Abstract
Intratumor heterogeneity (ITH) is a driver of tumor evolution and a main cause of therapeutic resistance. Despite its importance, measures of ITH are still not incorporated into clinical practice. Consequently, standard treatment is frequently ineffective for patients with heterogenous tumors as changes to treatment regimens are only made after recurrence and disease progression. More effective combination therapies require a mechanistic understanding of ITH and ways to assess it in clinical samples. The growth of technologies enabling the spatially intact analysis of tumors at the single cell level and the development of sophisticated preclinical models give us hope that ITH will not simply be used as a predictor of a poor outcome but will guide treatment decisions from diagnosis through treatment.
Keywords: Intratumor heterogeneity, single-cell technologies, treatment-resistance, tumor plasticity, clonal interaction
Definition and sources of intratumor heterogeneity
Heterogeneity is one of the defining features of cancer. Cancer cells and stromal cells within the same tumor can vary widely in their genetic, epigenetic, and phenotypic characteristics. Despite significant research efforts and improvements in treatment, most advanced cancers remain deadly, largely due to their high level of heterogeneity. The sources of intratumor heterogeneity (ITH) are numerous and include genetic alterations (single nucleotide and copy number), epigenetic changes (chromatin patterns and DNA methylation), and microenvironmental factors (drug treatment, hypoxia, stiffness, and cellular interactions) [1]. Tumors experience a range of microenvironmental stresses including hypoxia, nutrient deprivation, and immune surveillance leading to continuous selection for the fittest subclones (see Glossary), (Figure 1) [2,3]. Thus, ITH in combination with microenvironmental factors drive tumor progression and treatment responses. Recent technical and computational advancements have enabled in-depth analysis of tumors at single-cell resolution to evaluate ITH in clinical samples. Using these tools, numerous studies have demonstrated the clinical relevance of ITH and highlighted the importance of assessing ITH in the clinic to improve patient stratification. At the same time, novel experimental models have been developed to characterize the functional relevance of ITH, provide evidence for subclonal cooperation within tumors, and dissecting the underlying mechanism to serve as basis for the design of more effective treatment strategies for heterogeneous tumors. Despite the recognition of the clinical importance of ITH and the explosion of single-cell assays enabling characterization of ITH at unprecedented depths, ITH measures are still not used to guide treatment design in current standard-of-care. Consequently, patients with heterogeneous tumors commonly experience treatment failure and disease progression.
Figure 1. Subclonal evolution during cancer progression and therapy resistance.
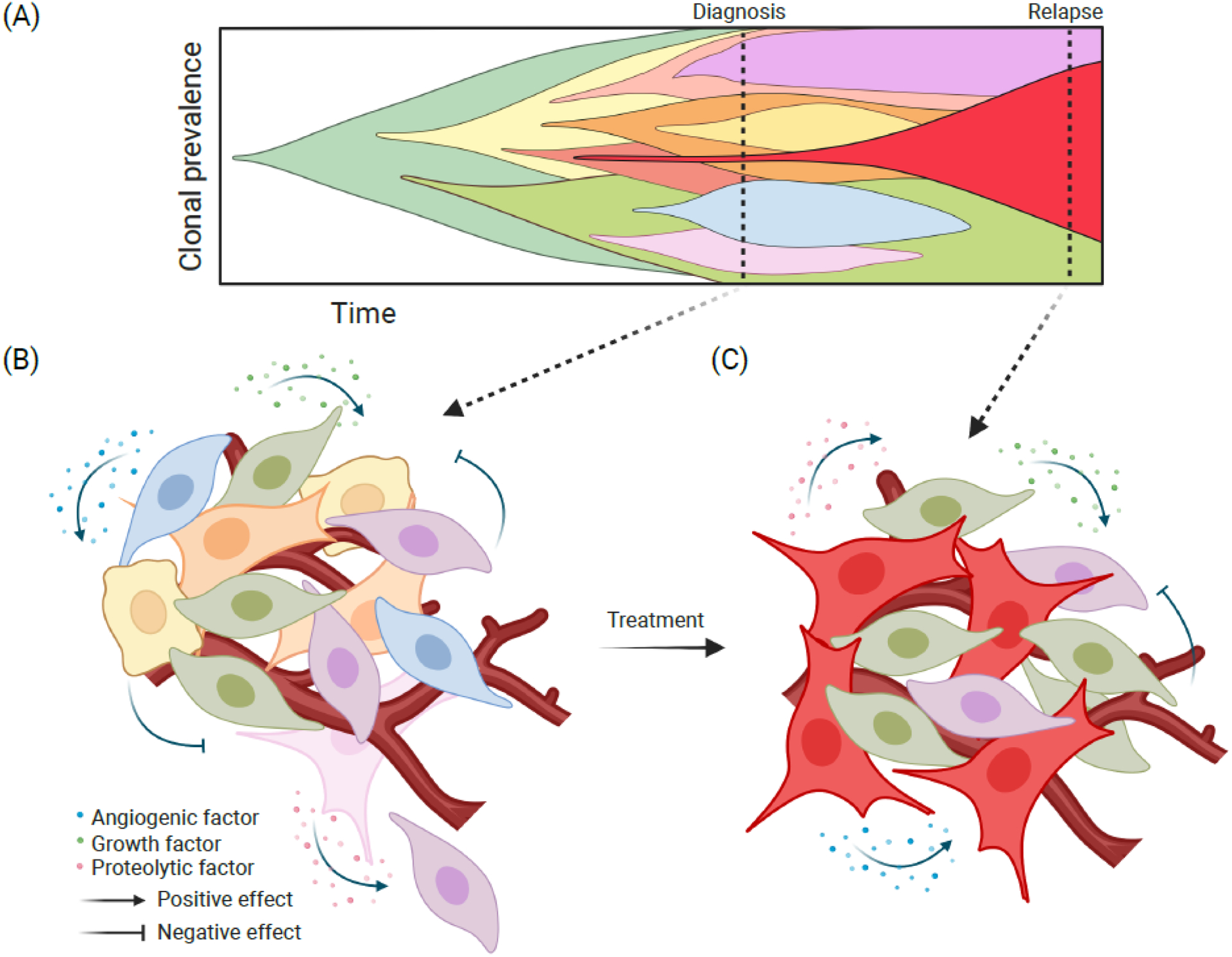
(A) Fish plot illustrating subclonal dynamics during tumor progression and selection by cancer therapies. Every color represents a different subclone matching cell colors in (B) at the time of diagnosis and in (C) at relapse. (B) During tumor development, an initiated neoplastic cell population, defined by genetic or epigenetic characteristics, will give rise to a variety of subclones, generating a heterogeneous tumor by the time of diagnosis. Interactions between the emerging subclones, either direct or indirect via the microenvironment, will also influence their phenotype and fitness for example via the secretion of different factors (e.g., angiogenic, growth, and proteolytic). Subclonal interactions can be negative where the different subclones are competing for limited resources and secrete factors that will limit the expansion of other subclones (i.e., clonal competition). In contrast, a subclonal cooperation can also be present where a subclone produces a factor that promotes the growth or invasive properties of other subclones. (C) Different subclones may also respond differently to treatment, leading to the selection for a limited set of subclones with preexisting or acquired therapeutic resistance. Subclones resistant to treatment (here depicted in red, purple, and green) will be selected leading to relapse. Created with BioRender.com
In this review we summarize recent advances in these areas focusing on assays used for the assessment of ITH, its clinical and functional relevance, along with the challenges to incorporate this accumulating knowledge into tools to guide clinical practice. Improvements in technologies and computational tools and the availability of combination therapies targeting both the cancer cells and the microenvironment make it likely that ITH will become a standard for individualized treatment design.
