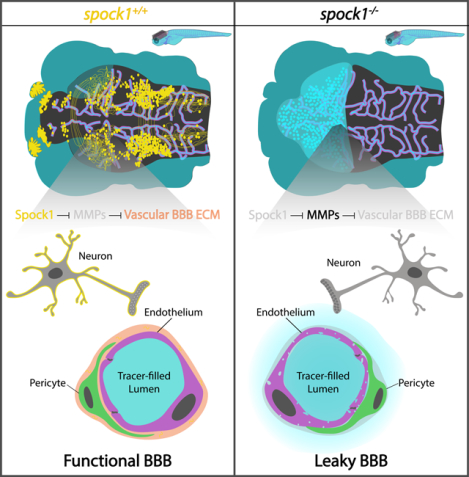
Abstract
The blood-brain barrier (BBB) is a unique set of properties of the brain vasculature which severely restricts its permeability to proteins and small molecules. Classic chick-quail chimera studies showed that these properties are not intrinsic to the brain vasculature but rather are induced by surrounding neural tissue. Here we identify Spock1 as a candidate neuronal signal for regulating BBB permeability in zebrafish and mice. Mosaic genetic analysis shows that neuronally-expressed Spock1 is cell non-autonomously required for a functional BBB. Leakage in spock1 mutants is associated with altered extracellular matrix (ECM), increased endothelial transcytosis, and altered pericyte-endothelial interactions. Furthermore, a single dose of recombinant SPOCK1 partially restores BBB function in spock1 mutants by quenching gelatinase activity and restoring vascular expression of BBB genes including mcamb. These analyses support a model in which neuronally secreted Spock1 initiates BBB properties by altering the ECM, thereby regulating pericyte-endothelial interactions and downstream vascular gene expression.
Graphical Abstract
One-Sentence Summary:
Spock1 is a signal secreted by neurons that induces barrier properties in the brain vasculature.
eTOC blurb
The blood-brain barrier (BBB) restricts movement of solutes across brain blood vessels and understanding BBB formation will help drug delivery and treatment of neurodegenerative disease where it often fails. Here, O’Brown et al. identify a secreted signal, Spock1, expressed in neurons that induces BBB properties in surrounding brain vasculature.
INTRODUCTION
The blood-brain barrier (BBB) maintains a tightly controlled homeostatic environment in the brain that is required for proper neural function. BBB breakdown has been implicated in multiple neurodegenerative diseases including Alzheimer’s, Parkinson’s, and Huntington’s Diseases.1 Conversely, the BBB also serves as an obstacle for effective drug delivery to the brain. Therefore, there is great interest in generating a better understanding of how to therapeutically regulate its permeability, both to restore BBB function in neurodegeneration and to transiently open it for improved chemotherapeutic access for brain tumors. The BBB is a specialized property of the brain vasculature, which is composed of a thin, continuous layer of non-fenestrated endothelial cells with uniquely restrictive properties. Brain endothelial cells create the barrier via two primary cellular mechanisms: 1) specialized tight junction complexes that block the transit of small water-soluble molecules between cells and 2) reduced levels of vesicular trafficking or transcytosis to restrict transit through endothelial cells.2 At a molecular level, functional barrier endothelial cells are differentiated from peripheral endothelial cells by high expression of the tight junction molecule Claudin-53 and the fatty acid transporter Mfsd2a,4,5 and an absence of the fenestra and vesicle associated protein Plvap.6 Expression of substrate-specific influx and efflux transporters such as Glut17 and Pgp,8 which dynamically regulate the intake of necessary nutrients and the removal of metabolic waste products,9 further enhances the selectivity of the BBB.
The restrictive properties of the BBB are not intrinsic to brain endothelial cells, but are induced and maintained by signals in the brain microenvironment during embryonic development.10 When avascularized quail neural tissue was transplanted into embryonic chicken gut cavities, the blood vessels ingressing from the non-barrier gut tissue obtained functional BBB properties, specifically both tight junctions and reduced levels of vesicles, resulting in Trypan blue tracer confinement within the blood vessels.10 Conversely, when avascularized quail somite tissue was transplanted into embryonic chick brain, the blood vessels ingressing from the neural tissue lost functional BBB properties, leaking tracer into the quail graft due to increased endothelial transcytosis and loss of tight junction function. These data show that signals from the neural microenvironment to the vasculature control barrier function. Furthermore, these microenvironmental signals are also required to actively maintain barrier properties throughout life.11–14
Mural cells called pericytes share the endothelial basement membrane and co-migrate with endothelial cells during neovascularization of the developing brain. This close interaction between pericytes and endothelial cells is required for both the initiation and continued maintenance of barrier properties.15–17 Astrocytes, glial cells found exclusively in the central nervous system (CNS), completely ensheath the vasculature with their endfeet in a polarized fashion.18 Astrocytes arise late in embryonic development, after neurogenesis is complete, with the vast majority of gliogenesis occurring postnatally in rodents. Mammalian astrocytes are potent inducers of barrier properties in vitro and in vivo, and are required for BBB maintenance.13,14,19 While the necessity of these two cell types has been known for decades, canonical Wnt signaling arising from both neuronal and astrocytic sources is the only microenvironmental signal known to induce and maintain BBB function.13,20–27 However, in addition to its role in regulating barrier properties, Wnt signaling also plays an important role in tip cell specification and angiogenesis in the developing brain.21,23,24 Furthermore, mouse endothelial cells are Wnt-responsive as early as E9.5,27 preceding the acquisition of functional barrier properties,22 suggesting that other signaling may be acting in conjunction with or in addition to Wnt signaling for the induction of BBB properties.
To further examine the molecular determinants of BBB development, we turned to the optically transparent zebrafish system, which allows for imaging of the intact BBB in toto throughout zebrafish development. Zebrafish brain endothelial cells express many of the same molecular markers as mammalian brain endothelial cells, including Glut1, Cldn5, ZO-1, and Mfsd2a.24,28–32 We previously characterized the molecular and subcellular mechanisms of functional BBB development in zebrafish, and determined that the zebrafish BBB becomes functionally mature by 5 days post fertilization (dpf) due to the suppression of transcytosis rather than through the acquisition of tight junction function, which was observed as early as 3 dpf.28 During these studies, we serendipitously discovered a recessive viable mutant with profound forebrain and midbrain barrier leakage which we identify here to be the neuronally produced and secreted proteoglycan Spock1. Using an array of perturbations and imaging techniques, we uncover the range of Spock1 signaling to the vasculature, and identify that mechanistically, Spock1 signaling acts through control of the pericyte-endothelial extracellular matrix (ECM) to control endothelial transcytosis.
RESULTS
Identification and mapping of a spontaneous leaky mutant to spock1
We found a spontaneous recessive mutant that leaks both an injected 1 kDa Alexa Fluor (AF) 405 NHS Ester dye and a transgenic 80 kDa serum protein DBP-EGFP29 into the forebrain and midbrain as early as 3 days post fertilization (dpf) with no improvement in BBB function throughout larval development (Figure 1A–E) or adulthood (Figure 1G). This increased BBB permeability in homozygous mutants occurs in the absence of hemorrhage or vascular patterning defects (Figure S1), apparent changes in Wnt signaling (Figure S1), or any reduction in viability or fertility. Time lapse imaging of injected 10 kDa Dextran revealed a steady accumulation of Dextran in the brain parenchyma of leaky mutants over the course of an hour (Figure S2). The rate of Dextran accumulation in the brain closely resembled the dynamics of Dextran leakage we previously reported in mfsd2aa mutants,28 which specifically display an increase in endothelial caveolae-mediated transcytosis without altered tight junction function.
Figure 1. Spock1 induces BBB functional development.
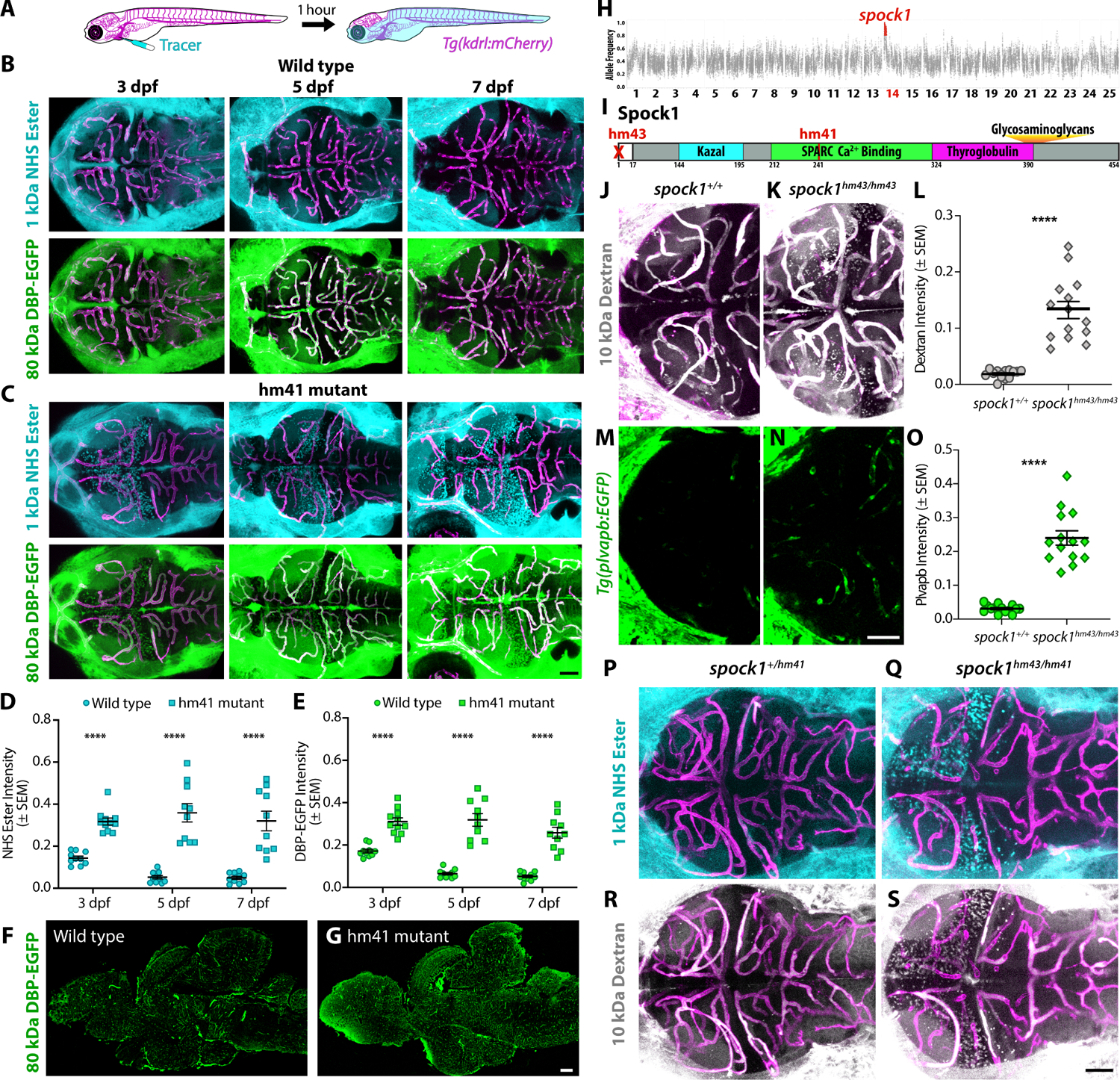
(A-E) Fluorescent tracer leakage assays in larval zebrafish reveal BBB leakage in the forebrain and midbrain of both injected 1 kDa Alexa Fluor 405 NHS Ester (turquoise) and 80 kDa DBP-EGFP (green) of hm41 homozygous larvae (C) compared to wild type controls (B) throughout larval development, as quantified in D and E. (F-G) Sagittal sections of adult brains show that the mutant leakage persists into adulthood (G). (H) Linkage mapping of the hm41 mutant phenotype reveals tight linkage to spock1 on chromosome 14. (I) Spock1hm41 has several point mutations (T241A and several silent mutations) in the SPARC domain. Spock1hm43 has a deletion of the 5’ UTR and start codon. (J-O) Dextran leakage assays show spock1hm43/hm43 mutants (K) have increased BBB leakage (L). spock1hm43/hm43 mutants also have increased expression of the leaky vessel marker plvapb (N and O). (P-S) Compound spock1hm43/hm41 heterozygotes also display increased NHS Ester (Q) and Dextran (S) leakage compared to spock1hm41/+ heterozygote siblings, which confine both injected tracers at 5 dpf (P and R). Scale bars represent 50 μm (C, N, S) and 200 μm (G). N = 10 (D and E) and 14 (L and O), with each point representing an individual fish. **** p<0.0001 by 2way ANOVA (D and E) and by t test (L and O).
To identify the mutation responsible for this leakage phenotype, we performed linkage mapping on 5 dpf mutant and wild type siblings using bulk segregant RNAseq33 and identified a single peak at chr14:2205271–3513919 (GRCz11; Figures 1H and S3). Of the 14 genes within the linkage region (Figure S3), 8 were expressed at 5 dpf. Two of these genes were differentially expressed between mutant and wild type, csf1ra and gstp2 (Table S1), and one had several mutations that segregated with the leakage phenotype, spock1. To test whether loss of any of these genes conferred the increased BBB permeability, we assessed tracer leakage in mosaic crispants (zebrafish larvae injected at the 1-cell stage with Cas9 protein and gene-specific sgRNAs) at 5 dpf and observed no BBB defects in 42 gstp2 or 35 csf1ra crispants. However, when we assessed BBB function in spock1 crispants, we observed a strong leaky phenotype in 26% of larvae (26/100 injected fish; Figure S3) and moderate leakage in 53% (53/100 injected fish; Figure S3) as well as a few that displayed BBB leakage restricted to one midbrain hemisphere (4/100 injected fish; Figure S3). Leakage in spock1 crispants corresponded with a loss of spock1 expression (Figure S3). Spock1 encodes a secreted protein of unclear function named for its conserved protein domains: SPARC (Osteonectin), Cwcv And Kazal Like Domains Proteoglycan 1 protein (also known as Testican-1). Spock1 has three predicted domains: 1) a Kazal-type serine protease inhibitor, 2) an extracellular SPARC calcium-binding region, and 3) a thyroglobulin type-1 repeat region, in addition to being decorated by both chondroitin sulfate (CS) and heparan sulfate (HS) glycosaminoglycan (GAG) chains (Figure 1I).34,35 Further sequencing of spock1 confirmed a few point mutations in the SPARC calcium-binding domain in the leaky fish (Figure 1I), including a T241A missense mutation and two silent mutations at Q249 and G250, hereafter referred to as spock1hm41/hm41 mutants.
