Abstract
Ninety-eight isolates of Cryptococcus neoformans were collected from 30 patients at the University of Iowa Hospitals and Clinics from December 1987 through December 1994. The susceptibility of each isolate was determined against fluconazole, itraconazole, amphotericin B, and flucytosine. Of the 98 isolates, 53 were recovered from blood, 19 were recovered from cerebrospinal fluid (CSF), and 26 were recovered from other sources. Although the strains were isolated from the same institution, DNA typing by electrophoretic karyotype (EK) revealed wide genetic variation. Overall, 23 different EK profiles were identified by computer-aided analysis. An isolate exhibiting a single EK was isolated from 24 of 30 patients (80%), whereas multiple strains with unique EKs were isolated from 6 of 30 (20%) patients. Of the six patients who had multiple strains recovered, only one individual had two strains isolated from unique body sites, one strain from the blood and the other from the CSF. Six strains were isolated from multiple patients. Nine patients had multiple sequential isolates recovered over periods of time ranging from 3 days to 4 months. EK analysis revealed persistence of the same genotype in six of the cases. Three patients, however, appeared to have an isolate with a second distinct EK emerge during therapy. Of the patients with sequential positive cultures, an increase in the MICs for test agents was observed in only one case. C. neoformans isolates were collected over a period of 7 years, during which time MICs at our institution remained stable.
Cryptococcus neoformans is a common cause of meningeal infection among AIDS patients. Untreated, cryptococcal meningitis is uniformly fatal among this patient population. Pharmacologic management of cryptococcal infections generally consists of primary therapy with amphotericin B, with or without flucytosine, for 14 days followed by lifelong suppressive therapy with an agent such as fluconazole. Despite seemingly appropriate antifungal therapy, positive cerebrospinal fluid (CSF) cultures persist in approximately 30% of patients (9). High rates of fungal persistence and frequent disease relapse have sparked a growing concern among clinicians regarding the potential for the emergence of resistance among cryptococci. In an effort to understand the evolving epidemology of cryptococcal infections, we collected C. neoformans isolates over a 7-year period and evaluated these isolates with respect to susceptibility profiles and genetic variability.
(Results presented at the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 28 September to 1 October 1997 [6].)
MATERIALS AND METHODS
Fungal isolates.
Ninety-eight isolates of C. neoformans were collected from 30 patients at the University of Iowa Hospitals and Clinics between December 1987 and December 1994. Isolates were identified by using the Vitek system (Vitek bioMerieux, Hazelwood, Mo.) and conventional methods. Following identification, isolates were banked in sterile water and stored at room temperature until being analyzed.
Antifungal agents.
Fluconazole (Pfizer, Inc., New York, N.Y.), itraconazole (Janssen Pharmaceutica, Inc., Piscataway, N.J.), amphotericin B (Sigma Chemical Co., St. Louis, Mo.), and flucytosine (Hoffmann-La Roche Laboratories, Nutley, N.J.) were used for in vitro susceptibility determinations.
Antifungal susceptibility testing.
The MICs of test agents were determined for each test isolate in accordance with National Committee for Clinical Laboratory Standards guidelines (7). RPMI 1640 medium (Sigma) buffered to a pH of 7.0 with morpholinepropane-sulfonic acid (MOPS) buffer (Sigma) served as the growth medium. The MIC of fluconazole, itraconazole, or flucytosine was defined as the lowest concentration of drug which resulted in an 80% reduction of fungal growth compared to control. The amphotericin B MIC was defined as the lowest concentration of drug which resulted in complete inhibition of visible growth.
Molecular typing.
