Abstract
A PCR-based assay was developed to amplify a conserved region of the pneumococcal autolysin gene. The amplified product was labelled with digoxigenin-labelled dUTP and was detected with a biotin-labelled probe in an enzyme immunoassay (EIA). The assay was initially tested with suspensions of various serotypes of Streptococcus pneumoniae and other gram-positive and gram-negative bacteria and was then applied to cerebrospinal fluid (CSF) specimens from patients with meningitis and those with other neurological disorders. The assay detected all the serotypes of S. pneumoniae tested, whereas all the other bacterial strains tested were negative. Seven of the 8 CSF specimens positive for pneumococcus by culture or latex agglutination (LA) were positive by PCR-EIA, whereas all 10 specimens positive for other organisms were negative. Among 11 patients with clinically diagnosed meningitis but with negative culture and LA results, 5 were positive by PCR-EIA. The assay was negative for all but one patient without meningitis; it was positive with the CSF from a child with immunodeficiency and pneumococcal abscesses on the scalp. PCR-EIA is a useful tool for the diagnosis of meningitis, especially when culture and LA are negative because of prior antibiotic treatment.
Except during an epidemic of meningococcal infection, Streptococcus pneumoniae (pneumococcus) is the commonest cause of acute bacterial meningitis in adults and in children over 5 years of age (11). The traditional methods of diagnosing pneumococcal infections are isolation in culture and antigen detection. In many developing countries isolation of organisms in culture is often hindered by the use of antibiotics by a large proportion of patients prior to their arrival at a center where culture facilities are available. The use of bacterial antigen detection is an alternative in such situations. However, the currently available methods such as latex agglutination (LA) and counterimmunoelectrophoresis require the presence of ≥103 CFU of organisms per ml for optimal sensitivity (3, 10). In recent years, commercial DNA probes have become available for the diagnosis of a number of infectious diseases (17). However, this test also requires the presence of ≥103 organisms to give a positive result (12). Such assays would therefore be of limited value in detecting pneumococci in the majority of patients for whom cultures are negative.
Vaccines for the control of Haemophilus influenzae type b infection have substantially reduced the incidence of invasive disease due to this organism in most developed countries (1). Conjugated pneumococcal polysaccharide vaccines are now undergoing evaluation and hold much promise (5). Adequate evaluation of these vaccines in field trials will require sensitive and specific tests for the diagnosis of these infections. The use of molecular methods of diagnosis of infections such as PCR would provide a useful tool, especially in regions where antibiotics are frequently used before performing bacterial culture.
Several methods for the diagnosis of pneumococcal and H. influenzae type b infections that use gene amplification have been described in the literature (7–9, 13, 14, 18, 19, 21). These assays have used a variety of targets for amplification and have been tested with a variety of clinical specimens. However, the value of these assays in detecting pneumococcal DNA in culture-negative cerebrospinal fluid (CSF) specimens has not been adequately evaluated. We have developed a sensitive and specific PCR-based assay which uses enzyme immunoassay (EIA) for specific detection of the product and have evaluated its use in diagnosing pneumococcal meningitis in specimens negative by culture and antigen detection. The preliminary data from this study are presented.
MATERIALS AND METHODS
Bacterial strains.
The following bacterial strains were obtained from the American Type Culture Collection (ATCC; Rockville, Md.): S. pneumoniae ATCC 10341, ATCC 10373, ATCC 6314, ATCC 33400, and ATCC 6301; Streptococcus agalactiae ATCC 13813; Streptococcus equi ATCC 33398; Streptococcus mutans ATCC 25175; and Streptococcus pyogenes ATCC 12344. The following strains were obtained from the microbiology laboratory of The Johns Hopkins Hospital, Baltimore, Md.: S. pneumoniae serotypes 4, 9, 18, 19, and 23; Enterococcus faecalis; H. influenzae type b; and Staphylococcus aureus. These strains were used to produce bacterial suspensions for the development and initial evaluation of the assay.
CSF specimens.
