Abstract
Background and Aims:
Surveillance of gastric intestinal metaplasia (GIM) may lead to early gastric cancer detection. Our purpose was to externally validate a predictive model for endoscopic GIM previously developed in a veteran population in a second U.S. population.
Methods:
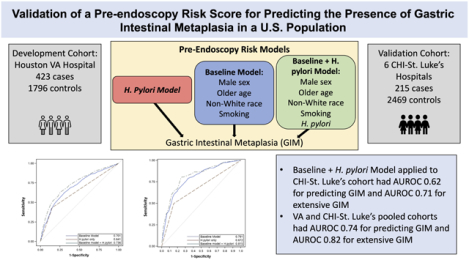
We previously developed a pre-endoscopy risk model for detection of GIM using 423 GIM cases and 1796 controls from the Houston VA Hospital. The model included sex, age, race/ethnicity, smoking, and H. pylori infection with an area under the receiver operating characteristic (AUROC) curve of 0.73 for GIM and 0.82 for extensive GIM. We validated this model in a second cohort of patients from six CHI-St. Luke’s hospitals (Houston, Texas) from January–December 2017. Cases were defined as having GIM on any gastric biopsy, and extensive GIM as involving both antrum and corpus. We further optimized the model by pooling both cohorts and assessing discrimination using AUROC.
Results:
The risk model was validated in 215 GIM cases (55 with extensive GIM) and 2469 controls. Cases were older than controls (59.8 vs. 54.7 years) with more non-whites (59.1% vs. 42.0%,) and H. pylori infection (23.7% vs. 10.9%). The model applied to the CHI-St. Luke’s cohort had AUROC 0.62 (95% confidence interval [CI] 0.57–0.66) for predicting GIM and AUROC 0.71 (95%CI 0.63–0.79) for extensive GIM. When the VA and CHI-St. Luke’s cohorts were pooled, discrimination of both models improved (GIM AUROC 0.74; extensive GIM AUROC 0.82).
Conclusion:
A pre-endoscopy risk prediction model was validated and updated using a second U.S. cohort with robust discrimination for endoscopic GIM. This model should be evaluated in other U.S. populations to risk stratify patients for endoscopic GIM screening.
Keywords: gastric intestinal metaplasia, risk prediction model, gastric cancer, Helicobacter pylori, cancer screening
Graphical Abstract
Introduction
Gastric cancer is the fourth leading cause of cancer-related death world-wide1. Although gastric cancer is decreasing in the overall U.S. population, the incidence rates are increasing among non-Hispanic white and Hispanic individuals <50 years old2–4. Most cases of gastric cancer are diagnosed at a late stage, when the 5-year survival is greatly diminished (i.e., 5%) as compared to those diagnosed with localized disease (up to 69%)5. Thus, diagnosis at an early stage is imperative to decreasing mortality from gastric cancer.
Gastric cancer is thought to develop via a progression from gastric atrophy and gastric intestinal metaplasia (GIM) mostly in the setting of Helicobacter pylori infection6. Thus, GIM is considered an important precursor of gastric cancer and a promising target for screening and endoscopic surveillance to facilitate detection and treatment of early gastric cancers7. Endoscopic screening programs for gastric cancer have been implemented in Korea and Japan8 with resultant increases in detection of early gastric cancers and reduced mortality rates9, 10. While U.S. societies have released guidance on treatment and surveillance after diagnosis of GIM11, there is no clear guidance on whom to screen for GIM, and this gap is in large part due to the absence of validated generalizable risk stratification tools.
We previously described several significant demographic and clinical risk factors for GIM among patients presenting for elective endoscopy as well as among patients in primary care settings12–16. Based on these risk factors, we developed pre-endoscopy risk prediction models for the presence of GIM in a U.S veteran population within Houston, Texas17. The predictive variables in our best performing risk model included H. pylori, sex, age, race/ethnicity, and smoking status. The weighted logistic regression model had good discrimination (area under the receiver operating characteristic [AUROC] 0.73) for detecting any GIM and slightly better discrimination for extensive GIM involving both antrum and corpus (AUROC 0.82)17. However, this risk prediction model needs to be externally validated in multiple populations to be generalizable and useful18. The purpose of this study was to externally validate and optimize pre-endoscopy risk prediction models for presence of GIM in a second non-veteran multi-ethnic U.S. population.
Methods
Study Populations
External Validation (CHI-St. Luke’s Population)
The external validation cohort consisted of a pooled cohort of patients from six Catholic Health Initiative-St. Luke’s (CHI-St. Luke’s) Hospital endoscopy units in Houston, Texas who underwent EGD with gastric biopsies from January to December 2017. We previously described the study population19. Patients underwent gastric biopsy for either symptomatic indication or endoscopic finding and were identified using Current Procedural Terminology (CPT) code 43,239 from the electronic endoscopic reporting software database (ProVation Medical; Minneapolis, MN). Only patients over 18 years of age were included, and we excluded patients with altered gastric anatomy. This study was approved by the Institutional Review Board at Baylor College of Medicine and the MEDVAMC. We used the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) guideline checklist20, which provides recommendations on developing and validating risk prediction models, in the reporting of our study.
