ABSTRACT
Klebsiella pneumoniae is an important human pathogen known for its resistance to carbapenem antibiotics, especially the increasing carbapenem-resistant hypervirulent variants. The carbapenem resistance is mainly caused by the carbapenemase gene blaKPC which was commonly found on the IncFII transferable plasmids in K. pneumoniae ST11 isolates in regions of China. However, the mechanisms of the plasmid-carrying blaKPC regulation by the host strain are not clear. To investigate the chromosome-encoded two-component system (TCS) that regulates the carbapenem resistance of K. pneumoniae caused by blaKPC, twenty-four TCSs of a carbapenem-resistant classical K. pneumoniae ST11 clinical isolate were knocked out. The deletion mutation of the TCS regulator cpxR exhibited increased sensitivity to carbapenem, which could be restored by complementation with cpxR in trans. Electrophoretic mobility shift, isothermal titration calorimetry and DNase I footprinting results revealed that CpxR directly bound to the promoter DNA of blaKPC and the binding was abolished by disrupting the DNA-binding domain in CpxR. The subsequent in vivo assays using the lacZ reporter system and qPCR showed that CpxR upregulates the transcription of blaKPC. Notably, CpxR was also found to activate the transfer of the blaKPC-carrying IncFII plasmid between the hypervirulent K. pneumoniae and E. coli isolates, in which CpxR promoted the transcription of the tra operon via binding to its promoter region. These results provide an important insight into the regulation of the host factor CpxR in the plasmid-carrying carbapenemase gene in the classical and hypervirulent K. pneumoniae.
KEYWORDS: Two-component system, CpxR, carbapenem resistance gene, plasmid conjugation, Klebsiella pneumoniae
GRAPHICAL ABSTRACT
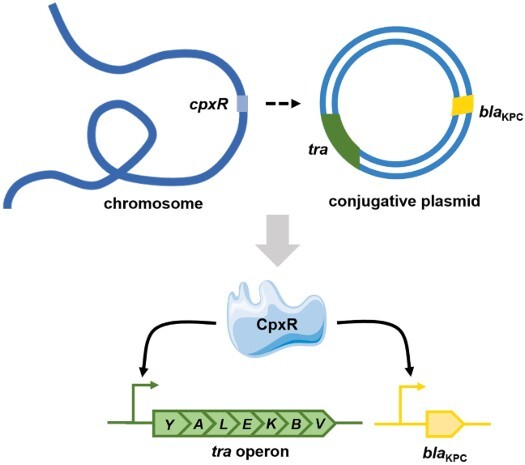
Highlights
CpxR contributes to the carbapenem antibiotics resistance of classical and hypervirulent K. pneumoniae.
CpxR binds to the promoter region of blaKPC and upregulates the blaKPC transcription.
CpxR binds to the promoter of the tra operon and activates the conjugation of the carbapenem-resistant plasmid.
Introduction
Klebsiella pneumoniae is one of the most prominent carbapenem-resistant Enterobacteriaceae (CRE). The carbapenem resistance rate of classical K. pneumoniae (cKP) increased over 8-fold from 2005 to 2018 in China [1]. Notably, the hypervirulent K. pneumoniae (hvKP) used to be sensitive to antimicrobial agents but now variants of hvKP with high degrees of carbapenem resistance have increasingly emerged [2]. The carbapenem resistance of K. pneumoniae is mainly caused by the carbapenemase gene blaKPC [3], which is much more frequently detected than other carbapenemase genes such as blaOXA-48, blaNDM-1 and blaIMP-1 [4, 5]. K. pneumoniae has the capability of acquiring new plasmids that code for antibiotic resistance determinants and/or virulence factors [6]. In the sequence type (ST) 11 carbapenem-resistant K. pneumoniae, the most prevalent K. pneumoniae strains in China, the blaKPC genes were commonly found in the IncFII transferable plasmids [7]. Since the acquisition of these resistant plasmids readily turns host bacteria resistant to carbapenems, blaKPC has been considered an autonomous gene that may be constitutively expressed [8]. However, little was known whether there exists any interaction of the blaKPC gene in the transferable plasmid with the regulation factors encoded by the host bacterial chromosome.
Bacterial two-component systems (TCSs) are important signal transduction systems that sense and respond to changes in the external environment. TCS usually consists of two components: a cell membrane protein histidine kinase (HK) and a cytosolic protein response regulator (RR). In response to extracellular changes, HK autophosphorylates at the conserved histidine residues and transfers the phosphoryl group to cognate RR. The phosphorylated RR binds to promoters and controls gene expression at the transcriptional level [9]. CpxAR is a TCS monitoring the inner-membrane homeostasis, with CpxA as HK and CpxR as RR [10]. Upon activation, CpxR strongly induces the transcription of downstream gene cpxP, whose mRNA is further processed into a small regulatory RNA CpxQ [11]. Notably, the CpxAR system has been reported to play a crucial role in antimicrobial resistance in a variety of bacterial pathogens [12–16]. Constitutive activation of CpxR in E. coli and Salmonella leads to intrinsic resistance to some but not all bactericidal drugs including aminoglycoside, hydroxyurea and β-lactam [17–19], despite the mechanisms remaining little understood. The inactivation of cpxAR in K. pneumoniae rendered bacteria more sensitive to cefepime and chloramphenicol [13]. The CpxAR system seems to regulate intrinsic resistance pathways by impacting drug transport and membrane porins [20–23]. However, it was unclear whether the chromosome-borne CpxAR directly regulates drug-resistance genes carried on the foreign, mobile plasmids.
We previously reported the carbapenem-resistant ST11 cKP clinical isolate HS11286, in which blaKPC was encoded on an IncFII-IncR conjugative plasmid called pKPHS2 [24]. The deletion of pKPHS2-borne blaKPC significantly decreased the level of meropenem resistance exhibited by the resultant mutant strains [25]. Here, we investigated the expression, regulation and interaction between the chromosome-coding TCS and the self-transferable plasmid-carrying blaKPC gene of K. pneumoniae.
Materials and methods
Strains, plasmids and primers
All strains and plasmids used in this study are listed in Table S1. All primers used in this study are listed in Table S2. All deletion mutants were generated by lambda red recombination and allelic exchange using the suicide vector pKOBEG-Apra as described in Bi et al. [25]. Polymerase chain reaction (PCR) was conducted using specific forward primer and reverse primer designed within the gene deletion region. If the DNA product was obtained from the wild-type but not from the mutant, it was considered successful construction of the mutant. The low-copy vector pXG10 with respective genes was used for complementation. All E. coli and K. pneumoniae strains were cultured in LB broth or on LB agar with appropriate antibiotics.
Antimicrobial susceptibility testing
The disc diffusion assay was performed according to the Clinical and Laboratory Standards Institute standard (CLSI, 2023). Fresh bacterial cells were made into bacterial suspensions with a McFarland turbidity of 0.5. After evenly spreading on Mueller-Hinton agar, a Meropenem Antimicrobial Susceptibility disc (Oxoid, 10 μg) was placed in the centre of the plate. After incubating at 37°C for 16 h, the diameter of the zone of inhibition was measured.