CSF specimens were obtained from patients admitted to the Christian Medical College Hospital, Vellore, India. To test the qualitative sensitivity and specificity of the assay in diagnosing pneumococcal meningitis, we used four sets of specimens: (i) specimens that were culture or LA positive for pneumococcus (positive controls); (ii) specimens that were culture positive for other organisms (negative controls); (iii) specimens from patients without meningitis, i.e., patients with five or fewer leukocytes per mm3 of CSF (negative controls); and (iv) culture-negative CSF specimens from patients with clinical meningitis and CSF pleocytosis, i.e., more than five leukocytes per mm3. Included among the negative controls (the third category) were CSF specimens from a patient with pneumococcal pneumonia with bacteremia as well as from a child with an uncharacterized immunodeficiency syndrome who had multiple abscesses on the scalp from which S. pneumoniae was isolated; neither patients had meningitis by CSF criteria. All CSF specimens were stored at −70°C till tested.
CSF cultures and the LA test were performed in the microbiology laboratory of the Christian Medical College Hospital, which is a 1,500-bed tertiary-care hospital. The microbiology laboratory is the reference laboratory for a multicenter study on invasive bacterial infections being conducted in six centers in India. External quality assurance of the pneumococcal identification and the results of serotyping in the laboratory is validated by blind comparisons with the results obtained by the Statens Seruminstitut, Copenhagen, Denmark. The laboratory offers 24-h service. Specimens are processed immediately on receipt in the laboratory at any time of the day. Smears are prepared, fixed, and stained with Gram stain and methylene blue. Turbid CSF specimens are smeared directly, whereas clear specimens are centrifuged at 1,500 rpm (International Centrifuge; International Equipment Co., Boston, Mass.) for 15 min and the sediment is used to prepare smears. All stained smears are counterchecked by a senior faculty member of the department. For culture, the specimens are inoculated onto culture plates containing Trypticase soy agar supplemented with 5% sheep blood or chocolate agar and into thioglycolate broth, and the cultures are incubated at 37°C in an atmosphere containing 5 to 10% CO2 (4). Isolates were identified by standard microbiological techniques (20). In addition, the LA test was also performed by using previously described reagents and techniques (16).
Sample preparation and DNA extraction.
Isolates to be tested were inoculated onto Trypticase soy blood agar plates, and the plates were incubated overnight at 37°C. When pure cultures were obtained, suspensions of the organisms were made in sterile normal saline.
DNA was extracted and purified from a 200-μl volume of a bacterial suspension or a CSF specimen with a QIAamp blood kit (QIAGEN, Chatsworth, Calif.) according to the manufacturer’s instructions.
Primers and probes.
A 413-bp fragment of the autolysin (lytA) gene of S. pneumoniae (base count, 625 to 1038) was used as the target for the pneumococcal PCR. The autolysin gene has been shown to be conserved among all tested pneumococcal serotypes (12). The following oligonucleotide primers were used: Lyt A1 (5′-GTC GGC GTG CAA CCA TAT AGG CAA-3′) and Lyt A2 (5′-GGA TAA GGG TCA ACG TGG TCT GAG-3′).
The following nested primers were used to prepare a biotinylated RNA probe as described previously (2): Lyt A3-T7 (5′-TTA ATA CGA CTC ACT ATA GGT GAA GCG GAT TAT CAC TGG-3′) and Lyt A4 (5′-AGC GTT TTC GGC AAA CCT GCT T-3′) (underscores indicate the T7 promoter sequence).
A 45-mer oligonucleotide DNA probe, which was biotinylated at the 5′ end, was also synthesized for detection of the PCR product by EIA. The nucleotide sequence of the probe was as follows: 5′-biotin-TGC ATC ATG CAG GTA GGA CCT GTT GAT AAT GGT GCC TGG GAC GTT-3′.
Neither the primers nor the intervening regions displayed any significant homology to nucleotides other than those derived from S. pneumoniae, as indicated by a computer search using the program Blast 2.0 (National Library of Medicine, Washington, D.C.).
PCR.
The DNA was amplified in a total volume of 100 μl with 0.5 μM primers, 0.2 mM (each) deoxynucleoside triphosphates, 1× PCR buffer, 2.5 mM MgCl2, and 2.5 U of Taq polymerase. The mixture was incubated in a thermal cycler for 30 cycles at 94, 55, and 72°C for 1 min at each temperature.
Detection of PCR product.
