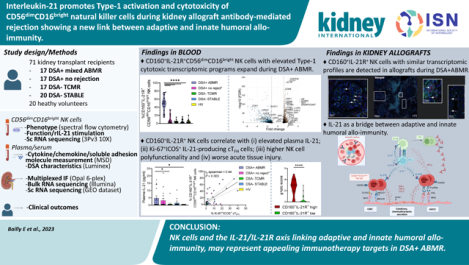
Abstract
The role of Natural killer (NK) cells during kidney allograft antibody-mediated rejection (ABMR) is increasingly recognized, but an in-depth characterization of mechanisms that contribute to such immune response is still under investigation. Here, we characterized phenotypic, functional, and transcriptomic profiles of peripheral blood circulating and allograft infiltrating CD56dimCD16bright NK cells during anti-HLA donor-specific antibody (DSA)+ ABMR. Cross- sectional analyses performed in 71 kidney transplant recipients identified a unique phenotypic circulating CD56dimCD16bright NK cell cluster expanded in DSA+ ABMR. This cluster co-expressed high levels of the interleukin-21 Receptor (IL-21R); Type-1 transcription factors T-bet and EOMES, CD160 and natural killer group 2D cytotoxic and activating co-stimulatory receptors. CD160+ IL-21R+ NK cells correlated with elevated plasma IL-21, Ki-67+ ICOS+ (CD278) IL-21-producing circulating T follicular helper cells, enhanced Type-1 pro-inflammatory cytokines, NK cell cytotoxicity, worse microvascular inflammation and graft loss. Single-cell transcriptomic analysis of circulating NK cells delineated an expanded cluster in DSA+ ABMR characterized by elevated pro-inflammatory/cytotoxic pathways, IL-21/STAT3 signaling, and leukocyte trans-endothelial migration pathways. Infiltration of CD160+ IL-21R+ NK cells with similar transcriptomic profile was detected in DSA+ ABMR allograft biopsies, potentially contributing to allograft injury. Thus, the IL-21/IL-21R axis, linking adaptive and innate humoral allo-immunity, or NK cells may represent appealing immunotherapy targets in DSA+ ABMR.
Keywords: Natural killer cell, antibody-mediated rejection, kidney transplantation, alloantibody, IL-21, cytotoxicity
Graphical Abstract
TRANSLATIONAL STATEMENT
While the involvement of NK cells during kidney allograft rejection has been increasingly described over the past decade, the role of potentially heterogeneous NK cell subsets involved in antibody-mediated rejection remains to be investigated. Here we report that IL-21, in addition to its known role during humoral B cell allo-responses, may also help NK cells differentiate into polyfunctional Type-1 activated cytotoxic effectors with a potential deleterious ability to infiltrate the kidney allograft during DSA+ ABMR and harm the vascular endothelium. Targeting the IL-21/IL-21R axis and NK cells could be valuable emerging therapeutic strategies during humoral rejection.
INTRODUCTION
Allorecognition and alloimmune effector mechanisms in solid organ transplantation are mediated by broad and diverse adaptive and innate cell types that show dynamic interactions1,2. Such alloimmune processes represent great hurdles for long-term allograft outcomes. In the field of solid-organ transplantation, there is still an unmet need for better characterization of both cellular and molecular mechanisms contributing to heterogeneous pathologic outcomes3. Gaining more insight into these processes will help advance personalized preventive or therapeutic strategies in the clinic4.
Despite robust immunosuppressive regimens, allorecognition of donor HLA molecules, ultimately reflected by anti-HLA donor-specific alloantibody (DSA) generation, can lead to antibody-mediated rejection (ABMR) with various phenotypes5,6. DSA characteristics such as MFI, complement-binding capacity and IgG subclass distribution proved to influence DSA pathogenicity7. An increased frequency of proliferating activated circulating T follicular helper cells (cTFH) with Th1/Th17 features and producing IL-21 has been observed during DSA+ ABMR8. cTFH cognate interaction with activated B cells results in the generation of pathogenic DSA with predominant IgG3 subclass profile8. DSA through the engagement with FcγR-expressing cellular effectors mediate antibody-dependent cellular cytotoxicity (ADCC) and cytokine release9–11. Among FcγR-expressing cells, NK cells infiltrating kidney allografts were observed in capillaries during ABMR and in the inflamed interstitium during T-cell mediated rejection (TCMR)12. NK cell infiltration has been associated with ABMR severity highlighting their importance in the humoral rejection pathogenesis13. DSA may interact with FcγR+ NK cells leading to their activation and to ADCC with release of lytic enzymes and pro-inflammatory mediators participating to microvascular inflammation (MVI). Transcripts indicative of FcγRIIIa (CD16a)-mediated NK cell activation have been identified during ABMR14,15. However, till date there is no comprehensive characterization of blood circulating NK cells profiles in DSA+ ABMR patients to corroborate the tissue transcriptomic findings.
Among circulating NK cells, CD56dimCD16bright NK cells are the most numerous and express high levels of CD16a that can bind DSA cross-linked to the endothelium. Here, we hypothesize that during alloimmune humoral responses, the altered cytokine environment could uniquely shape CD56dimCD16bright NK cell phenotype and function towards enhanced cytotoxicity and inflammation with direct consequences against the allograft endothelium. Detailed phenotypic, functional, and transcriptomic characterization of circulating and graft infiltrating CD56dimCD16bright NK cells were implemented to address our hypothesis. We identified a novel mechanistic link between adaptive and innate immunity that may be specifically targeted during DSA+ ABMR for improved long-term allograft outcomes.
METHODS
More detailed information is provided in the Supplementary Methods.
Experimental design
This observational study included patients who underwent kidney transplantation (KT) from January 2013 to December 2018 and participated in the Thomas E. Starzl Transplant Institute prospective biorepository initiative. Patients were screened in the three first year’s post transplantation for: occurrence of DSA, biopsy-proven clinical ABMR or TCMR. Diagnoses were established following the international Banff 2019 criteria5. Exclusion criteria comprised of absence of Thymoglobulin induction, positive flow cytometry crossmatch, BK virus nephropathy, HIV/HCV seropositivity, CMV viremia at the time of event, absence of protocol biopsies or available PBMC at the time of event. We identified patients with DSA with either mixed ABMR (“DSA+ ABMR”) or without pathology lesions (“DSA+ no rejection”). Age/sex-matched patients with no DSA during follow-up were identified with either TCMR (“DSA− TCMR”) or without MVI/rejection episodes during follow-up (“DSA− STABLE”) (Supplementary Figure S1A and S1B). Additionally, healthy volunteers (HV) were enrolled. The cohort characteristics are shown in Supplementary Table S1 and Supplementary Figure S1C. Samples were analyzed cross-sectionally at the time of rejection (DSA+ ABMR or DSA− TCMR), of the first DSA occurrence (DSA+ no rejection), or at a matched timepoint (DSA− STABLE group).
The study protocol was approved by the University of Pittsburgh institutional review board (IRB PRO12030552, PRO17020318).
Spectral flow cytometry and high-dimensional flow cytometry data analysis
PBMC were surface and intracellular stained (Supplementary Table S2), and subsequently acquired on Aurora spectral flow cytometer (Cytek). CD56dimCD16bright NK cells were gated on the CD3−CD19−CD14−CD7+CD56+ viable cells (Supplementary Figure S2A). Data were analyzed using the FlowJo software (BD Biosciences) and the Cytobank platform for t-distributed stochastic neighbor embedding (t-SNE) visualization, FlowSOM (Flow Self-Organizing Map) clustering16 and the CITRUS (cluster identification, characterization, and regression) predictive algorithm17.
Plasma cytokines measurement
Plasma cytokines and soluble adhesion molecules were measured by a Meso Scale Discovery V-PLEX Human Biomarker Kit.
Functional assays
Cell-mediated cytotoxicity (CMC) and ADCC
174xCEM.T2 TAP-deficient lymphoblastic cell line was used as NK cell target (Supplementary Figure S2B). For ADCC experiments, T2 cells were pre-coated with highly sensitized KT patient (HS) sera containing anti-HLA-A2 DSA with high MFI. PBMC and T2 cells (5:1 effector-to-target ratio) were incubated for 6 hours (37°C, 5% CO2). AB normal human serum that contains no anti-HLA Ab was used as control (Supplementary Figure S2C). CD56dim NK cell degranulation (CD107a) and cytokines (IFN-γ, TNF-α, IL-10) production was assessed by spectral flow cytometry.
IL-21 stimulation
PBMC were incubated for 5 days with rIL-21 (Gibco, 50 ng/mL). Some cocultures were supplemented with mouse IL-21R-Fc (R&D Systems, 10 μg/mL) to block IL-21 binding.
Single-cell RNA-sequencing analysis of circulating NK cells
NK cells from 4 DSA+ ABMR and 4 DSA− TCMR were FACS-sorted on a BD Biosciences Aria II cytometer. Illumina libraries were generated using the Chromium Single Cell 3′ Reagent Kit (v3 Chemistry; 10X Genomics) and loaded on a NovaSeq S2–100. Counts matrices were analyzed in the Partek Flow software.
Multispectral immunofluorescence of kidney allografts
Multispectral multiplexed immunofluorescence was performed on formalin-fixed paraffin-embedded (FFPE) tissue sections using the Opal 6-Plex Manual Detection Kit (Akoya Biosciences). One kidney allograft nephrectomy (chronic mixed ABMR), 2 DSA+ mixed ABMR and 2 DSA− TCMR biopsies were stained for CD31/CD3/CD56/CD16/CD160/IL-21R and DAPI. Imaging was performed on the PhenoImager HT and analysis on InForm v2.6 (Akoya Biosciences)18.
Bulk RNA-sequencing of kidney allograft biopsies
Total RNA extracted from twenty-eight FFPE renal allograft biopsies diagnosed as DSA+ ABMR (n=13), DSA− TCMR (n=9) or as normal (DSA− STABLE) (n=6), was analyzed (Supplementary Table S3) by bulk RNA-sequencing on an Illumina Next-Seq550. The differential gene expression analysis was performed with the DESeq2 algorithm in the Partek Flow software.
Single-cell RNA-sequencing analysis of kidney allograft biopsies
Published single-cell RNA-sequencing data (Illumina HiSeq) from 3 ABMR and 2 acute tubular necrosis (ATN) biopsies rejection-free were retrieved (GSE145927)19. After SCTransform integration, NK and NKT cells were annotated and analyzed for their transcriptomic signature, while endothelial cells were identified and assessed for apoptosis related pathways.
Statistical analysis
Mean, standard deviation and frequencies are provided for continuous and categorical variables, respectively. Means and proportions were compared using the t-test, χ2 test or the Mann-Whitney test and Fisher’s exact test if appropriate. Multiple groups were analyzed by 1-way ANOVA with the Tukey post-hoc test or Kruskal-Wallis test with Dunn’s test for multiple comparison adjustment. Correlations were evaluated by Spearman rank tests.
Death-censored allograft survival was assessed using the Kaplan-Meier method and compared with the log-rank test. Univariate Cox regressions were performed to investigate the association between survival and clinical/immunological/histological variables.
P < 0.05 were considered statistically significant. All tests were 2-sided. For the transcriptomic analysis, P values were adjusted by a Benjamini-Hochberg procedure (FDR < 5%). Figures were generated on GraphPad Prism version 9, R Studio version 2022.07.1 and BioRender.
