Abstract
Protein α-N-terminal (Nα) methylation is a post-translational modification catalyzed by N-terminal methyltransferase 1/2 (NTMT1/2) and METTL13. Nα methylation affects protein stability, protein-protein interaction, and protein-DNA interaction. Thus, Nα methylated peptides are essential tools to study the function of Nα methylation, generate specific antibodies for different states of Nα methylation, and characterize the enzyme kinetics and activity. Here we describe chemical methods of site-specific synthesis of Nα mono, di, and trimethylated peptides in the solid phase. In addition, we describe the purification of recombinant NTMT1 and the preparation of trimethylation peptides by NTMT1 catalysis.
Keywords: Nα methylation, solid phase synthesis, Nα methylated peptides, recombinant NTMT1
1. Introduction
Protein α-N-terminal (Nα) methylation is a post-translational modification featuring adding the methyl groups to the α-N-terminus of proteins. Nα methylation is an evolutionarily conserved modification across different species (Huang, 2019). Previous research showed that Nα methylation of cytochrome c-557 protected it from aminopeptidase degradation (Smith & Pettigrew, 1980). The Nα trimethylation or the dimethylation of proline produces a permanently positive charge on the Nα-amino group. Also, the nucleophilicity of the methylated Nα-amino group is lost in the form of a tertiary or quaternary amine (Stock et al., 1987). Due to hydrophobicity and charge state changes, Nα methylation affects the protein-protein interactions and protein-DNA interactions. For example, the Nα-trimethylated myosin alkali light chain A1 increased its binding affinity with actin compared to the unmethylated one (Hayashibara & Miyanishi, 1994). The Nα-methylated tail of RCC1 (regulator of chromosome condensation 1) stabilizes the dynamic interaction of RCC1 with chromatin (Hitakomate et al., 2010); Nα-trimethylation of CENP-B (centromere protein B) strengthens its interaction with CENP-B box (a 17-bp DNA motif) in centromeric DNA (Dai et al., 2013). Nα methylation of DDB2 (Damaged DNA-binding Protein 2) showed enhanced nuclear localization and recruitment of DDB2 to the DNA damage site (Cai et al., 2014). Besides the molecular interactions, Nα methylation also plays a vital role in regulating chromatin remodeling, DNA damage repair, mitosis, and breast cancer cell proliferation and migration (Bonsignore et al., 2015; Cai et al., 2014; Hao & Macara, 2008; Hitakomate et al., 2010).
To date, three mammalian enzymes were identified to use S-adenosyl-L-methionine (SAM) as the cofactor to catalyze the Nα methylation by transferring one to three methyl groups onto the Nα amine (mono, di, tri-methylation) (Petkowski et al., 2013; Schaner Tooley et al., 2010). Protein N-terminal methyltransferase 1 and 2 (NTMT1/2) recognize a specific N-terminal sequence X-P-K/R after methionine cleavage (X represents any amino acid other than D/E) Fig 1 (Petkowski et al., 2012). However, noncanonical sequences were also reported to be methylated by NTMT1/2. For instance, MYL9 (Myosin Regulatory Light Chain 9) with an SSK sequence was moderately methylated by NTMT1 with in vitro enzymatic assay (Nevitt et al., 2018). A similar result was found for a peptide derived from the N-terminus of ZHX2 (zinc fingers and homeoboxes 2), starting with ASK, in the in vitro enzymatic assay. Nα methylation level of ZHX2 was boosted by the addition of NTMT2 (Conner et al., 2022). Besides NTMT1/2, METTL13 was identified as a dual-specific methyltransferase METTL13 to catalyze the Nα methylation and Lys55 methylation on eukaryotic translation elongation factor 1 alpha (eEF1A). As eEF1A has an N-terminal GKEK motif, it provides an additional example of a noncanonical Nα methylation (Jakobsson et al., 2018).
Fig. 1.

General scheme for protein Nα methylation.
The substrate sequence analysis suggests more than 300 substrates for NTMT1/2. However, the number of proteins validated as substrates for Nα methylation is limited. Besides, the function of protein Nα methylation remains largely unknown compared to its widespread on possible substrates. The limited information for Nα methylation substrates and function presents an urgent need to study this modification and its enzymes. Hence, Nα methylated peptides are valuable tools for studying the role of Nα methylation. For example, Nα mono, di, and tri-methylated peptides can be used as antigens to generate and purify the antibodies specific to Nα-methylated proteins with different methylation states. Those specific antibodies help validate Nα-methylated proteins and evaluate the cellular inhibition activity of inhibitors for NTMTs. Meanwhile, recombinant Nα methylated proteins would be valuable tools in studying the function of Nα methylation. However, incubating a recombinant substrate with NTMTs imparts a mixture of methylated proteins with various methylation states. On the other hand, Nα methylated peptides would be necessary starting materials for generating site-specific Nα methylated proteins through the native chemical ligation (Agouridas et al., 2019). Synthesis of selectively Nα methylated peptides in the solid phase has been reported, but mainly for the monomethylation (Biron et al., 2006; Jensen, K. J., Shelton, P. T., Pedersen, 2013; Miller & Scanlan, 1997). All these methods start with o-NBS group protection of a primary amine to activate the N-terminal amine for alkylation. The protection group is then removed after methylation with DBU and dimethylsulfate. Our lab reported the methods of di- and trimethylation of Nα peptides in the solid phase. Specifically, we utilized sodium cyanoborohydride as the reducing agent and formaldehyde as the methyl donor for Nα dimethylation. In addition, potassium carbonate and methyl iodide were used for Nα trimethylation (Borch et al., 1971; Richardson et al., 2015).
