Abstract
Non-toxigenic Clostridioides difficile (NTCD) has been shown to decrease the risk of recurrent C. difficile infection (CDI) in patients following metronidazole or vancomycin treatment for CDI. Limited data on the prevalence of NTCD strains in symptomatic patients and their clinical characteristics are available. We conducted this study to investigate the prevalence of NTCD in diarrhoea patients and their clinical characteristics. Between July 2017 and June 2018, unduplicated stool specimens were collected from patients with diarrhoea. The characteristics and episodes of C. difficile infection in patients with NTCD and toxigenic strains were compared. Among the 1182 stool specimens collected, 236 (18.5%) were identified as growing C. difficile, and 19.5% of the identified isolates were found to be NTCD. Multivariate analysis showed that community-onset diarrhoea (OR = 4.13, 95% CI 1.07–15.97; p = 0.040), underlying diabetes (OR = 3.64, 95% CI 1.46–9.25; p = 0.006), previous use of glycopeptides (OR = 4.75, 95% CI 1.37–16.42; p = 0.014), and the lack of use of proton pump inhibitors (PPIs) (OR = 3.57, 95% CI 1.39–9.09; p = 0.009) were independently associated with the NTCD group. Although there was no statistical significance, the number of CDI episodes occurring after 90 days tended to be lower in the NTCD group (2.2%) than in the toxigenic group (11.2%). A considerable portion of the C. difficile strains isolated from patients with diarrhoea showed NTCD. Further, more extensive studies are needed to clearly define the protective effects of NTCD strains in patients with diarrhoea.
Keywords: Clostridioides difficile, non-toxigenic Clostridioides difficile, toxigenic Clostridioides difficile, diabetes, proton pump inhibitors, glycopeptides
1. Introduction
Clostridioides difficile is a Gram-positive, spore-forming anaerobic bacterium that accounts for 15–25% of all cases of antibiotic-associated diarrhoea [1]. C. difficile infection (CDI) remains an important cause of morbidity and mortality in healthcare-associated infections [2]. C. difficile produces toxins responsible for the disease, although not all strains produce toxins. Toxigenic C. difficile strains generally produce both toxins A (enterotoxin) and B (cytotoxin), and occasionally binary toxin (CDT); however, some are toxin A-negative due to mutations in the tcdA gene [3]. Ribotype (RT) 027 and other CDT-producing C. difficile strains have rarely been reported in Asian countries, whereas the most prevalent RTs are RT017, RT018, RT014, RT002, and RT001 [4,5]. No specific clinical features distinguish CDI from other causes of diarrhoea [6]. Therefore, rapid and accurate diagnosis of CDI is essential for the initiation of appropriate antibiotics and for controlling its spread [7]. The widely used assays are C. difficile toxin A and B enzyme immunoassays (toxin EIA), which detect free toxins in faeces; glutamate dehydrogenase (GDH) tests, which detect a common antigen produced by C. difficile; and nucleic acid amplification tests (NAATs), which detect toxin genes [6,8,9]. Toxin EIA correlates better with disease than GDH or NAAT, but this method has poor sensitivity, leading to missed cases [10,11]. In contrast, NAATs cannot differentiate between active infection and asymptomatic carriage because they detect toxin genes alone but not toxin production [11,12,13,14].
In contrast, non-toxigenic C. difficile (NTCD) strains cannot produce toxins and are usually not associated with symptomatic infections. NTCD strains do not contain the pathogenic island encoding toxins A and B (PaLoc genes) or exhibit dysregulation of the tcdA and tcdB genes, and they do not produce enough toxins to cause disease [15,16,17]. NTCD gastrointestinal colonisation has been shown to prevent CDI through exposure to a toxigenic strain [18,19]. NTCD-M3 has also been shown to be effective in the prevention of recurrent CDI in patients who have been treated with metronidazole or vancomycin for CDI [18]. Therefore, a promising preventive measure against CDI is the use of NTCD to colonise the destroyed gut after antibiotic treatment and prevent colonisation by toxigenic C. difficile.
Limited data on the prevalence of NTCD strains in symptomatic patients and their clinical characteristics are available. Therefore, we conducted a 1-year study of patients with diarrhoea to investigate the prevalence and of NTCD strains and patients’ characteristics.
