Abstract
Purpose:
Patients with neuroendocrine prostate cancer (NEPC) are often managed with immunotherapy regimens extrapolated from small cell lung cancer (SCLC). We sought to evaluate the tumor immune landscape of NEPC compared to other prostate cancer types and SCLC.
Experimental Design:
In this retrospective study, a cohort of 170 patients with 230 RNA sequencing and 104 matched whole exome sequencing data were analyzed. Differences in immune and stromal constituents, frequency of genomic alterations, and associations with outcomes were evaluated.
Results:
In our cohort, 36% of the prostate tumors were identified as CD8+ T-cell inflamed, while the remaining 64% were T-cell depleted. T-cell inflamed tumors were enriched in anti-inflammatory M2 macrophages and exhausted T-cells and associated with shorter overall survival relative to T-cell depleted tumors (HR=2.62, p<0.05). Among all prostate cancer types in the cohort, NEPC was identified to be the most immune depleted, wherein only 9 out of the 36 total NEPC tumors were classified as T-cell inflamed. These inflamed NEPC cases were enriched in interferon gamma signaling and PD-1 signaling compared to other NEPC tumors. Comparison of NEPC with SCLC revealed that NEPC had poor immune content and less mutations compared with SCLC, but expression of checkpoint genes PD-L1 and CTLA-4 was comparable between NEPC and SCLC.
Conclusions:
NEPC is characterized by a relatively immune-depleted tumor immune microenvironment compared with other primary and metastatic prostate adenocarcinoma except in a minority of cases. These findings may inform development of immunotherapy strategies for patients with advanced prostate cancer.
Keywords: immunogenomic, microenvironment, prostate cancer, neuroendocrine, NEPC
INTRODUCTION
Metastatic prostate cancer is a leading cause of cancer death for men globally [1]. While androgen receptor (AR)-targeted therapies have significantly improved outcomes for prostate cancer, prolonged and potent suppression of AR signaling has led to evolution of treatment resistant AR-independent variants of prostate cancer, such as neuroendocrine prostate cancer (NEPC), that rarely arises de novo but can develop in up to 15–20% of late-stage castration resistant prostate cancer (CRPC) [2–5]. NEPC is clinically aggressive with limited therapeutic options. NEPC is currently diagnosed by its morphologic features that are similar to other poorly differentiated small cell neuroendocrine (NE) carcinomas, though both pure NE carcinomas and mixed adeno/NE histologies may be observed within the spectrum of NEPC [6]. There have been very few trials dedicated to NEPC, and therefore treatment is often extrapolated from small cell lung cancer (SCLC) guidelines based on their clinical and pathologic similarities. Based on recent data in SCLC, platinum-based chemotherapy combined with anti-PDL1 immune checkpoint inhibitor therapy (atezolizumab or durvalumab) followed by immunotherapy maintenance is now used in the first line setting for patients with SCLC [7, 8]. Patients with NEPC are now often being managed based on updated SCLC regimens with chemotherapy plus immunotherapy despite a lack of strong supporting data [9]. A better understanding of the immune landscape is needed to inform how SCLC regimens are adopted for NEPC.
Prostate cancer generally has an immune ‘excluded’ tumor microenvironment (TME) with limited leukocyte infiltration dominated by immunosuppressive cells that promote tumor proliferation [10–12]. Strategies to prime the immune system to recognize tumors in prostate cancer involve combining conventional therapy with immunotherapy e.g., immune checkpoint blockade (ICB) [13]. However, success of these treatments has remained limited to rare cases of mismatch repair deficiency and microsatellite instability [14, 15]. Recent data from the IMbassador250 trial has suggested that a subset of advanced prostate cancer patients may still benefit from ICB if they expressed ICB related immune markers (e.g., PD-L1) [16]. Similarly, in a small clinical trial, one NEPC patient who showed exceptional durable response to avelumab had consistently high levels of Natural Killer and PD1+ helper T-cells coupled with increase in CXCR2+ T-cells over time[17]. Such findings emphasize the potential value of comprehensive TME profiling in advanced prostate cancer.
In this retrospective study, we profiled the immunogenomic landscape of 230 patient biopsies along the prostate cancer continuum. We report an immune-depleted profile of NEPC compared with prostate adenocarcinoma and SCLC pathologies suggesting an immune-evasive, tumor-promoting TME with potential implications for therapy.
METHODS
Patient cohort
We retrospectively analyzed data from previously published prostate cohorts for this study [2, 18–20]. A total of 230 tissue samples from 170 individuals with RNA sequencing (RNAseq) data were analyzed, comprised of benign prostate tissue (n=31), localized prostate cancer (PCa n=74), hormone naïve metastatic PCa (mPCa.HN, n=14), CRPC (n=75), NEPC (n=36). Metastatic patient characteristics (n=93) are reported in Suppl Table S1. NEPC was defined based on tumor morphology (6). 25 cases were pure small or large cell carcinoma and 11 were mixed morphology cases. Survival data was available for 82 patients and matched whole exome sequencing was available for 104 tumors. Data Availability Statement: Prostate cancer datasets used in this study are available within their corresponding publications [2, 18–20] or through dbGAP (phs000909.v.p1, phs001666.v1.p1). The gene expression data (quantified as FPKMs (fragments per kilobase of exon per million mapped fragments)) was obtained for the International Stand up to Cancer (SU2C)-Prostate Cancer Foundation (PCF) Dream Team from the cBioPortal (https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019) (RRID:SCR_014555) and for the TCGA cancer cohorts from the Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov) (RRID:SCR_014514). Data from SCLC (n=28) was procured from a controlled access dataset EGAS00001000334 [21].
RNA sequencing, data quantification and analysis
Sequenced reads were aligned to human reference genome hg19/GRC37 using STAR (RRID:SCR_004463) [22] and quantified as gene counts using HTSeq (RRID:SCR_005514) [23] and FPKMs using cufflinks (RRID:SCR_014597) [24] with GENCODE (RRID:SCR_014966) [25] v19 for Gene expression profiles (GEPs). Enrichment scores (ES) were calculated using single sample gene set enrichment analysis (ssGSEA) [26]. Cytolytic scores were calculated as geometric mean of genes PRF1 and GZMA [27]. Differentially expressed genes (DEGs) were identified using gene counts with DESeq2 (RRID:SCR_000154) [28] and pathway analysis of ranked DEGs was done using fgsea in R (false discovery rate (FDR) q < 0.25). Top and bottom DEGs selected based on log fold change threshold of 2 (q < 0.05) were annotated using the collection of ‘NCI-Nature_2016_pathways’ in Enrichr (RRID:SCR_001575) [29] (fdr < 0.25). All statistical tests done using Wilcoxon test or Fisher’s exact test in R.
