Abstract
Background:
Silent cerebral infarcts (SCI) in sickle cell anemia (SCA) are associated with future strokes and cognitive impairment, warranting early diagnosis and treatment. Detection of SCI, however, is limited by their small size, especially when neuroradiologists are unavailable. We hypothesized that deep learning may permit automated SCI detection in children and young adults with SCA, as a tool to identify the presence and extent of SCI in clinical and research settings.
Methods:
We utilized UNet, a deep learning model, for fully automated SCI segmentation. We trained and optimized UNet using brain MRIs from the Silent Infarct Transfusion (SIT) Trial. Neuroradiologists provided the ground truth for SCI diagnosis, while a vascular neurologist manually delineated SCI on FLAIR and provided the ground truth for SCI segmentation. UNet was optimized for the highest spatial overlap between automatic and manual delineation (Dice Similarity Coefficient, DSC). The optimized UNet was externally validated using an independent single-center prospective cohort of SCA participants. Model performance was evaluated through sensitivity and accuracy (% correct cases) for SCI diagnosis, DSC, intra-class correlation coefficient (ICC, metric of volumetric agreement), and Spearman’s correlation.
Results:
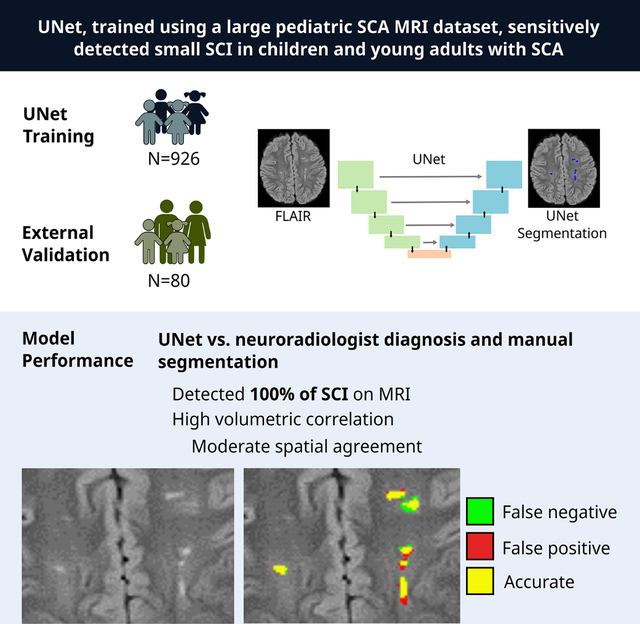
The SIT Trial (N=926, 31% with SCI, median age 6.8 y) and external validation (N=80, 50% with SCI, age 11.5 y) cohorts had small median lesion volumes of 0.40 mL and 0.25 mL, respectively. Compared to the neuroradiology diagnosis, UNet predicted SCI presence with 100% sensitivity and 74% accuracy. In MRIs with SCI, UNet reached a moderate spatial agreement (DSC 0.48) and high volumetric agreement (ICC 0.76, Rho=0.72, P<0.001) between automatic and manual segmentations.
Discussion:
UNet, trained using a large pediatric SCA MRI dataset, sensitively detected small SCI in children and young adults with SCA. While additional training is needed, UNet may be integrated into the clinical workflow as a screening tool, aiding in SCI diagnosis.
Graphical Abstract
INTRODUCTION
Individuals with sickle cell anemia (SCA) are at risk for developing silent cerebral infarcts (SCI), starting from infancy.1 By age 32, over 50% of adults with SCA demonstrate SCI on MRI.2,3 The presence and the absolute volume of SCI are associated with neurocognitive impairment,4 contributing to poor school performance in children and unemployment in adults.5,6 Additionally, children with SCI are at an elevated risk for overt strokes,7 which can cause even greater long-term disability and loss of independence. Early SCI detection may lead to an escalation in treatment strategy to prevent infarct recurrence and may prompt an assessment for individualized education plan for school-aged children.8,9
Several barriers prevent the timely detection of SCI in children with SCA. SCI are not associated with acute neurological symptoms or deficits detectable on bedside examinations. The majority of SCI can only be identified on MRIs, which may be difficult to obtain in young children due to the need for sedation and in certain geographical areas due to limited healthcare resources.10,11 Additionally, SCI are small and are defined as a minimum of 3 mm in diameter, measuring on average less than 0.50 mL.12 Thus, given their small size and varying degrees of signal intensity on MRI, SCI may be missed or misclassified by clinicians unfamiliar with MRI findings in individuals with SCA.11,13 Furthermore, in certain clinical settings with limited resources, while MRIs may be performed, neuroradiologists may not be on-site to review the scans.14 Recent American Society of Hematology guidelines recommend a minimum of one screening MRI in individuals with SCA to evaluate for the presence of SCI.11 As more clinicians incorporate screening MRIs as a part of routine care, clinicians may benefit from automated methods, which may sensitively and efficiently detect SCI.
Over the past decade, the role of machine learning in lesion detection has expanded across several clinical and research settings. Various semi-automated methods have been optimized for white matter hyperintensity (WMH) segmentation on T2 FLAIR brain MRI.15 These existing models, however, may not be suitable for SCI segmentation given SCI are typically markedly smaller than WMHs caused by sporadic, age-related cerebral small vessel disease. Thus, we utilized a deep learning benchmark model based on convolutional neural networks (UNet) and trained UNet on a large, existing pediatric SCA MRI dataset, which included patients with and without SCI. UNet has offered fully automated lesion segmentation in neurological diseases such as multiple sclerosis, ischemic stroke, and cerebral small vessel disease with success.16,17 We hypothesized that UNet could be optimized to detect the presence of SCI on MRI and to accurately delineate infarcts, serving as a potential clinical and research tool in children and young adults with SCA.
METHODS
We followed the reporting guideline on the use of machine learning predictive models in biomedical research.18
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Study populations
We utilized two independent study populations. The population used for UNet model training and optimization consisted of participants from the Silent Infarct Transfusion (SIT) Trial. The SIT Trial was a multicenter, international, randomized clinical trial (NCT00072761) aimed to determine the efficacy of regular blood transfusions for SCI prevention in children with SCA.8,12 Participants were recruited from December 2004 to May 2010 and underwent screening brain MRIs. Participants with a history of overt infarcts were excluded. Additional inclusion and exclusion criteria were listed elsewhere.8,12 The SIT Trial was approved by the institutional review board at each of the 29 participating sites.
The second independent study population was utilized for external UNet model validation. This cohort consisted of both children and young adults enrolled in a longitudinal brain MRI study at Washington University School of Medicine. Inclusion criteria were participants with hemoglobin SS (HbSS), HbS β-thalassemia, or hemoglobin SC (HbSC) genotype. Exclusion criteria were a history of stem cell transplantation, chronic transfusion therapy, overt stroke, or intracerebral hemorrhage. Institutional Review Board approved the study. Written informed consent was obtained from all participants.