2. Results
2.1. Characteristics of C. difficile Strains
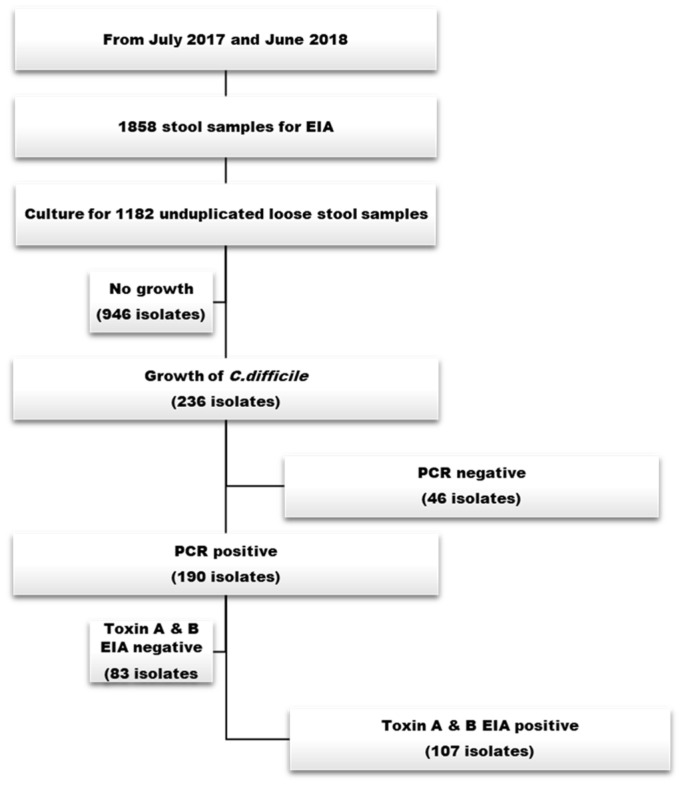
Between July 2017 and June 2018, 1182 unduplicated specimens from 1858 stool specimens submitted for C. difficile toxin EIA testing were cultured. Among the 1182 specimens, 236 (18.5%) were positive for C. difficile. NTCD strains (n = 46) were observed in 19.5% of the C. difficile strains isolated from patients with diarrhoea. Among the 190 toxin gene polymerase chain reaction (PCR) + strains, 160 (84.2%) isolates were identified as A+B+CDT-, 22 (13.8%) as A-B+CDT-, and 8 (4.2%) as A+B+CDT+ strains. In addition, 83 isolates were identified as toxin EIA negative/toxin gene PCR positive and 107 isolates were identified as toxin EIA positive/toxin gene PCR positive (Figure 1 and Table 1).
Figure 1.
Overall study scheme.
Table 1.
Distribution of toxin gene profiles in patients with diarrhoea.
| Toxin Gene Profile (n = 236) | Toxin EIA Results | N (%) |
|---|---|---|
| A+B+CDT- strain | positive | 92 (39.0) |
| negative | 68 (28.8) | |
| A-B+CDT- strain | positive | 11 (4.7) |
| negative | 11 (4.7) | |
| A+B+CDT+ strain | positive | 4 (1.7) |
| negative | 4 (1.7) | |
| A-B-CDT- strain | negative | 46 (19.5) |
EIA—enzyme immunoassays.
2.2. Comparison of Clinical Features between Non-Toxigenic and Toxigenic C. difficile
The characteristics of patients with NTCD strains were compared with those with toxigenic C. difficile (Table 2). Community onset was more common (19.6% vs. 11.2%, p = 0.006), and underlying diabetes was more prevalent in patients with NTCD strains (34.8% vs. 17.8%, p = 0.022). Patients with NTCD strains were less likely to have received antimicrobial therapy in the preceding month (78.3% vs. 90.6%, p = 0.039) but were more likely to have received glycopeptide therapy (18.2% vs. 5.9%, p = 0.031). The use of proton pump inhibitor (PPI) therapy was significantly lower in patients with NTCD strains than in patients with toxigenic C. difficile (21.7% vs. 40.6%, p = 0.025).
Table 2.
Comparison of characteristics between non-toxigenic and toxigenic Clostridium difficile.
| Non-Toxigenic (n = 46) |
Toxigenic (n = 107) |
p-Value | |
|---|---|---|---|
| Age ≥ 65 y | 23 (50.0) | 68 (63.6) | 0.117 |
| Hospital stays, days, median (IQR) | 7.5 (1.0–20.5) | 7.5 (1.0–25.3) | 0.562 |
| ICU | 9 (19.6) | 12 (11.2) | 0.169 |
| Male sex | 24 (52.2) | 53 (49.5) | 0.764 |
| Category of infection | |||
| Community onset | 9 (19.6) | 5 (4.7) | 0.006 |
| Community-onset healthcare facility associated | 8 (17.4) | 32 (29.9) | 0.106 |
| Hospital onset | 29 (63.0) | 70 (65.4) | 0.778 |
| Underlying disease | |||
| Diabetes | 16 (34.8) | 19 (17.8) | 0.022 |
| Cerebrovascular disease | 14 (30.4) | 44 (41.1) | 0.212 |
| Cardiovascular disease | 6 (13.0) | 21 (19.6) | 0.327 |
| Chronic lung disease | 5 (10.9) | 9 (8.4) | 0.760 |
| Liver cirrhosis | 2 (4.3) | 3 (2.8) | 0.637 |
| Chronic renal disease without dialysis | 8 (17.4) | 12 (11.2) | 0.299 |
| Dialysis | 4 (8.7) | 7 (6.5) | 0.735 |
| Solid tumour | 8 (17.4) | 22 (20.6) | 0.651 |
| Solid organ transplantation | 1 (2.2) | 2 (1.9) | 0.661 |
| Charlson’s score, median (IQR) | 2 (0–4) | 2 (1–5) | 0.175 |
| Previous medical history within 1 month | |||
| Operation | 11 (23.9) | 25 (23.4) | 0.942 |
| Immunosuppression | 5 (10.9) | 11 (10.3) | 0.559 |
| Antibiotic exposure | 36 (78.3) | 96 (90.6) | 0.039 |
| Extended spectrum cephalosporin | 9 (20.5) | 27 (26.5) | 0.439 |
| Quinolone | 8 (18.2) | 26 (25.5) | 0.338 |
| β-lactam/β-lactamases | 8 (18.2) | 20 (19.6) | 0.841 |
| Carbapenem | 11 (25.0) | 29 (28.4) | 0.670 |
| Glycopeptide | 8 (18.2) | 6 (5.9) | 0.031 |
| Gastrointestinal medication use at diagnosis | |||
| PPI | 10 (21.7) | 43 (40.6) | 0.025 |
| H2 receptor antagonist | 16 (34.8) | 35 (33.0) | 0.832 |
| Probiotics | 6 (13.0) | 19 (17.9) | 0.456 |
| Concurrent systemic infection | 27 (58.7) | 51 (47.7) | 0.211 |
| Antibiotics use at the time of diagnosis | 27 (58.7) | 65 (60.7) | 0.812 |
IQR—interquartile range; ICU—intensive care unit; PPI—proton pump inhibitor; Data are n (%) unless otherwise stated.