Assessing heterogeneity in clinical samples
Various methods have been developed to assess ITH in human clinical samples. Profiling of patient specimens enables the study of the complex and unique ecosystem of human tumors including tumor cell and tumor microenvironment (TME) heterogeneity.
Profiling of whole tumors
Omics analysis of bulk tumors using whole genome (WGS) or RNA-sequencing (RNA-seq) can be used to identify potential therapeutic targets including overexpressed or mutated genes and perturbed signaling pathways. Multi-region profiling can provide an additional level of resolution, but heterogeneity is hard to assess using this approach, because single-cell level information is lost. Nevertheless, novel computational analysis of bulk sequencing methods can help estimate cell type composition and trace back the evolutionary history of a tumor, thus retrieving important information about heterogeneity. A novel analytical tool reconstructing in silico subclonal composition based of somatic mutations can help decipher tumor cell heterogeneity and branching phylogenies [4]. The algorithm developed was able to follow the expansion of clones and permitted the identification of cancer-specific subclonal evolutionary patterns with driver mutations in a pan-cancer study.
Gene expression profiling of bulk tumor samples can also identify patients who may benefit from certain targeted therapies. An example of this is the designation of “BRCA-ness”, defined as molecular similarity to BRCA-mutant tumors in breast and ovarian cancer [5]. Tumors with loss of function mutation of BRCA1 and BRCA2 are deficient in homologous recombination repair and due to this highly sensitive to PARP inhibitors, and tumors with BRCA-ness signature may also respond to this treatment [5]. Similarly, the WINTHER trial tested the usefulness of DNA sequencing and expression profiling of bulk tissue samples to guide treatment decisions in patients who failed prior therapies [6]. Certain mutations identified led to change in treatment and exceptional responses in some patients. For example, mutant MSH6 DNA repair gene resulted in exceptional response to anti-PD-1 treatment, while overexpression of AKT2 and AKT3 identified cases that responded to mTOR inhibitors [6].
Subclonal evolution is strongly shaped by the immune environment [1]. Thus, numerous computational tools have been developed including CIBERSORT [7] and TIMER [8], to estimate the abundance of various immune cell types from bulk profiling data, which can predict the presence of active anti-tumor immune responses characterized by higher fraction of activated GZM+CD8+ T cells or identify an immunosuppressive environment enriched in FOXP3+ regulatory T cells (Treg) and myeloid derived suppressor cells (MDSCs). Understanding tumor immune cell composition can be used to guide treatment, for example by predicting the likelihood of response to immune checkpoint inhibitors. Other specialized methods, such as T cell receptor sequencing (TCR-seq), can measure the heterogeneity of the TCR clonotype repertoire of tumor-infiltrating or peripheral T cells. TCR clonotype diversity has been shown to be associated with response to immune checkpoint inhibitors such as the efficacy of PD-1 blockade in patients with metastatic melanoma [9]. These examples demonstrate that sequencing of whole tumor specimens is a cost-effective way to characterize ITH in clinical samples to predict treatment options and clinical outcomes in a more accessible and affordable way.
Single-cell profiling
In recent years numerous single-cell omics technologies have been developed to enable the study of cellular populations within clinical samples at a greater resolution as reviewed in [1]. These include single-cell genome, transcriptome (scRNA-seq), and assay for transposase-accessible chromatin (scATAC-seq) sequencing, imaging cytometry by time of flight (CYTOF), and more recently single-cell proteomics [10]. scRNA-seq, scATAC-seq or single-cell proteomics allow the more in-depth analysis of features in a non-bias way compared to CYTOF where only a limited number of markers can be accessed. On the other hand, CYTOF can give information on millions of cells versus a few thousand routinely used in scRNA-seq and related methods. scRNA-seq has been very useful for defining clinically and functionally relevant cell populations including identifying many different types of cancer-associated fibroblasts (CAFs) [11,12]. CAFs display high heterogeneity among and within tumors and also change dynamically during treatment like immunotherapy. Modulating the balance among the various CAFs could potentially be explored to improve therapies [12]. Beyond identifying cell types within tumors, scRNA-seq also enables exploring their functional relevance using algorithms developed to detect cell-cell interactions based on the expression of paired receptors and ligands. A systematic investigation of a single-cell transcriptional atlas of fibroblasts across cancer types was able to define distinct conserved interactions of the different CAFs subtypes with other cell components within tumors [11]. Predicted interactions between CAFs and various immune cells correlated with checkpoint inhibitor response and overall prognosis highlighting the clinical relevance of mapping cellular interactomes.
Significant advances have also been made into multiplexing single-cell omics assays that enable the assessment of different features (e.g., transcriptome, epigenome) on the same sample and provide an even more comprehensive assessment of ITH [13]. Multiplexed assays also help with dissecting the functional relevance of ITH, since they facilitate delineating underlying mechanisms. A nice example for this the discovery of why KMT2A-rearranged infant acute lymphoblastic leukemia (ALL) in younger patients is more likely to evade chemotherapy and immune-mediated control using multiplexed scRNA-seq and scATA-seq analysis [14]. The combined analysis of gene expression and chromatin profiles in the same cells revealed that leukemic cells in younger ALL patients have higher degree of lineage plasticity and a unique immunosuppressive leukemic blast population signaling to cytotoxic lymphocytes. By being able to track individual leukemic cells over time using this multi-omic approach allowed the visualization of the evolution of acute myeloid leukemia (AML) from preexisting myeloid blasts present even at initial ALL diagnosis. This study shows the power of multi-omic single cell assays by identifying tumor intrinsic and extrinsic factors linked to patient outcome and it gave insights that can be used to improve therapies for younger patients with ALL.