Whole-exome sequencing and data processing
WES data processing and alignment to human reference genome hg19/GRC37 was performed using established protocols described earlier [2, 30, 31]. Mutations were identified using Multi-Center Mutation Calling in Multiple Cancers (MC3) pipeline [32]. Tumor mutation burden (TMB) was calculated as non-silent mutation counts divided by the total n), modified by including Mutect2 (RRID:SCR_000559) and Strelka2 (RRID:SCR_005109) to existing tools VarScan2 (RRID:SCR_006849), Pindel (RRID:SCR_000560), SomaticSniper (RRID:SCR_005108), RADIA, and MuSE. All mutations calls were combined and only those identified by ≥2 tools were retained. Default MC3 read depth filters were applied and mutations were restricted to HaloPlex regions (umber of bases in the HaloPlex captured region per million. Copy number alterations were identified using CNVkit (RRID:SCR_021917) [33] and CNVSeeqer. The log2 copy ratio threshold of 0.10 was used to calculate copy number loads. Oncoprints were constructed with ComplexHeatmap in R (RRID:SCR_017270).
T-cell inflammation and deconvolution analysis
GEPs from 230 prostate samples were ordered by their median absolute deviation and top 65% most variable genes across the cohort were selected. The selected genes were median centered and used for unsupervised hierarchical clustering (wardD2 method), leading to identification of a 361-gene cluster with T-cell marker CD8A (Suppl Table S2). These 361 genes were used for supervised re-clustering of 230 samples with ConsensusClusterPlus [34] (pam clustering, 200 random selections, 80% re-sampling); 136 depleted and 94 inflamed samples were identified (Fig. 1A). To apply this T-cell signature to independent cancer cohorts, we used the signature for supervised consensus clustering with method pam and 1,000 random selections with 80% re-sampling, followed by removal of misclassified samples as identified by silhouette analysis (R package cluster). ssGSEA (RRID:SCR_003199) was applied to calculate a composite T-cell cluster score per sample for these 361 genes. ssGSEA was also applied to deconvolve GEPs based on 80 published immune and stromal signatures we considered representative of the TME (Suppl Table S3). Survival analysis was done using Cox Proportional-Hazards models and results visualized as forest plots (survival and forestplot, R). Plots were made with ggpubr or ComplexHeatmap R packages.
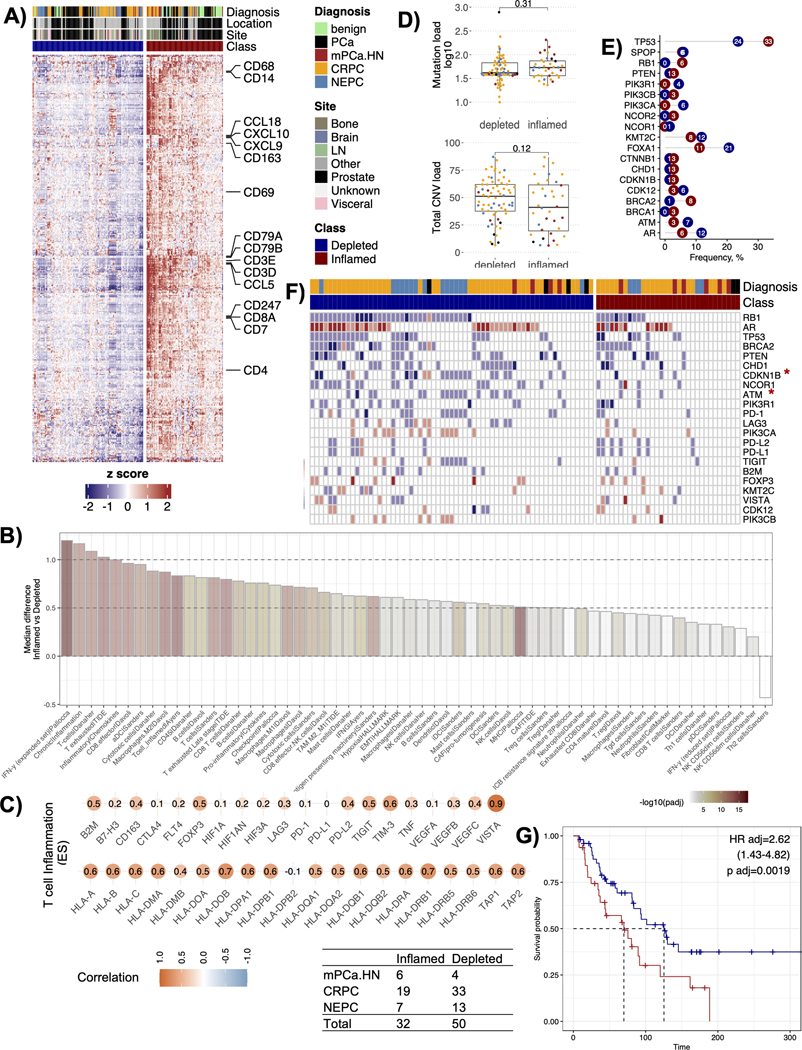
Figure 1.
Classification of prostate tumors based on T-cell immune status. A) Heatmap showing the result of consensus clustering using the 361 gene signature (derived in this study) to group prostate tumors into T-cell inflamed or depleted phenotypes. B) Barplot showing immune and stromal cell types and processes with significantly different median enrichment scores between the inflamed and the depleted T-cell groups (adjusted p value < 0.15, n=58). The x-axis shows the quantified features, and the y-axis shows the difference between the median enrichment scores from the inflamed versus the depleted clusters. The bars are colored by the adjusted p values on a -log10 scale. C) Pearson correlations between a composite T-cell cluster score (derived in this study) and the expression levels of a subset of genes related to immune checkpoints and functions. D) Boxplots showing the differences between the median tumor mutation burden (left panel) and the median total copy number variation (CNV) load (right panel) obtained from the two T-cell immune groups. p values reported are from the Wilcoxon signed-rank test, E) Plot showing the frequency of cases with mutations in known prostate driver genes separated by inflamed (red) versus depleted (blue) T-cell groups. F) Oncoprint plot showing the absolute copy number changes in 31 known prostate driver genomic alterations, MHC compels and common immune checkpoint genes. The oncoprint is segregated by T-cell immune groups and genes restricted to those with alterations in >=10 samples. Color red shows amplifications, blue shows deletions, and white shows a copy neutral status. Asterisk next to a gene denotes p-values significant at < 0.05 in a Fisher’s exact test for that gene. G) Kaplan-Meier survival curves showing a difference in overall survival (OS) between T-cell inflamed and the depleted groups (n=82). Hazard ratio (HR), 95% confidence intervals (upper and lower), and p values are reported from the Cox proportional-hazards regression model adjusted for the cancer types (adj).
