Abstract
The Genotype List (GL) String grammar for reporting HLA and Killer-cell Immunoglobulin-like Receptor (KIR) genotypes in a text string was described in 2013. Since this initial description, GL Strings have been used to describe HLA and KIR genotypes for more than 40 million subjects, allowing these data to be recorded, stored and transmitted in an easily parsed, text-based format. After a decade of working with HLA and KIR data in GL string format, with advances in HLA and KIR genotyping technologies that have fostered the generation of full-gene sequence data, the need for an extension of the GL String system has become clear. Here, we introduce the new GL String delimiter ”?”, which addresses the need to describe ambiguity in assigning a gene sequence to gene paralogs. GL Strings that do not include a “?” delimiter continue to be interpreted as originally described. This extension represents version 1.1 of the GL String grammar.
Keywords: genotype list string, killer-cell immunoglobulin-like receptor, HLA
INTRODUCTION
The HLA genes, on chromosome 6p21, are the most polymorphic, medically-relevant loci in the human genome[1–3]. As of January of 2023, 35,820 unique HLA nucleotide sequences, encoding 20,697 unique HLA proteins have been published. HLA genes are key elements of the adaptive immune system, and encode cell-surface antigens that present short, endogenously and exogenously derived peptides for inspection by T cells, permitting the discrimination of self from non-self by the adaptive immune system[4, 5]. The Killer-cell Immunoglobulin-like Receptor (KIR) genes, in the KIR gene cluster of the Leukocyte Receptor Complex on chromosome 19q13.4, expressed on Natural Killer (NK) cells, are key elements of the innate immune system that interact with class I HLA proteins to stimulate or inhibit NK cell activity[6–8]. The KIR region of chromosome 19 displays extensive haplotypic diversity, including many gene duplications and deletions[9]. As of December of 2022, 1617 unique KIR nucleotide sequences, encoding 703 unique proteins, have been published[10]. The key roles played by KIR and HLA molecules in innate and adaptive immunity motivate their routine genotyping for bone-marrow-, peripheral blood- and cord-blood-based hematopoietic progenitor cell therapies, as well as solid-organ transplantation efforts, and donor registries for these therapies store and share HLA and KIR genotypes for millions of potential donors[11–13].
Each unique HLA sequence is assigned a structured name by the World Health Organization Nomenclature Committee for Factors of the HLA System (WHO Nomenclature Committee), and each unique KIR sequence is assigned a similar name by the WHO Nomenclature Committee’s KIR subcommittee[14, 15]. Full-gene HLA and KIR sequencing has historically been difficult, and routine full-gene sequencing is only recently becoming cost-effective[16–19]. Given these historical limitations, HLA and KIR genotyping data have typically been ambiguous in that multiple distinct sets and combinations of allele names can be consistent with a pair of nucleotide sequences for a particular gene, when the complete gene sequence has not been determined for a subject. As shown in Table 1, allelic ambiguity occurs when only part of the gene sequence has been determined; alleles with identical nucleotide sequences in this region cannot be distinguished. Until recently, exons 2 and 3 have been commonly sequenced for class I HLA genes, and exon 2 has been commonly sequenced for class II HLA genes. For the KIR genes, though exons 3, 4, 5, 7, 8 and 9 may be sequenced, phase between each exon is not easily established. When the observed pair of allele sequences can appear in multiple pairs of alleles, genotype ambiguity occurs. For example, both the KIR2DS4*0010101 and KIR2DS4*002 genotype and the KIR2DS4*0040101 and KIR2DS4*0060102 genotype are consistent with identical sequences of exons 3, 4, 5, 7, 8 and 9, representing different phase options between exons.
Table 1.