For both study populations, laboratory studies including venous hemoglobin, and capillary gel electrophoresis to quantify the percentage of Hb isoforms and vital signs including oxygen saturation by pulse oximetry (SpO2) and systolic blood pressure were obtained before each MRI.
MRI protocol and image processing
The MRI protocol for the SIT Trial was described elsewhere.12 In brief, the majority of scans (>99%) were performed on 1.5 Tesla scanner, < 1% were performed on 3.0 Tesla scanner. Only the axial Fluid-attenuated Inversion Recovery (FLAIR) sequence was used for model training, with an in-plane resolution of ~1 × 1 mm and 5mm slice thickness. For the external validation cohort, all participants underwent a 3.0 Tesla brain MRI (Siemens, Erlangen, Germany). T1 MPRAGE (echo time [TE]/repetition time [TR] = 2.95/ 1,800 ms, inversion time [TI] = 1,000 ms, flip angle = 8°, resolution = 1.0 × 1.0 × 1.0 mm) and FLAIR (TE/TR=93/9000 ms, TI=2500 ms, resolution=1.0 × 0.9 × 5.0 mm for children, 1.0 × 0.9 × 3.0 mm for adults) were acquired.
FLAIR images were skull-stripped and corrected for gain variation using the FMRIB Software Library (fmrib.ox.ac.uk/fsl, v6.0).19 For deep learning-based SCI segmentation, the FLAIR image intensity was further normalized with z-score like transformation using the mean image intensity within the brain region and twice the standard deviation to normalize the image intensity within the range [−1, 1].
Defining ground truth for SCI presence and SCI segmentation
SCI adjudication and manual infarct delineation for the SIT Trial were described previously8,12 and served as the ground truth for UNet training. Briefly, SCI presence and SCI segmentation were defined through a two-step process. First, baseline MRIs were evaluated and adjudicated by the SIT neuroradiology committee. An agreement between two out of three study neuroradiologists was required to determine the presence of SCI within each MRI. A SCI was defined as a MRI signal abnormality at least 3 mm in one dimension and visible in two planes on FLAIR images. Second, in MRIs adjudicated to have SCI, a board-certified vascular neurologist (A.L.F) manually delineated SCI on axial FLAIR images using the Medical Image Processing Analysis and Visualization (https://mipav.cit.nih.gov/) application. The application offers a semi-automatic approach to delineate infarct lesions and create lesion masks for model training. For the external validation cohort, the ground truth for SCI presence was determined by a single neuroradiologist (M.R). Lesions were manually delineated by a board-certified vascular neurologist.
Deep learning segmentation model training
UNet consisted of a down-sampling contracting path and a symmetric up-sampling expansive path, allowing context capture and precise localization (Figure 1).20 For our study, the input of UNet was a 2D axial FLAIR image. The final output was a binary segmentation map indicating the probability of SCI per voxel for that axial image. A representative case of UNet SCI segmentation is shown in Figure 2.
Figure 1. Schematic of UNet architecture.
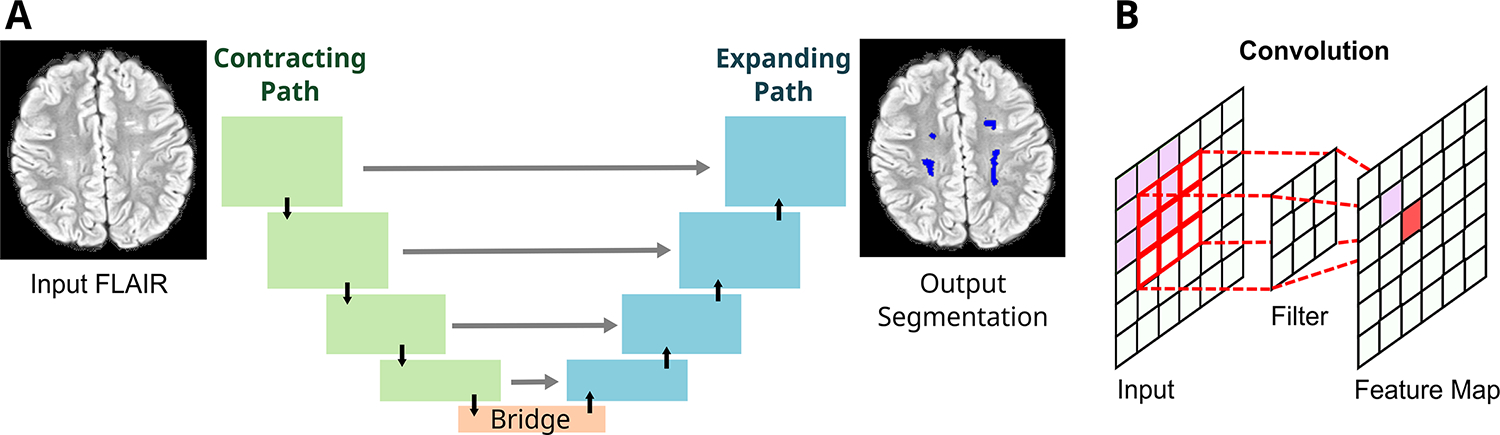
The UNet is a deep learning network that is increasingly utilized for automatic medical image segmentation (A). It is comprised of a contracting path (green boxes), an expanding path (blue boxes), and a bridge that connects the two paths (orange box). The contracting path is a feature extractor and learns an abstract representation of the input 2D axial FLAIR image through a sequence of the contraction layers. Four layers are depicted in the figure. Within each layer, UNet goes through convolutions, which are linear operations that multiplies a set of image pixel values to two-dimensional weights (filter, B). Repeated overlapping application of this filter creates a two-dimensional output array known as a feature map. Each value in the feature map is then passed through a nonlinearity activation function to improve network learning. Finally, the spatial dimensions (height and width) of the feature maps are reduced by half between each contraction layers to reduce the computational time by decreasing the number of trainable parameters.
The expanding path projects the abstract representation of the image in the form of lower resolution discriminative, abstract features learnt during the contracting path, onto the higher resolution voxel space to eventually generate an output segmentation mask. This is carried out within the expansion layers through upsampling which restores the condensed feature maps to the original size of the input image by expanding the feature dimensions, and combining with skip connection (gray arrow) feature maps from the corresponding expansion layers. These skip connections ensure conservation of features from earlier layers to preserve learning. This design allows the UNet to learn increasingly complex features at multiple scales without sacrificing spatial information. Finally, the feature maps are converted to an output probability map indicating the probability of each image voxel being a SCI, with spatial dimensions corresponding to the original FLAIR image. The UNet-estimated SCI volume was calculated as the product of [the number of SCI positive voxels with a probability >=0.5] and [the volume of each voxel].