Here, we describe the method to generate Nα-methylated peptides through chemical and enzymatic methodologies site-specifically. SPPS (solid phase peptide synthesis) was used to prepare the α-N-methylated peptides. Chemical synthesis of Nα mono, di, and the tri-methylated peptide was described respectively in this protocol. Recombinant His-TEV tagged NTMT1 was expressed in Escherichia coli and purified through the conventional Ni-NTA agarose column. Enzymatic catalysis of the trimethylated peptide by recombinant NTMT1 was also described as an alternative method.
2. Preparation of recombinant NTMT1
2.1. Equipment
Shaker (Thermo Scientific™ MaxQ™ 6000 Incubated/Refrigerated Stackable Shakers)
Cooling centrifuge (Thermo Sorvall Legend XTR Refrigerated Centrifuge)
Sonicator (Fisher Scientific, Model: FB505)
Electrophoresis system (PowerPac™ Basic Power Supply and Mini-Protean Tetra system, Bio-Rad)
Nanodrop spectrophotometer (NanoDrop 1000, Thermo Scientific)
Vivaspin™ protein concentrator spin columns (20 MWCO 10 kDa)
2.2. Material and buffer recipes
Expression Escherichia coli: BL21 (DE3) Competent E. coli (New England Biolabs, C2527I)
LB Broth, Miller (Fisher #BP97235)
Ampicillin sodium salt (Sigma #A0166–25G)
IPTG (CHEM-IMPEX INT’L INC. Cat #00194)
Halt™ Protease inhibitor Cocktail (100x, Thermo Scientific #78438)
Ni-NTA Agarose (QIAGEN #30210)
TEV protease (Expressed and purified with pRK793-TEV plasmid)
-
Buffers:
Lysis buffer: 20 mM Tris, pH 7.4, 500 mM NaCl, 20 mM imidazole, 5 mM BME, 1x Halt™ protease inhibitor cocktail
Elution buffer A: 20 mM Tris, pH 7.4, 500 mM NaCl and 20 mM imidazole, 5 mM BME
Elution buffer B: 20 mM Tris, pH 7.4, 500 mM NaCl and 500 mM imidazole, 5 mM BME
Dialysis buffer (Storage buffer): 20 mM Tris, pH 7.4, 50 mM KCl.
2.3. Expression
Expression vector pET28a encoding full-length NTMT1 with His6-TEV tag is expressed in Escherichia coli BL21 (DE3) codon plus RIL cells in LB medium in the presence of 100 μg/mL ampicillin. Glycerol stocks of Escherichia coli BL21 cells are prepared and stored at −80 oC.
Inoculate 10 mL LB medium and grow in a shaker at 37 oC with constant agitation (180 rpm) overnight.
Inoculate 1 L LB medium (with 100 μg/mL ampicillin) with 10 mL starter culture in step 2. Grow at 37 oC with shaking at 180 rpm. Check OD600 hourly until it reaches ~0.5 – 0.7, which usually takes 3–4 hours for 1 L LB medium.
Take 20 μL uninduced culture into a microcentrifuge tube containing 5 μL 5x SDS-PAGE loading dye. Label the sample as uninduced whole cell lysate and store it at −20 oC.
Protein overexpression is then induced with 1 mM IPTG (add 1 mL of 1 M IPTG stock solution to 1 L medium) and shaken overnight at 16 oC for 16 h. Take 20 μL induced culture and mix with 5 μL 5x SDS-PAGE loading dye. Label the sample as induced whole cell lysate and store it at −20 oC. (see Note 2.5.1)
Harvest cells by centrifugation at 3,000 g at 4 oC for 15 min. Decant supernatant and store cell pellets at −80 oC until ready to use.
2.4. Purification
2.4.1. Preparation of cell lysate
Resuspend cell pellets in a Lysis Buffer (40 mL for pellets per liter of LB culture) and transfer the suspension into a 50 mL centrifuge tube.
Lyse the cells via sonication on the ice at 50% amplitude, pulse: 5 seconds on, 15 seconds off for 15 min with intermittent 5 min pause to check the temperature of suspension and tip.
Centrifuge the total lysate at 15,000 g for 30 min at 4 oC, collect the supernatant, and centrifuge again at 15,000 g for 30 min at 4 oC. Take a tiny portion of the pellet and mix it with 25 μL of 1x SDS-PAGE loading dye. Label the sample as pellet and store it at −20 oC. (see Note 2.5.2)
Filter the supernatant using 0.45 μm filters. Save 20 μL of the supernatant and mix with 5 μL 5x SDS-PAGE loading dye. Label the sample as cell lysate and store it at −20 oC.
2.4.2. Enrichment of His-TEV-NTMT1 through Ni-NTA column
Wash 1 mL of the Ni-NTA agarose resin with 5 column volumes (CVs) of milli-Q grade water in a gravity flow column.
Equilibrate the column with 10 CVs of lysis buffer and drain the buffer by gravity flow.
Apply the supernatant from step 2.4.1.4 to the column twice, allowing it to pass through the resin by gravity. Collect the flow through and save 20 μL sample for SDS-PAGE gel. Label the sample as flowthrough and store it at −20 oC. (see Note 2.5.3)
Wash the column with at least 20 CVs of purification buffer A. Meanwhile, monitor the absorbance of the last washing buffer at 280 nm by NanoDrop and keep washing until the absorbance reaches baseline.