Examples of Allele Ambiguity for the HLA and KIR Genes
| Locus | Identical Gene Feature1 | Ambiguous Alleles | Group2 |
|---|---|---|---|
| HLA-DRB1 | Exon 2 | DRB1*01:01:01:01, DRB1*01:01:01:02, DRB1*01:01:01:03, DRB1*01:01:01:04, DRB1*01:01:01:05, DRB1*01:01:01:06, DRB1*01:01:01:07, DRB1*01:01:01:08, DRB1*01:01:01:09, DRB1*01:01:01:10, DRB1*01:01:01:11, DRB1*01:01:01:12, DRB1*01:01:01:13, DRB1*01:01:01:14, DRB1*01:01:01:15, DRB1*01:01:01:16, DRB1*01:01:01:17, DRB1*01:01:01:18, DRB1*01:01:01:19, DRB1*01:01:32, DRB1*01:01:33, DRB1*01:01:35, DRB1*01:01:37, DRB1*01:01:39, DRB1*01:01:41, DRB1*01:01:44, DRB1*01:50, DRB1*01:67, DRB1*01:77, DRB1*01:82, DRB1*01:91Q, DRB1*01:95, DRB1*01:98, DRB1*01:100, DRB1*01:103, DRB1*01:104, DRB1*01:107, DRB1*01:115, DRB1*01:117, DRB1*01:119, DRB1*01:120, DRB1*01:121, DRB1*01:128, DRB1*01:129, DRB1*01:132, DRB1*01:137 and DRB1*01:139 | HLA-DRB1*01:01:01G |
| HLA-A | Exons 2 & 3 | A*23:01:01:01, A*23:01:01:02, A*23:01:01:03, A*23:01:01:04, A*23:01:01:05, A*23:01:01:06, A*23:01:01:07, A*23:01:01:08, A*23:01:01:09, A*23:01:01:10, A*23:01:01:11, A*23:01:01:12, A*23:01:01:13, A*23:01:01:14, A*23:01:01:15, A*23:01:01:16, A*23:01:01:17, A*23:01:01:18, A*23:01:01:19, A*23:01:01:20, A*23:01:01:21, A*23:01:01:22, A*23:01:01:23, A*23:01:01:24, A*23:01:01:25, A*23:01:01:26, A*23:01:01:27, A*23:01:01:28, A*23:01:01:29, A*23:01:01:30, A*23:01:05, A*23:01:19, A*23:01:24, A*23:01:25, A*23:01:27, A*23:01:28, A*23:01:31, A*23:01:34, A*23:07N, A*23:17:01:01, A*23:17:01:02, A*23:17:01:03, A*23:17:01:04, A*23:17:02, A*23:18, A*23:20, A*23:58, A*23:85, A*23:86, A*23:87, A*23:88, A*23:91N, A*23:92, A*23:93, A*23:94, A*23:95, A*23:96, A*23:103N, A*23:115, A*23:116, A*23:117, A*23:118, A*23:121, A*23:123 and A*23:125 | HLA-A*23:01:01G |
| KIR2DL5 | Exons 3, 5, 7, 8 & 9 | KIR2DL5A*001, KIR2DL5B*006, KIR2DL5B *008 and KIR2DL5A*012 | KIR2DL5*0013 |
| KIR3DL2 | Exons 3, 4, 5, 7, 8 & 9 | KIR3DL2*002, KIR3DL2*101 and KIR3DL2*102 | KIR3DL2*0023 |
Gene Features are the structural elements of a gene – Introns, Exons and Untranslated Regions (UTRs)[27].
Groups are identifiers that represent sets of alleles sharing a predefined set of gene features. For the HLA alleles, “G groups” (e.g., HLA-DRB1*01:01:01G) represent all of the alleles with identical sequence for exons 2 and 3 for class I HLA alleles, and exon 2 for class II HLA alleles. G group membership described here is specific to IPD-IMGT/HLA Database Release 3.50.0.
For KIR alleles, the KIR2DL5*001 and KIR3DL2*002 groups represent all alleles that share identical sequence for KIR exons 3–5 and 7–9 with KIR2DL5A*001 and KIR3DL2*002, respectively[28]. The KIR allele group memberships described here are specific to the KIR typing approach described by Wagner et al., 2018[19]. The use of the KIR2DL5 designation without specifying the A or B ending is not consistent with the official KIR nomenclature.
