Abstract
A total of 199 Gram-negative bacterial isolates from urinary tract infections and 162 from bloodstream infections were collected from 12 healthcare systems throughout the United States between May 2021 and August 2022. The isolates, phenotypically non-susceptible to 2nd or 3rd generation cephalosporins or carbapenems, were characterized through antimicrobial susceptibility testing and whole genome sequence analysis to obtain a broad snapshot of beta-lactamase-mediated resistance among these two sample types. Overall, 23 different carbapenemase genes were detected among 13 species (20.5% of isolates). The blaKPC-3 and blaKPC-2 subtypes were the most common carbapenemase genes identified, followed by blaNDM and the co-carriage of two different blaOXA carbapenemases by Acinetobacter baumannii isolates. All carbapenemase-producing A. baumannii isolates were mCIM negative. Extended-spectrum beta-lactamase genes were identified in 66.2% of isolates; blaCTX-M-15 was the most common. AmpC genes, both plasmid and chromosomal, were detected in 33.2% of isolates. Importantly, 2.8%, 8.3%, and 22.2% of blaKPC-positive organisms were susceptible to ertapenem, imipenem, and meropenem, respectively. The correlation between broth microdilution and disk diffusion results was high for most drugs except cefepime, where the detection of resistance was statistically lower by disk diffusion. Thus, there were gaps in the accuracy of susceptibility testing for some mechanisms of resistance.
Keywords: carbapenemases, ESBL, AmpC, antimicrobial resistance, susceptibility testing
1. Introduction
Antimicrobial-resistant Gram-negative bacteria, especially those producing extended-spectrum beta-lactamases (ESBL) or carbapenemases, have been recognized as urgent global public health threats [1,2]. Such organisms increased in prevalence during the COVID-19 pandemic due to the broad use of antimicrobial agents and infection control measures that focused more on halting the spread of SARS-CoV-2 in hospitals than on traditional healthcare-associated pathogens [3,4]. In fact, the World Health Organization (WHO) has placed carbapenem-resistant (CR) Acinetobacter baumannii, CR Pseudomonas aeruginosa, CR Enterobacterales, and Enterobacterales resistant to third-generation cephalosporins on the critical priority list for antimicrobial development due to the limited therapeutic options for infections caused by these organisms [5]. Despite the impact on the selection of therapeutic choices for infections with antimicrobial-resistant organisms, surveillance for these organisms in various regions of the United States is often spotty [6]. Thus, there is a need for additional data about the prevalence, genotypes, and extended susceptibility profiles of ESBL- and carbapenemase-producing organisms [7]. Multicenter surveys of beta-lactam resistance in Gram-negative bacteria in the U.S. have largely been limited to specific resistance mechanisms [8], selected bacterial species [9,10], or the pre-COVID period [11]. This study is intended to address this gap.
The goals of this study were to use whole genome sequencing to identify the genes encoding ESBLs, carbapenemases, and other beta-lactamases in Gram-negative bacilli obtained from patients with urinary tract infections and bloodstream infections in the United States, to compare the resistance determinants in the organisms from the two specimen types, and to investigate emerging mechanisms of beta-lactam resistance.
2. Results
2.1. Description of the Isolates
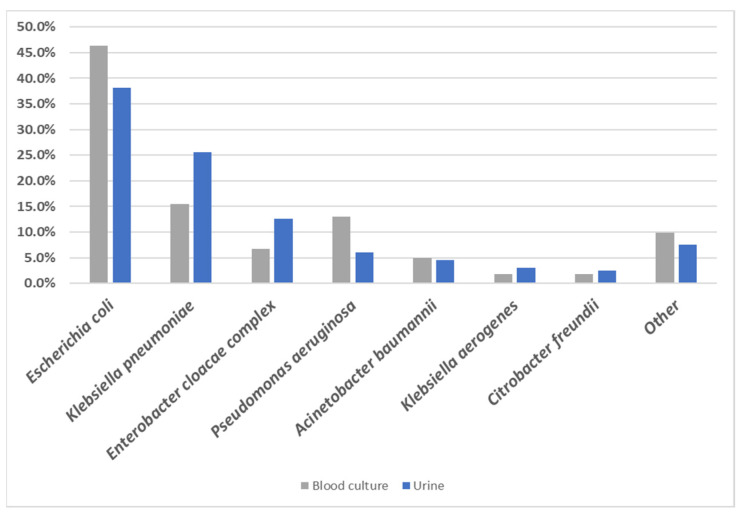
There were 361 isolates (199 urine isolates and 162 blood culture isolates) characterized in this study. E. coli represented 46.3% of isolates from blood cultures, compared to 38.2% of isolates from urine cultures, while K. pneumoniae represented only 15.4% of blood culture isolates but 25.6% of urine isolates (Figure 1). P. aeruginosa was isolated from 13.0% of blood cultures compared to only 6.0% from urine, while isolates of Enterobacter cloacae complex were isolated from 6.8% of blood culture samples but from 12.6% of urine cultures (Figure 1).
Figure 1.
Prevalence of the main bacterial species in blood and urine culture specimens. Isolates identified by sequence analysis to the subspecies level as Klebsiella pneumoniae subsp. pneumoniae and several Enterobacter spp. were grouped as K. pneumoniae and E. cloacae complex, respectively, for the purpose of this chart; see Table S1 for details.
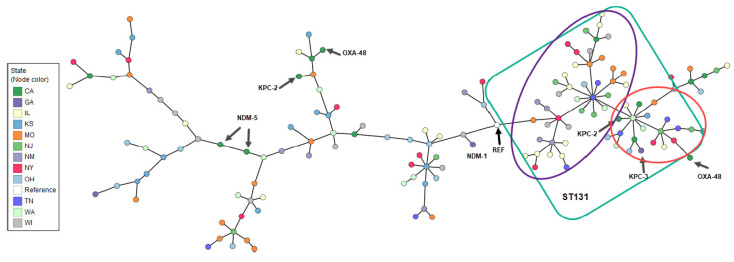
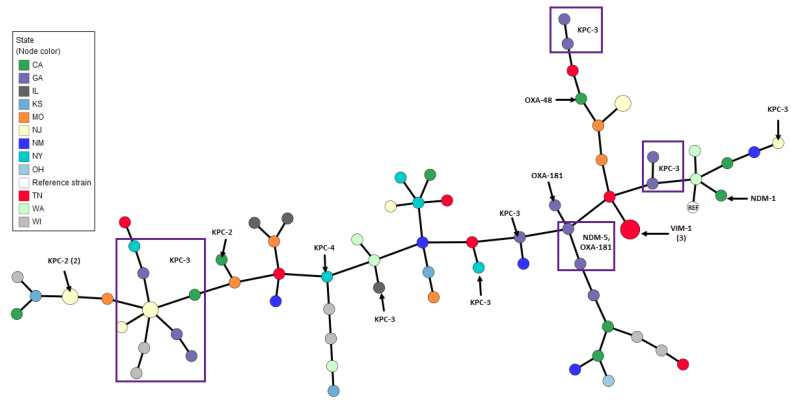
Considerable genetic diversity was observed among the bacterial species using multilocus sequence typing (MLST). Several high-impact lineages were identified in multiple states across the country. For example, among isolates of Acinetobacter baumannii, the ST2 clone, which represented 70.6% of all A. baumannii in this study, was present in New York, Tennessee, Illinois, Georgia, and California (Supplementary Materials Figure S1). Similarly, among E. coli isolates, the ST131 pandemic clone (ST43 by the Pasteur scheme) [12] was the most common strain type observed (37.8%) and was identified in eight U.S. states (Figure 2). Small sporadic clusters of related isolates, especially among the ST131 isolates, were seen in several states (Figure 2). Highly similar clusters of isolates were more evident among K. pneumoniae, some of which were indistinguishable when visualized by Minimum Spanning Tree (MST) analysis, including three clusters each containing two closely related strains from New Jersey and a cluster of three strains from Tennessee (Figure 3, larger nodes containing 2 and 3 isolates). Among the isolates from Georgia (purple nodes), a group of four closely related K. pneumoniae isolates were identified, with two carrying both blaNDM-5 and blaOXA-181, one harboring only blaOXA-181, and one with no carbapenemase gene. Isolated pairs of more closely related strains were also identified by MST, including one pair of isolates from Missouri, two distinct pairs from Washington (all harboring both blaCTX-M-15 and blaSHV), and one pair from Wisconsin (carrying blaKPC-3) (Figure 3). The most frequent sequence type among K. pneumoniae isolates was the emerging ST307 lineage (14.5%), which was present in eight states, followed by ST258 (10.5%), which was found in five states.