2.3. Predictors of NTCD Strains in Patients with Diarrhoea
A multivariate analysis of the potential predictors associated with NTCD strains is shown in Table 3. Variables with a p-value < 0.05 in the univariate analysis were included in the subsequent multivariate analysis. A logistic regression model revealed that community onset [OR = 4.13, 95% CI 1.07–15.97; p = 0.040)], underlying diabetes (OR = 3.64, 95% CI 1.46–9.25; p = 0.006), vancomycin therapy in the preceding month (OR = 4.75, 95% CI 1.37–16.42; p = 0.014), and non-concurrent use of PPIs (OR = 3.57, 95% CI 1.39–9.09; p = 0.009) were independent predictors of NTCD strains in patients with diarrhoea.
Table 3.
Univariable and multivariable logistic regression analyses for independent predictors of non-toxigenic C. difficile among patients with diarrhoea.
| OR (95% CI) | p-Value | Adjusted OR (95% CI) a | p-Value | |
|---|---|---|---|---|
| Age ≥ 65 y | 0.57 (0.29–1.15) | 0.119 | ||
| * Hospital stays | 1.02 (0.76–1.36) | 0.903 | ||
| ICU | 1.93 (0.75–4.95) | 0.174 | ||
| Male sex | 1.11 (0.56–2.22) | 0.765 | ||
| Category of infection | ||||
| Community onset | 4.96 (1.56–15.77) | 0.007 | 4.13 (1.07–15.97) | 0.040 |
| Community-onset healthcare facility associated | 0.49 (0.21–1.18) | 0.110 | ||
| Hospital onset | 0.90 (0.44–1.85) | 0.778 | ||
| Underlying disease | ||||
| Diabetes | 2.47 (1.13–5.41) | 0.024 | 3.64 (1.46–9.25) | 0.006 |
| Cerebrovascular disease | 0.63 (0.30–1.31) | 0.213 | ||
| Cardiovascular disease | 0.61 (0.23–1.64) | 0.331 | ||
| Chronic lung disease | 1.33 (0.42–4.20) | 0.630 | ||
| Liver cirrhosis | 1.58 (0.25–9.76) | 0.625 | ||
| Chronic renal disease without dialysis | 1.67 (0.63–4.40) | 0.302 | ||
| Dialysis | 1.36 (0.38–4.90) | 0.637 | ||
| Solid tumour | 0.81 (0.33–1.99) | 0.651 | ||
| Solid organ transplantation | 1.17 (0.10–13.20) | 0.901 | ||
| Charlson’s score * | 0.76 (0.42–1.35) | 0.347 | ||
| Previous medical history within 1 month | ||||
| Operation | 1.03 (0.46–2.32) | 0.942 | ||
| Immunosuppression | 1.06 (0.35–3.26) | 0.913 | ||
| Antibiotic exposure | 0.38 (0.14–0.98) | 0.044 | ||
| Extended spectrum cephalosporin | 0.71 (0.30–1.68) | 0.440 | ||
| Quinolone | 0.65 (0.27–1.58) | 0.340 | ||
| β-lactam/β-lactamases | 0.91 (0.37–2.26) | 0.841 | ||
| Carbapenem | 0.84 (0.38–1.88) | 0.670 | ||
| Glycopeptide | 3.56 (1.15–10.96) | 0.027 | 4.75 (1.37–16.42) | 0.014 |
| Gastrointestinal medication use at diagnosis | ||||
| No PPI | 0.41 (0.18–0.91) | 0.028 | 0.28 (0.11–0.72) | 0.009 |
| H2 receptor antagonist | 1.08 (0.52–2.24) | 0.832 | ||
| Probiotics | 0.69 (0.26–1.85) | 0.458 | ||
| Concurrent systemic infection | 1.56 (0.78–3.14) | 0.212 | ||
| Antibiotics use at the time of diagnosis | 0.92 (0.45–1.86) | 0.812 |
OR—odds ratio; CI—confidence interval; IQR—interquartile range; ICU—intensive care unit; PPI—proton pump inhibitor; * Log-transformation of the data is applied. a Variables with a p-value < 0.05 in the univariate analyses are included in the subsequent multivariate regression model. Hosmer–Lemeshow test, χ2 = 3.263, p = 0.917.