For the agar screening, a spot assay was used to determine bacterial sensitivity to meropenem. In brief, the bacterial suspensions with an OD600 nm value of 0.5 were diluted by a 10-fold gradient. Then 5 μl of all suspensions were carried out on LB agar with or without meropenem respectively. The concentration of meropenem used for K. pneumoniae HS11286 and its mutants was 2 μg/ml. And 1 μg/ml was used for K. pneumoniae 293Z and its mutants, 0.25 μg/ml was used for E. coli C600 and its mutants.
The minimal inhibitory concentrations (MICs) were determined using the broth microdilution method according to the CLSI standard (2023). The tested strains were inoculated at a final concentration of 0.5 McFarland standard into CAMHB (Cation-Adjusted Mueller-Hinton Broth) medium supplemented with meropenem at concentrations of 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.06, and 0.03 μg/ml. When determining the MIC of Meropenem-vaborbactam (MEV), a final concentration of 8 µg/ml vaborbactam was added to the mixture of the aforementioned bacterial cultures and meropenem. The cultures were incubated at 37°C under ambient air for 16 h, and bacterial growth was observed to determine the MIC. E. coli ATCC 29522 was used as the quality control strain.
Expression and purification of proteins
The wild-type cpxR gene and its mutation variants genes were cloned into vector pET28a(+) using primers listed in Table S2, and transferred into E. coli BL21(DE3). Cells containing plasmids were grown at 37°C with shaking. When an OD600 nm of 0.5 was reached, protein expression was induced overnight at 16°C with 0.2 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Subsequently, proteins were purified from cell lysis supernatant with cobalt metal affinity resin (Takara). Contaminating nucleic acid was removed by HiTrap Heparin HP (GE Healthcare) or HiTrap Capto Q (GE Healthcare).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were carried out by mixing wild-type or mutated CpxR at a range of concentrations with EMSA/Gel-Shift Binding Buffer (poly(dI-dC), DTT, Glycerol, EDTA, NaCl, MgCl2, Tris) (Beyotime) at room temperature for 10 min. Then, the 2 pmol promoter DNA probes with a 6-FAM modification at the 5-end were added and incubated for 20 min. The mixtures were then subjected to electrophoresis in native 6% polyacrylamide gel at 100 V for 60 min.
DNase I footprinting assay
DNase I footprinting assays were performed according to the method described by Wang et al. [26]. Four hundred ng promoter DNA probes with a 6-FAM modification at the 5-end were incubated with or without CpxR in a total volume of 40 µl in binding buffer (50 mM Tris-HCl pH 8.0, 100 mM KCl, 2.5 mM MgCl2, 0.2. mM DTT, 2 μg salmon sperm DNA and 10% glycerol) for 30 min at 25°C. Then 10 µl solution containing about 0.015 unit DNase I (Promega) and 100 nmol freshly prepared CaCl2 was added and further incubation was performed at 37°C for 1 min. The reaction was stopped by adding 140 µl stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA and 0.15% SDS). Samples were extracted with phenol/chloroform and precipitated with ethanol. Pellets were dissolved in 30 µl MiliQ water. The preparation of the DNA ladder, electrophoresis and data analysis were the same as described before, except that the GeneScan-LIZ600 size standard (Applied Biosystems) was used.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were performed on a TA NANO system. Genomic DNA was extracted from K. pneumoniae HS11286 and used as a template for PCR amplification of the promoter DNA fragment of blaKPC, using Pkpc403-F/R primers (listed in Table S2). The purified CpxR protein (0.1 μM) and the promoter DNA of blaKPC (0.01 μM) were dissolved in HEPES (4-(2-hydroxyethyl)−1-piperazineethanesulfonic acid) (pH 7.4). Sixty microliters purified CpxR protein was injected into the sample cell filled with 300 μl promoter DNA of blaKPC, and the injection was repeated 20 times with an equilibrium interval of 200 s. The experiment was conducted at 25°C. The equilibrium dissociation constant KD was obtained using the nanoAnalyzer software. 0.4 μM the CpxR-R195H protein and 0.04 μM promoter DNA of blaKPC were used.
Real-time quantitative PCR (qRT-PCR)
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). Then gDNA was removed and cDNA was produced by PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara). Finally, qPCR was performed using Hieff UNICON® qPCR SYBR Green Master Mix (YEASEN) and gapA was used as an internal control. The gene expression level was calculated by the 2-ΔΔCT method.
β-galactosidase activity assay
The β-galactosidase activity was quantified based on the hydrolysis of O-Nitrobenzene-β-D-galactopyranoside (ONPG). Bacteria were subcultured in LB broth to logarithmic phase (OD600 nm of 0.4 ∼ 1.0), then cells were pelleted and resuspended in an equal amount of Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol). One millilitre of cells (OD600 nm determined after blank against the Z buffer) were permeabilized by adding 100 μl chloroform and 50 μl 0.1% SDS and was equilibrated for 5 min in a 28°C water bath. The reaction was started by adding 0.2 ml O-nitropheny1-β-D-galactoside (ONPG, 4 mg/ml) at 28°C and was stopped after the yellow colour has developed by adding 0.5 ml 1 M Na2CO3. The time of reaction (T) was recorded precisely with a timer. Then the mixture was spined 5 min at maximum to remove debris and chloroform. OD420 nm and OD550 nm of supernatant were measured. Units of activity were calculated with the formula as follows. 1)
Conjugation assay
Bacteria were subcultured in LB broth to the logarithmic phase. Two millilitres of donor cells and recipient cells were washed with 10 mM MgSO4 for 3 times, resuspended in 40 μl of 10 mM MgSO4, mixed and then 20 μl was inoculated on LB agar for 12 h at 37°C. The mixtures of donors, recipients and transconjugants cells were resuspended in Phosphate Buffered Saline (PBS) and serially diluted. The serial dilutions were plated on LB agar with appropriate antibiotic (listed in Table S3) and the CFU of the transconjugants and donors cells were counted. The transconjugants were further validated by PCR. The gene blaKPC was located on pKPHS2 and used as a marker for the presence of plasmid. The gene khe exists only in K. pneumoniae but not in E. coli [27], which can distinguish between donors and recipients. The conjugation frequency was calculated as the ratio of transconjugants to donors.
Statistical analysis
The data of promoter activity and qRT-PCR were derived from three independent assays. Student’s t-test (unpaired, equal variance, two-sided) was performed using R package (https://www.r-project.org/). Statistical significance was considered when p ≤ 0.05. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001.