To validate that loss of Spock1 function causes the leakage phenotype in the spontaneous mutants, we generated a stable null allele designated spock1hm43 using CRISPR mutagenesis with two sgRNAs to completely delete the 5’ UTR and start codon of spock1 (Figure 1I). As expected based on the 550 bp genomic deletion, spock1hm43/hm43 fish do not transcribe spock1 unlike the original spontaneous spock1hm41/hm41 mutants, which maintain normal spock1 transcription (Figure S4). Functional tracer leakage assays using 10 kDa Dextran revealed the same increased BBB permeability specifically in the forebrain and the midbrain observed in the spontaneous spock1hm41/hm41 mutants (Figure 1J–L). This increased BBB permeability was accompanied with increased expression of the pathological marker of leaky endothelial cells plvapb:EGFP31 compared to the negligible levels observed in wild type siblings with a functional BBB (Figure 1M–O). Lastly, we crossed both lines to test for genetic complementation, and observed the leaky phenotype in spock1hm41/hm43 compound heterozygotes (Figure 1P–S), further indicating that the initial spontaneous mutant acts via loss of Spock1 function.
Neuronal Spock1 signals to the vasculature within a short range
Given the necessity of Spock1 in determining BBB function, we next wanted to assess where spock1 was expressed during BBB development. Using CRISPR we generated a stable spock1 knock in mCherry reporter (TgKI(spock1:mCherry)hm44) which revealed expression throughout the developing CNS, including the brain, retina and spinal cord (Figures 2A–B, S5). We validated spock1 expression in the brain by whole mount HCR fluorescent in situ hybridization, and saw that it colocalized with the neuronal marker elavl3 and never the vascular Tg(kdrl:mCherry) signal (Figure 2C–F, Figure S5), similar to its predominantly neural expression in the developing mouse CNS.36 This neuronal expression was confirmed further in live double transgenic TgKI(spock1:mCherry) and Tg(huc:GcAMP6s) animals (Figure S5).36 To determine whether spock1 is expressed by a particular subtype of neuron, we co-stained for spock1 and the glutamatergic marker slc17a6b and the GABAergic marker gad1b (Figure S5). These analyses revealed that while not every elavl3+ neuron expresses spock1, spock1 expression was not restricted to either neuronal subtype. However, spock1 expression did appear to be excluded from Tg(neurog1:egfp) neurons, indicating that spock1 is primarily expressed by post-mitotic neurons (Figure S5). Prior single-cell RNA sequencing (scRNA-seq) data37 and our own also showed expression of spock1 primarily in neurons and never in vascular endothelial cells (Figure S6). Taken together, these data indicate that we have identified a neuronal signal, Spock1, that plays a role in establishing endothelial BBB properties during development without altering vascular patterning.
Figure 2. Neuronal Spock1 regulates endothelial cells within a 10–20 μm range.
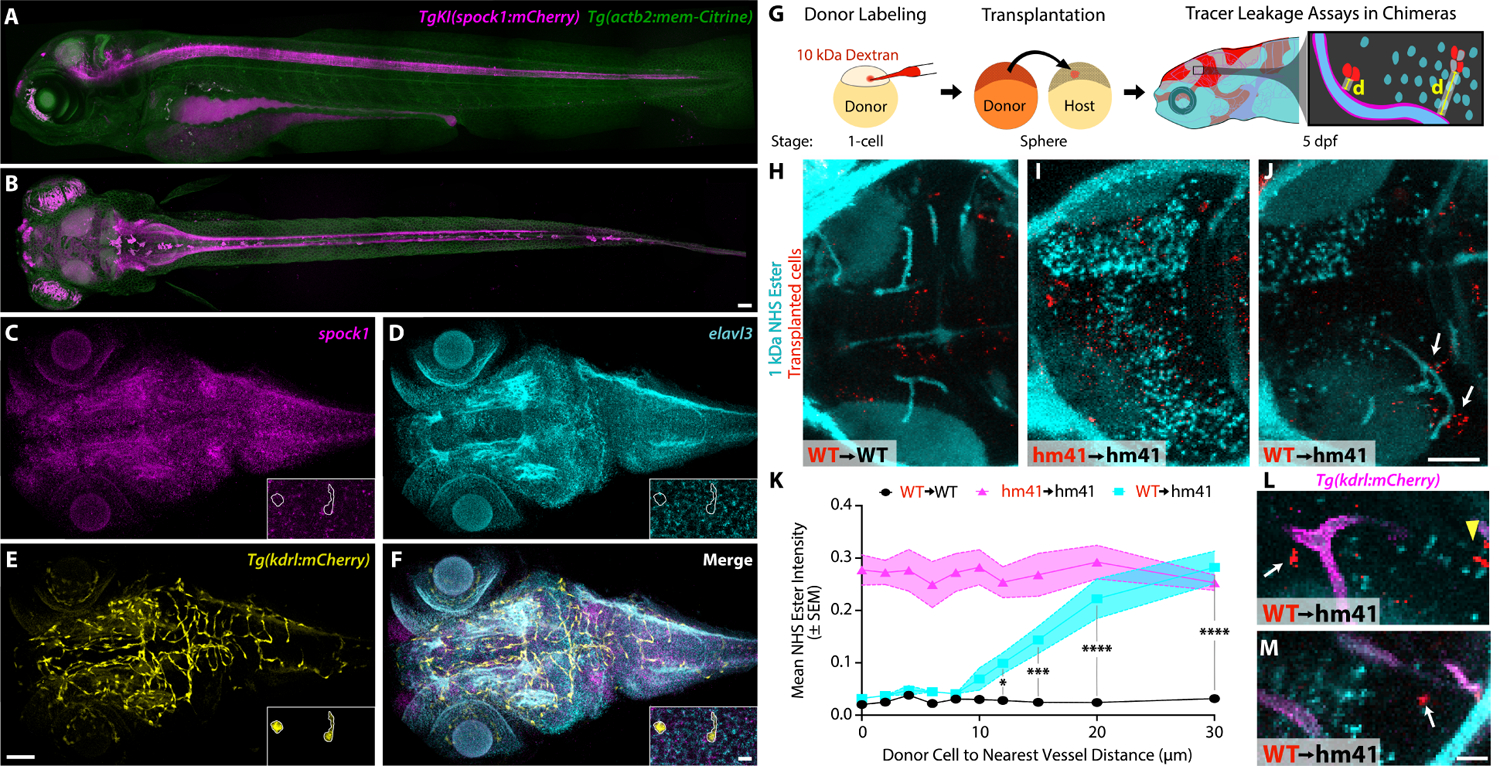
(A-B) Sagittal (A) and dorsal (B) maximum intensity projections (MIPs) of the TgKI(spock1:mCherry) (magenta) knock-in reveals expression of spock1 throughout the CNS, including the retina, brain and spinal cord with all cells in the fish labelled by Tg(actb2:memCitrine) (green). (C-F) Whole-mount HCR in situ hybridization for spock1 (C) confirms signal throughout the 5 dpf brain, in elavl3+ (D) neurons and absent from kdrl+ vasculature (E). Insets are zoomed in 10 μm thick MIPs from the midbrain region to further resolve the absence of spock1 signal in the vasculature (outlined in white). (G) Schematic of transplantation experiments. Cells from donor embryos labeled with 10 kDa Dextran (red) at the single cell stage are transplanted into unlabeled host embryos at sphere stage. Leakage of the injected 1 kDa AF 405 NHS Ester (turquoise) into the midbrain parenchyma is then measured in relationship to the distance (d) from the blood vessel to the nearest donor cell in the 5 dpf chimeric larvae. (H-J) Representative dorsal 20 μm thick MIP confocal image of a chimeric larva with transplanted wild type donor cells (red) into a wild type host (H, WT→WT), spock1hm41/hm41 donor cells into a spock1hm41/hm41 mutant host (I, hm41→hm41), and wild type donor cells into a spock1hm41/hm41 mutant host (J, WT→hm41). (K) Quantification of mean NHS leakage in WT→WT (black line, N=11) and hm41→hm41 (magenta line, N=8) reveals no change in tracer leakage in relationship to the nearest donor cell, with wild type fish confining the tracer and mutant fish leaking the tracer. However WT→hm41 (turquoise line, N=11) transplants reveal a full rescue of the leakage in the mutant background when the transplanted cell is within 10 μm of a blood vessel and no effect if the donor cell is further than 20 μm from the vessel. (L-M) Zoomed in images of WT→hm41 transplants. The white arrows point to instances of local rescue of tracer (turquoise) leakage when the wild type donor cell is close but not directly contacting the mutant vasculature (magenta). The yellow arrowhead marks wild type donor cells that fall outside the range of Spock1 signaling. Scale bars represent 50 μm (D and K) and 10 μm (G and N). * p=0.0495, *** p=0.0001, **** p<0.0001 by 2way ANOVA compared to WT→WT transplants.
To determine the range of the Spock1 signal, we performed cell transplantation experiments to make genetically mosaic zebrafish embryos and assessed tracer leakage in relationship to the closest transplanted cells at 5 dpf (Figure 2G). When we transplanted labeled wild type cells into wild type host embryos, we observed a negligible level of tracer accumulation in the brain parenchyma regardless of the proximity of the nearest donor cell (Figure 2H and 2K). In contrast, when we transplanted spock1hm41/hm41 mutant cells into mutant host embryos, we observed high levels of tracer accumulation in the parenchyma regardless of the proximity of the nearest donor cell (Figure 2I and 2K). Strikingly, when we transplanted wild type cells into spock1 mutant hosts, we observed a complete rescue of the mutant leakage if the wild type donor cell was within 10 μm of a blood vessel and no rescue if the donor cell was more than 20 μm away (Figure 2J–M), indicating that Spock1 can functionally signal at a distance of 10 to 20 μm. Importantly, wild type cells could rescue leakage in mutant hosts when they differentiate as neurons but not endothelial cells (Figure 2M).
Mutant leakage arises from increased transcytosis and altered pericyte-endothelial interactions
To assess the subcellular mechanism of increased BBB permeability in spock1hm41/hm41 mutants, we injected electron-dense NHS-gold nanoparticles (5 nm) into circulation, followed by transmission electron microscopy (TEM) imaging in 7 dpf spock1 mutant and wild type siblings. These TEM leakage assays revealed no alterations in the cellular composition of the neurovascular unit (NVU) in spock1 mutants, with endothelial cells and pericytes sharing a basement membrane that is surrounded by glia and neurons (Figure 3A and 3B). A modest impairment in tight junction function was observed in spock1 mutants (52/59 functional tight junctions; Figure 3D) compared to wild type siblings (57/57; Figure 3C). However, spock1 mutants had a significant increase in both small (<100 nm diameter) non-clathrin coated vesicles (Figure 3D and 3G) and large (>200 nm diameter) vesicles (Figure 3F and 3H) suggesting that the leakage results primarily from an increase in vesicular trafficking across endothelial cells. In addition to the increase in total large vesicular abundance in spock1 mutants, we also observed several of these large vesicles fused to the abluminal membrane in multiple larvae with a few examples of the gold nanoparticles visibly spilling into the endothelial basement membrane (Figure 3F). Although pericyte coverage of the endothelium was unaltered (Figure S1), spock1 mutants displayed overall thinner pericyte-endothelial basement membranes with several pericytes having long stretches of apparently direct contact on endothelial cells (Figure 3I–K), corresponding with observations by live confocal microscopy (Figure S1). These data suggest that loss of Spock1 function alters the critical pericyte extracellular interactions with endothelial cells, resulting in loss of BBB properties as observed in pericyte-deficient mice and zebrafish.16,17,38,39
Figure 3. Spock1 mutant leakage arises through increased endothelial vesicles.