Figure 2.
Minimum Spanning Tree based on MLST of Escherichia coli by U.S. state of isolation and carbapenemase gene identification. Arrows indicate isolates that harbor a carbapenemase gene. The green square shows the ST131 lineage. The purple circle delineates strains harboring blaCTX-M-15; the red circle shows the strains harboring mostly blaCTX-M-14 and blaCTX-M-27. Reference strain (REF): NZ_CP117235, Escherichia coli strain ATCC25922.
Figure 3.
Minimum Spanning Tree based on MLST of Klebsiella pneumoniae by U.S. state of isolation and carbapenemase gene identification. Node radius indicates the number of isolates. Squares indicate isolates that harbor the same carbapenemase gene. Reference strain (REF): NZ_CP035929 Klebsiella pneumoniae strain B31.
P. aeruginosa isolates exhibited considerable strain diversity, which included the pandemic clones ST235 (the most frequent), ST244, ST111, ST274, and ST357. The latter clone, ST357, harbored blaNDM-1. Additionally, a P. aeruginosa ST644 isolate carrying both blaIMP-62 and blaNDM-1 and an ST167 isolate that harbored a blaIMP-15 carbapenemase were identified (Figure S2).
2.2. Beta-Lactamase Gene Carriage by Specimen Type
One or more carbapenemase genes were detected in 20.5% of all isolates, alone or in conjunction with other beta-lactamase genes (Table 1 and Table S2). Among urine cultures, 23.6% of the isolates harbored a carbapenemase gene compared to 16.7% of blood culture isolates (p = 0.116) (Table 1).
Table 1.
Number and percentage of beta-lactamase genes identified in the two sample types.
| Blood Culture (N = 162) | Urine (N = 199) | All Isolates (N = 361) | |||||
|---|---|---|---|---|---|---|---|
| No. of Isolates | % | No. of Isolates | % | p-Value | No. of Isolates | % | |
| Carbapenemase genes | 27 | 16.7% | 47 | 23.6% | 0.116 | 74 | 20.5% |
| ESBL genes | 104 | 64.2% | 135 | 67.8% | 0.503 | 239 | 66.2% |
| AmpC genes | 58 | 35.8% | 62 | 31.2% | 0.370 | 120 | 33.2% |
| Other beta-lactamase genes detected 1 | 2 | 1.2% | 3 | 1.5% | 1.000 | 5 | 1.4% |
| No beta-lactamase genes detected | 2 | 1.2% | 6 | 3.0% | 0.304 | 8 | 2.2% |
1 One blaFONA-6 (Serratia fonticola), 3 blaTEM-1B (Escherichia coli) and 1 blaTEM-1A (Serratia nematodiphila).
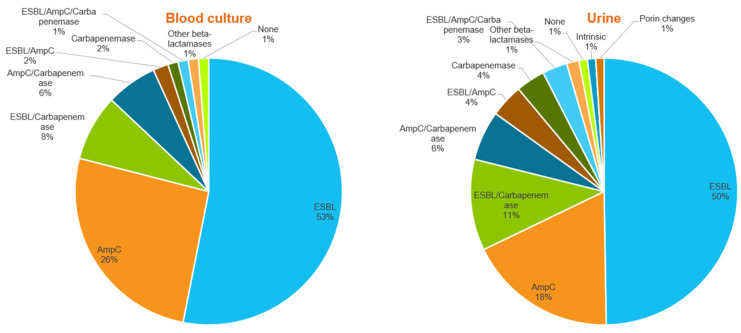
Overall, 66.2% of the isolates harbored ESBL genes, showing an even distribution between the two specimen types, with 67.8% of urine culture isolates carrying an ESBL gene compared to 64.2% of blood culture isolates (p = 0.656). AmpC genes were detected in 33.2% of all isolates and were equally distributed among the two sample types (35.8% of blood culture isolates vs. 31.2% of urine isolates; p = 0.370). Figure 4 shows the different combinations of beta-lactam resistance mechanisms observed among the blood culture and urine isolates. No significant difference was observed between the two sample types for any combination of beta-lactam resistance mechanisms, including the percentage of AmpC-only genes, which, although higher among blood cultures than among urine isolates, was not statistically significant (25.9% versus 18.1%; p = 0.094).
Figure 4.
Beta-lactam resistance mechanisms were identified among isolates from blood cultures and urine cultures.
2.3. Carbapenemase, ESBL, and AmpC Genes Detected by Whole Genome Sequencing
The predominant carbapenemase genes were blaKPC-2 and blaKPC-3, which were harbored by 9.7% of all study isolates and 47.3% of those isolates with carbapenemase genes (Table 2 and Figure S3). The blaNDM alleles were present in eight Enterobacterales isolates (two K. pneumoniae isolates also carried blaOXA-181) and two P. aeruginosa isolates, one of which also carried blaIMP-62. The remaining carbapenem-resistant P. aeruginosa isolate carried blaIMP-15. Co-carriage of two blaOXA-type carbapenemases was seen in 11 of 16 carbapenem-resistant A. baumannii isolates, including the combination of blaOXA-23 and blaOXA-66 in five isolates. Other carbapenemase genes in A. baumannii included blaOXA-24, blaOXA-71, blaOXA-78, blaOXA-82, blaOXA-95, blaOXA-237, and blaOXA-407. The remaining carbapenemase genes, including blaVIM, blaOXA-48, and blaSME, were detected sporadically.
Table 2.
