Abstract
Solute carrier (SLCs) transporters mediate the transport of a broad range of solutes across biological membranes. Dysregulation of SLCs has been associated with various pathologies, including metabolic and neurological disorders, as well as cancer and rare diseases. SLCs are therefore emerging as key targets for therapeutic intervention with several recently approved drugs targeting these proteins. Unlocking this large and complex group of proteins is essential to identifying unknown SLC targets and develop next generation SLC therapeutics. Recent progress in experimental and computational techniques has significantly advanced SLC research including drug discovery. Here, we review emerging topics in therapeutic discovery of SLCs, focusing on state-of-the-art approaches in structural, chemical, and computational biology, and discuss current challenges in transporter drug discovery.
Keywords: Membrane protein, drug design, protein structure prediction, ligand discovery
Biological importance of Solute Carrier (SLC) transporters
Membrane transporters play a critical role in communication between the cell and the environment. The largest membrane transport group in humans is the Solute Carrier (SLC) transporters that consists of 455 members classified into 66 families [1]. The SLCs transport a broad range of substrates (see Glossary), including nutrients, neurotransmitters, ions, and drugs, and take part in numerous biological processes, such as regulation of cell signaling and organization of the cellular organelles [2]. A large fraction of the SLCs are secondary active transporters that couple electrochemical gradient of ions and/or other molecules to transport substrates (eg symport, antiport) [1, 3]. Genetic variations in many SLC members have been linked to various diseases and disorders, such as neurological disorders, metabolic disease, and cancer [4, 5].
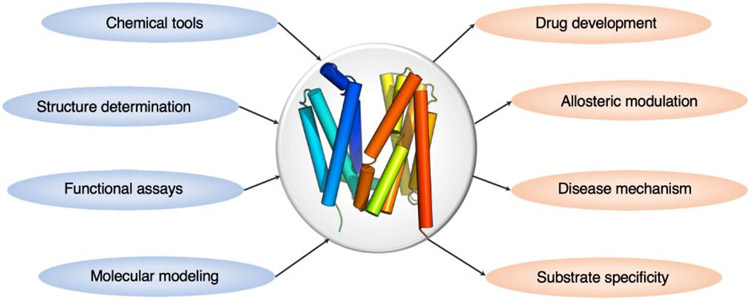
Notably, despite their biological importance, SLCs are still considered to be among the most understudied proteins relative to their family size, and many aspects of their biology remain unknown. Analyses by us and others highlight the gap in the current knowledge of human SLCs [2, 5-8]: many orphan transporters have no known substrates [5, 9] and established SLC transporters have no known chemical modulators [10], and a variety of disease-related mutations in SLCs have not been characterized [5]. Further, over ten protein families have been reclassified as SLCs in the past decade, such as the pyrophosphate exporter, progressive ankylosis protein homolog (ANKH; SLC62A1) [11], and the lysosomal cholesterol transporter Niemann-Pick disease type C1 (NPC1; SLC65A1) [12]. Notably, newly developed computational and experimental approaches discussed herein have addressed important questions in SLC biology, including long sought-after questions related to disease mechanisms and substrate specificity, as well as fast-growing fields in SLC research such as allosteric modulation and drug development (Key Figure, Figure 1).
Key Figure, Figure 1:
Overview of the emerging topics in SLC research. Approaches advancing the characterization of SLCs are shown on the left-hand side of the figure, while the biological questions currently addressed by the community are shown on the right-hand side.
Due to their broad physiological roles, the SLCs are proteins of the utmost pharmacological importance. Uptake and efflux SLC transporters are often localized in organs and tissues such as the kidney, liver, and blood-brain-barrier (BBB), where they control the absorption, distribution, and excretion of therapeutic drugs [13, 14]. For example, the peptide transporter PepT1 (SLC15A1) regulates the intestinal absorption of peptide-like drugs, such as β-lactam antibiotics (eg cefadroxil) and antiviral drugs (eg valacyclovir) across the cell membrane [15, 16]. Therefore, genetic variations in these transporters are often associated with differential drug response among patient populations (ie pharmacogenomics) [13]. In fact, genetic polymorphisms of the organic cation transporter OCT1 (SLC22A1), which is mainly expressed in the liver, impact the disposition, distribution, and toxicity of prescription drugs such as metformin, among different ethnic groups [17].
Moreover, multiple SLCs have been validated as important targets for therapeutic intervention [18]. Over the past 50 years, the monoamine transporters, including the norepinephrine transporter NET (SLC6A2), dopamine transporter DAT (SLC6A3), and serotonin transporter SERT (SLC6A4) have been targeted by drugs for treating of neurological and psychiatric disorders [19]. For instance, selective serotonin reuptake inhibitors (SSRIs, eg Escitalopram) and serotonin–norepinephrine reuptake inhibitors (SNRIs, eg Duloxetine), which increase neurotransmitter concentrations in synapses and control neurotransmission, are often prescribed to treat depression and anxiety disorders [20]. More recently, the SLC target space has been expanded to a variety of other indications such as metabolic disorders, cancer, and other pathologies [18]. For example, sodium-glucose cotransporter-2 SGLT2 (SLC5A2) inhibitors (eg canagliflozin) are used for the treatment of type 2 diabetes by lowering blood sugar levels [21]. Other novel SLC targets with drug candidates in clinical investigation include the creatine transporter CRTR (SLC6A8; gastrointestinal cancer, clinical trial number NCT03597581) and glycine transporter 1 GlyT1 (SLC6A9; schizophrenia) [22]. Finally, drugs targeting established SLC targets have been recently repurposed to address new indications. For example, the SGLT2 inhibitor Licogliflozin [23] and the apical sodium bile acid transporter ASBT (SLC10A2) inhibitor Elobixibat (clinical trial number NCT04006145 [24]) are currently in clinical trials for treating liver disease including nonalcoholic steatohepatitis (NASH).
In this review, we discuss emerging topics in SLC biology, including SLC structure and mechanism, the chemical space of SLC ligands, strategies to modulate SLC function, and how the most recent advances in computational modeling can be applied to characterize the SLCs. Finally, we discuss current challenges in SLC drug discovery.
SLC structures in rational drug design
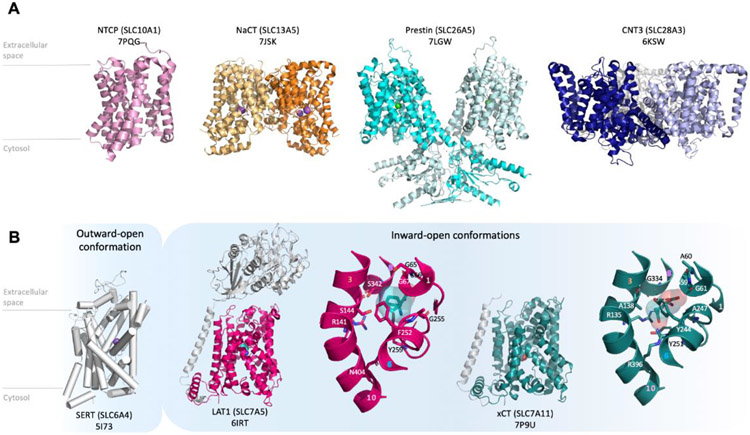
The three-dimensional structures of SLC transporters can help address fundamental questions related to their biology, including description of mechanisms of membrane transport and substrate specificity, as well as development of predictive models for the effect of disease-related mutations on SLC function. Over the past decade, there has been a surge in the number of experimentally determined structures of SLCs, primarily due to progress in cryo-electron micrography (cryo-EM) technologies for structure determination of membrane proteins [25]. These structures have shown that, unlike other functionally defined “Superfamilies” (eg ABC transporters, GPCRs), the SLCs are highly diverse in structure, consisting of several evolutionary unrelated, distinct structural classes or folds (Figure 2A) [6, 7, 26], where the Major Facilitator Superfamily (MFS; eg glucose transporters GLUTs (SLC2)) and the leucine transporter LeuT-like fold (eg neurotransmitters transporters of the SLC6 family) are the most common structural classes in the human SLCs [6] (Figure 2B). It was observed that conserved functionally important elements among members of distinct families (ie SLC6 and SLC7) within a structural class (ie LeuT-fold), allow for efficient functional annotation among SLC members [27]. Notably, it has been shown that transmembrane helices 1 and 6 (TM1 and TM6) are anchoring the ligands through interactions with backbone atoms of conserved residues (eg the GXG motif in TM1), while TM3, TM8, and TM10 consist of variable residues conferring the selectivity of the ligand for the transporter (Figure 2B).