Results
CpxR contributes to the resistance of K. pneumoniae to carbapenem antibiotics
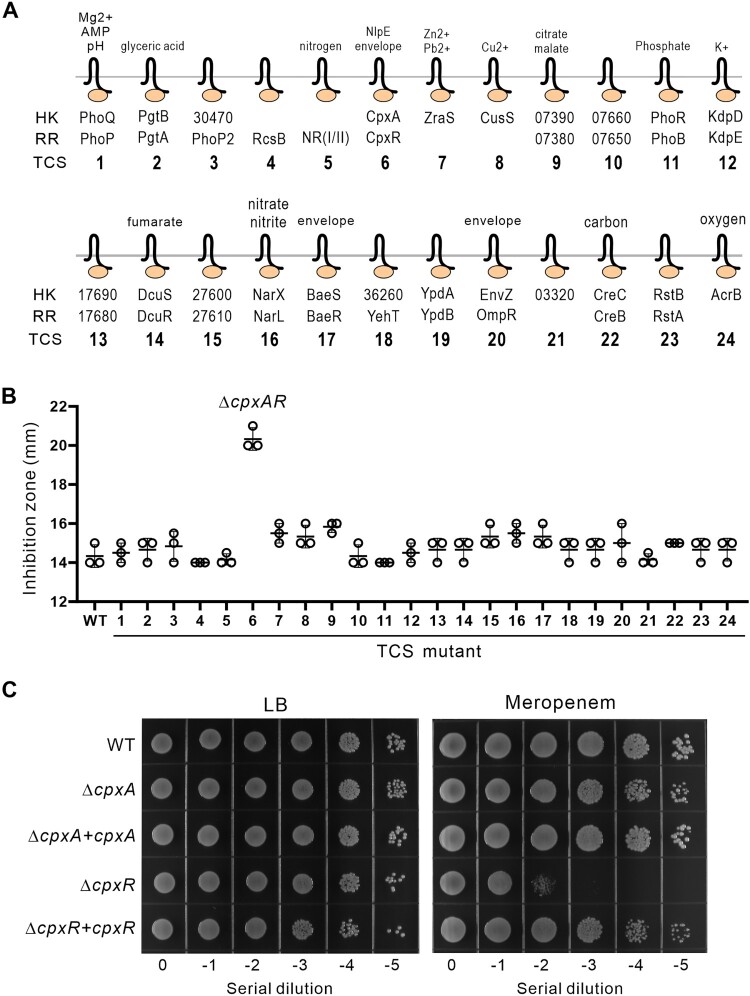
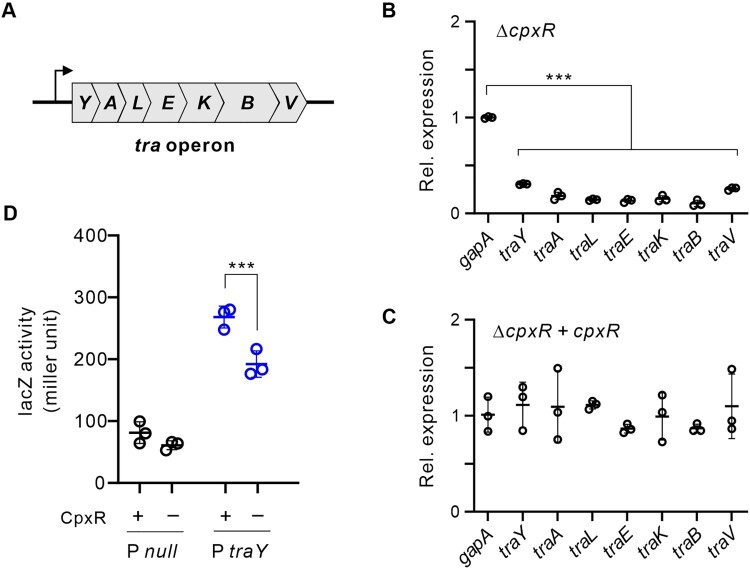
To better understand the involvement of TCS in antibiotic resistance, we first predicted all the potential TCSs encoded in the chromosome of ST11 cKP HS11286 using the Prokaryotic 2-Component Systems database (P2CS) [28]. Of all the TCSs that have been predicted and annotated (Table S4), we have successfully constructed mutants for 24 TCSs, each harbouring a chromosomal deletion of a TCS (Figure 1A, Figure S1). Using disk diffusion test, we discovered that the ΔTCS6 strain (K. pneumoniae HS11286 harbouring a deletion of cpxAR) showed an increased sensitivity to meropenem (a kind of carbapenem antibiotics) compared to the wild type (Figure 1B). According to the CLSI 2023, the ΔTCS6 strain exhibited intermediate resistance to meropenem, while the wild-type and other mutants were resistant. Further dissecting the responsible gene in the cpxAR bicistronic operon (Figure S2) confirmed the direct involvement of the cpxR gene, which encodes the response transcriptional regulator CpxR. A clean mutant of cpxR (ΔcpxR) showed impaired resistance to meropenem, which could be fully restored by complementation with a wild-type cpxR gene in trans (Figure 1C, Table S5).
Figure 1.
Identification of a two-component system that is required for resistance to β-lactam antibiotics in K. pneumoniae. (A) Twenty-four TCSs were individually deleted on the chromosome of K. pneumoniae HS11286. Abbreviations: HK, histidine kinase; RR, response regulator; TCS, two-component system; AMP, antimicrobial peptides. (B) Diameter of inhibition zone of 24 TCS deletion mutants to meropenem. (C) Growth of bacterial serial dilutions in the presence or absence of meropenem.
CpxR directly binds to the promoter region of blaKPC
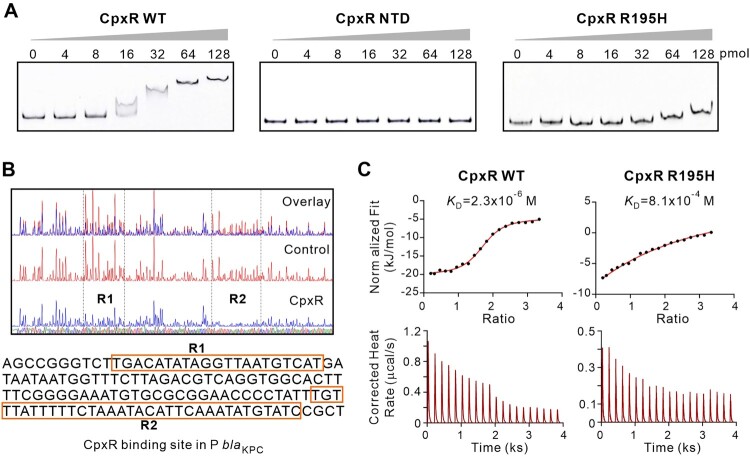
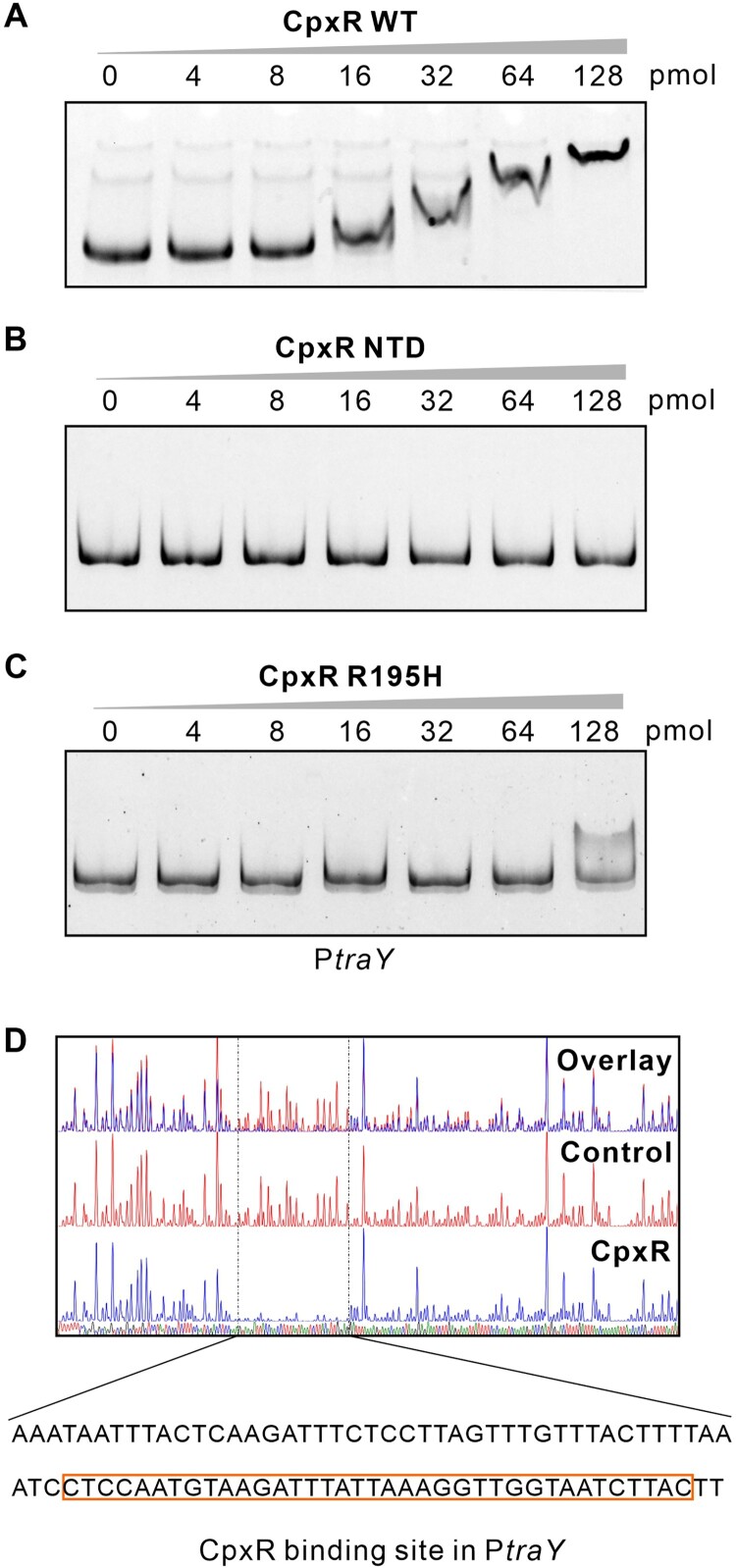
Meropenem resistance in K. pneumoniae is mainly due to the expression of β-lactamase K. pneumoniae carbapenemase (KPC, encoded by blaKPC gene), which efficiently hydrolyzes carbapenem-like drugs and mediates resistance to β-lactam antibiotics. Using vaborbactam [29], an inhibitor of β-lactamase KPC, carbapenem-resistant K. pneumoniae becomes susceptible to carbapenem (Table S5), supporting the alteration of carbapenemase as the cause for the change of meropenem resistances. We hypothesized that the impaired meropenem resistance in ΔcpxR may be due to transcriptional regulation of blaKPC by