Beta-lactamase gene carriage and phenotypic resistance profiles of carbapenemase-producing Enterobacterales isolates.
| Sample Type | Organism by K-mer Spectra |
MLST 1 | State | Carbapenemase Gene | AmpC Gene | ESBL Gene | ETP | IPM | MEM | Other Resistance Phenotypes | mCIM/eCIM Report |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BC | Citrobacter freundii | ST98 | TN | KPC-2 | CMY-109 | None | R | R | S | ATM, CTX, CAZ, TZP | serine carbapenemase detected |
| UC | Citrobacter freundii | ST415 | WI | KPC-2 | CMY-48 | SHV-12 | R | R | R | ATM, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Citrobacter freundii | ST344-like | WI | KPC-2 | CMY-108 | SHV-12 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Enterobacter asburiae | ST252 | CA | KPC-2 | ACT-3 | SHV-12 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Enterobacter cloacae | ST171 | NJ | KPC-3 | ACT-16 | None | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Enterobacter hormaechei | ST110 | CA | KPC-2 | ACT-15 | None | R | R | R | ATM, CTX, CAZ, CZA, CT, TZP | serine carbapenemase detected |
| UC | Enterobacter hormaechei | ST1377 | NM | NDM-1 | ACT-7 | None | R | R | R | FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| UC | Escherichia coli | ST721-like | CA | OXA-48 | DHA-1 | CTX-M-27 | R | I | S | ATM, FEP, CTX, CAZ, ETP | serine carbapenemase detected |
| UC | Escherichia coli | ST691-like | CA | KPC-2 | None | None | R | R | S | ATM, CTX, TZP | serine carbapenemase detected |
| UC | Escherichia coli | ST44 | CA | NDM-5 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| BC | Escherichia coli | ST2 | CA | NDM-5 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| BC | Escherichia coli | ST221 | CA | OXA-48 | None | None | S | I | S | TZP | serine carbapenemase detected |
| UC | Escherichia coli | ST721-like | CA | KPC-2 (truncated) | None | CTX-M-15 | S | S | S | ATM, FEP, CTX, CAZ | carbapenemase not detected |
| UC | Escherichia coli | ST43/ST131 2 | GA | KPC-3 | None | None | S | I | S | ATM, CTX, CAZ, TZP | serine carbapenemase detected |
| UC | Escherichia coli | ST33 | GA | NDM-1 | None | None | R | R | R | FEP, CTX, CAZ, CZA, CT, TZP | metallo beta-lactamase detected |
| UC | Klebsiella aerogenes | ST3-like | GA | KPC-3 | None | None | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella aerogenes | ST3-like | GA | KPC-3 | None | None | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella michiganensis | ST59-like | CA | NDM-7 | None | OXY-2-8 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| BC | Klebsiella michiganensis | ST85 | WI | KPC-2 | None | OXY-1-7 | R | R | R | ATM, CTX, TZP | serine carbapenemase detected |
| BC | Klebsiella michiganensis | ST311 | GA | KPC-3 | None | OXY-4-1, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella oxytoca | ST180 | GA | KPC-3 | None | OXY-1-4 | S | R | S | ATM, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST15 | CA | NDM-1 | CMY-6 | CTX-M, SHV-106 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| UC | Klebsiella pneumoniae | ST101 | CA | OXA-48 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CT, MEV, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST11 | CA | KPC-2 | None | CTX-M-65, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | NJ | KPC-3 | None | SHV-12 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST412 | NJ | KPC-2 | None | SHV-187 | R | R | R | ATM, CTX, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST412 | NJ | KPC-2 | None | SHV-187 | R | R | R | ATM, CTX, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | NJ | KPC-3 | None | SHV-12 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | NJ | KPC-3 | None | None | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST1683-like | NJ | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST379 | GA | KPC-3 | None | SHV-187, TEM-168 (trunc) | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST469 | GA | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST16 | GA | NDM-5, OXA-181 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| UC | Klebsiella pneumoniae | ST219 | GA | KPC-3 | None | CTX-M-15, SHV-187 | R | R | S | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST13 | GA | KPC-3 | None | SHV-197 | R | R | R | ATM, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST219 | GA | KPC-3 | None | CTX-M-15, SHV-187 | R | R | S | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST16 | GA | NDM-5, OXA-181 | None | CTX-M-15, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| UC | Klebsiella pneumoniae | ST258 | NY | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST348 | NY | KPC-4 | None | SHV-187 | R | I | S | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST307 | NY | KPC-3 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | CA | KPC-3 | None | SHV-187, TEM-168 (trunc) | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST405 | TN | VIM-1 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta-lactamase detected |
| BC | Klebsiella pneumoniae | ST405 | TN | VIM-1 | None | CTX-M-15, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo-beta lactamase detected |
| BC | Klebsiella pneumoniae | ST405 | TN | VIM-1 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CZA, CT, MEV, TZP | metallo beta lactamase detected |
| BC | Klebsiella pneumoniae | ST152 | IL | KPC-3 | None | CTX-M-15 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | WI | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST258 | WI | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST258 | GA | KPC-3 | None | SHV-12 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST13 | GA | KPC-3 | None | CTX-M-15, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Klebsiella pneumoniae | ST16 | GA | OXA-181 | None | CTX-M-15, SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| BC | Klebsiella pneumoniae | ST1199 | GA | KPC-3 | None | SHV-187 | R | R | R | ATM, FEP, CTX, CAZ, CT, TZP | serine carbapenemase detected |
| UC | Raoultella ornithinolytica | N/A | NM | OXA-181 | None | None | R | R | I | CTX, TZP | serine carbapenemase detected |
| BC | Serratia marcescens | N/A | CA | SME-2 | SRT-1 | None | R | N/R | R | ATM, CTX, CAZ, MEM | serine carbapenemase detected |
| BC | Serratia marcescens | N/A | TN | VIM-1 | SRT-1 | None | I | R | I | FEP, CTX, CAZ, CZA, CT, TZP | metallo beta lactamase detected |
1 Multi-locus sequencing typing was carried out using the 7-locus scheme present in CLC Genomic Workbench (CLC Type with MLST scheme 1.3). When multiple MLST schemes exist, such as for Acinetobacter baumannii and Escherichia coli, the Pasteur scheme is used. When all alleles cannot be identified, the closest sequence type is listed, followed by “-like”. N/A = no scheme available for that organism. 2 ST43 Pasteur/ST131 Achtman scheme ATM: aztreonam; FEP: cefepime; CTX: cefotaxime; CAZ: ceftazidime; CZA: ceftazidime/avibactam; CT: ceftolozane/tazobactam; ETP: ertapenem; IPM: imipenem; MEM: meropenem; MEV: meropenem/vaborbactam; TZP: piperacillin/tazobactam. The antimicrobial susceptibility testing results listed in this table were obtained using the broth microdilution method.
Among ESBL genes, blaCTX-M types were the most commonly identified (85.8% of all ESBLs) and included blaCTX-M-15 (58.2% of ESBL genes), blaCTX-M-27 (10.5%), and blaCTX-M-14 (7.5%) (Figure S4). The blaCTX-M genes, especially blaCTX-M-15, were detected with other ESBL genes, such as blaSHV-187, 24.9% of the time. All isolates harboring blaCTX-M genes displayed an ESBL phenotype (Figure S4). Among E. coli ST131 isolates in particular, 94.7% harbored an ESBL gene, of which 72.2% were blaCTX-M-15, 20.4% blaCTX-M-27, and 7.4% blaCTX-M-14. Isolates harboring the latter two blaCTX-M genes clustered in a separate branch on the E. coli Minimum Spanning Tree (MST) (Figure 2, red circle). Similarly, all ST307 K. pneumoniae isolates carried the blaCTX-M-15 genes (Table S2). The blaSHV beta-lactamase genes were the next most frequent ESBL family, and they were carried either alone or with other ESBL genes (31.4%). The blaSHV-187 gene was the most frequent (22.6%), followed by blaSHV-12 (3.3%).
A total of 120 AmpC genes were identified among 11 different species. The chromosomal blaPDC and blaADC genes, found in P. aeruginosa and A. baumannii, respectively, together constituted 42.5% of all the AmpC genes detected (Figure S5). Among the acquired AmpC genes, blaACT types were the most common (30.8% of all AmpC genes) and were identified in all E. cloacae complex isolates and in half of the K. aerogenes isolates. This is followed in frequency by blaCMY genes, which were identified in E. coli, C. freundii, K. aerogenes, P. mirabilis, and K. pneumoniae isolates (Figure S5).