To remove the endogenous histidine-rich proteins bound to the resin, wash the column with 10–20 CVs of 5% elution buffer B in buffer A (45 mM imidazole). Collect washing fractions, 5 mL each. (see Note 2.5.4)
Wash the column with 10–20 CVs of 40% elution buffer B in buffer A (200 mM imidazole) to elute the protein. Collect elution fractions, 3 mL each.
Wash the column with 20 CVs of 100% buffer B to remove any bound proteins. Wash the column with 10 CVs of milli-Q water and store it in 20% ethanol.
Take 20 μL from each fraction, mix with 5 μL of 5x SDS-PAGE loading dye, and run the gel for each fraction. The fractions containing the His-TEV-NTMT1 (~ 25 kDa) are collected. (Fig. 2 )
Fig. 2.

SDS-PAGE analysis of His-TEV-NTMT1 purification. Each well was loaded with 20 μL of samples. Lane 1: protein ladder; Lane 2: cell lysate; Lane 3: flow-through of cell lysate passed over the Ni-NTA column; Lane 4: 5% elution buffer B washing; Lane 5, 6, 7: elution samples obtained with 40% elution buffer B; Lane 8: collected protein fractions with TEV protease; Lane 9: flow-through of protein solution passed through Ni-NTA column to remove TEV protease; Lane 10: elution buffer A washing to remove the nonspecifically bound NTMT1 on the column.
2.4.3. Cleavage of His-TEV tag and removal of TEV protease
Transfer the collected protein solution to a 10 kDa MW cutoff dialysis bag. Save 20 μL for gel analysis.
Cleave the His-tag by adding TEV protease (1:50 molar ratio of TEV protease and NTMT1) to the dialysis bag.
Dialyze the protein mixture against the dialysis buffer overnight at 4 oC with constant stirring. Change the dialysis buffer three times.
Transfer the protein solution to a 50 mL centrifuge tube and spin at 10,000 g for 10 min at 4 oC to remove any insoluble. Decant the supernatant to a clean tube. Take 20 μL and save for gel analysis.
Wash the Ni-NTA column (1 mL) with 5 CVs of milli-Q water and equilibrate with 10 CVs of dialysis buffer.
To remove the His-tagged protease and the uncleaved His-NTMT1, the supernatant from step 4 is applied to the Ni-NTA column for the second affinity separation. Collect the flowthrough and save 20 μL for gel analysis.
To elute nonspecifically bound NTMT1, wash the column with 5 CVs of elution buffer A (20 mM imidazole), collect the fraction, and save 20 μL for gel analysis.
Run the gel for saved samples and pool fractions that contain cleaved NTMT1 based on the gel analysis. (Fig. 2)
The resulting solution is dialyzed in a storage buffer and concentrated to a desired concentration with a protein concentrator spin column (10 kDa cutoff MW) by centrifuging at 1,800 rpm under 4 oC. Aliquot, flash-frozen, and store at −80 oC.
2.5. Notes
Analyze the uninduced and induced cells through SDS-PAGE before protein purification to ensure the expression of the targeted protein.
For the first purified protein, it is recommended to save the cell pellet and analyze it through SDS-PAGE to check if the targeted protein is soluble or insoluble.
The flowthrough can be reloaded to the column if the gel indicates that binding is inefficient. So always run the sample for each step.
An elution buffer with an increasing imidazole gradient can be used when purifying the target protein for the first time to determine the appropriate imidazole concentration that can remove most impurities and retain the target protein on the column.
3. Synthesis of Nα methylated peptides
3.1. Equipment
CEM Automated Microwave Peptide Synthesizer (Liberty Blue)
Peptide reaction vessels (Torviq #SF-1000, #SF-0500)
Vac-Man® Laboratory Vacuum Manifold (Promega #A7231)
Vortex mixer (Corning #LSE 6776)
Tube revolver (Thermo Scientific #88881001)
Centrifuge (Eppendorf #5424R)
Bench Top Incubator (Thermo Scientific Heratherm #IMC18 50125590)
Voyager DE-Pro MALDI-TOF Mass Spectrometer
3.2. Materials
Resin: Rink amide resin (0.48 mmol/g, CHEM-IMPEX INT’L INC. #12662)
-
Organic solvent
N, N -Dimethylformamide (DMF, Honeywell #LP076–19))
Dichloromethane (DCM, Fisher #D37–20)
Methanol (Fisher #A412–20)
1-Methyl-2-pyrrolidine (NMP, Acros Organics #449180050)
-
Reagents
Standard Fmoc-protected amino acids (CEM): Fmoc-Ala-OH, Fmoc-Asn(Trt)-OH, Fmoc-Asp(OtBu)-OH, Fmoc-Arg(Pbf)-OH, Fmoc-Cys(Trt)-OH, Fmoc-Gln(Trt)-OH, Fmoc-Glu(OtBu)-OH, Fmoc-Gly-OH, Fmoc-His(Boc)-OH, Fmoc-Ile-OH, Fmoc-Leu-OH, Fmoc-Lys(Boc)-OH, Fmoc-Met-OH, Fmoc-Phe-OH, Fmoc-Pro-OH, Fmoc-Ser(tBu)-OH, Fmoc-Thr(tBu)-OH, Fmoc-Trp(Boc)-OH, Fmoc-Tyr(tBu)-OH, Fmoc-Val-OH
Oxyma (CEM, #S001-C)
DIC (N,N’-Diisopropylcarbodiimide, CHEM-IMPEX INT’L INC. #00110)
Piperidine (CHEM-IMPEX INT’L INC. #02351)
2-Nitrobenzenesulfonyl chloride (o-NBS-Cl, Alfa Aesar #B21522))
2,4,6-Trimethylpyridine (2,4,6-collidine, TCI #T0716)
1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU, CHEM-IMPEX INT’L INC. #12723)
Dimethyl sulfate (DMS, Sigma #D186309–100mL)
2-Mercaptoethanol (BME, Acros Organics #125472500)
Formaldehyde (HCHO, Fisher # BP531–500)
Acetic acid (AcOH, Supelco #AX0073–9))
Sodium cyanoborohydride (NaBH3CN, Alfa Aesar #87839)
Methyl iodide (MeI, Fisher #M212I-100))
18-crown-6 (Santa Cruz Biotechnology #sc-254005)
Cesium carbonate (Cs2CO3, Acros Organics #278020500)
Trifluoroacetic acid (TFA, CHEM-IMPEX INT’L INC. #02883)
3,6-Dioxa-1,8-octanedithiol (DODT, TCI #D2649)
Triisopropylsilane (TIPS, Oakwood chemical #S17975)
Diethyl ether (Et2O, Fisher #E138–4)
1,4-dithiothreitol (DTT, Thermo Scientific™ #409190010)
S-(5′-Adenosyl)-L-methionine chloride dihydrochloride (SAM, Sigma #A7007–100MG)
3.3. Synthesis of peptides
Peptides are synthesized on Rink amide resin using standard Fmoc chemistry with a CEM Liberty microwave peptide synthesizer. A peptide derived from protein SET (APKRQSPLPP, SET-10) is used as an example.