RESULTS
Patients’ demographics and clinical characteristics
We conducted a cross-sectional study on 71 KT patients: DSA+ ABMR (n=17), DSA+ no rejection (n=17), DSA− TCMR (n=17), DSA− STABLE (n=20) and 20 HV (Supplementary Figure S1A). Samples were analyzed cross-sectionally at the time of rejection/DSA occurrence or at a matched timepoint in the first 3 years post-Tx (Supplementary Figure S1B). All patients had a minimum follow-up of 3 years. Demographics are depicted in Supplementary Table S1. All patients had a low immunological risk (negative flow cytometry crossmatch) and were maintained on similar immunosuppressive regimens. DSA+ ABMR group displayed higher re-transplantation rate, calculated panel reactive antibody, preformed DSA frequency (41.2%), and number of HLA-DRB1 mismatches. At the time of sampling, DSA+ ABMR group presented higher immunodominant DSA MFI and number of DSA. In contrast, more living-donor transplantation and a lower number of HLA-A mismatches were observed in the DSA− STABLE group. DSA characteristics are depicted in Supplementary Figure S1C.
CD56dimCD16bright NK cells display increased cytotoxic molecule expression and Type-1 polarization during DSA+ ABMR
A decreased frequency of circulating CD56dimCD16bright NK cells was observed in KT recipients compared to HV with no differences among the patient groups (Supplementary Figure S3A). t-SNE and FlowSOM clustering revealed different CD56dimCD16bright NK cell density distribution and important phenotypic differences among groups (Figure 1A–1C). A unique cluster (cluster 2 in orange) expanded specifically in the DSA+ groups (Figure 1C). Highly prevalent in DSA+ ABMR, it was detected in only 3 DSA+ no rejection patients and was absent in all DSA− groups (Figure 1D). This cluster was characterized by a higher expression of the cytotoxic molecule CD160 (MHC class I molecule ligands). Notably, this cluster overexpressed the cytokine receptor IL-21R and Type-1 promoting transcription factors (TF) EOMES and Tbet (Figure 1B and 1E). In addition to its roles during humoral immunity, IL-21 was shown as physiologically involved in earlier stages of NK cell development and particularly relevant for triggering NK cell proliferation and cytotoxic activity20. NKG2D (MICA/B, ULBPs ligands) cytotoxic and activating receptor was also upregulated, while the activating receptor NKG2C was not. Those cells were not terminally differentiated, as indicated by the lack of CD57 expression. Another feature associated with this pro-inflammatory phenotype was the upregulation of CXCR3, suggesting a higher migration potential towards inflamed tissues. Interestingly, 8 out of 17 DSA+ ABMR patients (47.1%) also exhibited high expression of CD161/NK1.1 and of the ITIM-containing inhibitory receptor NKG2A (HLA-E ligand) (Figure 1B and 1E). CD161 expression marks pro-inflammatory NK cells with high cytokine responsiveness21. Its ligation with lectin-like transcript LLT1 maintains IFN-γ production but with a cytotoxicity inhibition21,22. Co-expression of CD161/NKG2A potentially acted as negative feedback to regulate NK cell activity. But in conditions of sufficient activation, in particular via cytokine activation, NKG2A may not reduce NK cell cytotoxicity23. Of note, PD1 was not co-expressed by this cluster. Variable levels of the proliferation marker Ki-67 were observed. Using supervised biaxial gating, we confirmed these observations (Supplementary Figure S3B). Thus, a unique CD56dimCD16bright NK cell subset responsive to IL-21 and displaying Type-1 and cytotoxic features was expanded during DSA+ ABMR.
Figure 1|. High-dimensional spectral flow cytometry analysis of CD56dimCD16bright natural killer cells from patients with donor-specific alloantibody-positive mixed antibody-mediated rejection (DSA + ABMR) reveals a cytotoxic, pro-inflammatory cell cluster with high interleukin-21 receptor (IL-21R) expression.
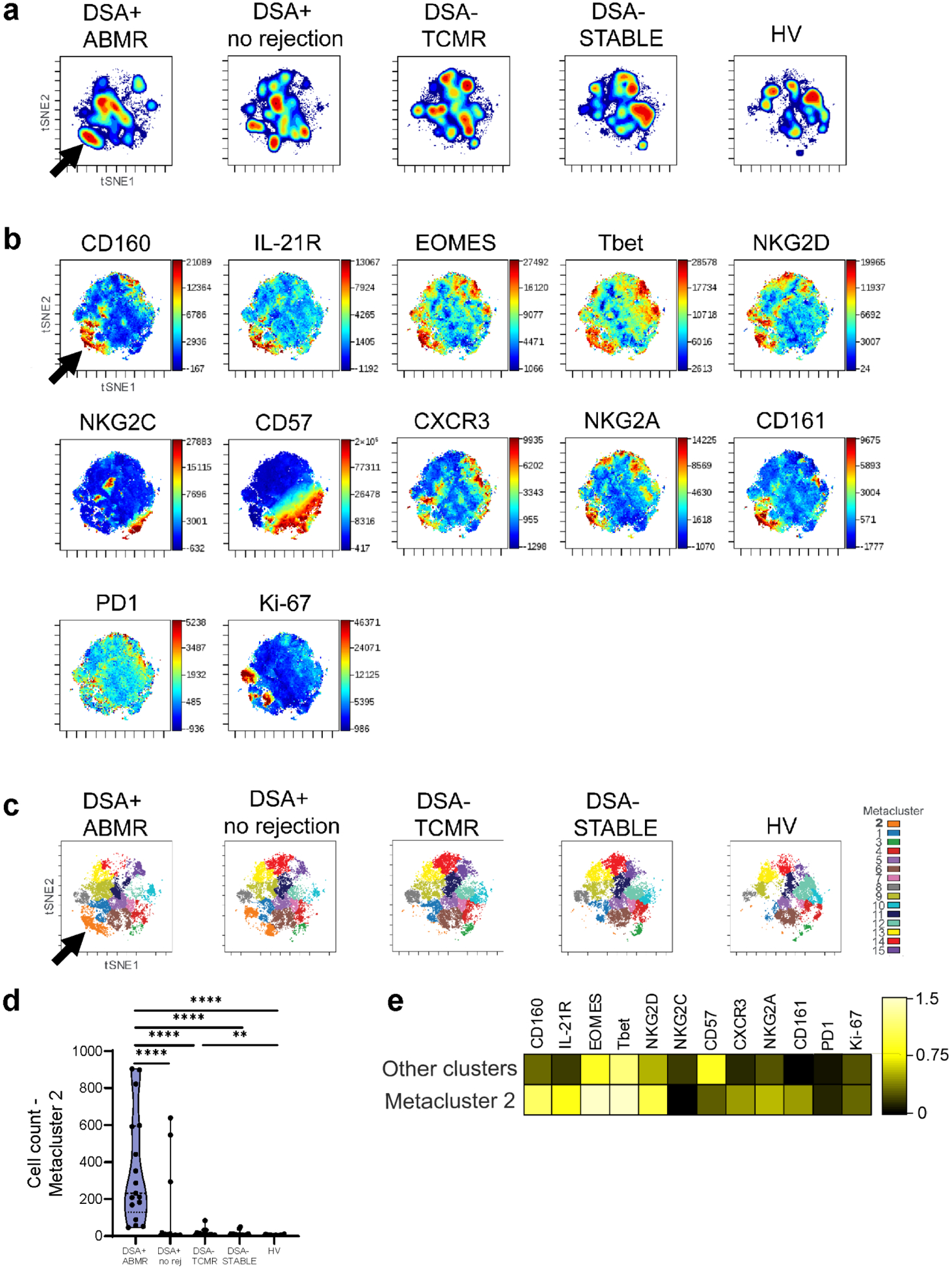
(a) t-Distributed stochastic neighbor embedding (t-SNE) projections generated from concatenated files for each group displaying cell density distribution, n = 71 patients and n = 20 healthy volunteers (HVs). (b) t-SNE projections displaying cell surface marker mean fluorescence intensity (MFI) gradients, (c) FlowSOM (Flow Self-Organizing Map) clustering for the 5 study groups, (d) Distribution of cluster 2 among the groups. Mann-Whitney test. **P < 0.01, ****p < 0.0001. (e) Heatmap with markers' MFI for cluster 2 versus the other clusters. Transformed ratio of the median MFI, displayed by column's minimum. CXCR3, CXC chemokine receptor 3; DSA+ no rejection, donor-specific alloantibody-positive without microvascular inflammation/rejection episodes; DSA− STABLE, donor-specific alloantibody-negative without microvascular inflammation/rejection episodes; DSA− TCMR, donor-specific alloantibody-negative T cell–mediated rejection; NKG2A, natural killer group 2A; NKG2C, natural killer group 2C; NKG2D, natural killer group 2D; PD1, programmed cell death protein 1.
CD160 and IL-21R co-expression on CD56dimCD16bright NK cells significantly distinguish DSA+ ABMR from DSA− TCMR phenotypes
To further parse phenotypic differences between CD56dimCD16bright NK cells from DSA+ ABMR and DSA− TCMR patients we used the CITRUS predictive algorithm to determine the lowest number of phenotypic markers that could significantly distinguish the two conditions. The model identified significant differential co-expression of CD160 and IL-21R on 10 different cell clusters (Supplementary Figure S4A and S4B). CD160 and IL-21R co-expression was able to differentiate the two rejection groups. Clusters 1 to 10 displayed higher levels of EOMES, Tbet, CXCR3 and NKG2A. NKG2D and Ki-67 were increased in proximal nodes, whereas CD161 in daughter nodes (Supplementary Figure S4C). Therefore, CD56dimCD16bright NK cells during DSA+ ABMR, show elevated CD160+IL-21R+ co-expression that uniquely distinguishes them from DSA− TCMR, reinforcing their differential inflammatory cytotoxic profile. High (>22.5% defined by the 75% percentile of the cohort) CD160+IL-21R+ expression was observed in 15/17 (88.2%) DSA+ ABMR and 3/17 (17.6%) DSA+ no rejection patients and was specific for those 2 DSA+ groups (Figure 2A).
Figure 2|. CD160+IL-21R+CD56dimCD16bright natural killer (NK) cells correlate with plasma interleukin-21 (IL-21) and with activated circulating T follicular helper (cTFH) cells.