2.4. Comparison of Phenotypic and Genotypic Resistance Profiles
Table 2 and Table 3 illustrate the most frequent carbapenemase genes identified and the correlation between the isolates’ genotype and phenotypic expression of resistance to ertapenem, imipenem, and meropenem for Enterobacterales, and imipenem and meropenem for A. baumannii and P. aeruginosa, respectively (see Figure S3 for additional information).
Table 3.
Beta-lactamase gene carriage and phenotypic resistance profiles of carbapenemase-producing Acinetobacter baumannii and Pseudomonas aeruginosa isolates.
| Sample Type | Organism by K-mer Spectra |
MLST 1 | State | Carbapenemase Gene | AmpC Gene | ESBL Gene | Imipenem Interpretation (BMD) | Meropenem Interpretation (BMD) | Other Resistance Phenotypes |
mCIM/eCIM Report |
|---|---|---|---|---|---|---|---|---|---|---|
| UC | Acinetobacter baumannii | ST2 | CA | OXA-23, OXA-66 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST2 | CA | OXA-237, OXA-66 | ADC-25 | None | I | R | CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST2 | CA | OXA-237, OXA-66 | ADC-25 | None | R | R | CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST235 | NJ | OXA-71 | ADC-25 | SHV-12 | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | GA | OXA-23 | ADC-25 | None | R | R | FEP, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | GA | OXA-23, OXA-66 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | GA | OXA-80 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST2 | GA | OXA-23, OXA-66 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST499 | NY | OXA-23, OXA-95 | ADC-25 | None | R | R | None | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | NY | OXA-23, OXA-82 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | NY | OXA-407 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST2 | NY | OXA-23, OXA-82 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST203 | NM | OXA-78 | ADC-25 | None | S | S | CTX (I) | carbapenemase not detected |
| UC | Acinetobacter baumannii | ST258 | CA | OXA-23, OXA-66 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST2 | TN | OXA-23, OXA-66 | ADC-25 | None | R | R | FEP, CTX, CAZ, TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST1525-like | TN | OXA-24, OXA-317 | ADC-25 | None | R | R | TZP | carbapenemase not detected |
| BC | Acinetobacter baumannii | ST2 | IL | OXA-24, OXA-66 | ADC-25 | None | R | R | CTX, CAZ, TZP | carbapenemase not detected |
| UC | Pseudomonas aeruginosa | ST644 | CA | IMP-62, NDM-1 | PDC-430 | PME-1 | R | R | ATM, FEP, CAZ, CZA, CT, TZP | serine carbapenemase detected |
| BC | Pseudomonas aeruginosa | ST357 | CA | NDM-1 | PDC-11 | VEB-1 | R | R | ATM, FEP, CAZ, CZA, CT, TZP | metallo beta-lactamase detected |
| BC | Pseudomonas aeruginosa | ST167 | CA | IMP-15 | PDC-445 | None | R | R | FEP, CAZ, CZA, CT, TZP | metallo beta lactamase detected |
1 Multi-locus sequencing typing was carried out using the 7-locus scheme present in CLC Genomic Workbench (CLC Type with MLST scheme 1.3). When multiple MLST schemes exist, such as for Acinetobacter baumannii and Escherichia coli, the Pasteur scheme is used. When all alleles cannot be identified, the closest sequence type is listed, followed by “-like”. N/A = no scheme available for that organism. ATM: aztreonam; FEP: cefepime; CTX: cefotaxime; CAZ: ceftazidime; CZA: ceftazidime/avibactam; CT: ceftolozane/tazobactam; ETP: ertapenem; IPM: imipenem; MEM: meropenem; MEV: meropenem/vaborbactam; TZP: piperacillin/tazobactam.
Only 61.3% of organisms harboring a blaSHV ESBL expressed the ESBL phenotype by disk diffusion (DD) and 66.7% by broth microdilution (BMD), when compared to 99.2% and 89.3% for isolates carrying blaCTX-M, respectively (p < 0.001 for both). This difference is likely due to the fact that 38.7% of blaSHV were present concurrently with a carbapenemase gene, while only 8.3% of blaCTX-M were associated with a carbapenemase gene, whose presence tends to confound phenotypic ESBL detection (Table S1). Other ESBL genes, like blaTEM-106 and blaTEM-168, blaVEB-1 and blaPME-1, and the chromosomal ESBLs blaOXY-1-1, blaOXY-1-7, and blaOXY-2-8, were seen sporadically (Table S2).
Eight isolates in our study, two from blood cultures and six from urine, concurrently harbored an ESBL, an AmpC, and a carbapenemase gene (Figure 4 and Table S1). These isolates, from California, New Jersey, and Wisconsin, included two C. freundii, two P. aeruginosa (one ST644 harboring two carbapenemase genes and epidemic clone ST357 carrying blaNDM-1), an E. coli, a K. pneumoniae (high-risk clone ST15), an Enterobacter asburiae, and an A. baumannii (ST235). All of the isolates were phenotypically resistant to the three carbapenems tested, except for the ST-721-like E. coli harboring blaOXA-48, blaCTX-M-27, and blaDHA-1, which was susceptible to meropenem and intermediate to imipenem by BMD (Table 2).
Overall, 90.3% of the 196 isolates with ESBL genes but no carbapenemase genes were resistant to cefepime by BMD (Minimum Inhibitory Concentration ≥16 µg/mL). (Table S1). The rate of cefepime resistance remained high (84.9%) even after excluding 70 isolates carrying blaOXA-1, which has been implicated in cefepime hydrolysis [2].
Among the 54 Enterobacterales that harbored a carbapenemase gene, 42 were resistant to ertapenem, imipenem, and meropenem. Five isolates were resistant to ertapenem and imipenem but intermediate or susceptible to meropenem, while an additional six isolates had variable patterns of resistance, intermediate, and susceptible results to the three antimicrobial agents. The final organism, which, was susceptible to all three drugs, harbored a blaKPC-2 gene, which on further analysis, was truncated and non-functional. The three P. aeruginosa isolates with carbapenemase genes were resistant to both imipenem and meropenem, while 15 of the 17 A. baumannii isolates were resistant to both antimicrobial agents. One A. baumannii isolate was intermediate to imipenem and resistant to meropenem, while the final isolate, which contained blaOXA-78, was susceptible to both carbapenem agents. Interestingly, all A. baumannii isolates were negative by the combined mCIM/eCIM method for detection of carbapenemase activity.
2.5. Resistance to Newer Beta-Lactam/Beta-Lactamase Inhibitor Combinations
Overall, 5.2% (16 of 309) Enterobacterales and 15.2% (5 of 33) P. aeruginosa isolates were resistant to ceftazidime–avibactam (CZA) by BMD (MIC ≥ 16/4 µg/mL) (Table S1). Of those, 12 Enterobacterales (including 6 K. pneumoniae, 3 E. coli, one E. cloacae hormaechei, one S. marcescens, and one K. michiganensis) and 3 P. aeruginosa contained metallo-beta-lactamases, two in combination with blaOXA-181. In addition, one E. hormaechei carried blaKPC-2, and five isolates, including P. aeruginosa, K. aerogenes, P. rettgeri, and S. nematodiphila (isolates #17,636, 17,708, 17,213, and 17,835 respectively in Table S1), were negative for carbapenemase genes and negative for mCIM and eCIM. Among Enterobacterales, 26.5% were resistant and 3.9% were intermediate to ceftolozane-tazobactam (CT), while resistance to the combination drug among P. aeruginosa was 12.1%, with an additional 3.0% of results reported as intermediate. Three of the four CT-resistant P. aeruginosa harbored a metallo-beta-lactamase gene, while the fourth carried only blaOXA-395 and blaPDC-471 (amino-acid mutations in blaPDC at R79Q and T105A have been linked to CT resistance) [13]. Resistance to meropenem-vaborbactam (MEV) among Enterobacterales was 3.9% (12 of 309). Ten of the resistant isolates harbored a metallo-beta-lactamase gene, one had a serine carbapenemase gene (blaOXA-48), and one P. rettgeri isolate did not carry a carbapenemase gene (Tables S1 and S2).