In the initial assay development part of the study, the PCR product was hybridized in solution at 78°C with the biotinylated RNA probe and was detected in an EIA by previously described methods (2). Subsequently, when the assay was performed with CSF specimens in the Indian laboratory, due to the nonavailability of some of the reagents used for the PCR-EIA in the U.S. laboratory, the assay was modified as follows: (i) Digoxigenin (DIG)-labelled PCR product was obtained by using 0.19 mM dTTP and 0.01 mM DIG-UTP in the reaction mixture with the Boehringer-Mannheim dig-labelling kit (Boehringer Mannheim GmbH), and (ii) the PCR product was hybridized in solution with the biotinylated 45-mer oligonucleotide DNA probe and the hybrid was detected by an EIA with the PCR dig-detection kit (Boehringer Mannheim GmbH) according to the manufacturer’s instructions. The EIA was performed in duplicate wells. A sample was considered positive if the mean optical density (OD) value for the duplicate samples was more than 3 standard deviations above the mean value for the negative controls run in the same batch.
Processing of specimens for PCR and EIA was performed in level 2 biosafety hoods. Separate workstations were used for DNA extraction, preparation of the PCR mixture, and EIA. Every fifth specimen in each batch tested was a reagent blank. The person doing the PCR-EIA was given coded specimens and was blinded to the results of culture and LA.
The PCR assay was developed in the U.S. laboratory, where testing of the ATCC and laboratory isolates and determination of the quantitative sensitivity were performed. The assay was then transferred to the Indian laboratory where the quantitative sensitivity was redetermined by the modified technique and was found to be the same as that when the assay was used in the U.S. laboratory. All CSF specimens from patients were tested in the Indian laboratory.
RESULTS
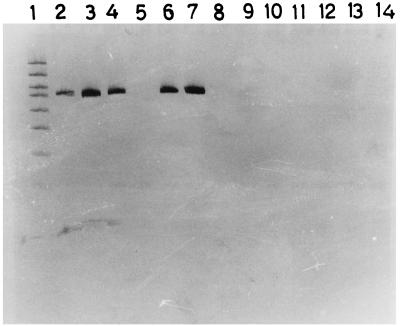
The assay detected all the serotypes of pneumococcus tested, whereas all other bacterial strains tested negative. The results of these tests are depicted in Fig. 1 and 2.
FIG. 1.
Polyacrylamide gel electrophoresis and silver staining showing gel marker (GelMarker I; Research Genetics, Huntsville, Ala.) (lane 1) and PCR products of S. pneumoniae ATCC 6301 (lane 2), S. pneumoniae ATCC 6314 (lane 3), S. pneumoniae ATCC 10341 (lane 4), S. pneumoniae ATCC 10341 (lane 6), S. pneumoniae ATCC 33400 (lane 7), S. mutans (lane 8), S. agalactiae (lane 9), S. pyogenes (lane 11), S. equi (lane 12), E. faecalis (lane 13), and reagent blanks (lanes 5, 10 and 14).
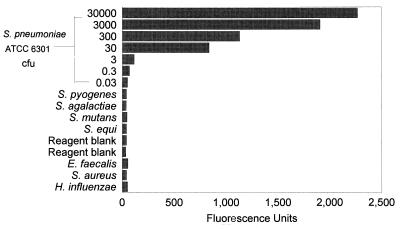
FIG. 2.
Quantitative sensitivity of the PCR-EIA showing fluorescence values by EIA produced by the PCR products of serial dilutions of S. pneumoniae ATCC 6301 DNA and of other bacteria.
To estimate the quantitative sensitivity of the assay, a suspension of S. pneumoniae ATCC 6301 was prepared. One aliquot was used for bacterial quantitation, while another aliquot was used for DNA extraction. The pneumococcal DNA content in the suspension was taken to be the DNA equivalent of the number of CFU of pneumococci in this suspension. PCR-EIA was performed with serial dilutions of this extracted DNA. The dilution of DNA which had the equivalent of 3 CFU of S. pneumoniae in the reaction mixture gave a reading which was more than 3 standard deviations above the mean value for the negative controls (Fig. 2).
The results of the PCR-EIA performed with CSF specimens are presented in Table 1. Eight specimens were positive for S. pneumoniae, including six that were positive by Gram staining, culture, and LA, one that was positive by Gram staining and culture but negative by LA, and one that was positive by Gram staining and LA but negative by culture. Seven of these eight specimens were positive by PCR-EIA; the specimen which was positive by LA and Gram staining but not by culture was also positive by PCR-EIA. Among four specimens positive for H. influenzae type b, two were positive by culture, Gram staining and LA, one was positive by culture alone, and one was positive by LA alone. All four specimens were negative for pneumococcus by PCR-EIA. Similarly, of the six specimens that were positive for other organisms, including four that were positive by Gram staining and culture and two that were positive by culture alone, none was positive for pneumococcus by PCR-EIA. Of the 11 CSF specimens from patients with meningitis for which Gram staining, culture, and LA for pneumococcus and H. influenzae type b were negative, five were positive for pneumococcus by PCR-EIA. The clinical characteristics of these five patients are presented in Table 2.