Model Optimization (MEDVAMC Population)
We previously identified 423 GIM cases and 1796 controls among patients recruited from primary care and endoscopy clinics at the Michael E. DeBakey VA Medical Center (MEDVAMC) in Houston, Texas between February 2008 and August 2013 who underwent esophagogastroduodenoscopy (EGD) with gastric mapping biopsies15. Patients were recruited from two sources: among asymptomatic patients undergoing research EGD along with their colonoscopy for colon cancer screening, and among symptomatic patients previously scheduled for EGD who then consented for additional gastric biopsies. This population was first used to develop the risk models for presence of GIM17 and used in the current study to further optimize the models after pooling with the CHI-St. Luke’s population.
Data Collection
We collected patient demographics (i.e., sex, age, race/ethnicity) and clinical data (i.e.,H. pylori, smoking history) from the electronic medical record (EPIC Hyperspace; Verona, WI). Presence of H. pylori infection was defined as positive test for H. pylori prior to or at the time of EGD based on stool antigen, urea breath test, serum antibody, gastric biopsy histopathology, or clinic notes indicating prior H. pylori infection.
Definition of GIM and Controls
Cases and controls were identified by reviewing consecutive endoscopies and corresponding pathology reports from all patients who underwent EGD with biopsy during the time period. Patients with GIM on any non-cardia gastric biopsy were defined as GIM cases, and controls were defined as patients not having GIM on any non-cardia gastric biopsy. Focal GIM was defined as GIM restricted to the antrum, and extensive GIM was defined as GIM involving both the antrum and corpus.
Statistical Analysis
We used chi-square test for categorical variables and student t-test for continuous variables to compare GIM cases and controls. For validation of the risk model, predictor variables previously included in the risk prediction model were evaluated: age (years), sex, race/ethnicity (non-Hispanic white, Hispanic, black), habitual smoking status (defined as usage of ≥1 cigarette per day), and H. pylori infection. We examined independent associations of each potential predictor variable with presence of GIM and extensive GIM in univariate and multivariate logistic regression analyses.
Risk Model Validation
To determine discrimination of the GIM model (i.e., ability of the model to correctly identify GIM), we applied the parameter estimates of the published model (i.e., the model/formula derived from the MEDVAMC development) to the CHI-St. Luke’s external validation cohort and reported the AUROC for 3 models: 1) H. pylori only model, 2) baseline model (sex, age, race/ethnicity [white, Hispanic, black], smoking status), and 3) combined model (H. pylori plus baseline model)17. We similarly determined predictive ability of the MEDVAMC models for presence of extensive GIM, defined as GIM in both antrum and corpus, in the CHI-St. Luke’s cohort. We also reported various probability thresholds for the combined model in the validation cohort with corresponding sensitivity, specificity, percentage of the population that would need to undergo screening EGD, and number of EGDs needed to detect one GIM case. We determined calibration (i.e., how closely the predicted risk correlates to the observed risk) by Hosmer-Lemeshow chi-square statistics. Furthermore, to optimize discrimination of the models, we pooled the MEDVAMC and CHI-St. Luke’s cohorts together and developed new risk estimates using the same variables and reported AUROC and 95% CI for the pooled cohorts. We additionally attempted to optimize the combined model by adding proton pump inhibitor (PPI) usage, which was the only significant variable independently associated with GIM in the validation cohort that was not originally included in the models19.
All analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC), and a 2-tailed p-value of < 0.05 was considered statistically significant.
Results
Study population
We included 215 GIM cases and 2469 controls in the CHI-St. Luke’s validation cohort, making the overall prevalence of GIM 8.0%. This was lower than the prevalence of GIM in the MEDVAMC development cohort (19.1%). Cases were older (mean age 59.8 vs. 54.7 years, p<0.01) with higher proportion of males (42.8% vs. 40.7%, p=0.54), non-white race/ethnicity (59.1% vs. 42.0%, p<0.01), smoking history (19.5% vs. 8.3%, p=0.03), and H. pylori infection (23.7% vs. 10.9%, p<0.01) compared to controls. Comparisons of the MEDVAMC development cohort and CHI-St. Luke’s validation cohort are in Table 1.
Table 1.