Program the synthesis method for peptides with the Liberty Blue software, including peptide sequence (APKRQSPLPP), synthesis scale (0.1 mmol), C-terminus (amide), resin type (not preloaded), deprotection cycle (final deprotection). Generate the usage report for reagent preparation through the usage calculator.
Weigh each Fmoc-AA-OH (5-fold excess, final concentration is 0.2 M in DMF) and activation reagents (DIC/Oxyma) according to the usage report and dissolve in the corresponding volume of DMF. Prepare 20% piperidine (v/v) in DMF for deprotection. Ensure DMF is enough for the synthesis.
Transfer 208 mg of resin (0.1 mmol, 0.48 mmol/g) to the reaction vessel and start synthesis. Fmoc deprotection is completed with a microwave at 75 °C for 15 seconds. Next, coupling reactions are performed in a microwave reactor for 5 minutes at 75 °C, 10 minutes at 50 °C for Cys and His, and double coupling for Arg. After the peptide synthesis, the Fmoc group at the N-terminus is deprotected for methylation.
Transfer the peptide-coupled resin to a clean vessel, wash the resin thoroughly with DCM and methanol three times alternatively, and drain the solvent by vacuum. Repeat the washing step twice. (see Note 3.7.1)
Dry the resin under a vacuum for 30 minutes and store it at −20 oC.
3.4. Nα Monomethylation of peptides
We follow a previously described method for the preparation of the monomethylated peptides in the solid phase (Chatterjee et al., 2008; Miller & Scanlan, 1997) with minor modifications. This procedure will first protect and activate the N terminus with o-NBS-Cl. The subsequent alkylation takes place with DBU and dimethylsulfate in NMP. This direct methylation is efficient for all the amino acids except His(Trt). (see Note 3.7.2) The N-terminal mono-methylated peptide is obtained after removing o-NBS protection with DBU and 2-mercaptoethanol. (see Fig. 3.1 for monomethylation synthetic scheme)
Fig. 3.
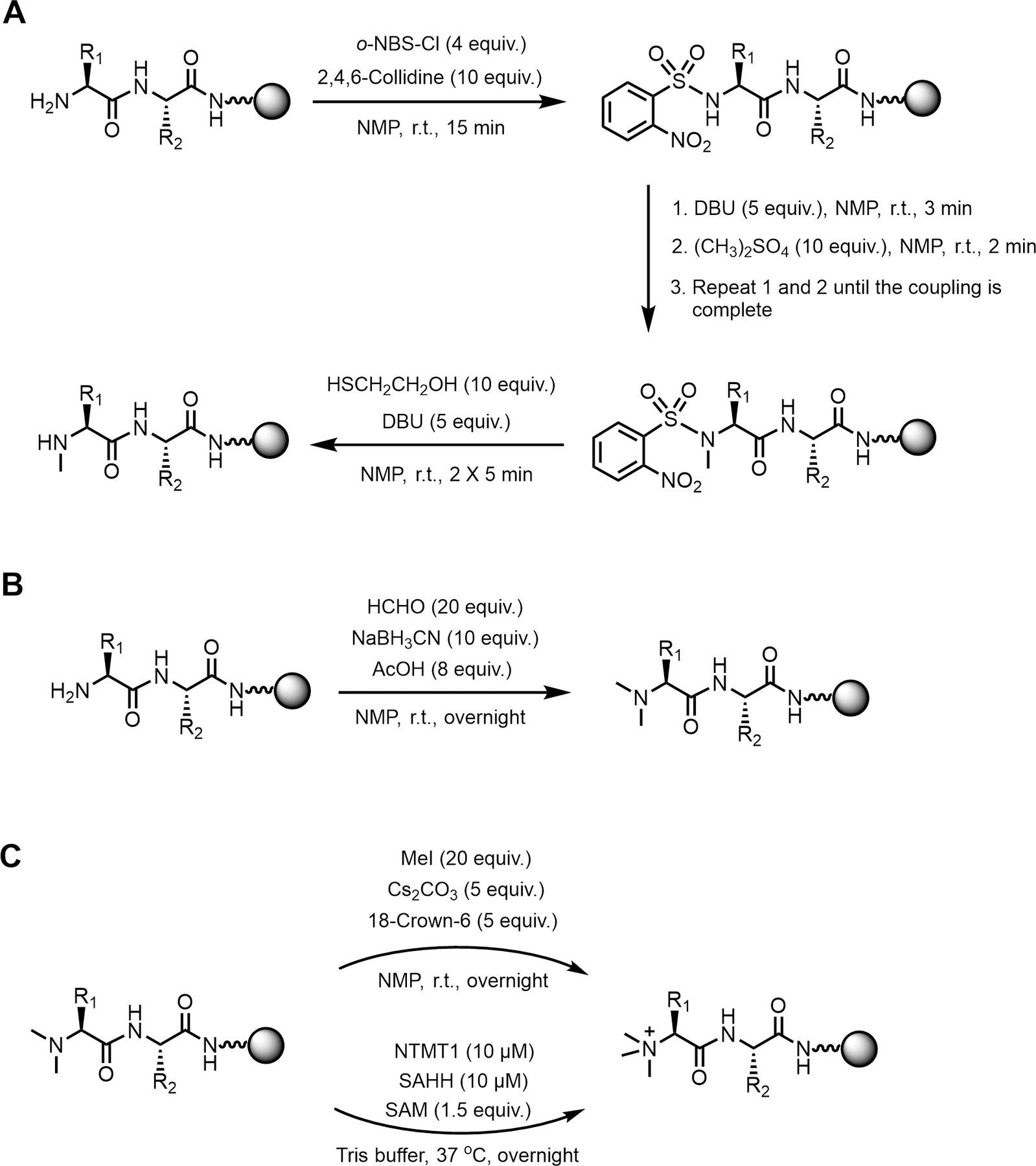
Synthetic scheme of the Nα mono-, di-, tri-methylation of peptides on a solid phase. 3.1 Nα mono-methylation of peptides. Three steps in total. Step one is o-NBS-Cl protection and activation. Step two, methylation can be achieved through direct alkylation. The final step is deprotection, which removes sulfonamide by 2-mercaptoethanol and DBU. 3.2 Nα di-methylation of peptides. A dimethylation reaction is performed with formaldehyde and sodium cyanoborohydride in the presence of acetic acid in one step. 3.3 Nα tri-methylation of peptides. Chemical trimethylation is achieved with MeI as the methylation reagent and Cs2CO3 as the base. Enzymatic trimethylation with NTMT1 and SAM is an alternative method with high efficiency but is limited to NTMT1 recognition peptides.