Figure 2. Representative UNet segmentation of SCI.
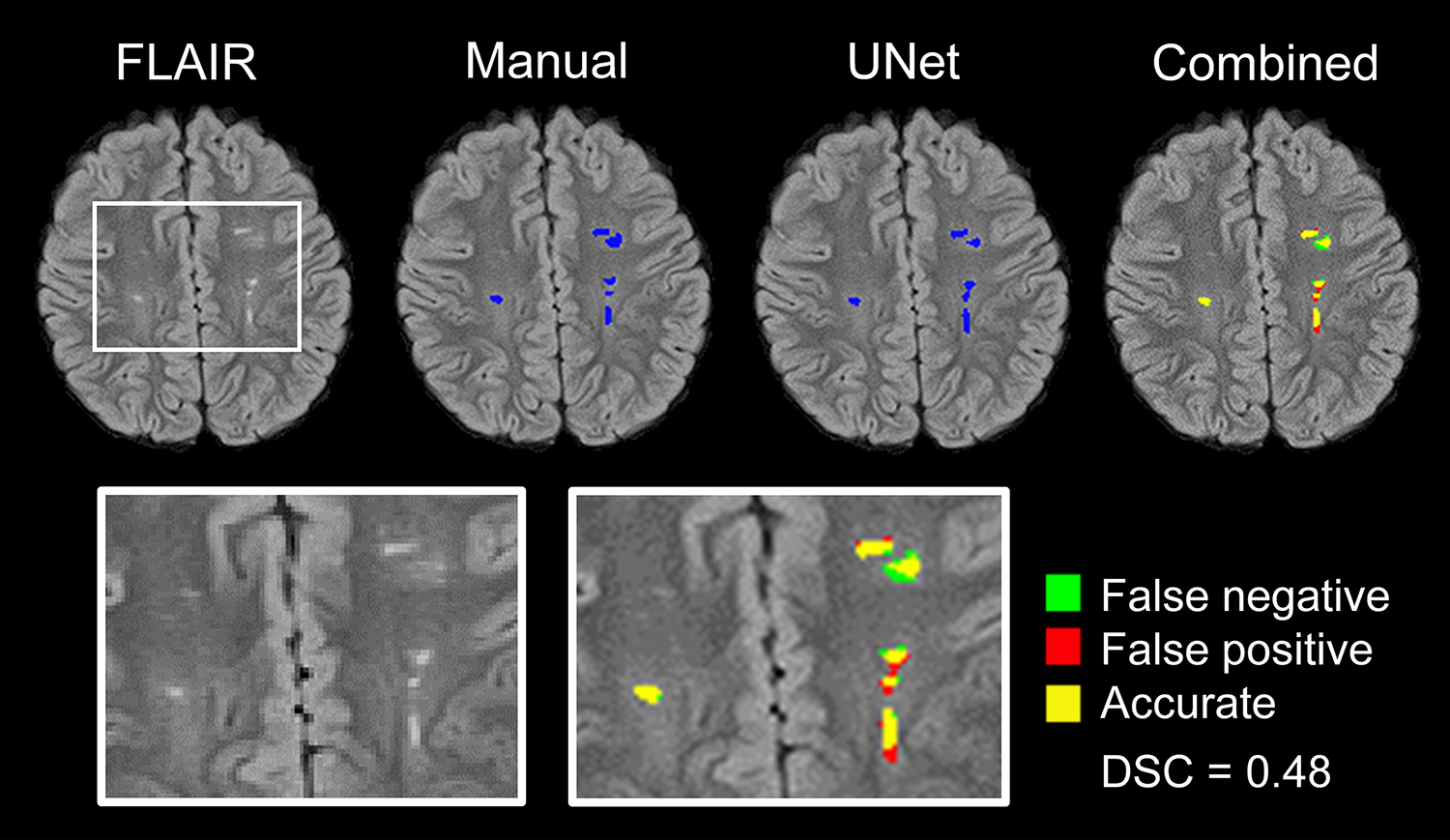
The original FLAIR map, manual segmentation mask, and UNet segmentation mask are shown. To determine spatial agreement, the manual segmentation was overlaid onto the UNet segmentation (combined). A region containing SCI, before and after segmentations, was enlarged for better visualization (boxes). Manual segmentation was defined as the “ground truth”. False negative, false positive, and accurate segmentations are shown in green, red, and yellow, respectively. In this example, UNet achieved a Dice Similarity Coefficient of 0.48.
Available screening MRIs from the SIT Trial were randomly divided into 90% for model training (“training” dataset) and 10% for model optimization (“optimization” dataset, Supplemental Figure 1). First, through the training dataset, we initiated model training using a pre-trained UNet model, previously optimized to detect white matter hyperintensities in adults with cerebral small vessel disease.17 Using the optimization dataset, we chose the UNet model that performed the best in lesion segmentation, achieving the maximal Dice Similarity Coefficient (DSC). DSC is a spatial overlap index that evaluates image similarity between UNet segmentation and manual delineation. It ranges from 0, indicating no spatial overlap, to 1, indicating perfect overlap. A DSC of 0.7 or greater is considered excellent agreement.21
Due to the relative scarcity of voxels positive for SCI compared to voxels negative for SCI, UNet could have easily achieved high accuracy by classifying all voxels as negative for SCI, yet at the expense of high false negatives. As a solution, using weighted binary cross entropy, we assigned a range of weights (1, 10, 100, and 1000) to SCI positive class to boost its weight relative to SCI negative class. A weight of 10, or SCI positive voxels set to 10 times the weight of SCI negative voxels, achieved the highest DSC in the optimization dataset.
Performance evaluation
The optimized UNet model was applied to our external validation cohort to assess model performance and generalizability. To assess UNet performance at detecting SCI presence, sensitivity, specificity, and accuracy of UNet performance were calculated. Accuracy is the proportion of correct cases, either true positive or true negative, identified by UNet. We categorized a MRI as positive for SCI if the UNet predicted volume was greater than 0 mL. UNet SCI detection was compared against the “ground-truth” SCI presence as adjudicated by a neuroradiologist.
To assess the ability of UNet to accurately delineate the location and the extent of SCI, DSC was used to evaluate spatial (i.e. voxel-wise) agreement, while intra-class correlation coefficient (ICC, two-way mixed effect, absolute agreement) and bivariate correlations were used to evaluate volumetric agreement between UNet and manual segmentations.
Statistical analysis
Data were reported as percentages for categorical data and median with interquartile range for continuous data. The normality of the distribution was assessed by the Shapiro-Wilk test. Clinical characteristics and laboratory measures were compared between the SIT Trial cohort and the external validation cohort using Mann Whitney U. Chi square or Fisher’s exact test was used for categorical variables.