DNA typing by electrophoretic karyotype (EK) was determined by using a contour-clamped homogeneous electrophoretic field system (CHEF-DRII; Bio-Rad, Richmond, Calif.). Isolates were obtained from the isolate bank and subcultured twice on potato dextrose agar (Remel, Lexana, Kans.). Fungi from a 48- to 72-h culture plate were suspended in 20 ml of YEPD (1% yeast extract, 2% glucose, 2% Bacto Peptone) and incubated at 35°C with agitation for 24 h. Cells were harvested by centrifugation and resuspended in 1 M sorbitol, and 200 μl of cells were mixed with 100 μl of lysing enzyme (20 mg of L2265 [Sigma]/ml from Trichoderma hazianum). The mixture was incubated at 37°C for 1 h. Cells were recovered by centrifugation and washed twice with 1 ml of SCE (1 M sorbitol, 0.6 M sodium citrate, and 0.06 M EDTA) (1). Following washes, cells were resuspended in 240 μl of SCE and mixed with 240 μl of 2% low-melting-temperature agarose (SeaPlaque GTG; FMC BioProducts, Rockland, Maine) and dispensed into molds to form plugs. Upon removal from molds, the plugs were incubated overnight at 50°C in 2 ml of NET (0.01 M Tris, 0.45 M EDTA, and 1% N-lauryl-sarcosine [pH 7.5]) containing 100 μl of proteinase K (1 mg/ml [Sigma]). Plugs were then washed four times with 5 ml of CHEF TE buffer (0.1 M Tris and 0.1 M EDTA [pH 7.5]) and stored at 5°C until being used.
Plugs, unknown samples and Saccharomyces cerevisiae chromosome DNA molecular weight markers, were placed on the teeth of a stationary comb, and liquid agarose (1% SeaKem GTG agarose [FMC BioProducts]) was poured into the mold. Gels were placed in a CHEF system containing 0.5× TBE buffer (0.5 M Tris, 0.5 M sodium borate, and 0.005 M Na2EDTA). The temperature of the system was maintained at 14°C and set at 4.5 V/cm with a switch time of 120 to 280 s. The run time of the system was 48 h. Once the gels were removed from the CHEF system, they were stained with ethidium bromide and photographed under UV light. Band migrations were compared among isolates and with the S. cerevisiae standards.
Analysis.
EK patterns were compared both visually and by using the Dendron software package version 2.1 (Solltech, Iowa City, Iowa). Visually, an isolate was classified as a unique EK if at least a one band difference from existing EKs was detected. For computer analysis, gel images were transformed into electronic images via a flatbed color scanner. Gels were normalized by using the S. cerevisiae chromosomal DNA standards as reference points. Band positions were identified by the software package, and similarity coefficients (SAB, number of shared bands × 2/total number of bands between the two isolates) were determined for each pair of isolates, following which SABs were compared among isolates. An SAB of 1.0 indicates that the isolates are identical (all bands match). Conversely, an SAB of 0.0 indicates that no band similarities were detected between isolates. Dendograms were generated based on SAB values by using an unweighted pair-group method. Isolate pairs exhibiting SABs of 1.0 were considered to represent the same EK and those with SABs ranging from 0.90 to 0.99 were considered to be highly related and were identified as subtypes. SABs of 0.90 to 0.99 are roughly equivalent to a one band difference among isolates. If SABs among isolates were <0.90 (usually representing ≥2 bands difference) they were considered to represent unique DNA types.
RESULTS
Fungal isolates.
A total of 98 cryptococcal isolates were recovered from 30 patients over the study period. Fifty-three isolates were recovered from blood, 19 were recovered from CSF, and 26 were recovered from other sources including bone marrow, bronchoalveolar lavage, ascitic fluid, and urine.
Antifungal susceptibility.
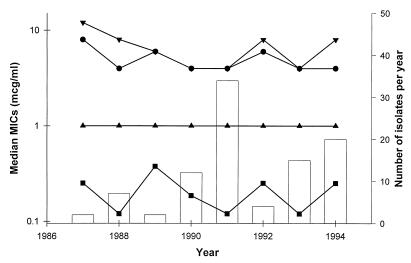
Antifungal susceptibility patterns are summarized in Table 1 and Fig. 1. All of the isolates were inhibited by clinically achievable concentrations of fluconazole (≤8 μg/ml) and itraconazole (≤0.5 μg/ml). Importantly, there was not a trend of increasing MIC with any of the agents over the study period.
TABLE 1.
Summary of C. neoformans (n = 98) susceptibility data
| Agent | MIC (μg/ml)a
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| Fluconazole | 4 | 8 | 0.5–8 |
| Itraconazole | 0.25 | 0.25 | 0.03–0.5 |
| Amphotericin B | 1 | 1 | 0.25–1 |
| Flucytosine | 8 | 8 | 0.5–>128 |
50% and 90%, MICs at which 50 and 90% of the isolates are inhibited, respectively.
