Abstract
Lung cancer heterogeneity is a major barrier to effective treatments and encompasses not only the malignant epithelial cell phenotypes and genetics, but also the diverse tumor-associated cell types. Current techniques used to investigate the tumor microenvironment (TME) can be time consuming, expensive, complicated to interpret, and often involve destruction of the sample. Here we utilize standard hematoxylin and eosin (H&E)-stained tumor sections and the HALO® Artificial Intelligence (AI) nuclear phenotyper software to characterize six distinct cell types (epithelial, mesenchymal, macrophage, neutrophil, lymphocyte, and plasma cells) in both murine lung cancer models and human lung cancer samples. CD3 immunohistochemistry and lymph node sections were used to validate lymphocyte calls, while F4/80 immunohistochemistry was used for macrophage validation. Consistent with numerous prior works, we demonstrate that macrophages predominate the adenocarcinomas while neutrophils predominate the squamous cell carcinomas in murine samples. In human samples, we show a strong negative correlation between neutrophils and lymphocytes as well as between mesenchymal cells and lymphocytes, and that higher percentages of mesenchymal cells correlate with poor prognosis. Taken together, we demonstrate utility of this AI software to identify, quantify and compare distributions of cell types on standard H&E-stained slides. Given the simplicity and cost-effectiveness of this technique, it may be widely beneficial for researchers designing new therapies, and clinicians working to select favorable treatments for their patients.
Keywords: Artificial intelligence, non-small cell lung cancer, tumor microenvironment, tumor immunology
INTRODUCTION
The complexity of the tumor microenvironment is vast, with numerous cellular and structural patterns that are distinct in each cancer. Even within non-small cell lung cancer (NSCLC), a wide variety of histological variants leads to numerous possible diagnoses, each with its own preferred treatment regimen 1,2. Among the different subtypes of NSCLC, adenocarcinoma (ADC) and squamous cell carcinoma (SCC) are the most commonly diagnosed. Pathological assessment of patient biopsies has long been the standard for diagnosis, and pathologists are skilled at identifying the multitude of cell types, in addition to the malignant epithelial cells, that can be found in carcinomas 3-5. While the role of the pathologist will never be replaced, a method for less highly trained researchers to quickly and accurately assess the types of cells within a histological sample would greatly benefit the field. As the NSCLC treatments move towards the use of immunotherapy as a first line therapy for most cancers 2, assessment of the tumor microenvironment (TME) is critical.
ADC and SCC TMEs are very heterogeneous and contain numerous types of cells that are often collectively referred to as ‘stroma’. Commonly identified cell types in NSCLC TME include mesenchymal cells, plasma cells, macrophages, neutrophils, and lymphocytes 3,4,6,7. Although cells such as CD8+ T cells in the TME can be tumor eliminating, it is generally accepted that tumors reprogram the microenvironment to favor tumor promoting cells. In particular, mesenchymal cells, such as cancer associated fibroblasts (CAFs), can produce a variety of cytokines, including TGF-β that can create an immunosuppressive TME by repressing CD8+ T cells and increasing regulatory T cells 8,9. Similarly, tumor-associated neutrophils are also thought to be predominately immunosuppressive, in particular through production of arginase and reactive oxygen species 10,11. A high neutrophil to lymphocyte ratio (NLR) typically predicts poor overall survival and poor responses to immunotherapies 12,13. However, current strategies to study the NLR of patients with cancer involves blood samples, but these may not represent the immune microenvironment at the site of the tumor.
ADC and SCC differ in their histology as well as genetic profiles, leading to divergent treatment options, with ADC possessing mutations allowing for more targeted therapies [1-3]. Among the many genetic alterations present in NSCLCs, mutations in genes such as KRAS, TP53, and EGFR are some of the most common. Mutations in KRAS and EGFR are more frequent in ADCs, with KRAS mutated in ~32% and EGFR in ~27% of tumors. However, mutations in these targetable genes are much less frequent in SCCs that instead have frequent mutations in TP53 (90%), PIK3CA and PTEN (15%) 1,14. In order to produce large amounts of histologically and genetically similar tumors, researchers have designed genetically engineered mouse models (GEMMs) of lung cancers to mimic patient genetics and have used these models for systematic characterization of lung cancer TMEs. Data have shown that murine lung adenocarcinomas tend to attract macrophages 15-17, but if there are alterations in EGFR, LKB1 or late stage KRAS, neutrophils may be recruited to these tumors 18-20. In murine squamous lung cancers neutrophils predominate and are thought to drive a transition from ADC to SCC states 15,17,21,22.
Current methods to characterize the TME include flow cytometry, single cell RNAseq, mass cytometry, and other multiplexed immunohistochemistry approaches 4,6,23,24. Flow cytometry and standard single cell RNAseq are robust methods, but require a relatively large amount of live starting material that must be dissociated prior to experimental assessment. Dissociating tissues can lead to loss of fragile cell populations and erases spatial information. Spatial scRNAseq can overcome this hurdle, but is expensive to implement. Other options include spatial profiling through multiplexed immunohistochemistry, including imaging mass cytometry techniques such as fluidigm or spatial proteomics25,26. These are powerful approaches, but require specialized equipment, a large investment of money for reagents, and trained personnel to interpret results 23. Therefore, developing a TME quantification method that is more cost effective, faster to implement, and retains the sample is imperative to further research efforts and to provide clinicians a tool to help guide treatment for their patients.
In this study, we utilize the HALO® Artificial Intelligence (AI) nuclear phenotyper software by Indica Labs to identify differences in the TME in both human and murine NSCLC tissues. We investigated five murine models, including KrasG12D/p53-null, Pik3caE545K/p53-null, EGFRT790M/L858R, KrasG12D/Lkb1-null, and Lkb1-null/Pten-null, to determine how the presence of these mutations alters the tumor microenvironment. We confirmed the accuracy of the algorithm with both correlation to IHC staining and by testing on lymphocyte-rich lymph node sections. We confirmed accumulations of macrophages in murine adenocarcinomas and neutrophils in murine squamous cell carcinomas in addition to further identifying differences in these tumors. In human samples, we uncovered extremely strong negative correlations between both mesenchymal cells and neutrophils with lymphocytes, and higher proportions of mesenchymal cells predicted poor overall survival.