Spatial profiling
Tumor heterogeneity is not only reflected in the presence of different subpopulations, the localization of the cells within tumors is also an important part of ITH. Relatively simple technologies like Fluorescence In Situ Hybridization (FISH) and immunostaining have been used for a long time to access spatial heterogeneity of particular genes/proteins and to guide treatment decision in the clinic. Testing for ERBB2 (encoding HER2) amplification by FISH and overexpression of HER2 by immunostaining in breast cancer is one of the best examples of this. More sophisticated spatial profiling methods like spatial transcriptomics and multiplexed ion beam imaging by time of flight (MIBI-TOF) enable the assessment of multiple markers on a single tissue slice at the single cell or even subcellular level, while preserving tissue topology [15]. The application of these technologies gives a more detailed view of the complexity of cancer cells and TME heterogeneity by assessing spatial cellular interaction networks and their associations with disease progression. For example, slide-TCR-seq allowed the mapping of T cell populations in renal cell carcinoma and melanoma demonstrating preferential localization of different T cell clones within tumors revealing immune niches and spatial interactions of T cells with tumor cells [16]. A spatial atlas of breast ductal carcinoma in situ (DCIS) using MIBI-TOF was also able to predict the risk of invasive progression based on changes in myoepithelial cells and extracellular matrix (ECM) [17]. Comparing normal breast tissues and DCIS with or without invasive recurrence the authors found that more “normal-like” myoepithelium with high E-cadherin expression and higher thickness in DCIS was associated with higher risk of invasive progression. Similarly, pairing spatial profiling with artificial intelligence (AI) can facilitate our understanding of heterogeneous tumor ecosystems and yield clinically relevant predictions based on a wide breadth of data. For example, imaging CYTOF-based characterization of spatial cellular phenotypes in combination with deep learning identified spatial neighborhoods in the tumor immune microenvironment that predicted clinical outcome from a single 1 mm2 tumor section [18,19].
Clinical relevance of tumor heterogeneity
ITH has been associated with poor outcomes in multiple solid tumor types such as lung, gastrointestinal, breast, and prostate cancers. The TRAcking Cancer Evolution through therapy (Rx), or TRACERx, is a prospective cohort study dedicated to investigating how cancer evolution impacts tumor biology and patient outcomes in lung and renal cancers, and now includes other cancers as well [20]. Through whole exome sequencing on over 300 tumor regions from 100 patients with early-stage non-small cell lung cancer (NSCLC), the TRACERx team found substantial intratumor heterogeneity both in somatic driver alterations and copy number alterations [21]. Among the 100 tumors, there was a median of 30% of somatic mutations identified as subclonal, and the number of subclonal mutations was significantly correlated with certain mutational signatures. Tumors with the most subclonal mutations were more likely to have APOBEC-mediated mutagenesis. While there was no significant association between the proportion of subclonal mutations and survival, there was a significant association between copy number heterogeneity and risk of recurrence or death. Among all patients, there was a median of 48% of copy number alterations that were identified as subclonal; having 48% or more subclonal copy number alterations was associated with a nearly five times higher risk of recurrence or death.
In addition to the clinical implications of heterogeneity in somatic alterations, heterogeneity in tumor subtype, such as in estrogen receptor (ER)-positive or HER2-positive breast cancers, is also known to affect clinical outcomes. Estrogen receptor status is typically ascertained pathologically via immunohistochemical staining. An analysis of 1,780 postmenopausal women with ER-positive breast cancer found that intratumor heterogeneity for ER-positive cells in a single tumor sample, as assessed by ten breast cancer pathologists, was significantly associated with worse breast cancer-specific survival [22]. Similarly, in HER2-positive breast cancers, characterized by amplification in ERBB2, intratumor HER2 heterogeneity has been associated with poor prognosis as well as resistance to HER2-directed treatments [23,24]. HER2 expression may vary between multiple sites within a single tumor and also between different metastatic sites in a single patient [25]. The impact of HER2 heterogeneity on pathologic treatment response has been prospectively studied in a phase II clinical trial of neoadjuvant HER2-directed therapy with trastuzumab emtansine (T-DM1) and pertuzumab: among 157 trial participants with evaluable tumors, 10% had heterogeneity in ERBB2 amplification. HER2 heterogeneity in this study was assessed with two biopsies from each patient’s tumor and defined following American Society of Clinical Oncology/College of American Pathologists (guidelines: as having either an area with ERBB2 amplification in more than 5% but less than 50% of tumor cells, or a HER2-negative area by FISH on at least one of two biopsies [26]. Of the 10% of participants with HER2 heterogeneity, none of these achieved a pathologic complete response, while over half of the patients with non-heterogeneous disease had a pathologic complete response [24]. Importantly, as was seen in this trial, HER2 heterogeneity in breast cancer seems to be more common in patients with ER-positive/HER2-positive tumors than in those with HER2-positive tumors that are ER-negative. Similarly, in the KRISTINE trial, patients with HER2 heterogeneity receiving pre-operative T-DM1 and pertuzumab were more likely to experience locoregional recurrence [27]. While next-generation antibody drug conjugates (ADCs) have specifically shown efficacy in tumors with low levels of HER2 expression, it is unclear whether these agents will be effective in HER2 heterogeneous tumors [28,29]. Ongoing clinical trials, such as the DESTINY-Breast11 and DESTINY-Breast05 trials studying trastuzumab deruxtecan (T-DXd) in the pre-operative and post-operative setting respectively, are actively investigating whether such novel ADCs may optimize therapy of early-stage HER2-positive breast cancers. It is possible that future strategies may overcome HER2 heterogeneity and improve treatment of patients with heterogeneous tumors (Box 1).
Box 1. Hypothetical case of HER2 heterogenous breast cancer.