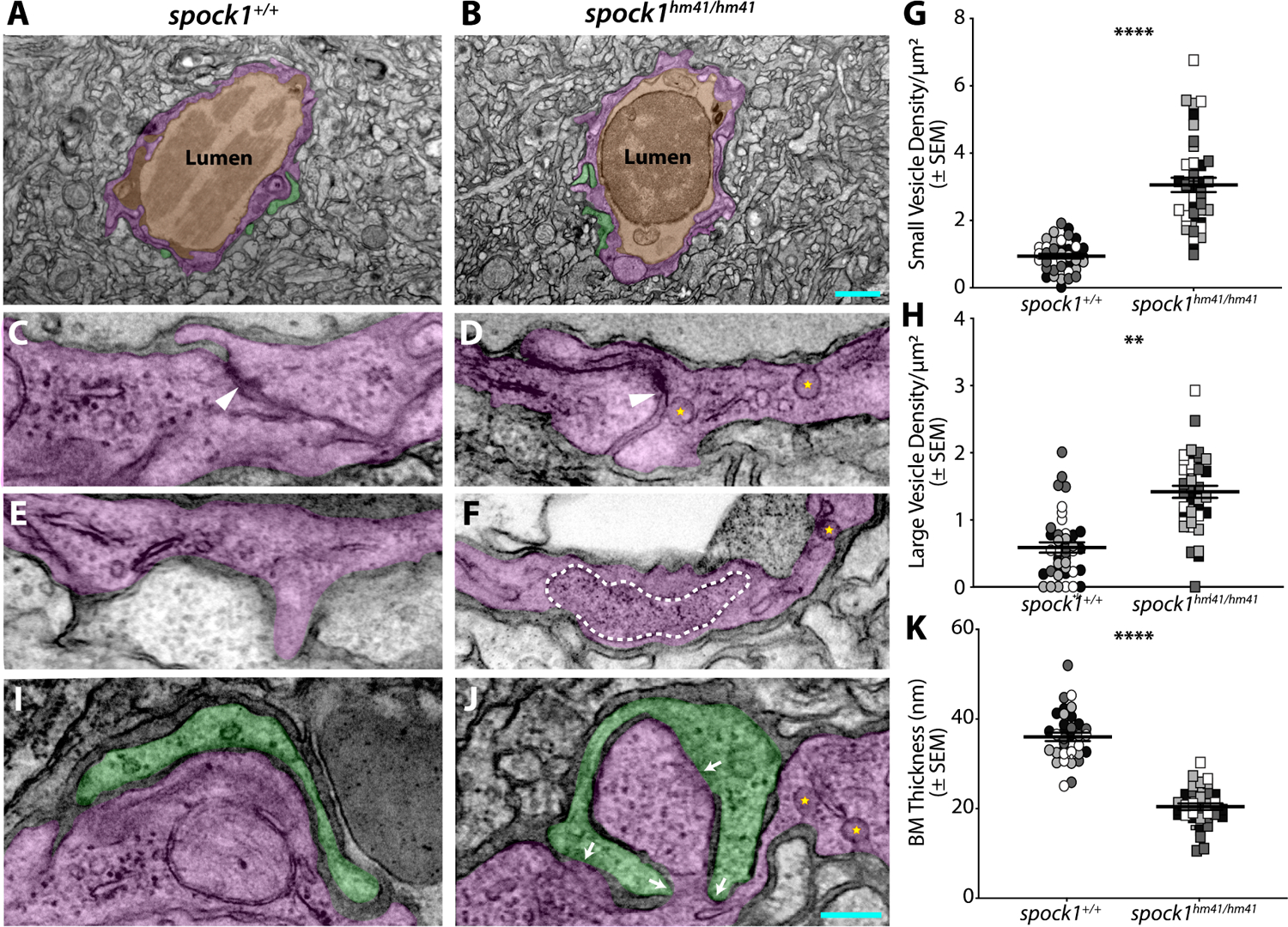
(A-B) The neurovascular unit remains intact in spock1hm41/hm41 mutants (B) with a continuous single layer of endothelial cells (pseudocolored magenta) enclosing the lumen (pseudocolored orange) and in close contact with pericytes (pseudocolored green). (C-H) The majority of tight junctions (white arrowheads) are functionally restrictive in the spock1hm41 mutant endothelial cells (88%). Mutant endothelial cells displayed a significant increase in vesicular density, including both small flask shaped vesicles (yellow stars, G) and larger vesicles greater than 200 nm in diameter (outlined by a white dashed line in F, H). (I-K) While pericyte coverage is unaltered in spock1hm41 mutants, the pericyte-endothelial cell interactions are altered in the mutants, with several instances of direct pericyte-endothelial cell contact (white arrows) and overall diminished average basement membrane thickness between the two vascular cells (K). Scale bars represent 1 μm (B) and 200 nm (J). N= 4 fish, each demarcated by a unique color, with 10 vessels analyzed per fish and shown as individual points. ** p=0.0029 (H), **** p<0.0001 by nested t test (G and K).
One of the proposed roles of Spock1 is to regulate matrix metalloproteinase (MMP) activation, specifically gelatinases MMP-2 and MMP-9.35,40–43 To assess whether spock1hm41/hm41 mutants had altered gelatinase activity that corresponded to observed changes in vascular cell interactions, we performed in vivo gelatin zymography by intracranial injections of highly quenched FITC-gelatin. Proteolytic cleavage of FITC-gelatin by either MMP-2 or −9 releases bright FITC peptides resulting in a non-reversible increase in fluorescence. This increase in fluorescence upon digestion is proportional to the proteolytic activity of the MMPs, allowing for the detection of gelatinase activity in vivo.44,45 Using this assay, we were able to see that in wild type 5 dpf fish, the majority of the signal strongly appears in the cerebral spinal fluid (CSF) and more subtly in many pericytes (Figure 4A). While spock1hm41/hm41 mutants also displayed strong gelatinase activity in the CSF, this was accompanied by a regional increase in gelatinase activity specifically in the forebrain and midbrain (Figure 4B and 4C), the same regions that were also prone to tracer accumulation (Figure 1). As MMPs are known to cleave many ECM substrates, we wondered which vascular ECM components were altered with the increased gelatinase activity in the spock1hm41/hm41 mutants. To investigate this, we performed immunofluorescence staining for ECM proteins with known roles in the vascular basement membrane, Fibronectin (Figure 4D–F) and Collagen IV (Figure 4G–I). Both proteins were present in the vascular basement membrane of wild type siblings, but at significantly diminished levels in the spock1hm41/hm41 mutants (Figure 4D–I). These data suggest that Spock1 helps induce barrier function by keeping gelatinase activity in check to allow proper deposition of necessary vascular basement membrane proteins.
Figure 4. The extracellular matrix is misregulated in spock1 mutants.
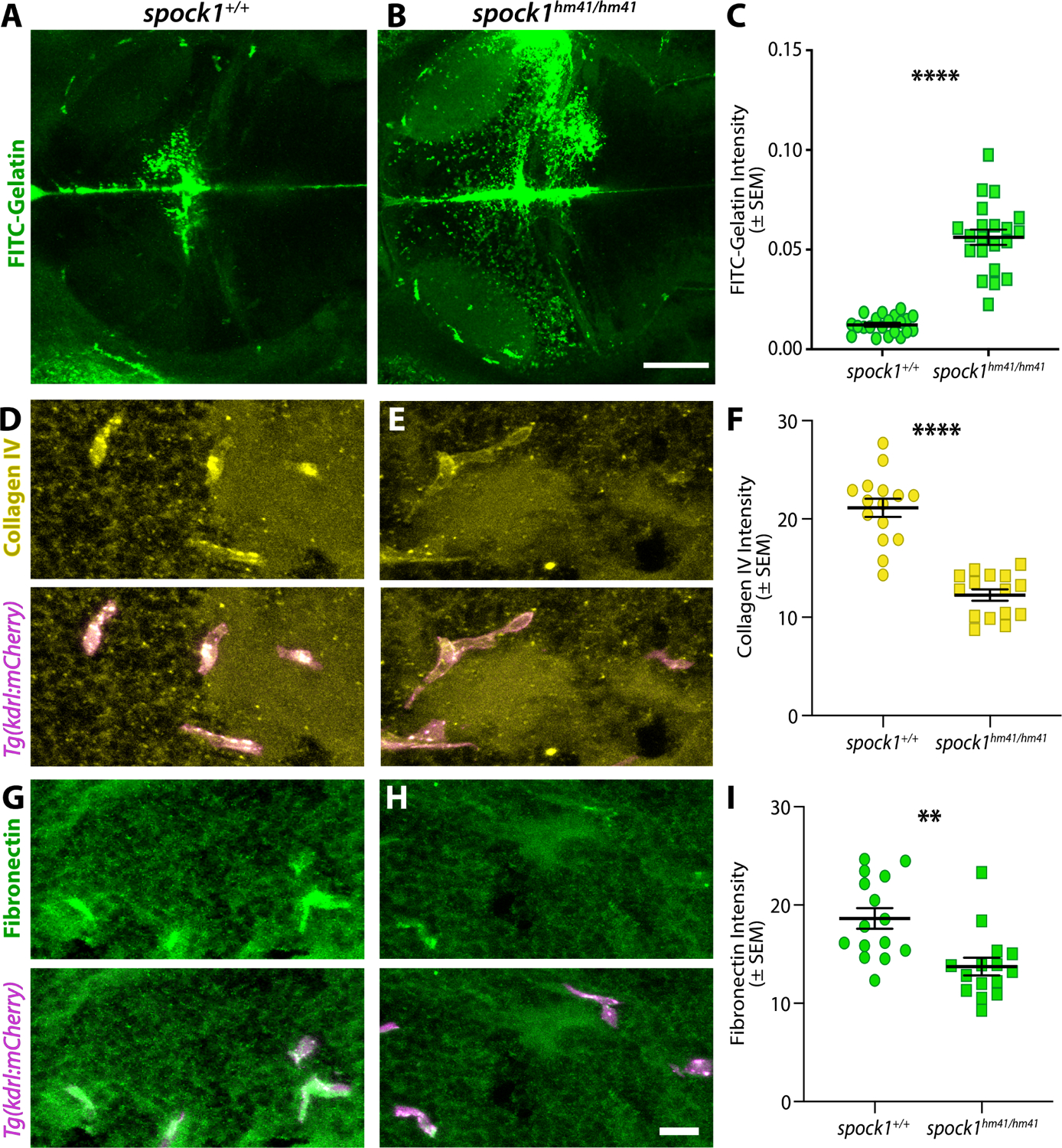
(A-C) In vivo gelatin zymography in wild type (A) and spock1hm41/hm41 mutants (B) reveals significantly increased gelatinase activity in the mutant midbrain compared to wild type siblings (C). (D-I) Immunofluorescence staining for vascular extracellular matrix proteins Collagen IV (yellow, D-F) and Fibronectin (green, G-I) that are required for vascular integrity reveal significantly reduced levels of all basement membrane proteins assayed within in the kdrl:mCherry labelled vasculature (magenta), quantified in F and I. N=21 (C) and 15 (F and I) fish analyzed for each genotype and depicted as individual points. Scale bars represent 50 μm (B) and 10 μm (H). ** p=0.0014, **** p<0.0001 by unpaired t test.
Spock1 plays a conserved role in inducing barrier function in mammals
To determine if Spock1 plays a conserved role in determining vertebrate BBB properties, we assessed BBB function in Spock1−/− mice46 on embryonic day 15.5 (E15.5),46 a stage when the cortex BBB is fully functional4 (Figure 5A). While wild type siblings confined both 550 Da Sulfo-NHS-Biotin (Figure 5B) and 10 kDa Dextran (Figure 5D) within the brain vasculature, Spock1−/− mice leaked both tracers into the cortex parenchyma (Figure 5C–G). This increased BBB permeability was also associated with increased expression of PLVAP (Figure 5H–L), as seen in the zebrafish mutants (Figure 1N). These data indicate that Spock1 plays a conserved role in inducing barrier properties during embryonic development. However, when we repeated these leakage assays in adult mice, we observed full recovery in BBB function in Spock1−/− mice (Figure S7). This contrasts with adult zebrafish spock1hm41/hm41 mutants, which have a defective BBB (Figure 1G), indicating species-specific differences in the adult BBB.
Figur 5. Spock1 plays a conserved role in inducing barrier function during embryonic development in mice.
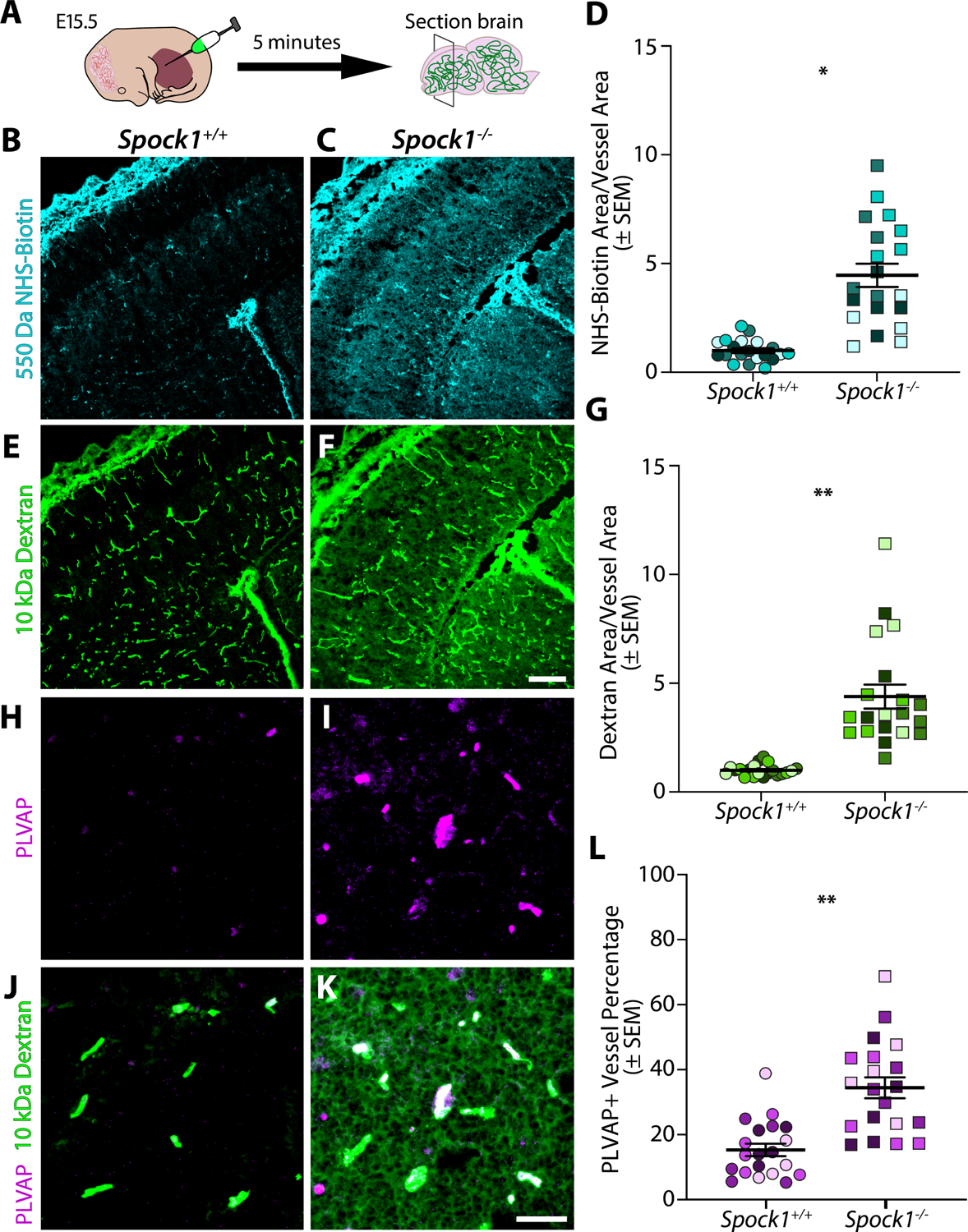
(A) Schematic of functional tracer leakage assays in embryonic day 15.5 (E15.5) mice. (B-G) Wild type mice confine both injected 550 Da NHS-Biotin (B) and 10 kDa Dextran (D) within the vasculature, as previously reported. However, Spock1−/− mice leak both NHS-Biotin (C) and Dextran (E) into the brain parenchyma. Quantification of the total area of NHS-Biotin leakage (D) and Dextran leakage (G) normalized to vessel area where a ratio of 1 indicates no leakage reveals a significant increase in extravasation of both tracers in the Spock1−/−embryos (p=0.0191 (F) and p=0.0048 (G) by nested t test). (H-L) Zoomed in view of 10 kDa Dextran (green) injected embryos immunostained for the leaky vessel marker PLVAP (magenta) in wild type (H,J) reveals that the increased BBB permeability in Spock1−/− mice is accompanied by an increase in PLVAP expression in the vasculature (I,K). Quantification of PLVAP expression within the vasculature (L) reveals a significant increase in PLVAP expression in the Spock1−/− embryos (p=0.0016 by nested t test). N=4 embryos for each genotype, marked by unique colors, with 5 sections analyzed per embryo and shown as individual points. Scale bars represent 100 μm (F) and 50 μm (K).