CpxR, a member of the OmpR/PhoB transcriptional regulator family with a C-terminal DNA-binding domain (DBD) (Figure S3A). Using electrophoretic mobility shift assay (EMSA), we indeed found that CpxR directly binds to the promoter region of blaKPC with high affinity (Figure 2A). As expected, this binding was reduced by disrupting the DNA-binding domain in CpxR (Figure 2A), either by removing the C-terminal domain (CTD) or by mutating the crucial Arg195 residue to a histidine [30]. The Isothermal titration calorimetry analysis, which measures the binding affinity between CpxR and the PblaKPC sequence, showed that the R195H mutation significantly reduced the DNA binding affinity of the CpxR by ∼100 fold (Figure 2C). We have further determined the CpxR-binding sites using DNase I footprinting assay and revealed two regions in the blaKPC promoter that were protected by CpxR (Figure 2B), i.e. 5’-TGACATATAGGTTAATGTCAT-3’ and 5’-TGTTTATTTTTCTAAATACATTCAAATATGTATC-3’. The sequence alignment of the obtained CpxR binding sites against the completely sequenced K. pneumoniae genomes available in the NCBI RefSeq database (Figure S4) revealed that the CpxR binding site sequences are present in 239 out of 416 (57%) of the promoter regions of blaKPC genes (Figures S5 and S6). Notably, this binding was not only observed for the blaKPC promoter region in K. pneumoniae Tn1721 of ST11, the dominant KPC-producing cKP clone in China, but also observed by EMSA for the promoter from Tn4401b of ST258, the dominant carbapenem-resistant K. pneumoniae clone in Europe and the United States (Figure S7). However, the CpxR binding sites to the blaKPC promoter region in Tn4401b were different from those of Tn1721 and require further investigation (Figures S5 and S6).
Figure 2.
CpxR recognizes and binds to the promoter region of blaKPC. (A) Electrophoretic mobility shift assay (EMSA) using increasing amounts of wild-type CpxR, CpxRNTD (CpxR missing C-terminal domain), and CpxRR195H with 2 pmol of FAM-labeled promoter of blaKPC. (Negative Controls, Figure S8) (B) DNase I footprinting assay. The FAM-labeled promoter region of blaKPC was incubated with or without CpxR protein, then digested by DNase I. Electropherograms indicated the sequence of protection region. The peaks at R1 and R2 were significantly decreased in the presence of CpxR, indicating direct binding sites of CpxR. (C) Isothermal titration calorimetry assay of the blaKPC promoter region with CpxR and CpxRR195H. The genetic organizations of CpxR, CpxRNTD, and CpxRR195H were depicted in Figure S3.