In current clinical practice, patients with early-stage or advanced HER2-positive breast cancer are treated uniformly regardless of heterogeneity in ERBB2 amplification (Figure I). As clinical trials investigate the efficacy of new therapies, such as novel antibody drug conjugates, treatments may be improved to more effectively treat tumors with HER2 heterogeneity.
Figure I.
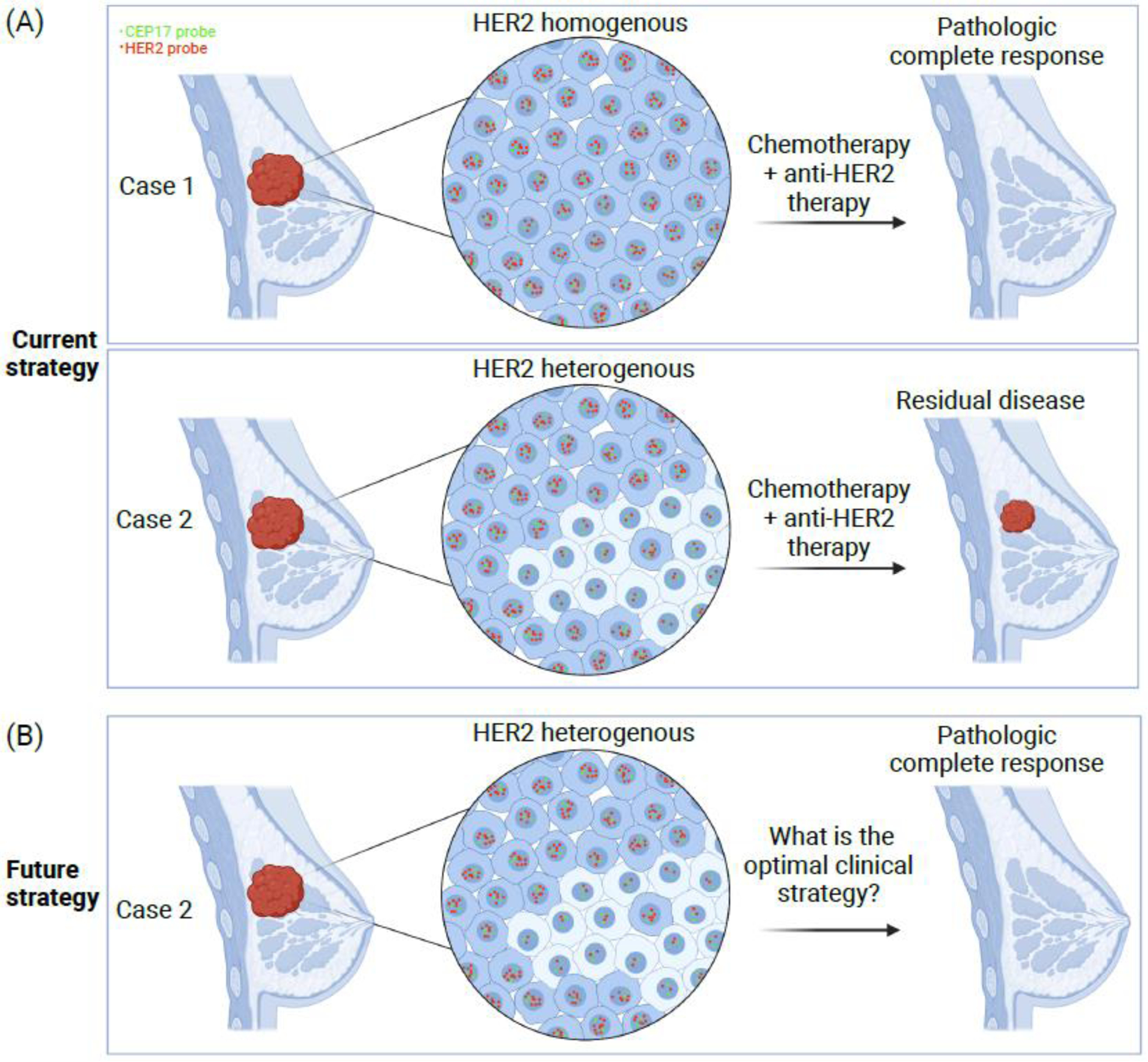
(A) In this hypothetical case study, the patient in Case 1 has a tumor that is homogenous in HER2 expression while the patient in Case 2 has HER2-heterogeneous disease. In current practice, both patients will be treated with neoadjuvant chemotherapy and HER2-targeting therapy. At the time of surgical evaluation, Case 1 is likely to have a pathologic complete response. However, Case 2, with HER2 heterogeneity, is more likely to have residual disease. (B) A potential future strategy may apply novel treatment regimens that are more effective for patients with HER2 heterogeneity. Created with BioRender.com
Heterogeneity in tumor subtype may indicate a path towards subtype switching. For example, the AURORA study, which analyzed primary and metastatic tumor pairs among 381 women with breast cancer, showed that intrinsic subtype switching occurred in 36% of cases [30]. In breast cancer, intrinsic subtype switching from primary tumor to distant metastasis appears to be most commonly switching from luminal A to luminal B subtype, though switching to basal or HER2-enriched subtypes also occurs [30,31]. The mechanism of subtype switching is unclear though may be related to pre-existing intratumor heterogeneity and selection for certain cell populations in particular microenvironments at distant sites [32]. Of note, while a switch from luminal A to luminal B subtype typically confers worse prognosis, this is not true for all cases of subtype switch. For example, in a series of 219 patients with metastatic breast cancer to the brain, HER2-positive status was gained in about 15% of cases and it was associated with improved survival [32].
Similarly, while most prostate cancers are adenocarcinomas (PACs) dependent on the androgen-receptor (AR) signaling pathway, acquired genomic and epigenetic alterations are thought to facilitate lineage switching such that tumors evolve into AR-independent neuroendocrine prostate cancers (NEPCs) [33]. Like other poorly differentiated neuroendocrine carcinomas, RB1 and TP53 loss is common in NEPCs and may play a role in this lineage switch. Epigenetic features, such as methylation changes and histone modifications, are also thought to contribute to evolution from PAC to NEPC [34,35]. The incidence of NEPC seems to be increasing, likely as a mechanism of resistance to newer AR-targeted therapies. While initial intratumor heterogeneity is likely required for the development of treatment-related NEPCs, recent research suggests that these NEPCs ultimately evolve to be more homogenous, perhaps via clonal selection [36]. Investigation of diagnostic approaches to identify treatment-related NEPC and tumors at risk for this lineage switching is important as this diagnosis has significant treatment implications: NEPCs, like neuroendocrine small cell lung cancers, are typically responsive to platinum-based chemotherapies and may similarly respond to immunotherapy.