To facilitate the lossless storage and transmission of HLA and KIR genotype data, while describing these genotyping ambiguities in a structured manner, the Genotype List (GL) String format was developed to standardize the encoding of genotyping results[20], using a set of case-defined “AND” and “OR” delimiters. While primarily developed for HLA and KIR data, the GL String format can be applied to any genetic system, regardless of its function or application. The Be The Match registry, DKMS, and other registries and donor centers use GL Strings to store and exchange HLA and KIR genotype data for over 40 million patients and volunteer donors[13, 21].
In the decade since the development and initial description of the GL String format, advances in HLA and KIR genotyping technologies have driven discovery and insight into the diversity of the HLA and KIR genes, as well as the large-scale haplotype structure of the KIR gene cluster. Challenges experienced in genotyping the paralogous KIR2DL5A and KIR2DL5B genes have resulted in the recognition of a new type of ambiguity in which nucleotide sequences cannot be assigned to either the centromeric KIR2DL5A gene or the telomeric KIR2DL5B gene.
Here, we introduce “?”, a new GL String delimiter that extends the original GL string standard to describe ambiguity in gene location. The extension of the GL String grammar to include the “?” delimiter will require the modification of GL String parsers to allow forward-compatibility, while maintaining backward-compatibility with original GL Strings. We define this extension of the grammar as Genotype List String version 1.1.
METHODS AND RESULTS
The five original GL String delimiters are evaluated in an order of precedence beginning with distinct genes (precedence 1, delimited by “^”), proceeding through ambiguous genotypes (precedence 2, delimited by “|”), gene copies (precedence 3, delimited by “+”), genes for which phase is known (precedence 4, delimited by “~”), and ending with ambiguous alleles (precedence 5, delimited by “/”). These grammatical rules have worked well for the last decade to accurately describe what is known and unknown about an HLA or KIR genotype. The application of these delimiters is further described in Table 2.
Table 2.
Genotype List String Delimiters
| Precedence1 | Delimiter | Identifies | Example |
|---|---|---|---|
| 0 | ? | Possible Gene Locations | KIR2DL5B*00201?KIR2DL5A*00501 |
| 1 | ^ | Unphased Genes | HLA-A*02:07+HLA-A*03:06^HLA-B*08:01+HLA-B*44:02 |
| 2 | | | Possible Genotypes | HLA-A*02:02:01G+A*03:01:01G | HLA-A*02:881+A*03:303 |
| 3 | + | Copies of Genes | HLA-A*02:02 + HLA-A*03:01 |
| 4 | ~ | Phased Genes | HLA-DRB1*03:01 ~ HLA-DRB3*01:01 |
| 5 | / | Possible Alleles | HLA-A*02:01 / HLA-A*02:02 |
Adapted from (Milius et al., 2013[20]).
GL String delimiters are evaluated in the numerical precedence order shown.
The order of delimited elements in a GL String does not inform how that string is parsed. For example, the order of slash-delimited alleles or pipe-delimited genotypes does not indicate the greater likelihood of one allele or one genotype over another. Similarly, the order of loci in a GL String need not correspond to the relative chromosomal position or chromosomal order of those loci. Further, when the “~” delimiter is used to identify chromosomal phase between genes encoding subunits of a heterodimeric protein (e.g., between an HLA-DQA1 allele and an HLA-DQB1 allele), the identified phase does not imply that those alleles will form a heterodimer.
When the examples with precedence 1 to 5 in Table 2 are assembled, the resulting GL String, “HLA-A*02:01/HLA-A*02:02+HLA-A*03:01|HLA-A*02:07+HLA-A*03:06^HLA-B*08:01+HLA-B*44:02/HLA-B*44:03^HLA-DRB1*03:01~HLA-DRB3*01:01+HLA-DRB1*03:01~HLA-DRB3*01:01”, accurately describes what is known and unknown about this HLA genotype.
It is possible to record the same genotype data with distinct but equivalent GL Strings. For example, “HLA-A*02:01/HLA-A*02:02+HLA-A*03:01” could also be written as “HLA-A*02:01+HLA-A*03:01|HLA-A*02:02+HLA-A*03:01”.