Figure 2: SLC structure and dynamics.
(A) Recent SLC structures revealed previously unknown structural classes or folds that operate via the alternating access mechanism, including, from left to right NTCP (SLC10A1) (PDB id 7PQG [120]), NaCT (SLC13A5) (PDB id 7JSK [50]), Prestin (SLC26A5) (PDB id 7LGW [121]) and CNT3 (SLC28A3) (PDB id 6KSW [122]). (B) Outward open structure of SERT (SLC6A4) (PDB id 5I73 [39]). Inhibitors-bound structures in inward-open conformations of the LeuT fold transporters LAT1 (SLC7A5) (PDB id 6IRT [34]), and, xCT (SLC7A11) (PDB id 7P9U [41]) as well as a close up view of their respective binding sites. Residues defining the binding site are labeled, with TM1 and TM6 labeled in white and blue, and TM3, TM8, and TM10 in orange, purple and pink, respectively.
Interestingly, despite their dissimilarity in structure, SLCs use a conserved ‘alternating access’ transport mechanism, in which the transporter interchangeably exposes its binding site at either side of the membrane [28]. The alternating access model is facilitated through the internal symmetry within the transporter structure, regardless of its structural class [29]. Notably, multiple structures were determined in different conformation of the transport cycle for only a small number human SLCs, where the majority of SLCs with known structures have only been solved in one or two conformations [18]. For example, proteins belonging to MFS such as the glucose transporters GLUTs (SLC2) use a rocker-switch mechanism, while members of the LeuT-like (eg neurotransmitters transporters of the SLC6 family) and GltPh-like (eg the amino acid transporters SLC1) structural classes use gated-pore and elevator transport mechanisms, respectively (reviewed in [8, 30]).
In addition to describing transport mechanisms, atomic resolution structures of SLC drug targets can be used for rational drug design. For example, structures of the cancer-related transporters of monocarboxylates (MCT1; SLC16A1) [31], glutamine (ASCT2; SLC1A5) [32], cystine (xCT, SLC7A11) [33], leucine (LAT1, SLC7A5) [34], and glucose (GLUT1, SLC2A1) [35] have been used to develop compounds targeting reprogrammed metabolic networks in cancer. Furthermore, the structures of SLCs related to drug transport and dynamics, such as the intestinal transporter PepT1 [36] and the liver and brain transporter OCT3 [37], have revealed previously unknown mechanisms of drug absorption, drug-drug interactions, and pharmacogenomics.
Notably, distinct conformations can be used to develop conformation-specific binders with unique scaffolds and specificity profiles [38]. Indeed, up until recently, it was thought that the most optimal transporter conformation for rational design would be outward-facing conformations, as observed in the SERT-escitalopram complex [39]. However, it was shown that small molecule inhibitors can also target inward conformations (Figure 2B) [34, 40-44]. In fact, SERT–ibogaine complexes were solved in multiple SERT states, including outward-facing, occluded, and inward-facing conformations [44]. Interestingly, one mechanism has been proposed in which an inhibitor targeting inward-facing conformation first diffuses across the membrane and subsequently binds the transporter within the cell [45]. This putative mechanism can potentially allow the compound to avoid competing with endogenous substrates found in the rich extracellular media. Taken together, these studies open new avenues for the design of novel small molecule inhibitors targeting particular conformations of the transport cycle.
Pharmacological space of SLCs
A tool compound can be defined as a chemical that selectively controls the function of a protein, allowing researchers to address fundamental and mechanistic questions about the target protein by using a range of experimental approaches, such as biochemical and cellular assays or in vivo methodologies [46]. Tool compounds can also potentially provide a starting point for the development of lead compounds for future therapeutics. Over the past decade, newly developed tool compounds targeting SLCs have advanced their structural and functional characterization. For example, a small molecule inhibitor of the cancer-related amino acid transporter ASCT2 (SLC1A5) called Lc-PBE was designed using a homology model based on the EAAT1 (SLC1A3) X-ray structure in a ligand bound outward-facing state; the compound facilitated the experimental structure determination of ASCT2 in a unique conformation, which allowed the further development of potent and selective ASCT2 inhibitors [45].
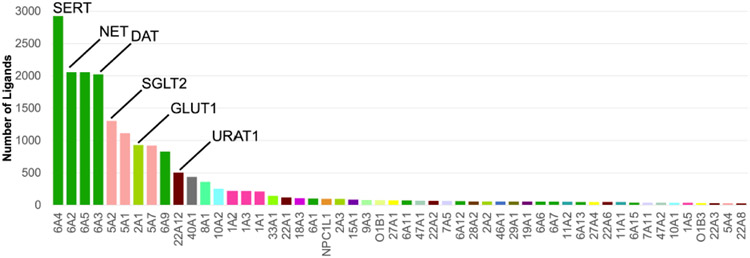
Notably, the current known space of SLC ligands is limited, where major barriers for effective discovery of useful chemical probes for SLCs include the limited availability of assays for this class of proteins [10] and the lack of SLC structures in distinct conformations available for rational drug design [18]. Analysis of the ChEMBL database [47] shows that small molecule ligands of SLCs, as measured by IC50 values of 1 mM or lower, exist for only 97 SLCs . Interestingly, there is a significant bias for well-established drug targets such as the neurotransmitter transporters, NET and DAT, and sugar transporters, SGLT1 and GLUT1, where each protein has more than 1,000 inhibitors (Figure 3), whereas hundreds of SLCs do not have any reported inhibitors. This highlights the need for potent and selective chemical tools for SLC transporters. In addition, analysis of relationships between proteins based on the chemical similarity of their small molecule ligands can reveal functional associations, as well as guide the deorphanization of proteins [48, 49] (Box 1).
Figure 3: Small molecule ligands of SLCs.
Each inhibitor is defined as having IC50 value of 1mM or lower. Analysis of ChEMBL [47] identified 97 SLCs with at least one inhibitor using this criterion (Supplementary Table 1). 52 SLCs had 25 or more ligands and are shown here. The colors correspond to the SLC family of each protein.
Box 1. Pharmacological space of SLCs.