Tissue assessment of tumor heterogeneity, via sampling at multiple tumor sites at once, is not typically included in routine clinical practice. However, some studies, including the prospective TRACERx cohorts, also study liquid biopsy, the testing of tumor biomarkers, including genetic material, in bodily fluids such as circulating peripheral blood (obtained via a blood test) or cerebrospinal fluid (obtained via a lumbar puncture). Liquid biopsy is a non-invasive and relatively quick method of assessing tumor genomic alterations – via digital droplet PCR, targeted next generation sequencing, or other methods – to identify targetable alterations for chemotherapeutics or to assess for known resistance mechanisms. Because tumor-specific alterations found in liquid biopsy often represent multiple tumor sites rather than an individual location from which a tumor biopsy was collected, liquid biopsy may be a helpful tool to assess ITH of multiple lesions at the same time, especially in metastatic settings where biopsies are not possible at every site (Figure 2). For example, in colorectal cancer, liquid biopsy detection and sequencing of cell-free DNA (cfDNA) more frequently detects important treatment resistance-associated alterations than tumor tissue sequencing [37]. Clinical trials are increasingly testing how this can be used to personalize treatment decisions: in the recent phase II CHRONOS trial, cfDNA assessment of resistance mutations was successfully used to guide treatment of heavily pretreated metastatic colorectal cancer [38]. In patients with central nervous system metastases or primary brain tumors, liquid biopsy testing of cfDNA from cerebrospinal fluid (CSF) may help to identify tumor genomic alterations from heterogeneous cancers [39]. Ongoing research will determine how to optimize CSF cfDNA detection to better inform prognosis, guide assessment of treatment resistance, and assess applicability of targeted therapies. In these ways, liquid biopsy is increasingly studied and used in clinical practice [40].
Figure 2. Liquid biopsy versus tissue biopsy: assessment of intratumor heterogeneity in genomic alterations.
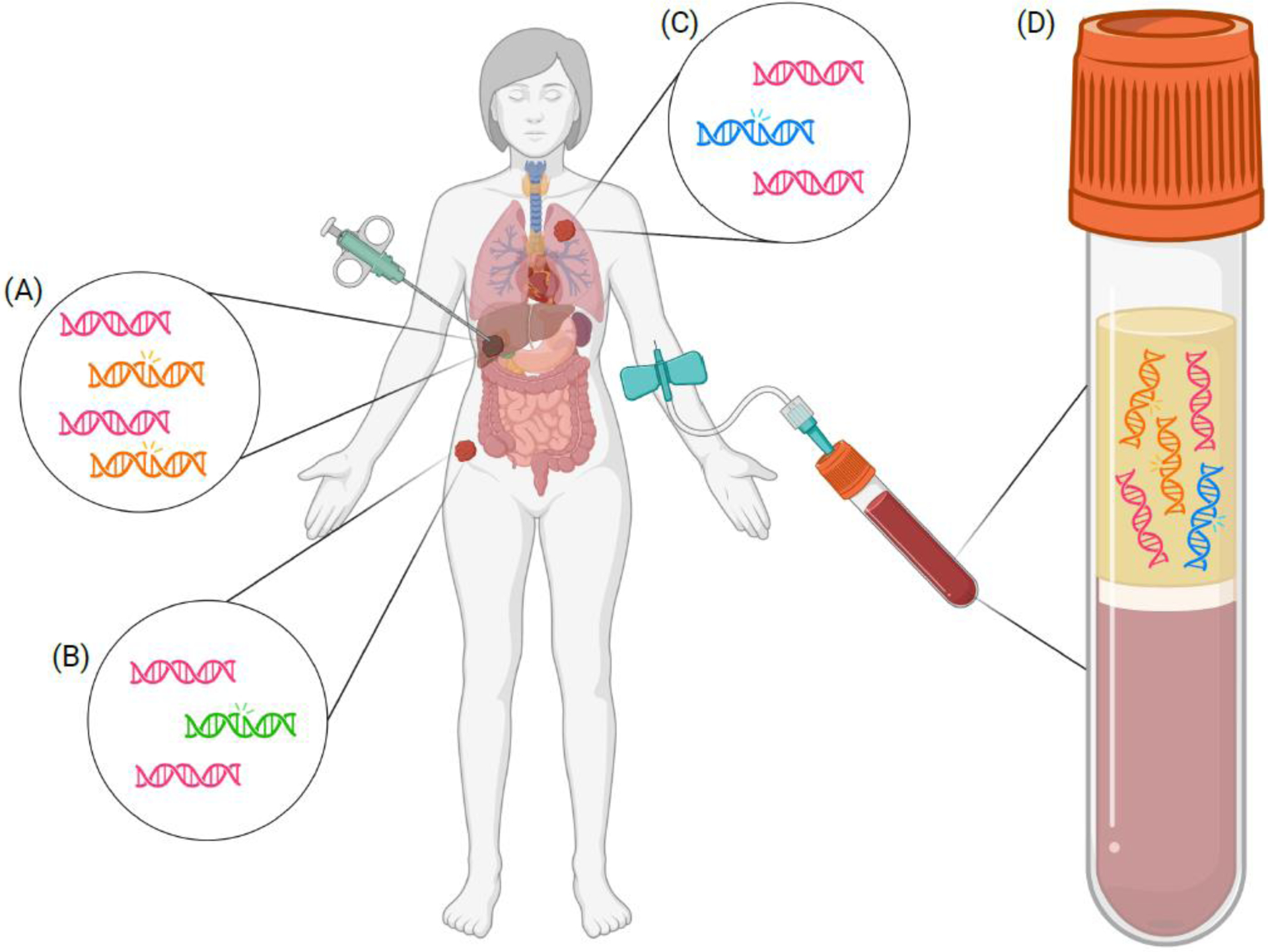
In this illustration of a patient with metastatic disease, both traditional tissue biopsy as well as liquid biopsy, obtained via peripheral blood testing, are depicted. (A) Tissue biopsy of the liver lesion may yield both DNA without alterations (indicated by pink helices) and tumor somatic mutations (as indicated by orange mutant helices). (B) Biopsy of the hip lesion may also reveal a tumor somatic mutation (indicated by green mutant helix) that may be different than the mutations seen at other metastatic sites. (C) Biopsy of a different metastatic site, such as a lung lesion, may reveal yet another unique tumor somatic mutation (indicated by the blue mutant helix). (D) In this liquid biopsy plasma sample, mutations from multiple metastatic sites are detected (as indicated by both the orange mutant helices and the blue mutant helix). This is an advantage over single-site metastatic biopsy, which would have captured only the somatic mutations in the one lesion that is biopsied. However, liquid biopsy may not detect genomic alterations from every metastatic site. For example, the mutation indicated by the green mutant helix is not detected in this patient’s plasma sample. Created with BioRender.com
Current oncologic treatment often includes moderating the host immune system, and heterogeneity in host immune function is also associated with clinical outcomes. In particular, peripheral blood TCR clonotype repertoire diversity has been associated with survival after immunotherapy treatment in melanoma, pancreatic ductal adenocarcinoma, and urothelial cancers [41]. One growing area of interest is the clinical implications of the presence and type of tumor infiltrating lymphocytes (TILs) [42]. The presence of stromal TILs has been associated with improved prognosis in multiple tumor types including longer overall survival in colorectal cancer and high grade serous ovarian cancer as well as longer disease-free survival in early-stage HER2-positive and triple negative breast cancers [43–46]. The presence and type of TILs are also thought to play a role in response to therapy with immune checkpoint inhibition. For example, tumors with mismatch repair deficiency acquire many point mutations, which is thought to lead to more tumor-associated neoantigens. This, in turn, is coupled with the increased presence of TILs and improved prognosis [42]. While high tumor mutation burden has been associated with response to treatment with immune checkpoint inhibitors, high subclonal heterogeneity itself has been associated with worse response to immune checkpoint inhibitors [47]. Additionally, some biomarkers that have been associated with response to immunotherapy are known to vary between metastatic sites, like for example PD-L1 expression [48]. In this way, heterogeneity in both tumor and host factors are important in prognosis and response to immune-modulating agents.
Functional relevance and experimental models
The functional relevance of ITH is difficult to decipher from profiling patient specimens since these most often offer a snapshot of tumor evolution as repeated sampling over time is rarely feasible. Therefore, experimental models of heterogenous tumors are needed for studying the impact of ITH on tumor progression and for developing more effective treatment strategies.