The need for an extension of the GL String grammar was initially raised as part of the ongoing Data Standards Hackathon for Next Generation Sequencing (DaSH for NGS) effort[22]. The advent of long-range, massively-parallel, single molecule sequencing has initiated a sea change in the Histocompatibility and Immunogenetics community, and larger genomics community, expanding and deepening the scope of knowledge of the structure, organization and polymorphism of HLA and KIR genes.
Since the 16th International HLA and Immunogenetics Workshop in 2012, research laboratories, stem cell donor registries, and donor centers have investigated, developed and adopted NGS technologies for routine genotyping[23]. A key outcome of these efforts is the acknowledgement that genotyping results that may once have been dismissed as unusual edge-cases are in fact legitimate polymorphisms that require new approaches for accurate description. Over the course of several DaSH meetings in 2020 and 2021, the extension to the GL String grammar described below was developed.
Describing Gene-Location Ambiguity
While the GL String grammar allows accurate descriptions of what is known and unknown about allele sequences and phasing information between those sequences, it cannot describe what is known or unknown about allele sequences for highly homologous gene paralogues.
As illustrated in Figure 1, the paralogous KIR2DL5A and KIR2DL5B genes share high sequence homology and are respectively located in the telomeric and centromeric regions of the KIR gene cluster[9, 24]. Recombination between the centromeric and telomeric regions of the KIR gene cluster defines four KIR gene cluster haplotypes by the presence and absence of the KIR2DL5A and KIR2DL5B genes -- those with KIR2DL5A and KIR2DL5B, those with only one of either the KIR2DL5A or KIR2DL5B gene, and those with no KIR2DL5A or KIR2DL5B gene (Figure 1), so that individuals can have between zero and four alleles of the KIR2DL5A and KIR2DL5B alleles. This is also the case for the KIR2DS3 and KIR2DS5 genes. KIR2DL5A and KIR2DL5B alleles are similar in sequence, and it can be difficult to distinguish centromeric and telomeric alleles without phased sequence across multiple exons and introns. In these cases, KIR2DL5A and KIR2DL5B ambiguity cannot be accurately described using the current GL String syntax, which does not permit alleles of paralogous loci to be included in a single “/” delimited string, and does not include an “OR” delimiter for gene location.
Figure 1.
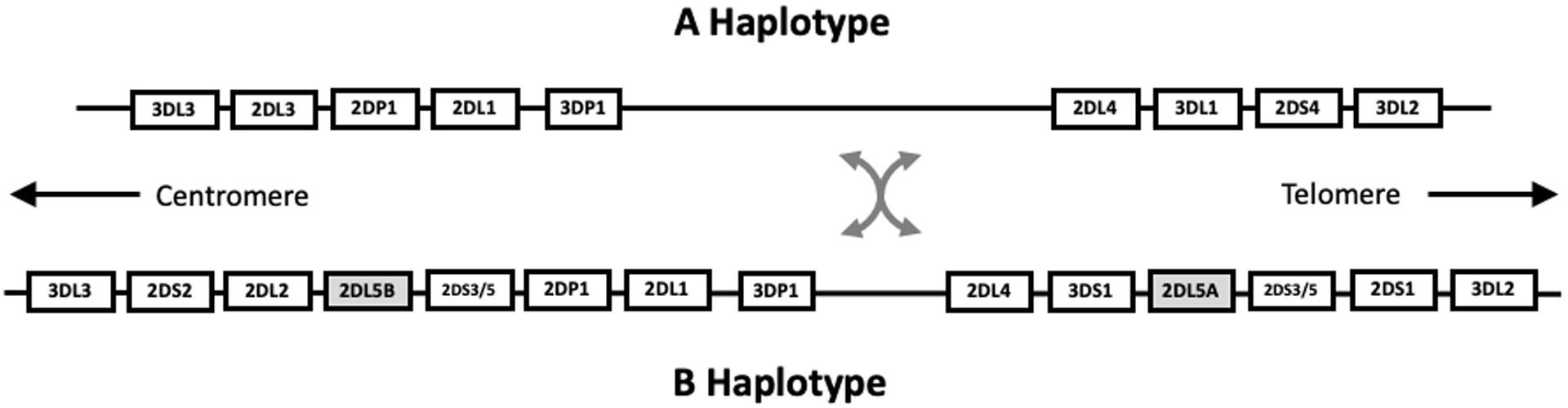
The KIR2LD5A and KIR2DL5B Genes in the KIR Gene Cluster
The relative organization of the KIR genes on the KIR A and B haplotypes is shown. Each KIR gene is identified with a rectangular box. Each haplotype can be divided into a centromeric and telomeric segment, between which recombination occurs, resulting in a centromeric A (cenA) haplotype, a centromeric B (cenB) haplotype, a telomeric A (telA) haplotype and a telomeric B (telB) haplotype[24]. The KIR2DL5A and KIR2DL5B genes are indicated in grey.