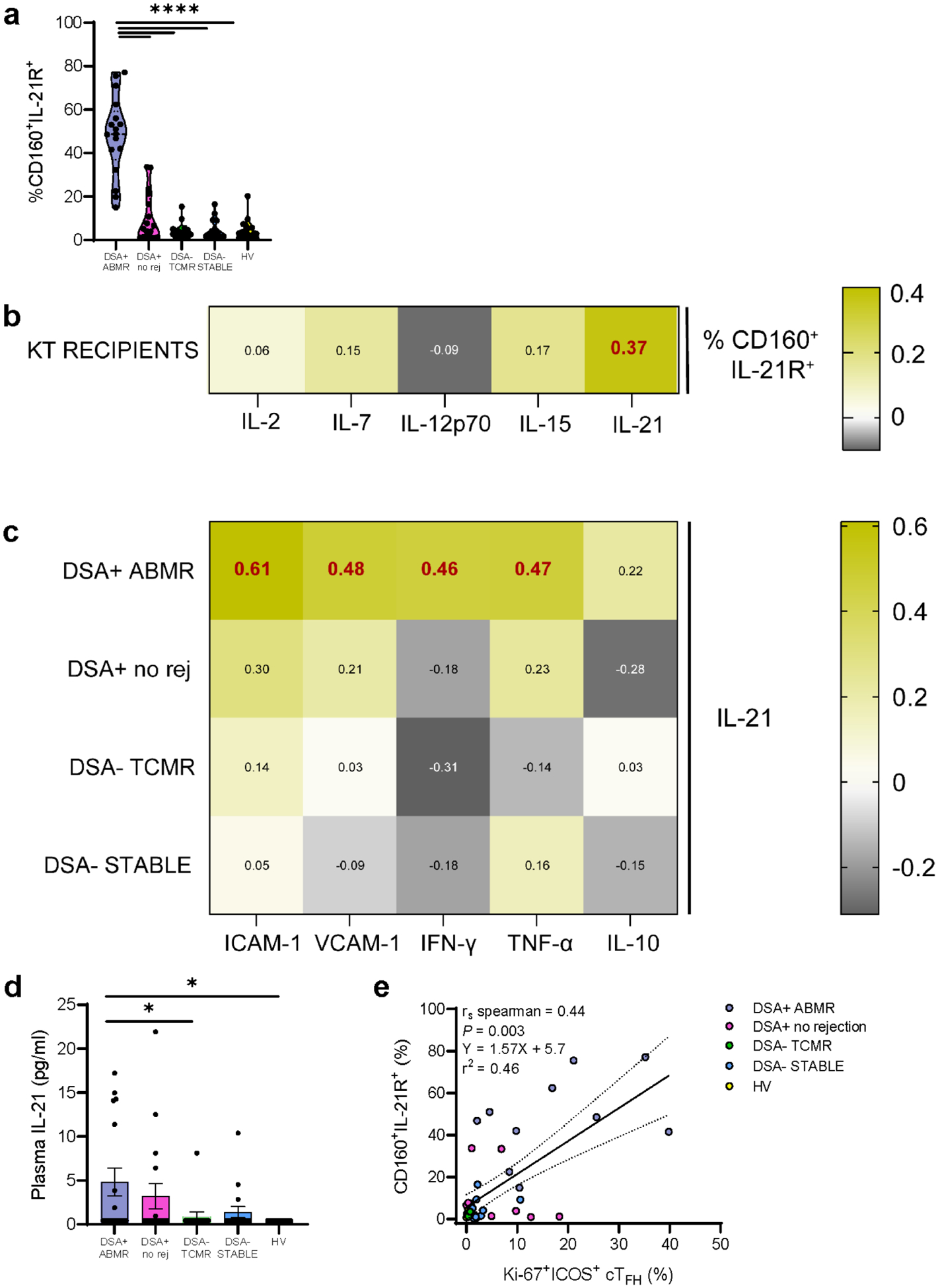
(a) Supervised quantification of CD160+IL-21R+CD56dimCD16bright NK cells in kidney transplant patients and healthy volunteers (HVs). Mann-Whitney test. ****p < 0.0001. (b) Matrix heatmap of Spearman rank correlations for the percentage of CD160+IL-21R+CD56dimCD16bright NK cells and plasma cytokine concentrations in kidney transplant recipients. rs coefficients in bold red have P < 0.05. (c) Matrix heatmap of Spearman rank correlations for the concentrations of plasma IL-21 and other cytokines as well as soluble adhesion molecules in the patient groups. rs coefficients in bold red have P < 0.05. (d) Plasma IL-21 concentration at the time of event. Mann-Whitney test. *P < 0.05. (e) Simple linear regression and Spearman rank correlation test for the percentage of CD160+IL-21R+CD56dimCD16bright NK cells and the percentage of Ki-67+ICOS+ cTFH cells in subjects phenotyped for both circulating NK cells and cTFH cells. n = 43 HVs/kidney transplant recipients. DSA+ ABMR, donor-specific alloantibody-positive mixed antibody-mediated rejection; DSA+ no rejection (no rej), donor-specific alloantibody-positive without microvascular inflammation/rejection episodes; DSA− STABLE, donor-specific alloantibody-negative without microvascular inflammation/rejection episodes; DSA− TCMR, donor-specific alloantibody-negative T cell-mediated rejection; ICAM-1, intercellular adhesion molecule-1; IFN-γ, interferon-γ; IL-21R, interleukin-21 receptor; TNF-α, tumor necrosis factor-α; VCAM-1, vascular cell adhesion molecule-1.
Plasma IL-21 correlates with CD160+IL-21R+CD56dimCD16bright NK cell phenotype and Type-1 pro-inflammatory cytokines released during DSA+ ABMR
We next inquired whether cytokines known to be involved in NK cell maturation and activation correlate with the observed NK cell phenotypic changes. Circulating CD160+IL-21R+CD56dimCD16bright NK cell frequency correlated with plasma IL-21 (Figure 2B). In DSA+ ABMR patients, plasma IL-21 also correlated with pro-inflammatory cytokines IFN-γ and TNF-α and with soluble ICAM-1 and VCAM-1, reflecting an elevated Type-1 inflammatory polarization and possible endothelial cell activation and injury24 (Figure 2C). Thus, phenotypic changes manifested by elevated CD160+IL-21R+ NK cells observed in DSA+ ABMR patients may occur in response to IL-21 and may subsequently contribute to endothelium injury via Type-1 effector mechanisms.
Activated cTFH correlate with CD160+IL-21R+CD56dimCD16bright NK cells and may represent a source of plasma IL-21 during DSA+ ABMR
We recently reported that Ki-67+ICOS+ cTFH cells produce elevated levels of IL-21 in DSA+ ABMR patients8. Here, plasma IL-21 was significantly elevated in DSA+ ABMR patients (Figure 2D). Moreover, a significant positive correlation between Ki-67+ICOS+ cTFH cell levels and CD160+IL-21R+CD56dimCD16bright NK cell frequency was observed (rs=0.44) (Figure 2E). These results suggest that Ki-67+ICOS+ cTFH cells may be a source of IL-21 for those DSA+ ABMR patients.
High levels of CD160+IL-21R+ expression associates with increased CD56dimCD16bright NK cell polyfunctionality
To further assess the functional potential of CD160+IL-21R+CD56dimCD16bright NK cells, we segregated KT recipients according to the percentage of CD160+IL-21R+ expression: 18 patients displayed high expression (≥75th percentile as described above) while 53 patients had low expression (Figure 3A). In CMC experiments, patients with high CD160+IL-21R+ displayed significantly higher polyfunctionality (e.g., degranulation (CD107a) + IFN-γ, IFN-γ + TNF-α) and with more single IFN-γ secretion (Figure 3B, Supplementary Figure S5A). In ADCC experiments, polyfunctional differences were also important (e.g., degranulation + IFN-γ and/or TNF-α) (Figure 3C, Supplementary Figure S5B). Among patients with a high CD160+IL-21R+CD56dimCD16bright NK cell frequency, 8 patients displayed high NKG2A expression. But despite NKG2A expression, high functionality was preserved (Supplementary Figure S5C and S5D).
Figure 3|. Polyfunctionaiity and higher pro-inflammatory cytokine release are observed in patients with a high percentage of CD160+IL-21R+CD56dimCD16bright natural killer (NK) cells.
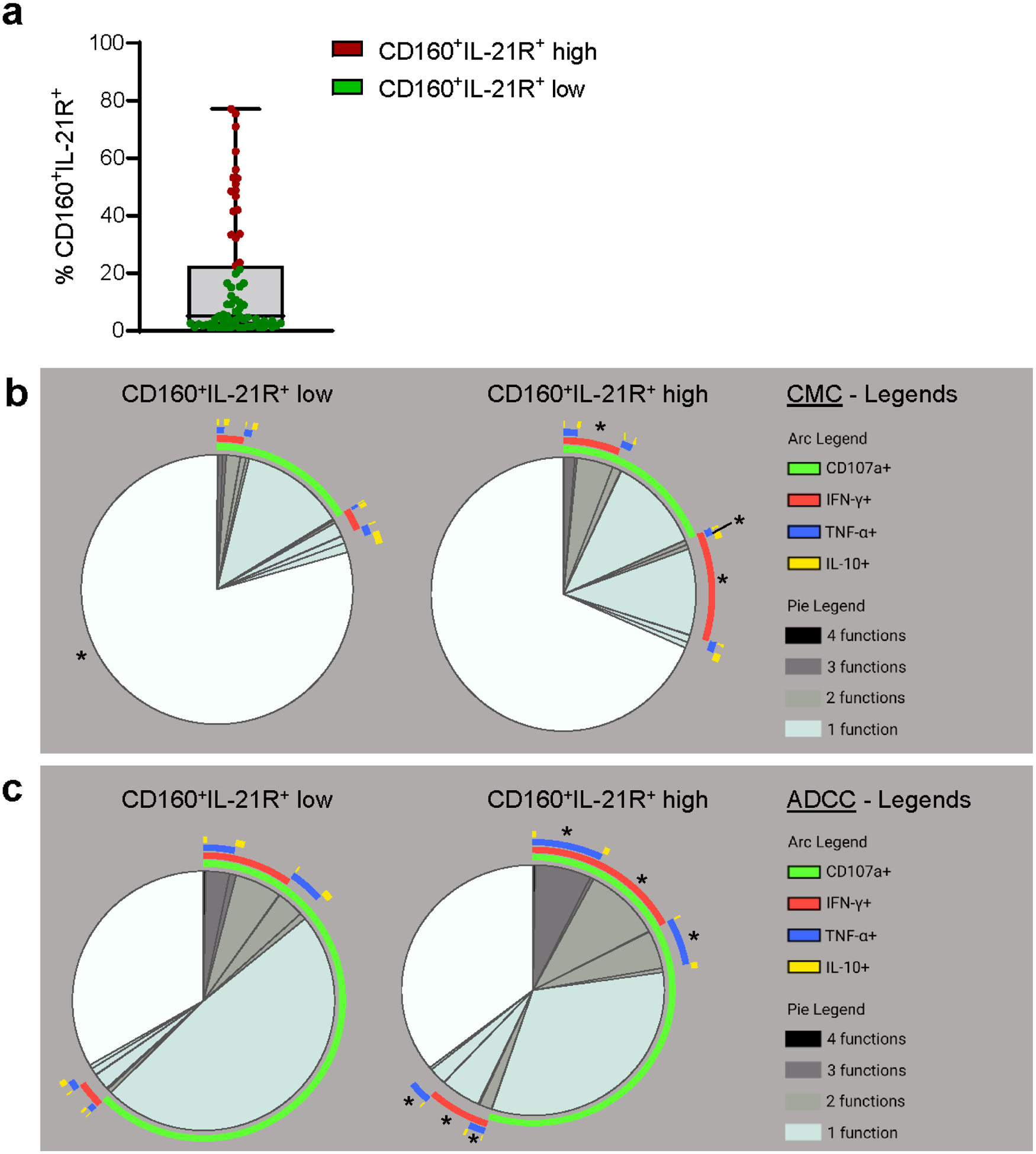
(a) Distribution of the percentage of CD160+IL-21+CD56dimCD16bright NK cells among the 71 kidney transplant recipients, identifying a group of 18 patients with high CD160+IL-21R+ expression (≥75th percentile) in red and 53 patients with low CD160+IL-21R+ expression in green. (b,c) CD56dimCD16bright NK cell degranulation and cytokine production in (b) cell-mediated cytotoxicity (CMC) and (c) antibody-dependent cellular cytotoxicity (ADCC) experiments according to CD160+IL-21R+ expression in 27 kidney transplant recipients with low (2 patients showing donor-specific alloantibody-positive mixed antibody-mediated rejection [DSA+ ABMR], 6 patients who were DSA-positive without microvascular inflammation/rejection episodes [DSA+ no rejection], 9 patients showing DSA-negative T cell-mediated rejection [DSA− TCMR], and 10 patients who were DSA-negative without microvascular inflammation/rejection episodes (DSA− STABLE]) versus 9 patients with high (6 DSA+ ABMR, 3 DSA+ no rejection) CD160+IL-21R+ expression. Boolean gates were created for CD 107a, interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and interleukin-10 (IL-10). Polyfunctional data were analyzed using the Simplified Presentation of Incredibly Complex Evaluations (SPICE) software (National Institutes of Health and National Institute of Allergy and Infectious Diseases). The pie charts display the mean frequencies of degranulating and/or cytokine-secreting CD56dimCD16bright NK cells for each possible combination of CD107a/IFN-γ/TNF-α/IL-10. Pie segments are color coded to represent monofunctionality to polyfunctionaiity. The arcs illustrate the analyte(s) produced by the pie segment underneath. Mann-Whitney tests were used to compare each pie segment between the 2 groups. Significance symbols were displayed on the pie segment with increased proportion. *P < 0.05.