2.4. Comparison of Clinical Signs and Subsequent CDI Episodes between Non-Toxigenic and Toxigenic C. difficile
Clinical signs were similar between non-toxigenic and toxigenic C. difficile. A body temperature > 38.0 °C and a white blood cell count of >15,000/µL were more common in those with toxigenic C. difficile. The number of subsequent CDI episodes following a period of 90 d tended to be lower in the NTCD group (2.2%) than in the toxigenic group (11.2%), although there was no statistical significance (Table 4).
Table 4.
Comparison of clinical signs and subsequent CDI episodes between non-toxigenic and toxigenic C. difficile.
| Non-Toxigenic (n = 46) |
Toxigenic (n = 107) |
p-Value | |
|---|---|---|---|
| Signs at diagnosis | |||
| Body temperature > 38.0 °C | 14 (30.4) | 51 (47.7) | 0.048 |
| Shock | 1 (2.3) | 9 (8.6) | 0.282 |
| Ileus | 0 (0) | 6 (5.8) | 0.179 |
| Laboratory finding | |||
| White blood cell count > 15,000/µL | 8 (18.6) | 35 (35.4) | 0.046 |
| Acute kidney injury | 2 (4.8) | 6 (6.2) | 0.545 |
| Albumin level, g/dL, mean ± SD | 3.5 ± 0.9 | 3.2 ± 0.7 | 0.073 |
| CRP, mmol/L, mean ± SD | 65.5 ± 78.7 | 71.7 ± 63.8 | 0.634 |
| CDI development within 90 days | 1 (2.2) | 12 (11.2) | 0.055 |
CRP—C-reactive protein; SD—standard deviation; CDI—C. Difficile infection; Data are n (%) unless otherwise stated.
3. Discussion
In this study, we investigated the prevalence and characteristics of NTCD in patients with diarrhoea. NTCD strains were observed in 19.5% of C. difficile strains isolated from patients with diarrhoea. The clinical features of patients with NTCD were significantly different from those with toxigenic C. difficile. NTCD was associated with community-onset diarrhoea, underlying diabetes, previous use of glycopeptides within 1 month, and non-concurrent use of PPIs. Although the difference was not statistically significant with this sample size, the amount of subsequent CDI episodes after 90 days tended to be lower in the NTCD group (2.2%) than in the toxigenic group (11.2%).
Little is known about the prevalence of non-toxigenic strains. A few studies on the epidemiology of NTCD strains among hospitalised patients with diarrhoea have been published in Asia [20,21,22]. A previous study in Malaysia showed that the prevalence of non-toxigenic strains was 12.4% (54/437) in inpatients aged 18–80 years who experienced diarrhoea [20], which was comparable to the reported prevalence in Indonesia (10.6%) [21] and Thailand (15.6%) [22]. While further extensive research is needed to determine the protective effects of NTCD strains in patients with diarrhoea, anecdotal reports and a recent multinational study indicate that in Southeast Asia, where the prevalence of NTCD is high, CDI generally presents as self-limiting diarrhoea, and recurrence is rare [23]. The prevalence of NTCD strains in our study (19.5%) was higher than that reported in previous studies [20,21,22], which could be due to differences related to the characteristics of patient populations, antimicrobial stewardship practices, or local animal or environmental reservoirs of C. difficile. Intriguingly, the amount of subsequent CDI episodes after 90 days tended to be lower in the NTCD group (2.2%) than in the toxigenic group (11.2%). This may be due to the protective role of non-toxigenic C. difficile. One study found that 88 (46%) of 192 asymptomatic patients with C. difficile colonisation carried NTCD, and the colonisation with C. difficile was associated with lower CDI rates than those who were not colonised [19]. In addition, NTCD-M3 has been demonstrated to be effective in the prevention of recurrent CDI in a Phase 2 clinical trial, where vancomycin and metronidazole were the treatment antibiotics [18]. Based on these data, NTCD-M3 is currently being developed as a novel live biotherapeutic to reduce recurrent CDI. A recent study also demonstrated the ability of NTCD-M3 to colonise hamsters after treatment with either fidaxomicin or vancomycin [24]. Therefore, a Phase 3 clinical trial is currently being developed to confirm this effect after treatment with current standard of care antibiotics, which will likely include fidaxomicin as well as vancomycin.
A total of 83 isolates (43.7%) among the 190 isolates with a positive toxin gene PCR test were shown to be toxin EIA negative. This finding is consistent with a previous study, which found that 55.3% (162 of 293) of patients with a positive C. difficile PCR test result lacked toxin using the clinical toxin immunoassay test [11]. Guerrero D.M. et al. also demonstrated that 43 (32%) patients who underwent EIA tested negative for toxin in a prospective study of 132 patients with a diagnosis of CDI using PCR [25]. Several studies provide evidence that C. difficile colonisation is frequently observed in hospitalised patients, and many cases of nosocomial diarrhoea are of non-infectious origin. In addition, it is possible that clinical toxin tests can miss the presence of toxin at low concentrations [26,27].