Comparison of demographic and clinical risk factors used in the risk prediction model for detection of gastric intestinal metaplasia for the development and validation cohorts
| Development Cohort (MEDVAMC) | Validation Cohort (CHI-St. Luke’s) | |||||
|---|---|---|---|---|---|---|
| Cases n=423(%) |
Controls n=1796 (%) |
p-value | Cases n=215 (%) |
Controls n=2469 (%) |
p-value | |
| Sex | <0.001 | 0.543 | ||||
| Male | 411 (97.16) | 1630 (90.76) | 92 (42.79) | 1004 (40.66) | ||
| Female | 12 (2.84) | 166 (9.24) | 123 (57.21) | 1465 (59.34) | ||
| Age | <0.001 | <0.001 | ||||
| <60 | 138 (32.62) | 761 (42.37) | 90 (41.86) | 1418 (57.43) | ||
| 60–69 | 218 (51.54) | 842 (46.88) | 72 (33.49) | 589 (23.86) | ||
| ≥70 | 67 (15.84) | 193 (10.75) | 53 (24.65) | 462 (18.71) | ||
| Race/Ethnicity | <0.001 | <0.001 | ||||
| White | 175 (41.37) | 1095 (60.97) | 88 (40.93) | 1431 (57.96) | ||
| Hispanic | 62 (14.66) | 148 (8.24) | 56 (26.05) | 425 (17.21) | ||
| Black | 178 (42.08) | 521 (29.01) | 44 (20.47) | 413 (16.73) | ||
| Asian | - | - | 22 (10.23) | 133 (5.39) | ||
| Other/Unknown | 8 (1.89) | 32 (1.78) | 5 (2.33) | 67 (2.71) | ||
| Habitual smoker | <0.001 | 0.031 | ||||
| No | 267 (63.12) | 1219 (67.87) | 173 (80.47) | 2120 (85.86) | ||
| Yes | 138 (32.62) | 473 (26.34) | 42 (19.53) | 206 (8.34) | ||
| Unknown/missing | 18 (4.26) | 104 (5.79) | 0 (0.00) | 0 (0.00) | ||
| Helicobacter pylori | <0.001 | <0.001 | ||||
| No | 199 (47.04) | 1377 (76.67) | 164 (76.28) | 2199 (89.06) | ||
| Yes | 219 (51.77) | 394 (21.94) | 51 (23.72) | 270 (10.94) | ||
| Unknown/missing | 5 (1.18) | 25 (1.39) | 0 (0.00) | 0 (0.00) | ||
| PPI use | 0.017 | 0.167 | ||||
| No | 205 (48.46) | 734 (40.87) | 172 (80.00) | 1865 (75.54) | ||
| Yes | 193 (45.63) | 935 (52.06) | 43 (20.00) | 595 (24.10) | ||
| Unknown/missing | 25 (5.91) | 127 (7.07) | 9 (0.36) | |||
PPI: proton pump inhibitor
Older age (ref age<60 years: age 60–69 years OR 2.18, 95% CI 1.56–3.04; age ≥70 years OR 2.29; 95% CI 1.58–3.30), non-White race/ethnicity (ref white race: Hispanic 2.26, 95% CI 1.56–3.26; black 1.65, 95% CI 1.12–2.43; Asian 2.67, 95% CI 1.59–4.47), habitual smoking (OR 1.60, 95% CI 1.11–2.30), and H. pylori infection (OR 2.30, 95% CI 1.62–3.28) were independently associated with GIM in the validation cohort after additionally adjusting for sex. The magnitudes of associations were stronger when comparing 55 cases with extensive GIM to 2469 controls without GIM (Table 2).
Table 2.
Associations of potential predictor variables with presence of gastric intestinal metaplasia (GIM) and extensive GIM (i.e., involving both antrum and corpus) among 215 GIM cases (of which 55 with extensive GIM) and 2469 controls in the CHI-St. Luke’s validation cohort
| GIM | Extensive GIM | |||
|---|---|---|---|---|
| Adjusted OR | 95% CI | Adjusted OR | 95% CI | |
| Sex | ||||
| Female | ref | ref | ref | ref |
| Male | 1.02 | 0.76–1.37 | 1.53 | 0.88–2.67 |
| Age | ||||
| <60 | ref | ref | ref | ref |
| 60–69 | 2.18 | 1.56–3.04 | 3.64 | 1.84–7.17 |
| ≥70 | 2.29 | 1.58–3.30 | 5.25 | 2.61–10.56 |
| Race/Ethnicity | ||||
| White | ref | ref | ref | ref |
| Hispanic | 2.26 | 1.56–3.26 | 4.20 | 2.06–8.58 |
| Black | 1.65 | 1.12–2.43 | 2.82 | 1.33–5.98 |
| Asian | 2.67 | 1.59–4.47 | 3.96 | 1.49–10.52 |
| Habitual Smoker | ||||
| No | ref | ref | ref | ref |
| Yes | 1.60 | 1.11–2.30 | 1.77 | 0.88–3.53 |
| Helicobacter pylori | ||||
| No | ref | ref | ref | ref |
| Yes | 2.30 | 1.62–3.28 | 2.62 | 1.39–4.96 |
CI: confidence interval; OR: odds ratio
Risk Model Validation
When the MEDVAMC models were applied to the CHI-St. Luke’s cohort, the combined model (i.e., H. pylori and baseline model) performed better (AUROC 0.62, 95% CI 0.57–0.66) than the model containing H. pylori alone (AUROC 0.57, 95% CI 0.53–0.60) or the baseline model (containing sex, age, race/ethnicity, smoking status; AUROC 0.59, 95% CI 0.54–0.64) (Table 3). Similarly, the combined risk model (H. pylori plus baseline model) for predicting extensive GIM performed better than the overall GIM model among 55 cases with extensive compared to 2469 controls (AUROC 0.71, 95% CI 0.63–0.79) (Table 3). The AUROCs were uniformly lower among the external validation cohort (the CHI-St. Luke’s cohort) compared with the MEDVAMC development cohort17 (Table 3).