Place 0.05 mmol of peptide-coupled resin (~160 mg) in a clean peptide reaction vessel. To swell the resin, add 2 mL NMP and agitate with constant shaking for 15 min.
Drain the solution by vacuum and wash the resin with 5 mL NMP.
Dissolve 0.2 mmol of o-NBS-Cl (4 equiv., 44.2 mg) in 1 mL NMP, add 0.5 mmol of 2,4,6-collidine (10 equiv. 66 μL) to the solution, and vortex to mix. Add the mixed solution to the resin and agitate for 15 min at room temperature.
Drain the reaction solution by vacuum. Add 5 ml of NMP to the resin, gently agitate it for 10 s and drain the solution. Repeat the washing step four more times to thoroughly wash the resin. (see Note 3.7.3)
Add 0.25 mmol of DBU (5 equiv.,38 μL) to 0.5 mL NMP, vortex to mix. Add the solution to the resin and agitate for 3 min.
Add 0.5 mmol of dimethyl sulfate (10 equiv., 48 μL) to 0.5 mL NMP and vortex to mix. Add the solution to the resin and agitate for 2 min.
Drain the solution and wash the resin once with 5 mL NMP.
Repeat steps 5–7.
A test cleavage is performed to check if the alkylation is complete (see Note 3.7.4). Repeat steps 5–7 if the methylation is incomplete. See figure 4A for the test cleavage result of complete methylation.
Wash the resin five times with 5 mL NMP.
Add 0.5 mmol of 2-mercaptoethanol (10 equiv., 35 μL) and 0.25 mmol of DBU (5 equiv., 38 μL) to 1 mL NMP, vortex to mix. Add the solution to the resin and agitate for 5 min.
Drain the solution by vacuum and wash the resin with 5 mL NMP.
Repeat steps 10 and 11.
Wash resin five times with 5 mL NMP.
Wash the resin thoroughly with DCM and methanol three times alternatively and drain the solvent by vacuum.
Dry the resin under a vacuum for 30 minutes and store it at −20 oC.
Fig. 4.

MALDI-MS spectrums of Nα methylated peptides. A. o-NBS-protected MeSET-10 peptide. A test cleavage was performed after dimethyl sulfate methylation to determine whether the reaction was completed. A methylation reaction should be performed again if starting material was observed. B. Monomethylated SET10 peptide. C. Dimethylated SET10 peptide. D. Trimethylated SET10 peptide.
3.4. Nα Dimethylation of peptides
We follow the method described in our previously published paper for the Nα dimethylation reaction of peptides or monomethylation of peptides starting with proline (Richardson et al., 2015b). The N-terminal dimethylated peptide is obtained by reacting with formaldehyde, sodium cyanoborohydride, and acetic acid. (see Fig. 3.2 for dimethylation synthetic scheme)
Place 0.05 mmol of peptide-coupled resin (~160 mg) in a clean peptide reaction vessel. To swell the resin, add 2 mL NMP and agitate with constant shaking for 15 min.
