataract and hypotrichosis due to compound heterozygous missense variants in LSS.16 The first variant c.1025T>G :p.(lle342Ser) localized to the N-terminal domain while the second variant c.1887G>T: p.(Trp629Cys) to the C-terminal domain.
Romano et al. described a number of cases from different ethnicity with the second Mendelian trait, hypotrichosis simplex, due to biallelic LSS variants.5 They hypothesized that the phenotype depends on the pathogenic variants localizing toward the N-terminus lead to hypotrichosis simplex (HS) while those near the C-terminus lead to congenital cataract.5 This assumption was later questioned after a family from China with both cataract and hypotrichosis and had biallelic LSS variants localized toward the N-terminus. 13 Several families with the HS phenotype have been described with the variants spread across both domains.8–11,14,18 Then the phenotypic spectrum of LSS was expanded to include a third and more severe neuro-ectodermal phenotype, APMR4, with a report of eleven patients from seven unrelated families having alopecia and intellectual disability secondary to LSS biallelic variants.2 The described LSS variants were distributed across both LSS domains thus providing no clear phenotype-genotype correlation (Figure 1d). Two more patients from Iraq and Egypt with APMR4 have been recently reported.10,12
Our proband displayed the core clinical features of APMR4 syndrome including alopecia, developmental delay, and early onset seizures in absence of cataract, hearing loss, nail dysplasia or skin lesions. Early-onset epilepsy is a common feature in reported subjects (8/15; 53%). Additional common features observed in our subject are absent or poor speech (10/13; 77%) and hypotonia (8/15; 53%).2,10,12
Our proband had micropenis, reported in four males with APMR4 and one male with the intermediate phenotype.2,16 It is proposed to result from the impairment in the cholesterol biosynthesis pathway due to defective LSS which is essential for the development of normal genitalia in early fetal life.20 Interestingly, normal plasma cholesterol level was reported in several patients with LSS-related disorders supporting an alternative pathway for cholesterol synthesis (Table 1S).2,5,12
Similar to our subject, patients of Wada et al. had partial agenesis of the corpus callosum; however, their motor and intellectual development was normal. Also, the brain MRI of both our subject and Elaraby et al. showed dilated lateral ventricles and thin corpus callosum.12 Nevertheless, no specific neuroimaging findings were reported by Besnard et al.2 Our patient had a small cerebellar vermis with prominent vermian folia, not described with APMR4.
Lately, a more severe end-of-spectrum phenotype, palmoplantar keratoderma with congenital alopecia (PPKCA2), was reported with biallelic LSS variants, with in addition, severe hand and feet skin involvement.15 Palmoplantar keratoderma was a finding in the family described by Ho et al. and initially thought to have “the intermediate phenotype”.13 Interestingly, the variant c.1025T>C;p.(Ile342Ser) is shared between four unrelated Han Chinese subjects with PPKCA2 and “the intermediate phenotype” with another different variant: c.818G>A ;p.(Trp273Ter), c.3G>A;p.(Met1?) and c.1887G>T;p.(Trp629Cys) respectively.13,15,16 P.(Ile342Ser) variant found to significantly reduce LSS protein expression and completely abolishes its enzymatic activity resulting in no lanosterol production.13,15 The p.(Ile342Ser) variant might represent a founder allele in the Han Chinese population possibly predisposing to the intermediate or PPKCA2 phenotypes. Recently, a Chinese subject has been reported with two novel missense LSS variants and solely mutilating palmoplantar keratoderma (PPK) confirming the association with PPK but also further adding to the complexity and plethora of the LSS-related phenotypic spectrum.17
Here, we report a novel splice site variant and a novel missense variant. The majority of the reported pathogenic variants were missense. Splicing, frameshift and nonsense variants have been also reported as pathogenic in 9/15 (60%) patients with APMR4, 2/3 (67%) with PPKCA2 and only 2/22 (9%) with hypotrichosis phenotype, with another missense variant. Murata et al. thought that a combination of mutations with their locations can be important for determining the severity of phenotypes.11 Hua et al. inferred that epigenetic and other modifier genes might be responsible for phenotypic heterogeneity of LSS.14 By reviewing the reported mutations in aggregate and their associated phenotypes in an attempt to find a phenotype-genotype correlation, we found that the map positions of the individual pathogenic variant alleles were not related to the observed clinical disease. However, cases with a more severe phenotype were more likely to have a biallelic combination of a LoF allele plus a missense. Future studies should evaluate the functional impact of the variants as it is still unclear whether the different phenotypes represent different diseases or a continuum of one disorder due to allelic combinations and allelic severity at the locus.13
Our report provides novel LSS variants to be added to the list of APMR4 and highlights the clinical heterogeneity of LSS-related disorders. Moreover, we suggest that some neuroimaging clues can be relevant for an early diagnosis. We also encourage further studies of the allelic series at this locus to better understand the mechanisms and functions of the altered proteins and the neurectoderm phenotypic variability that arises from LSS mutations.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded in part by research grant from US NIH/NHGRI, NHBLI, BHCMG/ UM1HG006542, BCM-GREGoR, U01 HG011758, NINDS/R35NS105078, MDA/512848, NIH/ T32 GM007526–42, NHGRI/ K08 HG008986, NINDS/ 1K23 NS125126–01A1, NIH/T32 NS043124–19, and from Egypt by STDF-33650.