Weighted gene co-expression network analysis (WGCNA) and module identification
We performed signed WGCNA using the WGCNA R package (RRID:SCR_003302) for automatic network identification and construction [35]. The best soft thresholding power of 12 was selected using the pickSoftThreshold function. Module-trait relationships correlation matrix and binomial generalized linear models were used to identify associations between modules and prostate cancer groups. The module eigengenes were gathered, and their ES per sample was calculated using ssGSEA. The modules were annotated for functional enrichment with ReactomePA and clusterProfiler and networks visualized by cnetplot in R.
Immunohistochemistry and pathological review
Tissue studies were conducted in accordance with the Declaration of Helsinki and after institutional review board approval (WCM IRB #1305013903) and with written informed consent from participants. Histopathology examination was performed through routine hematoxylin and eosin stain. NEPC was defined based on tumor morphology (6). Digital image analysis of immunohistochemistry (IHC) slides for CD8, CD68, GZMA and PD-L1 was performed using HALO® software (Indica Labs) [36]. Briefly, IHC slides were scanned at 40x with the Aperio AT2 slide scanner (Leica Biosystems, Buffalo Grove, IL). Annotation of tumor areas and desired regions of interest (ROIs) were selected through the HALO® software, directly. Using an automated algorithm with specific nuclear and membranous characteristics, staining was quantified (0 to 100%), excluding artifacts or nuclear staining (false positive).
RESULTS
Identification of T-cell inflamed and depleted phenotypes in prostate cancer
CD8+ T-cells are important mediators of anti-tumor immune responses [37], and their quantification in a sample can provide initial insights into the anti-tumor potential of the TME. To identify T-cell enriched samples in our cohort of 230 prostate biopsies, we clustered their gene expression profiles (GEPs) with unsupervised hierarchal clustering and identified a cluster of 361 genes enriched with known markers of T-cells and T-cell receptor signaling (Suppl Fig. 1A, Suppl Table S2). We re-clustered the samples using consensus clustering (supervised with these 361 genes) resulting in two groups: a T-cell inflamed group with relatively higher expression of CD8+ T-cell markers and cytolytic scores versus a T-cell depleted group (Fig. 1A, Suppl Fig. 1B). Two independent pathologists reviewed a subset of cases to confirm a positive lymphocyte content (10–20%) in inflamed (n=3) and a negative lymphocyte content (0%) in depleted cases (n=3) (Suppl Fig. 1C)
We quantified the immune and stromal related features to catalog the TME landscape in inflamed versus depleted groups (Suppl Table S3). In addition to T-cell inflammation (e.g., IFN-γ), the inflamed group harbored significantly higher anti-inflammatory M2 macrophages, and exhausted T-cells together with elevated levels of stromal cells e.g., cancer associated fibroblasts (CAFs) (Fig 1B). While most cell types were enriched in the inflamed group, T helper 2 (Th2) cells were significantly lower in the inflamed group (q < 0.15) (Fig 1B). Composite T-cell cluster score (quantified from the 361 genes used for clustering) correlated positively not only with genes from the antigen processing machinery, but also with the negative regulators of T-cell function (FOXP3, VEGFC) and immune checkpoints (VISTA, TIM-3, TIGIT, PD-L2, LAG3) (Fig 1C), with strongest positive correlations observed for VISTA (r=0.90), a gene shown to correlate with poor prostate cancer prognosis and increased in patients treated with ipilimumab likely due to increased M2 macrophage polarization and decreased IFN-γ production [38]. IHC analysis (CD8, GZMA, CD68 and PD-L1) of a subset of available cases (6 PCa, 6 CRPC, 3 NEPC) where tissue was available confirmed low immune infiltration in the deserted subtype (Suppl Fig. 1D).
To identify genomic alterations associated with T-cell inflammation, we used available WES data from 104 tumors. We did not find differences in median mutational loads between the inflamed and depleted groups (median TMB: inflamed = 1.46; depleted = 1.14 Mut/Mb) (Fig. 1D). Total copy number loads were also not different between the groups (Fig. 1D), but we noted significantly higher copy number gains in depleted versus inflamed tumors (Wilcox p value = 0.007) (Suppl Fig. 1E). The frequency of known prostate cancer mutations [2] did not differ significantly between the two groups, but we observed trends; TP53 (33% vs 24%) and BRCA2 (8% vs 1%) mutations were higher in inflamed, while FOXA1 (11% vs 21%), PIK3CA (0% vs 6%), ATM (3% vs 7%), and AR (6% vs 12%) mutations were higher in depleted tumors (Fig. 1E, Suppl Fig. 1F). For copy number changes (including those in genes related to antigen presentation and immune checkpoints), only CDKN1B and ATM had significantly higher copy number losses in the depleted compared to the inflamed groups (Fisher’s p = 0.012 (CDKN1B); p = 0.038 (ATM)) but did not remain significant after multiple hypothesis correction (q > 0.1) (Fig. 1F, Suppl Fig. 1G).
In many solid tumors, tumor immune infiltration correlates with clinical outcomes [39], but such correlations are not well established in prostate cancer [10, 40, 41]. In our cohort, 82 patients had overall survival (OS) data available. In this exploratory analysis, patients from the inflamed group associated with shorter OS compared to those from the depleted group (HR = 2.62, p < 0.05) (Fig. 1G). We reasoned that perhaps the elevated co-presence of pro-tumor factors (e.g., VISTA, M2 macrophages, CAFs, T-cell) with cytotoxic T-cells in the T-cell inflamed group antagonized anti-tumor T-cell function (Fig 1B–C). Further validation is required.