Table 3.
Discrimination of the models in predicting gastric intestinal metaplasia (GIM) and extensive GIM the in development and validation cohorts, reported as area under receiver operating characteristic (AUROC) curve and 95% confidence interval (CI) for 3 models: 1) baseline model containing sex, age, race/ethnicity, and smoking status, 2) H. pylori only model, and 3) expanded model that included H. pylori and baseline model
| MEDVAMC (development) | CHI-St. Luke’s (validation) | Pooled MEDVAMC & CHI-St. Luke’s | ||||
|---|---|---|---|---|---|---|
| AUROC | 95 % CI | AUROC | 95 % CI | AUROC | 95% CI | |
| GIM models | ||||||
| Baseline model | 0.67 | 0.64–0.70 | 0.59 | 0.54–0.64 | 0.70 | 0.68–0.72 |
| H. pylori only model | 0.66 | 0.63–0.68 | 0.57 | 0.53–0.60 | 0.64 | 0.62–0.66 |
| Baseline + H. pylori | 0.73 | 0.71–0.76 | 0.62 | 0.57–0.66 | 0.74 | 0.71–0.76 |
| Extensive GIM models | ||||||
| Baseline model | 0.77 | 0.72–0.81 | 0.69 | 0.62–0.78 | 0.79 | 0.75–0.83 |
| H. pylori only model | 0.69 | 0.64–0.73 | 0.59 | 0.52–0.65 | 0.67 | 0.63–0.71 |
| Baseline + H. pylori | 0.82 | 0.78–0.85 | 0.71 | 0.63–0.79 | 0.82 | 0.78–0.85 |
In order to optimize model performance and enhance transportability, we pooled the MEDVAMC and CHI-St. Luke’s cohorts and developed new risk estimates (Supplementary Table 1). The new combined risk model (including variables for sex, age, race/ethnicity, smoking, H. pylori) using the pooled cohorts had better predictive performance for GIM (AUROC 0.74, 95% CI 0.71–0.76; Figure 1) and extensive GIM (AUROC 0.82, 95% CI 0.78–0.85; Figure 2) than the CHI-St. Luke’s validation cohort using estimates from the MEDVAMC cohort (Table 3). The addition of PPI use did not significantly improve performance of the combined model for GIM overall (AUROC 0.74, 95% CI 0.71–0.76) and extensive GIM (AUROC 0.82, 95% CI 0.78–0.85) in the pooled cohort. Finally, the combined model was well-calibrated according to the Hosmer-Lemeshow test (p=0.08).
Figure 1.
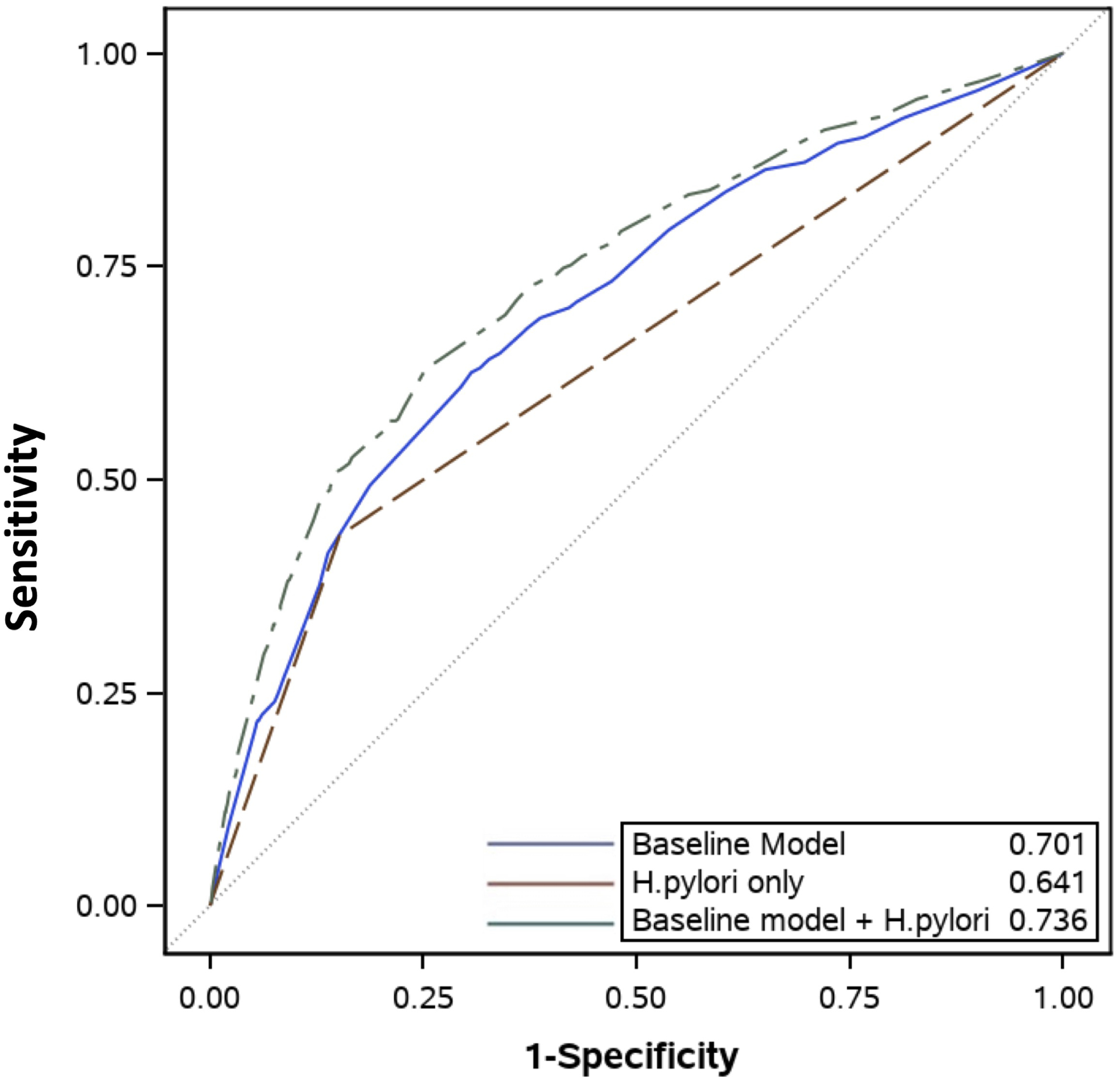