TABLE 1.
Results of PCR-EIA for detection of S. pneumoniae with CSF specimens from patients with meningitis
| CSF specimen source | No. of specimens tested | No. of specimens positive by PCR-EIA |
|---|---|---|
| Patients with meningitis | ||
| Culture or LA positive for S. pneumoniae | 8 | 7 |
| Culture or LA positive for H. influenzae | 4 | 0 |
| Culture positive for other organismsa | 6 | 0 |
| Culture and LA negative | 11 | 5 |
| Patients without meningitisb | 9 | 1 |
Other includes Proteus mirabilis (n = 1), pseudomonas (n = 1), coagulase-negative staphylococci (n = 1), alpha-hemolytic streptococci (n = 1), and micrococci (n = 2).
Febrile seizures (n = 2), congenital hydrocephalus (n = 1), neonatal sepsis without meningitis (n = 2), cytophagic panniculitis (n = 1), typhoid fever with encephalopathy (n = 1), pneumococcal pneumonia (n = 1), and pneumococcal scalp abscess (n = 1).
TABLE 2.
Selected characteristics of patients with meningitis for whom culture and LA were negative but PCR-EIA was positivea
| Patient no. | Age | Duration of illness (days) | Prior treatment | Mean OD value | OD cutoff | CSF findings | Treatment and clinical course |
|---|---|---|---|---|---|---|---|
| 1 | 5 yr | 7 | Unknown parenteral medication | 0.258 | 0.038 | WBC, 18/mm3; poly, 20%, glucose: 57 mg/dl; protein: 22 mg/dl | i.v. chloramphenicol and penicillin; recovered |
| 2 | 5 mo | 14 | Oral amoxycillin | 0.804 | 0.038 | WBC, 110/mm3; poly, 5%; glucose, 25 mg/dl; protein, 118 mg/dl | Also had pneumonia, subdural effusion, and hydrocephalus; i.v. cefotaxime; LAMA |
| 3 | 3 mo | 4 | Not known | 0.440 | 0.034 | WBC, 15/mm3; poly, 47%; glucose, 60 mg/dl; protein, 16 mg/dl | i.v. cefotaxime; recovered |
| 4 | 22 days | 3 | i.v. cefotaxime | 0.212 | 0.025 | WBC, 48/mm3; poly, 2%; glucose, 37 mg/dl; protein, 48 mg/dl | Also had pneumonia; i.v. cefotaxime; died |
| 5 | 7 yr | 5 | i.v. chloramphenicol | 0.160 | 0.032 | WBC, 120/mm3; poly, 30%; glucose, 63 mg/dl; protein, 22 mg/dl | i.v. cefotaxime; recovered |
| 0.174 | 0.027 |
Abbreviations: WBC, leukocyte count; Poly, polymorphonuclear leukocytes; i.v., intravenous; LAMA, left against medical advice.
CSF specimens from nine patients without clinical or CSF evidence of bacterial meningitis were tested. The specimens from eight patients including two neonates with sepsis in whom CSF cytology, protein, and glucose content were not suggestive of meningitis and one patient each with a bacteremic pneumococcal pneumonia, febrile seizure, congenital hydrocephalus, typhoid encephalopathy, cytophagic panniculitis, and seizure disorder were negative by PCR-EIA. CSF from a patient with an undiagnosed immunodeficiency disorder and pneumococcal abscesses on the scalp repeatedly tested positive by PCR-EIA, even though the CSF did not show pleocytosis or abnormality in the protein or glucose content.
DISCUSSION
We have adapted the PCR-EIA for amplification and detection of an autolysin gene fragment of the pneumococcus. The assay was found to have a quantitative sensitivity for the detection of DNA equivalent to the amount of DNA from three organisms. It detected all tested serotypes of pneumococcus, whereas other streptococci, H. influenzae type b, and S. aureus yielded a negative result.