Vascular cells are the only altered cell types in spock1 mutant brains
To determine how Spock1, a neuronally produced and secreted proteoglycan, signals to and regulates brain endothelial cell BBB properties, we turned to scRNA-seq of dissected 5 dpf spock1hm41/hm41 mutant and wild type brains, allowing us to globally assess cell type specific molecular changes occurring in the mutant brains. With Leiden clustering of the scRNA-seq data we defined cell clusters containing neuronal, glial, and vascular cell types (Figure S6). We performed differential gene expression (DGE) analysis for each cluster and did not observe any changes in the neuronal or glial populations, but did observe significant changes in cluster 13 (Table S2). When we subclustered cluster 13, we were able to resolve endothelial cells, pericytes and vascular smooth muscle cells (vSMCs) in addition to other cells in the brain, like the meninges, or contaminating pharyngeal arch remnants (Figure 6A). Due to the low vascular cell numbers present in the scRNA-seq data, we prioritized candidate genes that could potentially be involved in spock1 signaling to the vasculature for subsequent validation by their fold-change in gene expression rather than statistical significance. These analyses suggested molecular changes in the mutant endothelial cells indicative of a leaky phenotype, with increased expression of plvapb, which we already observed in both the mutant fish (Figure 1N–O) and embryonic knockout mice (Figure 5H–L), and decreased expression of the tight junction protein cldn5b (Figure 6B, Table S3). Interestingly, spock1 mutants had decreased expression of the melanoma cell adhesion molecule mcamb (also known as CD146) in both endothelial cells and pericytes (Figure 6B and 6C), as well as a decrease in pericyte expression of the forkhead transcription factor foxc1b (Figure 6C). This decreased expression of mcamb in the vasculature was validated by HCR FISH, with significantly reduced expression in mutant midbrain endothelial cells and pericytes but normal levels in hindbrain vascular cells, which maintain BBB function in spock1hm41/hm41 mutants (Figure 6D–G). CD146 has previously been shown to be required for BBB integrity, as CD146−/− mice exhibit BBB breakdown due to decreased pericyte coverage and downstream loss of endothelial expression of Cldn5.47 Pericyte expression of foxc1b was also down-regulated in the spock1hm41/hm41 mutant midbrain (Figure 6H–K). Pericyte specific loss of Foxc1 in mice also results in increased BBB leakage and micro-hemorrhage.48 Taken together, these results suggest that loss of Spock1 signaling alters pericyte identity without altering coverage.
Figure 6. Spock1hm41/hm41 mutant vascular cells have reduced expression of BBB regulators.
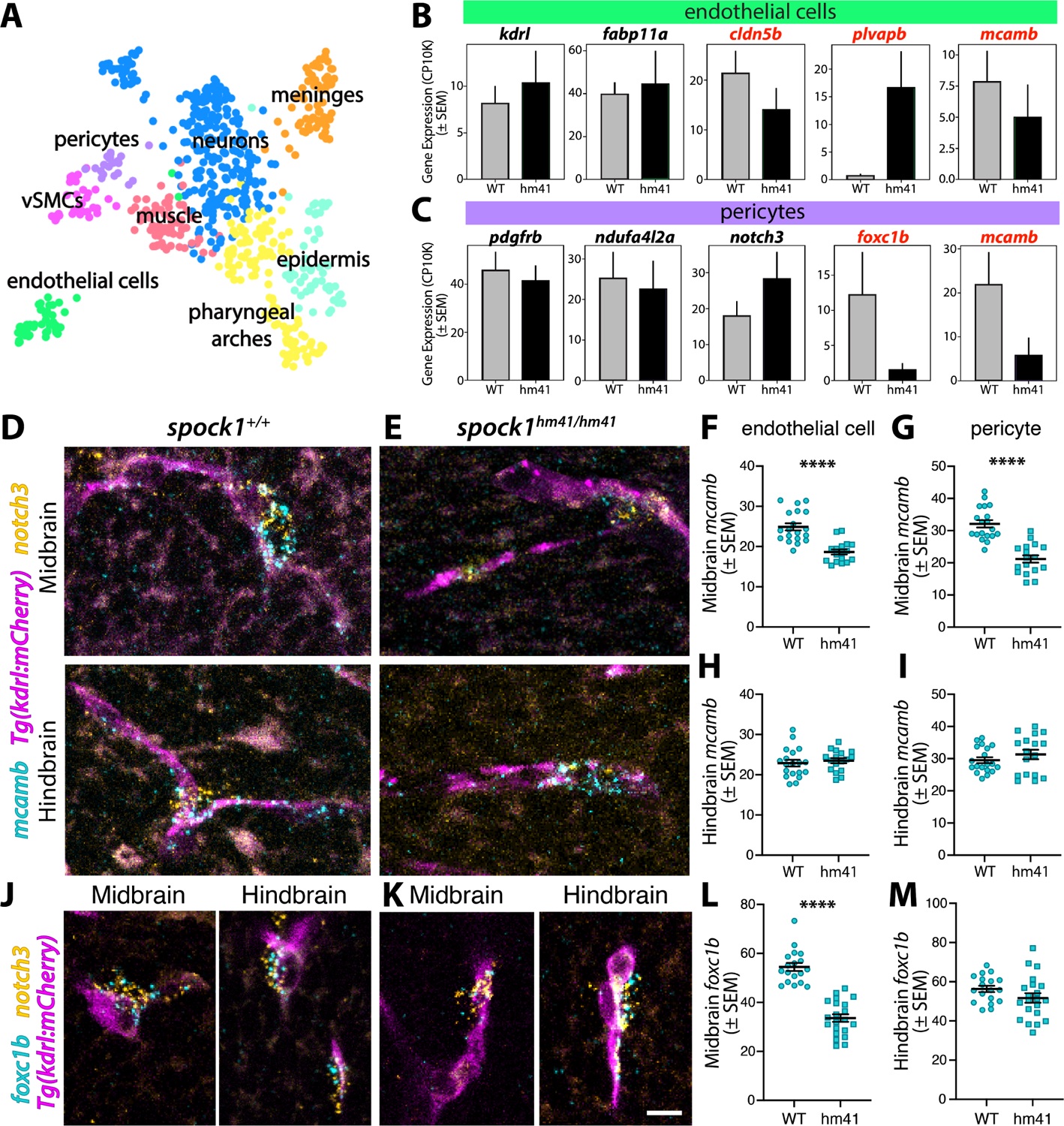
(A) UMAP of Leiden cluster 13 following subclustering and annotated by cell type, with pericytes (purple) separating from vascular smooth muscle cells (vSMCs, pink) and endothelial cells (green). (B-C) Mean gene expression in wild type (WT, grey bars) and spock1 mutant (hm41, black bars) endothelial cells (B) and pericytes (C). Error bars represent SEM. Mutants appear to have lower levels of mcamb in both pericytes and endothelial cells and reduced foxc1b expression in pericytes. (D-I) HCR FISH reveals strong expression of mcamb (turquoise) in wild type vessels (magenta) and notch3+ pericytes (orange, D), both in the midbrain (F-G) and hindbrain (H-I). Spock1 mutants have significantly reduced expression of mcamb in the midbrain (E-G), but normal levels in the hindbrain (H-I), where no leakage is observed. (J-M) HCR FISH reveals expression of foxc1b (turquoise) in wild type notch3+ pericytes (orange, J), both in the midbrain (L) and hindbrain (M). Spock1 mutants have significantly reduced expression of foxc1b (K) in the midbrain (L) but not in the hindbrain (H). Scale bar represents 10 μm. **** p<0.0001 by unpaired t test.
Exogenous SPOCK1 restores barrier function
To test the ability of Spock1 to rescue the mutant leakage phenotype, we injected recombinant human SPOCK1 (rSPOCK1) protein directly into the brain of spock1hm41/hm41 mutant larvae at 5 dpf and assessed brain permeability at 6 dpf. While control animals injected with PBS maintained high levels of brain permeability (Figure 7A), larvae that received at least 2.3 ng of rSPOCK1 per mg fish body weight showed a 50% reduction in brain permeability following a single dose (Figure 7B and 7C). These results further confirm that SPOCK1 is able to act cell non-autonomously to rescue the mutant BBB leakage, as these injections were targeted broadly to the neural tissue rather than the endothelial cells, and demonstrate functional conservation of Spock1 from human to zebrafish.
Figure 7. Spock1 induces BBB properties by modulating the brain microenvironment, altering vascular cell biology non-autonomously.
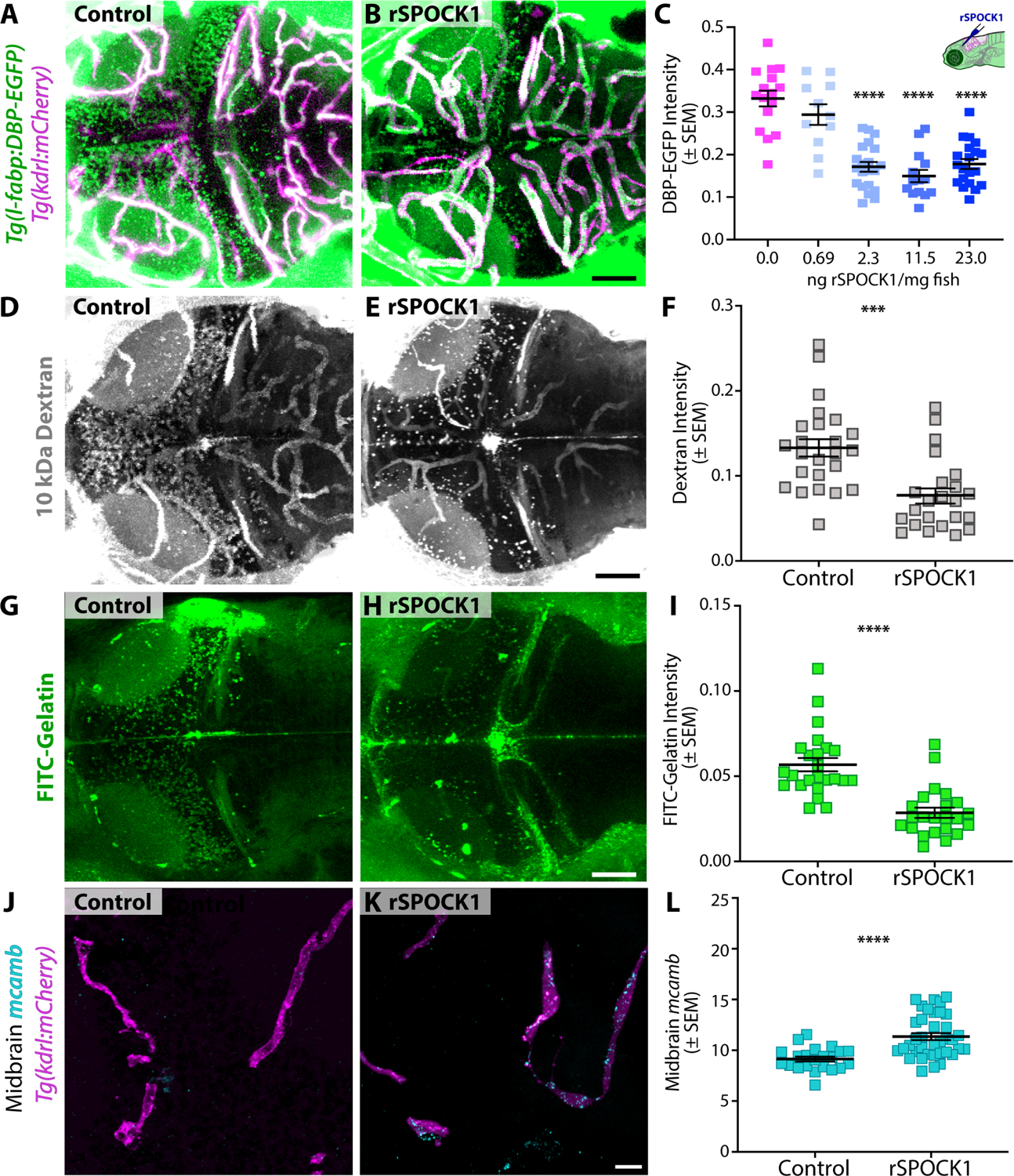
(A-C) A single intracranial injection of human rSPOCK1 into the brain at 5 dpf reduces mutant leakage of DBP-EGFP at 6 dpf about 50% (B) compared to controls injected with PBS alone (A), quantified in C. (D-F) Similarly, injection of rSPOCK1 at 4 dpf reduces mutant leakage of 10 kDa Dextran at 5 dpf (E), quantified in F. (F-I) The addition of rSPOCK1 reduces gelatinase activity in the mutant brain (H) compared to control injected mutants (F), quantified in I. (J-L) Vascular expression of mcamb in the midbrain is partially restored following rSPOCK1 intracranial injections (K), quantified in L. Scale bars represent 50 μm (C, E, F) and 10 μm (K). **** p<0.0001 by 2way ANOVA compared to PBS injected control spock1hm41/hm41 mutants in C and by t test in I and L, *** p<0.001 by t test in F.