Among all carbapenemase-producing E. coli, two isolates harboring blaNDM-5, one ST44 isolated from urine and one ST2 from a blood culture (isolates #17184 and 17198, respectively, in Table S2), both from the same institution in California, had the four amino acid YRIK insertion at position 333 in pbp3, associated with resistance to aztreonam-avibactam and decreased susceptibility to cefiderocol (phenotypic resistance to cefiderocol and aztreonam-avibactam were not assessed). The two isolates were obtained from different patients who were unlinked epidemiologically.
3. Discussion
The goal of this study was to investigate the prevalence of novel beta-lactamase-mediated resistance mechanisms in a convenience sample of cephalosporin- or carbapenem-resistant isolates from blood and urine specimens from patients in 12 geographically dispersed U.S. hospitals. A second goal was to identify potential gaps in susceptibility testing methods that may miss emerging resistance mechanisms. It is important to note that the mCIM and eCIM tests failed to detect carbapenemase production in every one of the carbapenem-resistant A. baumannii isolates in this study that carried a carbapenemase gene. Several new protocols with modified mCIM/eCIM conditions that may detect these isolates have been proposed but not yet standardized [14].
The correlation of broth microdilution results to disk diffusion results was high in this study, with the exception of the results for cefepime, for which recognition of resistance was statistically lower by the CLSI disk diffusion method when MIC results were interpreted using either CLSI or EUCAST breakpoints [15,16]. The CLSI and EUCAST MIC interpretations were highly correlated.
Minimum spanning tree analysis of the key bacterial species in this study indicated that, except for a few pairs of K. pneumoniae isolates and several E. coli ST131 isolates, the majority of the isolates, representing 15 species, did not appear to be associated with hospital outbreaks. Thus, this convenience sample provides us with a broad snapshot of beta-lactamase-mediated resistance among Gram-negative organisms in the United States, including ESBLs, AmpC enzymes, and carbapenemases. Not surprisingly, the blaCTX-M genes were the most common ESBLs detected; however, there were a number of SHV variants, including blaSHV-12, blaSHV-106 [17], and blaSHV-187, which were detected, often in combination with blaCTX-M genes. Newer blaSHV alleles, such as blaSHV-187, are emerging globally [18]. Plasmid-mediated AmpCs, such as blaACT, blaCMY, and blaDHA, were rare in the Enterobacterales, while chromosomal AmpC enzymes were found in all A. baumannii and P. aeruginosa isolates, where they likely mediated resistance to multiple beta-lactam agents.
The E. coli high-risk lineage ST131 was the predominant sequence type observed among the E. coli isolates from both blood and urine. ST131 has been described as the “quintessential example of an international multiresistant high-risk clone” [19], and in our convenience sample, both of the C subclades described in the literature, i.e., C1, characterized by carriage of blaCTX-M-14 and blaCTX-M-27, and C2, harboring blaCTX-M-15, were observed [20]. Strains belonging to ST131 were identified in isolates from all 12 participating laboratories, consistent with its broad geographic expansion during the last two decades [20]. Similarly, the two dominant clones among K. pneumoniae isolates, i.e., ST307 and ST258, are both high-risk international clones. ST307 isolates carried blaCTX-M-15 (similar to that of E. coli ST131 [20,21]), although only one ST307 was carbapenem-resistant and carried blaKPC-3. Similarly, most of the A. baumannii isolates characterized in our study belonged to the successful international clone ST2 (representative of global clone II) [22].
In our study, most ESBL isolates were resistant to cefepime by BMD. There have been mixed reports on the effectiveness of cefepime for the treatment of ESBL infections, ranging from cefepime therapy being comparable to carbapenem therapy to being inferior [23,24,25,26]. Higher cefepime MIC levels have been previously associated with CTX-M enzymes [27], which corroborates what was observed in our study.
The breadth of carbapenemases identified was surprising. A total of 23 different carbapenemase genes were seen among 13 different species. In addition to the blaKPC, blaNDM, blaOXA-48, and blaVIM variants detected among the Enterobacterales isolates, the A. baumannii isolates contained a variety of blaOXAAb variants, such as blaOXA-23, blaOXA-24, blaOXA-51, and several OXA variants not commonly reported from US isolates. These included the blaOXA-66 family (sometimes called the blaOXA-51/66 family) and its variants, blaOXA-71, blaOXA-78, and blaOXA-317. blaOXA-237 is another carbapenemase found in Acinetobacter species; however, one that is derived from the blaOXA-134 group. The proliferation of carbapenemase genes seen among Acinetobacter species in the United States is important for clinical microbiologists to recognize since many laboratories do not test Acinetobacter species for carbapenemases, presuming that the carbapenem resistance is efflux-mediated [28]. Furthermore, beta-lactam/beta-lactamase combination drugs with activity against Acinetobacter isolates producing class D beta-lactamases are now available [29].
As noted before, the mCIM/eCIM method was not reliable for detecting carbapenemases in A. baumannii in our study. Thus, it may be worth considering the use of genotypic methods for detecting and differentiating among serine and metallo-beta-lactamase genes among carbapenem-resistant Acinetobacter species to guide therapeutic strategies for serious infections.
There were relatively few P. aeruginosa isolates in this collection, and only three (two from Northern California and one from Southern California) contained a carbapenemase gene. Carbapenemase-producing strains of P. aeruginosa remain rare in the United States but are increasingly prevalent elsewhere and associated with higher mortality [30]. One isolate carried blaIMP-15; another, which belonged to the ST357 international lineage, harbored blaNDM-1; and the third, an ST644, contained both blaIMP-62 and blaNDM-1. An extensively drug-resistant P. aeruginosa ST644 isolate was recently reported from an infected footpad of a Magellanic penguin in Brazil [31]. Because metallo-carbapenemases are common in P. aeruginosa and especially since ceftolozane-tazobactam, which is often used to treat P. aeruginosa infections, is not active against metallo-enzymes, consideration should be given to testing isolates of P. aeruginosa, especially from positive blood cultures, for carbapenemase production [32]. As opposed to the problems noted for testing A. baumannii by mCIM and eCIM, all three P. aeruginosa isolates were positive for carbapenemase production with the mCIM test; however, one of the three isolates, the one that carried both blaIMP-62 and blaNDM-1, was incorrectly identified as a serine carbapenemase producer, likely because IMP enzymes are more resilient to zinc chelation and may cause a false-positive eCIM result unless EDTA concentration is increased [33].
All of the “big 5” carbapenemases are circulating in the United States [1]. The presence of a metallo-beta-lactamase (such as blaIMP, blaNDM, or blaVIM) in a bacterial isolate can compromise the use of novel beta-lactam/beta-lactamase inhibitor combinations, such as ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam, as these antimicrobial agents often are not effective in the presence of these enzymes [34]. It is also important to note that carbapenems may be ineffective for the treatment of carbapenemase-producing organisms even when the MIC is in the susceptible range [35,36]. One blaNDM-7 gene, which encodes an NDM variant with increased carbapenem-hydrolyzing activity [37], was observed in a K. michiganensis isolate from the urine of a patient in California. The blaNDM-7 gene was located on an IncX3-type plasmid, which is an important mechanism of resistance gene dissemination for blaNDM-5 determinants [38].