The qualitative sensitivity and specificity of the assay were estimated on the basis of the results of the assay for the positive controls (specimens for tests of sensitivity) and negative controls (specimens for tests of specificity) and the ability of the assay to detect additional cases of pneumococcal infection determined by the results of the assay with the culture-negative CSF specimens from patients with meningitis. Seven of the eight specimens that were culture or LA positive for pneumococcus were positive by PCR-EIA. The isolate from the only specimen that tested negative was the only one that showed intermediate resistance to penicillin. Since tolerance to penicillin is largely but not completely mediated by amidase activity (6), we suspected that the negative test result was the result of a mutation in the gene fragment that we were trying to amplify. However, when the isolate itself was subjected to PCR with the same primers, we were able to get a strong positive reaction. We suspected that the presence of inhibitors of PCR in the CSF specimen might have been responsible for the negative reaction. However, dilutions of the CSF also gave negative results; a sufficient volume of specimen was not available for a spike-back experiment to prove this hypothesis. All 10 specimens that were culture positive for other organisms were PCR-EIA negative, as were 8 of the 9 specimens from patients without meningitis. CSF from one child with multiple pneumococcal abscesses including abscesses on the scalp gave a positive result. We do not have a good explanation for this result. Either contamination of the CSF specimen with pneumococcus from the pneumococcal skin abscesses or the presence of pneumococci in the CSF in the absence of an inflammatory response are possible explanations.
These results suggest that the assay has a reasonably high sensitivity and specificity. Therefore, a positive reaction for 5 of 11 specimens from patients with meningitis for whom culture was negative for bacteria has a high degree of significance. This suggests that pneumococcal meningitis is more prevalent in India than was previously suspected. Beta-lactam antibiotics, including injectable preparations, are widely used by general practitioners for the treatment of febrile illnesses in children. This may render the CSF culture negative and account for the failure to diagnose many cases of pneumococcal meningitis. Since our study was done with stored CSF specimens, details of prior antibiotic treatment were not available for all patients. However, we were able to document prior antibiotic use by three patients for whom culture was negative but PCR-EIA was positive; a fourth patient had received injectable medications, but the nature of these medications was not known (Table 2). Prospective studies with specimens from patients for whom this information is available are being planned.
Previous studies evaluating PCR for the detection of pneumococcal DNA have used fragments of the pneumolysin gene (15, 19), autolysin gene (8, 9, 14), or PBP 2B gene (21). These assays were used with a variety of specimens including middle-ear fluid (19), serum (15), whole blood (14, 21), CSF (13), and culture supernatant (8, 9). In two of these studies, the PCR-based assays detected pneumococcal DNA in middle-ear fluid and blood culture supernatant when routine culture did not show bacterial growth. Several of these assays used a nested PCR to obtain optimal sensitivity (13–14, 15, 19). Nested PCR carries with it the risk of amplification product carryover, leading to false-positive results, especially when large numbers of samples are tested. In all the previous assays in which specific probes were used to identify the amplified product, radiolabelled oligonucleotide probes were used. This is a particular disadvantage in developing countries, where proper disposal of radioactive materials is difficult. We have overcome the problem of using radiolabelled probes by using an EIA to detect the PCR product. The use of EIA was found to have a substantially higher sensitivity in detecting DNA amplified by reverse transcription-PCR of influenza A virus compared to that of polyacrylamide gel electrophoresis with silver staining (2). The increased sensitivity of the assay when EIA instead of gel electrophoresis is used for detection of the product may obviate the need to use nested PCR (and therefore the risk of carryover of the amplification product) and also the problems of disposal of radioactive material.
Two previous studies have evaluated PCR for the detection of pneumococcal or H. influenzae type b DNA in CSF (13, 18). In the first assay (13), a seminested PCR was used. In the seminested PCR the first amplification resulted in a general bacterial amplification and the second resulted in specific amplification of meningococcus, H. influenzae type b, or streptococci (not specifically S. pneumoniae). This assay was found to have close to 90% sensitivity. The second study evaluated the use of PCR for the detection of H. influenzae, including the presence of genes conferring ampicillin resistance, in CSF samples (18). In both studies, sufficient numbers of culture-negative samples were not tested to determine the value of the assay with such specimens. CSF has fewer cellular and other potentially inhibitory components than serum or blood and therefore may be a preferred clinical specimen for DNA detection by PCR. A third study described the use of a set of a broad range of PCR primers for the 16S rRNA gene in bacteria with three series of probes to identify potentially pathogenic bacteria in CSF or common CSF contaminants (7). However, this assay was not evaluated with actual patient specimens.