Area Under Receiver Operating Characteristic (AUROC) curves of 3 models in predicting gastric intestinal metaplasia updated in the pooled MEDVAMC and CHI-St. Luke’s cohorts for: 1) baseline model without H. pylori (blue), 2) H. pylori only model (red), and 3) combined model that includes H. pylori and baseline model (green)
Figure 2.
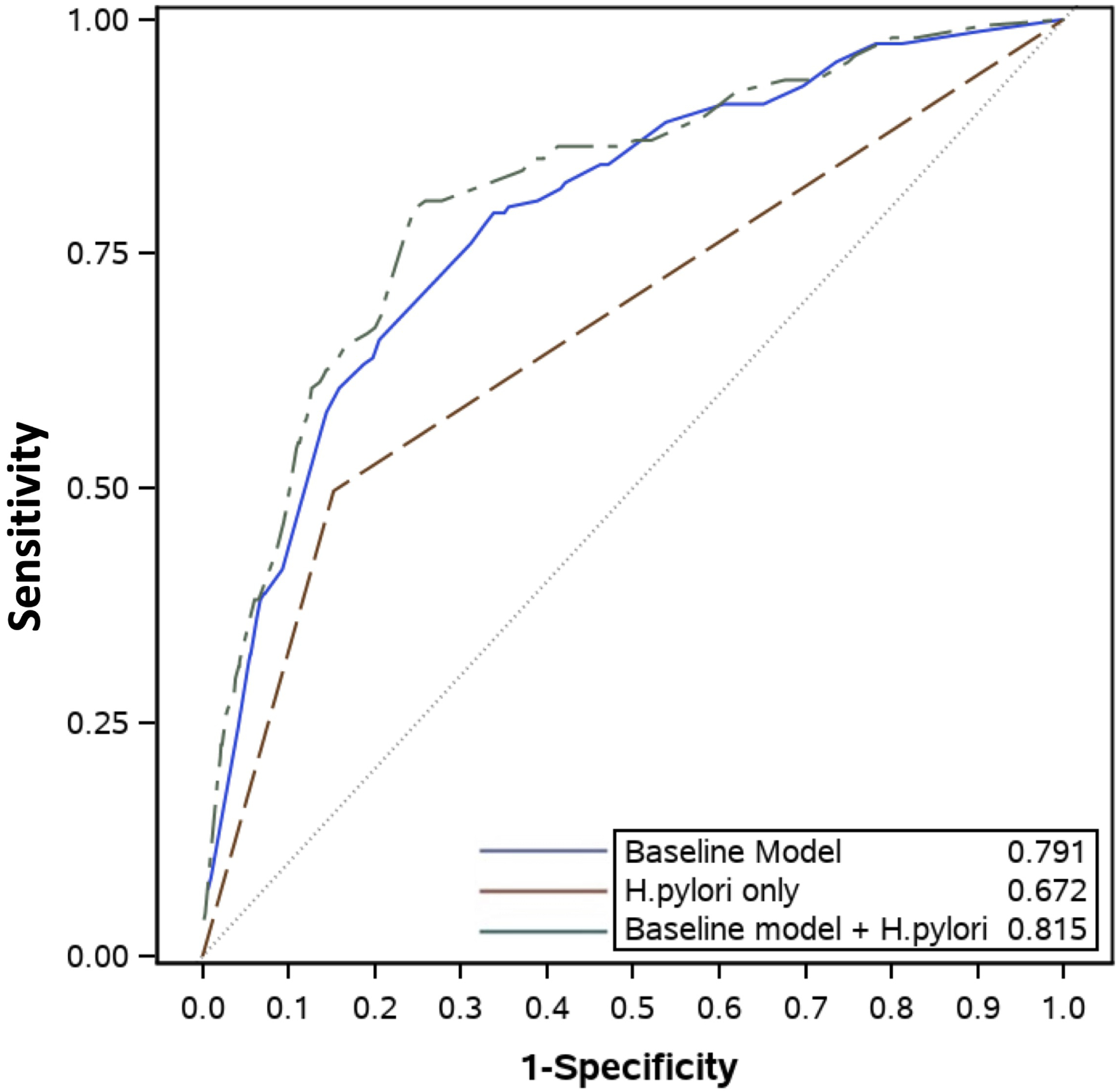
Area Under Receiver Operating Characteristic (AUROC) curves of 3 models in predicting extensive gastric intestinal metaplasia updated in the pooled MEDVAMC and CHI-St. Luke’s cohorts for: 1) baseline model without H. pylori (blue), 2) H. pylori only model (red), and 3) expanded model that includes H. pylori and baseline model (green)
We additionally examined different probability threshold cutoffs for the new combined model (H. pylori plus baseline model) in the pooled MEDVAMC and CHI-St. Luke’s cohorts and reported sensitivity, specificity, proportion of patients referred for EGD, and number needed to detect one GIM case at each cutoff threshold (Table 4). The first line represents screening all patients for GIM, thereby detecting all GIM cases (sensitivity 100%) but only 1 in 8 patients would have GIM diagnosed. If a probability threshold of 10% or more is selected to refer for endoscopy for example, the number of the population undergoing endoscopy would decrease to 45% but 26% of patients with GIM would be missed and not referred for endoscopy (74% sensitivity). As the probability cut-off increases, less patients are referred for endoscopy (and less costs) but more patients with GIM would be missed and not referred.
Table 4.
Performance of various probability thresholds for the expanded model in predicting gastric intestinal metaplasia in the pooled MEDVAMC and CHI-St. Luke’s cohorts.
| Probability Threshold | Sensitivity (%) | Specificity (%) | Patients undergoing endoscopy (%) | Number of EGDs needed to find one case |
|---|---|---|---|---|
| 0.00 | 100.0 | 0.0 | 100 | 8 |
| 0.05 | 92.6 | 17.0 | 84.2 | 7 |
| 0.10 | 74.0 | 59.5 | 44.83 | 5 |
| 0.15 | 55.7 | 78.2 | 26.16 | 4 |
| 0.20 | 50.5 | 85.4 | 19.27 | 3 |
| 0.30 | 31.6 | 92.9 | 10.23 | 3 |
| 0.40 | 11.6 | 98.1 | 3.19 | 2 |
| 0.50 | 4.5 | 99.3 | 1.18 | 2 |
| 0.60 | 0.0 | 100.0 | 0 | - |
| 0.70 | 0.0 | 100.0 | 0 | - |