To determine whether this restoration of barrier function corresponds to Spock1’s regulation of the MMP activity, we repeated the intracranial injections of 11.5 ng rSPOCK1/mg of fish in spock1hm41/hm41 mutants at 4 dpf and assessed barrier function and gelatinase activity at 5 dpf. While 5 dpf control injected spock1hm41/hm41 mutants continued to display high levels of BBB permeability and gelatinase activity, rSPOCK1 reduced mutant leakage of 10 kDa Dextran about 50% (Figure 7E and 7F), just as it did when we injected rSPOCK1 at 5 dpf and assessed BBB function at 6 dpf (Figure 7C). This restoration of BBB function was accompanied by a decrease in gelatinase activity (Figure 7H and 7I). Finally, to determine whether Spock1 lies upstream of vascular expression of the cell adhesion molecule mcamb, we assessed mcamb expression in the mutant midbrain vasculature at 5 dpf with or without the addition of rSPOCK1. While spock1hm41/hm41 mutants injected with PBS displayed negligible levels of mcamb expression in the midbrain (Figure 7J), mutants injected with rSPOCK1 had a 50% increase in mcamb expression in the vasculature (Figure 7K and 7L), with a noticeable increase in mcamb+ pericytes. Interestingly this 50% increase in mcamb expression directly correlates with the 50% reduction in leakage and gelatinase activity following rSPOCK1 injections (Figure 7F and 7I).
DISCUSSION
Together these data show that Spock1 is a secreted, neuronally expressed extracellular signal that regulates BBB permeability in zebrafish and mouse without altering vascular patterning during development (Figures 1 and 5). Mechanistically, this work suggests a model whereby Spock1 regulates the brain extracellular environment via its inhibition of gelatinase activity, thereby providing the appropriate chemical or mechanical signals for vascular pericytes and endothelial cells to communicate properly with one another. This critical pericyte-endothelial interaction thereby prompts a BBB transcriptional program, including increased expression of tight junction (cldn5b) and cell adhesion (mcamb) molecules, decreased expression of leaky endothelial genes (plvapb), and reduced transcytosis. While these vascular gene expression changes are reminiscent of those observed in Wnt mutants,20,49 we did not observe any alterations in Wnt signaling in our spock1 mutants (Figure S1). Thus, loss of Spock1 function contributes to increased BBB permeability by disruption of the complex extracellular scaffold and cell-cell interactions required for brain vasculature during development in a Wnt-independent manner. Together this work reveals how a secreted signal from the brain microenvironment can regulate the vasculature to give rise to the special properties of the BBB and provides targets for its therapeutic modulation.
Despite the expression of spock1 throughout the CNS, spock1 loss of function zebrafish mutants display regional leakage in the forebrain and midbrain and a completely intact hindbrain barrier. Spock1 is not alone in having region-specific regulation of barrier properties despite pan-CNS expression. A recent study demonstrated that the pericyte-produced ECM molecule vitronectin is required for functional retinal and cerebellar barriers despite its expression by all CNS pericytes.50 Even mutants in canonical Wnt signaling have varying levels or regionalized angiogenic or barrier defects despite wide-spread sources of Wnt signals and receptors throughout the brain.23,25,51–55 Interestingly, the regions with the highest susceptibility to single gene mutations in canonical Wnt signaling are the retina and the cerebellum, while the cortex can maintain a functional BBB even with mutations in several genes in the Wnt pathway.25 These data suggest that different regions of the brain are either molecularly or cellularly (or both) distinct, perhaps in nuanced ways that are often masked by current detection methods. For example, while the neurovascular unit of endothelial cells and pericytes in close contact with astrocytic endfeet appears uniform throughout the brain by immunostaining and EM, various scRNA-seq datasets have now shown that astrocytes are highly regionally distinct at a molecular level56–58 raising the possibility that they may respond to different signals. Pericytes in the brain, unlike in the rest of the body, arise from two discrete origins, mesodermal and neural crest.59,60 In zebrafish, the two origins have discrete domains, with the hindbrain being exclusively derived from mesodermal origins and the midbrain and forebrain being predominantly of neural crest origin,59 which interestingly matches the leakage boundaries in spock1 mutants.
Here we demonstrate that Spock1 regulates brain endothelial cell identity to initiate the barrier program during development, both at molecular and subcellular resolution. However, this does not preclude the fact that other brain barriers may be similarly affected by loss of this signal. In addition to the BBB, the neural tissue is also protected by the blood-cerebrospinal fluid (CSF) barrier, which is created by the choroid plexus epithelial cells lining the brain ventricles,61 and the arachnoid barrier, which is created by the epithelial cells beneath the dura that completely enwrap the brain.62 These various brain barriers mature on different developmental timelines, with the blood-CSF barrier becoming impermeable by 4 dpf,63 then the BBB by 5 dpf,28 and the arachnoid barrier maturing after 9 dpf32 in zebrafish. While the focus of our study has been the BBB, it is possible that these additional brain barriers also play a role in the observed leakage of various tracers into the brain in spock1 mutants. In fact, our scRNA-seq data suggest that the meninges of spock1hm41/hm41 mutants may also be impaired with decreased expression of the tight junction cldn11a (Table S3), which has recently been shown to be essential for arachnoid barrier functional development.64
The N-terminal region of Spock1 has previously been shown to regulate matrix metalloproteinase (MMP) activity, specifically gelatinases MMP-2 and −9.40 We show here that leaky spock1hm41/hm41 mutants have increased gelatinase activity in the midbrain occurring in the same place as the observed decrease in the endothelial-pericyte basement membrane (Figure 4 and S1), suggesting that this excessive MMP activity may be responsible for the breakdown of the basement membrane. Although all the cells in the neurovascular unit secrete MMP-2 and −9, pericytes have recently been shown to produce MMP-9 in response to ischemia and TGF-β1 exposure, leading to increased barrier breakdown and neuronal damage.44,65 For this reason, many have proposed gelatinases as a therapeutic target for several neurological diseases that are accompanied with barrier breakdown including stroke, multiple sclerosis, Alzheimer’s diseases, and cerebral hemorrhage.44,66–68 Given the ability of a single dose of rSPOCK1 to mitigate much of the leakage in the mutants and restore proper vascular expression of BBB genes, we propose that Spock1 may serve as a therapeutic target to reduce gelatinase activity in these neurological diseases and help restore barrier function.
Limitations of the Study
While we have thoroughly characterized where spock1 mRNA is expressed, we do not have an antibody that reliably labels Spock1 protein expression in zebrafish. This prevented us from following up further on identifying Spock1 binding partners. Furthermore, we turned to scRNA-seq to identify the mechanism of how Spock1 regulates BBB properties in an unbiased fashion and this revealed specific changes in vascular cells and no other brain cell types. When we subclustered this cluster, we were left with only a handful of endothelial cells and pericytes, and therefore focused on genes with large changes in gene expression, like mcamb and foxc1b. However, our list of differentially expressed genes in these vascular cells is not necessarily exhaustive, and could be further supplemented by sorting for cell types of interest. While there are point mutations, including one missense mutation, in hm41 mutants, it is not clear how these mutations abrogate Spock1 function.
STAR Methods
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Natasha O’Brown (natasha.obrown@rutgers.edu)
Materials availability
Zebrafish lines and plasmids generated in this study will be shared by the lead contact upon request.
Data and code availability
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available (GSE230236). All original code for the single-cell analysis is available as interactive Jupyter notebooks here: https://doi.org/10.5281/zenodo.7955159. Interactive exploration of the full single-cell data can be found here: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/2021_OBrown/2021_OBrown. Exploration of the cluster 13 subcluster cells can be found here: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/2021_OBrown_Vasculature/2021_OBrown_Vasculature. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Experimental model and study participant details
Zebrafish Strains and Maintenance
Zebrafish were maintained at 28.5°C following standard protocols.69 All zebrafish work was approved by the Harvard Medical Area Standing Committee on Animals under protocol number IS00001263–3. Adult fish were maintained on a standard light-dark cycle from 8 am to 11 pm and fed a mix of live artemia and tropical fish granules. Adult fish, age 3 months to 2 years, were crossed to produce embryos and larvae. For imaging live larvae, 0.003% phenylthiourea (PTU) was used beginning at 1 dpf to inhibit melanin production. These studies used the AB wild-type strains and the transgenic reporter strains Tg(l-fabp:DBP-EGFP)Iri500Tg abbreviated as Tg(l-fabp:DBP-EGFP),29 (Tg(kdrl:HRAS-mCherry)s896Tg abbreviated as Tg(kdrl:mCherry),70 TgBAC(pdgfrb:EGFP)ncv22Tg abbreviated as TgBAC(pdgfrb:EGFP),59 Tg(plvapb:EGFP)sj3Tg abbreviated as Tg(plvapb:EGFP),31 Tg(actb2:mem-citrine-citrine)hm30 abbreviated as Tg(actb2:mem-Citrine),71 Tg2(elavl3:GCaMP6s)a13203Tg abbreviated as Tg(elavl3:GCaMP6s),72 and Tg(−3.1neurog1:GFP)sb2Tg abbreviated as Tg(−3.1neurog1:GFP)73 in the text.
Spock1hm41 Mutants
Spock1hm41 mutants were maintained in the double transgenic Tg(l-fabp:DBP-EGFP; kdrl:mCherry) background. Heterozygous fish were intercrossed for all leakage assays, with the exception of the time lapse microscopy experiments, where a heterozygous fish was crossed to a homozygous mutant, and the cell transplantation experiments, where homozygous mutants were in-crossed. All larvae were imaged prior to genotyping to identify wild type and mutant fish. The spock1hm41 mutant line was genotyped using primers 1 and 2 (See Table S4) followed by a HaeIII restriction digest, which does not digest the wild-type product (327 bp).
Spock1hm43 Mutants
Spock1hm43 mutants were generated and maintained in the double transgenic Tg(plvapb:EGFP; kdrl:mCherry) background using CRISPR mutagenesis with two sgRNAs (spock1_1 and spock1_2, Table S4) to target the spock1 5’ UTR and start codon. Heterozygous fish were intercrossed for all leakage assays. All larvae were imaged prior to genotyping to identify wild type and mutant fish. The spock1hm43 mutant line was genotyped using primers 3 and 4 (Table S4) and KAPA HiFi Hotstart polymerase (Roche:KK2602) using a 58°C annealing temperature. The mutant PCR product of 364 bp was easily distinguishable by gel electrophoresis from the wild type product at 935 bp.
TgKI(Spock1-mCherry)hm44 Knock-ins
TgKI(Spock1-mCherry)hm44 knock-ins were generated and maintained in the AB background by injecting Cas9 mRNA (TriLink Biotechnologies; 100 ng/μl), crRNA:tracrRNA duplexes to target the first exon of spock1 (spock1_KI, Table S4) and the universal gRNA site in the donor plasmid (ugRNA, Table S4) (IDT DNA Technologies; 25 μM), and a donor plasmid with 30 bp homology arms to spock1 exon 1 (spock1-mCherry-KI; 25 ng/μl). F0 injected larvae with mCherry expression were raised to adulthood and outcrossed to find a founder with germ-line transmission as assessed by mCherry fluorescence. These F1 fish were then raised and outcrossed to other transgenic cell-type reporter lines as heterozygotes for the assessment of spock1 expression in vivo. Genomic integration in the correct locus was validated with PCR on genomic DNA using primers 5 and 6 (Table S4) followed by sequencing with primer 5.
Mouse Maintenance
Spock1−/− knockout mice46 were obtained from Ursula Hartmann’s lab and were backcrossed to and maintained on a C57Bl/6 background. Mice were maintained on 12 light/12 dark cycle and given LabDiet 5053 and water ad libitum. Adult mice were housed with a maximum of 5 mice per cage. Mice were genotyped with primers 7–9 (Table S4) using KAPA HiFi Hotstart polymerase (Roche:KK2602). This PCR reaction produced a 185 bp wild type band and a 750 bp knockout band in heterozygous animals. All animals were treated according to institutional and US National Institutes of Health (NIH) guidelines approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School under protocol IS00000045–6.
Method Details
Fluorescent Zebrafish Tracer Injections and Live Imaging
Larvae were immobilized with tricaine and placed in an agarose injection mold with their hearts facing upwards. 2.3 nl of Alexa Fluor 405 NHS Ester (Thermo Fisher: A30000) or Alexa Fluor 647 10 kDa Dextran (Thermo Fisher: D22914) fluorescently conjugated tracers (10 mg/ml) were injected into the cardiac sac using Nanoject II (Drummond Scientific, Broomall, PA). Larvae were then mounted with 1.5% low gelling agarose (Sigma: A9414) in embryo water on 0.17 mm coverslips and imaged live within 2 hours post injection on a Leica SP8 laser scanning confocal microscope using the same acquisition settings with 1 μm z-steps using a 25x water immersion objective with 0.75x magnification. Using these settings, we were able to image the entire brain of each fish. Pericyte and vascular double transgenics were similarly imaged on the Leica SP8 for quantification of vascular defects. Additional images were taken on a Zeiss LSM 980 with Airyscan 2 with 0.21 μm z-steps using a 20x water immersion objective for increased cellular resolution, allowing us to detect gaps between the two cell types in vivo.