Among the Enterobacterales, blaKPC, blaNDM, blaOXA-48, and blaVIM carbapenemases were detected, while blaIMP was observed in two P. aeruginosa isolates. The isolates carrying both metallo-beta-lactamases and ESBL genes were resistant to the entire panel of beta-lactam and beta-lactam–beta-lactamase inhibitor combinations tested, showing how this combination of resistance genes can limit therapeutic options. Two isolates of E. coli, both harboring blaNDM-5 and blaCTX-M-15, carried the pbp3 insertions that have been linked to decreased susceptibility to cefiderocol and to the beta-lactam–beta-lactamase inhibitor aztreonam-avibactam. The latter combination has shown promising results for the treatment of Enterobacterales co-harboring metallo-beta-lactamases and ESBLs or AmpC genes [39,40]. There have been increasing reports of blaKPC variants (including 77 out of 156 known blaKPC alleles according to the NCBI Reference Gene Catalog [41]) that can confer resistance to ceftazidime-avibactam [42]. We only identified one blaKPC-2-containing isolate (an E. hormaechei) with resistance to CZA. This organism also carried a blaACT-27-like ampC gene in addition to the carbapenemase gene. CZA resistance can be associated with mutations or with increased expression of ampC genes [43].
This study has several limitations, including the small number of participating laboratories and the limited number of isolates collected. Furthermore, the panel of beta-lactam and beta-lactam–beta-lactamase inhibitor combinations available for testing did not include imipenem-relebactam. However, the geographic diversity of the laboratories and the broad spectrum of beta-lactamases ultimately observed among the isolates indicate that the primary goal of our study, i.e., to obtain a snapshot of currently circulating beta-lactamase genes in the United States, was successful.
In summary, these data indicate that mechanisms of beta-lactamase-mediated resistance in bacterial isolates obtained from patients across the United States continue to evolve, with new mechanisms of carbapenem resistance emerging, especially in A. baumannii. Phenotypic detection of carbapenem resistance in this species proved difficult with the mCIM and eCIM methods. Clearly, carbapenemase genes are much more diverse than just blaKPC, and the number of metallo-carbapenemases, including blaIMP, blaNDM, and blaVIM, circulating could prove a challenge for the selection of effective antimicrobial therapy. This study also shows the importance of internationally recognized epidemic clones in spreading antimicrobial resistance.
4. Materials and Methods
4.1. Isolate Selection Criteria
Twelve geographically dispersed hospitals or hospital system laboratories from across the United States participated in this study. Each laboratory was invited to send 15 Gram-negative isolates from bloodstream infections and 15 Gram-negative organisms isolated from urinary tract infections, each from a unique patient, during 2021 and 2022. Isolates had to meet any of the following criteria: Enterobacterales that were non-susceptible to cefotaxime, ceftriaxone, ceftazidime, ertapenem, meropenem, or imipenem; Acinetobacter or Pseudomonas species that were non-susceptible to meropenem or imipenem; or any Gram-negative bacterial isolate that was positive by the modified carbapenem inactivation method (mCIM) or was confirmed to be a carbapenemase producer by nucleic acid amplification testing or other laboratory method.
4.2. Bacterial Identification and Antimicrobial Susceptibility Testing (AST)
Identification of the Gram-negative bacterial isolates was performed by MALDI-TOF MS (Bruker Daltonics GmbH, Bremen, Germany) according to the manufacturer’s instructions. Antimicrobial susceptibility testing was conducted using the Neg MIC 56 panel on the MicroScan WalkAway 40 Plus system (Beckman Coulter, Inc., West Sacramento, CA, USA) as described by the manufacturer. Minimal inhibitory concentration (MIC) results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines [15]. Quality control organisms included P. aeruginosa ATCC 27853, Escherichia coli ATCC 25922, and ATCC 35218, Klebsiella pneumoniae ATCC 700,603, and K. pneumoniae ATCC BAA-1705. A meropenem disk was added to agar plates in each subculture to maintain antimicrobial pressure and prevent loss of beta-lactam resistance determinants. Isolates were also tested for susceptibility to ertapenem, imipenem, meropenem, cefotaxime (with and without clavulanic acid), ceftazidime (with and without clavulanic acid), ceftriaxone, cefepime, aztreonam, and ceftazidime/avibactam by the disk diffusion method on Mueller-Hinton agar (Hardy Diagnostics, Santa Maria, CA, USA), as described by CLSI [44]. ESBL production was tested with the disk diffusion method using both cefotaxime (CTX) (30 mg) and ceftazidime (CAZ) (30 mg) disks alone and in combination with clavulanic acid (CA) (10 mg) (Becton, Dickinson, Sparks, MD, USA) as described by CLSI (CLSI M100). Carbapenemase production was detected with the modified carbapenem inactivation method (mCIM), which was employed in conjunction with the EDTA-modified carbapenem inactivation method (eCIM) to differentiate serine carbapenemases from metallo-carbapenemases, according to CLSI guidelines, with the following two exceptions: eCIM testing was extended to all isolates instead of only Enterobacterales, and mCIM/eCIM testing was also performed on Acinetobacter species [15].
4.3. Whole Genome Sequencing
Pure cultures of the organisms were grown overnight on blood agar plates with a meropenem disk (Hardy Diagnostics) added to the second streak area. Nucleic acids were extracted with the Qiagen DNeasy Blood and tissue kit using the Qiacube instrument (Qiagen, Valencia, Santa Clarita, CA, USA). Genomic libraries were prepared with the Illumina DNA Prep Kit (Illumina, San Diego, CA, USA) and sequenced on the Illumina MiSeq using Reagent Kit v2 chemistry (Illumina). De novo assemblies, multi-locus sequence typing (MLST), k-mer based prediction of species, construction of minimum spanning trees, and detection of acquired antimicrobial resistance genes and point mutations were performed with the CLC Genomics Workbench version 22.0.2 and CLC Microbial Genomics Module version 22.1.1 (QIAGEN Bioinformatics, Aarhus, Denmark). All procedures were performed in accordance with the manufacturers’ instructions. All nucleic acid sequence data obtained in this study have been deposited in the NCBI BioProject database at https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 8 June 2023) with links to BioProject Accession # PRJNA981469.
4.4. PBP3 Sequence Analysis
Polymorphisms in the penicillin-binding protein 3 (PBP3, encoded by the gene ftsI), specifically insertions of amino acids YRIN or YRIK at position 333, which have been associated with decreased susceptibility to aztreonam-avibactam as well as cefiderocol in E. coli, were determined as previously described [45,46,47].
