Abstract
We determined the nucleotide (nt) and amino acid (aa) heterogeneities of three distinct regions of the human cytomegalovirus (CMV) genome for 46 low-passage CMV isolates from four different patient populations (congenitally infected infants, children attending day-care centers, renal transplant recipients, and human immunodeficiency virus-infected individuals) and for two laboratory strains (CMV Ad169 and Towne). The gene regions for the major immediate-early (MIE) exon 4 gene (nt positions 1702 to 1982, aa positions 152 to 244), the DNA polymerase gene (nt positions 2797 to 3046, aa positions 713 to 795), and the glycoprotein B (gB) gene (nt positions 1698 to 1884, aa positions 567 to 628) were sequenced. The sequence information was used to design sets of nested PCR primers directed against the most highly conserved regions identified. MIE was the most variable gene region compared to the variability of the DNA polymerase and gB gene regions. Comparison of the sequences of all 46 isolates with that of Ad169 revealed nt and aa sequence homologies of 87.9 and 87.2%, respectively, within the MIE gene compared to 92.8 and 100% homologies, respectively, within the DNA polymerase gene and 93 and 95.2% homologies, respectively, within the gB gene. Within the MIE gene, compared to the Ad169 nt sequence the homology at the nt level among isolates obtained from children attending day-care centers was high (96.4%), while it was lower (90%) among isolates obtained from the other three patient populations. Preliminary results of a nested PCR with oligonucleotide primers selected from the DNA polymerase gene region with a low level of nt sequence variation indicates that primers selected from this region might be more powerful for use in PCR than primers selected from the MIE gene region.
The human cytomegalovirus (CMV) genome consists of 230 to 235 kbp of double-stranded DNA (4) and contains more than 200 predicted open reading frames (5). Like other herpesviruses, CMV induces latency, and the virus may become reactivated with immunosuppression. Reinfection with new genetically distinct CMV strains is prevalent, especially among transplant recipients (7). In immunocompromised hosts, primary or reactivated CMV infection or reinfection may result in a variety of clinical manifestations and may sometimes cause life-threatening CMV disease (2).
Early diagnosis of CMV infection is important if the development of CMV disease is to be prevented. Immunosuppressed patients might especially benefit from prompt institution of preemptive antiviral therapy (25). PCR has been widely used for the rapid diagnosis of CMV infection. Although PCR is rapid and sensitive in most cases, we have reported failure to detect CMV DNA from some CMV strains due to primer and target mismatches (27).
It has previously been shown by restriction fragment length polymorphism analysis that a number of CMV strains are circulating in the general population and that each of them displays its own restriction fragment length polymorphism pattern (12, 13). The extent to which the diversity found among human CMV strains is reflected in biological and functional differences is unknown at present. Little information about sequence variation within different CMV genes is available since only a few genes have been extensively studied. Previous studies indicate that sequence variation among CMV strains frequently occurs even in gene regions considered to be highly conserved (6, 8, 10).
We have studied three functionally important CMV genes, all of which are expected to be highly conserved: the IE1 region or the major immediate-early (MIE) gene region, the DNA polymerase gene region, and the glycoprotein B (gB) gene region. The MIE gene encodes the MIE protein (ppUL123), which already appears in infected cells after 1 h and has an important function in the regulation of CMV genes (19). The CMV DNA polymerase gene, encoded by the open reading frame UL54, plays an important role in DNA replication (19). The major envelope glycoprotein gB (gpUL55) is involved in cell-to-cell transmission of CMV and is the dominant target for neutralizing antibodies (19).
Our aims were to assess the nucleotide (nt) sequence variations in CMV isolates from both immunocompetent and immunocompromised individuals in comparison with the sequences of the laboratory strains Ad169 and Towne and to define genomic targets suitable for primer selection when constructing new diagnostic PCR systems for optimal detection of CMV in clinical samples from all patient populations.
MATERIALS AND METHODS
Clinical specimens.
Leukocytes separated from EDTA-anticoagulated blood from 12 renal transplant recipients and urine samples from a total of 34 patients, including 10 congenitally infected infants (ages, 0 to 14 days), 10 children (ages, 2 to 6 years) attending two different day-care centers, and 14 human immunodeficiency virus (HIV)-infected individuals without an obvious relation were inoculated onto human embryonic fibroblasts (HEFs). The CMV isolates were passaged one to four times in HEF cells and were stored at −70°C until PCR amplification for DNA sequencing.
Thirty selected specimens (23 EDTA-anticoagulated plasma samples, 5 bronchoalveolar lavage specimens, and 2 urine specimens) that had been stored at −70°C were used for a preliminary evaluation of a new nested PCR with primers from the CMV DNA polymerase gene region. The specimens were selected from a previous study and had been CMV culture positive but CMV PCR negative with our standard MIE primers and by alkaline treatment for DNA extraction (27).
Sample preparation for PCR.
For sequencing, 10 μl of tissue culture medium from infected fibroblasts was added directly to the PCR mixture. For a minority of samples, DNA extraction with phenol-chloroform and ethanol precipitation was performed to achieve successful CMV DNA amplification. For diagnosis, CMV DNA extraction was performed with the QIAamp blood kit (QIAGEN) according to the manufacturer’s instructions.
PCR amplification.