Bivariate correlations between UNet estimated volume and ground-truth volume were performed using Spearman’s rank correlation (ρ). ICC was also performed to assess Unet predicted and ground-truth volume agreement. Bland-Altman plots were generated to assess data variability and the agreement between estimated and ground-truth volumes. Mean bias, and lower and upper limits of agreement (LOA, ±1.96 *standard deviation), were calculated. One Sample t-Test was performed to evaluate if the mean bias was statistically different from zero. Linear regression analysis was performed to evaluate if the bias between the estimated and ground-truth volumes (Y-axis of Bland-Altman plot) was independent of the ground-truth SCI volume (X-axis of Bland-Altman plot). Statistical analyses were performed using SPSS version 26 (IBM Corp, Armonk, NY).
RESULTS
Clinical characteristics of the training, optimization, and external validation cohorts
The clinical characteristics of 926 pediatric participants who underwent MRI screening in the SIT Trial are shown in Table 1. The MRIs were partitioned into 90% for model training and 10% for model optimization. The 90% “training dataset” included N=833, of which 258 MRIs (31%) contained SCI. The 10% “optimization dataset” included N=93, of which 29 MRIs (31%) contained SCI. UNet was externally validated using an independent “external validation cohort” which consisted of 80 children and young adults with SCA, of whom 40 participants (50%) had SCI.
Table 1.
Baseline clinical characteristics of Silent Infarct Transfusion (SIT) Trial and external validation cohort
| SIT Trial | WashU Cohort | P value* | ||
|---|---|---|---|---|
|
|
||||
| Training dataset (N=833) | Optimization dataset (N=93) | External validation (N=80) | ||
| Median (IQR) or N (%) | Median (IQR) or N (%) | Median (IQR) or N (%) | ||
|
| ||||
| Age, y | 8.8 (6.8, 10.9) | 9.4 (7.1, 11.9) | 11.5 (8.3, 19) | <0.001 |
| Male, N (%) | 417 (50) | 49 (43) | 40 (50) | 1.0 |
| African American, N (%) | 788 (94.6) | 86 (92.5) | 80 (100) | 0.03 |
| Hemoglobin, g/dl | 8 (7.4, 8.9) | 8 (7.5, 8.7) | 8.4 (7.7, 9.5) | 0.002 |
| Hemoglobin F, % | 10 (6, 17) | 11 (5, 16) | 9.2 (3.2, 21.4) | 0.46 |
| Hemoglobin S, % | 84 (78, 89) | 85 (77, 88) | 74 (59.4, 84.0) | <0.001 |
| White blood cell, 106/μL | 12 (10, 14.8) | 12.9 (10, 14.7) | 10.5 (8.2, 13.0) | <0.001 |
| SpO2, % | 97 (95, 99) | 96 (94, 98) | 98 (95, 99) | 0.32 |
| Systolic blood pressure, mmHg | 108 (101, 115) | 107 (101, 114) | 116 (110, 122) | <0.001 |
| SCI present, N (%) | 258 (31) | 29 (31) | 40 (50) | <0.001 |
| SCI volume, mL | 0 (0, 0.13) | 0 (0, 0.23) | 0.01 (0, 0.25) | 0.005 |
| SCI volume, within participants with SCI, mL | 0.40 (0.18, 1.00) | 0.42 (0.29, 2.32) | 0.25 (0.13, 0.72) | 0.03 |
Abbreviations: SIT, Silent Infarction Transfusion; SCI, silent cerebral infarct; SpO2, peripheral oxygen saturation
Comparison between the SIT Trial cohort and the external validation cohort
Compared to the SIT Trial cohort, the external validation cohort was older in age (The SIT Trial median age: [IQR] 8.9 [6.8, 11.0] years; the external validation cohort median age: 11.5 [8.3, 19] years, p<0.001). Median infarct volume within participants with SCI was small at 0.40 mL and 0.25 mL for the SIT Trial and the external validation cohorts, respectively (p=0.03). Additional baseline clinical characteristics are presented in Table 1.
UNet performance for automated detection of silent cerebral infarcts
We aimed to use UNet as a screening tool for SCI with maximal sensitivity. To minimize false negative cases while allowing for false positive cases, we categorized a MRI as positive for SCI if UNet-predicted volume was greater than 0 mL. The presence of SCI as adjudicated by a neuroradiologist served as the ground-truth. In the optimization dataset, UNet had a sensitivity of 90%, specificity of 63%, and accuracy of 71%, with three false negative cases.
By comparison, in the external validation cohort, UNet had a sensitivity of 100%, specificity of 48%, and accuracy of 74%, with zero false negative cases and 21 false positive cases. While false negative cases were rare, occurring only in the optimization dataset, false positive cases were more common in both groups, and as expected given the goal to maximize sensitivity, at the expense of specificity. Representative examples of false negative cases from the optimization dataset and false positive cases for the external validation cohort are shown in Figure 3.
Figure 3. SCI misclassification by UNet.