To address this, the “?” delimiter describes locus ambiguity where the typing system cannot distinguish paralogous genes, in the same way that the “|” delimiter describes genotype ambiguity where the typing system cannot distinguish chromosomal phase, and the “/” delimiter describes allele ambiguity where the typing system cannot distinguish between alleles. The ”?” delimiter is assigned precedence 0, so that it is evaluated before all other GL String delimiters.
For example, the KIR2DL5A*0010101+KIR2DL5A*0010201?KIR2DL5A*0010301^KIR2DL5B*0010101?KIR2DL5B*0010201+KIR2DL5B*0010301” string describes three possible relationships between KIR2DL5 alleles for which partial sequences are known. It cannot be determined which alleles are at the KIR2DL5A and KIRDL5B loci, so all three possible combinations must be reported. In this case, sequence is known for both alleles, but the corresponding allele name depends on that gene’s location in the telomeric or centromeric section of the KIR region.
DISCUSSION AND CONCLUSION
The Histocompatibility and Immunogenetics community is in the “log phase” of the transition from established molecular genotyping methods to so-called third-generation sequencing approaches. The evaluation and application of these technologies is still in process. Extension of the GL String grammar to describe ambiguity in gene-location and gene-copy number will facilitate this transition by fostering more accurate, lossless communication and storage of HLA and KIR genotyping results.
Data for other gene systems that assign unique names to the nucleotide sequences of gene variants can be described using GL Strings, and this extension of the format could prove useful for gene systems characterized by large copy-number variation of highly homologous genes (e.g., class I MHC in rhesus macaques), and for describing novel or rare haplotype structures.
It should be noted that GL Strings, while designed to facilitate lossless storage and transmission of genotype data, do not inherently convey all aspects of the typing process itself. For instance, GL Strings of genotypes can be generated by interrogating different numbers of gene features for different subjects (e.g., HLA class I exons 2 and 3 for all subjects, and additionally exons 4 and 5 for a subset of subjects). Consequently, when GL Strings are compared directly, methodological details of a genotyping experiment need to be considered. These details do not typically accompany the reported genotype. DaSH efforts have focused on developing systems and services that formalize the description of immunogenomic genotyping methodologies[25, 26]. With this ongoing effort in mind, we propose that annotations of immunogenomic genotyping experiments include standardized descriptions of genotyping metadata pertinent to gene-feature targets and sequencing technology. As with any data recording format, knowledge of the assumptions and limitations of the format are key for informed data-analysis and application.
ACKNOWLEDGEMENTS
We thank the participants of the Data Standards Hackathons for NGS for their continued participation and for helpful discussion on the topics of data-representation and data-sharing. The work described here was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) grants R01AI128775 (SM, JH [UCSF], MM), R01AI173095 (LG), and U01AI152960 (LG), and by the US Office of Naval Research (ONR) grant N00014-20-1-2832 (MM, LG, SS, RM, JP, JS [NMDP]). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIAID, NIH, ONR or United States Government.
CONFLICT OF INTEREST STATEMENT
Loren Gragert has received research funding from National Marrow Donor Program (NMDP) and United Network for Organ Sharing (UNOS). The remaining authors have no conflicts of interest to declare.