FIG. 1.
Annual median MICs of fluconazole (•), itraconazole (■), amphotericin B (▴), and flucytosine (▾) against C. neoformans and the number of cryptococcal isolates recovered each year of the surveillance period (bars).
Among the nine patients from whom sequential positive cultures (defined as the collection of multiple isolates over a minimum of 3 days) were obtained, only one patient had isolates recovered for which MICs were elevated over time. Patient 12 had six isolates of the same EK (J2) and one isolate with a unique EK recovered over a 6-day period. Over this time, MICs of fluconazole, itraconazole, and amphotericin B increased four-, five-, and ninefold, respectively. Flucytosine MICs remained stable.
Molecular typing.
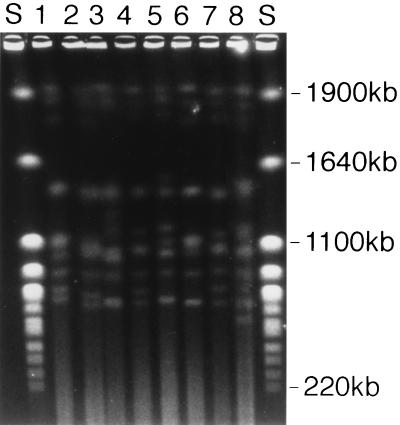
Of the 98 cryptococcal isolates recovered, 83 were able to be typed by EK. Among these 83 isolates, 30 unique EK profiles were noted upon visual inspection by using a one band difference as the discrimination criteria. By using computer-generated SAB values and a discrimination factor of ≥0.90, 23 different EKs were identified. Of these 23 EKs, seven had variant subtypes. Two subtypes were noted for four EKs (H, J, M, and V), three subtypes for two EKs (K and N), and four subtypes for one EK (O). As shown in Table 2, good agreement was noted between visual and computer-aided identification of EKs. The remainder of our analysis will focus on the EK data resulting from the computer-aided analysis. Representative EK from several isolates are presented in Fig. 2. Molecular types and antifungal susceptibilities, grouped by individual patients, are summarized in Table 2. Twenty-four patients had one strain represented by a single EK and six patients had two different strains recovered. Isolates with multiple subtype variants were isolated from four patients. Three patients had isolates of two subtypes, and one patient had isolates representing four different subtypes. Of the patients from whom strains with multiple unique EKs or subtypes were isolated, only two patients had these different isolates recovered from separate body sites. Patient 3 had two cryptococci with unique EKs recovered from separate body sites (CSF and blood), and patient 5 had two subtype variants isolated (CSF and blood). In both cases the different isolates shared similar susceptibility profiles. Six EKs were isolated from more than one patient: an isolate with a common EK was recovered from two pairs of patients, one cryptococcal strain with an identical EK was isolated from three patients, and three isolates with a common EK were each isolated from four patients. Nine patients had multiple sequential isolates recovered over periods ranging from 3 days to 4 months. Analysis of EKs revealed the persistence of a single genotype among six of the cases. Of these patients, two had multiple subtypes recovered. Three patients, however, had a strain with a second distinct genotype cultured during therapy (patients 11, 12, and 26). Three additional patients had multiple EKs cultured within the first 3 days of therapy (patients 3, 23, and 28). Thus, the majority of patients, 80%, were infected with a single cryptococcal strain.
TABLE 2.