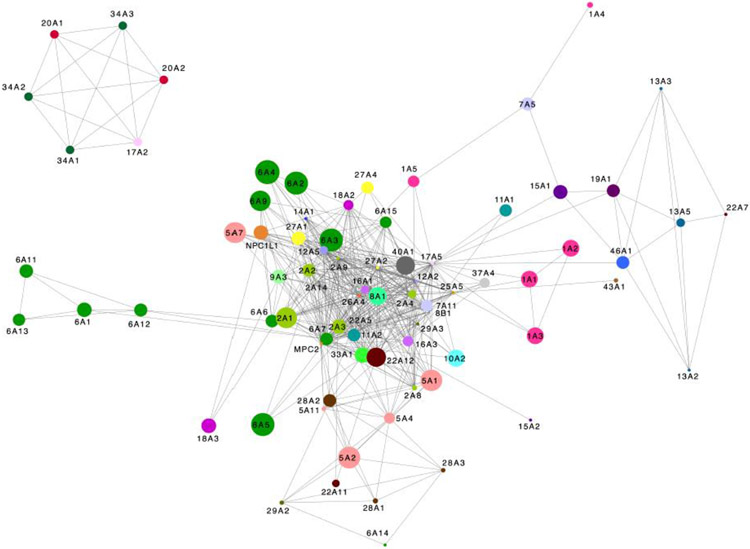
To demonstrate the relationships between proteins based on the chemical similarity of their small molecule ligands, we generated, as an example, a network in which the SLCs are connected to each other if they have chemically similar small molecule ligands in ChEMBL (Figure I). We observe that SLCs belonging to different, and often evolutionarily unrelated families, can be highly connected in the pharmacological space (Figure I). Some relationships among SLCs are expected from the overlap in their natural substrates. For example, members the evolutionarily unrelated sugar transporter families, including the SLC2 family of glucose transporters (GLUTs; light green; MFS fold) and the SLC5 family of sodium-dependent glucose transporters (SGLTs; salmon; LeuT-like fold) are highly interconnected based on the chemical similarity of their ligands. Similarly, phosphate transporters belonging to the distinct SLC families SLC34, SLC20, and SLC17 are also connected, forming a unique cluster. Interestingly, other connections among SLCs are not entirely obvious from their natural substrates, such as the subcluster formed by the sodium-dependent citric acid cycle (CAC) metabolites transporters of the SLC13 family (SLC13A2,3,5) and the unrelated folate transporters SLC19A1 and SLC46A1. Moreover, some members of the same SLC family are more closely associated with members of other SLC families than those of their own family. For example, SLC1A5, a neutral amino acid transporter, is linked only indirectly to SLC1 family members (SLC1A1,2,3) that transport acidic amino acids (eg glutamate), through SLC17A5, which transports sialic acids. Overall, filling the gap in the knowledge of the SLC ligands will allow us to connect more SLCs in the pharmacological space thereby guiding functional annotation and deorphanization of the SLCs.
Box 1, Figure I: Pharmacological space of SLCs.
The size of the circle in the map corresponds to the number of known small molecule inhibitors of the SLC according to the criteria described above. The distance between two SLCs is the inverse of the sum of the dice similarities based on the RDkit morgan fingerprint [123], with radius two, of every combination of pair of inhibitors from the two SLC inhibitor sets normalized by the number of comparisons. The arrangement of the SLCs is dictated by the Cytoscape edge weighted, force directed biolayout [124].
Chemical modulation of SLCs
Small molecule ligands of SLCs can control their function via different mechanisms. An SLC inhibitor can selectively inhibit the transport of substrates via the transporter across the cellular membrane. An inhibitor can bind the substrate binding site (‘orthosteric inhibitor’), competing with the substrate and blocking its binding and/or the conformational changes that are associated with transport. Over the past decade, many orthosteric inhibitors have been developed for a range of biomedically important transporters including the Na+/Citrate Cotransporter NaCT (SLC13A5) [50], glycine transporter-1 GlyT1 (SLC6A9) [42], and the GABA transporter 1 GAT1 (SLC6A1) [40]. For example, orthosteric inhibitors of SLCs that are associated with reprogrammed metabolic networks in cancer can deprive the tumor cell of nutrients. A recent study analyzed structure-activity relationship (SAR) around the xCT (SLC7A11) inhibitor sulfasalazine, allowing for the investigation of the toxicity of this drug when administered to patients. As a result, several potent xCT inhibitors were designed and validated in various cancer cell lines and presented minimal toxicity profiles in normal human astrocytes [51]. Interestingly, another recent study showed that inhibition of GLUT1 (SLC2A1) and/or GLUT3 (SLC2A3) resulted in disulfidptosis, a unique mechanism of cell death in SLC7A11high cancer cells. This work provides a novel strategy for treating numerous cancers with high xCT expression [52]. Moreover, it was shown that drugs targeting the orthosteric binding site in distinct SERT [53] and DAT [38] conformations can lead to differential pharmacological effect. This suggests that SLC transporters can modulate biased signaling potentially allowing for fine tuning pharmacological effects of drugs.
Alternatively, an inhibitor can bind a site distant from the substrate binding site or the substrate transport pathway (‘allosteric inhibitor’). Allosteric binding sites are often less conserved within a protein family and can thus potentially be targeted with more selective inhibitors than the substrate binding site inhibitors [54]. Moreover, the allosteric sites are more likely to be targeted by molecules chemically different from the endogenous substrates that bind the substrate binding site, and thus avoid off-target binding to proteins with similar substrate specificity. Thus, one significant advantage of allosteric inhibitors is that they can potentially improve drug-like properties and selectivity, which are often not seen in transporters’ substrate-like compounds such as amino acid-like or sugar-like ligands. Allosteric inhibitors in transporters have so far been described in members of the SLC6 family [55] (eg SERT/SLC6A4 [56], DAT/SLC6A2 [57], and GlyT2/SLC6A5 [58]), and the SLC1 family (eg in EAAT1/SLC1A3 [59]). For example, one mechanism of allosteric inhibition is binding the interface between the scaffold and mobile domains of the SLC1 family of elevator transporters, hence hindering the conformational change that is needed for transport, as observed for the allosteric EAAT1 inhibitor UCPH101 [60] (Figure 4A). Interestingly, the UCPH101 binding site overlaps with equivalent lipid binding sites in its homologs EAAT3 (Figure 4A) and ASCT2 (SLC1A5), where cholesterol was proposed to be important for ASCT2 function [32]. Interestingly, a recent study revealed that this allosteric mechanism is conserved among SLC1 members and that subtle differences in the allosteric binding site allow the identification of selective allosteric inhibitors, thereby providing an avenue for future drug development for members of this highly important SLC family [61].