Thus, through CD16a crosslinking by DSA and activating cytotoxic receptors trigger, CD160+IL-21R+CD56dimCD16bright NK cells are likely poised to augmented deleterious cytotoxic and pro-inflammatory activity in vivo during DSA+ ABMR.
Recombinant (r)IL-21 can induce CD160+IL-21R+CD56dimCD16bright NK cells to elevate Type-1 phenotype and function in vitro
We next inquired whether rIL-21 could promote in vitro phenotypic and functional changes to CD56dimCD16bright NK cells, similar to those observed ex vivo in DSA+ ABMR patients. rIL-21 triggered a significant up-regulation of IL-21R, Type-1 TF EOMES and Tbet as well as CD160 and NKG2D in CD56dimCD16bright NK cells (Figure 4A). NKG2A was increased but CXCR3 and CD161 were not enhanced by rIL-21. These phenotypic changes were inhibited by IL-21R-Fc antagonist treatment (Figure 4B).
Figure 4|. Recombinant interleukin-21 (rlL-21) activates CD56dimCD16bright natural killer (NK) cells in vitro, enhancing their type 1 polarization and cytotoxicity.
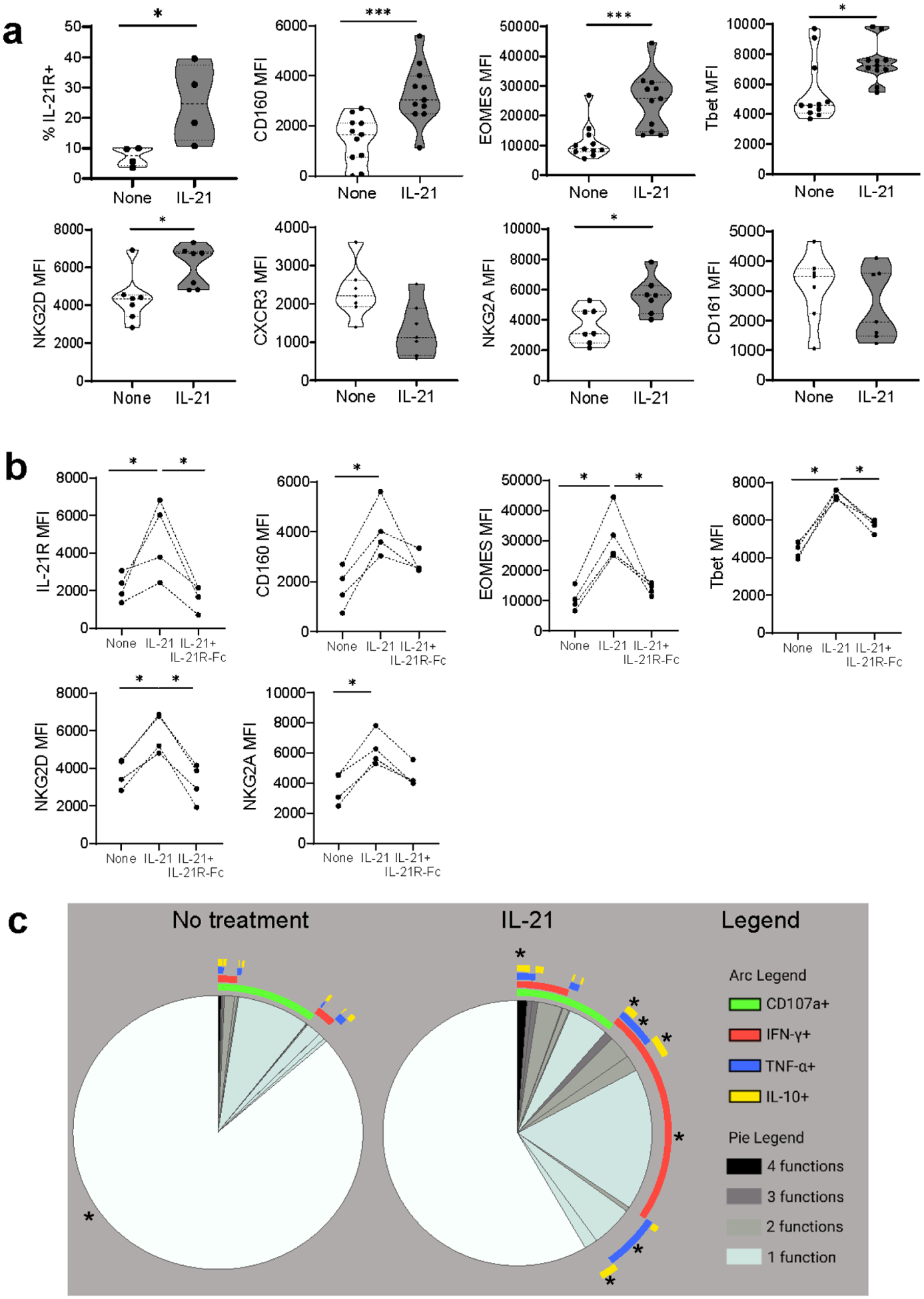
(a) Phenotypic NK cell changes after 5-day in vitro stimulation with rlL-21. n = 4–11 healthy volunteers (HVs). Mann-Whitney test, (b) Phenotypic modifications after 5-day rlL-21 stimulation ± IL-21 receptor (IL-21 R) antagonist (IL-21 R-Fc). n = 4 HVs. Mann-Whitney test, (c) CD56dimCD16bright NK cell degranulation and cytokine production after 5-day rlL-21 stimulation, n = 12 HVs. Boolean gates were created for CD107a, interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and interleukin-10 (IL-10). Polyfunctional data were analyzed using the Simplified Presentation of Incredibly Complex Evaluations (SPICE) software. The pie charts display the mean frequencies of degranulating and/or cytokine-secreting CD56dimCD16bright NK cells for each possible combination of CD107a/IFN-γ/TNF-α/IL-10. Pie segments are color coded to represent monofunctionality to polyfunctionaiity. The arcs illustrate the analyte(s) produced by the pie segment underneath. Mann-Whitney tests were used to compare each pie segment between the 2 groups. Significance symbols were displayed on the pie segment with increased proportion. *P < 0.05, ***P < 0.001. CXCR3, CXC chemokine receptor 3; MFI, mean fluorescence intensity; NKG2A, natural killer group 2A; NKG2D, natural killer group 2D.
Moreover, rIL-21 stimulation enhanced polyfunctionality and elevated single or multiple pro-inflammatory cytokine release (Figure 4C, Supplementary Figure S6A), all inhibited by IL-21R-Fc treatment (Supplementary Figure S6B). Also, rIL-21-pretreated CD56dimCD16bright NK cells performed better in ADCC (Supplementary Figure S6C and S6D).
These results support the critical role of IL-21 in inducing CD160+IL-21R+CD56dimCD16bright NK cell phenotypic changes and augmenting their Type-1 cytotoxic and pro-inflammatory activity during DSA+ ABMR.
Transcriptomic modifications of blood circulating CD56+CD16bright NK cells during DSA+ ABMR
To further scrutinize the transcriptional programs underlying the expansion of the cytotoxic IL-21-responsive subset during DSA+ ABMR, we analyzed FACS-sorted circulating CD56+ NK cells by 3Pv3 single-cell RNA-sequencing (21,888 NK cells from 4 DSA+ ABMR and 4 DSA− TCMR). All 4 DSA+ ABMR displayed the flow cytometry phenotypic cluster 2 described above. Several gene expression patterns were noted from the UMAP with rejection-specific areas (Figure 5A). Unsupervised clustering revealed 2 clusters with high FCGR3A transcript (inferring their CD56+CD16bright phenotype) expanded in DSA+ ABMR patients: cluster 2 (red) and 4 (green) (Figure 5B, 5C and 5D, Supplementary Table S4, and S5). Cluster 2, present in 3/4 DSA+ ABMR patients was over-represented as compared to DSA− TCMR patients. It was characterized by up-regulated transcripts and impacted pathways related to FcγR signaling, cell activation, cytotoxicity, and pro-inflammatory state (Figure 5E and 5F, Supplementary Figure S7A). CD160, KLRK1 (NKG2D) and their signaling, IFNG signaling as well as other Type-1-related cytotoxic/pro-inflammatory effector genes: EOMES, TBX21, IL-21-induced GZMA/B, PRF1 (perforin), CCL3, CCL4 were noted elevated, with an inactivated IL-10 signaling pathway. Importantly, IL-21 signaling related genes/pathways (IL2RG encoding for the cytokine receptor common subunit gamma that is part of IL-21R and STAT3 pathway) were upregulated. We discerned common genes for IL-21, FcγR and NKG2D signaling (JAK1/3, AKT1/2, MAP2K1-MAPK1). Some features related to activation regulation were expressed (KLRB1 (CD161)). Leukocyte trans-endothelial migration, integrins, actin cytoskeleton regulation for focal adhesion pathways were also upregulated suggesting high capacity for diapedesis and migration (Figure 5F).
Figure 5|. Three prime v3 single-cell RNA sequencing of fluorescence-activated cell sorting (FACS)–sorted natural killer (NK) cells from patients with donor-specific alloantibody-positive mixed antibody-mediated rejection (DSA+ ABMR) identifies the expansion of a transcriptomic signature with cytotoxicity features and interleukin-21 (IL-21)/signal transducer and activator of transcription 3 (STAT3) pathway as compared with patients with DSA-negative T cell-mediated rejection (DSA− TCMR).