Lineage tracing and molecular barcoding
Single-cell tracing methods using genetic or optical barcodes track the spatiotemporal fate of cells and their progeny mapping tumor evolution over time [49]. The combination of these methods with omics technologies also yields information on phenotypic features that can be used to predict the functional properties of tumor cell clones. The ClonMapper system utilizing expressed DNA barcodes even goes a step beyond this and in addition to tracking and characterizing clones by scRNA-seq it enables the retrieval of specific barcoded cell populations for functional analysis [50]. The application of this technology to a chronic lymphocytic leukemia cell line demonstrated the presence of distinct clones with different survivorship trajectories during chemotherapy. Retrieving clones before, during, and after treatment facilitated the study of therapy-induced cellular diversification and durable transcriptional changes.
A recently developed Cas9-based barcoding system introduces indels on target sites resulting in a heritable barcode sequence that generates unique barcodes during each cell division enabling lineage tracing at a single cell level [51]. This approach enabled the construction of phylogenetic trees during tumor growth and metastatic progression in a xenograft model of lung cancer. Animals were followed for over two months unveiling significant diversity in metastatic capacities originating from preexisting, heritable variations in gene expression patterns and identifying candidate drivers of distant metastases.
Topologic tracking of cell populations in vivo also permits deeper analysis of tumor subclonal behavior and interactions. Spatial epitope tagging combining barcoding and MIBI (EpicMIBI) characterized patches of tumor clones in an experimental model of small cell lung cancer deciphering their phenotype and microenvironment [52]. This analysis revealed that the presence of PTEN null cells impacts the growth of PTEN wild-type cells in mixed tumor cell patches suggesting a non-cell autonomous role for PTEN loss.
Barcoding technologies require engineering the cells prior to tumor initiation limiting their use to experimental models. However, recent studies have described that tracking mitochondrial DNA mutations can be used as a natural barcoding system and can be used to study subclonal dynamics and tumor evolution not only in murine models [53], but also in human cancers [54].
Genetically engineered mouse models (GEMMs)
The development of more effective treatment for heterogeneous tumors requires experimental models that reproduce the ITH of human tumors. GEMMs have been widely used to study tumor initiation and progression and the validation of novel therapeutic targets. Some GEMMs were developed to model the genetic ITH of human cancers, like for example ones that combine mutations of cancer-causing genes. In a mouse model of lung cancer, the introduction of mutant Kras with or without Trp53 deletion recapitulated different stages of cancer development and scRNA-seq analysis of the resulting tumors showed a dramatic increase of ITH during progression [55]. The capacity of cells to modify their characteristics and acquire new biological traits is known as plasticity and can be responsible for increased of heterogeneity and acquired resistance to treatment. This study highlighted the importance of a highly plastic cell state in ITH and treatment resistance and demonstrated the presence of these plastic cells in human cancer samples validating the functional relevance of this experimental model. In prostate cancer, a model was developed that recapitulates the adenocarcinoma-to-neuroendocrine prostate cancer progression by combining mutations in Pten, Rb, and Trp53 leading to plasticity and resistance to antiandrogen therapy [56]. Using organoids derived from this model, the authors identified the JAK2-STAT3 signaling pathway as a driver of plasticity and multilineage transcriptional programs in stem-like clones, suggesting that JAK/STAT inhibition could be used to overcome resistance to antiandrogen therapy [56,57].
Ex vivo models
Cell culture models that retain ITH could offer relatively easy, high throughput approaches to study subclonal interactions and test new treatment strategies. Based on recent studies, patient-derived organoid models may have these characteristics. Pediatric high-grade gliomas display high subclonal heterogeneity and have dismal outcomes. Organoids derived from distinct glioma clones in combination with spatial computational modelling of cellular interactions have been used to explore the effects of subclonal interactions on tumorigenicity, invasion, and therapy resistance [58,59]. Analysis of single-cell-derived clones of diffuse midline glioma (DIPG) in organoid cultures revealed that clonal interactions increased tumor invasiveness where a non-invasive clone becomes invasive in the presence of a specific clone [58]. A rare tumor cell population with a mutation in KMT5B (SUV420H1) encoding histone H4 lysine 20 methyltransferase was shown to increase the invasive capacity of the neighboring cells [59]. These studies highlight the importance of subclonal interactions in tumor evolution and demonstrate that dissecting the underlying mechanisms can identify novel therapeutic targets.