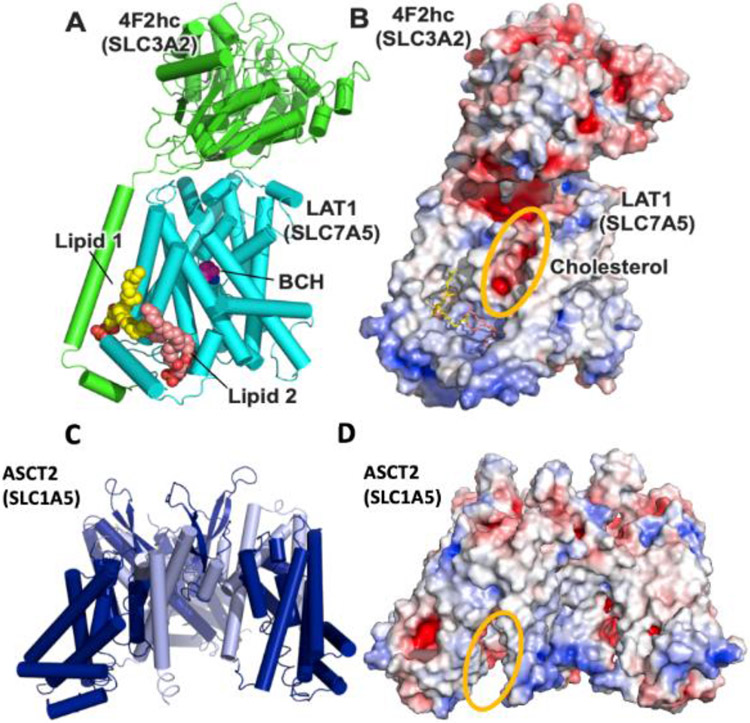
Figure 4: Allosteric modulatory surfaces in amino acid transporters.
(A) Trimeric structure of Excitatory amino acid transporter 3 EAAT3 (SLC1A1) shown as cylinder (left; PDB id 6S3Q) where each protomer is shown in a different color. Phospholipids in the interface between protomer 1 and 2 are shown as purple and yellow spheres. Also shown is surface representation of protomer 1 of EAAT1 (SLC1A3) (right; PDB id 5MJU) with the allosteric inhibitor UCPH101 (orange sticks), whose location overlaps with the location of the phospholipids colored in purple in EAAT3. (B) Heterodimeric structure of LAT1 (SLC7A5; cyan) and 4F2hc (SLC3A2; green) (PDB id 6IRT) in complex with two lipid molecules (yellow and pink), and the substrate binding site inhibitor BCH (magenta; left). Also shown is surface representation of LAT1 structure colored based on electrostatic potential (right). The putative cholesterol binding site is highlighted in orange.
Additionally, allosteric binding sites can be targeted by small molecule activators – compounds enhancing the transport efficiency of natural or synthetic substrates across biological membranes. To the best of our knowledge, only one allosteric activator has been rationally designed for an SLC, the glutamate transporter EAAT2 (SLC1A2) [62]. This activator is localized at the interface between the trimerization and transport domain, another region proposed to be important for enabling the conformational change required for transport. Binding sites of sterols and other lipids in other transporters have also been shown to be amenable for allosteric modulation, further demonstrating the importance of these molecules for function, similarly to other membrane protein families such as ion channels [63, 64], where cholesterol can interact with its target, triggering conformational changes that are associated with activation or inhibition. For example, recent structural, biochemical, and pharmacological data suggest that the transport of substrates by the LeuT-fold transporter LAT1 (SLC7A5) depends on its interaction with SLC3A2 and is mediated by two lipid molecules, as well as by cholesterol which serves as a LAT1 activator [34] (Figure 4B). Further, a variety of related SLCs with a LeuT-fold are modulated by lipids, such as the dopamine transporter DAT (SLC6A3) [65, 66], serotonin transporter SERT (SLC6A4) [67], and glycine transporter GlyT2 (SLC6A5) [58].
Another type of activators are pharmacochaperones, compounds that rescue folding and/or trafficking of misfolded proteins. Pharmacochaperones improve the activity of proteins carrying disease-causing mutations that affect their folding, stability, or localization to the membrane [68]. For example, pharmacochaperones have been recently developed for the treatment of cystic fibrosis, caused by a specific mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) [69, 70]. These drugs represent a novel strategy of rescuing the function of a malfunctioned transporter through small molecule binding. Particularly, the CFTR modulators can be classified into two groups: i) ‘potentiators’ (i.e., Ivacaftor) are compounds that increase the channel conductance; and ii) ‘correctors’ (i.e., Elexacaftor and Tezacaftor) are molecules that rescue CFTR folding and trafficking to the plasma membrane [70]. The necessity of combining both types of modulators for trafficking and functional rescue shows the complexity of pharmacochaperones development. Pharmacocharperoning development has recently been explored for the monoamine transporters of the SLC6 family [71, 72]. For example, derivatives of the psychedelic drug ibogaine were able to correct folding deficient DAT (SLC1A3), thus paving the way for rational design of pharmacochaperones targeting SLC transporters.
Importantly, SLCs have different regions that control other aspects of their function, including post-translational modifications and protein-protein interactions [73]. For example, DAT physically and functionally interacts with the voltage-gated K+ channel Kv2.1 to modulate dopamine neurotransmission [74]. Moreover, the C-terminus of DAT binds Ca2+/calmodulin-dependent protein kinase II α (CaMKIIα) to facilitate phosphorylation of the N-terminus of DAT, thereby modulating amphetamine-induced dopamine efflux [75]. Interestingly, the termini of the SLC6 members are divergent in sequence and size and are thought to play a critical role in functional differences among these proteins, including their interactions with substrates and lipids [73, 76]. Some of these interacting regions can be highly dynamic, or even involve unstructured regions; thus, developing small molecule compounds targeting these regions is highly challenging.
Finally, over the past decade, new approaches have emerged to control proteins through their selective degradation [77]. For instance, PROteolysis-TArgeting ChimeraS (PROTACS) are bifunctional molecules that consist of a small molecule protein ligand that is connected via a linker to an E3 ubiquitin ligase (E3) ligand that recruits the protein degradation machinery [78]. PROTACs as well as other degraders have shown promise on a variety of targets, including tau [79] and KRAS [80], as well as multiple compounds currently tested in clinical trials. Designing a PROTAC for SLCs is particularly challenging. It requires the formation of a tertiary complex from a large variety of warheads, linkers, and E3 ligase ligand. Moreover, it needs to bind the target from the intracellular domain, which might not include an accessible, druggable site. It was recently shown that selected SLCs representing distinct SLC families are amenable for degradation by PROTACS, providing encouraging data for using this approach to modulate transporters [81]. For example, a degrader with broad specificity for the SLC9 family that showed promising data on cancer cell line was developed [81].
Computational modeling approaches
The SLCs adopt distinct conformational states to allow for alternating access transport of substrates. Despite advancements in structure determination of membrane proteins with experimental techniques, using these approaches to characterize different conformations of membrane can be costly and time consuming. Computational protein structure prediction aims to bridge the gap between the sequence space of the SLCs and their structural coverage (Figure 5). Traditional modeling approaches include homology or template-based modeling (TBM), in which the target protein is modeled based on its sequence alignment to one or more known experimentally determined structure(s) of homolog protein(s) that serve as modeling templates. Conversely, in de novo or ab initio modeling, the target is modeled directly from its amino acid sequence [82]. The accuracy of these methods has significantly improved over the years, partly due to the integration of spatial restraints derived from the analysis of sequence co-evolution [83] or experimental data [84].