Footnotes
CONFLICT OF INTEREST:
JRL has stock ownership in 23andMe and is a consultant for Genome International.
DATA AVAILABILITY
All data are available upon request to corresponding author.
REFERENCES
- 1.Muzammal M, Ahmad S, Ali MZ, Khan MA. Alopecia-mental retardation syndrome: Molecular genetics of a rare neuro-dermal disorder. Ann Hum Genet 2021;85(5):147–154. [DOI] [PubMed] [Google Scholar]
- 2.Besnard T, Sloboda N, Goldenberg A, et al. Biallelic pathogenic variants in the lanosterol synthase gene LSS involved in the cholesterol biosynthesis cause alopecia with intellectual disability, a rare recessive neuroectodermal syndrome. Genet Med 2019;21(9):2025–2035. [DOI] [PubMed] [Google Scholar]
- 3.Bloch K The biological synthesis of cholesterol. Science 1965;150(3692):19–28. [DOI] [PubMed] [Google Scholar]
- 4.Thoma R, Schulz-Gasch T, D’Arcy B, et al. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 2004;432(7013):118–122. [DOI] [PubMed] [Google Scholar]
- 5.Romano MT, Tafazzoli A, Mattern M, et al. Bi-allelic Mutations in LSS, Encoding Lanosterol Synthase, Cause Autosomal-Recessive Hypotrichosis Simplex. Am J Hum Genet 2018;103(5):777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reza Sailani M, Jahanbani F, Nasiri J, et al. Association of AHSG with alopecia and mental retardation (APMR) syndrome. Hum Genet 2017;136(3):287–296. [DOI] [PubMed] [Google Scholar]
- 7.Zhao L, Chen XJ, Zhu J, et al. Lanosterol reverses protein aggregation in cataracts. Nature 2015;523(7562):607–611. [DOI] [PubMed] [Google Scholar]
- 8.Wada Y, Kikuchi A, Kaga A, et al. Metabolic and pathologic profiles of human LSS deficiency recapitulated in mice. PLoS Genet 2020;16(2):e1008628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Liao C, Li R, et al. A novel and a known mutation in LSS gene associated with hypotrichosis 14 in a Chinese family. J Dermatol 2019;46(11):e393–e395. [DOI] [PubMed] [Google Scholar]
- 10.Cesarato N, Wehner M, Ghughunishvili M, et al. Four hypotrichosis families with mutations in the gene LSS presenting with and without neurodevelopmental phenotypes. Am J Med Genet A 2021;185(12):3900–3904. [DOI] [PubMed] [Google Scholar]
- 11.Murata M, Hayashi R, Kawakami Y, Morizane S, Shimomura Y. Two cases of severe congenital hypotrichosis caused by compound heterozygous mutations in the LSS gene. J Dermatol 2021;48(3):392–396. [DOI] [PubMed] [Google Scholar]
- 12.Elaraby NM, Ahmed HA, Ashaat NA, et al. Expanding the Phenotypic Spectrum of APMR4 Syndrome Caused by a Novel Variant in LSS Gene and Review of Literature. J Mol Neurosci 2022;72(11):2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S, Lo I, Luk H. Expansion of Phenotype of Lanosterol Synthase-related Disease: A Case Report and Literature Review. J Paediatr (New Series) 2022(27):37–41. [Google Scholar]
- 14.Hua S, Ding Y, Zhang J, Qian Q, Li M. Novel mutations in Chinese hypotrichosis simplex patients associated with LSS gene. J Dermatol 2021;48(3):408–412. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Jiang X, Zhu Y, et al. Biallelic Variants in Lanosterol Synthase (LSS) Cause Palmoplantar Keratoderma-Congenital Alopecia Syndrome Type 2. J Invest Dermatol 2022;142(10):2687–2694.e2682. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Liu L. Congenital cataract with LSS gene mutations: a new case report. J Pediatr Endocrinol Metab 2017;30(11):1231–1235. [DOI] [PubMed] [Google Scholar]
- 17.Zhou S, Jiang X, Zhu Y, et al. Biallelic mutations in LSS in autosomal-recessive mutilating palmoplantar keratoderma. Exp Dermatol 2023. [DOI] [PubMed]
- 18.Zhao B, Tang Y, Chen W, Wan H, Yang J, Chen X. A novel homozygous mutation in LSS gene possibly causes hypotrichosis simplex in two siblings of a Tibetan family from the western Sichuan province of China. Front Physiol 2022;13:992190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori M, Li G, Abe I, et al. Lanosterol synthase mutations cause cholesterol deficiency-associated cataracts in the Shumiya cataract rat. J Clin Invest 2006;116(2):395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fakheri RJ, Javitt NB. Autoregulation of cholesterol synthesis: physiologic and pathophysiologic consequences. Steroids 2011;76(3):211–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request to corresponding author.