Concluding remarks and future perspectives
The prognostic and therapeutic implications of ITH is an intense area of investigation. However, many clinical and translational questions remain (see Outstanding questions). What is the best way to assess ITH in standard clinical practice? Even though scRNA-seq, spatial transcriptomics, and related omic technologies are highly informative, these are often not practical to use in standard clinical care due to the complexity of data acquisition and analyses as well as lack of data regarding the impact on clinical outcomes. While tissue biopsy of multiple sites at the time of progression may prove helpful, this is often not feasible. Liquid biopsy may provide a relatively non-invasive way to assess alterations at multiple tumor sites and evaluate ITH based on genomic alterations, as well as changes in transcription and methylation. However, blood or cerebrospinal fluid sampling may not capture variants from all potential metastatic sites and may not always provide a full picture of the extent of ITH within a patient. What type of ITH (i.e., genetic, epigenetic, phenotypic) is the most helpful to test? Multiomic technologies give the most information, but many findings are not currently actionable in the clinic. Thus, assays would have to be selected based on tumor type, clinical data, and available treatment options. Improved biomarker identification and novel diagnostic technologies are needed to enable assessment of ITH in routine oncology care. Further data, in clinical trials, is needed to determine how to incorporate such diagnostic technologies and biomarkers into clinical practice and whether these assessments improve survival outcomes. This is particularly important as contemporary therapies allow patients to live longer and receive more lines of treatment than ever before. As chemotherapies, immunotherapies, and targeted therapies improve, so too will tumor mechanisms of resistance evolve and likely contribute to increasing intratumor heterogeneity. The effect of new therapeutics on treatment of tumors with substantial heterogeneity is a critical area of future work, both in the laboratory and in the clinic, to improve outcomes for patients with heterogeneous tumors.
Outstanding questions.
Awareness of the implications of ITH on treatment response and overall prognosis is emerging. What are the best methods for assessing ITH in standard clinical practice?
Incorporating ITH measures as part of routine oncologic care may improve treatment design to increase efficacy of response and reduce the risk of relapse. How and at what stage should ITH be used for tailoring and sequencing existing treatment strategies and assigning patients to new experimental therapies?
Heterogeneity of the immune system has an impact on the response to immunotherapy. How can we incorporate host heterogeneity assessment into clinical practice?
Highlights.
High ITH is associated with poor outcome in many human cancer types, and it may include heterogeneity for genetic, epigenetic, and phenotypic features.
The assessment of intratumor heterogeneity (ITH) in clinical samples using deep-resolution single-cell multi-omics approaches facilitate the understanding of its role in tumor evolution and therapeutic resistance.
Sampling of multiple tumor or metastatic sites is challenging, thus, the development of less invasive diagnostic methods to assess ITH and monitor tumor evolution, including the use of circulating biomarkers, is an area of active investigation.
Recent clinical trials have incorporated assessment of ITH, as well as heterogeneity in host immune factors, in order to better understand the response and resistance to specific targeted therapies.
Experimental models of ITH have been used to investigate the functional relevance of ITH and for the development of novel therapeutic strategies to improve the treatment of heterogeneous tumors.
Acknowledgements
We express our gratitude to members of our laboratory and Dr. Sara Tolaney for their insightful feedback on the manuscript and valuable suggestions. The authors were supported by the National Cancer Institute R35 CA197623 (K.P.) and the Canadian Institute of Health Research Postdoctoral Fellowship (M.A.G.).
Glossary
- APOBEC
apolipoprotein B mRNA-editing enzyme that edits mRNA and/or deaminates DNA thereby driving genomic and/or transcriptomic changes. Dysregulation of APOBEC can induce tumor mutagenesis, the acquisition of somatic mutations in cancers that may facilitate tumor heterogeneity.
- Barcoding
identification method based on genetic or optical marks of cells that allows the tracking of cells through space and time.
- Cell subpopulation
a group of cells defined by shared phenotypic or genetic characteristics but not necessarily sharing clonal ancestry.
- CYTOF
cytometry by time of flight (CYTOF) is an application of mass cytometry to quantify multiple targets in individual cells using metal-isotope-tagged antibodies combined with time-of-flight detection.
- MIBI-TOF
multiplexed ion beam imaging by time of flight (MIBI-TOF) is mass spectrometry imaging of metal-tagged antibodies at subcellular resolution in a tissue section.
- scRNA-seq
single-cell RNA sequencing is a technic to investigate the transcriptome at the single-cell level in a large number of cells.
- scATAC-seq
single-cell ATAC-seq is the application of assay for transposase-accessible chromatin for single cells enabling the assessment of open chromatin at the single-cell level.
- Subclone
cancer cells derived from the same clone that have additional shared genetic and/or epigenetic characteristics.
- Subtype switching
the process by which tumors may change over time from one tumor subtype to another via genetic or epigenetic alterations, microenvironmental changes, or other mechanisms.
- Tumor subtype
classification of tumors based on certain shared characteristics that reflects biological and clinical differences (e.g., HER2-positive breast cancer is a subtype of breast adenocarcinoma).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
K.P. serves on the Scientific Advisory Board of Novartis, Vividion Therapeutics, Ideya Biosciences, and Scorpion Therapeutics, holds equity options in Scorpion Therapeutics, received honorarium from Astra-Zeneca, New Equilibrium Biosciences, and Roche in the past 12 months, and receives sponsored research funding from Novartis. The remaining authors have no interests to declare.