Summary of EK subtypes and antifungal susceptibility profiles
| Patient | Source of isolatea | Collection date | EK determination by:
|
MIC (μg/ml)b
|
||||
|---|---|---|---|---|---|---|---|---|
| Visual inspection | SAB ≥ 0.9 | FLU | ITRA | AMB | 5-FC | |||
| 1 | Urine | 12/16/87 | I | Q | 8 | 0.25 | 1 | 8 |
| Blood | 12/19/87 | I | Q | 8 | 0.25 | 1 | 16 | |
| 2 | NR | 7/14/88 | Y | O4 | 8 | 0.12 | 1 | 4 |
| 3 | Blood | 10/10/88 | A | H1 | 8 | 0.25 | 1 | 8 |
| CSF | 10/10/88 | J | F | 8 | 0.25 | 1 | 8 | |
| 4 | CSF | 11/20/88 | B | H2 | 4 | 0.25 | 1 | 8 |
| 5 | CSF | 9/24/89 | C | N1 | 8 | 0.5 | 1 | 8 |
| Blood | 9/25/89 | D | N2 | 4 | 0.25 | 1 | 4 | |
| 6 | CSF | 1/12/90 | E | N2 | 4 | 0.25 | 1 | 8 |
| 7 | Blood | 4/30/90 | F | N3 | 4 | 0.25 | 1 | 8 |
| CSF | 5/1/90 | F | N3 | 4 | 0.25 | 1 | 16 | |
| 8 | CSF | 6/28/90 | F | N3 | 4 | 0.12 | 1 | 4 |
| BAL | 6/28/90 | H | N3 | 4 | 0.12 | 1 | 4 | |
| CSF | 7/1/90 | F | N3 | 4 | 0.12 | 1 | 4 | |
| Blood | 7/1/90 | F | N3 | 4 | 0.25 | 1 | 4 | |
| Blood | 7/2/90 | F | N3 | 4 | 0.06 | 1 | 4 | |
| Blood | 7/3/90 | F | N3 | 8 | 0.06 | 1 | 4 | |
| 9 | Blood | 12/23/90 | K | W | 8 | 0.25 | 1 | 4 |
| Blood | 12/24/90 | K | W | 8 | 0.25 | 1 | 4 | |
| 10 | CSF | 1/25/91 | L | P | 4 | 0.25 | 1 | 4 |
| 11 | Blood | 1/31/91 | O | V1 | 2 | 0.06 | 1 | 4 |
| Blood | 2/4/91 | N | U | 2 | 0.06 | 1 | 4 | |
| Blood | 2/6/91 | O | V2 | 2 | 0.03 | 1 | 4 | |
| Blood | 2/6/91 | O | V2 | 2 | 0.03 | 1 | 4 | |
| 12 | Blood | 3/26/91 | P | J2 | 8 | 0.5 | 1 | 4 |
| Blood | 3/28/91 | P | J2 | 0.25 | 0.015 | 0.12 | 8 | |
| Blood | 3/28/91 | P | J2 | 1 | 0.03 | 0.12 | 4 | |
| Blood | 3/29/91 | P | J2 | 4 | 0.12 | 1 | 8 | |
| Blood | 3/29/91 | Q | R | 4 | 0.12 | 1 | 8 | |
| Blood | 3/31/91 | P | J2 | 4 | 0.25 | 1 | 8 | |
| Blood | 3/31/91 | P | J2 | 4 | 0.12 | 1 | 8 | |
| 13 | Lung bx | 4/12/91 | R | T | 8 | 0.5 | 1 | 4 |
| 14 | Blood | 5/9/91 | S | C | 4 | 0.03 | 0.5 | 4 |
| CSF | 5/10/91 | S | C | 4 | 0.12 | 0.5 | 4 | |
| CSF | 5/13/91 | S | C | 8 | 0.06 | 0.5 | 4 | |
| CSF | 5/13/91 | S | C | 2 | 0.12 | 0.25 | 4 | |
| CSF | 5/15/91 | S | C | 4 | 0.12 | 0.25 | 4 | |
| CSF | 5/15/91 | S | C | 2 | 0.12 | 0.25 | 0.5 | |
| 15 | Blood | 6/12/91 | J | O1 | 4 | 0.12 | 1 | 4 |
| Blood | 6/12/91 | J | O2 | 0.5 | 0.03 | 0.25 | 8 | |