NEPC is predominantly T-cell depleted
Tumors in the T-cell inflamed, and depleted groups differed by their localization and aggressiveness (X-squared = 20.48, p-value = 0.0004) (Fig. 2A, Suppl Fig. 2A). We found that prostate tumors were relatively immune depleted (n=127/199; 64%) compared to the benign prostate tissues (n=9/31; 29%) (Fig. 2A). To understand how this classification compared to other prostate cancers, we applied our signature to the SU2C-PCF metastatic CRPC and TCGA localized prostate cancer cohorts (Suppl Fig. 2 B,C). Overall, we found that the majority of samples classified as T-cell depleted (56%) in the SU2C-PCF cohort (Suppl Fig. 2B). Similarly, 78% of the samples classified as depleted in the primary prostate cohort of the TCGA (Suppl Fig. 2C). Analysis of other cancer types using the same method identified 59% primary lung adenocarcinomas and 51% of metastatic melanomas as T-cell inflamed (Suppl Fig. 2C).
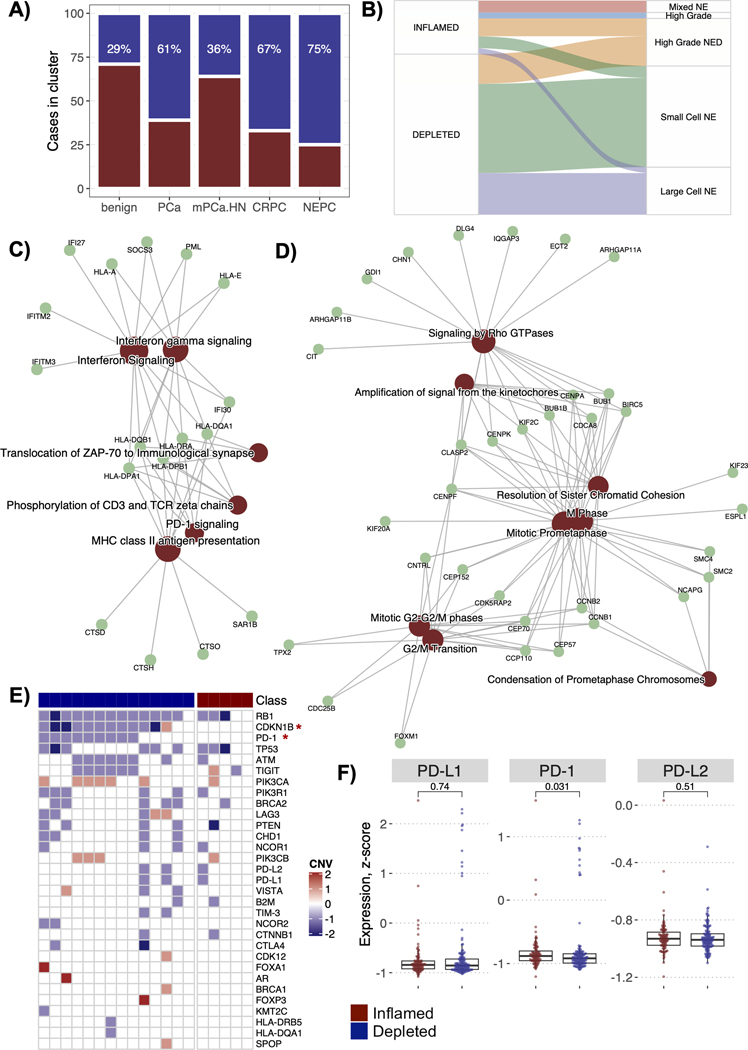
Figure 2.
T-cell groups by prostate cancer diagnoses. A) Barplot showing the percentage of cases among each prostate cancer diagnosis that belongs to T-cell inflamed or depleted groups, B) Sankey plot for the NEPC cases showing which NEPC histological subtype do the T-cell inflamed and depleted cases belong to, C) Enrichment maps for WGCNA module (MEmagenta) enriched in the inflamed NEPC cases. The map shows the network of key module genes and overrepresented pathways from the REACTOME database. D) Enrichment maps for WGCNA module (MEpurple) enriched in the depleted NEPC cases. E) Oncoprint plot for the NEPC cases showing the absolute copy number changes in 31 known prostate driver genomic alterations, MHC complex and common immune checkpoint genes. The oncoprint is segregated by the NEPC inflamed and depleted categories. Color red shows amplifications, blue shows deletions, and white shows a copy neutral status. F) Boxplots to show differences in the gene expression levels of the checkpoint gene PD-1 and its ligands PD-L1 and PD-L2 among the NEPCs, grouped by their inflamed or depleted status. p values reported are from the Wilcoxon signed-rank test.
In our cohort, metastatic prostate cancers were mostly T-cell depleted, apart from mHN.PCa (Fig. 2A). Among the prostate cancers (PCa, mPCa.HN, CRPC, NEPC), we found the highest frequency of T-cell depleted samples in the most aggressive NEPCs (n=27/36, 75%), with only 9 NEPC tumors classified as T-cell inflamed (iNEPC) (Fig. 2A). Analysis of the SU2C-PCF dataset [5] also confirmed T cell depletion in NEPC cases (Supp Fig. 1D). In our cohort, iNEPC cases had the lowest median expression of AR and were of both pure and mixed morphology (p < 0.05) (Suppl Fig. 3A, Fig. 2B). These observations revealed a minority of NEPC cases with inflammation in the otherwise T-cell depleted NEPC.
We sought to identify additional biological processes associated with inflammation in the subset of iNEPC. We applied WGCNA to NEPC GEPs and found a gene module (MEmagenta) with strongest positive correlation with iNEPC compared with non-inflamed NEPCs (r=0.68). This module enriched in genes of IFN-γ signaling, major histocompatibility complex (MHC) Class II antigen presentation, and PD-1 signaling (Fig. 2C, Suppl Fig. 3B). Similarly, we identified that the extracellular matrix organization and collagen degradation processes also correlated positively with iNEPC (r=0.59) (Suppl Fig. 3C). Conversely, a gene module (MEpurple) with strongest negative correlation with iNEPC (r = −0.52) was enriched in pathways associated with cell cycle (e.g., M phase, G2/M transition) (Fig. 2D).