The results of this preliminary study indicate that PCR-based assays are useful adjuncts to conventional bacterial culture and antigen detection tests in establishing the bacterial etiology in meningitis in settings where substantial numbers of specimens are culture negative.
ACKNOWLEDGMENTS
This work was supported by research grants from the International Clinical Epidemiology Network and the Rockefeller Foundation.
REFERENCES
- 1.Adams W G, Deaver K A, Cochi S L, et al. Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA. 1993;269:221–226. [PubMed] [Google Scholar]
- 2.Cherian T, Bobo L, Steinhoff M C, Karron R, Yolken R H. Use of PCR-enzyme immunoassay for identification of influenza A matrix RNA in clinical samples negative for cultivable virus. J Clin Microbiol. 1994;32:623–628. doi: 10.1128/jcm.32.3.623-628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis T E, Fuller D D. Direct identification of bacterial isolates in blood cultures by using DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes B A, Granato P A. Processing specimens for bacteria. In: Murray P R, Baron E J, Pfaffer M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 265–281. [Google Scholar]
- 5.Giebink G S. Immunology: promise of new vaccines. Pediatr Infect Dis J. 1994;13:1064–1068. [PubMed] [Google Scholar]
- 6.Gray B M. Pneumococcal infections in an era of multiple antibiotic resistance. Adv Pediatr Infect Dis. 1996;11:55–99. [PubMed] [Google Scholar]
- 7.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan-King M, Baldeh I, Secka O, Falade A, Greenwood B. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J Clin Microbiol. 1994;32:1721–1724. doi: 10.1128/jcm.32.7.1721-1724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan-King M, Baldeh I, Adegbola R, Omosigho C, Usen S O, Oparaugo A, Greenwood B M. Detection of Haemophilus influenzae and Streptococcus pneumoniae DNA in blood cultures by a single PCR assay. J Clin Microbiol. 1996;34:2030–2032. doi: 10.1128/jcm.34.8.2030-2032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan S L. Rapid diagnostic techniques. In: Feigin R D, Cherry J D, editors. Textbook of pediatric infectious diseases. 3rd ed. Philadelphia, Pa: The W. B. Saunders Co.; 1992. pp. 2384–2395. [Google Scholar]
- 11.Luby J P. Infections of the central nervous system. Am J Med Sci. 1992;304:379–391. doi: 10.1097/00000441-199212000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Pozzi G, Oggioni M R, Tomasz A. DNA probe for identification of Streptococcus pneumoniae. J Clin Microbiol. 1989;27:370–372. doi: 10.1128/jcm.27.2.370-372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radstrom P A, Backman N Y, Qian N Y, Kragsbjerk P, Pahlson C, Olcen P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudolf K M, Parkinson A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salo P, Ortqvist A, Leinonen M. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J Infect Dis. 1995;171:479–482. doi: 10.1093/infdis/171.2.479. [DOI] [PubMed] [Google Scholar]
- 16.Singhal A, Lalitha M K, John T J, Thomas K, Raghupathy P, Jacob S, Steinhoff M C. Modified latex agglutination test for rapid detection of Streptococcus pneumoniae and Haemophilus influenzae in cerebrospinal fluid and direct typing of Streptococcus pneumoniae. Eur J Microbiol Infect Dis. 1996;15:472–477. doi: 10.1007/BF01691314. [DOI] [PubMed] [Google Scholar]
- 17.Tenover F C. Diagnostic deoxyribonucleic acid probes for infectious disease. Clin Microbiol Rev. 1988;1:82–101. doi: 10.1128/cmr.1.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover F C, Huang M B, Rasheed J K, Persing D H. Development of PCR assays to detect ampicillin resistance gene in cerebrospinal fluid samples containing Haemophilus influenzae. J Clin Microbiol. 1994;32:2729–2737. doi: 10.1128/jcm.32.11.2729-2737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virolainen A, Salo P, Jero J, Karma P, Eskola J, Leinonen M. Comparison of PCR assay with bacterial culture for detecting Streptococcus pneumoniae in middle ear fluid of children with acute otitis media. J Clin Microbiol. 1994;32:2667–2670. doi: 10.1128/jcm.32.11.2667-2670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Manual for national surveillance of antimicrobial resistance of S. pneumoniae and H. influenzae: epidemiology and microbiological methods. Geneva, Switzerland: World Health Organization; 1991. [Google Scholar]
- 21.Zhang Y, Isaacman D J, Wadowsky R M, Rydquist-White J, Post J C, Ehrlich G D. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]