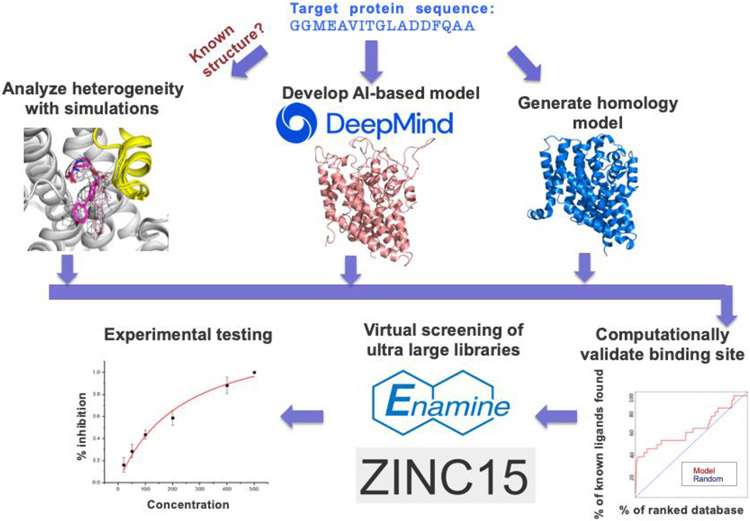
Figure 5: Workflow for structure-based virtual screening of SLC transporters.
For transporters with known structures, MD simulations can be applied to sample different druggable conformations. For SLCs with unknown structure, or for those SLCs that lack structure in a desirable conformation, structural models can be generated using the AI-based method AlphaFold2 or through homology modeling. All models can then be evaluated on the basis of their ability to predict known ligands (when applicable) using enrichment, and subsequently refined based on these results. Virtual screening of large purchasable compound libraries is then performed where top scoring compounds are tested experimentally.
Molecular Dynamics (MD) simulations have played a central role in describing dynamic properties of membrane proteins [85], and are particularly useful when combined with experimental methods [86]. For example, MD simulations were used to describe different aspects of SERT mechanism, including domain movement [87], ion binding [88], substrate specificity [89], oligomerization [90], mechanism of inhibitor binding and the resulting conformational changes [44], as well as the effect of mutations on its structure/function [91]. Additionally, advances made in atomistic and coarse-grained (CG) force-fields have allowed for improved modelling of protein-lipid interactions and the role lipids play on protein dynamics and function (reviewed in [92, 93]). MD has also been used to gain insights on lipid interactions with transporters affect conformation stability and transitions. For example, in DAT, PIP2 is involved in regulating the transition to the inward facing state by interacting with DAT’s N-term and intracellular loop 4 [94]. Additionally, simulations aided in identifying a conserved cholesterol binding site in SERT and cholesterol binding stabilizes SERT in an outward facing conformation [67].
Notably, atomic-level simulations were employed to characterize the full transport cycle of the bacterial glucose transporter SemiSWEET, including transitions from the transporter’s outward-facing to its inward-facing conformation [95]. This analysis was performed on one of the smallest known transporters with a simple geometry and energy coupling mechanism; however, MD simulations’ ability to model the translocation cycle in human SLCs is more challenging due to the long timescales required to overcome the high free-energy barriers that separate distinct states, as well as the inaccuracies of the force fields used.
One way to address some of the challenges in unguided all-atom simulations is by using biased or guided simulation techniques (reviewed in [96]), which often provide novel mechanistic insights on SLC transporters, especially when combined with experimental testing. For example, accelerated MD (aMD) applied to the human DAT revealed unknown insights into the sequential gating and transport events of this protein [97].
Alternatively, artificial intelligence (AI) and machine learning (ML) based methods have recently emerged as powerful and accurate approaches for structure prediction. Specifically, the recent release of the two open-source structure prediction methods AlphaFold2 (AF2) [98] and RosettaFold [99] launched a new era in protein structure prediction, providing tremendous support for addressing fundamental scientific questions using structural biology insights. For example, AF2 has been applied to a variety of challenging problems, such as structure modeling of protein complexes [100] and large assemblies [101], as well as experimental structure determination of particularly challenging targets [102] and identification of protein disordered regions [103].
A current limitation of modern AI based modeling is that there is no straightforward one-size-fits-all procedure that accurately captures the conformational diversity of proteins [104-106]. Moreover, it is unclear whether these methods can accurately model the protein’s amino acid sidechains [107], binding pockets or point mutations effect on structure [108], which are critical for rational drug design. Therefore, model generation should be performed and evaluated judiciously. One way to evaluate the relevance of the model for rational design is its ability to enrich for known ligands as compared to a data set of the ligands and likely non-binders or decoys, using docking [109] (Figure 5). By iteratively generating models and evaluating their binding site with enrichment the model’s binding site is optimized for protein-small molecule ligand complementarity and structure-based ligand discovery [110, 111]. Sampling biologically relevant conformations to be evaluated with enrichment can be done with a range of modeling approaches such as sidechain modeling on a fixed backbone, MD simulations, as well as other approaches [111]. In addition, using integrated approaches that include data derived from low resolution experimental data can guide rational drug design. For example, metainference MD simulations generate an ensemble of conformations that are consistent with available cryo-EM data, accounting for the concurrent presence of data ensemble-averaging, structural heterogeneity, and noise level variability in different regions of the experimental map [112]. In a recent study, metainference has advanced the discovery of a unique binding site and inhibitor conformation that was useful for the design of potent inhibitors [45].
Overall, computer guided ligand discovery campaigns have been applied to characterize a range of SLC transporters, representing different families and mechanisms, including PepT1 (SLC15A1) [113], NaCT [114] LAT1 (SLC7A5) [115], BGT1 (SLC6A12) [116], and GLUT3 (SLC2A3) [117]. Recently, a virtual screen of an ultra-large compound library (200 million compounds) on a membrane transporter was conducted using the inward-open SERT structure, leading to the discovery of selective SERT inhibitors with potencies up to 200 times better than the SSRI fluoxetine as well as improved efficacy in various mouse behavioral models. This study also demonstrated that even for a highly studied membrane transporter target, new structural information and improved computational approaches can inform the development novel lead compounds [53].
Concluding remarks
Recent advancements in chemical and structural biology methodologies, as well as in our conceptual understanding of the importance of the transport process, have allowed emergence of the SLCs as a major drug target family. The wealth of structural information and newly established computational methods can also allow us to rationalize how polymorphisms cause disease or differential drug response among individuals. Can we predict which mutation would be neutral, loss-of-function, or gain-of-function [118]? Describing mutation’s effect on structure / function is expected to improve the understanding of disease mechanisms, identify novel drug targets, and advance precision medicine.
Furthermore, structures of biomedically important transporters revealed previously unknown mechanisms of transport modulation that can be harnessed for the development of next generation of transporter drugs. For instance, transporters utilizing elevator-like transport mechanisms can be targeted with small molecules physically blocking the movement of the mobile domain (eg in EAAT1; Figure 4). Moreover, substrate binding site inhibitors targeting specific conformations, such as those of SERT lead to compounds with improved in vivo efficacy, further refining our understanding of pharmacological control of transporter function. Fine-tuning transporter function with chemical tools can lead to new areas in transporter pharmacology and drug design, such as pharmacology of biased signaling in GPCRs [119].
In addition, the CFTR drugs showed that small molecule activators can correct disease phenotype caused by transporter malfunctions. There are hundreds of diseases associated mutations causing defective transporters, which in principle, could potentially be targeted with approaches similar to those taken in the development of the CFTR drugs. However, key questions remain: are there general rules that determine which compound will be a substrate that goes through the transporter, an inhibitor that binds the transporter and blocks transport, or an activator that improves the transport of a defective transporter? Generalized strategies to develop each compound type are expected to allow the development of chemical tools that facilitate the characterization of SLCs as well as future SLC drugs. Finally, transporters often contain other regions that mediate other aspects of their function, including protein-protein interactions and post-translational modification. Description of the structure / function of these regions is expected to reveal unknown modulatory surfaces that are amenable for drug design.