CpxR upregulates the transcription of blaKPC and carbapenem resistance
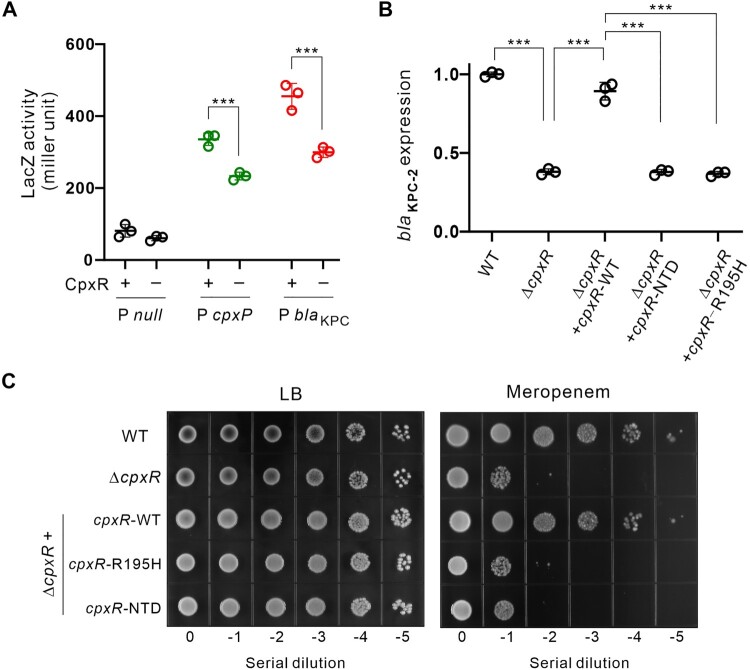
Having established a direct binding of CpxR to the blaKPC promoter, we next thought to analyze the impact of CpxR on blaKPC transcription using a lacZ transcriptional reporter system. The 403 bp region of the blaKPC promoter was cloned and fused to a promoterless lacZ gene, and the β-galactosidase activity was examined. As shown in Figure 3A, the transcription of blaKPC-lacZ was significantly reduced when cpxR was absent. A similar reduction was also observed for the PcpxP promoter, a known CpxR-dependent promoter as a positive control [11]. Further analysis at the mRNA level showed consistent results as above (Figure 3B). The expression of blaKPC mRNA was only about one-third of the wild-type (−2.631 fold, p < 0.001) when cpxR was deleted. And the expression of KPC was decreased by half when cpxR was deleted (Figure S9). This reduction was restored by a trans-complementation of the wild-type cpxR gene, but not by the CpxR mutants without DNA-binding capacity (NTD, R195H). As expected, these two CpxR mutants also failed to restore the carbapenem resistance phenotype, whereas the WT CpxR did (Figure 3C, Table S5). Together, these results confirm that CpxR upregulates the transcription of carbapenem-hydrolyzing class A β-lactamase KPC from the large conjugative plasmid pKPHS2 and promotes carbapenem resistance in cKP.
Figure 3.
CpxR promotes the transcription of blaKPC. (A) The activity of lacZ reporter fused to respective promoter was determined in the presence or absence of cpxR. The promoter of cpxP was used as a positive control [11]. The null promoter was used as a negative control. This experiment was performed in a heterologous system using E. coli DH5ɑΔcpxR as a surrogate host, in which the K. pneumoniae cpxR gene was provided on plasmid. (B) The expression level of blaKPC in the wild-type K. pneumoniae, ΔcpxR mutant, complementation of ΔcpxR with a wild-type cpxR, complementation of ΔcpxR with cpxRNTD (CpxR N-terminal domain), complementation of ΔcpxR with cpxRR195H. (C) Bacterial growth in the presence or absence of meropenem using 10-fold serial dilutions.
CpxR activates the transcription of tra operon and plasmid conjugation
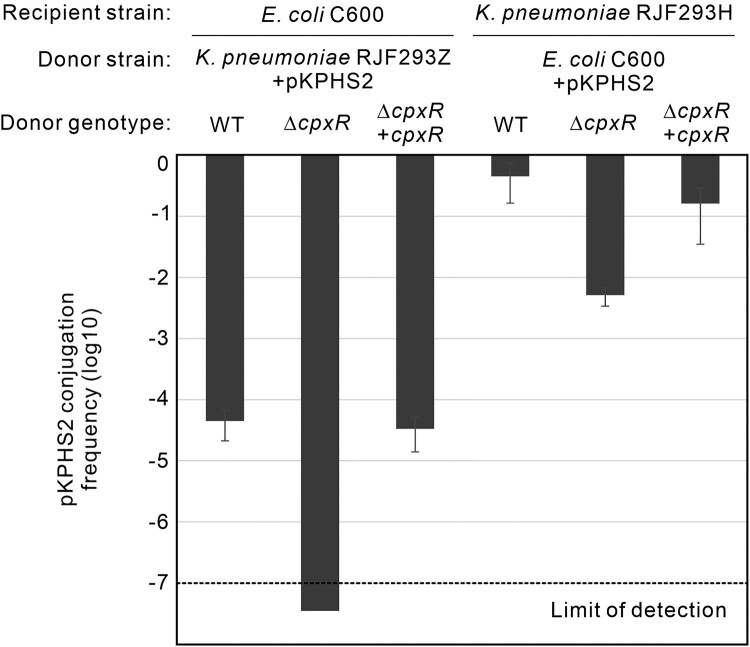
Initially identified as a major regulator of the conjugative plasmid expression (cpx), CpxR has been established to inhibit the conjugation of F plasmid in E. coli [31, 32]. We wondered whether CpxR regulates the conjugation of the blaKPC-carrying IncFII self-transferable plasmid between the cKP, hvKP and E. coli isolates. First, we transferred pKPHS2, the blaKPC-carrying IncFII plasmid of cKP HS11286, to RJF293Z as the donor to test the conjugation frequency of pKPHS2 with E. coli C600 as the recipient (Figure S12A). Strikingly, the conjugation frequency of pKPHS2 was strongly reduced to below the limit of detection in the ΔcpxR donor strain of K. pneumoniae (Figure 4). Then, in the second round of conjugation, E. coli C600 + pKPHS2 was employed as the donor, and the hvKP strain RJF293H was used as the recipient (Figure S12B). Again, we observed a strong reduction (∼100-fold) in conjugation efficiency with the ΔcpxR mutant compared to the WT donor (Figure 4), supporting that CpxR is necessary for the efficient transfer of the IncFII blaKPC-carrying pKPHS2 plasmid by conjugation.
Figure 4.
CpxR is required for the efficient transfer of the IncFII blaKPC-carrying pKPHS2 plasmid. Conjugation frequency of the pKPHS2 was determined using K. pneumoniae RJF293Z strains as donor and E. coli C600 as recipient; or using E. coli C600 strains as donor and RJF293H as recipient. (Verification of the pKPHS2 conjugation between E. coli C600 and K. pneumoniae RJF293, Figure S13)
The pKPHS2 plasmid of K. pneumoniae HS11286 encodes the tra operon required for conjugation (Figure 5A). This operon expresses a long polycistronic mRNA starting with traY as the first gene (Figure S10). To understand how CpxR regulates the conjugation of pKPHS2, we analyzed the expression of tra genes using qRT-PCR. The results showed that the expression of all seven tra genes was significantly down-regulated when cpxR was deleted (Figure 5B), and was again restored to the WT level in the CpxR complementation strain (Figure 5C). Using the traY-lacZ transcriptional reporter assay, we have confirmed that CpxR acts at the transcriptional level to promote the expression of tra operon (Figure 5D). To further establish a direct binding of CpxR to the traY promoter, we performed EMSA assay with the traY promoter DNA fragment. The results showed that the WT CpxR protein directly binds to the traY promoter (Figure 6A), whereas the CpxR mutants did now show any productive interaction (Figure 6BC). The CpxR binding site in the traY promoter has been determined using a DNase I protection assay (Figure 6D), i.e. 5’-CTCCAATGTAAGATTTATTAAAGGTTGGTAATCTTAC-3’. Altogether, our results showed that CpxR promotes conjugation by transcriptional activation of the tra operon on pKPHS2 in cKP.
Figure 5.
CpxR promotes the transcription of tra operon on the IncFII blaKPC-carrying plasmid pKPHS2. (A) Genetic organization of the tra operon. The genes traY, traA, traL, traE, traK, traB and traV form a polycistron that is transcribed from a promoter upstream (bended arrow). (B) The expression of tra genes in ΔcpxR mutant, using gapA as a control. (C) The expression level of tra genes in ΔcpxR complemented with a wild-type cpxR. (D) The beta-galactosidase activity of lacZ fused to the traY promoter was determined in the presence or absence of cpxR. The null promoter was used as a negative control. This experiment was performed in a heterologous system using E. coli DH5ɑΔcpxR as a surrogate host, in which the K. pneumoniae cpxR gene was provided on plasmid.