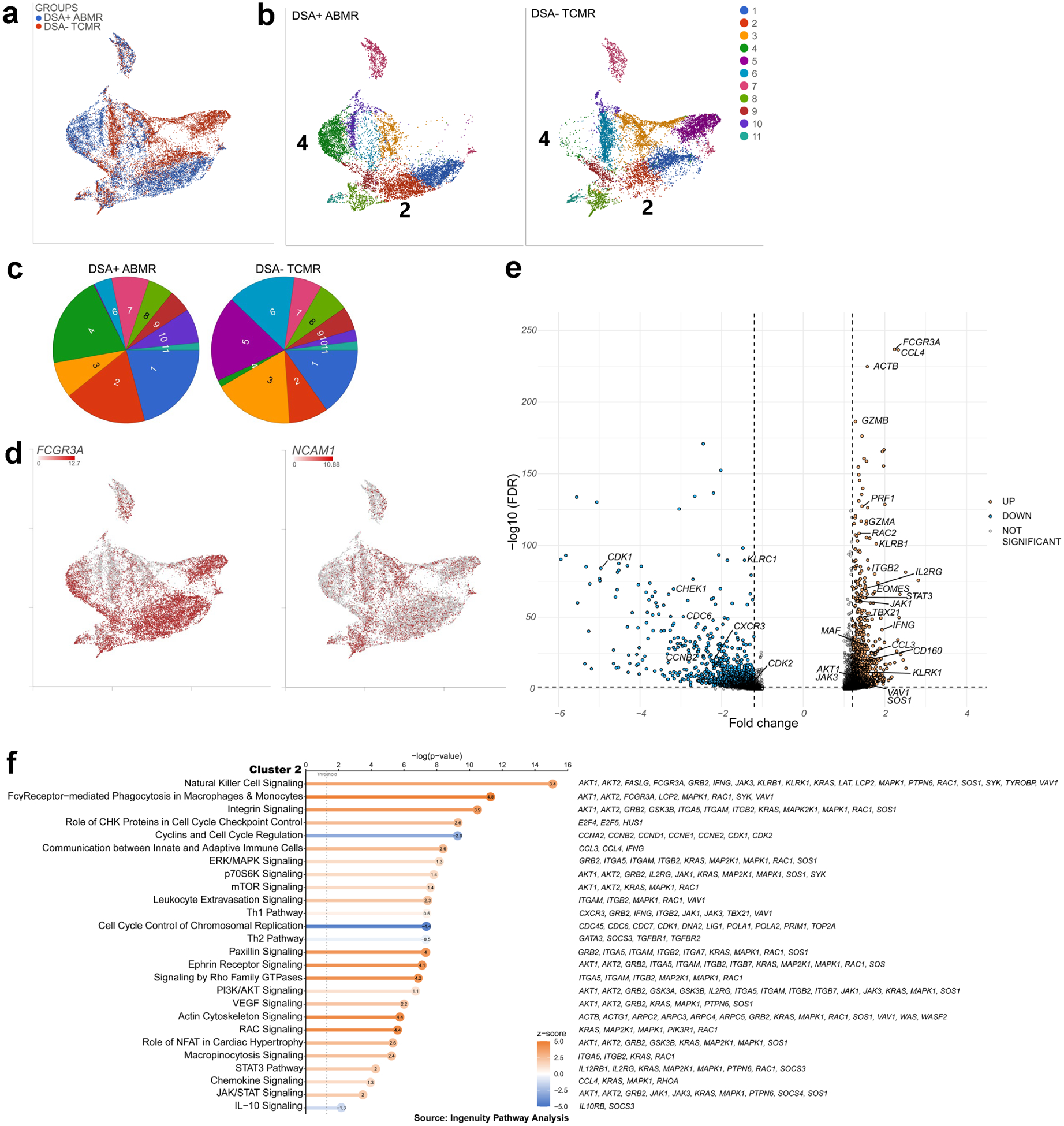
(a) Uniform manifold approximation and projection (UMAP) displaying the DSA+ ABMR and DSA− TCMR groups, n = 4 in each group, (b) UMAP displaying the graph-based clustering split by group, (c) Cluster frequency in the 2 groups, (d) UMAP featuring expression of Fc gamma receptor 3 A (FCGR3A) and neural cell adhesion molecule 1 (NCAM1) transcripts, (e) Volcano plot of selected differentially expressed genes from the analysis of variance of cluster 2 versus the other graph-based clusters. Significance was defined as fold change ≤−1.2 or ≥1.2 and false discovery rate (FDR) ≤0.05. (f) Pathway analysis (Ingenuity Pathway Analysis) of cluster 2 differentially expressed genes. Pathways are sorted by P value, and z-scores are displayed by color, with the z-score values inside the dots. Selected genes participating in each pathway are displayed on the right. AKT, serine/threonine-protein kinase AKT; CHK, checkpoint kinase; ERK, extracellular signal-regulated kinase; IL-10, interleukin-10; JAK, Janus kinase; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; NFAT, nuclear factor of activated T cells; PI3K, phosphatidylinositol-3'-kinase; RAC, rac family small GTPase; Th1, T helper cell 1; Th2, T helper cell 2; VEGF, vascular endothelial growth factor.
Cluster 4, expanded in one DSA+ ABMR patient, had common features with cluster 2 with FCGR3A signaling, a pro-inflammatory profile (Th1 pathway, IFN-γ-induced IRF1, CD160, GZMB, CCL3/5, and STAT3). But it displayed more inhibitory features: LAG3, CTLA4, KLRB1, KLRC1 (NKG2A) (Supplementary Figure S7B and S7C), compatible with signs of NK cell exhaustion in a context of chronic inflammation and antigen exposure25. Clusters 5 and 6 expanded in DSA− TCMR patients did not share those features listed above.
In summary, the transcriptional state of CD56+CD16bright NK cells during DSA+ ABMR is distinguishable from DSA− TCMR by the enrichment of cells with uniquely activated programs confirming the phenotypic observations and identifying novel features of high ability migration to the inflamed tissue.
A high CD160+IL-21R+CD56dimCD16bright NK cell frequency is associated with severe graft outcomes
We next evaluated the clinical relevance of the CD160+IL-21R+CD56dimCD16bright NK cell subset. Patients with a high percentage (≥75th percentile, n=18) of CD160+IL-21R+CD56dimCD16bright NK cells manifested significantly more severe ABMR lesions [worse MVI (g+ptc) and tubulointerstitial inflammation (i+t)] than patients with a low cell percentage (n=53) (Figure 6A). Among ABMR patients, those with a high percentage (n=15) of CD160+IL-21R+CD56dimCD16bright NK cells had a trend for more severe acute lesions. Moreover, those patients had the worse death-censored graft survival at 80 months post-KT (Figure 6B). In a Cox univariate analysis, the percentage of CD160+IL-21R+CD56dimCD16bright NK cells significantly increased the risk of graft loss (HR 4.46 (1.66–11.97), P = 0.003).
Figure 6|. High CD1 60+IL-21R+CD56dimCD16bright natural killer (NK) cell frequency is associated with severe graft outcomes.
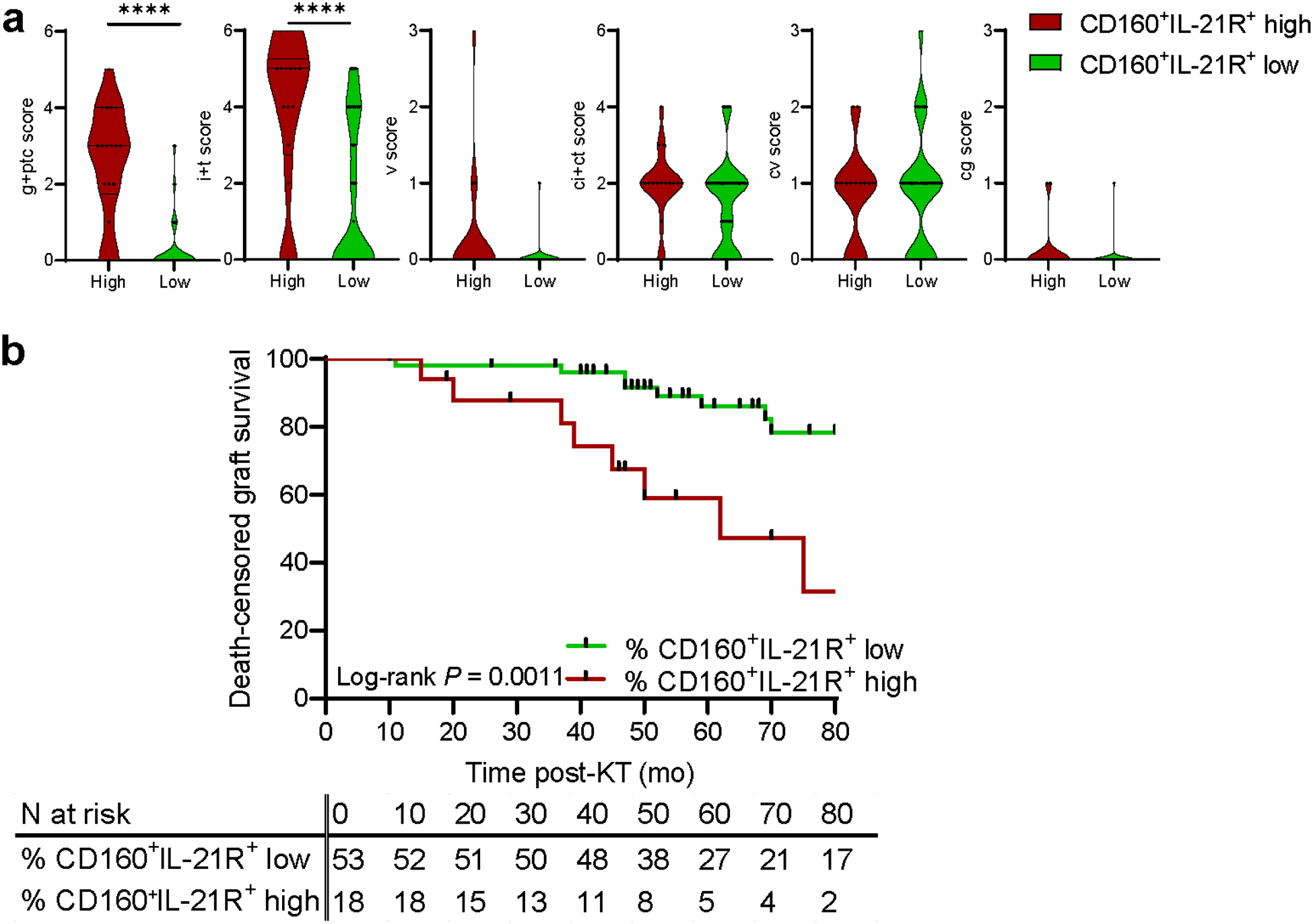
(a) Banff lesions severity according to low or high CD160+IL-21R+CD56dimCD16bright NK cell percentage (based on the 75th percentile = 22.5%) in the 71 kidney transplant recipients. Mann-Whitney test. ****P < 0.0001. (b) Death-censored allograft survival of the 71 patients stratified into 2 subgroups of CD160+IL-21R+CD56dimCD16bright NK cells (based on the 75th percentile = 22.5%). Kaplan-Meier curve of death-censored graft survival and log-rank test, cg, transplant glomerulopathy; ci + ct, interstitial fibrosis and tubular atrophy; cv, chronic vasculopathy; g + ptc, glomerulitis + peritubular capillaritis (microvascular inflammation); i + t, interstitial inflammation + tubulitis; v, arteritis.
Thus, the magnitude of CD160+ IL-21R+CD56dimCD16bright NK cell expansion is associated with tissue injury and long-term graft survival.
CD160+IL-21R+CD56+CD16+ NK cells infiltrate kidney allografts during DSA+ ABMR
To gain insight whether the described NK cell subset may be detected within the graft, we performed multispectral immunofluorescent staining and bulk RNA-sequencing on kidney allograft biopsies. NK cell infiltration was defined as CD3−CD56+CD16+ cells and analyzed for IL-21R and CD160 co-expression in the intravascular compartment (glomeruli, peritubular capillaries, arteries defined by CD31) or the extravascular compartment (the rest of the tissue) (Supplementary Figure S8A). As surface CD16 can be shed after NK cell activation, phenotype quantification independently of CD16 expression was also scrutinized. Overall, NK cell infiltration was more frequent in DSA+ ABMR (allograft nephrectomy with chronic mixed ABMR or mixed ABMR needle biopsies) compared to DSA− TCMR biopsies, in both compartments (Supplementary Figure S8B). Cell phenotyping confirmed the presence of NK cell CD16+ with various co-expression of CD160 and/or IL-21R (Figure 7A and 7B). CD160+ and IL-21R+ densities were increased during DSA+ ABMR (Figure 7B). CD16+CD160+IL-21R+ NK cells were observed intravascularly in the graft nephrectomy and infiltrated the interstitium in both the graft nephrectomy and the DSA+ ABMR biopsies but not during DSA+TCMR (Figure 7A and 7C). A potential for interstitial infiltration with cell adhesion and diapedesis activated pathways was anticipated transcriptionally as described above (Figure 5F).
Figure 7|. CD160+IL-21R+CD56+CD16+ natural killer (NK) cells infiltrate kidney allografts during donor-specific alloantibody-positive antibody-mediated rejection (DSA+ ABMR).