METHODS
Mouse Models
Mouse models were: KRASG12D/p53-null lung adenocarcinoma27,28, KRASG12D/p53-null/Ezh2-heterozygous and KRASG12D/p53-null/Ezh2-null lung adenocarcinoma 29, PIK3CAE545K/p53-null lung adenocarcinoma 30, EGFRT790M/L858R lung adenocarcinoma 31, KRASG12D/Lkb1-null mixed lineage tumors 17,32, and Lkb1-null/Pten-null squamous cell carcinoma 15. All animal work was approved by University of Kentucky, Dana-Farber Cancer Center or Boston Children’s Hospital IACUC. Adult mice were allowed to inhale Cre- or FlpO-encoding virus to initiate autochthonous lung tumors. Mice housed at University of Kentucky all received 2.9x106 pfu of adenoviral Cre (University of Iowa). Historical banked tissues from previous studies were also used.
Patient Samples
Patient tissue was obtained from the Markey Cancer Center Biospecimens Procurement and Translational Pathology Shared Resource Facility (BPTP SRF). A tissue microarray was prepared from patient biopsies from de-identified excess tissue. The array consists of three core biopsies from each patient, and there are a total of 83 adenocarcinoma, 102 squamous cell carcinoma, 14 adenosquamous and mixed histology tumors and 17 poorly differentiated tumors, including 2 large cell carcinoma, 1 giant cell carcinoma, 1 pleomorphic carcinoma, and 1 sarcomatoid carcinoma.
Histology and immunohistochemistry
All tissues were fixed with 10% neutral-buffered formalin overnight. They were then transferred to ethanol, embedded in paraffin, and sectioned at 4μm. CD3 immunohistochemistry was performed by the Biospecimens Procurement and Translational Pathology Shared Resource Facility (BPTP SRF) at the Markey Cancer Center. Antigen retrieval was performed in Biocare Medical decloaking chamber at 95°C for 30 minutes with Dako Target Retrieval Solution, pH 9 (S236784-2). Endogenous peroxidase activity was quenched with Dako peroxidase block (K800021-5). Primary antibody for CD3 (Dako IR503 ready-to-use) was added and incubated at room temperature for 45 minutes. Amplification was performed with Dako Envision anti-rabbit-HRP for 30 minutes at room temperature (K4003). Detection was performed with DAB for 10 minutes (Dako K346711-2). Slides were then counterstained with Harris’s hematoxylin. F4/80 staining was also carried out by the BPTP SRF. Antigen retrieval was performed on the Ventana Discovery Ultra system using the CC2 mild protocol (citrate pH 6 buffer, 32 minutes at 91°C). Primary antibody, anti-F4/80 (Cell Signaling, #70776) was used at 1:200 for 1 hour at 37°C. Detection with performed with Ventana OmniMap anti-Rabbit-HRP for 20 minutes, followed by Ventana DAB. Slides were then counterstained with Mayer’s hematoxylin and blued with ammonia water. Dehydrated mounted slides were scanned with the Aperio slide scanner at 40x and 6 tumor containing regions of 355mm2 were selected and the HALO® classifier Multiplex IHC v3.0.4 was used to quantify the positively stained cells within the regions.
HALO® AI nuclear phenotyper algorithm
Histology slides were scanned at 20-40x with an Aperio AT2 scanner in brightfield mode. The scanner had an Olympus 20x objective with an optional 2x magnifier, with a resolution of 0.5μm/pixel at 20x and 0.25μm/pixel at 40x. The file format was .SVS, which is a tiled TIFF, that was then compressed to JPEG. Z-stacking was not used. The images were loaded into the HALO® program, and the AI nuclear phenotyper algorithm was trained for 556,930 iterations using a total of 34,427 nuclei from 52 different samples. The accuracy of the final nuclear phenotyper algorithm was verified by pathologists at the Markey Cancer Center. For each murine lung sections, the tumors areas with approximately 50μm surrounding area were hand-annotated for analysis. For the KRAS/Lkb1-null tumors, adenocarcinoma and squamous cell carcinoma regions were manually sub-annotated by an experienced researcher.
Statistics and reproducibility
Statistics were performed with GraphPad Prism version 9.2.0 and the statistical tests used and exact n are listed in the figure legends. The HALO software versions were: HALO v3.5.3577.214 and HALO AI 3.5.3577. For murine data, all tumors of a given subtype present on a slide were analyzed together. For the human data, results from three independent core biopsies were averaged. The n listed in the figures are biological replicates, which were individual mice or humans.