The nested PCR systems used for sequencing and diagnosis are listed in Table 1, including the optimal conditions for buffers, the primers, and the times and temperatures during the cyclings in the different assays (3, 27). For the second primer pair, which was used for sequencing, one primer (MIE4, POL2, or gBF1) was biotinylated, while the other primer (MIE3, POL1, or gBE1) contained a 21-nt handle sequence complementary to the sequencing primer. After amplification, 10 μl of the amplified product was electrophoresed on a 3% agarose gel containing ethidium bromide, and the gels were subsequently photographed under UV illumination. False-positive results due to contamination were excluded by analyzing water as every second sample. Crude lysates of diluted HEF cells infected with human CMV Ad169 were used as positive controls.
TABLE 1.
Characteristics of nested PCR assays used for detection of CMV DNA for sequencing and diagnosis of CMV infection using standard and new primer sequences selected from conserved gene regions within MIE exon 4, DNA polymerase, and gB genes
| PCR assay and gene | PCR buffera
|
Sense primer (5′→3′) | Sense primer start–stop nt positions | Antisense primer (5′→3′) | Antisense primer start–stop nt positions | Amplicon length (bp) | Temp (°C)-duration (s)/no. of cyclesb | |||
|---|---|---|---|---|---|---|---|---|---|---|
| MgCl2 concn (mM) | dNTP concn (mM each) | pH | Primer (concn [μM]) | |||||||
| Sequencing | ||||||||||
| CMV MIE gene | 7 | 0.8 | 9.6 | MIE1, MIE2 (0.2) | GGT GCG GCA TAG AAT CAA GG | 1534–1553 | CCC GTA CAT GGT CAT CAT AC | 2130–2149 | 616 | 94, 55, 72-45, 45, 45/20 |
| 10 | 1 | 9.6 | MIE3, MIE4 (0.4) | #c-TTG CAG AAT GCC TTA GAT | 1623–1642 | Yd-GCG TGA GCA CCT TGT CTC TC | 2029–2048 | 426 | 94, 55, 72-45, 45, 45/40 | |
| CMV POL system 1 | 3 | 0.4 | 9.6 | POL3, POL4 (0.3) | TAA CAG TAG TAG CAG CGT CG | 2639–2658 | TCA TAA GAG AGA CGA AGA CC | 3515–3534 | 895 | 94, 55, 72-60, 45, 120/20 |
| 3 | 0.4 | 9.6 | POL1, POL2 (0.3) | #-GTA CTG CGG CGG TTT CGT AC | 2728–2747 | Y-AAC TCC AGC TTG ACG GGC TC | 3489–3508 | 781 | 94, 64, 72-60, 45, 120/35 | |
| CMV gB gene | 4.5 | 0.8 | 9.6 | gBG, gBH (0.13) | AAC CCG TCA GCT ATT CTC TC | 1570–1589 | AGA TGC TGC TGA GGT CAA TC | 1908–1927 | 358 | 94, 48, 72-30, 30, 30/20 |
| 5.5 | 0.8 | 9.6 | gBE1, gBF1 (0.5) | #-GAT GTC CTG GGT CTG GCC | 1630–1647 | Y-ATG CGT TTG AAG AGG TAG TCC ACG | 1884–1907 | 278 | 94, 52, 72-30, 30, 30/40 | |
| Diagnostic | ||||||||||
| CMV MIE gene (standard) | 10 | 0.5 | 9.6 | C, D (0.15) | TGA GGA TAA GCG GGA GAT GT | 1729–1748 | ACT GAG GCA AGT TCT GCA GT | 1951–1970 | 242 | 92, 53, 72-30, 30, 30/20 |
| 10 | 1 | 9.6 | A, B (0.3) | AGC TGC ATG ATG TGA GCA AG | 1767–1786 | GAA GGC TGA GTT CTT GGT AA | 1893–1912 | 146 | 92, 53, 72-30, 30, 30/40 | |
| New CMV MIE gene (unvalidated) | NDe | ND | ND | MIE1, MIE2 | TTG TAC CTG AGG ATA AGC G | 1722–1740 | GGG GAG CAC TGA GGC AAG T | 1959–1977 | 256 | ND |
| ND | ND | ND | MIE3, MIE4 | CGG GAG ATG TGG ATG GCT TG | 1739–1758 | GCC ATT GGT GGT CTT AGG GA | 1911–1930 | 192 | ND | |
| New CMV POL gene (validated) | 4 | 1 | 8.3 | CMVP1, CMVP2 (0.4) | TGG CCG TGT TCG ACT TTG C | 2797–2815 | GAG CGC CAT CTG TTC CTT G | 3071–3089 | 293 | 94, 50, 72-45, 45, 45/20 |
| 4 | 1 | 8.3 | CMVP3, CMVP4 (0.4) | CAG CCT CTA CCC TTC CAT CA | 2816–2835 | GCA CCG AGA CGC GCA CCG AA | 2957–2976 | 161 | 94, 50, 72-45, 45, 45/40 | |
| New CMV gB gene (unvalidated) | ND | ND | ND | gB1, gB3 | TGA AGG AAT CGC CAG GAC GC | 1700–1719 | TGA GGC TGG GAA GCT GAC AT | 1830–1849 | 150 | ND |
| ND | ND | ND | gB2, gB4 | ACG ACC CGT GGT CAT CTT TA | 1728–1747 | GCG GTG GTT GCC CAA CAG GA | 1802–1821 | 94 | ND | |
The first and second lines for each assay refer to first and second PCR rounds, respectively. Note that 10 mM Tris HCl and 50 mM NaCl were included in all assays in a total volume of 50 μl. dNTP, deoxynucleoside triphosphate.