| 0.80 | 0.0 | 100.0 | 0 | - |
| 0.90 | 0.0 | 100.0 | 0 | - |
| 1.00 | 0.0 | 100.0 | 0 | - |
Discussion
In the field of gastroenterology, many risk prediction models are developed to predict or stratify an individual’s risk of having a disease. While these models are often internally validated, the overwhelming majority never go on to be externally validated (and optimized), which is the gold standard for modeling and a necessity for usefulness in clinical practice18. Our study aimed to externally validate a risk prediction model that used measurable pre-endoscopy risk factors (i.e., age, sex, race/ethnicity, smoking status, H. pylori infection) to predict an individual’s risk of GIM and especially extensive GIM17. The model was originally developed in the MEDVAMC population and was tested here in a separate CHI-St. Luke’s population (geographic validation). The under performance of the model in the validation cohort was likely due to several factors including the differing prevalence of GIM among the two populations (8% in CHI-St. Luke’s, 19% in MEDVAMC), different distribution of risk factors (more females and Hispanics in the CHI-St. Luke’s cohort in addition to lower rates of smoking and H. pylori), and possibly selection bias (MEDVAMC cohort included asymptomatic patients undergoing EGD for screening and CHI-St. Luke’s cohort only included patients undergoing EGD for symptomatic indications). On further evaluation, we found that male sex was a predictor of GIM in the MEDVAMC cohort, but not in the CHI-St. Luke’s cohort. Given these limitations with generalizability, we updated and optimized the original model by adding in the CHI-St. Luke’s cohort and adjusted the variable coefficients in the risk model which greatly improved the discrimination for GIM (AUROC 0.74) and especially extensive GIM (AUROC 0.82). These are frequently encountered limitations when validating risk models in differing populations and explain the limited number of external validation studies. Overcoming these limitations by fine-tuning risk models to better accommodate multiple populations is necessary for transportability and generalizable use in clinical practice.