In addition to the biological processes differentially enriched among the NEPC subtypes, we found that the CDKN1B loss was exclusive to non-inflamed NEPC (Fisher’s p = 0.0025), as was PD-1 loss (Fisher’s exact p = 0.03) (Fig. 2E). The expression of PD-1 was significantly higher in iNEPC (p = 0.03) while expression of its ligand PD-L1 was comparable to other NEPC cases (Fig. 2F). Taken together, higher CD8+ T-cells, IFN-γ signaling and higher expression of PD-1 in a subset of NEPC suggests a potential window of opportunity for ICB [42].
NEPC has higher PD-L1 expression and cell proliferation compared to other prostate cancers
We surveyed shifts in TME profiles of samples (comprised of 80 immune and stromal features) along the prostate cancer continuum. A principal component analysis (PCA) of the TME profiles showed that advanced tumors (e.g., CRPC, NEPC) had higher heterogeneity in their TME profiles while the localized PCa and benign tissues clustered more tightly (Fig. 3A). We identified distinctions within the TME profiles of NEPC by T-cell immune class. Clustering of TME profiles averaged by cancer diagnosis showed that immune-depleted (dNEPC) clustered independently while iNEPC clustered with the PCa tumors and benign prostate tissues (Fig. 3A).
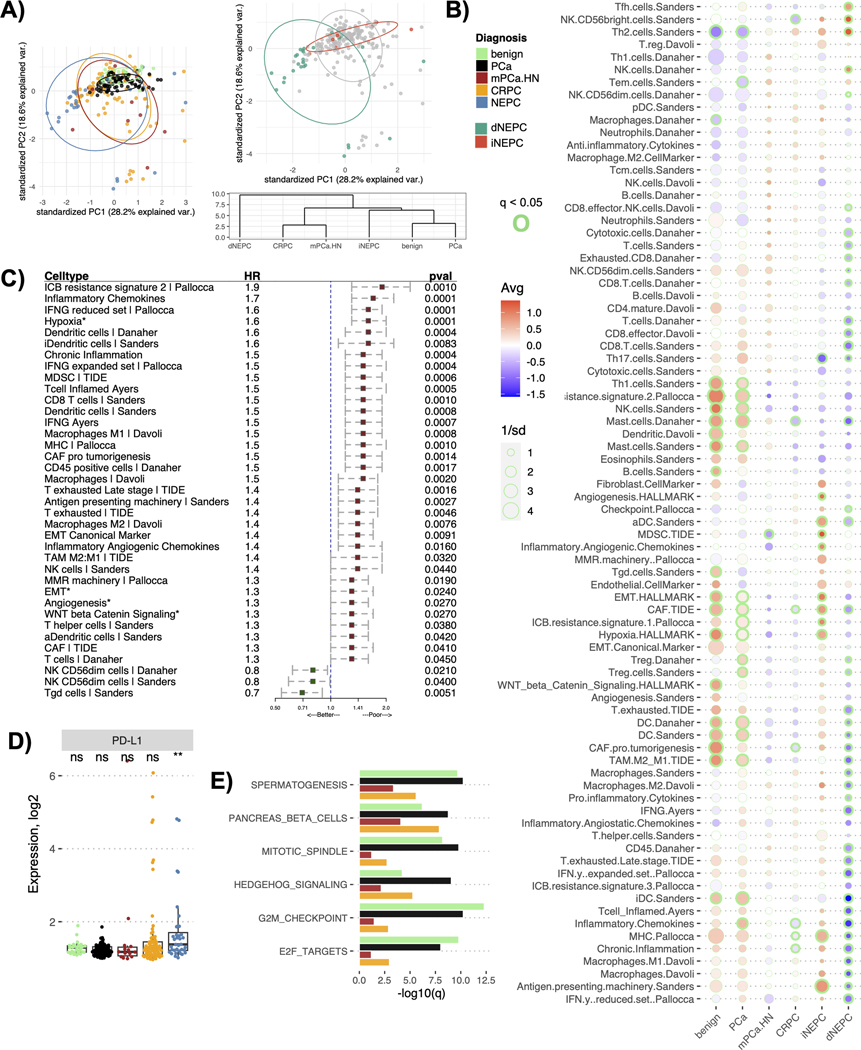
Figure 3.
TME profile of distinct prostate cancer types in the cohort. A) Plot for the principal component analysis applied to the TME profiles (comprised of 80 gene signatures) of the prostate cancer cohort colored either by diagnosis (left panel) or by NEPC immune classes (right panel top). The right panel bottom shows a cluster dendrogram for the prostate cancer diagnosis based on their averaged TME profiles. The dendrogram was constructed using the hierarchical clustering (distance= euclidean, method=ward D2), B) Bubble plot showing differences in the averaged TME profiles among the prostate cancer diagnosis in the cohort. The color of the circles indicates the mean (Avg) enrichment score by diagnosis for that feature, with red denoting high values and blue denoting low values on a color gradient scale. The size of the circle is determined by 1/standard deviation (sd) of the scores within that diagnosis, larger the circle smaller the sd among the tumors for that feature scores. The enrichment scores significantly different from at least four other cancers diagnoses being compared at a q < 0.05 are outlined with green, the thickness of the circle denotes the number of diagnoses it is significantly different from, C) Forest plots showing associations between the OS and individual features of the TME profile. Asterisk indicates that the corresponding feature is a cancer hallmark pathway. Hazard ratio (HR) and p-values (pval) are reported from the Cox proportional-hazards regression models adjusted for cancer type. The plot only shows associations that were significant at p < 0.05, D) Boxplot showing the differences in the gene expression levels of PD-L1 among the prostate cancer types. Asterisk denotes levels significantly different from all other cancer types compared (p < 0.05), while ns denotes not significant p-values from Wilcoxon signed-rank test, E) Barplot showing the cancer hallmark pathways significantly overexpressed in NEPCs compared to all other prostate diagnosis. The x-axis shows q values (-log10 scale) for the differences between the enrichment scores for the corresponding pathways in NEPC versus the cancer type being compared to.
The TME profiles averaged by cancer type showed an overall decrease in immune and stromal features from benign prostate tissues towards PCA, CRPC and finally NEPC, especially in dNEPC (Fig. 3B). The only cell types enriched in dNEPC compared to other cancers were Natural Killer (NK), T follicular helper (Tfh) and Th2 cells (Fig. 3B, Suppl Fig. 4A). Median levels of Th2 cells increased progressively with cancer aggressiveness (Suppl Fig. 4A). Consistent with our WGCNA, we found significantly higher expression of MHC presentation pathway and antigen presenting machinery genes in iNEPC compared to dNEPC (Fig. 3B).