Figure 6.
CpxR binds to the traY promoter. (A-C) EMSA using 2 pmol FAM-labeled traY promoter with increasing amount of wild-type CpxR (A), CpxRNTD (CpxR missing C-terminal domain) (B), or CpxRR195H (C) (Negative Control, Figure S11). (D) DNase I footprinting assay. The FAM-labeled traY promoter was incubated with or without CpxR protein, then digested by DNase I. Electropherograms indicated the CpxR protection region. The peaks in the dashed box were significantly decreased, indicating the binding site of CpxR. The organizations of CpxR, CpxRNTD, CpxRR195H were depicted in Figure S3.
CpxR is the host factor promoting carbapenem resistance in the recipient strain
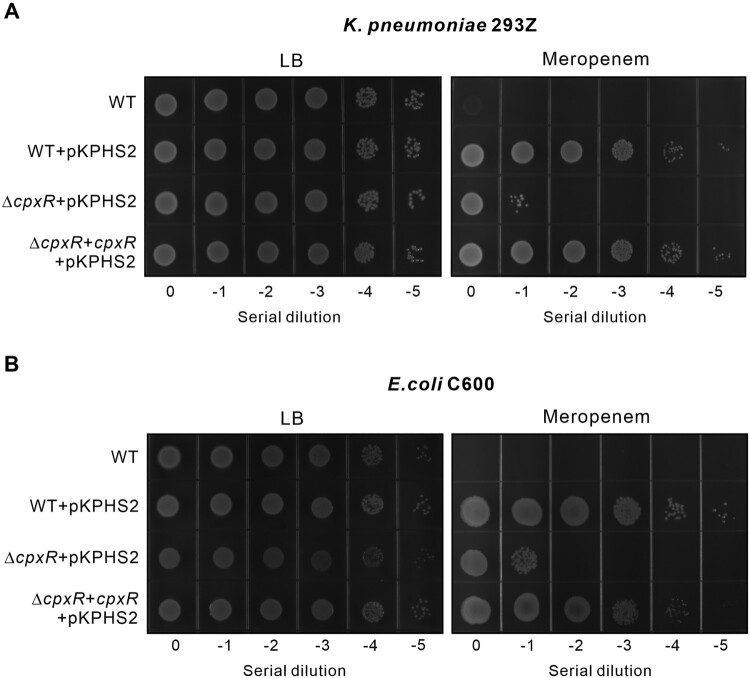
The transfer of blaKPC-carrying plasmids is a major reason for the spread of antibiotic resistance among Gram-negative bacteria, most of which encode a conserved cpxR gene on the chromosomes. To determine the contribution of CpxR in the recipient bacteria to meropenem resistance, we transferred the pKPHS2 plasmid into the hvKP strain 293Z and E. coli and analysed their resistance to meropenem (Figure 7, Table S5). Both hvKP and E. coli gained full resistance to meropenem after receiving the pKPHS2 plasmid. The resistance level was reduced by 4 logs when cpxR was deleted in the recipient strains, and it was restored by the complementation of cpxR in trans. Therefore, our results showed that CpxR is a conserved host factor crucial for carbapenem resistance mediated by the plasmid-borne blaKPC gene.
Figure 7.
CpxR regulates meropenem resistance in recipient strains containing an acquired blaKPC gene. (A) Growth of K. pneumoniae 293Z, 293Z + pKPHS2, 293ZΔcpxR + pKPHS2, 293ZΔcpxR + cpxR + pKPHS2 in the presence or absence of meropenem. (B) Growth of E. coli C600, C600 + pKPHS2, C600ΔcpxR + pKPHS2, C600ΔcpxR + cpxR + pKPHS2 in the presence or absence of meropenem.
Discussion
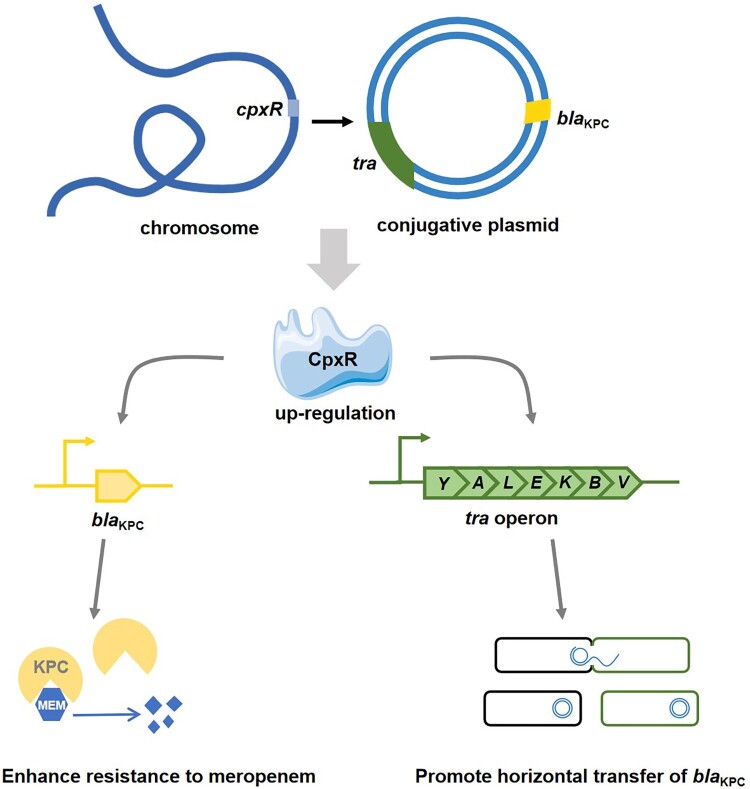
Carbapenem antibiotics are one of the most important antibacterial drugs for the treatment of serious bacterial infections. Their effectiveness is compromised by the recent emergence of CRE in clinics, especially the problematic carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) [33]. The carbapenem resistance is acquired commonly due to a conjugative plasmid carrying the blaKPC gene, but the intrinsic resistance mechanism in K. pneumoniae has been little understood. In this study, we discovered that the chromosome-encoded TCS regulator CpxR directly regulates the expression and the mobilization of blaKPC gene on the IncFII conjugative plasmid, playing a crucial role in the growing problem of carbapenem resistance in the classical and hypervirulent K. pneumoniae strains (Figure 8).
Figure 8.
Proposed model for CpxR-mediated transcriptional regulation of blaKPC and tra operon on the conjugative resistance plasmid.