The genes tcdA and tcdB that encode toxins A and B, respectively, are located near other genes that control their expression (tcdC and tcdR) and the release of biologically active forms of the toxin (tcdE). This region of the C. difficile genome is referred to as the pathogenicity locus (PaLoc). NTCD isolates lack an intact PaLoc, which means they do not produce toxin A or B and are typically not associated with symptomatic infections [15,16,28]. In addition to these strains, some NTCD strains have altered regulation of the tcdA and tcdB genes, resulting in insufficient expression of the bioactive toxins that cause disease. These strains are considered ‘phenotypically’ non-toxigenic. For instance, the M90 strain carries a PaLoc but fails to produce detectable toxin levels, possibly due to poor gene transcription [29]. In our study, 83 isolates were toxin EIA negative/toxin gene PCR positive, which could be ‘phenotypically’ non-toxigenic or ‘genotypically’ toxigenic. Toxin EIA positivity best defines true cases of C. difficile infections. Therefore, we excluded 83 toxin EIA negative/toxin gene PCR positive strains from the study and compared the NTCD strain and toxin-producing toxigenic C. difficile groups to evaluate the characteristics of NTCD in patients with diarrhoea. NTCD was associated with community-onset diarrhoea, underlying diabetes, previous use of glycopeptides within 1 month, and the lack of concurrent use of PPIs in our study. Diabetes increases the risk of recurrent CDI; however, metformin treatment seems to have a protective effect against the development of CDI by altering the gut microbiota composition [30]. Our study also showed that the proportion receiving metformin was higher in the NTCD group than in the toxigenic group (37.0% vs. 12.1%, p = 0.001). On the other hand,, previous use of glycopeptides within 1 month and no concurrent use of PPIs were also more frequent in the NCTD group. Approximately 80–90% of each intravenous vancomycin dose is excreted in urine and has little effect on intestinal microbiota, and intravenous vancomycin is classified as a low-risk antibiotic for CDI. In addition, pooled analysis of 50 studies showed a significant association between PPI use and risk of developing CDI (OR = 1.26, 95% CI, 1.12–1.39) compared with non-users [31]. Although the mechanism underlying the association between the aforementioned factors and NTCD has not been investigated, these factors may not affect the gut microbiota, including NTCD, allowing NTCD to retain its protective role against CDI.
The present study had some limitations. First, all the strains included in this study were clinical isolates from patients with diarrhoea, but NTCD-positive patients may still be carriers, and their diarrhoea is probably due to an alternative aetiology. In addition, NTCD may be involved in mixed infections with toxigenic strains. Second, environmental contamination by C. difficile could be an important source of transmission [32]. Therefore, subsequent CDI development may be affected by differences in cleaning and disinfection practices. In a study of a large patient cohort using whole-genome sequencing, the researchers were able to determine an association with previous CDI cases in only 55% of newly developed cases [33]. Third, although the value of leukocytes in the faeces of patients with diarrhoea is considered significant, we did not measure leukocyte counts in the faeces. Finally, this study was conducted at a single centre and had a small sample size. Subsequent CDI episodes after 90 days exhibited differences between the NTCD group (2.2%) and the toxigenic group (11.2%); however, this difference did not reach statistical significance. It is important to acknowledge that a small sample size can often yield statistically insignificant results. This is due to the increased variability within the data associated with limited sample sizes, resulting in wider confidence intervals and larger p-values. Consequently, even if a true difference or effect exists within the population, a small sample size may not provide sufficient evidence to establish its statistical significance. In addition, while the odds ratios have statistically significant p-values (p < 0.05), the confidence intervals still encompass a broad range of values. The calculated odds ratios provide valuable insights, but the wide confidence intervals caution us to interpret the results cautiously and with awareness of the potential variability and uncertainty in the estimates. Therefore, the results need to be validated through a relatively larger study or a more refined study design, in which an appropriate sample size is calculated beforehand, particularly focusing on providing more detailed information on the characteristics of patients with diarrhoea.
4. Materials and Methods
Study Design and Population
This cohort study was conducted among patients hospitalised at Samsung Changwon Hospital between July 2017 and June 2018. Eligible specimens were prospectively identified by reviewing the stool specimens submitted for toxin EIA during the study period. Only unduplicated specimens from patients with at least three loose or watery stools within 24 h were included. Clinical data were retrospectively obtained from the medical records of each patient.