Denaturation, annealing, and extension temperatures-respective durations.
#, specific handle sequence (CGT TGT AAA ACG ACG GCC AGT) complementary to the sequencing primer.
Y, biotin.
ND, has still not been determined.
Solid-phase DNA sequencing.
The CMV DNA in PCR-positive samples was sequenced as described by Wahlberg et al. (24) by using an AutoRead T7 sequencing kit (Pharmacia Biotech). Briefly, the double-stranded PCR templates were immobilized on magnetic beads (Dynabeads M-280 Streptavidin; Dynal AS, Oslo, Norway) and were subsequently converted into single strands by alkaline treatment. The annealing buffer and a fluorescing sequencing primer were added, and after heating to 65°C and cooling to room temperature, the beads with the immobilized single-stranded template DNA were transferred to a 96-well plate. Extension buffer and T7 DNA polymerase were added, and the mixture was dispensed into four wells, whereupon the dideoxynucleotides (ddATP, ddCTP, ddGTP, and ddTTP) were added. After incubation for 5 min at 37°C, stop solution was added. The mixture was heated to 92°C just prior to loading of the gel. The fluorescing bands were read on a 6% polyacrylamide gel with an automated laser fluorescence sequencing apparatus (Pharmacia LKB), and the nt and amino acid (aa) sequences were deduced.
Comparison of CMV gene sequences.
The length of the MIE exon 4 gene is 1,218 bp (nt positions 1505 to 2722) (1), and it encodes 406 aa. The sequence of nt positions 1702 to 1982 (281 bp encoding aa positions 152 to 244) was determined for each of the 46 isolates and the laboratory strains (Ad169 and Towne). The CMV DNA polymerase gene consists of 3,726 bp (1,242 aa) (16). The CMV DNA polymerase has eight conserved regions (A and I to VII) (11, 14, 26) with high degrees of homology to other DNA polymerases (22). The sequence from bases 2797 to 3046 (250 bp encoding aa 713 to 795) (16) including the major part of region II (aa 696 to 742) and the whole of region VI (aa 771 to 790) was determined. In total, 50 of the 83 aa sequenced within the DNA polymerase are located in conserved regions. The gB gene consists of 2,721 bp (907 aa) (9, 20), and the sequence of gB from bases 1698 to 1884 (187 bp encoding aa 567 to 628) was analyzed. The major neutralizing epitope of gB consists of aa 552 to 635 (23).
Phylogenetic tree analysis.
Phylogenetic tree analysis of the nucleotide sequences was conducted with the GCG package by using the distance matrix and Growtree based Neighbor-joining tree building method and the Jukes-Cantor one-parameter substitution model.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the reported sequences are AF099506 to AF099643.
RESULTS
nt variation within CMV isolates.
Sequencing of the 46 clinical isolates and 2 laboratory strains (CMV Ad169 and Towne) revealed DNA sequences of 250 to 500 bp in length. It was possible to assess for all isolates the nt sequence variations of 281-bp fragments within the MIE exon 4 gene (nt positions 1702 to 1982 encoding aa positions 152 to 244), 250-bp fragments within the DNA polymerase gene (nt positions 2797 to 3046 encoding aa positions 713 to 795), and 187-bp fragments within the gB gene (nt positions 1698 to 1884 encoding aa positions 567 to 628). No deletions or insertions were detected in the 46 isolates.
The sequences of fragments of Ad169 and Towne that were sequenced were identical to previously published sequences (1, 9, 15, 16, 20). The published sequence differences between the Ad169 and Towne strains within the MIE exon 4 gene region at a single nt position (nt position 1952) (1), 6 nt positions within the DNA polymerase gene (nt positions 2810, 2868, 2891, 2900, 2954, and 2978) (15, 16), and 4 nt positions within the gB gene (nt positions 1698, 1734, 1794, and 1866) (9, 20) were confirmed.
Analyses of the sequences of the three gene fragments in the different isolates revealed a number of nt changes at several positions. Table 2 presents the number of nt substitutions in the sequences of all 46 isolates as well as in those of isolates from each of the four patient populations compared to the sequence of strain Ad169. We found that the MIE gene was the most variable gene region, and 34 of 281 bp sequenced (12.1%) had nt variations in one or more isolates. In comparison, nt variations occurred in the DNA polymerase gene at 18 of 250 positions (7.2%) and in the gB gene at 13 of 187 positions (7%). Comparisons between the 46 isolates and the Towne strain revealed nt variations of the same magnitude as those for Ad169, and variations were found at 12.1, 8, and 7.5% of the positions of the MIE, DNA polymerase, and gB genes, respectively.
TABLE 2.