By performing an unbiased screen of twenty-four TCS knockout mutants, we discovered that CpxAR is the sole system in Klebsiella contributing to carbapenem resistance. CpxR, known as a key regulator of envelope stress, has been linked to the resistance of multiple classes of antibiotics in gut bacteria, including aminoglycosides [12, 34] beta-lactams [21], Fosfomycin [12] and cationic antimicrobial peptides [35]. To our knowledge, this is the first report that CpxR influences resistance to carbapenems, one of the most effective antibiotics in clinical treatments. In many previous studies, the contribution of CpxR was studied using artificially engineered variants of CpxR that are constitutively activated [18], casting some doubts on the true involvement of CpxR in antibiotic resistance. Here in this study, a clean deletion of cpxR gene from its native chromosomal locus exhibits a clear deficit in meropenem resistance, which was fully rescued by CpxR provided in trans. Our results unequivocally demonstrated that the endogenous CpxR plays an indispensable role in antibiotic resistance in bacteria.
CpxR promotes carbapenem resistance by a direct regulation of resistance genes blaKPC in Klebsiella. Our data showed that CpxR binds to the promoter region of blaKPC (Figures 2 and 3), and activates the transcription of blaKPC in the IncFII plasmid of K. pneumoniae ST11 (Figure 3). It was previously reported that CpxR mainly acts on the envelope permeability to limit intracellular antibiotic concentrations, either by activating efflux pumps [21–23], or regulating major membrane porins [20, 21]. The direct regulation of blaKPC gene provides a new mechanism for how CpxR influences resistance to antibiotics. Such direct regulations have been documented for other TCS in different bacteria. For example, the VbrKR system in V. parahaemolyticus was shown to activate the expression of class A β-lactamase (blaAVP, vpa0477) to confer ampicillin resistance [36].
Different from the chromosomally encoded blaAVP, the blaKPC gene is located on a mobile genetic element (i.e. the IncFII conjugative plasmid) that may circulate in numerous enteric bacterial species. It was anticipated that the regulation of mobile blaKPC would be conserved in different bacterial hosts in which CpxR is conserved [11]. Investigating this hypothesis, we have proven that 1) the blaKPC can be transferred to new bacterial species via conjugation, 2) the newly acquired blaKPC gene is regulated by the host CpxR in the recipient bacteria (Figure 7, Table S5). Strikingly, we found that CpxR facilitates the transmission of pKPHS2 between cKP, hvKP and E. coli (Figure 4), promoting the dissemination of blaKPC genes via horizontal transfer. The IncFII conjugative plasmid pKPHS2 encodes an entire F-type Type IV secretion system (T4SS), which makes the blaKPC highly susceptible to strain-to-strain transmission by conjugation. CpxR activates the expression of tra genes that are required for conjugative transfer. CpxR directly binds to the promoter of the tra operon (PtraY), upregulates the transcription of tra genes, and facilitates the mobilization of pKPHS2 and the plasmid-encoded blaKPC (Figures 4–6). When pKPHS2 is transferred to other Enterobacterales, CpxR acts as the key host factor promoting carbapenem resistance in recipient bacteria (Figure 7, Table S5). CpxAR is a highly conserved TCS in the core genomes in Enterobacterales. Our analysis of 1280 K. pneumoniae complete genome sequences currently available on GenBank confirmed the presence of intact cpxAR genes (Figure S14). Thus, the cross-regulation of mobile blaKPC by host CpxR may likely represent a ubiquitous regulatory mechanism, which contributes to the ongoing emergence of the carbapenem antibiotics resistance of classical and hypervirulent K. pneumoniae.
CpxR regulates transcription by binding directly to the promoter region of target genes. The canonical binding sites were thought to resemble a sequence motif GTAAA-(N)4-8-GTAAA [11, 37–39]. However, in recent years, an increasing number of different CpxR motifs have been identified, such as GTATT-N5-GAAAA [35], GAAAT-N5-GTAAA [35], TTAAA-N6-GTAAA [40], TTGAC-N5-CTTGC [14] and TGAAA-N3-TTTAT [16] among others (Table S5). In this study, three CpxR binding sites were identified by DNaseI footprinting, each containing the new motifs TTAAT-N6-ATAAT, AAATA-N5-AAATA, and TTAAA-N5-GTAAT, respectively. These results expand our understanding of the CpxR binding sites and various genes directly regulated by CpxR.
Summary
By screening twenty-four putative TCS encoded on the chromosome of K. pneumoniae HS11286, we have discovered that the CpxAR TCS is required in the carbapenem resistance. CpxR not only upregulated the expression of the plasmid-carrying gene blaKPC but also promoted the conjugation of blaKPC-carrying plasmid pKPHS2 between the cKP, hvKP and E. coli strains. As a highly conserved regulatory system in many bacteria, the CpxR may contribute to the transmission of carbapenem antibiotics resistance among Enterobacteriaceae.
Supplementary Material
Acknowledgements
We thank Dr. Yingzhou Xie from Shanghai Pulmonary Hospital for help with experiments design and Dr. Heng Li from Soochow University for help with experiments operation.
Funding Statement
This work was supported by the National Key Research and Development Program of China [grant no 2018YFE0102400, 2022YFE0111800, 2022YFC2303200], the Natural Science Foundation of China [grant no 32070572, 32270064], the Science and Technology Commission of Shanghai Municipality [grant no 21ZR1471300, 2019SHZDZX02], the Open Funding Project of State Key Laboratory of Microbial Metabolism [grant no MmlKF22-06], the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University [grant no XK2019025], and the Chinese Academy of Sciences [grant no 176002GJHZ2022022MI].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Zhiyuan Liu designed the study, performed experiments and revised the manuscript. Jiahao Guan analyzed and processed the data. Zhaoyan Chen participated in experiment performance. Cui Tai and Zixin Deng revised the manuscript. Yanjie Chao, Hong-Yu Ou and Zhiyuan Liu drafted the manuscript. Yanjie Chao and Hong-Yu Ou designed the study, supervised the project and revised the manuscript. All authors read and approved the final manuscript.