Laboratory Testing
All stool samples were subjected to a C. difficile toxin EIA (RIDASCREEN 43 Clostridium difficile toxin A/B, R-Biopharm AG, Darmstadt, Germany) and reported clinically. Formed stools were rejected. Stool specimens were cultured anaerobically on C. difficile selective media (chromID C. difficile, bioMérieux, Lyon, France) for 48 h at 37 °C as previously described [34]. Putative C. difficile colonies were confirmed via colony analysis, odour, and Gram staining. These colonies were subcultured on a universal anaerobic culture medium (Brucella agar plate). DNA was extracted from the colonies grown on Brucella agar plates. C. difficile isolates were identified using 16S rRNA sequencing of the extracted DNA. PCR for tcdA, tcdB, cdtA, and cdtB genes was performed using the previously described method [3,35]. Two primer sets were used to detect the toxin A gene; primers NK3 and NK2 were derived from the nonrepeating portion of the C. difficile toxin A gene, and primers NK11 and NK9 were derived from the repeating portion of the C. difficile toxin A gene. A segment of the toxin B gene was amplified by using primer NK104 and primer NK105, which were derived from the nonrepeating sequence of the C. difficile toxin B gene. Probe NK106 was used and was 3′ end labelled with digoxigenin with a digoxigenin labelling kit. The thermal profile for primer pairs NK3-NK2 and NK104-NK105 was 35 cycles of 95 °C for 20 s, 55 °C for 120 s and 74 °C for 5 min. PCR amplification with primer pair NK11-NK9 was performed for 35 cycles, consisting of 95 °C for 20 s, 62 °C for 120 s and 74 °C for 5 min. Primers designed to amplify regions of cdtA and cdtB were as follows: cdtApos 5′-TGAACCTGGAAAAGGTGATG-3′ (position, cdtA 507–526); cdtArev 5′-AGGATTATTTACTGGACCATTTG-3′ (position, cdtA 882–860); cdtBpos 5′-CTTAATGCAAGTAAATACTGAG-3′ (position, cdtB 368–389); and cdtBrev 5′-AACGGATCTCTTGCTTCAGTC-3′ (position, cdtB 878–858). Reactions were subjected to 30 cycles of 94 °C for 45 s, 52 °C for 1 min and 72 °C for 1 min 20 s. The positive controls were ATCC 43594, 43598, and 9689, representing the A+B+CDT-, A-B+CDT-, and A+B+CDT+ ribotypes, respectively. Laboratory parameters were obtained 2 days before or 1 day after the diagnosis of diarrhoea. Albumin and CRP were measured as part of the automated chemistry analysis using a Roche Modular D2400 system (Roche Diagnostics, Indianapolis, IN), and the reference ranges of our institution are 3.1–5.2 g/dL for albumin and 0–5.0 mmol/L for CRP. White blood cell counts were obtained using a Sysmex XN-10 hematology analyzer.
Clinical Data Collection
Diarrhoea was defined as the passage of at least three loose or watery stools within 24 h. The case definition of CDI included patients with documented diarrhoea and a positive C. difficile EIA toxin assay or a positive toxin gene PCR. Clinical and laboratory characteristics, including age, sex, ward of acquisition, underlying comorbidities, recent medical history within 30 days of diarrhoea, concurrent infection, and concomitant medication were obtained from the medical records of each patient. To determine the severity of the illness, the modified Charlson’s comorbidity index was used for all patients [36]. We traced the development of a CDI patient over a 90-day period by reviewing medical records. For patients who were discharged within this timeframe, we confirmed the development of CDI through telephone records. A toxigenic C. difficile strain was defined as a case with positive PCR results for tcdA, tcdB, cdtA, or cdtB genes of an anaerobically cultured colony of the C. difficile strain with toxin production. An NTCD strain was defined as a case with negative toxin gene PCR results for tcdA, tcdB, cdtA, or cdtB genes of an anaerobically cultured colony of the C. difficile strain.
Statistical Analysis
The characteristics of NTCD and toxigenic C. difficile strains with toxin production were compared. The discrete data are expressed as frequencies and percentages, while continuous variables are presented as either mean ± standard deviation or median and interquartile range based on their distribution, which was determined using the Shapiro–Wilk normality test. Characteristics were compared using appropriate statistical tests such as the χ2 test, Fisher’s exact test, the two-sample t-test, or the Mann–Whitney U-test. A multivariable logistic regression model was employed to identify predictors of NTCD. In cases where the continuous data exhibited a skewed distribution, log transformations were performed during univariable analyses. Variables with a p-value < 0.10 in the bivariate analysis were considered candidates for multivariate analysis. The Hosmer–Lemeshow statistic was used to evaluate the goodness of fit of the final model. The Statistical Package for the Social Sciences for Windows (version 18.0; SPSS Inc., Chicago, IL, USA) was used to perform all analyses.
5. Conclusions
A considerable portion of C. difficile strains isolated from patients with diarrhoea showed NTCD. Community onset, underlying diabetes, previous use of glycopeptides, and non-concurrent usage of PPI were associated with NTCD strains. Further, more extensive studies are needed to clearly define the protective effects of NTCD strains in patients with diarrhoea.
Author Contributions
Conceptualization, Y.M.W.; methodology, Y.M.W.; validation, Y.M.W., C.-H.J. and S.-H.K.; formal analysis, Y.M.W.; investigation, Y.M.W., C.-H.J. and S.-H.K.; resources, Y.M.W., C.-H.J. and S.-H.K.; data curation, Y.M.W., C.-H.J. and S.-H.K.; writing—original draft preparation, Y.M.W.; writing—review and editing, Y.M.W., C.-H.J. and S.-H.K.; supervision, Y.M.W.; project administration, Y.M.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in agreement with the principles of the Declaration of Helsinki and was approved by the ethics committee of Institutional Review Board of Samsung Changwon Hospital (SCMC 2017-04-003).