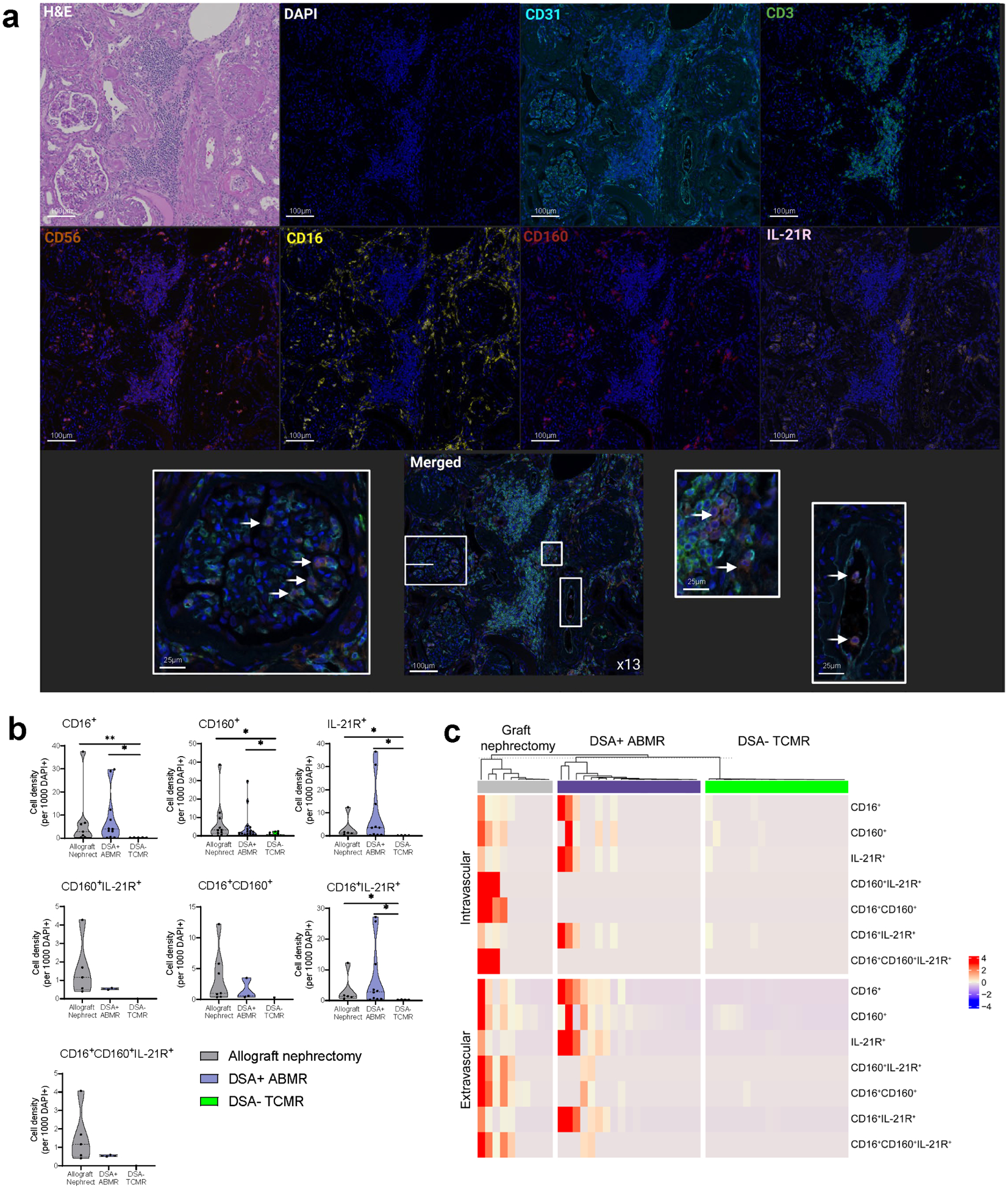
(a) Multispectral immunofluorescence of an allograft nephrectomy (nephrect) biopsy diagnosed with chronic mixed ABMR. Infiltrating NK cells were defined as CDB−CDS6+ cells and analyzed for CD16, CD160, and/or interleukin-21 receptor (IL-21R) expression, (b) Cell density of infiltrating NK cells with different coexpression of CD16, CD160, and/or IL-21R calculated in each 10 regions of interest (ROIs) per biopsy. Five biopsies were analyzed: an allograft nephrectomy biopsy with chronic mixed ABMR, 2 donor-specific alloantibody-positive mixed antibody-mediated rejection (DSA-f mixed ABMR) biopsies, and 2 DSA-negative T cell-mediated rejection (DSA− TCMR) needle biopsies. ROIs with no cells were excluded. Mann-Whitney t test. *P < 0.05, **P < 0.01. (c) Heatmap representing normalized cell density for different NK cell phenotypes (CD3−CD56+ with different coexpression of CD16, CD160, and/or IL-21R) in each ROI of the intravascular compartment (glomerular, peritubular capillaries, and arteries) and the extravascular compartment (the rest of the tissue) from all biopsies. Cell density was calculated for 1000 4',6-diamidino-2-phenylindole-positive (DAPI+) cells in each tissue compartment. ROIs with no cells were excluded. Unsupervised clustering with Euclidean distance (R, ComplexHeatmaps package26). H&E, hematoxylin and eosin. To optimize viewing of this image, please see the online version of this article at www.kidney-international.org.
To confirm these observations, 13 DSA+ ABMR, 9 DSA− TCMR and 6 DSA− STABLE (rejection-free) allograft biopsies were analyzed by RNA-sequencing. The transcriptomic profile associated with DSA+ ABMR notably diverged from both DSA− TCMR (Figure 8A, Supplementary Table S6) and DSA− STABLE (Figure 8A, Supplementary Table S7). Significant canonical pathways implied NK cell involvement with pro-cytotoxic molecular features (e.g., FCGR3A, TBX21, IL21R and IL2RG, Interferon signaling, KLRK1, CXCR3, PRF1) (Supplementary Table S7). Transcripts for the described circulating NK cell subset features were increased in DSA+ ABMR biopsies (FCGR3A, CD160, IL21R, EOMES, TBX21, KLRK1, CXCR3) (Figure 8B).
Figure 8|. Transcriptomic profile of allograft infiltrating CD160+IL-21R+FCGR3A+ natural killer (NK) cells shows pro-inflammatory and cytotoxic features.
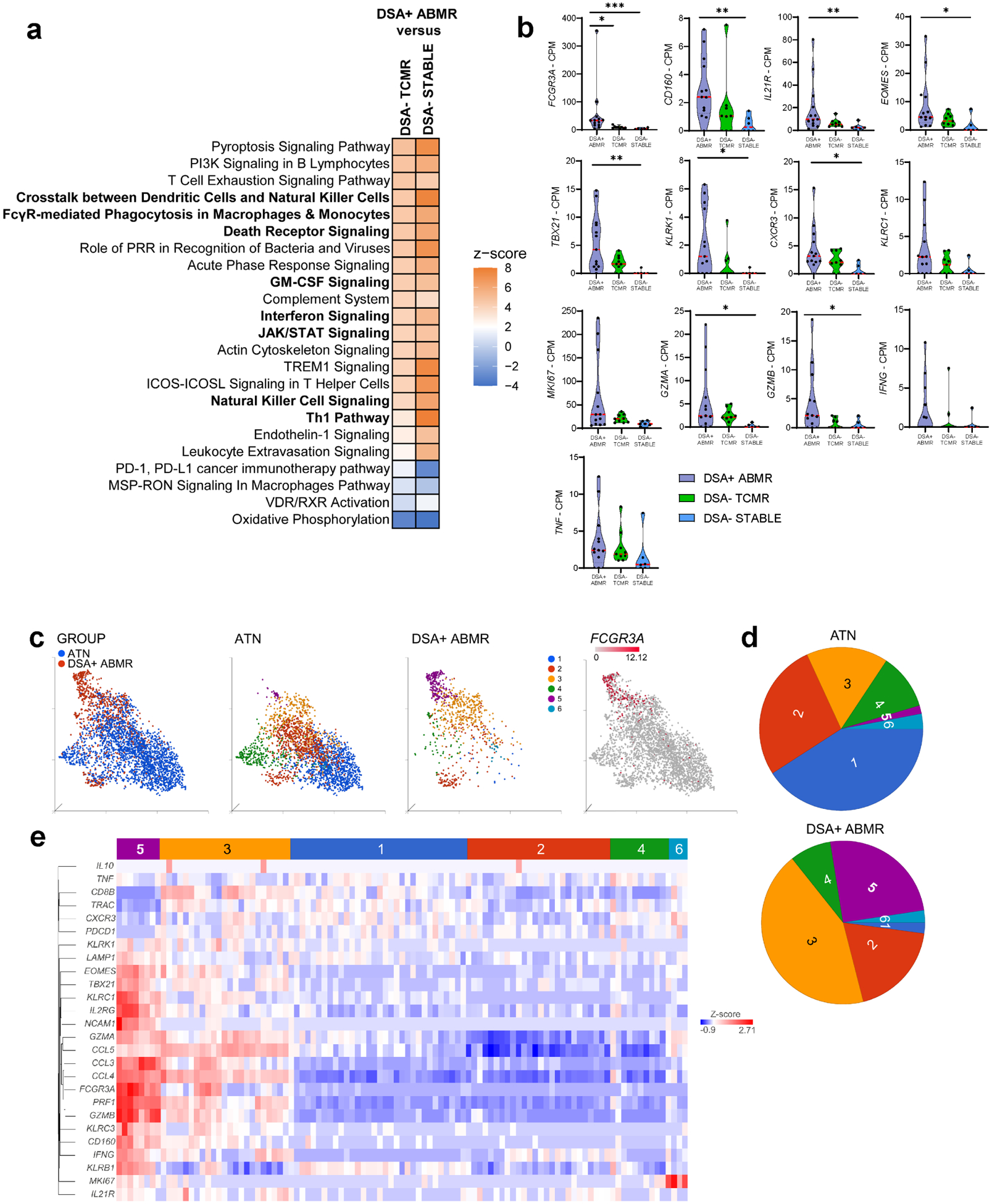
(a,b) Bulk RNA-sequencing analysis comparing donor-specific ailoantibody-positive mixed antibody-mediated rejection (DSA+ mixed ABMR; n = 13) biopsies with DSA-negative T cell-mediated rejection (DSA− TCMR; n = 9) biopsies or with DSA− negative without microvascular inflammation/rejection episodes (DSA− STABLE; n = 6) biopsies, (a) Transcriptomic analysis of infiltrating NK cell-related signatures in kidney allograft biopsies. All pathways are significantly modified and are ordered by z score in the comparison of DSA+ ABMR and DSA− TCMR. Canonical pathways displayed in bold suggest NK cell involvement, (b) Quantification of NK cell–related transcripts in kidney allograft biopsies: TBX21 (Tbet), MKI67 (Ki-67), KLRK1 (natural killer group 2D [NKG2D]), and KLRC1 (natural killer group 2A [NKG2A]). Kruskal-Wallis test with the Dunn post hoc test. *P < 0.05, **P < 0.01, ***p < 0.001. (c-e) Single-cell RNA sequencing of acute tubular necrosis (ATN; n = 2) and DSA+ ABMR (n = 3) biopsies from the Gene Expression Omnibus repository (GSE145927). (c) Uniform manifold approximation and projection plots representing the 2-group distribution (left), the graph-based clustering split by group (middle), and the expression value of FCGR3A transcript (right), (d) Graph-based cluster frequency in the 2 groups, (e) Heatmap representing scaled expression values of NK cell–related transcripts described in the phenotypic analysis. CPM, count per million; CXCR3, CXC chemokine receptor 3; FCGR3A, Fc gamma receptor 3 A; GM-CSF, granulocyte-macrophage colony-stimulating factor; ICOS, inducible T-cell costimulator; ICOSL, inducible T-cell costimulator ligand; IL-21 R, interleukin-21 receptor; JAK, Janus kinase; MSP-RON, macrophage-stimulating protein-récepteur d'origine nantais; PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1; PI3K, phosphatidylinositol-3’-kinase; PRR, pattern recognition receptor; RXR, retinoid X receptor; STAT, signal transducer and activator of transcription; TREM1, triggering receptor expressed on myeloid cells 1; VDR, vitamin D receptor.