We recently additionally evaluated risk prediction models for GIM in the Harris Health System in Houston, Texas, which provides healthcare to mostly uninsured and underinsured populations with high number of immigrants and racial minorities21. While this model included many of the same variables as our current study (age, sex, race/ethnicity, smoking, H. pylori infection), it also included birthplace (AUROC 0.67, 95% CI 0.64–0.71 for discriminating GIM risk). In the Harris Health study, birthplace outside of the U.S. was found to be an important risk factor for GIM (adjusted OR 1.75, 95% CI 1.16–2.66) even after adjusting for race/ethnicity. While our current model did not include birthplace, all other risk factor variables overlapped with the Harris Health model. The Harris Health model only included patients undergoing EGD for symptomatic indications and did not reflect a true screening population. If risk models are to be used to risk stratify patients at high risk for precancerous lesions (GIM) who should be referred for EGD (i.e., secondary prevention of gastric cancer), they should be developed and validated among asymptomatic screening populations.
Other limitations of our study included the retrospective electronic medical record review to ascertain the data, which limited the availability of important risk factors such as birthplace and family history of gastric cancer. Furthermore, Asian race was a risk factor in the CHI-St. Luke’s validation cohort, but it was not included in the risk model due to small number of Asians in the MEDVAMC development cohort. This model cannot be applied to populations with large number of Asians without first adapting and calibrating the model among these populations.
We believe that the probabilities of finding GIM from our model can be used for shared decision making between patients and providers. Clinical use of risk models depend on choosing a cut-point threshold to stratify risk. Examples include determination of liver transplantation based on the model for end-stage liver disease (MELD) score22 or a discriminant function score of 32 for medical treatment of alcoholic hepatitis23. The selection of a cut-point from our model to predict GIM will depend on a balance between maximizing GIM case finding and costs of negative EGDs. At a probability threshold of 5%, we would find 93% of all GIM cases, but there would be 6 unnecessary EGDs for every case of GIM diagnosed (number needed to diagnose one case: 7). At a probability threshold of 10%, only 4 unnecessary EGDs are done for every GIM case diagnosed (number need to diagnose: 5), but 26% of GIM cases would be missed. Ultimately, the number of unnecessary EGDs (false positives) would depend on the likelihood of finding GIM (i.e., prevalence) in the population. These models should be further validated in other U.S. populations to further fine-tune model performance, thus achieving better prediction of GIM pre-endoscopy and reducing the number of EGDs required to find 1 case of GIM. Cost effectiveness analyses that incorporate different thresholds within the context of GIM clinical course, effect of GIM surveillance, and costs are eventually needed for a more comprehensive guidance of shared decision making.
In summary, we externally validated a previously developed risk prediction model which used multiple pre-endoscopy factors, including sex, age, race/ethnicity, smoking, and H. pylori, to determine endoscopic risk of GIM and extensive GIM. While the original model under performed in the CHI-St. Luke’s validation cohort due to differences in distribution of risk factors, the updated model that pooled the MEDVAMC and CHI-St. Luke’s cohorts had high discrimination for predicting endoscopic GIM. By validating these models in multiple populations, we expand upon the generalizability and help their advancement towards clinical application in U.S. populations.
Supplementary Material
Conflicts of interest:
Mohamed O. Othman is consultant for Olympus, Boston Scientific Corporation, Conmed, Apollo, Lumendi and Abbvie. Mohamed O. Othman had received research grants from Abbvie, Lumendi and Lucid Diagnostics. The other authors report no competing interests for this publication.
Grant support:
Research reported in this publication was supported by the National Institute Of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under Award Number K23DK129776. This work was supported in part by National Institutes of Health grant P30 DK056338 (Study Design and Clinical Research Core), which supports the Texas Medical Center Digestive Diseases Center, and by Cancer Prevention and Research Institute of Texas (CPRIT) grant RP220127. This research was supported in part with resources at the VA HSR&D Center for Innovations in Quality, Effectiveness and Safety (#CIN 13–413), at the Michael E. DeBakey VA Medical Center, Houston, TX. The opinions expressed reflect those of the authors and not necessarily those of the Department of Veterans Affairs, the National Institutes of Health, the U.S. government or Baylor College of Medicine.