Nucleotide variation in CMV DNA sequences compared to CMV Ad169 sequence
| Patient population | No. (%) of nt substitutions compared to Ad169 sequence
|
Total no. of nt substitutions compared to Ad169 sequence/total no. of bases (%)
|
||||
|---|---|---|---|---|---|---|
| MIE gene (281 bp) | DNA polymerase gene (250 bp) | gB gene (187 bp) | MIE gene (281 bp) | DNA polymerase gene (250 bp) | gB gene (187 bp) | |
| Total (n = 46) | 34 (12.1) | 18 (7.2) | 13 (7.0) | 423/12,926 (3.3) | 318/11,500 (2.8) | 93/8,602 (1.1) |
| Congenitally infected infants (n = 10) | 28 (10.0) | 13 (5.2) | 8 (4.3) | 83/2,810 (3.0) | 71/2,500 (2.8) | 32/1,870 (1.7) |
| Children in day-care centers (n = 10) | 10 (3.6) | 12 (4.8) | 9 (4.8) | 55/2,810 (2.0) | 62/2,500 (2.5) | 22/1,870 (1.2) |
| Renal transplant recipients (n = 12) | 28 (10.0) | 15 (6.0) | 8 (4.3) | 136/3,372 (4.0) | 90/3,000 (3.0) | 23/2,244 (1.0) |
| HIV-infected individuals (n = 14) | 28 (10.0) | 12 (4.8) | 7 (3.7) | 149/3,934 (3.8) | 95/3,500 (2.7) | 16/2,618 (0.6) |
The degrees of homology between the sequence of the single most variable patient strain and that of strain Ad169 for each gene fragment were 97.9% for the MIE gene, 96.4% for the CMV DNA polymerase gene, and 96.3% for the gB gene.
Differences in the frequencies of nt sequence variations in isolates from the different patient populations were seen (Table 2). Thus, isolates from children attending two different day-care centers had the least nt sequence variation, with nt sequence homology within the MIE gene of 96.4% compared to the sequence of Ad169. The nt sequence homology compared to the sequence of Ad169 for the isolates from the three other groups was only 90% for this gene region. A greater nt sequence homology and no significant differences in sequence homologies between isolates from the different groups were found for the other two gene regions.
The total number of nt sequence variations within each gene was determined as the total number of nt substitutions occurring within the sequenced isolates divided by the total number of nt positions sequenced. The total nt sequence homology for all 46 isolates compared to the sequence of the Ad169 strain was 96.7% for the MIE gene, 97.2% for the CMV DNA polymerase gene, and 98.9% for the gB gene (Table 2).
Definition of new reference sequences.
The laboratory strains Ad169 and Towne displayed greater nt sequence variations than the wild-type isolates, which might indicate that Ad169 and Towne are less representative of the CMV strains that are now circulating in society. Reference sequences were determined by using the sequence data for the wild-type isolates obtained in the present study. For nt substitutions occurring in ≥50% of the CMV isolates, the sequences were considered the consensus sequence. The defined sequences are suggested to be used as new reference sequences (Fig. 1). By this definition, 4 of the 34 nt substitutions found within the MIE gene (when compared to the sequence of the Ad169 strain) were now considered to represent the consensus sequence. In the same manner, 8 of the 18 nt substitutions and 1 of the 13 nt substitutions within the DNA polymerase and gB genes, respectively, were considered to represent the consensus sequence. When the sequences of the isolates were compared to that of the Towne strain instead, 3 of the 34 nt substitutions in the MIE gene, 6 of the 20 nt substitutions in the DNA polymerase gene, and 1 of the 14 nt substitutions in the gB gene were considered to represent the consensus sequence.
FIG. 1.
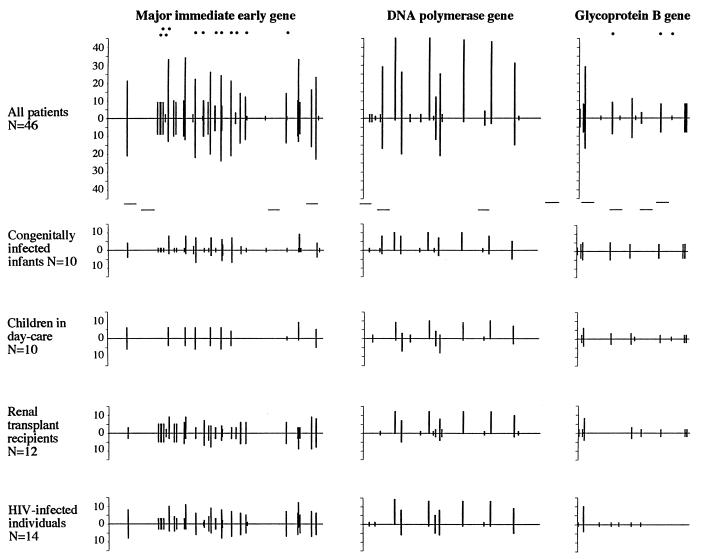
Distributions of nt substitutions and aa alterations within MIE exon 4 (nt positions 1702 to 1982) (1), DNA polymerase (nt positions 2797 to 3046) (13), and gB (nt positions 1698 to 1884) (16) gene sequences compared to the CMV Ad169 and new reference sequences among 46 CMV isolates obtained from four patient populations. The long horizontal lines represent the sequence of each gene region analyzed. The vertical lines above the horizontal lines represent the number of nucleotide substitutions occurring at certain positions of the sequences among the sequenced isolates compared to the sequence of CMV Ad169. The vertical lines below the horizontal lines represent nt substitutions compared to the new reference sequence. Nucleotide substitutions resulting in aa alterations are indicated (•). Variation at the aa level occurred within MIE exon 4 at aa positions 175 (H→Y), 176 (D→N), 177 (V→L), 178 (S→T), 190 (Q→K), 193 (A→V), 198 (D→E), 201 (R→K, N, or T), 205 (M→I), 207 (M→I), 212 (I→V or M), and 230 (Q→B) and within gB at positions 585 (A→V), 613 (L→F), and 619 (F→S). The new PCR primer sequences selected from highly conserved regions within all three gene regions are indicated by short horizontal lines.