References
- 1.Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019 Dec;38(12):2275–2281. doi: 10.1007/s10096-019-03673-1 [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020 Feb 1;75(2):327–336. doi: 10.1093/jac/dkz446 [DOI] [PubMed] [Google Scholar]
- 3.Logan LK, Weinstein RA.. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017 Feb 15;215(suppl_1):s28–s36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng X, Yang J, Duan J, et al. Assessing molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in central China. Sci Rep. 2019 Feb 19;9(1):2271, doi: 10.1038/s41598-018-38295-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu MC, Tang HL, Chiou CS, et al. Clonal dissemination of carbapenemase-producing Klebsiella pneumoniae: two distinct sub-lineages of sequence type 11 carrying blaKPC-2 and blaOXA-48. Int J Antimicrob Agents. 2018 Nov;52(5):658–662. doi: 10.1016/j.ijantimicag.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Zhang J, Wang M, et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021 Jul 22;13(1):119, doi: 10.1186/s13073-021-00936-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Goh YX, Tai C, et al. Vrprofil(e2). detection of antibiotic resistance-associated mobilome in bacterial pathogens. Nucleic Acid Res. 2022 May 7;50(W1):W768–W773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian D, Liu X, Chen W, et al. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect. 2022 Dec;11(1):1936–1949. doi: 10.1080/22221751.2022.2103454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stock AM, Robinson VL, Goudreau PN.. Two-component signal transduction. Annu Rev Biochem. 2000 Jul;69:183–215. doi: 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 10.Mitchell AM, Silhavy TJ.. Envelope stress responses: balancing damage repair and toxicity. Nat Rev Microbiol. 2019 Jul;17(7):417–428. doi: 10.1038/s41579-019-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao Y, Vogel J.. A 3’ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell. 2016 Feb 4;61(3):352–363. doi: 10.1016/j.molcel.2015.12.023 [DOI] [PubMed] [Google Scholar]
- 12.Kurabayashi K, Hirakawa Y, Tanimoto K, et al. Role of the CpxAR two-component signal transduction system in control of fosfomycin resistance and carbon substrate uptake. J Bacteriol. 2014 Jan 15;196(2):248–256. doi: 10.1128/JB.01151-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharathi SV, Vasanth V, Amitabha M, et al. Role of the Two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. Plos One. 2012 Apr 4;7(4):e33777, doi: 10.1371/journal.pone.0033777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weatherspoon-Griffin N, Yang D, Kong W, et al. The CpxR/CpxA two-component regulatory system Up-regulates the multidrug resistance cascade to facilitate escherichia coli resistance to a model antimicrobial peptide. J Biol Chem. 2014 Nov 21;289(47):32571–32582. doi: 10.1074/jbc.M114.565762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng-Ke Z, Meng-Yao Z, Shuo-Bo L, et al. Double deletion of cpxR and tolC significantly increases the susceptibility of Salmonella enterica serovar Typhimurium to colistin. J Antimicrob Chemother. 2021 Nov 12;76(12):3168–3174. doi: 10.1093/jac/dkab332 [DOI] [PubMed] [Google Scholar]
- 16.Zhai YJ, Sun HR, Luo XW, et al. CpxR regulates the colistin susceptibility of Salmonella Typhimurium by a multitarget mechanism. J Antimicrob Chemother. 2020 Oct 1;75(10):2780–2786. doi: 10.1093/jac/dkaa233 [DOI] [PubMed] [Google Scholar]
- 17.Jing W, Liu J, Wu S, et al. Role of cpxA mutations in the resistance to aminoglycosides and β-lactams in salmonella enterica serovar typhimurium. Front Microbiol. 2021 Feb 4;12:604079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahoney TF, Silhavy TJ.. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol. 2013 May 1;195(9):1869–1874. doi: 10.1128/JB.02197-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirakawa H, Nishino K, Yamada J, et al. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in escherichia coli. J Antimicrob Chemother. 2003 Oct 1;52(4):576–582. doi: 10.1093/jac/dkg406 [DOI] [PubMed] [Google Scholar]
- 20.Hu WS, Chen HW, Zhang RY, et al. The expression levels of outer membrane proteins STM1530 and OmpD, which Are influenced by the CpxAR and BaeSR two-component systems, play important roles in the ceftriaxone resistance of salmonella enterica serovar typhimurium. Antimicrob Agents Chemother. 2011 Aug;55(8):3829–3837. doi: 10.1128/AAC.00216-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masi M, Pinet E, Pagès J.. Complex response of the CpxAR two-component system to β-lactams on antibiotic resistance and envelope homeostasis in enterobacteriaceae. Antimicrob Agents Chemother. 2020 May 21;64(6):e00291–20. doi: 10.1128/AAC.00291-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian ZX, Yi XX, Cho A, et al. CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 2016 Oct 13;12(10):e1005932. doi: 10.1371/journal.ppat.1005932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao XL, Chen ZG, Yang TC, et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci Transl Med. 2021 Dec 22;13(625):eabj0716. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Li P, Jiang X, et al. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012 Apr 1;194(7):1841–1842. doi: 10.1128/JB.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi D, Jiang X, Sheng ZK, et al. Mapping the resistance-associated mobilome of a carbapenem-resistant klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J Antimicrob Chemother. 2015 Oct;70(10):2770–2774. doi: 10.1093/jac/dkv204 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Cen XF, Zhao GP, et al. Characterization of a new GlnR binding box in the promoter of amtB in streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol. 2012 Oct 1;194(19):5237–5244. doi: 10.1128/JB.00989-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Guo X, Xiang S, et al. Comparative analyses of phenotypic methods and 16S rRNA, khe. rpoB genes sequencing for identification of clinical isolates of Klebsiella pneumoniae. Antonie van Leeuwenhoek. 2016 Jul;109(7):1029–1040. [DOI] [PubMed] [Google Scholar]
- 28.Ortet P, Whitworth DE, Santaella C, et al. P2cs: updates of the prokaryotic two-component systems database. Nucleic Acids Res. 2015 Jan 28;43(Database issue):D536–D541. doi: 10.1093/nar/gku968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hecker SJ, Reddy KR, Totrov M, et al. Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem. 2015 May 14;58(9):3682–3692. doi: 10.1021/acs.jmedchem.5b00127 [DOI] [PubMed] [Google Scholar]
- 30.Mechaly AE, Haouz A, Sassoon N, et al. Conformational plasticity of the response regulator CpxR, a key player in gammaproteobacteria virulence and drug-resistance. J Struct Biol. 2018 Nov;204(2):165–171. doi: 10.1016/j.jsb.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 31.Gubbins MJ, Lau I, Will WR, et al. The positive regulator, TraJ, of the escherichia coli F plasmid is unstable in a cpxA* background. J Bacteriol. 2002 Oct 15;184(20):5781–5788. doi: 10.1128/JB.184.20.5781-5788.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau-Wong IC, Locke T, Ellison MJ, et al. Activation of the Cpx regulon destabilizes the F plasmid transfer activator, TraJ, via the HslVU protease in escherichia coli. Mol Microbiol. 2008 Feb;67(3):516–527. doi: 10.1111/j.1365-2958.2007.06055.x [DOI] [PubMed] [Google Scholar]
- 33.Yang X, Sun Q, Li J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent klebsiella pneumoniae in China. Emerg Microbes Infect. 2022 Dec;11(1):841–849. doi: 10.1080/22221751.2022.2049458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohanski MA, Dwyer DJ, Wierzbowski J, et al. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008 Nov 14;135(4):679–690. doi: 10.1016/j.cell.2008.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weatherspoon-Griffin N, Zhao G, Kong W, et al. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J Biol Chem. 2011 Feb 18;286(7):5529–5539. doi: 10.1074/jbc.M110.200352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Wang Q, Zhang H, et al. Sensor histidine kinase is a β-lactam receptor and induces resistance to β-lactam antibiotics. Proc Natl Acad Sci U S A. 2016 Feb 9;113(6):1648–1653. doi: 10.1073/pnas.1520300113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Wulf P, McGuire AM, Liu X, et al. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J Biol Chem. 2002 Jul 19;277(29):26652–26661. doi: 10.1074/jbc.M203487200 [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto K, Ishihama A.. Characterization of copper-inducible promoters regulated by CpxA/CpxR in escherichia coli. Biosci Biotechnol Biochem. 2006 Jul;70(7):1688–1695. doi: 10.1271/bbb.60024 [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Xu Y, Jiang B, et al. Systematic identification of CpxRA-regulated genes and their roles in escherichia coli stress response. mSystems. 2022 Oct 26;7(5):e0041922. doi: 10.1128/msystems.00419-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Liu F, Peng W, et al. The CpxA/CpxR two-component system affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front Cell Infect Microbiol. 2018 Mar 20;8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.