Abbreviations
- AUROC
area under the receiver operating characteristic
- CHI-St. Luke’s
Catholic Health Initiative-St. Luke’s
- CI
confidence interval
- CPT
Current Procedural Terminology
- EGD
esophagogastroduodenoscopy
- GIM
gastric intestinal metaplasia
- MEDVAMC
Michael E. DeBakey VA Medical Center
- MELD
model for end-stage liver disease
- OR
odds ratio
- PPI
proton pump inhibitor
- TRIPOD
Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Graham DY, Khan A, et al. Incidence of gastric cancer in the USA during 1999 to 2013: a 50-state analysis. Int J Epidemiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, El-Serag HB, Thrift AP. Increasing Incidence of Advanced Non-cardia Gastric Cancers Among Younger Hispanics in the USA. Dig Dis Sci 2020. [DOI] [PubMed] [Google Scholar]
- 4.Liu KS, Raza SA, El-Serag HB, et al. Recent Trends in the Incidence of Gastric Cancer in the United States. J Clin Gastroenterol 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol 2019;14:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correa P, Haenszel W, Cuello C, et al. A model for gastric cancer epidemiology. Lancet 1975;2:58–60. [DOI] [PubMed] [Google Scholar]
- 7.Tan MC, Graham DY. Screening for Gastric Cancer: Focus on the Ants Instead of the Ant Hill. Clin Gastroenterol Hepatol 2021;19:1990–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim GH, Liang PS, Bang SJ, et al. Screening and surveillance for gastric cancer in the United States: Is it needed? Gastrointest Endosc 2016;84:18–28. [DOI] [PubMed] [Google Scholar]
- 9.Lee H, Min BH, Lee JH, et al. Survival outcome associated with the screening interval for gastric cancer in Korea. Digestion 2011;84:142–8. [DOI] [PubMed] [Google Scholar]
- 10.Jun JK, Choi KS, Lee HY, et al. Effectiveness of the Korean National Cancer Screening Program in Reducing Gastric Cancer Mortality. Gastroenterology 2017;152:1319–1328.e7. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Li D, El Serag HB, et al. AGA Clinical Practice Guidelines on Management of Gastric Intestinal Metaplasia. Gastroenterology 2020;158:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jove AG, Holmes HM, Tan MC, et al. Inverse Association Between Gluteofemoral Obesity and Risk of Non-Cardia Gastric Intestinal Metaplasia. Clin Gastroenterol Hepatol 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes HM, Jove AG, Tan MC, et al. Alcohol consumption and the risk of gastric intestinal metaplasia in a U.S. Veterans population. PLoS One 2021;16:e0260019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thrift AP, Jove AG, Liu Y, et al. Associations of Duration, Intensity, and Quantity of Smoking With Risk of Gastric Intestinal Metaplasia. J Clin Gastroenterol 2022;56:e71–e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan MC, Mallepally N, Liu Y, et al. Demographic and Lifestyle Risk Factors for Gastric Intestinal Metaplasia Among US Veterans. Am J Gastroenterol 2020;115:381–387. [DOI] [PubMed] [Google Scholar]
- 16.Tan MC, Mallepally N, Ho Q, et al. Dietary Factors and Gastric Intestinal Metaplasia Risk Among US Veterans. Dig Dis Sci 2021;66:1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan MC, Ho Q, Nguyen TH, et al. Risk Score Using Demographic and Clinical Risk Factors Predicts Gastric Intestinal Metaplasia Risk in a U.S. Population. Dig Dis Sci 2022;67:4500–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thrift AP, Kanwal F, El-Serag HB. Prediction Models for Gastrointestinal and Liver Diseases: Too Many Developed, Too Few Validated. Clin Gastroenterol Hepatol 2016;14:1678–1680. [DOI] [PubMed] [Google Scholar]
- 19.Kligman E, Ali H, Chen E, et al. Ethnicity Is an Important Consideration in Screening for Gastric Intestinal Metaplasia. Dig Dis Sci 2022;67:4509–4517. [DOI] [PubMed] [Google Scholar]
- 20.Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): the TRIPOD Statement. Br J Surg 2015;102:148–58. [DOI] [PubMed] [Google Scholar]
- 21.Tan MC, Jamali T, Nguyen TH, et al. Race/Ethnicity and Birthplace as Risk Factors for Gastric Intestinal Metaplasia in a Multiethnic United States Population. Am J Gastroenterol 2022;117:280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamath PS, Wiesner RH, Malinchoc M, et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001;33:464–70. [DOI] [PubMed] [Google Scholar]
- 23.Akriviadis E, Botla R, Briggs W, et al. Pentoxifylline improves short-term survival in severe acute alcoholic hepatitis: a double-blind, placebo-controlled trial. Gastroenterology 2000;119:1637–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