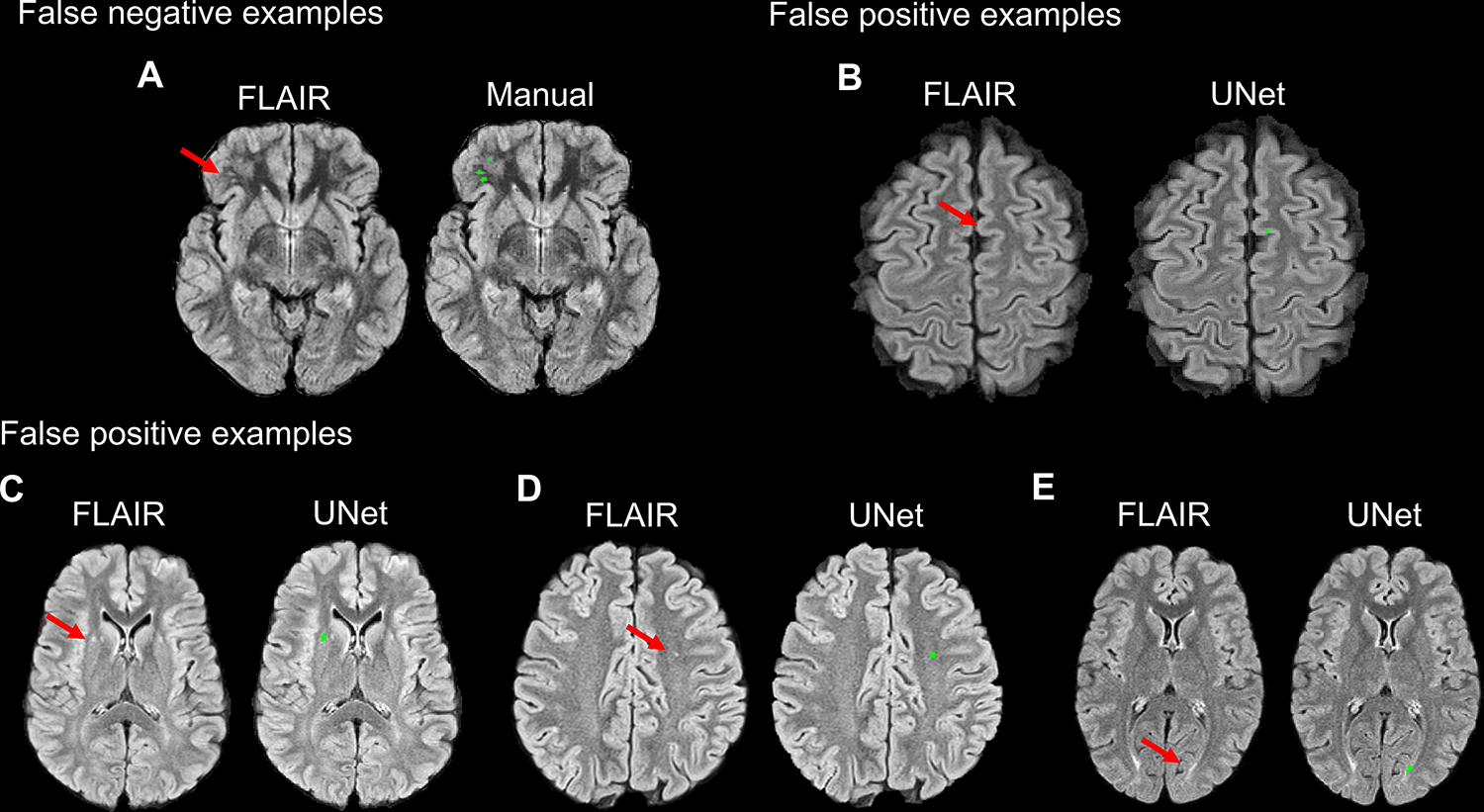
For demonstration purposes, false negative SCI case examples were selected from the optimization dataset as no false negatives were present in the external validation cohort. In the optimization dataset, UNet failed to segment small and faint SCI lesions in the subcortical white matter (arrow, A). In false positive SCI case examples from the external validation cohort, UNet erroneously segmented portions of cortical gray matter (arrow, B) and CSF pulsation artifact adjacent to the lateral ventricle (arrow, C) as SCI lesions. UNet also segmented white matter FLAIR hyperintensities smaller than 3mm in diameter (arrow, D), which fell below the size threshold for radiological definition of SCI, and symmetrical periventricular white matter changes (arrow, E) that are non-specific in adults and of unknown significance in children.
Of the false positive cases in the external validation cohort, UNet erroneously segmented portions of cortical or deep gray matter (8 cases, Figure 3B) and CSF pulsation artifact (2 cases, Figure 3C) as SCI lesions. UNet also segmented white matter FLAIR hyperintensities smaller than 3mm in diameter (3 cases, Figure 3D), which fell below the size threshold for radiological definition of a SCI, and non-specific symmetrical periventricular white matter hyperintensities (4 cases, Figure 3E). Head motion accounted for the rest of the false positive segmentations.
UNet performance for spatial and volumetric accuracy of silent cerebral infarcts
UNet performance for spatial agreement was evaluated on a subset of MRIs positive for SCI, excluding MRIs without SCI. The median infarct volume for the optimization dataset and external validation cohort was small at 0.42 mL and 0.25 mL, respectively. In the optimization dataset, UNet achieved moderate spatial agreement on MRIs with SCI only (median DSC [IQR]: 0.48 [0.25, 0.60]). In the external validation cohort, UNet achieved similar spatial agreement on MRIs with SCI only (0.48 [0.30, 0.58]). Notably, DSC was not measured for all MRI scans. On a SCI negative MRI, DSC could only be 1 or 0, depending on whether UNet correctly segmented zero voxels (DSC=1) or incorrectly segmented one or more voxels (DSC=0). Thus, inclusion of MRIs without SCI may falsely over- or under-estimate median DSC values, since these two extreme values would be included.
We next examined the volumetric correlation of estimated volume relative to ground-truth volume (Figure 4A, B). In the optimization dataset, median UNet estimated and ground-truth volumes were 0.30 (0.09, 1.76) mL and 0.42 (0.29, 2.31) mL, respectively. In the external validation cohort, median UNet and ground-truth volumes were 0.33 (0.16, 0.57) mL and 0.25 (0.13, 0.72) mL, respectively. UNet volume was highly correlated with ground-truth volume within the optimization dataset (ρ = 0.70, p < 0.001, Figure 4A) and within the external validation cohort (ρ = 0.74, p < 0.001, Figure 4B). UNet also demonstrated high volumetric agreement in the optimization dataset (ICC 0.73 [98% CI, 0.43–0.87]) and the external validation cohort (ICC 0.76 [98% CI, 0.55–0.87]).
Figure 4. UNet SCI volume estimation in optimization and external validation cohort.