In a survival analysis, we found that higher levels of individual TME features correlated with poor OS, a finding consistent with the outcomes observed in the T-cell inflamed versus depleted groups (Fig. 1G, Fig. 3C). Top features with negative associations with OS included ICB resistance signature (HR = 1.9) and hypoxia (HR = 1.6). Features with significantly positive associations with OS were T gamma delta (Tgd) and the NK CD56-dim cells (Fig. 3C).
In context of immunotherapy options for patients in our cohort, we reviewed the expression of prominent checkpoint genes such as the PD1 - PD-L1 axis for cytotoxic T-cell regulation [43]. We identified PD-L1 as the only checkpoint gene which had a significantly higher expression in NEPC compared to other prostate cancers (p < 0.05) (Fig. 3D, Suppl Fig. 4B). PD-L1 expression was higher in tumors that metastasized to lymph nodes or visceral organs compared to localized or other metastatic sites (Suppl Fig. 4C). Despite high PD-L1, NEPCs had significantly lower expression of B2M (key for neoantigen presentation) compared to other cancers, possibly indicative of immune evasion in NEPC (Suppl Fig. 4B).
Since NEPC tumors were broadly immune and stroma depleted compared to other prostate cancers, we sought to identify pathways that were specifically upregulated in NEPC. We quantified 50 cancer hallmark pathways (MSigDB RRID:SCR_016863) [44] from GEPs using ssGSEA (Suppl Fig. 4D). Pathways significantly upregulated in NEPC (compared to all other prostate cancers) were functionally related to cell proliferation and cancer invasiveness i.e., cell cycle G2/M checkpoint, E2F targets, Hedgehog signaling (q < 0.1) (Fig. 3E, Suppl Fig. 4D). These findings concluded that NEPC, the most aggressive variant of prostate cancer, has actively proliferating tumor cells coupled with an immune desert.
NEPC has distinct immune profile compared to SCLC
NEPC shares histological and molecular features with SCLC and because patients with SCLC are treated with ICB [7, 8, 45], we compared the immune features of NEPC and SCLC. A PCA plot of GEPs showed that NEPC tumors were clearly distinguishable from SCLC and were more heterogenous among themselves compared to PCa and SCLC tumors (Fig. 4A, Suppl Fig. 5A). This may be, in part, a reflection of the heterogeneity observed within the spectrum of NEPC, with both small cell carcinoma and mixed small cell/adenocarcinoma histologies [46]. Globally, SCLC had significantly higher expression of most immune features tested compared to NEPC, except the expression of two checkpoints PD-L1 (proposed predictor of response to pembrolizumab in SCLC [47] ) and CTLA-4 which was not distinguishable between SCLC and NEPC tumors (PD-L1 p = 0.93, CTLA-4 p = 0.11) (Fig 4B, Suppl Fig. 5B–D).
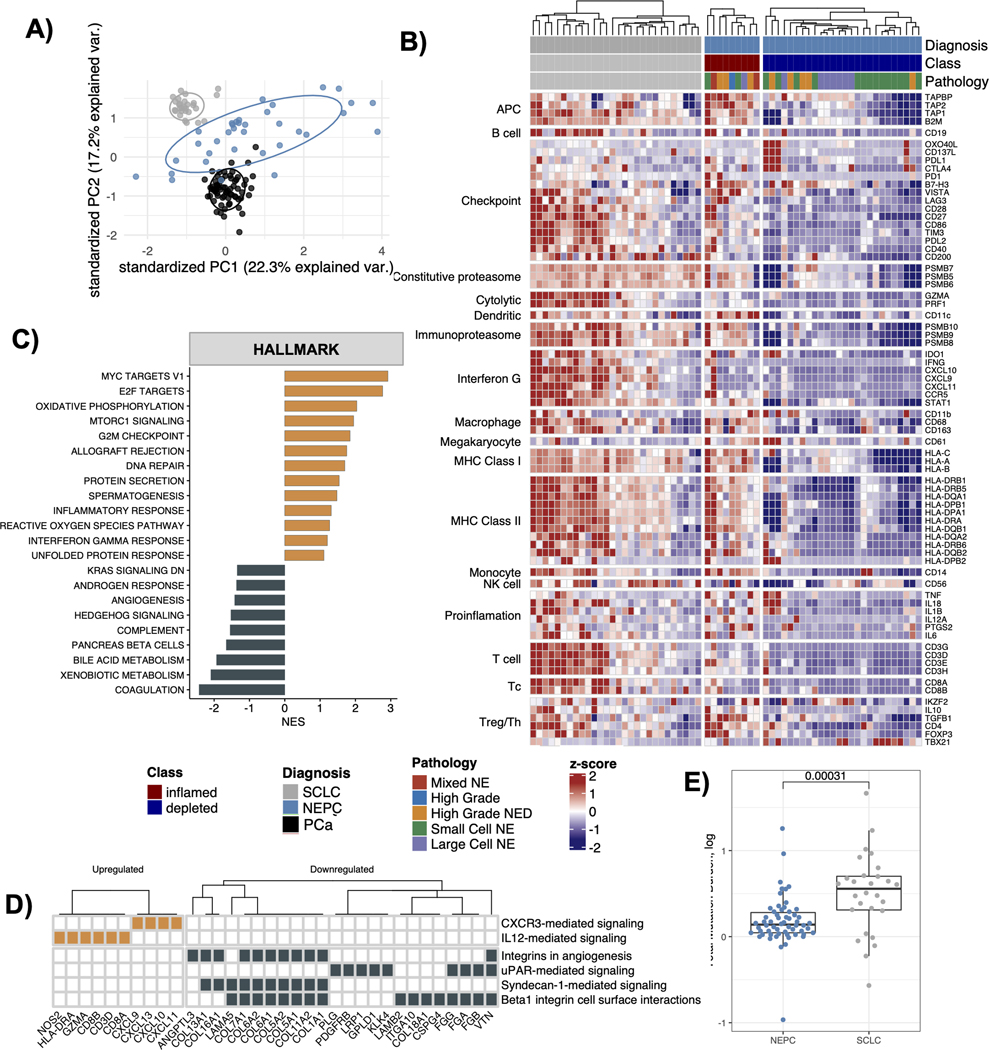
Figure 4.