RESULTS
Training and Validation of Nuclear Phenotyper Algorithm for Human and Murine Lung Cancer
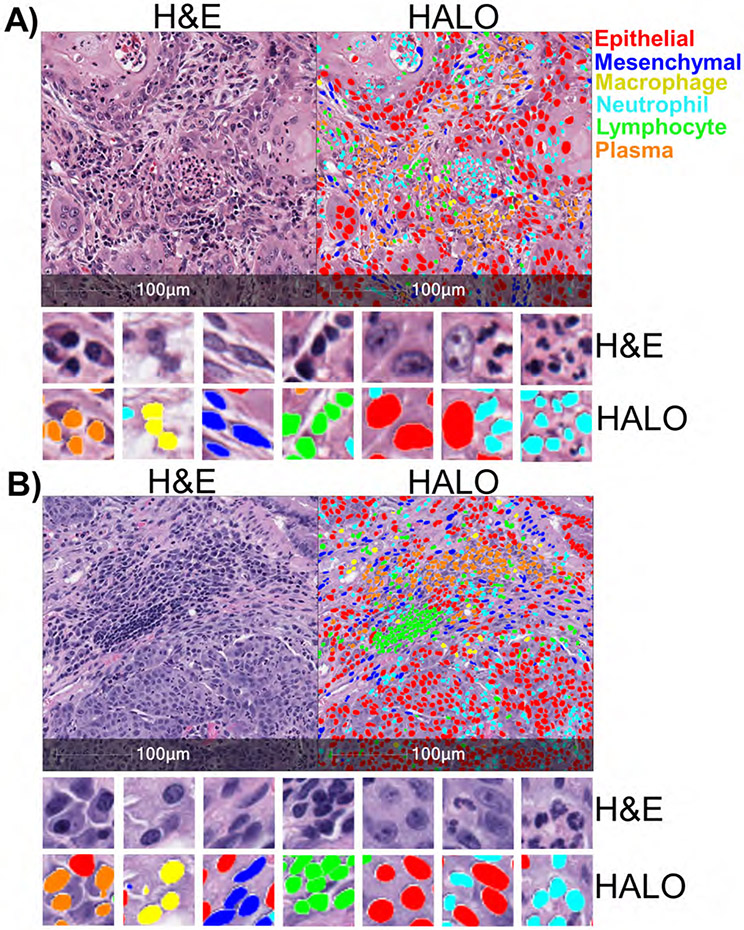
In order to profile lung cancer TME components using H&E-stained sections, we developed an algorithm that could reliably identify cell types in both patient and murine samples based on nuclear morphology. We used the HALO® AI nuclear phenotyper software and trained the software in iterations to identify six distinct cell types: epithelial cells, mesenchymal cells, plasma cells, macrophages, neutrophils and lymphocytes. Overall, 556,930 iterations were run, using 34,427 nuclei from 19 human samples and 33 murine samples. Representative images of human squamous cell carcinoma (Figure 1A) and murine squamous cell carcinoma (Figure 1B) show all six subtypes of cells identified by the algorithm. Neutrophils, overlaid in light blue, were identified by their polymorphonuclear or segmented nuclei. Plasma cells, shown in orange, contained a characteristic perinuclear “hot” that is a light area adjacent to the nucleus. Tumor cells, or epithelial cells, overlaid in red, were identified by their generally large cell size, pleomorphic nucleus and dark nucleoli. Mesenchymal cells, in dark blue, had elongated and spindle-shaped nuclei resemble endothelial cells, smooth muscle, fibroblasts and malignant epithelial cells that have undergone an epithelial-mesenchymal transition (EMT). Lymphocytes, in green, were characterized by small cytoplasm containing a small but often dark and rounded nucleus. Lastly, macrophages, in yellow, were identified by their round nuclei and diffuse cytoplasmic area.
Figure 1: Artificial Intelligence identifies nuclear phenotypes in human and mouse lung cancers.
A) H&E-stained sample from a patient with squamous cell carcinoma (left). Same H&E-stained image with different cell types identified by the HALO AI nuclear phenotyping algorithm overlaid with corresponding colors (right). Zoomed images show more detail of the different cell types identified in the tumor immune microenvironment. B) H&E-stained sample from KrasG12D/Lkb1-null mouse with squamous cell carcinoma (left), with the cell types identified by the HALO AI algorithm (right). Cell types are color-coded: epithelial/tumor cells (red), mesenchymal cells (dark blue), neutrophils (light blue), plasma cells (orange), lymphocytes (green), and macrophages (yellow).
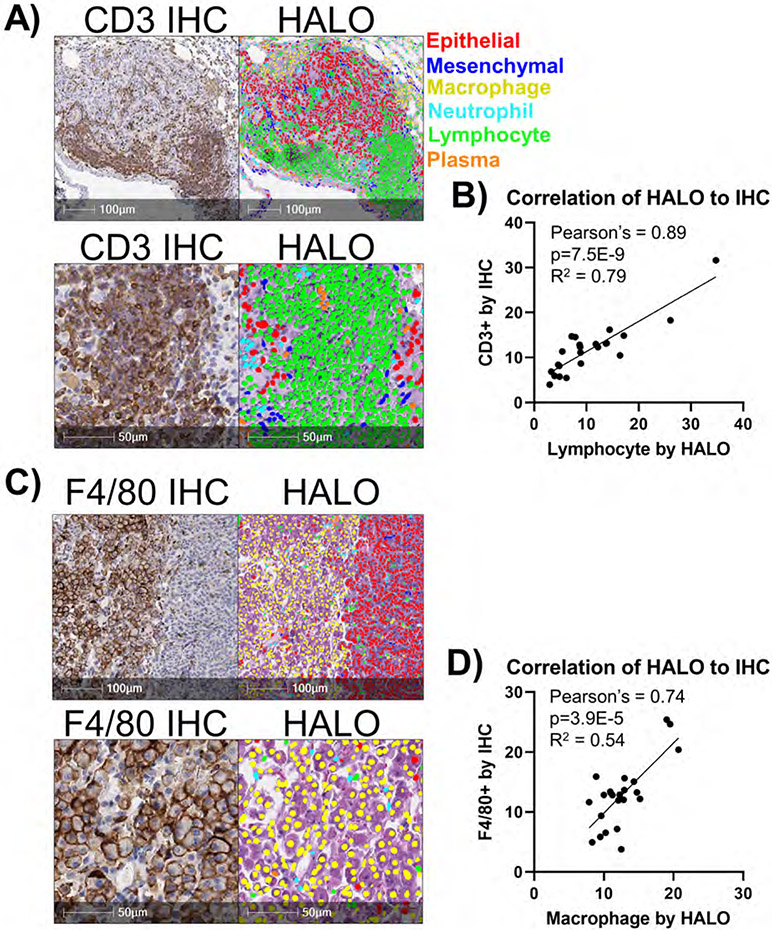
We next examined the concordance of the nuclear phenotyper algorithm with a traditional immunohistochemistry (IHC) approaches. First, we compared identification of lymphocytes using the algorithm to IHC staining of the T cell marker CD3 and observed a strong overlap in cell identification by the two methods (Figure 2A). We recently reported that when one copy of the gene encoding the histone methyltransferase EZH2 is deleted in KrasG12D/p53-null murine tumors, there are more abundant lymphocytes in the tumors29. Using tumors that were EZH2 wild-type, heterozygous and null, we compared directly the abundance of lymphocytes in the tumors as measured by the nuclear phenotyper algorithm and by CD3 IHC quantification (Supplementary Figures 1A+B). With both methods, we observed that EZH2 heterozygous tumors had statistically more lymphocytes as a percentage of total cells. The concordance of the two methods was also strong (Figure 2B). To further validate the ability of the algorithm to accurately identify lymphocytes and plasma cells, we examined murine lymph nodes. The lymph nodes of a murine sample can be seen with an abundance of lymphocytes, and lower numbers of the other cell types, which is expected for typical lymph node (Supplementary Figure 1C). In contrast, the nuclear phenotyer identifies numerous other cell types, including abundant epithelial cells when a metastatic tumor is present in the lymph node tissue (Supplementary Figure 1D). Lastly, we used F4/80 IHC and compared to macrophages identified by the phenotyper and again observed a strong concordance between the methods (Figure 2C+D). Taken together, these data indicate that the HALO® AI nuclear phenotyper is an easy and reproducible way to identify changes to the tumor microenvironment.
