Abstract
We have developed a PCR-based method to detect macrolide resistance and the virulence gene cagA in Helicobacter pylori within 24 h, thereby improving the lengthy process of culture-based approaches. Total DNA was prepared directly from stomach biopsy specimens. The procedure proved to be rapid and reliable and could be utilized for diagnostic purposes.
Helicobacter pylori plays a significant role in the pathogenesis of gastritis and peptic ulcer disease. In addition, carriage of H. pylori increases the risk for gastric adenocarcinoma (6). The factors that differentiate virulent bacteria causing severe illness from bacteria infecting asymptomatic carriers are not yet well defined. However, one marker for virulence is well known, the cytotoxin-associated gene A, cagA, which codes for a 120-kDa protein of unknown function. The cagA gene has been shown to be more prevalent in strains infecting patients with ulcer disease than in strains infecting patients with gastritis alone (1, 5, 15). If host and bacterial virulence factors associated with gastric pathology can be defined, it may help in focusing therapy on individuals infected with pathogenic strains or harboring endogenous constituents predisposing for disease.
Treatment of H. pylori-associated peptic ulcer disease with triple therapy practically always heals the mucosal lesion but fails to eradicate the microbe in about 10% of all cases (16). Antimicrobial resistance may, in part, account for treatment failures (9). Clarithromycin, an intracellularly active antibiotic, is one of the cornerstones in present ulcer therapy (3, 11, 16). With increasing use and thereby increased risk of bacterial resistance, there is an explicit need for more rapid methods to detect resistance among H. pylori strains (4). More than 90% of macrolide resistance in H. pylori is mediated by either of two transition mutations (A→G) at adjacent positions 2143 and 2144 in one or both of the bacterium’s two 23S rRNA genes (9, 12, 17). A transversion mutation (A→C) at position 2143 has been reported to be the cause of resistance in 7% of the resistant isolates (12). The mutations are stable, and resistance to one macrolide leads to cross-resistance to other members of this group of antibiotics (9). Standard methods for antibiotic susceptibility testing use culture-based approaches and therefore take about 2 weeks.
Methods to detect H. pylori by PCR directly on DNA derived from biopsy specimens or dental plaque have been evaluated previously (7, 8, 18, 19). Detection of macrolide resistance mutations has been performed only on DNA derived from cultured strains (13, 14). Recently, a method to type H. pylori strains with restriction fragment length polymorphism directly on biopsy specimens was evaluated (10).
The aim of this study was to develop and evaluate a PCR-based method for rapid detection of macrolide resistance-associated mutations and the virulence factor cagA of H. pylori directly on biopsy material.
Biopsy samples were collected from 69 patients (39 females and 30 males; mean age, 57 years) who were referred to endoscopy because of dyspeptic symptoms. Sixty-three were included consecutively; four of these had received treatment against H. pylori prior to sample collection. Six patients were chosen for inclusion separately as possible cases of infection with clarithromycin-resistant strains. These patients had unsuccessfully been treated with omeprazol, clarithromycin, and amoxicillin or metronidazole. Patients were initially tested for H. pylori infection with the CLOtest (Delta West Limited, Bentley, Australia) at the endoscopy unit. The study was approved by the local ethics committee at the Karolinska Institute, Stockholm, Sweden.
Two biopsy specimens were analyzed; total DNA was prepared from one biopsy specimen by using the QIAamp Tissue Kit (Qiagen, Hilden, Germany), and the other was used for culture of H. pylori according to the method described previously (9). DNA was prepared from one single colony of H. pylori (2) and analyzed as described above to evaluate the sensitivity of the method. Approximately 105 CFU and an Etest strip (AB Biodisk, Solna, Sweden) were applied onto chocolate agar plates and incubated for 4 days before the MICs were read. The 23S rRNA PCR followed by detection of resistance mutations and the cagA PCR were performed as previously described (2, 9, 17).
Of 69 patients, 29 were positive for H. pylori according to the CLOtest (2 were not analyzed) and 34 were positive by culture. Seven of the 10 treated patients were positive for H. pylori by both culture and CLOtest. To mimic the expected clinical situation, only biopsy specimens from the 29 patients positive by CLOtest were analyzed further. All of these patients were also positive by culture.
Of 29 biopsy samples, 17 were positive for cagA. When DNA from the cultures was tested, 19 of 29 samples were positive. There were no false-positive biopsy DNA samples compared to the DNA from the cultured strains.
The 23S rRNA gene fragment was amplified from 28 of the 29 culture- and CLOtest-positive samples. The only sample where the gene was not detected was obtained from a patient infected with a clarithromycin-resistant strain as determined by Etest.
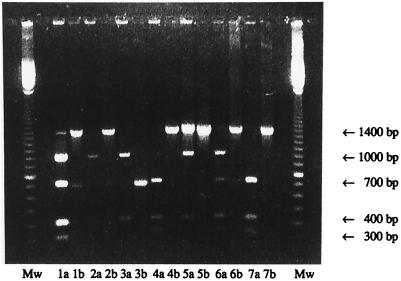
Seven patients were carrying clarithromycin-resistant H. pylori strains, and six of these had received treatment against H. pylori. In addition, one treated patient was infected with a sensitive strain (Fig. 1, patient 5), probably representing a reinfection. All the resistant strains were found to have either of the two resistance mutations (Fig. 1). One strain had the A2143G mutation but was found to be susceptible to clarithromycin, which may be due to a mixed H. pylori infection.
FIG. 1.