Abbreviations
- DaSH
Data Standards Hackathon
- GL
Genotype List
- KIR
Killer-cell Immunoglobulin-like Receptor
- MHC
Major Histocompatibility Complex
- NGS
Next Generation Sequencing
- NK
Natural Killer
- UTR
Untranslated Region
- WHO
World Health Organization
REFERENCES
- 1.Welter D, et al. , The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res, 2014. 42(Database issue): p. D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson J, et al. , Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles. PLoS Genet, 2017. 13(6): p. e1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker DJ, et al. , The IPD-IMGT/HLA Database. Nucleic Acids Res, 2023. 51(D1): p. D1053–d1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parham P, et al. , Nature of polymorphism in HLA-A, -B, and -C molecules. Proceedings of the National Academy of Sciences of the United States of America, 1988. 85(11): p. 4005–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lechler R and Warrens A, eds. HLA in Health and Disease. 2 ed. 2000, Academic Press Limited: London. [Google Scholar]
- 6.Martin AM, et al. , The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics, 2000. 51(4–5): p. 268–80. [DOI] [PubMed] [Google Scholar]
- 7.Vales-Gomez M, et al. , Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors. Immunity, 1998. 9(3): p. 337–44. [DOI] [PubMed] [Google Scholar]
- 8.Moretta A, et al. , P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med, 1993. 178(2): p. 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Lozano N, et al. , Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics, 2002. 54(5): p. 314–9. [DOI] [PubMed] [Google Scholar]
- 10.Robinson J, et al. , The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res, 2015. 43(Database issue): p. D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggeri L, et al. , Natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Int J Hematol, 2005. 81(1): p. 13–7. [DOI] [PubMed] [Google Scholar]
- 12.Petersdorf EW, et al. , Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med, 2001. 345(25): p. 1794–800. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt AH, et al. , Immunogenetics in stem cell donor registry work: The DKMS example (Part 1). Int J Immunogenet, 2020. 47(1): p. 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh SG, et al. , Nomenclature for factors of the HLA system, 2010. Tissue Antigens, 2010. 75(4): p. 291–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh SGE, et al. , Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens, 2003. 62(1): p. 79–86. [DOI] [PubMed] [Google Scholar]
- 16.Ehrenberg PK, et al. , High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics, 2014. 15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin WM, et al. , High-throughput Interpretation of Killer-cell Immunoglobulin-like Receptor Short-read Sequencing Data with PING. PLoS Comput Biol, 2021. 17(8): p. e1008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schöfl G, et al. , 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics, 2017. 18(1): p. 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner I, et al. , Allele-Level KIR Genotyping of More Than a Million Samples: Workflow, Algorithm, and Observations. Front Immunol, 2018. 9: p. 2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milius RP, et al. , Genotype List String: a grammar for describing HLA and KIR genotyping results in a text string. Tissue Antigens, 2013. 82(2): p. 106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker T, et al. , The National Marrow Donor Program: improving access to hematopoietic cell transplantation. Clin Transpl, 2011: p. 55–62. [PubMed] [Google Scholar]
- 22.Matern BM, et al. , Standard reference sequences for submission of HLA genotyping for the 18th International HLA and Immunogenetics Workshop. Hla, 2021. 97(6): p. 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Santis D, et al. , 16th IHIW : Review of HLA typing by NGS. Int J Immunogenet, 2013. 40(1): p. 72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roe D, et al. , Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun, 2017. 18(3): p. 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack SJ, et al. , Minimum information for reporting next generation sequence genotyping (MIRING): Guidelines for reporting HLA and KIR genotyping via next generation sequencing. Hum Immunol, 2015. 76(12): p. 954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milius RP, et al. , Histoimmunogenetics Markup Language 1.0: Reporting next generation sequencing-based HLA and KIR genotyping. Hum Immunol, 2015. 76(12): p. 963–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack SJ, A gene feature enumeration approach for describing HLA allele polymorphism. Hum Immunol, 2015. 76(12): p. 975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solloch UV, et al. , Estimation of German KIR Allele Group Haplotype Frequencies. Front Immunol, 2020. 11: p. 429. [DOI] [PMC free article] [PubMed] [Google Scholar]