Figure 2: HALO AI nuclear phenotyper algorithm accurately identifies cell types.
A) Representative CD3 IHC staining of lung tumor with abundant tumor-infiltrating lymphocytes. Lymphocytes in the IHC are stained with anti-CD3 antibody and stained brown (left) with HALO AI nuclear phenotyping algorithm overlaid on the H&E-stained section (right). B) Correlation of percentages of CD3+ cells versus lymphocytes identified by HALO AI are graphed with Pearson’s correlation coefficient and R2 values indicated on graph, n= 24 mice total. C) Representative F4/80 IHC staining of lung tumor with abundant tumor-infiltrating lymphocytes. Lymphocytes in the IHC are stained with anti-F4/80 antibody and stained brown (left) with HALO AI nuclear phenotyping algorithm overlaid on the H&E-stained section (right). D) Correlation of percentages of F4/80+ cells versus macrophages identified by HALO AI are graphed with Pearson’s correlation coefficient and R2 values indicated on graph, n= 24 mice total.
Identification of Predominant Cell Types within Lung Cancer GEMMs
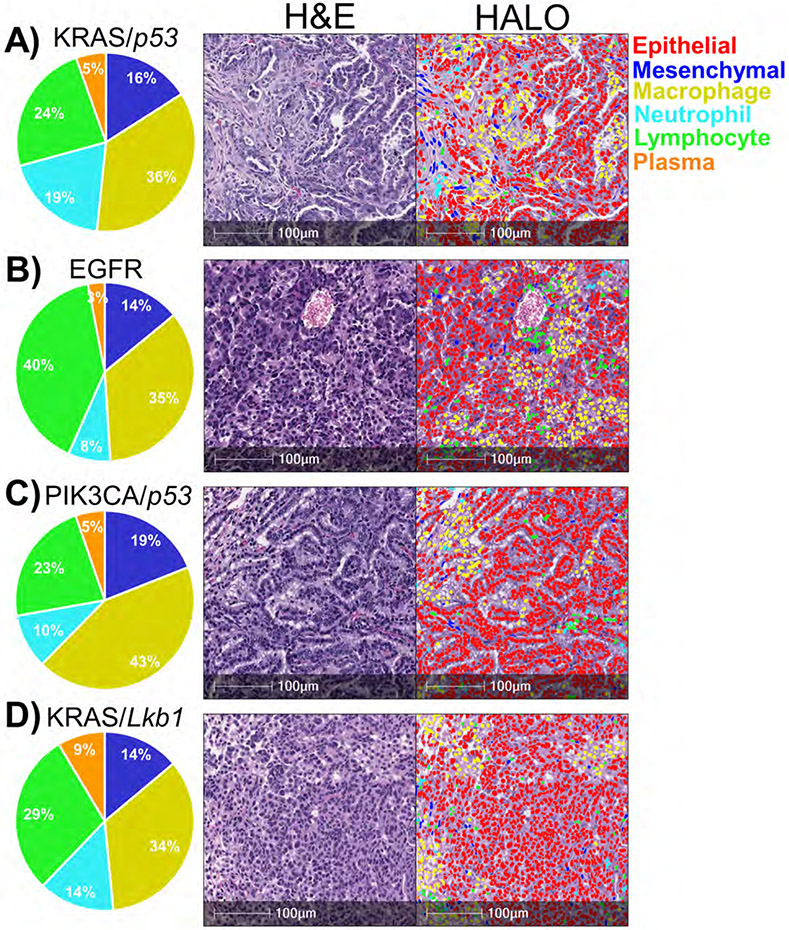
To further investigate the capabilities of the HALO® AI nuclear phenotyper algorithm, we examined H&E-stained tumors from four distinct lung cancer models: KRASG12D/p53-null lung adenocarcinoma 27,28, PIK3CAE545K/p53-null lung adenocarcinoma 30, EGFRT790M/L858R lung adenocarcinoma31, KRASG12D/Lkb1-null mixed lineage tumors 17,32. Consistent with numerous previous reports 15-17, all four models had large populations of macrophages at the site of the tumors. When considering the other abundant populations, the KRASG12D/p53-null model possessed the highest percentage of neutrophils relative to the other two models (Figure 3A); the PIK3CAE545K/p53-null model had the highest percentage of mesenchymal cells (Figure 3B); the EGFRT790M/L858R mutant model had the largest proportion of lymphocytes present (Figure 3C); and the KRASG12D/Lkb1-null had the most plasma cells (Figure 3D).
Figure 3: Diverse genotypes of murine lung adenocarcinomas have predominant macrophage infiltration.