Linkage Mapping
Larvae from 2 separate crosses were screened for leakage of DBP-EGFP at 5 dpf and pooled into 3 groups of 5 to 6 leaky or wild type heads. RNA was extracted from the pools using the RNeasy mini kit and ribo-depleted. RNA sequencing libraries were prepared using Wafergen Directional RNA-Seq kits and sequenced on NextSeq High-Output sequencers producing 75 bp paired end reads. Reads were mapped to the GRCz11 genome using tophat and bowtie2. Linkage mapping was performed on the mapped reads using RNAmapper33 with the following specifications: zygosity=25, coverage=1, linkedRatio=0.96, neighbors=10. Differential gene expression analysis was performed on these libraries using rsem.74
CRISPR Mutants
Gstp2 crispant fish were generated by injection of Cas9 protein and 3 guide RNAs (gstp2_1–3; Table S4) into 1-cell fertilized double transgenic Tg(l-fabp:DBP-EGFP; kdrl:mCherry) embryos. Csf1ra crispant fish were generated similarly with 4 guide RNAs (csf1ra_1–4; Table S4), as were spock1 crispants (spock1_3–6; Table S4). F0 crispants were analyzed for leakage of the DBP-EGFP tracer outside of the kdrl:mCherry labeled vasculature at 5 dpf.
HCR Fluorescent In Situ Hybridization (FISH)
Slide:
HCR RNA in situ hybridization (Molecular Instruments) experiments on 14 μm cryosections of fixed 5 dpf larvae were performed as previously described.28,75 Briefly, sections were air dried and re-fixed in 4% paraformaldehyde (PFA) in PBS for 10 minutes at room temperature. Following fixation, slides were washed in PBS and then permeabilized using 1 μg/ml Proteinase K (ThermoFisher) for 5 minutes, followed by PBS washes and refixation. Tissues were further permeabilized by an ethanol dehydration series of 50%, 70% and two rounds of 100% ethanol and air dried for 5 minutes. Dried slides were put into probe hybridization solution for at least 10 minutes at 37°C. Subsequently, the probes for spock1and mcamb (Table S4, IDT DNA oPools) were added to the slides at a final concentration of 4 nM to hybridize overnight at 37°C. The next day, slides were washed with a series of wash buffer to 5x SSCT (5x SSC with 0.1% Tween 20). Excess liquid was then removed and samples were immersed in amplification buffer at room temperature for 30 minutes prior to hairpin amplification, which occurred overnight at room temperature. Slides were then washed in 5x SSCT, washed in 5x SSC and mounted with Fluoromount-G (Electron Microscopy Sciences).
Whole mount:
Larvae were fixed in 4% PFA overnight and then stored in methanol at −20°C until staining was performed and within 6 months post fixation. All staining was performed in PCR strip tubes. Larvae were rehydrated in a series of methanol/PBST (0.1% Tween) washes and then permeabilized with 30 μg/ml Proteinase K for 45 minutes at room temperature. Larvae were then refixed in freshly made 4% PFA for 20 minutes and treated with probe hybridization buffer at 37°C. Subsequently, the spock1, elavl3, mcamb, notch3, foxc1b, slc17a6b, or gad1b probes (Table S4, IDT DNA oPools and Molecular Instruments) were added at a final concentration of 4 nM to hybridize 24–72 hours rocking at 37°C. The next day, larvae were washed with a series of wash buffer to 5x SSCT (5x SSC with 0.1% Tween 20). Excess liquid was then removed and samples were immersed in amplification buffer at room temperature for 30 minutes prior to hairpin amplification, which occurred overnight rocking at room temperature at concentration of 15 pmol. Larvae were then washed and imaged in 5x SSCT.
Transplantation
Donor embryos were injected with 2.3 nl of 10 mg/ml Alexa Fluor 647 10 kDa Dextran (Thermo Fisher: D22914) at the 1-cell stage to distinguish them from host cells. Following injection, embryos were incubated at 28.5°C until transplantation. Host and donor embryos were dechorionated with 1 mg/ml Pronase (Roche:11459643001) at oblong stage and transferred to transplantation agarose dishes in 1/3 Ringer’s buffer. Unfertilized or injured embryos were discarded. To generate clonal sources secreting wild type or mutant Spock1, approximately 40–80 cells were transplanted from sphere stage dextran labeled donor embryos into sphere stage wild type and mutant hosts (similar to previous studies).76 Embryos recovered overnight in 1/3 Ringer’s buffer and were subsequently transferred to Danieau Buffer with PTU. Tracer injections were performed at 5 dpf, as described above, with 1 kDa Alexa Flour 405 NHS Ester.
Transmission Electron Microscopy (TEM)
Larvae (7 dpf) were anesthetized with tricaine and injected with 2.3 nl of 5 nm NHS-activated gold nanoparticles (Cytodiagnostics: CGN5K-5–1, ~1.114 particles/ml in PBS) just as for the fluorescent tracer injections. After 5 minutes of circulation, the larvae were initially fixed by immersion in 4% paraformaldehyde (VWR:15713-S) /0.1M sodium cacodylate (VWR:11653). Following this initial fixation, larvae were further fixed for 7 days in 2% glutaraldehyde (Electron Microscopy Sciences: 16320)/ 4% paraformaldehyde/ 0.1M sodium cacodylate at room temperature. Following fixation, larvae were washed overnight in 0.1M sodium cacodylate. Entire larval heads were post-fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide, dehydrated, and embedded in epoxy resin. Ultrathin sections of 80 nm were then cut from the block surface and collected on copper grids.
Intracranial rSPOCK1 Injections
Larvae were anesthetized with tricaine and placed in an agarose injection mold with their heads facing upwards. 2.3 nl of recombinant human SPOCK1 (rSPOCK1; R&D Systems: 2327-PI-050) at a range of 0.001–10 mg/ml was injected directly into the midbrain using Nanoject II (Drummond Scientific, Broomall, PA). Following injections, larvae recovered in normal embryo water for 1 day. The effects of rSPOCK1 were either assessed in live fish using functional tracer leakage assays and in vivo gelatin zymography or in fixed samples followed by slide HCR in situ hybridization, as described above.
In vivo Gelatinase Zymography
Larvae were anesthetized and intracranially injected as for rSPOCK1 with 2.3 nl of FITC-Gelatin (Biovision: M1303) at 5 mg/ml. After one hour, larvae were also injected with 10 kDa Dextran in the cardiac sac to assess BBB function and then mounted with 1.5% low gelling agarose (Sigma: A9414) in embryo water on 0.17 mm coverslips and imaged live within 2 hours post injection on a Leica SP8 laser scanning confocal microscope using the same acquisition settings with 1 μm z-steps and a 25x water immersion objective.
Zebrafish Immunofluorescence Staining
Immunofluorescence staining was performed on 14 μm thick cryosections of paraformaldehyde (4% in PBS) fixed 5 dpf wild type and mutant larvae. Slides were air dried for at least 20 minutes at room temperature and then washed twice with PBS. Sections were then blocked in 10% normal goat serum (NGS) in PBST (0.1% Tween) for 2 hours at room temperature. Antibodies against Fibronectin (Abcam: ab6328) and Collagen IV (Abcam: ab6586) were used at 1:200 in the same blocking solution and applied overnight at 4°C. Sections were then washed in PBST several times followed by treatment with fluorescently-conjugated secondary antibodies (Jackson Immunoresearch) at 1:500 in the same blocking buffer at room temperature.
Mouse Tracer Injections and Immunofluorescence Staining
Heterozygous Spock1+/− mice were intercrossed and used for tracer leakage assays performed at embryonic day 15.5 (E15.5) as previously described4. In brief, 5 μl of tracer cocktail (10 mg/ml EZ-Link NHS-Biotin (Thermo Fisher: 20217) and 5 mg/ml 10 kDa Dextran Alexa Fluor 488 (Thermo Fisher: D22910)) was injected into the liver of each embryo and allowed to circulate for 5 minutes. Embryonic heads were fixed by immersion in 4% paraformaldehyde overnight at 4°C and then frozen in TissueTek OCT (Sakura). 20 μm thick sections were then collected and immunostained with Streptavidin Alexa Fluor 405 (1:200; Thermo Fisher: S32351) and rat anti-PLVAP (1:200; BD Biosciences:553849). All embryos were injected and fixed for processing blind before genotyping. Adult mice were retroorbitally injected with 0.5 mg of EZ-Link NHS-Biotin per g of mouse body weight. The NHS-Biotin circulated for 30 minutes prior to brain dissections and fixation with 4% PFA. 30 μm thick cryosections were then collected and immunostained with Streptavidin Alexa Fluor 568 (1:200; Thermo Fisher: S32351) and goat anti-CD31 (1:100; R&D Systems: AF3628).
Single-cell RNA-sequencing
Larval zebrafish brains were dissected and split along the midbrain-hindbrain boundary in DMEM. Brains were dissociated using a modified protocol.77 Briefly, chemical dissociations were performed at 30.5°C using a mixture of 0.25% Trypsin-EDTA, Collagenase/Dispase (8 mg/mL) and DNaseI (20 μg/mL) for 15–20 minutes with gentle pipetting every few minutes and quenched with 10% fetal bovine serum in DMEM. Samples were hashed using Multi-seq as previously described with slight modifications.78 For each sample, 80 pmoles of lipid modified oligos (LMOs) were used to hash every 500k cells. The hashing reaction was quenched using 1% BSA in PBS and barcoded samples were subsequently pooled and washed with 1% BSA. The pooled cell mixture was resuspended in PBS + 0.1%BSA + 18% Optiprep at a final concentration of ~300k cells/mL prior to single-cell capture with inDrops. The Single-cell Core (SCC) at Harvard Medical School captured single-cell transcriptomes and prepared NGS libraries as previously described79 with a target capture of 45k cells per experiment.
Gene expression and hashtag libraries were mixed (9:1 ratio) and sequenced on an Illumina Nova-seq 6000 with the NovaSeq S2 kit. Reads were mapped onto the Zebrafish GRCz11 Release 101 genome assembly using previously described methods.80 Hashtags were identified using custom code relying largely on SCANPY,81 and available on: https://doi.org/10.5281/zenodo.7955159.
Transcriptomes with greater than 350 UMIs were further filtered for viability by removing cells with >20% mitochondrial reads. Cell demultiplexing was performed manually by applying thresholds to delineate single cells from background and multiple populations. The resulting counts matrix was normalized to the mean UMIs per cell in the dataset.
To visualize the data, we first mean-normalized the data and identified highly variable genes from the wild type AB and RNF datasets (minimum of 3 transcripts per cell, minimum of 3 cells expressing gene, minimum V-score percentile of 85%). We then Z-scored counts for each gene and performed principal component analysis (PCA) with 50 components. Cells from the spock1 mutant libraries were projected into the same principal component subspace. A k-nearest neighbor graph (k=10) was generated based on the Euclidean distance in gene expression between cells within this subspace. The Leiden algorithm was used to cluster cells into subgroups.82 The neighborhood graph was embedded and visualized using UMAP and the data was explored interactively with SPRING83 to aid in cell cluster annotation.
Cells belonging to the liver, pharyngeal arches, skin, and muscle were removed prior to differential gene expression (DGE) analysis. The filtered dataset was reanalyzed using the methods described above. The Wilcoxon rank-sum test was used to generate a list of potential genes that are differentially regulated (fold change > 2) between genotypes across each Leiden cluster (Table S2). Genes were only considered for DGE analysis if they were expressed in at least 5% of cells in a given cluster and had a minimum mean of 10 transcripts per cell. Cells belonging to the AB and RNF background were treated as a single wild type genotype for the analysis. The vascular cluster (Leiden 13) was subclustered and similarly analyzed for DGE (Table S3).
Quantification and Statistical Analysis
Zebrafish Permeability Quantification
All quantification was performed on blinded image sets. For static images, parenchymal fluorescent tracer intensity was measured using Fiji84 in the entire regional parenchyma outside of the vasculature in 60 μm thick maximum intensity projections (MIPs) of the larval brains. These projections began on average 15 μm below the mesencephalic vein to reduce the effects of potential leakage diffusion from the surface vessels and had the vasculature masked and removed for average intensity quantification. These parenchymal tracer intensity values of the entire larval brains were then background subtracted and normalized to the tracer intensity within the vasculature to account for differential amounts of circulating tracer between fish. The average midbrain parenchymal tracer intensity for each individual fish was plotted as a single point for all tracer leakage assays. For time lapse imaging, Dextran intensity was measured in six parenchymal regions of average intensity projections of the time lapse videos and averaged as a single value per fish and similarly normalized to the average blood vessel luminal fluorescence intensity at each time point.
Vascular Coverage
80 μm thick MIPs of the entire larval brain were measured for vascular and pericyte coverage using Fiji84 as total masked vascular or pericyte area divided by total brain area.
HCR Quantification
To measure levels of mcamb expression in endothelial cells and pericytes, we made 20 μm thick MIPs from the whole mount larvae and manually traced kdrl+ endothelial cells and notch3+ pericytes and measured average intensity within each cell type using Fiji on blinded image sets. On average, 4 vessels and pericytes were measured for each region (midbrain and hindbrain) and averaged into a single value for each individual fish. Similar measurements were performed for foxc1b expression in notch3+ pericytes. Quantification of spock1 co-localization with neuronal markers (elavl3, slc17a6b, and gad1b) was performed on 2.5 μm thick MIPs of the dorsal midbrain. We first used Ilastik85 to segment individual cells in the brain based on the transgenic mem-Citrine signal. Taking this cell mask, we then measured average spock1 and other neuronal gene levels per segmented cell using the Fiji BioImaging and Optics Platform (BIOP) plugin. Additional images were taken on a Zeiss LSM 980 with Airyscan 2 with 0.21 μm z-steps using either a 20x or a 40x water immersion objective to further enhance the resolution of the mcamb signal.