Furthermore, we interrogated independently published single-cell RNA-sequencing data from 5 kidney allograft biopsies (3 DSA+ ABMR, 2 ATN rejection-free). Among NK and NKT cells, cluster 5 identified as NK cell-FCGR3A enriched was unique to ABMR (Figure 8C and 8D). It exhibited higher expression of IL2RG, CD160, TBX21, EOMES transcripts, transcripts for activating/cytotoxic genes (KLRK1, IFNG, GZMA/B, PRF1, CCL3/4/5) as well as inhibitory receptor genes (KLRB1, KLRC1) features similar to our own findings (Figure 8E). Furthermore, endothelial cells displayed significantly increased transcripts related to caspases (CASP6, CASP7, CASP8, CASP10) and to mediators of cell apoptotic signals (TNFSF10, TNFRSF10A/B, FADD, TP53) in DSA+ ABMR biopsies, reflecting ongoing apoptosis processes (Supplementary Figure S8C). Gene set enrichment analysis confirmed the strong increase of apoptotic processes (Supplementary Figure S8D). Thus, at the transcriptomic level, CD16+ NK cells with elevated Type-1 cytotoxic programs and IL-21 responsiveness signature together with endothelial cells enriched in apoptosis-related genes were concomitantly present in DSA+ ABMR kidney biopsies.
Thence, this novel circulating cytotoxic CD160+IL-21R+CD56dimCD16bright NK cell subset could infiltrate the allograft during DSA+ ABMR, inferring that it may directly be deleterious to the endothelium by CMC, ADCC and by pro-inflammatory molecule release (Supplementary Figure S9).
DISCUSSION
While the involvement of NK cells during kidney allograft rejection has been increasingly described in animal models26 and more recently in humans, the role of potentially heterogeneous NK cell subsets involved in different rejection types remains to be investigated. Missing self-induced NK cell activation proved to play a role in complement-independent chronic ABMR outcome27. Since not all patients with predicted killer Ig-like receptors mismatch develop pathological lesions, other mechanisms, yet to be identified, could contribute to NK cell cytotoxicity towards the vascular endothelium. Modulation of NK cell inhibitory receptors were observed in patients with anti-HLA antibodies (DSA or non-DSA) compared to patients with no antibodies, suggesting NK cell phenotypic modifications during a humoral response28.
Our data demonstrate that circulating CD56dimCD16bright NK cells from DSA+ ABMR patients uniquely overexpress IL-21R, correlate with elevated plasma IL-21, and upregulate IL2RG and STAT3 transcripts/pathways. Interestingly, we reported that donor-reactive cTFH cells were detected in KT recipients who further develop DSA29. While those cTFH can help activated memory B cells differentiate into DSA-producing plasma cells in an IL-21-dependent manner30, here we report for the first time that IL-21 may also help NK cells differentiate into polyfunctional Type-1 activated NK cells with a potential deleterious ability to harm the vascular endothelium. Indeed, rIL-21 induced Type-1 TF and cytotoxic receptors expression in vitro, and promoted pro-inflammatory polyfunctionality, supporting its pivotal role in NK cell changes during DSA+ ABMR.
IL-21 has been described to augment NK cell cytotoxic activity with enhanced Type-1-like cytotoxicity and ADCC in healthy subjects20,31,32. In vitro-generated NK cells overexpressing T-bet or EOMES improved functionality and target cell killing33. Interestingly, the IL-21R+CD56dimCD16bright subset identified here also showed increased cytotoxic molecules co-expression and elevated CMC. CD160 transcript has been previously observed in ABMR kidney biopsies14. Its engagement with MHC molecules was shown to trigger cytotoxicity and pro-inflammatory mediator production34 as we observed here. NKG2D, co-expressed by the same phenotypic cluster, may also participate in increased cytotoxicity. NKG2D proved necessary for chronic cardiac allograft vasculopathy in a mouse model35. DSA-dependent ADCC potential was also increased in CD160+IL-21R+CD56dimCD16bright NK cells, certainly with a role of IL-2132. In KT, a higher ADCC responsiveness has been associated with reduced graft function36. Here, we report that high percentages of circulating CD160+IL-21R+CD56dimCD16bright NK cell were associated with worse graft survival.
Our data provide novel evidence that during humoral alloimmune response, a potential crosstalk between IL-21-secreting cTFH cells and IL-21-responsive Type-1 CD56dimCD16bright NK cells occurs, bridging innate to adaptive immunity against donor. Activated proliferating cTFH cells could play a key role in influencing NK cell phenotype and function. We further postulate that these NK cells are directly deleterious against the kidney allograft vascular endothelium via CMC, ADCC and release of pro-inflammatory mediators, and in turn may have the potential to sustain pathogenic cTFH cell activity through IFN-γ and TNF-α feedback help (Supplementary Figure S9). Similarly, NK cells from patients diagnosed with myasthenia gravis were found to promote the differentiation and activation of TFH cells37.
The overexpression of CXCR3 in this phenotype, likely not augmented by IL-21, may facilitate attraction to inflamed tissue, as it has been described an accumulation of CXCR3+ immune cells in kidney acute vascular rejection38. Moreover, pathways related to leukocyte trans-endothelial migration and diapedesis were upregulated in our DSA+ ABMR cases and such CD160+/IL-21R+ NK cells were observed in our DSA+ ABMR KT biopsies, strongly supporting their potentially detrimental role locally.
While our observational pilot study encompassed a relatively small cohort, these patients were clinically, histologically and immunologically carefully characterized, and with a long-term follow-up. It was reported that NK cell phenotype can be modified by maintenance immunosuppressive drugs39. However, their usage was not different among our groups. Other confounding factors (demographical and histological) were found to participate in the poorer graft survival aside high CD160+IL-21R+ circulating NK cell phenotype in the univariate Cox analysis, but a multivariate analysis was not appropriate to run due to multicollinearity with the DSA characteristics and pathology. Nonetheless, the presence of this newly described circulating NK cell subset in DSA+ ABMR patients was confirmed by single-cell transcriptional analysis of circulating NK cells. Importantly, cells with similar phenotypic and transcriptional profiles were found infiltrating DSA+ ABMR kidney biopsies. Interestingly, such cells were observed in a failed allograft removed for severe mixed ABMR (Figure 7) suggesting their persistence in the tissue despite repeated therapeutic strategies.
Future multi-center studies on larger populations, with longitudinal analysis and comparison with other clinical presentations (pure versus mixed ABMR) will be needed to further confirm the potential of this NK cell subset in immune monitoring.
Targeting IL-21 signaling could be an emerging therapeutic strategy for interfering with both cTFH cell and NK cell pathogenesis during humoral rejection. A fully human monoclonal IgG1 anti-IL-21R Ab (ATR-107) proved to delay allograft rejection in a humanized mouse skin transplant model40, but its development was stopped due to high immunogenicity in healthy volunteers41. An early safety PK/PD trial with a human anti-rIL-21 monoclonal Ab (NNC0114-0005) was conducted in rheumatoid arthritis patients with preliminary encouraging signs of reduced disease activity42. Yet, no therapeutic agent blocking the IL-21/IL-21R axis is ready for clinical use and more data may be needed to elucidate the optimal administration schedule. In a BXSB-Yaa mouse model of systemic lupus erythematosus, IL-21 blockade by a soluble IL-21R-Fc showed a biphasic effect with disease exacerbation in the early phase but benefits at later timepoints43. Moreover, safety data will be essential in transplant patients already at higher risk of infection and neoplastic complications.
Supplementary Material
Supplementary Methods.
Supplementary Figure S1 | Flow chart, study outline and description of DSA characteristics in the DSA+ groups.
Supplementary Figure S2 | Gating strategy for CD56dimCD16bright NK cells and T2 cell line characterization.
Supplementary Figure S3 | CD56dimCD16bright NK cell frequency, count and phenotypic modifications.
Supplementary Figure S4 | CITRUS predictive analysis integrating the 9 NK cell markers and comparing DSA+ ABMR and DSA− TCMR patients.
Supplementary Figure S5 | Increased CD160+ IL-21R+ CD56dimCD16bright NK cell polyfunctionality during CMC and ADCC.
Supplementary Figure S6 | rIL-21 enhances CD56dimCD16bright NK cell polyfunctionality in vitro.
Supplementary Figure S7 | Pathway analyses of circulating NK cell single-cell RNA-sequencing clusters.
Supplementary Figure S8 | NK cells infiltrate kidney allografts and endothelial cells undergo apoptosis during DSA+ ABMR
Supplementary Figure S9 | Schematic representation of a hypothetical model based on our study findings.
Supplementary Table S1 | Cohort’s demographics (for the NK cell phenotypic, functional, transcriptomic analysis).
Supplementary Table S2 | Fluorochrome-tagged Abs used with flow cytometry.
Supplementary Table S3 | Patients’ demographics for bulk RNA-sequencing of allograft biopsies.
Supplementary References.
Supplementary Table S4 | Pathway-related genes for the circulating NK cell single-cell RNA-sequencing analysis of cluster 2.
Supplementary Table S5 | Pathway-related genes for the circulating NK cell single-cell RNA-sequencing analysis of cluster 4.
Supplementary Table S6 | Pathway-related genes for kidney allograft biopsy bulk RNA-sequencing analysis. Comparison DSA+ ABMR versus DSA− TCMR.
Supplementary Table S7. Pathway-related genes for kidney allograft biopsy bulk RNA-sequencing analysis. Comparison DSA+ ABMR versus DSA− STABLE.
ACKNOWLEDGMENTS
We thank Dr Douglas Landsittel (currently at the Epidemiology and Biostatistics department, Indiana University, Bloomington IN, USA) and Avantika Srivastava for the advice on biostatistics, Dr Chris Fleming (Cytek) for the help in the designing of the spectral flow cytometry NK cell panel; Dr Srilakshmi Chaparala and Dr Ansuman Chattopadhyay (Health Sciences Library System, University of Pittsburgh), Dr Xiaowen Wang (Partek) and Tracy Tabib (Single Cell Core, University of Pittsburgh) for the help in the generation and analysis of single-cell RNA-sequencing data; Michelle Lucas and David McMichael (Thomas E. Starzl Transplantation Institute) for the help with clinical data collection. We are grateful to Dr Walter Storkus (Department of Dermatology, University of Pittsburgh) for kindly providing the 174xCEM.T2 cell line. We thank Dr Marion Joy (Translational Pathology Imaging Lab, UPMC Hillman Cancer Center) for the performance of the kidney allograft tissue staining, the imaging, and the generation of the phenotyping dataset. We also thank the Unified Flow Core, Department of Immunology, University of Pittsburgh as well as the Center for Research Computing and the Health Sciences Library System, University of Pittsburgh.
Sources of support:
This work was supported by a National Institutes of Health grant [R01AI130010 (DM)]; the Human Immunology Program at the Thomas E. Starzl Transplantation Institute and a Thomas E. Starzl Postdoctoral Fellowship in Transplantation Biology at the University of Pittsburgh (EB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE
None of the authors has relevant disclosures and conflicts of interest.