A-D) Representative H&E-stained sections with different cell types identified by the HALO® AI nuclear phenotyping algorithm. Pie charts represent percentages of non-epithelial cells. A) KRASG12D/p53-null lung adenocarcinomas, n=8. B) PIK3CAE545K/p53-null lung adenocarcinomas, n=6. C) EGFRT790M/L858R lung adenocarcinomas, n=6. D) KRASG12D/Lkb1-null adenocarcinomas, n=9.
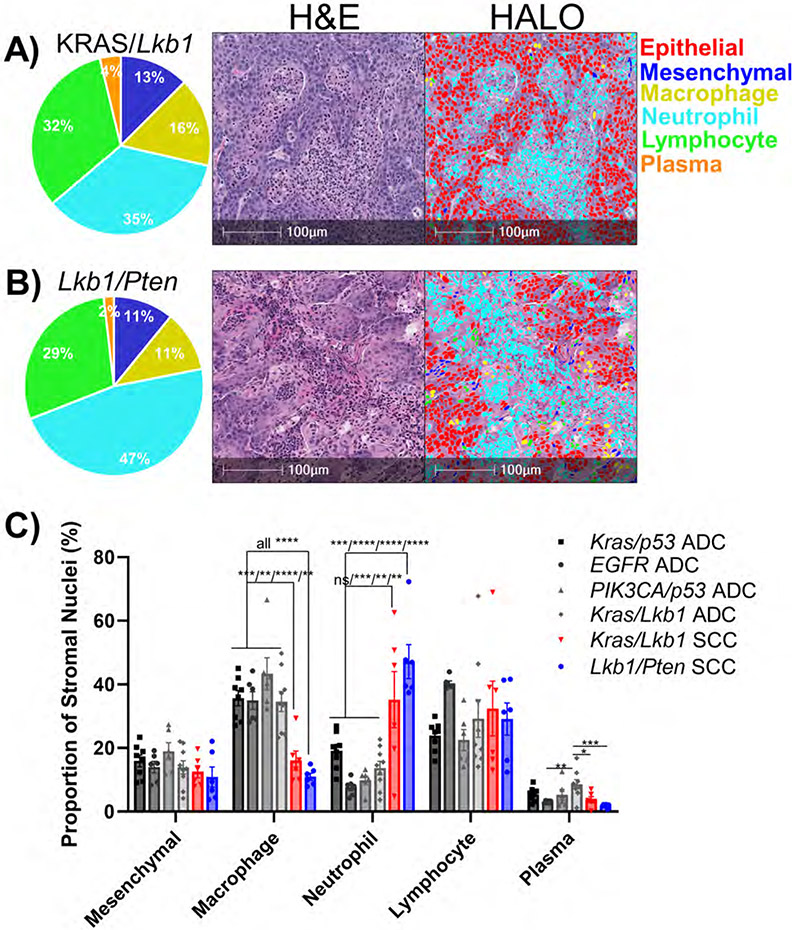
In addition to the KRASG12D/Lkb-null mouse model generating adenocarcinomas, it is also capable of creating tumors of adenosquamous and fully squamous histology 17,32. Again, consistent with several reports 17,21,22, the algorithm showed that squamous areas from the KRASG12D/Lkb-null model predominately recruit neutrophils (Figure 4A). We then explored the SCO model generated through biallelic deletion of the tumor suppressors Lkb1-null and Pten-null 15. Tumors from this model recruit a considerable number of neutrophils, indicated by the presence of large pockets of polymorphonuclear cells (Figure 4B). Likewise, when we compared immune cell composition from squamous tumor models, neutrophils are predominating; however, among all the adenocarcinoma models it is macrophages that predominate (Figure 4C). The distinct tropisms of the adenocarcinoma and squamous cell carcinomas were particularly evident when the two tumors were juxtaposed, showing macrophages recruited toward the adenocarcinoma histology and neutrophils toward the squamous tumor (Supplementary Figure 2).
Figure 4: Neutrophil infiltration predominates in genetically-defined murine lung squamous cell carcinomas.
A+B) Representative H&E-stained sections with different cell types identified by the HALO® AI nuclear phenotyping algorithm. Pie charts represent percentages of non-epithelial cells. A) KRASG12D/Lkb1-null squamous cell carcinomas, n=6. B) Lkb1-null/Pten-null squamous cell carcinoma, n=6. C) Bar graph represents proportion of non-epithelial nuclei for the indicated genetic mouse models of NSCLC, plotted as mean +/− SEM. * indicates p<0.02, ** p<0.006, *** p<0.001, **** p<0.0001 by one-way ANOVA with multiple comparisons and Holm-Šídák's multiple comparisons test.
Characterization of Human Non-Small Cell Lung Cancer Cell Profiles
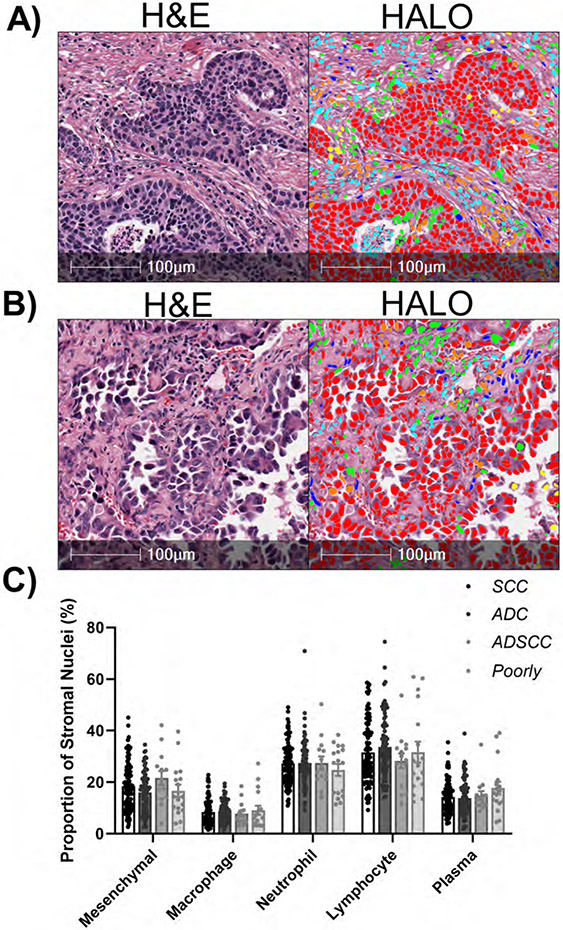
To examine the TME heterogeneity in human samples, we used a tissue microarray (TMA) that was generated from 216 NSCLC tumor samples. Using this TMA, we were able to recapitulate an accurate identification of the cells within the TME using the HALO® nuclear phenotyper algorithm in both ADC and SCC (Figure 5A+B). In contrast to our observations in the mouse models of NSCLC, we did not see any significant differences between the cell types present within these tumors (Figure 5C). The most commonly identified TME cell type was lymphocytes, in agreement with other work 4,7. The fact that the histotype did not predict cell infiltrations suggests that there are more heterogeneous factors contributing to the recruitment of immune cells within the tumor stroma in human samples than are present in our genetically-defined mouse models.