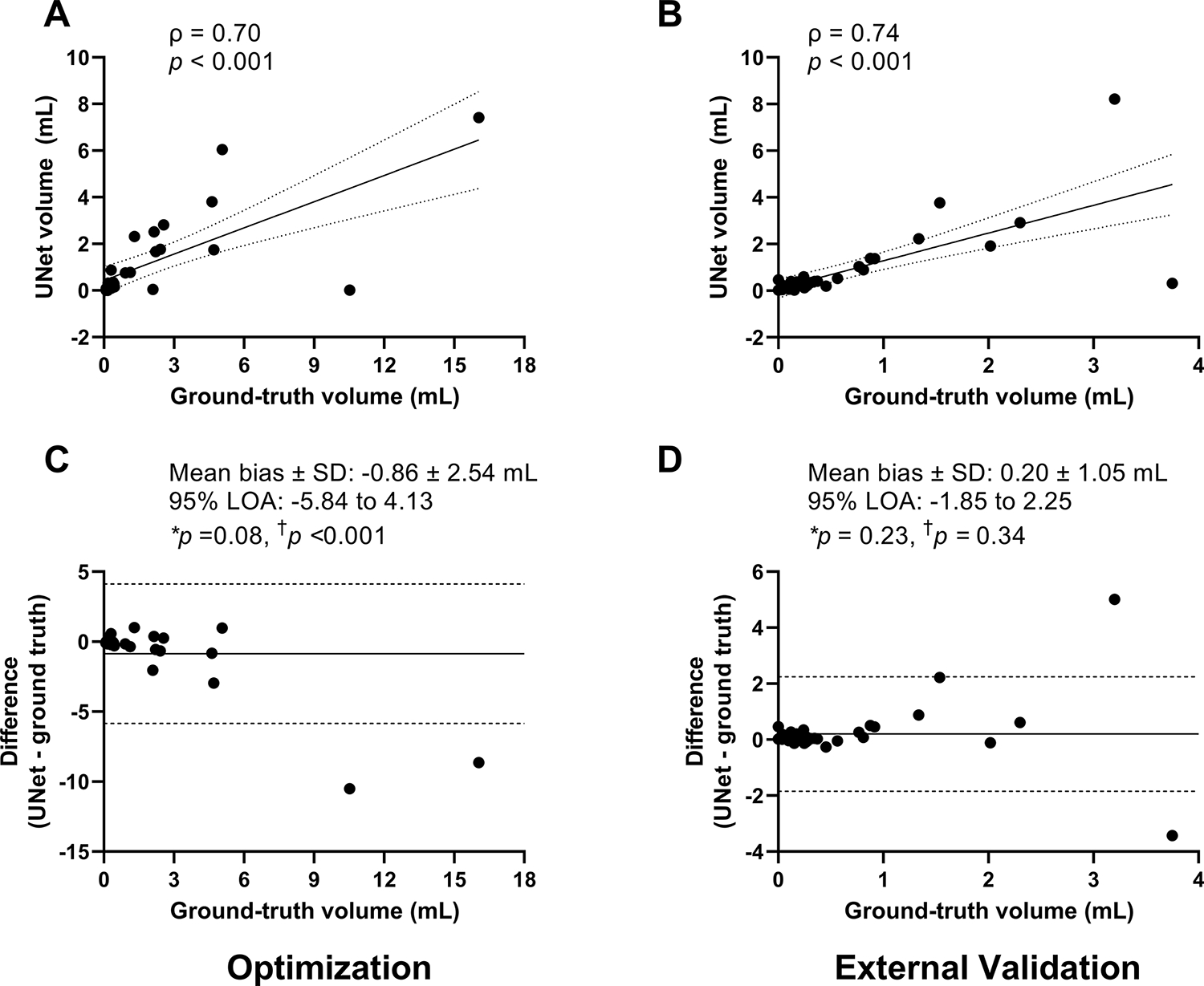
Simple correlations between UNet volume vs ground-truth volume in the optimization dataset (A) and external validation cohort (B) are shown. UNet volume was strongly and highly correlated with ground-truth volume. Linear regression line (solid line) with 95% confidence interval (dashed lines) are shown. On Bland-Altman plots, in the optimization dataset, there was a proportional bias where UNet systematically underestimated SCI volume (p < 0.001, C). On average, UNet underestimated SCI volume by 0.86 mL, trending significance (p = 0.08, C). In contrast, in the external validation cohort, there was no proportional bias in UNet predicted volumes compared to ground-truth volumes (p = 0.34, D). Its averaged difference between UNet and ground-truth volumes was not statistically different from zero (mean bias 0.13 mL, p = 0.11). The mean bias (solid line) and the lower and upper limits of agreement (LOA, ±1.96 *standard deviation, dashed lines) are shown. A negative difference on the Y-axis represents under-estimation by machine learning. A positive difference represents over-estimation by machine learning. *One Sample t-Test. † Linear regression.
To evaluate for the presence and direction of bias between estimated and ground-truth SCI volumes, Bland-Altman plots were created (Figure 4C, D). In the optimization dataset, UNet, on average, underestimated SCI volume by 0.86 mL, trending significance (p = 0.08). There was a proportional bias where UNet had greater underestimation of SCI volume as ground-truth volume increased (p < 0.001). In contrast, in the external validation cohort, there was no proportional bias in UNet predicted volumes as ground-truth volume increased (p = 0.34). The averaged difference between UNet and ground-truth was not statistically different from zero (mean bias 0.13 mL, p = 0.11).
DISCUSSION
Our results demonstrate proof-of-concept that deep learning may serve as a tool for SCI identification and spatial delineation in children and young adults with SCA. Traditionally, a major barrier for accurate SCI detection has been their small size and varying signal intensity and contrast with surrounding white matter.11,13 Infarcts are typically singular and less than 0.5 mL.12 Thus, to non-neuroradiologists, identifying SCI on MRI may be likened to “finding a needle in a haystack” and can be complicated by differentiating SCI from radiological mimics of SCI.13 To aid in SCI detection, we trained and optimized UNet using one of the largest pediatric SCA MRI datasets available, including nearly 1000 children with and without SCI. We applied our best performing UNet model to an independent cohort and found that, not only did UNet offer high sensitivity at diagnosing SCI, but also UNet had excellent volume correlation and moderate spatial agreement between automated and manual segmentations. By demonstrating external validity with high sensitivity for detecting SCI, deep learning may be considered as a tool to augment clinical workflow for the general SCA population.
American Society of Hematology has recently recommended a minimum of one screening MRI in individuals with SCA to evaluate for SCI.11 This guideline will likely increase the need for neuroradiologists, specifically pediatric subspecialists, who are trained to recognize SCI and its mimics,11 as well as certain MRI findings such as focal white matter hyperintensities that are considered “normal” in older adults, but not in children. In underserved clinical care settings, where pediatric neuroradiologists are scarce, incorporating deep learning into clinical workflow may alleviate demand and assist in SCI detection. This UNet model yielded excellent sensitivity and good accuracy in pediatric and young adult SCA populations. By maximizing sensitivity, at the expense of yielding false positive cases, we aimed to minimize false negative cases on initial screening (Figure 3A), as a missed diagnosis might delay a patient’s access to important medical therapy for future SCI prevention and neuropsychological testing.8,9,11 To prevent a patient from being falsely diagnosed with SCI, radiologists would still be needed to review MRIs that initially screen positive for SCI.
Some literature has supported separate radiological definitions of SCI for children and adults with SCA.11 The pediatric definition of SCI, ≥ 3 mm in diameter and visible in two planes on FLAIR, was set by the SIT Trial12 and correlated with cognitive impairment and increased risk of new SCI and future overt strokes. The adult definition of SCI was based on, now outdated, imaging characteristics of lacunar stroke, a hyperintense lesion ≥ 5 mm on FLAIR with corresponding T1 hypointensity.22,23 As image resolution and signal-to-noise improve, with 3D acquisition becoming increasingly available, we are likely to visualize smaller and more subtle FLAIR hyperintensities.11 However, we need to evaluate if these subtle FLAIR hyperintensities are predictive of new cerebral infarcts or cognitive impairment before attributing them as clinically important. The association of SCI burden with cognitive impairment is dependent on lesion size.24 For example, the association between SCI presence and increased cognitive impairment was no longer detected when the more restrictive adult definition of SCI was applied to children from the SIT Trial.24 In the false positive SCI cases from our validation cohort, some cases were erroneous segmentations of gray matter (Figure 3B) and ventricular CSF pulsation artifact (3C), while other cases were segmentations of FLAIR hyperintensities < 3 mm (3D) and symmetrical periventricular hyperintensities that are considered non-specific in adults but of unclear clinical significance in children (3E). Further studies are needed to examine the clinical significance of these FLAIR hyperintensities that are not categorized as SCI under current radiological definitions. When implementing automated SCI detection in older adults with SCA, additional training would be required to ensure detection of additional MRI findings associated with aging and non-SCA cerebrovascular risk factors, such as lacunar strokes.