Acknowledgments
We thank the following members of the HAI Consortium for providing the isolates for this study: N. Banaei, Stanford University School of Medicine, Palo Alto, CA; R.M. Humphries, Vanderbilt University Medical Center, Nashville, TN; J. Mortensen, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH; E. Beck, ACL Laboratories, Advocate Aurora Health, West Allis, WI; A.D. Rojtman, Hackensack-Meridian, Jersey Shore University Medical Center, Neptune, NJ; E.M. Burd, Emory University Hospital, Atlanta, GA; F.C. Fang, Harborview Medical Center, Seattle, WA; K. Culbreath, TriCore Reference Laboratories, Albuquerque, NM; M.A. Henthorne, Lawrence Memorial Hospital, Lawrence, KS; M. Morgan, Cedars-Sinai, Los Angeles, CA; P. Granato, Laboratory Alliance of Central New York, Syracuse, NY; R. Selvarangan and N. Kanwar, Childrens Mercy Hospital, Kansas City, MO.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12091386/s1, Figure S1: Minimum Spanning Tree of Acinetobacter baumannii color-coded by sequence type; Figure S2: Minimum Spanning Tree of Pseudomonas aeruginosa by sequence type; Figure S3: Number of isolates with carbapenemase genes identified and the non-susceptibility of the isolates to carbapenems (ertapenem, imipenem, and meropenem) by BMD; Figure S4: Most frequent ESBL genes identified and their phenotypic identification by BMD and DD; Figure S5: Most frequent AmpC genes identified and their phenotypic resistance to cefepime by BMD and DD (grouped by AmpC gene family); Table S1: Summary of phenotypic results result; Table S2: Summary of genotypic results.
Author Contributions
Conceptualization, F.C.T., D.K. and I.A.T.; methodology, F.C.T., D.K. and I.A.T.; software, A.E.O.; validation, A.E.O. and I.A.T.; formal analysis, I.A.T. and F.C.F.; investigation, A.E.O. and I.A.T.; resources, F.C.T.; data curation, I.A.T.; writing—original draft preparation, F.C.T. and I.A.T.; writing—review and editing, F.C.T., D.K., F.C.F., A.E.O. and I.A.T.; visualization, I.A.T.; supervision, F.C.T.; project administration, D.K.; funding acquisition, F.C.T. The Healthcare Associated Infections Consortium contributed the bacterial isolates. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article and in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 8 June 2023) with links to BioProject Accession # PRJNA981469.
Conflicts of Interest
Isabella A. Tickler and Diane Kawa are employees of Cepheid, and Fred C. Tenover is a former employee of Cepheid. Ferric C. Fang has received consulting fees, educational grants, and speaker honoraria from Cepheid. Anne E. Obradovich has received research funding from Cepheid.
Funding Statement
This research was funded by Cepheid.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Logan L.K., Weinstein R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanheira M., Simner P.J., Bradford P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021;3:dlab092. doi: 10.1093/jacamr/dlab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. [(accessed on 19 December 2022)]; Available online: http://www.cdc.gov/DrugResistance/Biggest-Threats.html.
- 4.Centers for Disease Control and Prevention. National Center for Emerging and Zoonotic Infectious Diseases. Division of Healthcare Quality Promotion . COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. Centers for Disease Control and Prevention; Atlanta, Georgia: 2022. [Google Scholar]
- 5.Shrivastava S., Shrivastava P., Ramasamy J. World Health Organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018;32:76–77. doi: 10.4103/jms.jms_25_17. [DOI] [Google Scholar]
- 6.Livorsi D.J., Chorazy M.L., Schweizer M.L., Balkenende E.C., Blevins A.E., Nair R., Samore M.H., Nelson R.E., Khader K., Perencevich E.N. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control. 2018;7:55. doi: 10.1186/s13756-018-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asokan G.V., Ramadhan T., Ahmed E., Sanad H. WHO Global Priority Pathogens List: A Bibliometric Analysis of Medline-PubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Med. J. 2019;34:184–193. doi: 10.5001/omj.2019.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamma P.D., Smith T.T., Adebayo A., Karaba S.M., Jacobs E., Wakefield T., Nguyen K., Whitfield N.N., Simner P.J. Prevalence of blaCTX-M Genes in Gram-Negative Bloodstream Isolates across 66 Hospitals in the United States. J. Clin. Microbiol. 2021;59:e00127-21. doi: 10.1128/JCM.00127-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castanheira M., Kimbrough J.H., DeVries S., Mendes R.E., Sader H.S. Trends of β-Lactamase Occurrence Among Escherichia coli and Klebsiella pneumoniae in United States Hospitals During a 5-Year Period and Activity of Antimicrobial Agents against Isolates Stratified by β-Lactamase Type. Open Forum Infect. Dis. 2023;10:ofad038. doi: 10.1093/ofid/ofad038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sader H.S., Mendes R.E., Duncan L., Kimbrough J.H., Carvalhaes C.G., Castanheira M. Ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam activities against multidrug-resistant Enterobacterales from United States Medical Centers (2018–2022) Diagn. Microbiol. Infect. Dis. 2023;106:115945. doi: 10.1016/j.diagmicrobio.2023.115945. [DOI] [PubMed] [Google Scholar]
- 11.Sader H.S., Castanheira M., Streit J.M., Flamm R.K. Frequency of occurrence and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bloodstream infections in United States medical centers (2015–2017) Diagn. Microbiol. Infect. Dis. 2019;95:114850. doi: 10.1016/j.diagmicrobio.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Cummins E.A., Snaith A.E., McNally A., Hall R.J. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur. J. Clin. Microbiol. Infect. Dis. 2021 doi: 10.1007/s10096-021-04359-3. [DOI] [PubMed] [Google Scholar]
- 13.Rada A.M., De La Cadena E., Agudelo C.A., Pallares C., Restrepo E., Correa A., Villegas M.V., Capataz C. Genetic Diversity of Multidrug-Resistant Pseudomonas aeruginosa Isolates Carrying blaVIM-2 and blaKPC-2 Genes that Spread on Different Genetic Environment in Colombia. Front. Microbiol. 2021;12:663020. doi: 10.3389/fmicb.2021.663020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitteregger D., Wessely J., Barišić I., Bedenić B., Kosak D., Kundi M. A Variant Carbapenem Inactivation Method (CIM) for Acinetobacter baumannii Group with Shortened Time-to-Result: rCIM-A. Pathogens. 2022;11:482. doi: 10.3390/pathogens11040482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 33rd ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2023. CLSI Supplement M100. [Google Scholar]
- 16.The European Committee on Antimicrobial Susceptibility Testing EUCAST: Clinical Breakpoints and Dosing of Antibiotics. [(accessed on 19 June 2023)]. Available online: https://www.eucast.org/clinical_breakpoints/