Figure 5: HALO AI nuclear phenotyper illustrates heterogeneity in patient NSCLC samples.
A) Representative human squamous cell carcinoma. B) Representative human adenocarcinoma. C) Proportions of the different cell types within the tumor micro-environment, SCC n=102, ADC n=83, ADSCC n=14, Poorly Differentiated n=17.
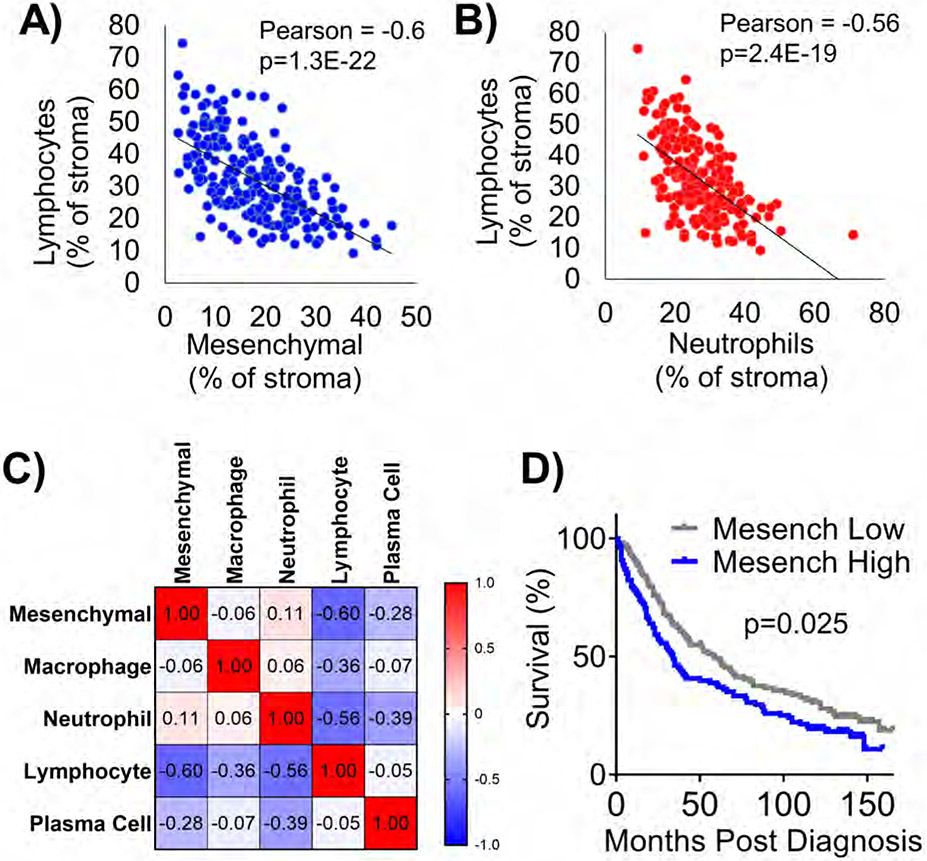
When comparing the percentages of mesenchymal cells to lymphocytes, we observed a very significant negative correlation between the two populations (Figure 6A). Furthermore, we observed a strongly significant negative correlation between tumor infiltrating neutrophils and lymphocytes (Figure 6B). This finding may indicate that mesenchymal cells and neutrophils present in the tumor stroma are phenotypically detrimental to lymphocyte survival or recruitment. To further interrogate the relationship between all the immune infiltrates identified by the HALO® AI nuclear phenotyper we created a correlation plot (Figure 6C). Positive correlations were observed between mesenchymal cells and neutrophils, which suggests that the mesenchymal cells could be phenotypically attracting neutrophils and together supporting a highly immunosuppressed environment. A negative correlation was also observed between macrophages and lymphocytes, but the degree of the correlation was lower.
Figure 6: Lymphocytes negatively correlate with neutrophils and mesenchymal cells, and mesenchymal cells predict poor prognosis in human samples.
A) Correlation plot between percentages of mesenchymal cells and lymphocytes, Pearson’s correlation coefficient and p value indicated on graph. B) Correlation plot between percentages of neutrophils and lymphocytes, Pearson’s correlation coefficient and p value indicated on graph. C) Correlation matrix depicts relationships between all tumor-associated cell types. D) Kaplan-Meier survival plot between mesenchymal low vs mesenchymal high tumors split at median, p value shown is Mantel-Cox LogRank test, n=216 patients for all plots.