A comparison of the new reference sequences and the sequences of the 46 isolates whose sequences were determined showed that the MIE gene remained the most variable gene region since nt substitutions occurred at 30 of 281 positions (10.7%), whereas substitutions occurred at 11 of 250 positions (4.4%) within the DNA polymerase gene and 12 of 187 positions (6.4%) within the gB gene (Table 3).
TABLE 3.
Nucleotide variation in CMV DNA sequences compared to new reference sequences
| Patient population | No. (%) of nt substitutions compared to the new reference sequences
|
Total no. of nt substitutions compared to the new reference sequences/total no. of bases (%)
|
||||
|---|---|---|---|---|---|---|
| MIE gene (281 bp) | DNA polymerase gene (250 bp) | gB gene (187 bp) | MIE gene (281 bp) | DNA polymerase gene (250 bp) | gB gene (187 bp) | |
| Total (n = 46) | 30 (10.7) | 11 (4.4) | 12 (6.4) | 297/12,926 (2.3) | 30/11,500 (0.3) | 66/8,602 (0.8) |
| Congenitally infected infants (n = 10) | 24 (8.5) | 5 (2.0) | 7 (3.7) | 51/2,810 (1.8) | 5/2,500 (0.2) | 27/1,870 (1.4) |
| Children in day-care centers (n = 10) | 6 (2.1) | 4 (1.6) | 8 (4.3) | 28/2,810 (1.0) | 9/2,500 (0.4) | 18/1,870 (1.0) |
| Renal transplant recipients (n = 12) | 24 (8.5) | 7 (2.8) | 7 (3.7) | 111/3,372 (3.3) | 8/3,000 (0.3) | 15/2,244 (0.7) |
| HIV-infected individuals (n = 14) | 24 (8.5) | 4 (1.6) | 6 (3.2) | 107/3,934 (2.7) | 8/3,500 (0.2) | 6/2,618 (0.2) |
When the nt sequences for all isolates were compared to the new reference sequences, the homologies within the MIE, CMV DNA polymerase, and gB genes were 97.7, 99.7, and 99.2%, respectively (Table 3).
Phylogenetic tree analysis.
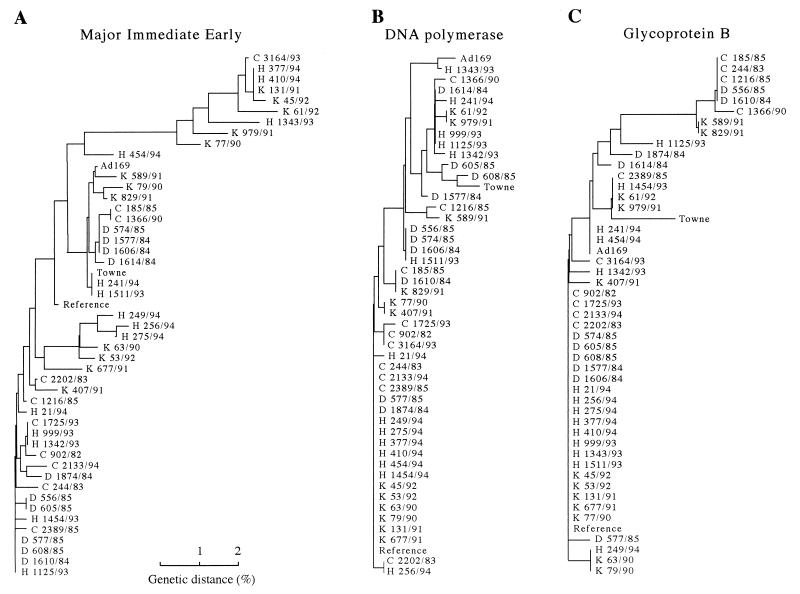
Phylogenetic tree analysis was used to compare the genetic distances for all 46 isolates, the 2 laboratory strains (Ad169 and Towne), and the suggested new reference sequences. Figure 2 shows the trees generated by using sequences from the MIE, DNA polymerase, and gB gene regions. The trees demonstrate, like the nt sequence variation data, that the most genetic drift has occurred in the MIE gene sequences in comparison with the CMV DNA polymerase and gB gene sequences. No conclusive difference in genetic distance between the 46 CMV isolates obtained from the different patient populations was found. A comparison of the 10 genetically most distant CMV strains within each tree reveals that, for the MIE gene, drift had occurred in 10 isolates mainly obtained from two patient populations, kidney transplant recipients (n = 5) and HIV-infected individuals (n = 4), as well as an isolate obtained from one congenitally infected infant (Fig. 2A). In contrast, for the DNA polymerase gene the 10 most distant strains consisted of both the Ad169 and Towne laboratory strains together with eight isolates obtained from the four different patient populations (HIV-infected individuals [n = 3], kidney transplant recipients [n = 2], children attending day-care centers [n = 2], and congenitally infected infants [n = 1]) (Fig. 2B). For the gB gene, the Towne strain was also found among the 10 most distantly related strains, together with 9 isolates obtained from all patient populations (congenitally infected infants [n = 4], children attending day-care centers [n = 2], kidney transplant recipients [n = 2], and HIV-infected individuals [n = 1]) (Fig. 2C).
FIG. 2.