Comparative analysis between NEPC and SCLC. A) Principal component analysis plot for the gene expression profiles obtained from NEPC, PCa and SCLC tumors, B) Heatmap showing expression of marker genes for immune cell types and immune checkpoint among the NEPC and SCLC tumors. The gene expression values are scaled by row and the plot is segregated by cancer diagnosis and the immune status of NEPCs tumors, C) Barplot showing cancer hallmark pathways identified as significantly upregulated (orange) or downregulated (black) in the SCLC versus the NEPCs (fdr < 0.25). x-axis shows normalized enrichment scores (NES) from the GSEA analysis applied to the list of genes differentially expressed between SCLC versus NEPC, ranked by their log2 fold change values, D) Heatmap showing 6 out of the 12 total enriched NCI-Nature_2016 pathways (enrichR) in the DEGs for SCLC versus NEPC, E) Boxplot showing the difference in tumor mutation burden between the NEPC and the SCLC tumors. p values reported are from the Wilcoxon signed-rank test.
Differential gene expression analysis between SCLC and NEPC tumors revealed that the cancer hallmark pathways associated with immune processes (e.g., IFN-γ, inflammatory response, allograft rejection) were significantly upregulated in SCLC tumors (q < 0.25) compared to immune depleted NEPC (Fig 4C, Suppl Table S4, Suppl Fig. 5E). In addition, cell cycle processes were upregulated while angiogenesis and hedgehog signaling genes were downregulated in SCLC compared to NEPC (Fig 4C). The top DEGs in SCLC versus NEPC (LFC > 2, q < 0.05) contained canonical T-cell signaling markers (CD8A, CD8B, CD3D, GZMA), while the bottom DEGs (LFC < −2, q < 0.05) were enriched with integrin and collagen genes, and uPAR mediated signaling (which regulates extracellular matrix degradation and cancer cell migration and metastasis) (Suppl Table S4–S5, Fig 4D).
Higher mutation burden is a predictor of ICB response [48], and in that context we found that the median mutation load in SCLC was significantly higher than NEPC as well as other prostate cancers (Fig 4E, Suppl Fig. 5F). Taken together these findings suggest that despite similarities in tumor morphology and comparable PD-L1 expression, highly immune depleted TME and relatively lower mutation load in NEPC could make it challenging to extend the clinical success of immunotherapies in SCLC to patients diagnosed with NEPC.
DISCUSSION
Transcriptome analysis of a large cohort of patients with prostate cancer comprising localized PCa, mHN.PCa, CRPC and NEPC for signatures of T-cell infiltration delineated a significant lack of immune infiltration in prostate cancer. While immune infiltration was limited across the different of stages of prostate cancer progression, mHN.PCa was an outlier with more than half displaying an inflamed phenotype. Further analysis of TME profiles across our cohort demonstrated a loss of reactive immune cells across the malignant castration resistant subgroups, indicative of a poor immune response.
Of specific interest was the observation that a small subset of NEPC had increased immune infiltration. When interrogated independently, NEPC represents the most immune-depleted TME, with an absence of antigen presenting machinery or cytotoxic T-cells. The immunosuppressive Tregs observed in NEPC cases are perhaps indicative of prior therapeutic interventions. Cases of iNEPC display a significantly higher expression of PD1, PDL-2 and cytolytic activity compared to other NEPC subtypes and upregulation of antigen presentation pathways. While the number of cases was small and validation is required, this finding indicates that there might be a subset of NEPC cases that respond to immunotherapy that warrants further study.
We further characterized our NEPC cohort by comparing the immunogenomic landscape to another small-cell NE carcinoma, SCLC. Clinically, NEPC is often treated with SCLC therapies given a lack of prospective trials showing efficacy to support NEPC-specific therapeutics, and this approach is supported by National Comprehensive Cancer Network (NCCN) guidelines. We discovered NEPC to be significantly less immunogenic than SCLC, an observation that may be due, in part, to the higher mutation load associated with SCLC and/or a higher MHC 1 expression in a subset. When we examine the checkpoint marker PD-L1 and CTLA-4, NEPC tumors had expression levels comparable to SCLC. The implication of checkpoint inhibition in this context is significant and currently being tested in ongoing clinical trials (NCT03179410, NCT04848337, NCT04709276, NCT03582475). Our data suggests that while NEPC is an immune-depleted tumor subtype, the expression profile of a limited subset of clinically relevant immune-related genes is closely associated with other small-cell malignancies.
There are several limitations to this study. Our findings are based on GEPs, and further studies that include multiplexed IHC or spatial- omics analyses are needed to confirm expression of identified immune markers within the tumor and stromal milieu as well as distinguish those expressed by tumor and immune cells. NEPC is a heterogenous disease including pure small cell carcinoma and mixed features [6]; moreover, there are other emerging phenotypes within CRPC (e.g., AR-negative, NE-negative double-negative CRPC and amphicrine CRPC) that have not been addressed here [6]. The study is retrospective and furthermore, treatment response data for patients treated with ICB was not available. Finally, these analyses were not stratified by race or ethnicity, and it is possible that the TME and immune milieu differs across diverse patient populations.
By assessing the expression profiles of NEPC within the paradigms of prostate malignancies and small-cell lung carcinoma, we have revealed distinct immune-related gene expression profiles related to NEPC. These findings may have implications for future immunotherapy strategies and for biomarker development.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Histologic transformation from prostate adenocarcinoma to small cell neuroendocrine prostate cancer (NEPC) is an emerging resistance mechanism seen in up to 15% of advanced prostate cancers. There are no approved therapies for NEPC and treatment, including immunotherapy, is often extrapolated from small cell lung cancer, a tumor type with similar histological features. We characterized the immunogenomic landscape of the tumor microenvironment in NEPC compared to prostate adenocarcinoma and small-cell lung carcinoma (SCLC). We found that NEPC is relatively the most T-cell immune depleted, except for a subset of inflamed tumors. Tumor microenvironment profiling, as reported in this study, may inform patient selection and immunotherapy strategies for patients with advanced prostate cancer.
Funding:
H.B. is supported by the Prostate Cancer Foundation, Department of Defense (W81XWH-17–1-0653), and NIH/NCI (R37CA241486, P50-CA211024).