We last sought to determine if abundance of any cell types within the tumors predicted poor overall survival in this cohort. We observed that patients whose tumors were classified as mesenchymal low survived longer than patients whose tumors were classified mesenchymal high (Figure 6D). This result was similar whether we queried all causes of death or if we limited the cohort to lung cancer-specific death (Supplementary Figure 3A). However, with multivariate analysis, confounding variables of sex, age at diagnosis, and tumor histology reduced the significance of correlation of mesenchymal cells with poor survival (Supplementary Figure 3B+C, Supplementary Table 1). Although not significant, high lymphocyte abundance predicted better prognosis, while a higher number of plasma cells predicted worse prognosis (Supplementary Figure 3B+C).
DISCUSSION
While there are many methods to investigate the TME, some of these methods are expensive, time consuming, and often destroy the sample in the process of data generation. We present utilizing a HALO® AI nuclear phenotyper algorithm to examine the TME in NSCLC. This method boasts low overall costs, quick turnaround, and the ability to retain the original sample. Using both human and mouse histology samples, we were able to demonstrate the algorithm’s ability to identify varied TME cell types, validating it as a tool for researchers, and as a potential clinical tool for the selection of appropriate treatments. Using GEMMS that have known immune cell tropisms, we confirmed that the nuclear phenotyping algorithm works very well to define the neutrophil and macrophage populations in lung cancer. This is one of the many ways in which this software can be used to differentiate between the microenvironments of these subtypes of NSCLC. We further confirmed the efficacy of the algorithm by assessing lymph node sections, comparing the algorithm’s results to a more traditional IHC staining for CD3. The algorithm performed very well with each of these known data-sets and was able to provide additional information about the abundant cell types in the GEMMs examined.
In a patient sample tissue microarray, the AI nuclear phenotyper provided a rapid and simple way to study the correlation of the different cell types at the tumor site. Our data demonstrate that neutrophils and lymphocytes are highly negatively correlated in human NSCLCs. This finding is in agreement with fresh patient tissue flow cytometry, which demonstrated that neutrophils are negatively correlated to CD8+ T cells 24. Given that CD8+ T cells are responsible for the elimination of malignant epithelial cells in immunotherapy contexts, a high neutrophil/low lymphocyte tumor may not respond well to immunotherapy, and this appears to be the case in patient samples 33. Furthermore, we show a strong negative correlation between mesenchymal cells and lymphocytes, and this is an additional avenue that could be explored for immunotherapy response. Given that the mesenchymal cell population also correlated with poor prognosis, further characterizing this heterogenous will be an important next step. Lastly, we observed numerous plasma cells in human NSCLCs, and recent reports have demonstrated that plasma cells may have a negative correlation to survival in lung cancer patients 34,35. These findings emphasize the importance of this program as a tool for researchers to understand the function of the TME, and the potential of this method to help determine patient treatment strategies. This easy-to-implement approach allows for specific understanding of the TME at the site of the tumor itself, and has potential to allow researchers to further investigate how abundance of different cell types influences the efficacy of therapies.
The data described here do not represent the first instance where AI has been suggested as a potential prognostic technique for histopathology. In fact, it has been predicted that AI will become a useful, if not necessary, tool for pathologists to triage slide analysis, quantify phenotypes and even to predict genetic alterations 36,37. However, there remain concerns about the ability of AI techniques to accurately differentiate between histopathologies, particularly if the algorithm was not trained on certain distinct patterns. Among proponents of the AI, it is widely accepted that the rigor of these algorithms will need to be tested abundantly to prove that their efficiency is equal to that of a pathologist before they are implemented as a prognostic technique 37. Therefore, the immediate implications of this technique are to allow translational research to begin to adopt this technique while further clinical validation is ongoing. While we specifically study NSCLC, we believe this artificial intelligence-based cell detection algorithm will have wider utility in many other diseases characterized by heterogenous chronic inflammation.
Supplementary Material
FUNDING ACKNOWLEDGEMENTS
This work was supported in part by the American Cancer Society Grant 133123-RSG-19-081-01-TBG, the American Institute for Cancer Research, NIGMS P20 GM121327, and NCI R01 CA237643. This research was also supported by the Cancer Research Informatics and Biospecimen Procurement & Translational Pathology Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30 CA177558). We thank Dana Napier at the BPTP for extensive support with the software and immunohistochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ETHICS APPROVAL/CONSENT TO PARTICIPATE
All animal work was approved by and performed in accordance with University of Kentucky, Dana-Farber Cancer Center or Boston Children’s Hospital Institutional Animal Care and Use Committees guidelines. A tissue microarray was prepared from patient surgical specimens. The samples were leftover clinical specimens collected under an IRB-approved protocol with informed consent or waiver. The University of Kentucky Biospecimens Procurement and Translational Pathology Shared Resource Facility acts as an honest broker and de-identifies the tissue. The Cancer Research Informatics Shared Resource Facility provides the necessary clinical data on the de-identified specimens. Because the investigators do not have patient identifying information, this is considered IRB-exempt research.
DATA AVAILABILITY STATEMENT
All data presented in this manuscript are available from the corresponding author upon reasonable request. The HALO nuclear phenotyer is available on Zenodo (DOI: 10.5281/zenodo.7883644). An ONNX (open neural network file) is available upon request with the appropriate MTA.