| Blood | 6/12/91 | J | O3 | 4 | 0.12 | 1 | 4 | |
| BM | 6/13/91 | J | O2 | 4 | 0.25 | 1 | 8 | |
| Blood | 6/17/91 | V | O4 | 4 | 0.25 | 1 | 4 | |
| 16 | BAL | 7/9/91 | T | S | 4 | 0.25 | 1 | 4 |
| 17 | Blood | 8/14/91 | H | K3 | 4 | 0.25 | 1 | 8 |
| Blood | 8/14/91 | H | K3 | 4 | 0.25 | 1 | 8 | |
| Catheter | 8/14/91 | H | K3 | 4 | 0.25 | 1 | 4 | |
| Blood | 8/15/91 | H | K3 | 4 | 0.12 | 0.5 | 8 | |
| AF | 8/15/91 | H | K3 | 4 | 0.12 | 1 | 8 | |
| 18 | Blood | 9/19/91 | GG | K2 | 4 | 0.12 | 1 | 8 |
| Blood | 9/24/91 | GG | K2 | 8 | 0.12 | 1 | 8 | |
| Blood | 9/25/91 | H | K3 | 4 | 0.25 | 1 | 8 | |
| BAL | 9/25/91 | H | K3 | 8 | 0.25 | 1 | 8 | |
| 19 | T. aut | 2/11/92 | J | O2 | 8 | 0.25 | 1 | 8 |
| 20 | CSF | 7/29/92 | W | J1 | 4 | 0.25 | 1 | 8 |
| 21 | Blood | 4/2/93 | M | M2 | 4 | 0.12 | 1 | 4 |
| Blood | 4/7/93 | M | M2 | 8 | 0.25 | 1 | 8 | |
| Blood | 4/7/93 | M | M2 | 4 | 0.25 | 1 | 4 | |
| 22 | Blood | 5/14/93 | H | K3 | 8 | 0.12 | 1 | 8 |
| Blood | 5/14/93 | H | K3 | 4 | 0.12 | 1 | 4 | |
| 23 | Blood | 8/14/93 | Z | L | 4 | 0.12 | 1 | 4 |
| Blood | 8/14/93 | D | M1 | 4 | 0.25 | 0.5 | 8 | |
| 24 | CSF | 8/17/93 | P | D | 4 | 0.25 | 1 | 8 |
| AF | 8/17/93 | P | D | 4 | 0.25 | 1 | 8 | |
| 25 | Blood | 9/26/93 | GG | A | 4 | 0.12 | 1 | 4 |
| CSF | 9/26/93 | GG | A | 4 | 0.12 | 1 | 4 | |
| CSF | 9/30/93 | GG | A | 4 | 0.12 | 0.5 | 4 | |
| 26 | Blood | 7/21/94 | BB | H2 | 4 | 0.25 | 1 | 8 |
| Blood | 7/25/94 | AA | I | 4 | 0.25 | 1 | 8 | |
| Blood | 12/14/94 | AA | I | 4 | 0.25 | 1 | 8 | |
| CSF | 12/17/94 | AA | I | 8 | 0.5 | 1 | 8 | |
| Blood | 12/18/94 | AA | I | 8 | 0.5 | 1 | 8 | |
| 27 | Blood | 7/28/94 | H | G | 4 | 0.12 | 0.5 | 4 |
| 28 | CSF | 11/13/94 | CC | B | 4 | 0.25 | 1 | 4 |
| Blood | 11/13/94 | EE | E | 4 | 0.25 | 1 | 8 | |
| Blood | 11/13/94 | CC | B | 4 | 0.25 | 1 | 2 | |
| Blood | 11/13/94 | CC | B | 0.5 | 0.12 | 1 | 4 | |
| 29 | Blood | 11/14/94 | FF | K1 | 4 | 0.25 | 1 | 8 |
| 30 | Blood | 11/17/94 | DD | H2 | 4 | 0.25 | 1 | 8 |
| Blood | 11/17/94 | DD | H2 | 4 | 0.25 | 1 | 8 | |
| Blood | 11/17/94 | DD | H2 | 4 | 0.25 | 1 | 8 | |
| Blood | 11/17/94 | DD | H2 | 4 | 0.5 | 1 | 8 | |
| Blood | 11/18/94 | DD | H2 | 4 | 0.25 | 1 | 8 | |
NR, not recorded; BAL, bronchoalveolar lavage; Lung bx, lung biopsy; T. aut, tissue autopsy; BM, bone marrow; AF, ascitic fluid.