Automated SCI segmentation and volume quantification in patients with silent infarcts have potential to revolutionize research applications, especially within large, multi-center datasets in which manual lesion delineation is time-prohibitive, taking hundreds of hours and specialized training, and at risk for variability in lesion delineation due to user error and differing imaging quality. In contrast, deep learning algorithms can obtain both the volume and the location of a lesion in minutes. Thus, detection of new lesions and quantification of progressive lesion growth can be easily performed and serve as valuable imaging endpoints for many trials designed to prevent SCI and understand their cognitive ramifications. Additionally, unlike human raters, machine learning does not produce variability on repeated measurements. Thus, longitudinal assessments of SCI growth may be more accurate through automated approaches.
Our current UNet model demonstrated high volumetric agreement (ICC 0.76) and moderate spatial agreement (DSC 0.48) in the external validation cohort. The accuracy of UNet estimated volume, however, may be influenced by lesion size. UNet was mainly trained on SCI less than 0.5 mL from the SIT Trial training dataset. As a result, UNet markedly underestimated lesions greater than 10 mL in the optimization dataset and was overall less accurate at measuring lesions over 3 mL (Figure 4C, D). In the optimization dataset, UNet was less accurate at segmenting the larger lesions, thus skewing the mean bias, resulting in the mean bias to exceed the average ground-truth infarct volume. Additional model training is needed to improve SCI volume quantification over a wide range of lesion sizes.
Direct comparison of the UNet model with other machine learning models is limited by a lack of published research on automated SCI segmentation in SCA. In WMH segmentation of participants with non-SCA cerebral small vessel disease, various deep learning models have obtained similar to higher spatial agreement, with DSC ranging from 0.5 to 0.8.16 Variations in model performance may be explained by the significant size difference between SCI and cerebral small vessel disease-related WMH, which is often 30–40 times larger than SCI, but recent algorithms have improved detection of even smaller WMH lesions.17 A previous study has shown that spatial agreement was positively associated with lesion volume in a large MRI database of WMH caused by various neurological diseases.16 As the quality of image acquisition improves over time, minimizing the effects of artifacts, additional model development will likely allow improved automated segmentation of very small lesions.
Our study has several strengths. We used the largest pediatric SCA MRI database of nearly 1000 children to train our deep learning model. Next, we externally validated our trained model in a cohort independent from the SIT Trial. Despite key demographic, clinical, and MRI protocol differences between the SIT Trial and the external validation cohort, we achieved an excellent sensitivity and good accuracy at identifying SCI in our validation cohort, which had a very small median SCI volume of 0.25 ml. Thus, deep learning has the potential to be applied to a broader SCA population in clinical practice and in research settings. Our study has several limitations. First, the prevalence of SCI in our external validation cohort was higher than previously reported.3 Potential causes include having defined SCI as ≥ 3 mm in diameter on 2D axial FLAIR, without confirmation on coronal FLAIR. We also utilized 3 Tesla MR for all participants in the external validation cohort, whereas previous studies, including the SIT Trial, utilized 1.5 Tesla for the majority of the scans. Due to differences in SCI definition and better image quality, we detected smaller SCIs and a higher proportion of SCI-positive MRIs. To accommodate the differences in SCI definition, we only utilized 2D axial FLAIR images when training and optimizing UNet, thus potentially lowering model performance. Additional studies are needed to evaluate if improving spatial resolution or signal to noise ratio by incorporating coronal sections or 3D FLAIR will improve model performance. Secondly, given the difficulty at times visualizing small and faint SCI that are around 3 mm and the user subjectivity in identifying and outlining SCI, the “ground-truth” may not always be correct. False positive cases predicted by UNet may be true positives in some instances. As a result, the “true” specificity and accuracy of our UNet model may be higher than reported. Furthermore, not all hyperintensities on FLAIR are SCI. Radiographical mimics of SCI include terminal zones of myelination and periventricular leukomalacia.13 Likewise, head motion and CSF pulsation produce artifacts on FLAIR also lower model specificity. Additional model training is needed to differentiate SCI from SCI mimics and artifacts. Thirdly, we optimized UNet to be very sensitive in detecting voxels positive for SCI in order to avoid false negative classifications, but at the expense of producing false positive ones. A tiered training approach, first focusing on SCI detection and then lesion segmentation, may be needed to improve overall accuracy. Lastly, while UNet is currently the deep learning benchmark model for medical image segmentation, we will explore other network structures in the future for better segmentation performance.
In conclusion, we developed and tested a deep learning approach to automate SCI detection and delineation in children and young adults with SCA. Our findings demonstrate that, with additional model development, deep learning may aid clinical SCI diagnosis and imaging endpoint quantifications in a research setting, improving the overall care of individuals with SCA.
Supplementary Material
SOURCES OF FUNDING
NIH, National Center for Advancing Translational Science (KL2TR002346 [Y.W.]); NIH, National Heart, Lung, and Blood Institute (R01 HL129241 [A.L.F.], K23HL136904 and R01HL157188 [M.E.F.]); and the NIH, National Institute of Neurological Disorders and Stroke (K23NS099472 and R01NS121065 [K.P.G.], RF1NS116565 [A.L.F, H.A.]).).
ABBREVIATIONS
- DSC
Dice Similarity Coefficient
- ICC
Intra-class correlation coefficient
- SCI
Silent cerebral infarcts
- SCA
Sickle cell anemia
- SIT
Silent Infarct Transfusion
- UNet
Convolutional neural network
- WMH
White matter hyperintensity
Footnotes
DISCLOSURES
Dr. Fields reports salary support in Global Blood Therapeutics, Inc, Monteore Medical Center, and Proclara Biosciences and from Washington University in St. Louis. Dr. An reports compensation from Pfizer Inc. for consultant services and grants from the National Institute of Health. Dr. Binkley reports salary support from CNS Consultants LLC and OpenCell Technologies. Dr. Lee reports salary support from Biogen. Dr. McKinstry reports salary support from NOUS Imaging Inc, Philips, and Siemens. Dr. Jordan reports compensation for expert witness and salary support from National Institutes of Health and Vanderbilt University. Dr. DeBaun reports salary support from Forma Therapeutics, Global Blood Therapeutics, Graphite Bio, Novartis, and Vanderbilt University. Y. Chen, Y. Wang, C-L Phuah, K.P. Guilliams, S. Fellah, M. Reis, and A.L. Ford report no disclosures relevant to the manuscript.