Outstanding Questions.
Are there general structural determinants of SLC modulators that discriminate between substrates, inhibitors, and activators?
Can we accurately predict mutational effect on SLC structure / function (neutral, gain-of-function, loss-of-function), to improve the understanding of disease mechanisms and identify novel SLC drug targets?
Can we harness emerging AI technologies and superior computational power to improve our understanding of transport mechanisms and guide the development of future SLC drugs?
Highlights.
Solute carrier (SLC) transporters are a highly understudied class of proteins that transport ions, nutrients, and drugs, across biological membranes, and are often mutated in disease.
Unlocking this large and complex group of proteins is essential to identify unknown SLC targets and develop next generation SLC drugs.
Recent advances in chemical, structural, and computational biology have allowed to develop innovative strategies to modulate their function as well as unique tool compounds and future drugs.
Acknowledgments
We thank Enrico Girardi (Solgate, GmbH) for valuable comments on the manuscript. We also thank Peter Man-Un Ung and Rachel-Ann Garibsingh (Mt. Sinai), Christof Grewer (Binghamton University), Allen Thomas (UNK-Kearny), Kathleen Giacomini (UCSF), and Gerhard Ecker (University of Vienna) for useful discussions. This work was supported in part by the National Institutes of Health grant R01 GM108911 to A. S.
Glossary
- Allosteric inhibitor
an inhibitor that binds to a site different from the orthosteric site of the transporter
- Allosteric modulation
mechanism of regulation in which a molecule that binds at a different site than the substrate binding site and enhances (allosteric activators) or inhibits (allosteric inhibitor) SLC transport
- Alternating access
a process in which membrane transporters undergo conformational changes to alternate between different states such as outward-facing, occluded, and inward-facing, enabling selective movement of substrates across the membrane
- Antiport
two or more different substrates, such as molecules or ions, are concurrently transported across the cell membrane in opposite directions
- Disulfidptosis
regulated cell death arising from disulfide stress induced by elevated SLC7A11 expression combined with glucose starvation
- Elevator
a type of alternating access transport where the substrate binds the transport domain and is then moved across the membrane via a significant rigid-body movement of the transport domain against the scaffold domain, which is typically involved in oligomerization
- Gated-pore or rocking-bundle
a type of alternating access transport mechanism in which a static scaffold domain and a mobile bundle domain that alternatively opens and close during the transport mechanism
- Molecular dynamics (MD) simulation
predicting the positions of atoms in a biomolecular system over time by applying Newton’s equations using a force field to specify the system’s parameters
- Orphan transporter
a transporter whose endogenous substrate or physiological function has not yet been characterized
- Orthosteric inhibitor
an inhibitor that binds to the substrate or orthosteric site of the transporter
- Orthosteric binding site
the binding site of the transporter’s substrate or other competitive ligands
- Pharmacochaperone
a small molecule that aids in the correct folding, stabilization, and/or trafficking of misfolded or unstable proteins, thereby rescuing their functional activity
- Pharmacogenomics
the study of how individual’s response to drugs is influenced by their genetic makeup
- PROteolysis-TArgeting Chimera (PROTAC)
bifunctional molecule consisting of a small molecule protein ligand that is connected via a linker to an E3 ubiquitin ligase (E3) ligand that recruits the protein degradation machinery
- Rocker-switch
a type of alternating access transport mechanism in which two symmetrically related domains shift around a central substrate-binding site, with the protein essentially moving around the substrate to alternately expose the binding site to either of the membrane
- Substrate
a molecule or an ion that gets transported across the membrane by a transporter
- Symport
two or more different substrates, such as molecules or ions, are concurrently transported across the cell membrane in the same direction
- Transport mechanism
the process of mediating substrate movement across the membrane with three commonly described states, including outward-facing, occluded, and inward-facing conformations
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hediger MA et al. (2004) The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch 447 (5), 465–8. [DOI] [PubMed] [Google Scholar]
- 2.Cesar-Razquin A et al. (2015) A Call for Systematic Research on Solute Carriers. Cell 162 (3), 478–87. [DOI] [PubMed] [Google Scholar]
- 3.Forrest LR (2013) Structural biology. (Pseudo-)symmetrical transport. Science 339 (6118), 399–401. [DOI] [PubMed] [Google Scholar]
- 4.Vander Heiden MG et al. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324 (5930), 1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin L et al. (2015) SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov 14 (8), 543–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlessinger A et al. (2013) SLC Classification: An Update. Clin Pharmacol Ther 94 (1), 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie T et al. (2022) Rational exploration of fold atlas for human solute carrier proteins. Structure. [DOI] [PubMed] [Google Scholar]
- 8.Colas C et al. (2016) SLC Transporters: Structure, Function, and Drug Discovery. Medchemcomm 7 (6), 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meixner E et al. (2020) A substrate-based ontology for human solute carriers. Mol Syst Biol 16 (7), e9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casiraghi A et al. (2021) Recent developments in ligands and chemical probes targeting solute carrier transporters. Curr Opin Chem Biol 62, 53–63. [DOI] [PubMed] [Google Scholar]
- 11.Haferkamp S et al. (2020) Extracellular Citrate Fuels Cancer Cell Metabolism and Growth. Front Cell Dev Biol 8, 602476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huizing M and Gahl WA (2020) Inherited disorders of lysosomal membrane transporters. Biochim Biophys Acta Biomembr 1862 (12), 183336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacomini KM et al. (2010) Membrane transporters in drug development. Nature Reviews Drug Discovery 9 (3), 215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardi E et al. (2020) A widespread role for SLC transmembrane transporters in resistance to cytotoxic drugs. Nat Chem Biol 16 (4), 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzel U et al. (1995) Stereoselective uptake of beta-lactam antibiotics by the intestinal peptide transporter. Br J Pharmacol 116 (7), 3021–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamai I et al. (1997) The predominant contribution of oligopeptide transporter PepT1 to intestinal absorption of beta-lactam antibiotics in the rat small intestine. J Pharm Pharmacol 49 (8), 796–801. [DOI] [PubMed] [Google Scholar]
- 17.Yee SW et al. (2018) Influence of Transporter Polymorphisms on Drug Disposition and Response: A Perspective From the International Transporter Consortium. Clin Pharmacol Ther 104 (5), 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garibsingh RA and Schlessinger A (2019) Advances and Challenges in Rational Drug Design for SLCs. Trends Pharmacol Sci 40 (10), 790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen AS et al. (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63 (3), 585–640. [DOI] [PubMed] [Google Scholar]
- 20.Andersen J et al. (2009) Recent advances in the understanding of the interaction of antidepressant drugs with serotonin and norepinephrine transporters. Chem Commun (Camb) (25), 3677–92. [DOI] [PubMed] [Google Scholar]
- 21.Koepsell H (2017) The Na(+)-D-glucose cotransporters SGLT1 and SGLT2 are targets for the treatment of diabetes and cancer. Pharmacol Ther 170, 148–165. [DOI] [PubMed] [Google Scholar]
- 22.Rosenbrock H et al. (2023) Development of the novel GlyT1 inhibitor, iclepertin (BI 425809), for the treatment of cognitive impairment associated with schizophrenia. Eur Arch Psychiatry Clin Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison SA et al. (2022) Licogliflozin for nonalcoholic steatohepatitis: a randomized, double-blind, placebo-controlled, phase 2a study. Nat Med 28 (7), 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.https://www.clinicaltrials.gov/, (accessed).