Transplantation Quantification
Blinded image sets were analyzed using Fiji. We took each z-stack of an individual larval brain and generated 10 μm thick MIPs to span the entire brain, with an average of 10 MIPs analyzed per individual fish. Using these MIPs, we then measured average NHS tracer intensity in an 8 μm wide rectangle between donor cells and the nearest blood vessel and normalized to average blood vessel tracer intensity. We then binned tracer measurements by distance (D) to the nearest donor cell (0, 2, 4, 6, 8, 10, 12, 15, 20, and 30+ μm) and took the median tracer intensity value for each bin per individual fish. All analyzed fish had at least 1 donor cell per each distance bin. Statistics were calculated using 2way ANOVA comparing to WT→WT at every distance bin. Mean values per each bin were plotted per genotype with standard error bars.
Gelatinase Zymography
All quantification was performed on blinded 80 μm thick MIPs of the entire larval brain. Average parenchymal FITC-Gelatin probe intensity was measured using Fiji in the entire midbrain tectum and normalized to the average intensity in the ventricles, which have high gelatinase activity, to account for fish to fish injection variation. Regions of obvious damage at the injection site were excluded for quantification.
Zebrafish Immunofluorescence
Images were collected with a Leica SP8 and the same settings were used across samples. Vessels were masked with the transgenic kdrl:mCherry signal in MIPs of the entire 14 μm thick z-stacks and used to quantify average fluorescence intensity of each ECM component within the vasculature in Fiji. Background levels of fluorescence in the surrounding brain tissue were subtracted from the average vessel intensity for each fish.
TEM Quantification
Grids were imaged using a 1200EX electron microscope (JEOL) equipped with a 2k CCD digital camera (AMT) and quantified using Fiji.84 Vesicular density values were calculated from the number of non-clathrin coated small vesicles less than 100 nm in diameter or large vesicles greater than 200 nm in diameter per μm2 of endothelial area for each image collected. Average pericyte basement membrane (BM) thickness was quantified by measuring the total BM area divided by the length of the pericyte-endothelial contact. All images for analysis were collected at 12000x magnification. 10–15 vessels were quantified for each fish, with each color representing a different fish.
Mouse Permeability and PLVAP Quantification
Using a Leica SP8 with a 25x water immersion objective, we collected z-stacks spanning 20–30 μm thick sections using the same settings across samples. We collected 5 separate images for each sample. Quantification of tracer permeability into the brain parenchyma was performed on blinded MIPs using a previously reported Fiji permeability macro86 that measures the total area of thresholded tracer signal and divides that by vascular area for each image. The same macro was used to quantify area of PLVAP+ vessels and divided by total vessel area
Supplementary Material
Table S1: Differential gene expression analysis of mutant and wild type larvae by bulk RNAseq, related to Figure 1.
Table S2: Differential gene expression analysis of whole brain scRNA-seq, related to Figure 6.
Table S3: Gene expression in Leiden cluster 13 subclusters, related to Figure 6.
Table S4: Primer, gRNA, and HCR oligonucleotide sequences, related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Fibronectin | Abcam | ab6328 |
| Rabbit Anti-Collagen IV | Abcam | ab6586 |
| Rat Anti-PLVAP | BD Biosciences | 553849 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FITC-Gelatin | Biovision | Cat# M1303 |
| recombinant human SPOCK1 protein | R&D Systems | Cat# 2327-PI-050 |
| NHS-activated gold nanoparticles | Cytodiagnostics | Cat# CGN5K-5–1 |
| Paraformaldehyde 20% Solution, EM Grade | VWR | Cat# 15703-S |
| Sodium Cacodylate Buffer, 0.2M | VWR | Cat# 11653 |
| Aqueous Glutaraldehyde EM Grade 50% | Electron Microscopy Sciences | Cat# 16320 |
| Dextran, Alexa Fluor 647; 10,000 MW, Anionic, Fixable | Thermo Fisher | Cat# D22914 |
| Alexa Fluor 405 NHS Ester (Succinimidyl Ester) | Thermo Fisher | Cat# A30000 |
| HCR v3.0 Amplifiers and Buffers | Molecular Instruments | https://www.molecularinstruments.com/hcr-rnafish-products |
| Agarose, low gelling temperature | Sigma | A9414 |
| EZ-Link NHS-Biotin | Thermo Fisher | 20217 |
| Dextran, Alexa Fluor 488; 10,000 MW, Anionic, Fixable | Thermo Fisher | D22910 |
| Streptavidin, Alexa Fluor 405 Conjugate | Thermo Fisher | S32351 |
| N-Phenylthiourea (PTU) | Sigma | 222909 |
| Deposited Data | ||
| bulk and single cell RNA-sequencing | We collected this data | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE230236 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: AB wildtype strain | Zebrafish International Resource Centre | ZDB-GENO-960809–1 |
| Zebrafish: spock1hm41 | This study | hm41 |
| Zebrafish: spock1hm43 | This study | hm43 |
| Zebrafish: Tg(kdrl:HRAS-mCherry)s896Tg | Chi et al, 200870 | ZDB-ALT-081212–4 |
| Zebrafish: Tg(l-fabp:DBP-EGFP)Iri500Tg | Xie et al, 201029 | ZDB-ALT-120118–1 |
| Zebrafish: TgBAC(pdgfrb:EGFP)ncv22Tg | Ando et al, 201659 | ZDB-ALT-160609–1 |
| Zebrafish:Tg(plvapb:EGFP)sj3Tg | Umans et al, 201731 | ZDB-ALT-170816–3 |
| Zebrafish: Tg(actb2:memcitrine-citrine)hm30 | Xiong et al, 201471 | ZDB-TGCONSTRCT-130114–2 |
| Zebrafish: Tg(elavl3:GCaMP6s)a13203Tg | Kim et al, 201772 | ZDB-ALT-180502–2 |
| Zebrafish: Tg(−3.1neurog1:GFP)sb2Tg | Blader et al, 200373 | ZDB-ALT-030904–2 |
| Mouse: Spock1tm1Htmn | Röll et al, 200646 | MGI:3665280 |
| Zebrafish: TgKI(Spock1-mCherry)hm44 | This study | hm44 |
| Oligonucleotides | ||
| See Table S4 | ||
| Recombinant DNA | ||
| spock1-mCherry-KI | This study | |
| Software and Algorithms | ||
| SCANPY | Wolf et al, 201881 | https://github.com/scverse/scanpy |
| Fiji | Shindelin et al, 201284 | https://imagej.net/software/fiji/ |
| Ilastik | Berg et al, 201985 | https://www.ilastik.org/ |
| SPRING | Weinreb et al, 201783 | https://kleintools.hms.harvard.edu/tools/spring.html |
| RNAmapper | Miller et al, 201333 | http://www.rnamapper.org/ |
Highlights.
Spock1, secreted from neurons, induces BBB functional development
Spock1 regulates vascular basement membrane composition
Exogenous SPOCK1 can partially restore BBB function in leaky mutants
Acknowledgments
We thank members of the Megason laboratory for data discussion and comments on the manuscript; Dr. Zach O’Brown for discussions and comments on the manuscript; Ignas Mazelis for single cell demultiplexing software; the HMS Single Cell Core for single cell library preparation; Dr. Bela Anand-Apte (Cleveland Clinic) for providing the transgenic l-fabp:DBP-EGFP fish line and Dr. Leonard Zon for providing the transgenic Tg(kdrl:HRAS-mCherry) line; and the HMS Electron Microscopy Core Facility, with special thanks to Louise Trakimas for all of her assistance in preparing the TEM samples. This work was supported by the Damon Runyon Cancer Foundation (N.M.O.), NIH K99HD103911 (N.M.O.), NIH R01HD096755 (S.G.M), an Allen Distinguished Investigator Award (C.G.), NIH R35NS116820 (C.G), and partially supported by Faculty Scholar grant from the Howard Hughes Medical Institute (C.G.). C.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
We declare no competing interests.
References:
- 1.Sweeney MD, Sagare AP, and Zlokovic BV (2018). Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nature Publishing Group 14, 133–150. 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reese TS, and Karnovsky MJ (1967). Fine structural localization of a blood-brain barrier to exogenous peroxidase. J Cell Biology 34, 207–217. 10.1083/jcb.34.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M, Kiang A-S, Kenna PF, Kerskens C, Blau C, O’Dwyer L, Tivnan A, Kelly JA, Brankin B, Farrar G-J, et al. (2008). RNAi-mediated reversible opening of the blood-brain barrier. The Journal of Gene Medicine 10, 930–947. 10.1002/jgm.1211. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, and Gu C (2014). Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511. 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreone BJ, Chow BW, Tata A, Lacoste B, Ben-Zvi A, Bullock K, Deik AA, Ginty DD, Clish CB, and Gu C (2017). Blood-Brain Barrier Permeability Is Regulated by Lipid Transport-Dependent Suppression of Caveolae-Mediated Transcytosis. Neuron 94, 581–594.e5. 10.1016/j.neuron.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallmann R, Mayer DN, Berg EL, Broermann R, and Butcher EC (1995). Novel mouse endothelial cell surface marker is suppressed during differentiation of the blood brain barrier. Dev Dynam 202, 325–332. 10.1002/aja.1002020402. [DOI] [PubMed] [Google Scholar]
- 7.Harik S, Kalaria R, Andersson L, Lundahl P, and Perry G (1990). Immunocytochemical localization of the erythroid glucose transporter: abundance in tissues with barrier functions. J Neurosci 10, 3862–3872. 10.1523/jneurosci.10-12-03862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schinkel AH (1999). P-Glycoprotein, a gatekeeper in the blood–brain barrier. Adv Drug Deliver Rev 36, 179–194. 10.1016/s0169-409x(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez-Covarrubias L, Slosky LM, Thompson BJ, Davis TP, and Ronaldson PT (2014). Transporters at CNS barrier sites: obstacles or opportunities for drug delivery? Current pharmaceutical design 20, 1422–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart PA, and Wiley MJ (1981). Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: A study using quail-chick transplantation chimeras. Dev Biol 84, 183–192. 10.1016/0012-1606(81)90382-1. [DOI] [PubMed] [Google Scholar]
- 11.Lyck R, Ruderisch N, Moll AG, Steiner O, Cohen CD, Engelhardt B, Makrides V, and Verrey F (2009). Culture-induced changes in blood-brain barrier transcriptome: implications for amino-acid transporters in vivo. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 29, 1491–1502. 10.1038/jcbfm.2009.72. [DOI] [PubMed] [Google Scholar]
- 12.Urich E, Lazic SE, Molnos J, Wells I, and Freskgård P-O (2012). Transcriptional Profiling of Human Brain Endothelial Cells Reveals Key Properties Crucial for Predictive In Vitro Blood-Brain Barrier Models. PloS one 7, e38149–16. 10.1371/journal.pone.0038149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérit S, Fidan E, Macas J, Czupalla CJ, Figueiredo R, Vijikumar A, Yalcin BH, Thom S, Winter P, Gerhardt H, et al. (2021). Astrocyte-derived Wnt growth factors are required for endothelial blood-brain barrier maintenance. Prog Neurobiol 199, 101937. 10.1016/j.pneurobio.2020.101937. [DOI] [PubMed] [Google Scholar]
- 14.Heithoff BP, George KK, Phares AN, Zuidhoek IA, Munoz‐Ballester C, and Robel S (2021). Astrocytes are necessary for blood–brain barrier maintenance in the adult mouse brain. Glia 69, 436–472. 10.1002/glia.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armulik A, Genové G, Mäe M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. (2010). Pericytes regulate the blood-brain barrier. Nature 468, 557–561. 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 16.Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566. 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell RD, Winkler EA, Sagare AP, Singh I, LaRue B, Deane R, and Zlokovic BV (2010). Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 68, 409–427. 10.1016/j.neuron.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbott NJ, Rönnbäck L, and Hansson E (2006). Astrocyte–endothelial interactions at the blood–brain barrier. Nature reviews. Neuroscience 7, 41–53. 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 19.Janzer RC, and Raff MC (1987). Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 325, 253–257. 10.1038/325253a0. [DOI] [PubMed] [Google Scholar]
- 20.Benz F, Wichitnaowarat V, Lehmann M, Germano RF, Mihova D, Macas J, Adams RH, Taketo MM, Plate K-H, Guérit S, et al. (2019). Low wnt/β-catenin signaling determines leaky vessels in the subfornical organ and affects water homeostasis in mice. Elife 8, e43818. 10.7554/elife.43818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, and McMahon AP (2008). Canonical Wnt Signaling Regulates Organ-Specific Assembly and Differentiation of CNS Vasculature. Science 322, 1247–1250. 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 22.Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, and Barres BA (2009). Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proceedings of the National Academy of Sciences of the United States of America 106, 641–646. 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Rattner A, Zhou Y, Williams J, Smallwood PM, and Nathans J (2012). Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell 151, 1332–1344. 10.1016/j.cell.2012.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanhollebeke B, Stone OA, Bostaille N, Cho C, Zhou Y, Maquet E, Gauquier A, Cabochette P, Fukuhara S, Mochizuki N, et al. (2015). Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. eLife 4. 10.7554/elife.06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Cho C, Williams J, Smallwood PM, Zhang C, Junge HJ, and Nathans J (2018). Interplay of the Norrin and Wnt7a/Wnt7b signaling systems in blood–brain barrier and blood–retina barrier development and maintenance. Proceedings of the National Academy of Sciences of the United States of America 115, E11827–E11836. 10.1073/pnas.1813217115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin M, Vermeiren S, Bostaille N, Eubelen M, Spitzer D, Vermeersch M, aci CP, Pozuelo E, Toussay X, Raman-Nair J, et al. (2022). Engineered Wnt ligands enable blood-brain barrier repair in neurological disorders. Science 375. 10.1126/science.abm4459. [DOI] [PubMed] [Google Scholar]