Restriction enzyme digestions of samples from patients treated for H. pylori. Numbers denote patient samples; “a” lanes show digestion of the 23S rRNA gene with BsaI, and “b” lanes show digestion with MboII. Mw, 100-step basepair ladder (Pharmacia Biotech, Sollentuna, Sweden). The 700-bp band indicates a resistant strain, since the mutation site is in the middle of the 1.4-kb PCR fragment. Sample 1 was digested at 700 bp by both MboII and BsaI and thus mutated in both positions. Samples 2 and 3 were digested at the mutation by MboII; i.e., they had the A2143G mutation. Samples 4, 6, and 7 were digested by BsaI; i.e., they had the A2144G mutation. Sample 5 was not resistant.
Macrolides, especially clarithromycin, are widely used for the eradication of H. pylori. Consequently, the number of resistant strains will inevitably increase. This increases the demand for diagnostic tools to guide the care provider in choosing the appropriate therapy. In its present design, the method described here detects only the two most common resistance mutations, A2143G and A2144G.
Virulence markers, such as cagA, can also be detected with this method. As our understanding of the molecular determinants that are associated with H. pylori pathology increases, our method could also be helpful in directing treatment to specific at-risk patients or groups. The analyses may be performed immediately after gastroscopy, and the treatment can immediately be adjusted for the particular patient.
PCR-based methods should not be undertaken unless it is known that the patient is infected with H. pylori, i.e., infection is verified with a urea breath test or a rapid urease test on biopsy specimens. We suggest that it be used after unsuccessful treatment with clarithromycin or when some genetic marker of the strain needs to be identified.
Acknowledgments
We thank Per Falk and Helena Enroth for excellent assistance with proofreading of the manuscript.
This work was supported with grants from the Swedish Medical Research Council (project no. 10848), The Swedish Cancer Society (project no. 3523-B97-03XBC), the Nanna Svartz Foundation, and the Swedish Foundation for Strategic Research (project no. A4J 16:5).
REFERENCES
- 1.Crabtree J E, Taylor J D, Wyatt J I, Heatley R V, Shallcross T M, Tompkins D S, Rathbone B J. Mucosal IgA recognition of Helicobacter pylori 120 kDa protein, peptic ulceration, and gastric pathology. Lancet. 1991;338:332–335. doi: 10.1016/0140-6736(91)90477-7. [DOI] [PubMed] [Google Scholar]
- 2.Enroth, H., O. Nyrén, and L. Engstrand. One stomach—one strain: does Helicobacter pylori strain variation influence disease outcome? Dig. Dis. Sci., in press. [DOI] [PubMed]
- 3.The European Helicobacter Pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Gut. 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falk P. Helicobacter pylori—can the mechanisms of pathogenesis guide us towards novel strategies for treatment and prevention? J Intern Med. 1996;240:319–332. doi: 10.1046/j.1365-2796.1996.75881000.x. [DOI] [PubMed] [Google Scholar]
- 5.Figura N, Guglielmetti P, Rossolini A, Barberi A, Cusi G, Musmanno R A, Russi M, Quaranta S. Cytotoxin production by Campylobacter pylori strains isolated from patients with peptic ulcers and from patients with chronic gastritis only. J Clin Microbiol. 1989;27:225–226. doi: 10.1128/jcm.27.1.225-226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham D Y. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology. 1997;113:1983–1991. doi: 10.1016/s0016-5085(97)70019-2. [DOI] [PubMed] [Google Scholar]
- 7.Hammar M, Tyszkiewicz T, Wadström T, O’Toole P W. Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J Clin Microbiol. 1992;30:54–58. doi: 10.1128/jcm.30.1.54-58.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho S-A, Hoyle J A, Lewis F A, Secker A D, Cross D, Mapstone N P, Dixon M F, Wyatt J I, Tompkins D S, Taylor G R, Quirke P. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543–2549. doi: 10.1128/jcm.29.11.2543-2549.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hultén K, Gibreel A, Sköld O, Engstrand L. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob Agents Chemother. 1997;41:2550–2553. doi: 10.1128/aac.41.11.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Ha T, Chi D S, Ferguson D A, Jiang C, Laffa J J, Thomas E. Differentiation of Helicobacter pylori strains directly from gastric biopsy specimens by PCR-based restriction fragment length polymorphism analysis without culture. J Clin Microbiol. 1997;35:3021–3025. doi: 10.1128/jcm.35.12.3021-3025.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind T, Velhuyzen van Zanten S, Unge P, Spiller R, Bayerdörffer E, O’Morain C, Bardhan K D, Bradette M, Chiba N, Wrangstadh M, Cederberg C, Idström J-P. Eradication of Helicobacter pylori using one week triple therapies combining omeprazol with two antimicrobials: the MACH I study. Helicobacter. 1996;1:138–144. doi: 10.1111/j.1523-5378.1996.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 12.Stone G G, Shortridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka S K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 13.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczebara F, Dhaenens L, Vincent P, Husson M O. Evaluation of rapid molecular method for detection of clarithromycin resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1997;16:162–164. doi: 10.1007/BF01709478. [DOI] [PubMed] [Google Scholar]
- 15.Tummuru M K, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Hulst R W M, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 17.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahlfors J, Meurman J H, Toskala J, Korhonen A, Alakuijala P, Janatuinen E, Kärkkäinen U M, Nuutinen P, Jänne J. Development of a rapid PCR method for identification of Helicobacter pylori in dental plaque and gastric biopsy specimens. Eur J Clin Microbiol Infect Dis. 1995;14:780–786. doi: 10.1007/BF01690993. [DOI] [PubMed] [Google Scholar]
- 19.Wang J T, Lin J T, Sheu J C, Yang J C, Chen D S, Wang T H. Detection of Helicobacter pylori in gastric biopsy tissue by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1993;12:367–371. doi: 10.1007/BF01964436. [DOI] [PubMed] [Google Scholar]