Unrooted neighbor-joining trees comparing the MIE (A), CMV DNA polymerase (B), and gB (C) gene sequences of isolates from 46 individuals from four patient populations, the Ad169 and Towne laboratory strains, and the newly defined reference strains. The CMV isolates are indicated by patient population (C, congenitally infected infants; D, children attending day-care centers; K, kidney transplant recipients; and H, HIV-infected individuals). The phylogenetic tree analysis was conducted with sequences corresponding to nt positions 1702 to 1982 within the MIE exon 4 gene, nt positions 2797 to 3046 within the DNA polymerase gene, and nt positions 1698 to 1884 within the gB gene with the GCG package by using the distance matrix and Growtree-based neighbor-joining tree building method and the Jukes-Cantor one-parameter substitution model.
aa alteration within CMV isolates.
In this study 93 aa (aa positions 152 to 244) within the MIE exon 4 gene, 83 aa (aa positions 713 to 795) within the DNA polymerase gene, and 62 aa (aa positions 567 to 728) within the gB gene were deduced from the nt sequences for all isolates. We found aa alterations at 12 positions (13%) within the MIE gene and at three positions (5%) within the gB gene, while the CMV DNA polymerase gene was highly conserved at the aa level since the sequences of all 46 isolates whose sequences were determined were identical to the sequence of Ad169 (Fig. 1). The aa alterations within MIE exon 4 occurred at positions 175 (H→Y), 176 (D→N), 177 (V→L), 178 (S→T), 190 (Q→K), 193 (A→V), 198 (D→E), 201 (R→K, N, or T), 205 (M→I), 207 (M→I), 212 (I→V or M), and 230 (Q→B). Within the gB gene aa alterations occurred at positions 585 (A→V), 613 (L→F), and 619 (F→S). When the sequences of the CMV isolates were compared to the new reference sequences instead of the sequence of Ad169, a single aa alteration at position 178 (S→T) within the MIE gene was found in ≥50% of the isolates and was therefore considered the consensus sequence, while the other aa alterations within the MIE and gB genes occurred at the same positions compared with the sequence of Ad169.
A total of 35 of 46 isolates (76%) displayed a total of 143 aa alterations at 12 positions in the MIE gene compared to the sequence of Ad169 (Fig. 1). The majority of alterations, 88 of 143 (62%), were found among 10 renal transplant patients and 11 HIV-infected individuals (Fig. 1). The number of aa alterations found in the individual isolates varied between two and five (average, 3.1).
Sequencing of the gB gene revealed that 12 of 46 isolates (26%) possessed a total of 18 aa alterations at three positions within the major neutralizing epitope compared to the sequence of Ad169 (Fig. 1). The majority, 15 of 18 (83%), of aa alterations occurred in isolates from five congenitally infected infants (50%) and in four isolates from children attending day-care centers (40%) (Fig. 1).
Conserved CMV DNA regions suitable for use as PCR primer sequences.
The sequence data obtained in this study were used to design sets of primers for nested PCR directed against the most highly conserved MIE, CMV DNA polymerase, and gB gene regions identified. With the use of the computer program OLIGO, we selected four primer sequences from each of the three gene regions for potential use in a diagnostic nested PCR (Table 1 and Fig. 1). The selected primer sequences were carefully analyzed for optimal energy, GC content, and the risk of primer dimer formations. Examination for nt homology to the human herpesviruses, herpes simplex virus types 1 and 2, human herpesvirus 6, Epstein-Barr virus, and varicella-zoster virus excluded cross-reactivity with the new primers.
The selected primer sequences from the CMV DNA polymerase gene region (new CMV POL) (Table 1) were validated for their potential use in a nested PCR assay for the detection of CMV DNA in clinical specimens. Thirty specimens (23 EDTA-anticoagulated plasma specimens, 5 bronchoalveolar lavage specimens, and 2 urine specimens) were analyzed with our standard MIE primers and the new POL primers. The QIAamp blood kit was used for DNA extraction. CMV DNA was demonstrated in 13 of 30 specimens (10 plasma specimens, 2 bronchoalveolar lavage specimens, and 1 urine specimen) with the MIE primers and in 24 of 30 specimens (20 plasma specimens, 3 bronchoalveolar lavage specimens, and 1 urine specimen) with the POL primers. The detection limit of the standard PCR with the MIE primers is at least 5 to 10 fg of CMV DNA, whereas the detection limit of the new PCR with the POL primers is 1 to 10 fg of CMV DNA. These findings indicate that the new primer sequences from the CMV DNA polymerase gene region might be more powerful than the standard MIE primers for diagnostic purposes. However, their sensitivity and specificity need to be further evaluated. The selected new gB and MIE primers still remain unvalidated.
DISCUSSION
In this study independent clinical CMV isolates selected from four clinically different patient populations were analyzed for their degrees of sequence variation and for their phylogenetic relationships. Regions from the following conserved genes with functional properties were selected: the MIE gene (MIE exon 4), the DNA polymerase gene, and the gB gene. So far, only a few CMV genes have been sequenced, and most studies have concentrated on isolates from immunosuppressed patient populations (3, 6, 8, 17). The MIE and gB gene regions have been studied previously (1, 3, 6, 8–10, 17, 20), while little information is available for the DNA polymerase gene (15, 16, 18). New reference CMV gene sequences, based on the sequencing data that have been obtained, are proposed as substitutes for the high-passage laboratory CMV strains Ad169 and Towne. It was also possible to identify new primer sites within highly conserved gene regions for the identification of CMV DNA from infected patients.
The MIE gene was the most variable gene region, and the frequent nt substitutions were scattered at several positions throughout the whole gene region, especially among isolates from kidney transplant recipients and HIV-infected individuals (Fig. 2). Within the DNA polymerase gene region, the nt substitutions appeared to be clustered at fewer positions within isolates from all patient populations, and within the gB gene a clustered distribution of nt substitutions at only a few distinct positions was found, especially among isolates from congenitally infected infants and children attending day-care centers (Fig. 1).