- 25.de Oliveira TM et al. (2021) Cryo-EM: The Resolution Revolution and Drug Discovery. SLAS Discov 26 (1), 17–31. [DOI] [PubMed] [Google Scholar]
- 26.Perland E and Fredriksson R (2017) Classification Systems of Secondary Active Transporters. Trends Pharmacol Sci 38 (3), 305–315. [DOI] [PubMed] [Google Scholar]
- 27.Colas C and Laine E (2021) Targeting Solute Carrier Transporters through Functional Mapping. Trends Pharmacol Sci 42 (1), 3–6. [DOI] [PubMed] [Google Scholar]
- 28.Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211 (5052), 969–70. [DOI] [PubMed] [Google Scholar]
- 29.Forrest LR et al. (2011) The structural basis of secondary active transport mechanisms. Biochimica et biophysica acta 1807 (2), 167–88. [DOI] [PubMed] [Google Scholar]
- 30.Drew D and Boudker O (2016) Shared Molecular Mechanisms of Membrane Transporters. Annu Rev Biochem 85, 543–72. [DOI] [PubMed] [Google Scholar]
- 31.Wang N et al. (2021) Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell 184 (2), 370–383 e13. [DOI] [PubMed] [Google Scholar]
- 32.Garaeva AA et al. (2019) A one-gate elevator mechanism for the human neutral amino acid transporter ASCT2. Nat Commun 10 (1), 3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan R et al. (2022) The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis. Cell Res 32 (7), 687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan R et al. (2019) Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 568 (7750), 127–130. [DOI] [PubMed] [Google Scholar]
- 35.Deng D et al. (2014) Crystal structure of the human glucose transporter GLUT1. Nature 510 (7503), 121–5. [DOI] [PubMed] [Google Scholar]
- 36.Killer M et al. (2021) Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Sci Adv 7 (45), eabk3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khanppnavar B et al. (2022) Structural basis of organic cation transporter-3 inhibition. bioRxiv, 2022.07.14.499921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanda G et al. (2021) Modafinil and its structural analogs as atypical dopamine uptake inhibitors and potential medications for psychostimulant use disorder. Curr Opin Pharmacol 56, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman JA et al. (2016) X-ray structures and mechanism of the human serotonin transporter. Nature 532 (7599), 334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motiwala Z et al. (2022) Structural basis of GABA reuptake inhibition. Nature 606 (7915), 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker JL et al. (2021) Molecular basis for redox control by the human cystine/glutamate antiporter system xc(). Nat Commun 12 (1), 7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahsavar A et al. (2021) Structural insights into the inhibition of glycine reuptake. Nature 591 (7851), 677–681. [DOI] [PubMed] [Google Scholar]
- 43.Kapoor K et al. (2016) Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides. Proc Natl Acad Sci U S A 113 (17), 4711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coleman JA et al. (2019) Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport. Nature 569 (7754), 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garibsingh RA et al. (2021) Rational design of ASCT2 inhibitors using an integrated experimental-computational approach. Proc Natl Acad Sci U S A 118 (37). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arrowsmith CH et al. (2015) The promise and peril of chemical probes. Nat Chem Biol 11 (8), 536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendez D et al. (2019) ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47 (D1), D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlessinger A et al. (2010) Comparison of human solute carriers. Protein Science 19 (3), 412–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoichet BK and Kobilka BK (2012) Structure-based drug screening for G-protein-coupled receptors. Trends in pharmacological sciences 33 (5), 268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauer DB et al. (2021) Structure and inhibition mechanism of the human citrate transporter NaCT. Nature 591 (7848), 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cirillo D et al. (2021) Structure-Activity-Relationship-Aided Design and Synthesis of xCT Antiporter Inhibitors. ChemMedChem 16 (17), 2650–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X et al. (2023) Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol 25 (3), 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh I et al. (2023) Structure-based discovery of conformationally selective inhibitors of the serotonin transporter. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Changeux JP and Christopoulos A (2016) Allosteric Modulation as a Unifying Mechanism for Receptor Function and Regulation. Cell 166 (5), 1084–1102. [DOI] [PubMed] [Google Scholar]
- 55.Niello M et al. (2020) Allosteric Modulation of Neurotransmitter Transporters as a Therapeutic Strategy. Trends Pharmacol Sci 41 (7), 446–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plenge P et al. (2020) The mechanism of a high-affinity allosteric inhibitor of the serotonin transporter. Nature Communications 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aggarwal S et al. (2019) Identification of a Novel Allosteric Modulator of the Human Dopamine Transporter. ACS Chem Neurosci 10 (8), 3718–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mostyn SN et al. (2019) Identification of an allosteric binding site on the human glycine transporter, GlyT2, for bioactive lipid analgesics. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Canul-Tec JC et al. (2017) Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erichsen MN et al. (2010) Structure-activity relationship study of first selective inhibitor of excitatory amino acid transporter subtype 1: 2-Amino-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-5,6,7,8-tetrahydro-4H-chrom ene-3-carbonitrile (UCPH-101). J Med Chem 53 (19), 7180–91. [DOI] [PubMed] [Google Scholar]
- 61.Dong Y et al. (2022) Conserved allosteric inhibition mechanism in SLC1 transporters. bioRxiv, 2022.09.21.508810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kortagere S et al. (2018) Identification of Novel Allosteric Modulators of Glutamate Transporter EAAT2. ACS Chem Neurosci 9 (3), 522–534. [DOI] [PubMed] [Google Scholar]
- 63.Levitan I et al. (2014) Cholesterol binding to ion channels. Front Physiol 5, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenhouse-Dantsker A (2017) Insights Into the Molecular Requirements for Cholesterol Binding to Ion Channels. Curr Top Membr 80, 187–208. [DOI] [PubMed] [Google Scholar]
- 65.Jones KT et al. (2012) Importance of cholesterol in dopamine transporter function. J Neurochem 123 (5), 700–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeppelin T et al. (2018) A direct interaction of cholesterol with the dopamine transporter prevents its out-to-inward transition. PLoS Comput Biol 14 (1), e1005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laursen L et al. (2018) Cholesterol binding to a conserved site modulates the conformation, pharmacology, and transport kinetics of the human serotonin transporter. J Biol Chem 293 (10), 3510–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marinko JT et al. (2019) Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem Rev 119 (9), 5537–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fiedorczuk K and Chen J (2022) Mechanism of CFTR correction by type I folding correctors. Cell 185 (1), 158–168.e11. [DOI] [PubMed] [Google Scholar]
- 70.Gramegna A et al. (2021) Elexacaftor-tezacaftor-ivacaftor: The new paradigm to treat people with cystic fibrosis with at least one p.Phe508del mutation. Curr Opin Pharmacol 57, 81–88. [DOI] [PubMed] [Google Scholar]
- 71.Bhat S et al. (2020) A tropane-based ibogaine analog rescues folding-deficient SERT and DAT. bioRxiv, 2020.07.14.202325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhat S et al. (2019) How to rescue misfolded SERT, DAT and NET: targeting conformational intermediates with atypical inhibitors and partial releasers. Biochem Soc Trans 47 (3), 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Razavi AM et al. (2018) How structural elements evolving from bacterial to human SLC6 transporters enabled new functional properties. BMC Biol 16 (1), 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lebowitz JJ et al. (2019) Clustered Kv2.1 decreases dopamine transporter activity and internalization. J Biol Chem 294 (17), 6957–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fog JU et al. (2006) Calmodulin kinase II interacts with the dopamine transporter C terminus to regulate amphetamine-induced reverse transport. Neuron 51 (4), 417–29. [DOI] [PubMed] [Google Scholar]