References
- 1.Li Z et al. (2022) Untangling the web of intratumour heterogeneity. Nat Cell Biol 24, 1192–1201. [DOI] [PubMed] [Google Scholar]
- 2.Marine JC et al. (2020) Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 20, 743–756 [DOI] [PubMed] [Google Scholar]
- 3.Vendramin R et al. (2021) Cancer evolution: Darwin and beyond. EMBO J 40, e108389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dentro SC et al. (2021) Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell 184, 2239–2254 e2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord CJ and Ashworth A (2016) BRCAness revisited. Nat Rev Cancer 16, 110–120. [DOI] [PubMed] [Google Scholar]
- 6.Rodon J et al. (2019) Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med 25, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman AM et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nature Methods 12, 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T et al. (2017) TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 77, e108–e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valpione S et al. (2021) The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nature Communications 12, 4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett HM et al. (2023) Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat Methods 20, 363–374 [DOI] [PubMed] [Google Scholar]
- 11.Luo H et al. (2022) Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nat Commun 13, 6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster DS et al. (2022) Multiomic analysis reveals conservation of cancer-associated fibroblast phenotypes across species and tissue of origin. Cancer Cell 40, 1392–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandereyken K et al. (2023) Methods and applications for single-cell and spatial multi-omics. Nature Reviews Genetics. 10.1038/s41576-023-00580-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C et al. (2022) Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood 139, 2198–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keren L et al. (2019) MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Science Advances 5, eaax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S et al. (2022) Spatial maps of T cell receptors and transcriptomes reveal distinct immune niches and interactions in the adaptive immune response. Immunity 55, 1940–1952 e1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risom T et al. (2022) Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell 185, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karimi et al. , Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature 614, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorin M et al. (2023) Single-cell spatial landscapes of the lung tumour immune microenvironment. Nature 614, 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey C et al. (2021) Tracking Cancer Evolution through the Disease Course. Cancer Discov 11, 916–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamal-Hanjani M et al. (2017) Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 376, 2109–2121 [DOI] [PubMed] [Google Scholar]
- 22.Lindström LS et al. (2018) Intratumor Heterogeneity of the Estrogen Receptor and the Long-term Risk of Fatal Breast Cancer. JNCI: Journal of the National Cancer Institute 110, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pernas S and Tolaney SM (2020) Targeting HER2 heterogeneity in early-stage breast cancer. Curr Opin Oncol 32, 545–554 [DOI] [PubMed] [Google Scholar]
- 24.Filho OM et al. (2021) Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov 11, 2474–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geukens T et al. (2023) Abstract HER2–16: HER2–16 Inter-lesion heterogeneity of HER2-status in metastatic breast cancer: possible implications for treatment with anti-HER2 antibody-drug conjugates. Cancer Research 83, HER2–16-HER12–16. [Google Scholar]
- 26.Vance GH et al. (2009) Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med 133, 611–612 [DOI] [PubMed] [Google Scholar]
- 27.Hurvitz SA et al. (2019) Neoadjuvant Trastuzumab Emtansine and Pertuzumab in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer: Three-Year Outcomes From the Phase III KRISTINE Study. J Clin Oncol 37, 2206–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarantino P et al. (2020) HER2-Low Breast Cancer: Pathological and Clinical Landscape. J Clin Oncol 38, 1951–1962 [DOI] [PubMed] [Google Scholar]
- 29.Modi S et al. (2022) Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N Engl J Med 387, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aftimos P et al. (2021) Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative. Cancer Discov 11, 2796–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klebe M et al. (2020) Frequent Molecular Subtype Switching and Gene Expression Alterations in Lung and Pleural Metastasis From Luminal A–Type Breast Cancer. JCO Precision Oncology, 848–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulsbergen AFC et al. (2020) Subtype switching in breast cancer brain metastases: a multicenter analysis. Neuro-Oncology 22, 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada Y and Beltran H (2021) Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr Oncol Rep 23, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beltran H and Demichelis F (2021) Therapy considerations in neuroendocrine prostate cancer: what next? Endocr Relat Cancer 28, T67–t78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cejas P et al. (2021) Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat Commun 12, 5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltran H et al. (2020) Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 130, 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikh AR et al. (2019) Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nature Medicine 25, 1415–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sartore-Bianchi A et al. (2022) Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med 28, 1612–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoane J et al. (2019) Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Annals of Oncology 30, 211–218. [DOI] [PubMed] [Google Scholar]
- 40.Choucair K et al. (2022) Liquid Biopsy-based Precision Therapy in Patients with Advanced Solid Tumors: A Real-world Experience from a Community-based Oncology Practice. The Oncologist 27, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi K et al. (2019) Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat Med 25, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paijens ST et al. (2021) Tumor-infiltrating lymphocytes in the immunotherapy era. Cell Mol Immunol 18, 842–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs TL et al. (2020) Assessment of Tumor-infiltrating Lymphocytes Using International TILs Working Group (ITWG) System Is a Strong Predictor of Overall Survival in Colorectal Carcinoma: A Study of 1034 Patients. Am J Surg Pathol 44, 536–544 [DOI] [PubMed] [Google Scholar]
- 44.Hwang C et al. (2019) Stromal tumor-infiltrating lymphocytes evaluated on H&E-stained slides are an independent prognostic factor in epithelial ovarian cancer and ovarian serous carcinoma. Oncol Lett 17, 4557–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim RS et al. (2019) Stromal Tumor-infiltrating Lymphocytes in NRG Oncology/NSABP B-31 Adjuvant Trial for Early-Stage HER2-Positive Breast Cancer. J Natl Cancer Inst 111, 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García-Teijido P et al. (2016) Tumor-Infiltrating Lymphocytes in Triple Negative Breast Cancer: The Future of Immune Targeting. Clin Med Insights Oncol 10, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf Y et al. (2019) UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 179, 219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozenblit M et al. (2020) Comparison of PD-L1 protein expression between primary tumors and metastatic lesions in triple negative breast cancers. J Immunother Cancer 8. 10.1136/jitc-2020-001558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano A et al. (2022) Mastering the use of cellular barcoding to explore cancer heterogeneity. Nature Reviews Cancer 22, 609–624 [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez C et al. (2021) Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat Cancer 2, 758–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinn JJ et al. (2021) Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rovira-Clave X et al. (2022) Spatial epitope barcoding reveals clonal tumor patch behaviors. Cancer Cell 40, 1423–1439 e1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penter L et al. (2023) Mitochondrial DNA Mutations as Natural Barcodes for Lineage Tracing of Murine Tumor Models. Cancer Res 83, 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ludwig LS et al. (2019) Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell 176, 1325–1339.e1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marjanovic ND et al. (2020) Emergence of a High-Plasticity Cell State during Lung Cancer Evolution. Cancer Cell 38, 229–246 e213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan JM et al. (2022) Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 377, 1180–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng S et al. (2022) Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat Cancer 3, 1071–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tari H et al. (2022) Quantification of spatial subclonal interactions enhancing the invasive phenotype of pediatric glioma. Cell Rep 40, 111283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinci M et al. (2018) Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat Med 24, 1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]