These findings might be of importance for diagnostic purposes since, according to the literature, most laboratories use primer sequences from the MIE gene region in their diagnostic PCR assays (3, 6, 17, 27). Also, the majority of samples analyzed in routine viral diagnostic laboratories are obtained from immunocompromised patients, since these patients benefit most from a rapid diagnosis of CMV infection and subsequent preemptive antiviral therapy. Furthermore, we have previously reported the failure to detect CMV DNA in clinical samples by PCR using our standard primers from the MIE exon 4 gene region (27). Analysis of the sequence of this gene region showed primer and target mismatches at four nt positions within one of the inner MIE primers and at six nt positions and one nt position, respectively, within the outer primers. These mismatches might explain the previously reported failure to detect CMV DNA.
Considering these facts, we found it necessary to define new primer sequences based on the sequence data obtained for the wild-type isolates. The selection of primer sequences from within the MIE gene sequence was hampered by the frequent nt substitutions occurring throughout the gene. Since the nt substitutions within the DNA polymerase and gB genes were fewer and were more clustered at distinct positions, it was easier to find conserved regions of DNA suitable as primer targets for PCR amplification. The outer reverse CMV POL primer CMVP2 was chosen from a region (nt positions 3071 to 3089) which is located outside the sequenced regions of all isolates whose sequences were compared. The selected region was considered to be more optimal according to the OLIGO analysis. Furthermore, the sequences of the majority of isolates had been sequenced throughout this region. Mutations in the CMV DNA polymerase gene associated with antiviral resistance have been described and should be considered at primer selection. However, only a few mutations conferring antiviral resistance have been mapped to region II (21).
The primers from the more conserved DNA polymerase gene region were validated, and we found that the new PCR with POL primers was more sensitive for the detection of CMV DNA in clinical specimens than our standard PCR with MIE primers. In fact, use of the new PCR with POL primers and QIAamp DNA extraction instead of PCR with MIE primers reduced the proportion of previously nonidentifiable CMV strains by nearly 50%.
New reference strain sequences based on the data obtained for the wild-type isolates are suggested in the present study. These sequences are more closely related to those of CMV isolates circulating in society today compared to those of the old and highly passaged Ad169 and Towne strains. Comparison of the nt sequence variations within the MIE genes from the small number of isolates obtained from the different patient populations with the sequence of the Ad169 strain and the sequences of the new reference strain revealed that the least genotypic variation was found in isolates obtained from children attending day-care centers compared to that in isolates from the other patient populations. For the DNA polymerase and gB genes, no major difference in nt sequence homology between isolates from the patient populations and the Ad169 and the new reference strains was found (Tables 2 and 3).
The phylogenetic tree analysis also supported the findings that the MIE gene sequences are more heterogeneous than the DNA polymerase and gB gene sequences and the fact that no close genetic relationship exists between wild-type isolates and laboratory strains Ad169 and Towne. Subdivision of the CMV strains from the different patient populations on the basis of the phylogenetic tree analysis was not possible.
The fact that none of the nt substitutions within the DNA polymerase gene led to aa alterations might reflect the strong functional conservation within this gene due to its importance during viral replication. nt substitutions within the gB and MIE exon 4 gene regions led to three (5%) and 12 (13%) aa alterations, respectively. The aa alterations at positions 585 and 613 within the gB gene have been described previously by Chou and Dennison (8), while the aa alteration F→S at position 619 found in a single isolate in the present study has not been reported previously. Within the MIE exon 4 gene region the aa alterations at positions 175, 176, 178, and 198 and the aa alteration I→V at position 212 have previously been reported by Chou (6), while the alterations at positions 190 and 205 and the R→K or N substitution at position 201 were previously described by Brytting et al. (3). In this study another six aa alterations that have not been reported previously were found, namely, V→L at position 177, A→V at position 193, R→T at position 201, M→I at position 207, I→M at position 212, and Q→B at position 230. gB is continuously subjected to the immune response, and the variation in the gB gene sequence may be of functional interest since gB induces a strong neutralizing antibody response. Congenitally infected infants had the highest degree of aa variability (14.5%) in this restricted fragment, while HIV-infected individuals had the lowest (1.6%), which allows us to speculate that a poor immune response might allow the dominance of few viral variants or that a high degree of selection pressure allows the survival of only a few strains. The functional importance of the frequent aa alterations within the regulatory MIE gene found in isolates from all patient populations remains to be determined.
In summary, optimal diagnosis of CMV infection by PCR in all patients can be achieved only if primer sequences with the potential to detect all CMV strains are used. The strategy of this study was to gather sequence information by assessing the nt sequence variations in independent clinical CMV isolates selected from four clinically different patient populations in order to identify conserved CMV sequences that can act as targets for primer selection. The DNA polymerase and gB gene regions proved to be more conserved and therefore presumably more suitable for primer selection for diagnostic PCR than the more variable MIE exon 4 gene region. The new PCR with POL primers is being evaluated with clinical samples obtained from different patient populations sent to the laboratory for diagnosis.
ACKNOWLEDGMENTS
We thank Marianne Brundin and Mehrdad Mousavi-Jazi for helpful contributions.
This study was supported by grants from the Swedish Medical Research Council (project K97-06X-12180-01A).