- 76.Khelashvili G and Weinstein H (2015) Functional mechanisms of neurotransmitter transporters regulated by lipid-protein interactions of their terminal loops. Biochim Biophys Acta 1848 (9), 1765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dale B et al. (2021) Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer 21 (10), 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alabi SB and Crews CM (2021) Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J Biol Chem 296, 100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu M et al. (2018) Discovery of a Keap1-dependent peptide PROTAC to knockdown Tau by ubiquitination-proteasome degradation pathway. Eur J Med Chem 146, 251–259. [DOI] [PubMed] [Google Scholar]
- 80.Bond MJ et al. (2020) Targeted Degradation of Oncogenic KRAS(G12C) by VHL-Recruiting PROTACs. ACS Cent Sci 6 (8), 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bensimon A et al. (2020) Targeted Degradation of SLC Transporters Reveals Amenability of Multi-Pass Transmembrane Proteins to Ligand-Induced Proteolysis. Cell Chemical Biology 27 (6), 728–739.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baker D and Sali A (2001) Protein structure prediction and structural genomics. Science 294 (5540), 93. [DOI] [PubMed] [Google Scholar]
- 83.Morcos F et al. (2011) Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA 108 (49), E1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rout MP and Sali A (2019) Principles for Integrative Structural Biology Studies. Cell 177 (6), 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang T et al. (2020) Computational Dissection of Membrane Transport at a Microscopic Level. Trends Biochem Sci 45 (3), 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chavent M et al. (2016) Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr Opin Struct Biol 40, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fenollar-Ferrer C et al. (2014) Structure and regulatory interactions of the cytoplasmic terminal domains of serotonin transporter. Biochemistry 53 (33), 5444–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szollosi D and Stockner T (2021) Investigating the Mechanism of Sodium Binding to SERT Using Direct Simulations. Front Cell Neurosci 15, 673782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sweeney CG et al. (2017) Dopamine Transporter Amino and Carboxyl Termini Synergistically Contribute to Substrate and Inhibitor Affinities. J Biol Chem 292 (4), 1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderluh A et al. (2017) Direct PIP2 binding mediates stable oligomer formation of the serotonin transporter. Nat Commun 8, 14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ponleitner M et al. (2022) Thermal Unfolding of the Human Serotonin Transporter: Differential Effect by Stabilizing and Destabilizing Mutations and Cholesterol on Thermodynamic and Kinetic Stability. Mol Pharmacol 101 (2), 95–105. [DOI] [PubMed] [Google Scholar]
- 92.Leonard AN et al. (2019) Developing and Testing of Lipid Force Fields with Applications to Modeling Cellular Membranes. Chem Rev 119 (9), 6227–6269. [DOI] [PubMed] [Google Scholar]
- 93.Muller MP et al. (2019) Characterization of Lipid-Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation. Chem Rev 119 (9), 6086–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khelashvili G et al. (2015) Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem Neurosci 6 (11), 1825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Latorraca NR et al. (2017) Mechanism of Substrate Translocation in an Alternating Access Transporter. Cell 169 (1), 96–107 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harpole TJ and Delemotte L (2018) Conformational landscapes of membrane proteins delineated by enhanced sampling molecular dynamics simulations. Biochim Biophys Acta Biomembr 1860 (4), 909–926. [DOI] [PubMed] [Google Scholar]
- 97.Cheng MH and Bahar I (2015) Molecular Mechanism of Dopamine Transport by Human Dopamine Transporter. Structure 23 (11), 2171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jumper J et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baek M et al. (2021) Accurate prediction of protein structures and interactions using a three-track neural network. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Evans R et al. (2021) Protein complex prediction with AlphaFold-Multimer. bioRxiv, 2021.10.04.463034. [Google Scholar]
- 101.Mosalaganti S et al. (2021) Artificial intelligence reveals nuclear pore complexity. bioRxiv, 2021.10.26.465776. [Google Scholar]
- 102.McCoy AJ et al. (2022) Implications of AlphaFold2 for crystallographic phasing by molecular replacement. Acta Crystallogr D Struct Biol 78 (Pt 1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tunyasuvunakool K et al. (2021) Highly accurate protein structure prediction for the human proteome. Nature 596 (7873), 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.del Alamo D et al. (2021) Sampling the conformational landscapes of transporters and receptors with AlphaFold2. bioRxiv, 2021.11.22.469536. [Google Scholar]
- 105.Schlessinger A and Bonomi M (2022) Exploring the conformational diversity of proteins. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mitrovic D et al. (2022) Reconstructing the transport cycle in the sugar porter superfamily using coevolution-powered machine learning. bioRxiv, 2022.09.24.509294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jumper J et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akdel M et al. (2022) A structural biology community assessment of AlphaFold2 applications. Nat Struct Mol Biol 29 (11), 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fan H et al. (2009) Molecular docking screens using comparative models of proteins. J Chem Inf Model 49 (11), 2512–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schlessinger A et al. (2011) Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. Proc Natl Acad Sci U S A 108 (38), 15810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carlsson J et al. (2011) Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nature Chemical Biology 7 (11), 769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bonomi M et al. (2018) Simultaneous Determination of Protein Structure and Dynamics Using Cryo-Electron Microscopy. Biophys J 114 (7), 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Colas C et al. (2017) Chemical Modulation of the Human Oligopeptide Transporter 1, hPepT1. Mol Pharm 14 (12), 4685–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Colas C et al. (2015) Structure-Based Identification of Inhibitors for the SLC13 Family of Na(+)/Dicarboxylate Cotransporters. Biochemistry 54 (31), 4900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chien HC et al. (2018) Reevaluating the Substrate Specificity of the L-Type Amino Acid Transporter (LAT1). J Med Chem 61 (16), 7358–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kickinger S et al. (2021) Molecular Determinants and Pharmacological Analysis for a Class of Competitive Non-transported Bicyclic Inhibitors of the Betaine/GABA Transporter BGT1. Front Chem 9, 736457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iancu CV et al. (2022) GLUT3 inhibitor discovery through in silico ligand screening and in vivo validation in eukaryotic expression systems. Sci Rep 12 (1), 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stein D et al. (2022) Genome-wide prediction of pathogenic gain- and loss-of-function variants from ensemble learning of diverse feature set. bioRxiv, 2022.06.08.495288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kenakin T and Christopoulos A (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov 12 (3), 205–16. [DOI] [PubMed] [Google Scholar]
- 120.Goutam K et al. (2022) Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606 (7916), 1015–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ge J et al. (2021) Molecular mechanism of prestin electromotive signal amplification. Cell 184 (18), 4669–4679 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou Y et al. (2020) Cryo-EM structure of the human concentrative nucleoside transporter CNT3. PLoS Biol 18 (8), e3000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.RDKit: Open-source cheminformatics, version Q3_2013, http://www.rdkit.org (accessed Nov 2013). [Google Scholar]
- 124.Shannon P et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 (11), 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]