Weigh 0.5 mmol of NaBH3CN (10 equiv., 32 mg) and dissolve in 1 mL NMP. Add 1 mmol of HCHO (20 equiv., 37 μL), 0.4 mmol of AcOH (8 equiv., 32 μL) to the solution, and vortex to mix.
Add the solution to the resin and agitate with constant shaking at room temperature overnight.
A test cleavage can be performed to check if the dimethylation is complete. (see Note 3.7.4) Repeat steps 2–3 if the methylation is incomplete.
Wash resin five times with 5 mL NMP.
Wash the resin thoroughly with DCM and methanol three times alternatively and drain the solvent by vacuum. Repeat the washing step twice.
Dry the resin under a vacuum for 30 minutes and store it at −20 oC.
3.5. Nα Trimethylation of peptides
3.5.1. Solid phase synthesis with MeI
Chemical Nα trimethylation of peptides or the dimethylation of peptides starting with proline is achieved by following the method described in our previously published paper (Richardson et al., 2015). MeI is used as the methyl donor, and Cs2CO3 is used as the base. (see Fig. 3.3 for trimethylation synthetic scheme)
Place 0.05 mmol of Nα dimethylated peptide coupled resin (~160 mg) in a clean peptide reaction vessel. To swell the resin, add 2 mL NMP and agitate with constant shaking for 15 min. (see Note 3.7.4)
Weigh 0.25 mmol of Cs2CO3 (5 equiv., 80 mg) and 0.25 mmol of 18-Crown-6 (5 equiv., 66 mg) and dissolve in 1 mL NMP. Add 1 mmol of MeI (20 equiv., 63 μL) to the solution and vortex to mix.
Add the solution to the resin and agitate with constant shaking at room temperature overnight.
A test cleavage is performed to check if the trimethylation is complete. (see Note 3.7.5) Repeat steps 2–3 if the methylation is incomplete.
Wash resin five times with 5 mL NMP.
Wash the resin thoroughly with DCM and methanol three times each alternatively and drain the solvent by vacuum. Repeat the washing step twice.
Dry the resin under a vacuum for 30 minutes and store it at −20 oC.
3.5.2. NTMT1 catalyzed trimethylation of peptides
Chemical Nα trimethylation reaction can be applied to all the peptides with different sequences. However, the methylation efficiency is not as high as mono- and di-methylation typically needs two or more coupling cycles to complete the trimethylation. So, here we also describe the enzymatic Nα trimethylation method. This method requires a shorter time and yields higher purity than chemical methylation, though it is limited to peptides with NTMT1 recognition motif (XPK/R).
Prepare reaction buffer (NTMT1 storage buffer, 20 mM Tris, pH 7.4, 50 mM KCl), and reagent stock solution (100 mM DTT, 100 mM SAM).
Purify Nα dimethylated peptide and prepare peptide solution (see section 4 for details of peptide cleavage and purification). Dissolve the desired amount in the reaction buffer to make 10 mM peptide solution.
Take NTMT1, SAHH from −80 oC and thaw on ice. (see Note 3.7.6)
For 2 mL reaction, add 100 μL of 10 mM peptide solution, 15 μL of 100 mM SAM (1.5 equiv.), 20 μL of 100 mM DTT to 1 mL reaction buffer. Adjust the pH of the reaction buffer to around 7.4. (see Note 3.7.4)
Add 340 μL of NTMT1 (1.5 mg/mL, 60 μM) and 70 μL of SAHH stock (20 mg/mL, 300 μM) to the reaction mixture to make 10 μM as the final concentration (the concentration ratio of peptide to NTMT1 is 50:1). (see Note 3.7.7)
Add reaction buffer to make the volume of 2 mL.
Place the reaction in a 37 °C incubator and incubate the reaction overnight.
Quench the reaction with 20 mL of cold methanol (10 times the reaction volume).
Keep the reaction mixture solution at −20 oC for 30 min to precipitate the proteins.
Centrifuge at 10,000 g for 10 min at 4 oC. Transfer the supernatant to a clean flask, and concentrate through the rotary evaporator to the desired volume. Purify through HPLC or store at −20 oC.
3.6. Notes
Alternately wash the resin with DCM and methanol to swell and shrink the resin to eliminate the solvent NMP in the resin.
Nα monomethylation of peptides starting with His(Trt) showed unexpected loss of the trityl protecting group (Biron et al., 2006), yielding products with methylation of the side chain. To solve this problem, the Mitsunobu reaction can be used. Methylation is performed with 5 equiv. of triphenylphosphine, 10 equiv. of methanol, and 5 equiv. of diisopropyl azodicarboxylate (DIAD) in THF for 10 min (Jensen, K. J., Shelton, P. T., Pedersen, 2013).
A Kaiser test can be performed to check if the protection with o-NBS is complete.
Unmethylated peptides can also be the starting material to synthesize trimethylated peptides directly. In the case of an unmethylated peptide as starting material for chemical synthesis, the amount of reagents can keep the same, as a sufficient extra amount has been used. However, for enzymatic synthesis, the amount of SAM needs to increase to 4.5 equiv., as three methyl groups are needed for unmethylated peptide.