SUPPLEMENTAL MATERIAL
REFERENCES
- 1.Kwiatkowski JL, Zimmerman RA, Pollock AN, Seto W, Smith-Whitley K, Shults J, Blackwood-Chirchir A, Ohene-Frempong K. Silent infarcts in young children with sickle cell disease. British journal of haematology. 2009;146:300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Fellah S, Fields ME, Guilliams KP, Binkley MM, Eldeniz C, Shimony JS, Reis M, Vo KD, Chen Y, et al. Cerebral Oxygen Metabolic Stress, Microstructural Injury, and Infarction in Adults With Sickle Cell Disease. Neurology. 2021;97:e902–e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kassim AA, Pruthi S, Day M, Rodeghier M, Gindville MC, Brodsky MA, Debaun MR, Jordan LC. Silent cerebral infarcts and cerebral aneurysms are prevalent in adults with sickle cell anemia. Blood. 2016;127:2038–2040. [DOI] [PubMed] [Google Scholar]
- 4.van der Land V, Hijmans CT, de Ruiter M, Mutsaerts HJMM, Cnossen MH, Engelen M, Majoie CBLM, Nederveen AJ, Grootenhuis MA, Fijnvandraat K. Volume of white matter hyperintensities is an independent predictor of intelligence quotient and processing speed in children with sickle cell disease. British Journal of Haematology. 2015;168:553–556. [DOI] [PubMed] [Google Scholar]
- 5.Sanger M, Jordan L, Pruthi S, Day M, Covert B, Merriweather B, Rodeghier M, DeBaun M, Kassim A. Cognitive deficits are associated with unemployment in adults with sickle cell anemia. Journal of Clinical and Experimental Neuropsychology. 2016;38:661–671. [DOI] [PubMed] [Google Scholar]
- 6.Schatz J, Brown RT, Pascual JM, Hsu L, DeBaun MR. Poor school and cognitive functioning with silent cerebral infarcts and sickle cell disease. Neurology. 2001;56:1109–1111. [DOI] [PubMed] [Google Scholar]
- 7.Pegelow CH, Macklin EA, Moser FG, Wang WC, Bello JA, Miller ST, Vichinsky EP, DeBaun MR, Guarini L, Zimmerman RA, et al. Longitudinal changes in brain magnetic resonance imaging findings in children with sickle cell disease. Blood. 2002;99:3014–3018. [DOI] [PubMed] [Google Scholar]
- 8.DeBaun MR, Gordon M, McKinstry RC, Noetzel MJ, White DA, Sarnaik SA, Meier ER, Howard TH, Majumdar S, Inusa BPD, et al. Controlled Trial of Transfusions for Silent Cerebral Infarcts in Sickle Cell Anemia. New England Journal of Medicine. 2014;371:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King AA, White DA, McKinstry RC, Noetzel M, DeBaun MR. A pilot randomized education rehabilitation trial is feasible in sickle cell and strokes. Neurology. 2007;68:2008–2011. [DOI] [PubMed] [Google Scholar]
- 10.Barkovich MJ, Xu D, Desikan RS, Williams C, Barkovich AJ. Pediatric neuro MRI: tricks to minimize sedation. Pediatric radiology. 2018;48:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBaun MR, Jordan LC, King AA, Schatz J, Vichinsky E, Fox CK, McKinstry RC, Telfer P, Kraut MA, Daraz L, et al. American Society of Hematology 2020 guidelines for sickle cell disease: prevention, diagnosis, and treatment of cerebrovascular disease in children and adults. Blood Advances. 2020;4:1554–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford AL, Ragan DK, Fellah S, Binkley MM, Fields ME, Guilliams KP, An H, Jordan LC, McKinstry RC, Lee JM, et al. Silent infarcts in sickle cell disease occur in the border zone region and are associated with low cerebral blood flow. Blood. 2018;132:1714–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeBaun MR, Armstrong FD, McKinstry RC, Ware RE, Vichinsky E, Kirkham FJ. Silent cerebral infarcts: a review on a prevalent and progressive cause of neurologic injury in sickle cell anemia. Blood. 2012;119:4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kija EN, Saunders DE, Munubhi E, Darekar A, Barker S, Cox TCS, Mango M, Soka D, Komba J, Nkya DA, et al. Transcranial Doppler and Magnetic Resonance in Tanzanian Children with Sickle Cell Disease. Stroke. 2019;50:1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinen R, Steenwijk MD, Barkhof F, Biesbroek JM, van der Flier WM, Kuijf HJ, Prins ND, Vrenken H, Biessels GJ, de Bresser J, et al. Performance of five automated white matter hyperintensity segmentation methods in a multicenter dataset. Scientific Reports. 2019;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Duan Y, Wang X, Zhuo Z, Haller S, Barkhof F, Liu Y. A deep learning algorithm for white matter hyperintensity lesion detection and segmentation. Neuroradiology. 2021;1:3. [DOI] [PubMed] [Google Scholar]
- 17.Phuah CL, Chen Y, Strain JF, Yechoor N, Laurido-Soto OJ, Ances BM, Lee JM. Association of Data-Driven White Matter Hyperintensity Spatial Signatures With Distinct Cerebral Small Vessel Disease Etiologies. Neurology. 2022;99:e2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, Shilton A, Yearwood J, Dimitrova N, Ho TB, et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res. 2016;18:e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. NeuroImage. 2012;62:782–790. [DOI] [PubMed] [Google Scholar]
- 20.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: IEEE Access. 2015:234–241. [Google Scholar]
- 21.Anbeek P, Vincken KL, Van Osch MJP, Bisschops RHC, Van Der Grond J. Probabilistic segmentation of white matter lesions in MR imaging. NeuroImage. 2004;21:1037–1044. [DOI] [PubMed] [Google Scholar]
- 22.Vichinsky EP, Neumayr LD, Gold JI, Weiner MW, Rule RR, Truran D, Kasten J, Eggleston B, Kesler K, McMahon L, et al. Neuropsychological dysfunction and neuroimaging abnormalities in neurologically intact adults with sickle cell anemia. JAMA - Journal of the American Medical Association. 2010;303:1823–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, Lindley RI, O’Brien JT, Barkhof F, Benavente OR, et al. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. The Lancet Neurology. 2013;12:822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhury NA, DeBaun MR, Rodeghier M, King AA, Strouse JJ, McKinstry RC. Silent cerebral infarct definitions and full-scale IQ loss in children with sickle cell anemia. Neurology. 2018;90:E239–E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