Conflict of Interest
H.B. has served as consultant/advisory board member for Janssen, Astellas, Merck, Pfizer, Foundation Medicine, Blue Earth Diagnostics, Amgen, Bayer, Oncorus, LOXO, Daicchi Sankyo, Sanofi, Curie Therapeutics, Novartis, Astra Zeneca; H.B. has received research funding from Janssen, AbbVie/Stemcentrx, Eli Lilly, Astellas, Millennium, Bristol Myers Squibb, Circle Pharma, Daicchi Sankyo, Novartis. O.E. is supported by Janssen, J&J, Astra-Zeneca, Volastra and Eli Lilly research grants; O.E. is scientific advisor and equity holder in Freenome, Owkin, Pionyr Immunotherapeutics, Volastra Therapeutics and One Three Biotech and a paid scientific advisor to Champions Oncology; V.C. has served as a consultant/advisory board member for Janssen, Astellas, Merck, AstraZeneca, Amgen, and Bayer; V.C. has received speaker honoraria or travel support from Astellas, Janssen, Ipsen, Bayer and Bristol.
REFERENCES
- 1.Sung H, et al. , Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin, 2021. 71(3): p. 209–249. [DOI] [PubMed] [Google Scholar]
- 2.Beltran H, et al. , Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med, 2016. 22(3): p. 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluemn EG, et al. , Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell, 2017. 32(4): p. 474–489 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aggarwal R, et al. , Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol, 2018. 36(24): p. 2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abida W, et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A, 2019. 116(23): p. 11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein JI, et al. , Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol, 2014. 38(6): p. 756–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn L, et al. , First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med, 2018. 379(23): p. 2220–2229. [DOI] [PubMed] [Google Scholar]
- 8.Paz-Ares L, et al. , Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet, 2019. 394(10212): p. 1929–1939. [DOI] [PubMed] [Google Scholar]
- 9.Beltran H. and Demichelis F, Therapy considerations in neuroendocrine prostate cancer: what next? Endocr Relat Cancer, 2021. 28(8): p. T67–T78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitkin N, et al. , The Tumor Immune Contexture of Prostate Cancer. Front Immunol, 2019. 10: p. 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calcinotto A, et al. , IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature, 2018. 559(7714): p. 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stultz J. and Fong L, How to turn up the heat on the cold immune microenvironment of metastatic prostate cancer. Prostate Cancer Prostatic Dis, 2021. 24(3): p. 697–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall CH and Antonarakis ES, Emerging treatments for metastatic castration-resistant prostate cancer: Immunotherapy, PARP inhibitors, and PSMA-targeted approaches. Cancer Treat Res Commun, 2020. 23: p. 100164. [DOI] [PubMed] [Google Scholar]
- 14.Goswami S, Aparicio A, and Subudhi SK, Immune Checkpoint Therapies in Prostate Cancer. Cancer J, 2016. 22(2): p. 117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sena LA, et al. , Tumor Frameshift Mutation Proportion Predicts Response to Immunotherapy in Mismatch Repair-Deficient Prostate Cancer. Oncologist, 2021. 26(2): p. e270–e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powles T, et al. , Atezolizumab with enzalutamide versus enzalutamide alone in metastatic castration-resistant prostate cancer: a randomized phase 3 trial. Nat Med, 2022. 28(1): p. 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown LC, et al. , A phase 2 trial of avelumab in men with aggressive-variant or neuroendocrine prostate cancer. Prostate Cancer Prostatic Dis, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beltran H, et al. , Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov, 2011. 1(6): p. 487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltran H, et al. , Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol, 2015. 1(4): p. 466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran H, et al. , A phase II trial of the aurora kinase A inhibitor alisertib for patients with castration resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin Cancer Res, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudin CM, et al. , Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2013. 29(1): p. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, and Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics, 2015. 31(2): p. 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc, 2012. 7(3): p. 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derrien T, et al. , The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res, 2012. 22(9): p. 1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbie DA, et al. , Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature, 2009. 462(7269): p. 108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney MS, et al. , Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell, 2015. 160(1–2): p. 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, and Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov MV, et al. , Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res, 2016. 44(W1): p. W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rennert H, et al. , Development and validation of a whole-exome sequencing test for simultaneous detection of point mutations, indels and copy-number alterations for precision cancer care. NPJ Genom Med, 2016. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson BD, et al. , Upper tract urothelial carcinoma has a luminal-papillary T-cell depleted contexture and activated FGFR3 signaling. Nat Commun, 2019. 10(1): p. 2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellrott K, et al. , Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst, 2018. 6(3): p. 271–281 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talevich E, et al. , CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol, 2016. 12(4): p. e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson MD and Hayes DN, ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics, 2010. 26(12): p. 1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P. and Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 2008. 9: p. 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao F, et al. , The neurotrophic tyrosine kinase receptor TrkA and its ligand NGF are increased in squamous cell carcinomas of the lung. Sci Rep, 2018. 8(1): p. 8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskov H, et al. , Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer, 2021. 124(2): p. 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, et al. , VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med, 2017. 23(5): p. 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giraldo NA, et al. , The clinical role of the TME in solid cancer. Br J Cancer, 2019. 120(1): p. 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ness N, et al. , Infiltration of CD8+ lymphocytes is an independent prognostic factor of biochemical failure-free survival in prostate cancer. Prostate, 2014. 74(14): p. 1452–61. [DOI] [PubMed] [Google Scholar]
- 41.Davidsson S, et al. , CD4 helper T cells, CD8 cytotoxic T cells, and FOXP3(+) regulatory T cells with respect to lethal prostate cancer. Mod Pathol, 2013. 26(3): p. 448–55. [DOI] [PubMed] [Google Scholar]
- 42.Tumeh PC, et al. , PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature, 2014. 515(7528): p. 568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun C, Mezzadra R, and Schumacher TN, Regulation and Function of the PD-L1 Checkpoint. Immunity, 2018. 48(3): p. 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberzon A, et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst, 2015. 1(6): p. 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conteduca V, et al. , Clinical features of neuroendocrine prostate cancer. Eur J Cancer, 2019. 121: p. 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labrecque MP, et al. , Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest, 2019. 129(10): p. 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott PA, et al. , Pembrolizumab in Patients With Extensive-Stage Small-Cell Lung Cancer: Results From the Phase Ib KEYNOTE-028 Study. J Clin Oncol, 2017. 35(34): p. 3823–3829. [DOI] [PubMed] [Google Scholar]
- 48.Chan TA, et al. , Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol, 2019. 30(1): p. 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.