A test cleavage should be performed to check if the methylation is complete before the final cleavage. For the test cleavage, prepare 1 mL of the TFA cleavage cocktail (TFA/DODT/ddH2O/TIPS = 94/2.5/2.5/1 v/v). Remove a small amount of resin and add to the cleavage cocktail; shake the mixture at room temperature for 1 h. The TFA is removed under a nitrogen line, and the peptide is precipitated with precooled diethyl ether. Centrifuge and carefully decant the supernatant, dissolve the peptide pellet with H2O, and analyze by MALDI-TOF MS.
SAHH (S-adenosylhomocysteine hydrolase) is used to hydrolyze SAH, the product of SAM methylation. SAH is a general inhibitor for methyltransferase, which may decrease NTMT1 methylation efficiency. Thus, SAHH is added to the reaction to prevent the accumulation of SAH.
To achieve the complete methylation of peptides, a high concentration (10 μM) of NTMT1 was used.
4. Cleavage and purification of Nα methylated peptides
4.1. Equipment
Peptide reaction vessels (Torviq #SF-1000, #SF-0500)
Tube revolver (Thermo Scientific #88881001)
Cooling centrifuge (Thermo Sorvall Legend XTR Refrigerated Centrifuge)
Analytical and semipreparative HPLC system
Rotary evaporators (BUCHI R-100, B-100)
Oil vacuum pump (Fisher brand MaximaDry)
Voyager DE-Pro MALDI-TOF Mass Spectrometer
4.2. Reagents
-
Solvent
Acetonitrile (ACN, HPLC grade, Sigma-Aldrich #34851)
Deionized water with 0.1% (v/v) TFA
-
Reagents
Trifluoroacetic acid (TFA, CHEM-IMPEX INT’L INC. #02883)
3,6-Dioxa-1,8-octanedithiol (DODT, TCI #D2649)
Triisopropylsilane (TIPS, Oakwood chemical #S17975)
Diethyl ether (Et2O, Fisher #E138–4))
4.3. Cleavage of peptides
Prepare cleavage solution (4 mL, 94% TFA (3.76 mL), 2.5% DODT (100 μL), 2.5% water (100 μL), 1% TIPS (40 μL)) in a clean centrifuge tube. Add it to the dried peptide-coupled resin (0.05 mmol).
Gently agitate the reaction with constant shaking at room temperature for 4 h. Filter the cleavage mixture into a 15 mL centrifuge tube.
Wash the resin with 1 mL of cleavage solution twice and collect the filtrate.
Remove TFA by blowing nitrogen over the solution in a fume hood; the remaining solution volume is less than 1 mL.
Place diethyl ether at −20 oC for 30 min, then add 10 mL of precooled diethyl ether to the concentrated peptide cleavage solution (the volume of the diethyl ether is at least 10 times the concentrated cleavage mixture).
Remove supernatant by centrifugation at 4,000 g for 10 min at 4 oC.
Add 10 mL of precooled diethyl ether to wash the pellet and triturate the product. Decant the solvent by centrifugation.
Air-dry the peptide pellet for about 10 min. Store the pellet at −20 oC for purification.
4.4. Purification of peptides
Dissolve the crude peptide pellet with 5 mL of milli-Q water containing 0.1% (v/v) TFA. Filter peptide solution using 0.45 μm filters. (see Note 4.5.1)
Edit semipreparative HPLC general method. Solvent A: deionized H2O with 0.1% (v/v) TFA, solvent B: HPLC grade ACN, running time:30 min, gradient: 5–50% of ACN, wavelength: 214 nm and 254 nm.
Analyze 100 μL of the crude peptide solution via semipreparative HPLC according to the conditions described in step 2. Collect the fractions corresponding to each peak and confirm the target peptide mass by MALDI-TOF MS.
Modify the purification method, and purify the rest of the crude peptide.
Collect the pure fractions in a round-bottom flask. Remove the solvent with a rotary evaporator (keep the temperature below 45 oC).
Freeze the concentrated solution at −80 oC and then lyophilize it overnight to obtain purified peptide. Confirm the identity of the peptide by MALDI-TOF MS.
4.5. Notes
Filtering the crude peptide solution before HPLC injection is critical for purification, as the insoluble/invisible impurities may block the HPLC lines.
5. Summary and prospects
Protein Nα methylation as an evolutionarily conserved modification began to show emerging importance in mitosis, DNA-damage repair, stem cell regulation, and cervical cancer cell proliferation and migration. NTMT1, the primary enzyme responsible for Nα methylation, is essential to study this modification. In this chapter, we described detailed protocols for the expression and purification of recombinant NTMT1. Recombinant NTMT1 can be used in different in vitro assays, such as fluorescent assay, mass spec assay, and inhibitor evaluation. We also outlined the protocols for synthesizing Nα methylated peptides, including mono, di, and trimethylation. Nα-methylated peptides play important roles in studying Nα methylation, such as the kinetic study of NTMT1 with peptides of different methylation states, peptide pull-down studies to identify the interactor, and antibody purification for Nα methylation. Nα monomethylated peptides have one N-H bond left so that the amino acid coupling can continue after the methylation. Peptides containing N-methylated residues in the middle can also be synthesized using the described monomethylation method. Unlike aliphatic side-chain amines with a pKa value around 10.5, Nα amine will be ionized under lower pH as its pKa is 6~8. Thus, it is possible to control the ionization of Nα amine with a low pH buffer and achieve the Nα dimethylation in aqueous conditions. Though this method needs further validation, it provides another thought of Nα methylated peptide synthesis. We hope that these techniques will be helpful for labs to start working on Nα methylation and would be utilized in elucidating the function of Nα methylation.