- 27.Liebner S, Corada M, Bangsow T, Babbage J, Taddei A, Czupalla CJ, Reis M, Felici A, Wolburg H, Fruttiger M, et al. (2008). Wnt/β-catenin signaling controls development of the blood–brain barrier. J Cell Biology 183, 409–417. 10.1083/jcb.200806024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brown NM, Megason SG, and Gu C (2019). Suppression of transcytosis regulates zebrafish blood-brain barrier function. eLife 8, 41. 10.7554/elife.47326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J, Farage E, Sugimoto M, and Anand-Apte B (2010). A novel transgenic zebrafish model for blood-brain and blood-retinal barrier development. BMC developmental biology 10, 76. 10.1186/1471-213x-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guemez-Gamboa A, Nguyen LN, Yang H, Zaki MS, Kara M, Ben-Omran T, Akizu N, Rosti RO, Rosti B, Scott E, et al. (2015). Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nature Genetics 47, 809–813. 10.1038/ng.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umans RA, Henson HE, Mu F, Parupalli C, Ju B, Peters JL, Lanham KA, Plavicki JS, and Taylor MR (2017). CNS angiogenesis and barriergenesis occur simultaneously. Developmental Biology 425, 101–108. 10.1016/j.ydbio.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong J-Y, Kwon H-B, Ahn J-C, Kang D, Kwon S-H, Park JA, and Kim K-W (2008). Functional and developmental analysis of the blood–brain barrier in zebrafish. Brain research bulletin 75, 619–628. 10.1016/j.brainresbull.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 33.Miller AC, Obholzer ND, Shah AN, Megason SG, and Moens CB (2013). RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome research 23, 679–686. 10.1101/gr.147322.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnet F, Périn JP, Charbonnier F, Camuzat A, Roussel G, Nussbaum JL, and Alliel PM (1996). Structure and cellular distribution of mouse brain testican. Association with the postsynaptic area of hippocampus pyramidal cells. The Journal of biological chemistry 271, 4373–4380. 10.1074/jbc.271.8.4373. [DOI] [PubMed] [Google Scholar]
- 35.Edgell C-JS, BaSalamah MA, and Marr HS (2004). Testican-1: a differentially expressed proteoglycan with protease inhibiting activities. International review of cytology 236, 101–122. 10.1016/s0074-7696(04)36003-1. [DOI] [PubMed] [Google Scholar]
- 36.Charbonnier F, Chanoine C, Cifuentes-Diaz C, Gallien CL, Rieger F, Alliel PM, and Périn JP (2000). Expression of the proteoglycan SPOCK during mouse embryo development. Mechanisms of development 90, 317–321. 10.1016/s0925-4773(99)00255-5. [DOI] [PubMed] [Google Scholar]
- 37.Raj B, Farrell JA, Liu J, Kholtei JE, Carte AN, Acedo JN, Du LY, McKenna A, Relić Đ, Leslie JM, et al. (2020). Emergence of Neuronal Diversity during Vertebrate Brain Development. Neuron, 1–24. 10.1016/j.neuron.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armulik A, Genové G, and Betsholtz C (2011). Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Developmental cell 21, 193–215. 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Pan L, Moens CB, and Appel B (2013). Notch3 establishes brain vascular integrity by regulating pericyte number. Development (Cambridge, England) 141, 307–317. 10.1242/dev.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Z, Lin Z, Wang Z, Liu D, Tian D, and Xia L (2020). SPOCK1 overexpression induced by platelet-derived growth factor-BB promotes hepatic stellate cell activation and liver fibrosis through the integrin α5Î21/PI3K/Akt signaling pathway. Laboratory Investigation, 1–15. 10.1038/s41374-020-0425-4. [DOI] [PubMed] [Google Scholar]
- 41.Ye Z, Chen J, Hu X, Yang S, Xuan Z, Lu X, and Zhao Q (2020). SPOCK1: a multi-domain proteoglycan at the crossroads of extracellular matrix remodeling and cancer development. Am J Cancer Res 10, 3127–3137. [PMC free article] [PubMed] [Google Scholar]
- 42.Váncza L, Tátrai P, Reszegi A, Baghy K, and Kovalszky I (2022). SPOCK1 with unexpected function. The start of a new career. Am J Physiol-cell Ph. 10.1152/ajpcell.00033.2022. [DOI] [PubMed] [Google Scholar]
- 43.Nakada M, Yamada A, Takino T, Miyamori H, Takahashi T, Yamashita J, and Sato H (2001). Suppression of membrane-type 1 matrix metalloproteinase (MMP)-mediated MMP-2 activation and tumor invasion by testican 3 and its splicing variant gene product, N-Tes. Cancer research 61, 8896–8902. [PubMed] [Google Scholar]
- 44.Underly RG, Levy M, Hartmann DA, Grant RI, Watson AN, and Shih AY (2017). Pericytes as Inducers of Rapid, Matrix Metalloproteinase-9-Dependent Capillary Damage during Ischemia. The Journal of Neuroscience 37, 129–140. 10.1523/jneurosci.2891-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia‐Alloza M, Prada C, Lattarulo C, Fine S, Borrelli LA, Betensky R, Greenberg SM, Frosch MP, and Bacskai BJ (2009). Matrix metalloproteinase inhibition reduces oxidative stress associated with cerebral amyloid angiopathy in vivo in transgenic mice. J Neurochem 109, 1636–1647. 10.1111/j.1471-4159.2009.06096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Röll S, Seul J, Paulsson M, and Hartmann U (2006). Testican-1 is dispensable for mouse development. Matrix Biology 25, 373–381. 10.1016/j.matbio.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Luo Y, Hui H, Cai T, Huang H, Yang F, Feng J, Zhang J, and Yan X (2017). CD146 coordinates brain endothelial cell–pericyte communication for blood–brain barrier development. Proc National Acad Sci 114, E7622–E7631. 10.1073/pnas.1710848114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegenthaler JA, Choe Y, Patterson KP, Hsieh I, Li D, Jaminet S-C, Daneman R, Kume T, Huang EJ, and Pleasure SJ (2013). Foxc1 is required by pericytes during fetal brain angiogenesis. Biol Open 2, 647–659. 10.1242/bio.20135009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam SJ, Richmond DL, Kaminker JS, Modrusan Z, Martin-McNulty B, Cao TC, Weimer RM, Carano RAD, van Bruggen N, and Watts RJ (2012). Death receptors DR6 and TROY regulate brain vascular development. Developmental cell 22, 403–417. 10.1016/j.devcel.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 50.Ayloo S, Lazo CG, Sun S, Zhang W, Cui B, and Gu C (2022). Pericyte-to-endothelial cell signaling via vitronectin-integrin regulates blood-CNS barrier. Neuron. 10.1016/j.neuron.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Wang Y, Tischfield M, Williams J, Smallwood PM, Rattner A, Taketo MM, and Nathans J (2014). Canonical WNT signaling components in vascular development and barrier formation. J Clin Invest 124, 3825–3846. 10.1172/jci76431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, et al. (2004). Vascular Development in the Retina and Inner Ear Control by Norrin and Frizzled-4, a High-Affinity Ligand-Receptor Pair. Cell 116, 883–895. 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 53.Cho C, Smallwood PM, and Nathans J (2017). Reck and Gpr124 Are Essential Receptor Cofactors for Wnt7a/Wnt7b-Specific Signaling in Mammalian CNS Angiogenesis and Blood-Brain Barrier Regulation. Neuron 95, 1056–1073.e5. 10.1016/j.neuron.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, and Ye W (2009). TSPAN12 Regulates Retinal Vascular Development by Promoting Norrin-but Not Wnt-Induced FZD4/β-Catenin Signaling. Cell 139, 299–311. 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, and Nathans J (2014). Gpr124 Controls CNS Angiogenesis and Blood-Brain Barrier Integrity by Promoting Ligand-Specific Canonical Wnt Signaling. Dev Cell 31, 248–256. 10.1016/j.devcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Q, Shah S, Dries R, Cai L, and Yuan G-C (2018). Identification of spatially associated subpopulations by combining scRNA-seq and sequential fluorescence in situ hybridization data. Nat Biotechnol, 10.1038/nbt.4260. 10.1038/nbt.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herrero-Navarro Á, Puche-Aroca L, Moreno-Juan V, Sempere-Ferràndez A, Espinosa A, Susín R, Torres-Masjoan L, Leyva-Díaz E, Karow M, Figueres-Oñate M, et al. (2021). Astrocytes and neurons share region-specific transcriptional signatures that confer regional identity to neuronal reprogramming. Sci Adv 7, eabe8978. 10.1126/sciadv.abe8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohlig S, Clavreul S, Thorwirth M, Simon-Ebert T, Bocchi R, Ulbricht S, Kannayian N, Rossner M, Sirko S, Smialowski P, et al. (2021). Molecular diversity of diencephalic astrocytes reveals adult astrogenesis regulated by Smad4. Embo J 40, e107532. 10.15252/embj.2020107532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ando K, Fukuhara S, Izumi N, Nakajima H, Fukui H, Kelsh RN, and Mochizuki N (2016). Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development (Cambridge, England) 143, 1328–1339. 10.1242/dev.132654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trost A, Lange S, Schroedl F, Bruckner D, Motloch KA, Bogner B, Kaser-Eichberger A, Strohmaier C, Runge C, Aigner L, et al. (2016). Brain and Retinal Pericytes: Origin, Function and Role. Frontiers in cellular neuroscience 10, 3171–13. 10.3389/fncel.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolburg H, and Paulus W (2010). Choroid plexus: biology and pathology. Acta neuropathologica 119, 75–88. 10.1007/s00401-009-0627-8. [DOI] [PubMed] [Google Scholar]
- 62.Abbott NJ (2004). Evidence for bulk flow of brain interstitial fluid: significance for physiology and pathology. Neurochem Int 45, 545–552. 10.1016/j.neuint.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Henson HE, Parupalli C, Ju B, and Taylor MR (2014). Functional and genetic analysis of choroid plexus development in zebrafish. Frontiers in neuroscience 8, 364. 10.3389/fnins.2014.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derk J, Como CN, Jones HE, Joyce LR, Kim S, Spencer BL, Bonney S, O’Rourke R, Pawlikowski B, Doran KS, et al. (2023). Formation and function of the meningeal arachnoid barrier around the developing mouse brain. Dev Cell 58, 635–644.e4. 10.1016/j.devcel.2023.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takahashi Y, Maki T, Liang AC, Itoh K, Lok J, Osumi N, and Arai K (2014). p38 MAP kinase mediates transforming-growth factor-β1-induced upregulation of matrix metalloproteinase-9 but not −2 in human brain pericytes. Brain Res 1593, 1–8. 10.1016/j.brainres.2014.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avolio C, Ruggieri M, Giuliani F, Liuzzi GM, Leante R, Riccio P, Livrea P, and Trojano M (2003). Serum MMP-2 and MMP-9 are elevated in different multiple sclerosis subtypes. J Neuroimmunol 136, 46–53. 10.1016/s0165-5728(03)00006-7. [DOI] [PubMed] [Google Scholar]
- 67.Kook S-Y, Hong HS, Moon M, and Mook-Jung I (2013). Disruption of blood-brain barrier in Alzheimer disease pathogenesis. Tissue Barriers 1, e23993. 10.4161/tisb.23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosell A, Cuadrado E, Ortega-Aznar A, Hernández-Guillamon M, Lo EH, and Montaner J (2008). MMP-9–Positive Neutrophil Infiltration Is Associated to Blood–Brain Barrier Breakdown and Basal Lamina Type IV Collagen Degradation During Hemorrhagic Transformation After Human Ischemic Stroke. Stroke 39, 1121–1126. 10.1161/strokeaha.107.500868. [DOI] [PubMed] [Google Scholar]
- 69.Westerfield M (1993). The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio).
- 70.Chi NC, Shaw RM, Val SD, Kang G, Jan LY, Black BL, and Stainier DYR (2008). Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes & Development 22, 734–739. 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong F, Ma W, Hiscock TW, Mosaliganti KR, Tentner AR, Brakke KA, Rannou N, Gelas A, Souhait L, Swinburne IA, et al. (2014). Interplay of Cell Shape and Division Orientation Promotes Robust Morphogenesis of Developing Epithelia. Cell 159, 415–427. 10.1016/j.cell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim DH, Kim J, Marques JC, Grama A, Hildebrand DGC, Gu W, Li JM, and Robson DN (2017). Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat Methods 14, 1107–1114. 10.1038/nmeth.4429. [DOI] [PubMed] [Google Scholar]
- 73.Blader P, Plessy C, and Strähle U (2003). Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Develop 120, 211–218. 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- 74.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development (Cambridge, England) 145. 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pomreinke AP, Soh GH, Rogers KW, Bergmann JK, Bläßle AJ, and Müller P (2017). Dynamics of BMP signaling and distribution during zebrafish dorsal-ventral patterning. Elife 6, e25861. 10.7554/elife.25861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bresciani E, Broadbridge E, and Liu PP (2018). An efficient dissociation protocol for generation of single cell suspension from zebrafish embryos and larvae. Methodsx 5, 1287–1290. 10.1016/j.mex.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, et al. (2019). MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat Methods 16, 619–626. 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell 161, 1187–1201. 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 108, eaar4362–13. 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf FA, Angerer P, and Theis FJ (2017). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep-uk 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinreb C, Wolock S, and Klein AM (2017). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16, 1226–1232. 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 86.Chow BW, and Gu C (2017). Gradual Suppression of Transcytosis Governs Functional Blood-Retinal Barrier Formation. Neuron 93, 1325–1333.e3. 10.1016/j.neuron.2017.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Differential gene expression analysis of mutant and wild type larvae by bulk RNAseq, related to Figure 1.
Table S2: Differential gene expression analysis of whole brain scRNA-seq, related to Figure 6.
Table S3: Gene expression in Leiden cluster 13 subclusters, related to Figure 6.
Table S4: Primer, gRNA, and HCR oligonucleotide sequences, related to STAR Methods.
Data Availability Statement
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available (GSE230236). All original code for the single-cell analysis is available as interactive Jupyter notebooks here: https://doi.org/10.5281/zenodo.7955159. Interactive exploration of the full single-cell data can be found here: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/2021_OBrown/2021_OBrown. Exploration of the cluster 13 subcluster cells can be found here: https://kleintools.hms.harvard.edu/tools/springViewer_1_6_dev.html?client_datasets/2021_OBrown_Vasculature/2021_OBrown_Vasculature. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