FLU, fluconazole; ITRA, itraconazole; AMB, amphotericin B; 5-FC, flucytosine.
FIG. 2.
Representative EK profiles (patient number, EK). Lanes S, S. cerevisiae standard; lane 1, 21, M2; lane 2, 8, N3; lane 3, 11, V2; lane 4, 24, D; lane 5, 15, O1; lane 6, 18, K3; lane 7, 12, J2; lane 8, 26, I.
DISCUSSION
Infections secondary to fungi are being recognized with increased frequency, and in fact yeasts currently rank as the fourth most commonly encountered nosocomial bloodstream pathogen (5). Along with the heightened awareness of fungal infections has come a corresponding increase in the overall use of antifungal agents. At our institution, total gram usage of antifungals increased 83% from 1988 to 1992 (unpublished data). Increased antifungal use coupled with the use of prolonged treatment regimens, long-term maintenance therapies, and routine antifungal prophylaxis has spurred concern over development of drug resistance. Infection secondary to C. neoformans is an excellent example of a clinical situation necessitating prolonged antifungal usage. Following typical treatment guidelines, anticryptococcal therapy generally consists of an active induction treatment period lasting approximately 2 to 3 weeks followed by lifelong suppressive therapy. Therefore, we desired to describe the susceptibility patterns and genetic diversity of cryptococci at our institution over time.
No significant changes in the in vitro susceptibilities of C. neoformans isolates recovered at our institution were noted over the 7-year period evaluated (Fig. 1). Over this time frame, the gram usage of amphotericin B remained relatively constant; however, there was a concurrent 154% increase in the amount of azole antifungals and a 26% reduction in the amount of flucytosine prescribed. This suggests that institutional antifungal usage patterns had little impact on the overall susceptibility of C. neoformans at our hospital.
Despite being isolated from the same institution, cryptococcal isolates demonstrated wide genetic variability. Molecular typing revealed 23 unique EKs among our 30 infected patients. Twenty-four patients were infected with a single EK; however, six strains were isolated from multiple patients. Among patients infected with the same EK no commonalities among clustered patients other than the date of specimen collection were detected. With respect to the date of collection, isolates with similar EKs were isolated from temporally clustered (date of collection separated by less than 1 year) patients. For example, EK N was isolated from four patients, all of whom had their specimens collected between September 1989 and July 1990. Similarly, strains with EKs H, K, M, and O were each isolated within a year’s time from two separate patients. These findings suggest the possibility of a common environmental exposure for these groups of individuals.
Multiple, positive, sequential cultures were collected from nine patients. Despite anti-cryptococcal therapy, persistence of the same EK subtype was observed in eight of the individuals. Changes in the MICs for the recovered isolates were noted in only one patient, patient 12. Patient 12 was an end-stage AIDS patient who was admitted for treatment of Toxoplasma encephalitis and pneumonia. In addition to several broad-spectrum antibiotics, this individual also received nystatin and clotrimazole for oral thrush prophylaxis. On the 25th day following admission, C. neoformans was isolated from this patient’s blood. This initial isolate exhibited MICs of fluconazole, itraconazole, amphotericin B, and flucytosine of 8, 0.5, 1, and 4 μg/ml, respectively. Amphotericin B therapy was initiated 2 days following the collection date of the first culture. Two cultures were obtained on this date. Both cultures were positive for C. neoformans; however, the MICs of each of the antifungals were severalfold lower than had been observed for the initial isolate. Subsequent cultures were obtained while the patient was on amphotericin B, and several of these cultures were also positive for C. neoformans. The susceptibility profiles exhibited by these final cultures were similar to that noted for the original culture and were stable over several days. The patient appeared to respond to amphotericin B therapy. No cultures were obtained after the fourth day of therapy; however, the patient became and remained afebrile. Following 27 days of inpatient therapy, the patient was discharged on home amphotericin B therapy.