REFERENCES
- 1.Akrigg A, Wilkinson G W G, Oram J D. The structure of the major immediate early gene of human cytomegalovirus strain Ad169. Virus Res. 1985;2:107–121. doi: 10.1016/0168-1702(85)90242-4. [DOI] [PubMed] [Google Scholar]
- 2.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2493–2523. [Google Scholar]
- 3.Brytting M, Wahlberg J, Lundberg J, Wahren B, Uhlen M, Sundqvist V-A. Variations in the cytomegalovirus major immediate-early gene found by direct genome sequencing. J Clin Microbiol. 1992;30:955–960. doi: 10.1128/jcm.30.4.955-960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 6.Chou S. Effect of interstrain variation on diagnostic DNA amplification of the cytomegalovirus major immediate-early gene region. J Clin Microbiol. 1992;30:2307–2310. doi: 10.1128/jcm.30.9.2307-2310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou S. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N Engl J Med. 1986;314:1418–1423. doi: 10.1056/NEJM198605293142205. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Dennison K M. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229–1234. doi: 10.1093/infdis/163.6.1229. [DOI] [PubMed] [Google Scholar]
- 9.Cranage M P, Kouzarides T, Bankier A T, Satchwell S C, Weston K, Tomlinson P, Barrell B G, Hart H, Bell S E, Minson A C, Smith G L. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darlington J, Super M, Patel K, Grundy J, Griffiths P, Emery V C. Use of the polymerase chain reaction to analyse sequence variation within a major neutralizing epitope of glycoprotein B (gp58) in clinical isolates of human cytomegalovirus. J Gen Virol. 1991;72:1985–1989. doi: 10.1099/0022-1317-72-8-1985. [DOI] [PubMed] [Google Scholar]
- 11.Gibbs J S, Chiou H C, Bastow K F, Cheng Y C, Coen D M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci USA. 1988;85:6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grillner L, Strangert K A. A prospective molecular epidemiological study of cytomegalovirus infection in two day-care centres in Sweden. No evidence for horizontal transmission within the centres. J Infect Dis. 1988;157:1080–1083. doi: 10.1093/infdis/157.5.1080. [DOI] [PubMed] [Google Scholar]
- 13.Ho M. Cytomegalovirus: biology and infection. 2nd ed. New York, N.Y: Plenum Medical Book Co.; 1991. pp. 229–300. [Google Scholar]
- 14.Hwang C B, C, Ruffner K L, Coen D M. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J Virol. 1992;66:1774–1776. doi: 10.1128/jvi.66.3.1774-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ihara S, Takekoshi M, Mori N, Sakuma S, Hashimoto J, Watanabe Y. Identification of mutation sites of a temperature-sensitive mutant of HCMV DNA polymerase activity. Arch Virol. 1994;137:263–275. doi: 10.1007/BF01309474. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T, Bankier A T, Satchwell S C, Weston K, Tomlinson P, Barrell B G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987;61:125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehner R, Stamminger T, Mach M. Comparative sequence analysis of human cytomegalovirus strains. J Clin Microbiol. 1991;29:2494–2502. doi: 10.1128/jcm.29.11.2494-2502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lurain N S, Thompson K D, Holmes E W, Read G S. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J Virol. 1992;66:7146–7152. doi: 10.1128/jvi.66.12.7146-7152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2447–2492. [Google Scholar]
- 20.Spaete R, Thayer R M, Probert W S, Masiarz F R, Chamberlain S H, Rasmussen L, Merigan T C, Pachl C. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology. 1988;167:207–225. doi: 10.1016/0042-6822(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 21.Tatti K M, Smith I L, Schinazi R F. Mutations in human cytomegalovirus (HCMV) DNA polymerase associated with antiviral resistance. Int Antivir News. 1998;6:1. [Google Scholar]
- 22.Teo I A, Griffin B E, Jones M D. Characterization of the DNA polymerase gene of human herpesvirus 6. J Virol. 1991;65:4670–4680. doi: 10.1128/jvi.65.9.4670-4680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner B, Kropff B, Kalbacher H, Britt W, Sundqvist V-A, Östberg L, Mach M. A continuous sequence of more than 70 amino acids is essential for antibody binding to the dominant antigenic site of glycoprotein gp58 of human cytomegalovirus. J Virol. 1992;66:5290–5297. doi: 10.1128/jvi.66.9.5290-5297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahlberg J, Lundeberg J, Hultman T, Uhlén M. General colorimetric method for DNA diagnostics allowing direct solid phase genomic sequencing of the positive samples. Proc Natl Acad Sci USA. 1990;87:6569–6573. doi: 10.1073/pnas.87.17.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winston D J. Prevention of cytomegalovirus disease in transplant recipients. Lancet. 1995;346:1380–1381. doi: 10.1016/s0140-6736(95)92401-9. [DOI] [PubMed] [Google Scholar]
- 26.Wong S W, Wahl A F, Yuan P-M, Arai N, Pearson B E, Arai K, Korn D, Hunkapiller J W, Wang T W-F. Human DNA polymerase α gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988;7:37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zweygberg Wirgart B, Claesson K, Eriksson B-M, Brundin M, Tufvesson G, Tötterman T, Grillner L. Cytomegalovirus (CMV) DNA amplification from plasma compared with CMV pp65 antigen (ppUL83) detection in leucocytes for early diagnosis of symptomatic CMV infection in kidney transplant patients. J Clin Diagn Virol. 1996;7:99–110. doi: 10.1016/s0928-0197(96)00258-9. [DOI] [PubMed] [Google Scholar]