REFERENCES
- 1.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. [DOI] [PubMed] [Google Scholar]
- 2.Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. The Lancet. 2021;398(10299):535–554. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong K-K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stankovic B, Bjørhovde HAK, Skarshaug R, et al. Immune Cell Composition in Human Non-small Cell Lung Cancer. Frontiers in immunology. 2018;9:3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadota K, Nitadori J-i, Ujiie H, et al. Prognostic Impact of Immune Microenvironment in Lung Squamous Cell Carcinoma. Journal of Thoracic Oncology. 2015;10(9):1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zilionis R, Engblom C, Pfirschke C, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50(5):1317–1334.e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Yu Q, Song T, et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduction and Targeted Therapy. 2022;7(1):289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koppensteiner L, Mathieson L, O'Connor RA, Akram AR. Cancer Associated Fibroblasts - An Impediment to Effective Anti-Cancer T Cell Immunity. Frontiers in immunology. 2022;13:887380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker AT, Abuwarwar MH, Poly L, Wilkins S, Fletcher AL. Cancer-Associated Fibroblasts and T Cells: From Mechanisms to Outcomes. J Immunol. 2021;206(2):310–320. [DOI] [PubMed] [Google Scholar]
- 10.Aarts CEM, Hiemstra IH, Béguin EP, et al. Activated neutrophils exert myeloid-derived suppressor cell activity damaging T cells beyond repair. Blood advances. 2019;3(22):3562–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez PC, Quiceno DG, Zabaleta J, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64(16):5839–5849. [DOI] [PubMed] [Google Scholar]
- 12.Cupp MA, Cariolou M, Tzoulaki I, Aune D, Evangelou E, Berlanga-Taylor AJ. Neutrophil to lymphocyte ratio and cancer prognosis: an umbrella review of systematic reviews and meta-analyses of observational studies. BMC medicine. 2020;18(1):360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J, Yang L, Liu D, Li W. Association of the neutrophil to lymphocyte ratio and clinical outcomes in patients with lung cancer receiving immunotherapy: a meta-analysis. BMJ open. 2020;10(6):e035031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48(6):607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Fillmore Christine M, Koyama S, et al. Loss of Lkb1 and Pten Leads to Lung Squamous Cell Carcinoma with Elevated PD-L1 Expression. Cancer Cell. 2014;25(5):590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D-H, Lee H-S, Yoon D, et al. Progression of EGFR-Mutant Lung Adenocarcinoma is Driven By Alveolar Macrophages. Clinical Cancer Research. 2017;23(3):778–788. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Fillmore Brainson C, Koyama S, et al. Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nature communications. 2017;8:14922–14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Research. 2016;76(5):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfirschke C, Engblom C, Gungabeesoon J, et al. Tumor-Promoting Ly-6G(+) SiglecF(high) Cells Are Mature and Long-Lived Neutrophils. Cell Rep. 2020;32(12):108164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang KH, Li S, Khodadadi-Jamayran A, et al. Combined Inhibition of SHP2 and CXCR1/2 Promotes Antitumor T-cell Response in NSCLC. Cancer Discovery. 2022;12(1):47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaraj AS, Lahtela J, Hemmes A, et al. Cell of Origin Links Histotype Spectrum to Immune Microenvironment Diversity in Non-small-Cell Lung Cancer Driven by Mutant Kras and Loss of Lkb1. Cell Reports. 2017;18(3):673–684. [DOI] [PubMed] [Google Scholar]
- 22.Mollaoglu G, Jones A, Wait SJ, et al. The Lineage-Defining Transcription Factors SOX2 and NKX2-1 Determine Lung Cancer Cell Fate and Shape the Tumor Immune Microenvironment. Immunity. 2018;49(4):764–779.e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffitt JR, Lundberg E, Heyn H. The emerging landscape of spatial profiling technologies. Nature reviews. Genetics. 2022;23(12):741–759. [DOI] [PubMed] [Google Scholar]
- 24.Kargl J, Busch SE, Yang GHY, et al. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nature Communications. 2017;8:14381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Lin Y, Wang Y, et al. Characterization of the Tumor Immune Microenvironment in Lung Squamous Cell Carcinoma Using Imaging Mass Cytometry. Frontiers in oncology. 2021;11:620989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Maldegem F, Valand K, Cole M, et al. Characterisation of tumour microenvironment remodelling following oncogene inhibition in preclinical studies with imaging mass cytometry. Nature Communications. 2021;12(1):5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Development. 2001;15(24):3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson EL, Olive KP, Tuveson DA, et al. The Differential Effects of Mutant p53 Alleles on Advanced Murine Lung Cancer. Cancer Research. 2005;65(22):10280–10288. [DOI] [PubMed] [Google Scholar]
- 29.Chen F, Byrd AL, Liu J, et al. Polycomb deficiency drives a FOXP2-high aggressive state targetable by epigenetic inhibitors. Nature Communications. 2023;14(1):336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, Liu J, Song X, et al. EZH2 inhibition confers PIK3CA-driven lung tumors enhanced sensitivity to PI3K inhibition. Cancer letters. 2022;524:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F, Liu J, Flight RM, et al. Cellular Origins of EGFR-Driven Lung Cancer Cells Determine Sensitivity to Therapy. Advanced science (Weinheim, Baden-Wurttemberg, Germany). 2021;8(22):e2101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–810. [DOI] [PubMed] [Google Scholar]
- 33.Kargl J, Zhu X, Zhang H, et al. Neutrophil content predicts lymphocyte depletion and anti-PD1 treatment failure in NSCLC. JCI Insight. 2019;4(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurebayashi Y, Emoto K, Hayashi Y, et al. Comprehensive Immune Profiling of Lung Adenocarcinomas Reveals Four Immunosubtypes with Plasma Cell Subtype a Negative Indicator. Cancer Immunology Research. 2016;4(3):234–247. [DOI] [PubMed] [Google Scholar]
- 35.Lee HE, Luo L, Kroneman T, et al. Increased Plasma Cells and Decreased B-cells in Tumor Infiltrating Lymphocytes are Associated with Worse Survival in Lung Adenocarcinomas. Journal of clinical & cellular immunology. 2020;11(1). [PMC free article] [PubMed] [Google Scholar]
- 36.Colling R, Pitman H, Oien K, et al. Artificial intelligence in digital pathology: a roadmap to routine use in clinical practice. The Journal of pathology. 2019;249(2):143–150. [DOI] [PubMed] [Google Scholar]
- 37.Acs B, Hartman J. Next generation pathology: artificial intelligence enhances histopathology practice. The Journal of pathology. 2020;250(1):7–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this manuscript are available from the corresponding author upon reasonable request. The HALO nuclear phenotyer is available on Zenodo (DOI: 10.5281/zenodo.7883644). An ONNX (open neural network file) is available upon request with the appropriate MTA.