Informed Consent Statement
Informed consent was waived by the Institutional Review Board of Samsung Changwon Hospital because of the retrospective nature of the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ananthakrishnan A.N. Clostridium difficile infection: Epidemiology, risk factors and 322 management. Nat. Rev. Gastroenterol. Hepatol. 2011;8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 2.Magill S.S., Edwards J.R., Bamberg W., Beldavs Z.G., Dumyati G., Kainer M.A., Lynfield R., Maloney M., McAllister-Hollod L., Nadle J., et al. Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H., Kato N., Watanabe K., Iwai N., Nakamura H., Yamamoto T., Suzuki K., Kim S.M., Chong Y., Wasito E.B. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 1998;36:2178–2182. doi: 10.1128/JCM.36.8.2178-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins D.A., Hawkey P.M., Riley T.V. Epidemiology of Clostridium difficile infection in Asia. Antimicrob. Resist. Infect. Control. 2013;2:21. doi: 10.1186/2047-2994-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo M.R., Kim J., Lee Y., Lim D.G., Pai H. Prevalence, genetic relatedness and antibiotic resistance of hospital-acquired Clostridium difficile PCR ribotype 018 strains. Int. J. Antimicrob. Agents. 2018;51:762–767. doi: 10.1016/j.ijantimicag.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Crobach M.J., Planche T., Eckert C., Barbut F., Terveer E., Dekkers O., Wilcox M., Kuijper E. European Society of Clinical Microbiology and Infectious Diseases: Update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2016;22((Suppl. 4)):S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Tschudin-Sutter S., Kuijper E.J., Durovic A., Vehreschild M.J.G.T., Barbut F., Eckert C., Fitzpatrick F., Hell M., Norén T., O’Driscoll J., et al. Guidance document for prevention of Clostridium difficile infection in acute healthcare settings. Clin. Microbiol. Infect. 2018;24:1051–1054. doi: 10.1016/j.cmi.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 8.McDonald L.C., Gerding D.N., Johnson S., Bakken J.S., Carroll K.C., Coffin S.E., Dubberke E.R., Garey K.W., Gould C.V., Kelly C., et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin. Infect. Dis. 2018;66:e1–e48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly C.R., Fischer M., Allegretti J.R., LaPlante K., Stewart D.B., Limketkai B.N., Stollman N.H. ACG clinical guidelines: Prevention, diagnosis, and treatment of Clostridioides difficile infections. Am. J. Gastroenterol. 2021;116:1124–1127. doi: 10.14309/ajg.0000000000001278. [DOI] [PubMed] [Google Scholar]
- 10.Planche T.D., Davies K.A., Coen P.G., Finney J.M., Monahan I.M., Morris K.A., O’Connor L., Oakley S.J., Pope C.F., Wren M.W., et al. Differences in outcome according to Clostridium difficile testing method: A prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect. Dis. 2013;13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polage C.R., Gyorke C.E., Kennedy M.A., Leslie J.L., Chin D.L., Wang S., Nguyen H.H., Huang B., Tang Y.-W., Lee L.W., et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern. Med. 2015;175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox M.H. Overcoming barriers to effective recognition and diagnosis of Clostridium difficile infection. Clin. Microbiol. Infect. 2012;18:13–20. doi: 10.1111/1469-0691.12057. [DOI] [PubMed] [Google Scholar]
- 13.Kelly S.G., Yarrington M., Zembower T.R., Sutton S.H., Silkaitis C., Postelnick M., Mikolajczak A., Bolon M.K. Inappropriate Clostridium difficile testing and consequent overtreatment and inaccurate publicly reported metrics. Infect. Control Hosp. Epidemiol. 2016;37:1395–1400. doi: 10.1017/ice.2016.210. [DOI] [PubMed] [Google Scholar]
- 14.Saade E., Deshpande A., Kundrapu S., Sunkesula V.C., Guerrero D.M., Jury L.A., Donskey C.J. Appropriateness of empiric therapy in patients with suspected Clostridium difficile infection. Curr. Med. Res. Opin. 2013;29:985–988. doi: 10.1185/03007995.2013.803956. [DOI] [PubMed] [Google Scholar]
- 15.Brouwer M.S., Allan E., Mullany P., Roberts A.P. Draft Genome Sequence of the Nontoxigenic Clostridium difficile Strain CD37. J. Bacteriol. 2012;194:2125–2126. doi: 10.1128/JB.00122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S.H., Tang Y.J., Silva J., Jr. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 2000;181:659–663. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 17.Fluit A.C., Wolfhagen M.J., Verdonk G.P., Jansze M., Torensma R., Verhoef J. Nontoxigenic strains of Clostridium difficile lack the genes for both toxin A and toxin B. J. Clin. Microbiol. 1991;29:2666–2667. doi: 10.1128/jcm.29.11.2666-2667.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerding D.N., Meyer T., Lee C., Cohen S.H., Murthy U.K., Poirier A., Van Schooneveld T.C., Pardi D.S., Ramos A., Barron M.A., et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: A randomized clinical trial. JAMA. 2015;313:1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 19.Shim J.K., Johnson S., Samore M.H., Bliss D.Z., Gerding D.N. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351:633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 20.Riley T.V., Collins D.A., Karunakaran R., Kahar M.A., Adnan A., Hassan S.A., Zainul N.H., Rustam F.R.M., Wahab Z.A., Ramli R., et al. High Prevalence of Toxigenic and Nontoxigenic Clostridium difficile Strains in Malaysia. J. Clin. Microbiol. 2018;56:e00170-18. doi: 10.1128/JCM.00170-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins D.A., Gasem M.H., Habibie T.H., Arinton I., Hendriyanto P., Hartana A., Riley T. Prevalence and molecular epidemiology of Clostridium difficile infection in Indonesia. New Microbes New Infect. 2017;18:34–37. doi: 10.1016/j.nmni.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putsathit P., Maneerattanaporn M., Piewngam P., Kiratisin P., Riley T.V. Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand. New Microbes New Infect. 2017;15:27–32. doi: 10.1016/j.nmni.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins D.A., Sohn K.M., Wu Y., Ouchi K., Ishii Y., Elliott B., Riley T.V., Tateda K., for the Clostridioides difficile Asia-Pacific Study Group Clostridioides difficile infection in the Asia-Pacific region. Emerg. Microbes Infect. 2019;9:42–52. doi: 10.1080/22221751.2019.1702480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambol S.P., Skinner A.M., Serna-Perez F., Owen B., Gerding D.N., Johnson S. Effective Colonization by Nontoxigenic Clostridioides difficile REA Strain M3 (NTCD-M3) Spores following Treatment with Either Fidaxomicin or Vancomycin. Microbiol. Spectr. 2023;11:e0051723. doi: 10.1128/spectrum.00517-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero D.M., Chou C., Jury L.A., Nerandzic M.M., Cadnum J.C., Donskey C.J. Clinical and infection control implications of Clostridium difficile infection with negative enzyme immunoassay for toxin. Clin. Infect. Dis. 2011;53:287–290. doi: 10.1093/cid/cir361. [DOI] [PubMed] [Google Scholar]
- 26.Alasmari F., Seiler S.M., Hink T., Burnham C.A., Dubberke E.R. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin. Infect. Dis. 2014;59:216–222. doi: 10.1093/cid/ciu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polage C.R., Solnick J.V., Cohen S.H. Nosocomial diarrhea: Evaluation and treatment of causes other than Clostridium difficile. Clin. Infect. Dis. 2012;55:982–989. doi: 10.1093/cid/cis551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wren B.W., Heard S.R., al-Saleh A.I., Tabaqchali S. Characterisation of Clostridium difficile strains by polymerase chain reaction with toxin A- and B-specific primers. J. Med. Microbiol. 1993;38:109–113. doi: 10.1099/00222615-38-2-109. [DOI] [PubMed] [Google Scholar]
- 29.Mathis J.N., Pilkinton L., McMillin D.E. Detection and transcription of toxin DNA in a nontoxigenic strain of Clostridium difficile. Curr. Microbiol. 1999;38:324–328. doi: 10.1007/PL00006811. [DOI] [PubMed] [Google Scholar]
- 30.Eliakim-Raz N., Fishman G., Yahav D., Goldberg E., Stein G.Y., Zvi H.B., Barsheshet A., Bishara J. Predicting Clostridium difficile infection in diabetic patients and the effect of metformin therapy: A retrospective, case-control study. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:1201–1205. doi: 10.1007/s10096-015-2348-3. [DOI] [PubMed] [Google Scholar]
- 31.Cao F., Chen C.X., Wang M., Liao H.R., Wang M.X., Hua S.Z., Huang B., Xiong Y., Zhang J.Y., Xu Y.L. Updated meta-analysis of controlled observational studies: Proton-pump inhibitors and risk of Clostridium difficile infection. J. Hosp. Infect. 2018;98:4–13. doi: 10.1016/j.jhin.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Walker A.S., Eyre D.W., Wyllie D.H., Dingle K.E., Harding R.M., O’Connor L., Griffiths D., Vaughan A., Finney J., Wilcox M.H., et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med. 2012;9:e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eyre D.W., Cule M.L., Wilson D.J., Griffiths D., Vaughan A., O’Connor L., Ip C.L., Golubchik T., Batty E.M., Finney J.M., et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 2013;369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park K.S., Ki C.S., Lee N.Y. Isolation and Identification of Clostridium difficile Using 375 ChromID C. difficile Medium Combined with Gram Staining and PRO Disc Testing: A 376 Proposal for a Simple Culture Process. Ann. Lab. Med. 2015;35:404–409. doi: 10.3343/alm.2015.35.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbs S., Rupnik M., Gibert M., Brazier J., Duerden B., Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 2000;186:307–312. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 36.Charlson M.E., Carrozzino D., Guidi J., Patierno C. Charlson Comorbidity Index: A Critical Review of Clinimetric Properties. Psychother. Psychosom. 2022;91:8–35. doi: 10.1159/000521288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.