DATA SHARING
All data associated with this study are present in the paper and Supplementary Materials. The single-cell RNA-sequencing dataset from FACS-sorted circulating NK cell for DSA+ ABMR and DSA− TCMR patients has been deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE216203 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216203). The RNA-sequencing of kidney allograft biopsies dataset has also been deposited and is accessible through GEO Series accession number GSE214703 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214703). The dataset of single-cell RNA-sequencing of kidney allograft biopsies used in this manuscript was downloaded from the GEO database (GSE145927, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145927).
REFERENCES
- 1.Callemeyn J, Lamarthée B, Koenig A, et al. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022;101:692–710. [DOI] [PubMed] [Google Scholar]
- 2.Lebraud E, Eloudzeri M, Rabant M, et al. Microvascular Inflammation of the Renal Allograft: A Reappraisal of the Underlying Mechanisms. Front Immunol. 2022;13:864730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mengel M, Loupy A, Haas M, et al. Banff 2019 Meeting Report: Molecular diagnostics in solid organ transplantation-Consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am J Transplant. 2020;20:2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis K, Macedo C, Lefaucheur C, et al. Adaptive immune cell responses as therapeutic targets in antibody-mediated organ rejection. Trends Mol Med. 2022;28:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loupy A, Haas M, Roufosse C, et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transplant. 2020;20:2318–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tambur AR, Campbell P, Chong AS, et al. Sensitization in transplantation: Assessment of risk (STAR) 2019 Working Group Meeting Report. Am J Transplant. 2020;20:2652–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefaucheur C, Viglietti D, Bentlejewski C, et al. IgG Donor-Specific Anti-Human HLA Antibody Subclasses and Kidney Allograft Antibody-Mediated Injury. J Am Soc Nephrol. 2016. 2016;27:293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis K, Macedo C, Bailly E, et al. Coordinated Circulating T Follicular Helper and Activated B Cell Responses Underlie the Onset of Antibody-Mediated Rejection in Kidney Transplantation. J Am Soc Nephrol. 2020;31:2457–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Taeye SW, Bentlage AEH, Mebius MM, et al. Fcγr binding and ADCC activity of human igg allotypes. Front Immunol. 2020;11:740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valenzuela NM, Hickey MJ, Reed EF. Antibody subclass repertoire and graft outcome following solid organ transplantation. Front Immunol. 2016;7:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvani J, Terada M, Lesaffre C, et al. In situ multiplex immunofluorescence analysis of the inflammatory burden in kidney allograft rejection: A new tool to characterize the alloimmune response. Am J Transplant. 2019;20:942–953. [DOI] [PubMed] [Google Scholar]
- 13.Yazdani S, Callemeyn J, Gazut S, et al. Natural killer cell infiltration is discriminative for antibody-mediated rejection and predicts outcome after kidney transplantation. Kidney Int. 2019. 2019;95:188–198. [DOI] [PubMed] [Google Scholar]
- 14.Parkes MD, Halloran PF, Hidalgo LG. Evidence for CD16a-Mediated NK Cell Stimulation in Antibody-Mediated Kidney Transplant Rejection. Transplantation. 2017;101:e102–e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamarthée B, Callemeyn J, Herck YV, et al. Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection. [published online July 10, 2022] medRxiv. 2022. 10.1101/2022.07.07.22276374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Gassen S, Callebaut B, Van Helden MJ, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. [DOI] [PubMed] [Google Scholar]
- 17.Polikowsky HG, Drake KA. Supervised Machine Learning with CITRUS for Single Cell Biomarker Discovery. Methods Mol Biol. 2019;1989:309–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Scientific Reports. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone AF, Wu H, Fronick C, et al. Harnessing expressed single nucleotide variation and single cell RNA sequencing to define immune cell chimerism in the rejecting kidney transplant. J Am Soc Nephrol. 2020;31:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nat Rev Drug Discov. 2014;13:379–395. [DOI] [PubMed] [Google Scholar]
- 21.Kurioka A, Cosgrove C, Simoni Y, et al. CD161 Defines a Functionally Distinct Subset of Pro-Inflammatory Natural Killer Cells. Front Immunol. 2018;9:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathew PA, Chuang SS, Vaidya SV, et al. The LLT1 receptor induces IFN-gamma production by human natural killer cells. Mol Immunol. 2004;40:1157–1163. [DOI] [PubMed] [Google Scholar]
- 23.Mahaweni NM, Ehlers FAI, Sarkar S, et al. NKG2A Expression Is Not per se Detrimental for the Anti-Multiple Myeloma Activity of Activated Natural Killer Cells in an In Vitro System Mimicking the Tumor Microenvironment. Front Immunol. 2018;9:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Videm V, Albrigtsen M. Soluble ICAM-1 and VCAM-1 as markers of endothelial activation. Scand J Immunol. 2008;67:523–31. [DOI] [PubMed] [Google Scholar]
- 25.Myers JA, Schirm D, Bendzick L, et al. Balanced engagement of activating and inhibitory receptors mitigates human NK cell exhaustion. JCI Insight. 2022;7: e150079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyairi S, Baldwin WM, Valujskikh A, et al. Natural Killer Cells: Critical Effectors During Antibody-Mediated Rejection of Solid Organ Allografts. Transplantation. 2020; 105:284–290. [DOI] [PubMed] [Google Scholar]
- 27.Koenig A, Mezaache S, Callemeyn J, et al. Missing Self-Induced Activation of NK Cells Combines with Non-Complement-Fixing Donor-Specific Antibodies to Accelerate Kidney Transplant Loss in Chronic Antibody-Mediated Rejection. J Am Soc Nephrol. 2020;32:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crespo M, Yelamos J, Redondo D, et al. Circulating NK-cell subsets in renal allograft recipients with anti-HLA donor-specific antibodies. Am J Transplant. 2015;15:806–814. [DOI] [PubMed] [Google Scholar]
- 29.Macedo C, Hadi K, Walters J, et al. Impact of Induction Therapy on Circulating T Follicular Helper Cells and Subsequent Donor-Specific Antibody Formation After Kidney Transplant. Kidney Int Rep. 2019;4:455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis K, Bailly E, Macedo C, et al. T-bet+CD27+CD21− B cells poised for plasma cell differentiation during antibody-mediated rejection of kidney transplants. JCI Insight. 2021;6: e148881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strengell M, Sareneva T, Foster D, et al. IL-21 up-regulates the expression of genes associated with innate immunity and Th1 response. J Immunol. 2002;169:3600–3605. [DOI] [PubMed] [Google Scholar]
- 32.Skak K, Frederiksen KS, Lundsgaard D. Interleukin-21 activates human natural killer cells and modulates their surface receptor expression. Immunology. 2008;123:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiekens L, Van Loocke W, Taveirne S, et al. T-BET and EOMES Accelerate and Enhance Functional Differentiation of Human Natural Killer Cells. Front Immunol. 2021;12:732511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Bouteiller P, Tabiasco J, Polgar B, et al. CD160: a unique activating NK cell receptor. Immunol Lett. 2011;138:93–96. [DOI] [PubMed] [Google Scholar]
- 35.Lin CM, Gill RG, Mehrad B. The natural killer cell activating receptor, NKG2D, is critical to antibody-dependent chronic rejection in heart transplantation. Am J Transplant. 2021;21:3550–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legris T, Picard C, Todorova D, et al. Antibody-Dependent NK Cell Activation Is Associated with Late Kidney Allograft Dysfunction and the Complement-Independent Alloreactive Potential of Donor-Specific Antibodies. Front Immunol. 2016;7:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R-T, Li W, Guo D, et al. Natural killer cells promote the differentiation of follicular helper T cells instead of inducing apoptosis in myasthenia gravis. Int Immunopharmacol. 2021;98:107880. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann U, Segerer S, Rümmele P, et al. Expression of the chemokine receptor CXCR3 in human renal allografts--a prospective study. Nephrol Dial Transplant. 2006;21:1373–1381. [DOI] [PubMed] [Google Scholar]
- 39.Neudoerfl C, Mueller BJ, Blume C, et al. The Peripheral NK Cell Repertoire after Kidney Transplantation is Modulated by Different Immunosuppressive Drugs. Front Immunol. 2013;4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leur K, Luk F, van den Bosch TPP, et al. The Effects of an IL-21 Receptor Antagonist on the Alloimmune Response in a Humanized Mouse Skin Transplant Model. Transplantation. 2019;103:2065–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua F, Comer GM, Stockert L, et al. Anti-IL21 receptor monoclonal antibody (ATR-107): Safety, pharmacokinetics, and pharmacodynamic evaluation in healthy volunteers: a phase I, first-in-human study. J Clin Pharmacol. 2014;54:14–22. [DOI] [PubMed] [Google Scholar]
- 42.Ignatenko S, Skrumsager BK, Mouritzen U. Safety, PK, and PD of recombinant anti-interleukin-21 monoclonal antibody in a first-in-human trial. Int J Clin Pharmacol Ther. 2016;54:243–252. [DOI] [PubMed] [Google Scholar]
- 43.Bubuer J, Bennett S, Sproule T et al. Treatment of BXSB-Yaa mice with IL-21R-Fc fusion protein minimally attenuates systemic lupus erythematosus. Ann N Y Acad Sci. 2007;1110:590–601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods.
Supplementary Figure S1 | Flow chart, study outline and description of DSA characteristics in the DSA+ groups.
Supplementary Figure S2 | Gating strategy for CD56dimCD16bright NK cells and T2 cell line characterization.
Supplementary Figure S3 | CD56dimCD16bright NK cell frequency, count and phenotypic modifications.
Supplementary Figure S4 | CITRUS predictive analysis integrating the 9 NK cell markers and comparing DSA+ ABMR and DSA− TCMR patients.
Supplementary Figure S5 | Increased CD160+ IL-21R+ CD56dimCD16bright NK cell polyfunctionality during CMC and ADCC.
Supplementary Figure S6 | rIL-21 enhances CD56dimCD16bright NK cell polyfunctionality in vitro.
Supplementary Figure S7 | Pathway analyses of circulating NK cell single-cell RNA-sequencing clusters.
Supplementary Figure S8 | NK cells infiltrate kidney allografts and endothelial cells undergo apoptosis during DSA+ ABMR
Supplementary Figure S9 | Schematic representation of a hypothetical model based on our study findings.
Supplementary Table S1 | Cohort’s demographics (for the NK cell phenotypic, functional, transcriptomic analysis).
Supplementary Table S2 | Fluorochrome-tagged Abs used with flow cytometry.
Supplementary Table S3 | Patients’ demographics for bulk RNA-sequencing of allograft biopsies.
Supplementary References.
Supplementary Table S4 | Pathway-related genes for the circulating NK cell single-cell RNA-sequencing analysis of cluster 2.
Supplementary Table S5 | Pathway-related genes for the circulating NK cell single-cell RNA-sequencing analysis of cluster 4.
Supplementary Table S6 | Pathway-related genes for kidney allograft biopsy bulk RNA-sequencing analysis. Comparison DSA+ ABMR versus DSA− TCMR.
Supplementary Table S7. Pathway-related genes for kidney allograft biopsy bulk RNA-sequencing analysis. Comparison DSA+ ABMR versus DSA− STABLE.
Data Availability Statement
All data associated with this study are present in the paper and Supplementary Materials. The single-cell RNA-sequencing dataset from FACS-sorted circulating NK cell for DSA+ ABMR and DSA− TCMR patients has been deposited in NCBI’s Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE216203 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE216203). The RNA-sequencing of kidney allograft biopsies dataset has also been deposited and is accessible through GEO Series accession number GSE214703 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214703). The dataset of single-cell RNA-sequencing of kidney allograft biopsies used in this manuscript was downloaded from the GEO database (GSE145927, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145927).