- 17.Mendonça N., Ferreira E., Louro D., ARSIP Participants. Caniça M. Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int. J. Antimicrob. Agents. 2009;34:29–37. doi: 10.1016/j.ijantimicag.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Legese M.H., Asrat D., Aseffa A., Hasan B., Mihret A., Swedberg G. Molecular Epidemiology of Extended-Spectrum Beta-Lactamase and AmpC Producing Enterobacteriaceae among Sepsis Patients in Ethiopia: A Prospective Multicenter Study. Antibiotics. 2022;11:131. doi: 10.3390/antibiotics11020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathers A.J., Peirano G., Pitout J.D.D. Escherichia coli ST131: The quintessential example of an international multiresistant high-risk clone. Adv. Appl. Microbiol. 2015;90:109–154. doi: 10.1016/bs.aambs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Pitout J.D.D., Chen L. The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli. Infect. Dis. Ther. 2023;12:1029–1041. doi: 10.1007/s40121-023-00791-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Núñez-Samudio V., Pimentel-Peralta G., Herrera M., Pecchio M., Quintero J., Landires I. Molecular Genetic Epidemiology of an Emerging Antimicrobial-Resistant Klebsiella pneumoniae Clone (ST307) Obtained from Clinical Isolates in Central Panama. Antibiotics. 2022;11:1817. doi: 10.3390/antibiotics11121817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamidian M., Nigro S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 2019;5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benanti G.E., Brown A.R.T., Shigle T.L., Tarrand J.J., Bhatti M.M., McDaneld P.M., Shelburne S.A., Aitken S.L. Carbapenem versus Cefepime or Piperacillin-Tazobactam for Empiric Treatment of Bacteremia Due to Extended-Spectrum-β-Lactamase-Producing Escherichia coli in Patients with Hematologic Malignancy. Antimicrob. Agents Chemother. 2019;63:10–1128. doi: 10.1128/AAC.01813-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaiskos I., Giamarellou H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics. 2020;9:61. doi: 10.3390/antibiotics9020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S.A., Altshuler J., Paris D., Fedorenko M. Cefepime versus carbapenems for the treatment of urinary tract infections caused by extended-spectrum β-lactamase-producing enterobacteriaceae. Int. J. Antimicrob. Agents. 2018;51:155–158. doi: 10.1016/j.ijantimicag.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Tamma P.D., Aitken S.L., Bonomo R.A., Mathers A.J., van Duin D., Clancy C.J. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa) Clin. Infect. Dis. 2022;75:187–212. doi: 10.1093/cid/ciac268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W.L., Pfaller M.A., Winokur P.L., Jones R.N. Cefepime MIC as a predictor of the extended-spectrum beta-lactamase type in Klebsiella pneumoniae, Taiwan. Emerg. Infect. Dis. 2002;8:522–524. doi: 10.3201/eid0805.010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover F.C. Using Molecular Diagnostics to Develop Therapeutic Strategies for Carbapenem-Resistant Gram-Negative Infections. Front. Cell Infect. Microbiol. 2021;11:715821. doi: 10.3389/fcimb.2021.715821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papp-Wallace K.M., McLeod S.M., Miller A.A. Durlobactam, a Broad-Spectrum Serine β-lactamase Inhibitor, Restores Sulbactam Activity Against Acinetobacter Species. Clin. Infect. Dis. 2023;76:S194–S201. doi: 10.1093/cid/ciad095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes J., Komarow L., Chen L., Ge L., Hanson B.M., Cober E., Herc E., Alenazi T., Kaye K.S., Garcia-Diaz J., et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): A prospective cohort study. Lancet Microbe. 2023;4:e159–e170. doi: 10.1016/S2666-5247(22)00329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sellera F.P., Fernandes M.R., Moura Q., Souza T.A., Nascimento C.L., Cerdeira L., Lincopan N. Draft genome sequence of an extensively drug-resistant Pseudomonas aeruginosa isolate belonging to ST644 isolated from a footpad infection in a Magellanic penguin (Spheniscus magellanicus) J. Glob. Antimicrob. Resist. 2018;12:88–89. doi: 10.1016/j.jgar.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Tenover F.C., Nicolau D.P., Gill C.M. Carbapenemase-producing Pseudomonas aeruginosa—An emerging challenge. Emerg. Microbes Infect. 2022;11:811–814. doi: 10.1080/22221751.2022.2048972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill C.M., Lasko M.J., Asempa T.E., Nicolau D.P. Evaluation of the EDTA-Modified Carbapenem Inactivation Method (eCIM) for Detecting Metallo-β-lactamase-producing Pseudomonas aeruginosa. J. Clin. Microbiol. 2020;58:10–1128. doi: 10.1128/JCM.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamma P.D., Aitken S.L., Bonomo R.A., Mathers A.J., van Duin D., Clancy C.J. Infectious Diseases Society of America Antimicrobial-Resistant Treatment Guidance: Gram-Negative Bacterial Infections. [(accessed on 14 March 2023)]. Available online: https://www.idsociety.org/practice-guideline/amr-guidance/ [DOI] [PubMed]
- 35.Wiskirchen D.E., Nordmann P., Crandon J.L., Nicolau D.P. In vivo efficacy of human simulated regimens of carbapenems and comparator agents against NDM-1-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2014;58:1671–1677. doi: 10.1128/AAC.01946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiskirchen D.E., Nordmann P., Crandon J.L., Nicolau D.P. Efficacy of humanized carbapenem exposures against New Delhi metallo-β-lactamase (NDM-1)-producing enterobacteriaceae in a murine infection model. Antimicrob. Agents Chemother. 2013;57:3936–3940. doi: 10.1128/AAC.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Q., Zhou J., Wu S., Yang Y., Yu D., Wang X., Wu M. Characterization of the IncX3 Plasmid Producing blaNDM-7 from Klebsiella pneumoniae ST34. Front. Microbiol. 2020;11:1885. doi: 10.3389/fmicb.2020.01885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Fu Y., Shen M., Huang D., Du X., Hu Q., Zhou Y., Wang D., Yu Y. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control. 2018;7:59. doi: 10.1186/s13756-018-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sader H.S., Mendes R.E., Carvalhaes C.G., Kimbrough J.H., Castanheira M. Changing Epidemiology of Carbapenemases Among Carbapenem-Resistant Enterobacterales from United States Hospitals and the Activity of Aztreonam-Avibactam Against Contemporary Enterobacterales (2019–2021) Open Forum Infect. Dis. 2023;10:ofad046. doi: 10.1093/ofid/ofad046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sader H.S., Castanheira M., Kimbrough J.H., Kantro V., Mendes R.E. Aztreonam/avibactam activity against a large collection of carbapenem-resistant Enterobacterales (CRE) collected in hospitals from Europe, Asia and Latin America (2019–2021) JAC Antimicrob. Resist. 2023;5:dlad032. doi: 10.1093/jacamr/dlad032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Center for Biotechnology Information Reference Gene Catalog—Pathogen Detection. [(accessed on 8 June 2023)]; Available online: https://www.ncbi.nlm.nih.gov/pathogens/refgene/
- 42.Hobson C.A., Pierrat G., Tenaillon O., Bonacorsi S., Bercot B., Jaouen E., Jacquier H., Birgy A. Klebsiella pneumoniae Carbapenemase Variants Resistant to Ceftazidime-Avibactam: An Evolutionary Overview. Antimicrob. Agents Chemother. 2022;66:e0044722. doi: 10.1128/aac.00447-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong L., Wang X., Wang Y., Yu W., Zhou Y., Chi X., Xiao T., Xiao Y. Molecular mechanisms underlying bacterial resistance to ceftazidime/avibactam. WIREs Mech. Dis. 2022;14:e1571. doi: 10.1002/wsbm.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk Susceptibility Tests. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. Approved Standard-Thirteenth Edition M2-A13. [Google Scholar]
- 45.Alm R.A., Johnstone M.R., Lahiri S.D. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: Role of a novel insertion in PBP3. J. Antimicrob. Chemother. 2015;70:1420–1428. doi: 10.1093/jac/dku568. [DOI] [PubMed] [Google Scholar]
- 46.Livermore D.M., Mushtaq S., Vickers A., Woodford N. Activity of aztreonam/avibactam against metallo-β-lactamase-producing Enterobacterales from the UK: Impact of penicillin-binding protein-3 inserts and CMY-42 β-lactamase in Escherichia coli. Int. J. Antimicrob. Agents. 2023;61:106776. doi: 10.1016/j.ijantimicag.2023.106776. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q., Jin L., Sun S., Yin Y., Wang R., Chen F., Wang X., Zhang Y., Hou J., Zhang Y., et al. Occurrence of High Levels of Cefiderocol Resistance in Carbapenem-Resistant Escherichia coli before Its Approval in China: A Report from China CRE-Network. Microbiol. Spectr. 2022;10:e0267021. doi: 10.1128/spectrum.02670-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in this article and in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 8 June 2023) with links to BioProject Accession # PRJNA981469.