Acknowledgments
This work was funded by NIH grants R01GM117275 (RH).
References
- Agouridas V, El Mahdi O, Diemer V, Cargoët M, Monbaliu JCM, & Melnyk O (2019). Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chemical Reviews, 119(12), 7328–7443. 10.1021/acs.chemrev.8b00712 [DOI] [PubMed] [Google Scholar]
- Biron E, Chatterjee J, & Kessler H (2006). Optimized selective N-methylation of peptides on solid support. Journal of Peptide Science, 12(3), 213–219. 10.1002/psc.711 [DOI] [PubMed] [Google Scholar]
- Bonsignore LA, Sergesketter Butler J, Klinge CM, Schaner Tooley CE, & Tooley CES (2015). Loss of the N-terminal methyltransferase NRMT1 increases sensitivity to DNA damage and promotes mammary oncogenesis. Oncotarget, 6(14), 12248–12263. www.impactjournals.com/oncotarget/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borch RF, Bernstein MD, & Durst HD (1971). The Cyanohydridoborate Anion as a Selective Reducing Agent. Journal of the American Chemical Society, 93(12), 2897–2904. 10.1021/ja00741a013 [DOI] [Google Scholar]
- Cai Q, Fu L, Wang Z, Gan N, Dai X, & Wang Y (2014). α-N-methylation of damaged DNA-binding protein 2 (DDB2) and its function in nucleotide excision repair. The Journal of Biological Chemistry, 289(23), 16046–16056. 10.1074/jbc.M114.558510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee J, Gilon C, Hoffman A, & Kessler H (2008). N-methylation of peptides: A new perspective in medicinal chemistry. Accounts of Chemical Research, 41(10), 1331–1342. 10.1021/ar8000603 [DOI] [PubMed] [Google Scholar]
- Conner MM, Parker HV, Falcone DR, Chung G, & Schaner Tooley CE (2022). Novel regulation of the transcription factor ZHX2 by N-terminal methylation. Transcription, 13(1–3), 1–15. 10.1080/21541264.2022.2079184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Otake K, You C, Cai Q, Wang Z, Masumoto H, & Wang Y (2013). Identification of novel α-N-methylation of CENP-B that regulates its binding to the centromeric DNA. Journal of Proteome Research, 12(9), 4167–4175. 10.1021/pr400498y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, & Macara IG (2008). Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. The Journal of Cell Biology, 182(5), 827–836. 10.1083/jcb.200803110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibara T, & Miyanishi T (1994). Binding of the Amino-Terminal Region of Myosin Alkali 1 Light Chain to Actin and Its Effect on Actin-Myosin Interaction. Biochemistry, 33(43), 12821–12827. 10.1021/bi00209a013 [DOI] [PubMed] [Google Scholar]
- Hitakomate E, Hood FE, Sanderson HS, & Clarke PR (2010). The methylated N-terminal tail of RCC1 is required for stabilisation of its interaction with chromatin by Ran in live cells. BMC Cell Biology, 11(1), 43–53. 10.1186/1471-2121-11-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R (2019). Chemical Biology of Protein N-Terminal Methyltransferases. ChemBioChem, 20(8), 976–984. 10.1002/cbic.201800615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson ME, Małecki JM, Halabelian L, Nilges BS, Pinto R, Kudithipudi S, Munk S, Davydova E, Zuhairi FR, Arrowsmith CH, Jeltsch A, Leidel SA, Olsen JV, & Falnes P (2018). The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nature Communications, 9(1), 3411. 10.1038/s41467-018-05646-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KJ, Shelton PT, Pedersen SL (2013). Peptide synthesis and applications. Methods in Molecular Biology (2nd ed.). Humana Press, (Chapter 10). 10.1007/978-1-62703-544-6 [DOI] [Google Scholar]
- Miller SC, & Scanlan TS (1997). Site-selective N-methylation of peptides on solid support. Journal of the American Chemical Society, 119(9), 2301–2302. 10.1021/ja9635443 [DOI] [Google Scholar]
- Nevitt C, Tooley JG, & Schaner Tooley CE (2018). N-terminal acetylation and methylation differentially affect the function of MYL9. The Biochemical Journal, 475(20), 3201–3219. 10.1042/BCJ20180638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, Merchant ML, Macara IG, & Schaner Tooley CE (2013). NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. The Biochemical Journal, 456(3), 453–462. 10.1042/BJ20131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J, Hunt DF, Minor W, & MacAra IG (2012). Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT. Biochemistry, 51(30), 5942–5950. 10.1021/bi300278f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SL, Mao Y, Zhang G, Hanjra P, Peterson DL, & Huang R (2015). Kinetic mechanism of protein N-terminal methyltransferase 1. Journal of Biological Chemistry, 290(18), 11601–11610. 10.1074/jbc.M114.626846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, & MacAra IG (2010). NRMT is an α-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature, 466(7310), 1125–1128. 10.1038/nature09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GM, & Pettigrew GW (1980). Identification of N,N‐Dimethylproline as the N‐Terminal Blocking Group of Crithidia oncopelti Cytochrome c557. European Journal of Biochemistry, 110(1), 123–130. 10.1111/j.1432-1033.1980.tb04847.x [DOI] [PubMed] [Google Scholar]
- Stock A, Clarke S, Clarke C, & Stock J (1987). N-terminal methylation of proteins: structure, function and specificity, 220(1), 8–14. https://pubmed.ncbi.nlm.nih.gov/3301412/ [DOI] [PubMed] [Google Scholar]