Six patients had two or more isolates with distinctly different EKs recovered during their hospitalization. The observation of such genetic diversity arising from individual patients may be due to coinfection with or exposure to and infection with multiple strains of C. neoformans and is consistent with previous observations (1, 2). As discussed by Brandt et al. (2), the importance of minor variations in EK among cryptococci is unclear; however, such differences have been shown to be stable upon repeat testing and may be reflective of polyclonal infection due to multiple exposures for these highly immunocompromised individuals. It should be noted that chromosomal rearrangements are common in C. neoformans and karyotype instability could explain the apparent emergence of new strains within a given individual; however, differences in EK due to karyotype instability are usually indicated by a single band change in EK profile, whereas differences of two bands or more suggest different strains (2, 3).
Our data are similar to those reported by several other investigators and indicate that recurrent cryptococcal infection is generally due to persistence of the same infecting strain (1, 2, 4, 8, 10). Failure to adequately treat or control the initial infection is largely due to inadequate host defenses and not to the development of antifungal resistance or reinfection with a new strain of C. neoformans.
Cryptococcal infections among immunocompromised individuals remain a significant concern. This study describes the epidemiology of cryptococcal infections at a single institution over a 7-year period. From this surveillance we determined that MICs for C. neoformans have remained stable over this study period. Furthermore, it appears that approximately 80% of infections result from infection with a single cryptococcal strain and that recurrent infections are largely due to persistence of the original infecting strain. Lastly, since the MICs determined for sequential isolates increased significantly in only one of nine patients (11%), emergence of drug resistance during therapy is probably an infrequent cause of treatment failure.
REFERENCES
- 1.Barchiesi F, Hollis R J, Messer S A, Scalise G, Rinaldi M G, Pfaller M A. Electrophoretic karyotype and in vitro antifungal susceptibility of Cryptococcus neoformans isolates from AIDS patients. Diagn Microbiol Infect Dis. 1995;23:99–103. doi: 10.1016/0732-8893(95)00169-7. [DOI] [PubMed] [Google Scholar]
- 2.Brandt M E, Pfaller M A, Hajjeh R A, Graviss E A, Rees J, Spitzer E D, Pinner R W, Mayer L W the Cryptococcal Disease Active Surveillance Group. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated cryptococcosis. J Infect Dis. 1996;174:812–820. doi: 10.1093/infdis/174.4.812. [DOI] [PubMed] [Google Scholar]
- 3.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haynes K A, Sullivan D J, Coleman D C, Clarke J C, Emilianus R, Atkinson C, Cann K J. Involvement of multiple Cryptococcus neoformans strains in a single episode of cryptococcosis and reinfection with novel strains in recurrent infection demonstrated by random amplification of polymorphic DNA and DNA fingerprinting. J Clin Microbiol. 1995;33:99–112. doi: 10.1128/jcm.33.1.99-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones R N, Marshall S A, Pfaller M A, Wilke W W, Hollis R J, Erwin M E, Edmond M B, Wenzel R P the SCOPE Hospital Study Group. Nosocomial enterococcal blood stream infections in the SCOPE program: antimicrobial resistance, species occurrence, molecular testing results, and laboratory testing accuracy. Diagn Microbiol Infect Dis. 1997;29:95–102. doi: 10.1016/s0732-8893(97)00115-6. [DOI] [PubMed] [Google Scholar]
- 6.Klepser M E, Pfaller M A. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Variation in electrophoretic karyotype (EK) and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987-84, abstr. C-14; p. 48. [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. Wayne, Pa: National Committee for Laboratory Standards; 1997. [Google Scholar]
- 8.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 9.van der Horst C M, Saag M S, Cloud G A, Hamil R J, Graybill J R, Sobel J D, Johnson P C, Tuazon C U, Kerkering T, Moskovitz B L, Powderly W G, Dismukes W E the National Institute of Allergy and Infectious Diseases Mycoses Study Group and AIDS Clinical Trials Group. Treatment of cryptococcal meningitis associated with the acquired immunodeficiency syndrome. N Engl J Med. 1997;337:15–21. doi: 10.1056/NEJM199707033370103. [DOI] [PubMed] [Google Scholar]
- 10.Varma A, Swinne D, Staib F, Bennett J E, Kwon-Chung K J. Diversity of DNA fingerprints in Cryptococcus neoformans. J Clin Microbiol. 1995;33:1807–1814. doi: 10.1128/jcm.33.7.1807-1814.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]