Abstract
Construction of user-defined long circular single stranded DNA (cssDNA) and linear single stranded DNA (lssDNA) is important for various biotechnological applications. Many current methods for synthesis of these ssDNA molecules do not scale to multi-kilobase constructs. Here we present a robust methodology for generating user-defined cssDNA employing Golden Gate assembly, a nickase, and exonuclease degradation. Our technique is demonstrated for three plasmids with insert sizes ranging from 2.1–3.4 kb, requires no specialized equipment, and can be accomplished in five hours with a yield of 33–43 % of the theoretical. To produce lssDNA, we evaluated different CRISPR-Cas9 cleavage conditions and reported a 52 ± 8% cleavage efficiency of cssDNA. Thus, our current method does not compete with existing protocols for lssDNA generation. Nevertheless, our protocol can make long, user-defined cssDNA readily available to biotechnology researchers.
Introduction
Single-stranded DNA (ssDNA) is increasingly becoming important for novel techniques and advancements in biotechnology, synthetic biology, and biomaterial research. Long ssDNA molecules have a wide range of potential applications including genome engineering and recombineering, DNA storage, DNA origami, and gene therapy [1]. There is a need for an accessible method of synthesis of user-defined ssDNA that is cost-effective, simple, robust, and scalable.
One major application of ssDNA is its use as a template for genome engineering applications. RNA-programmable gene targeting nucleases like CRISPR-Cas9 have transformed genome engineering in animals [2]. Site-specific modification of genes and full allelic replacement can occur through double stranded breaks (DSB) at defined loci, followed by Homology Directed Repair (HDR) with a donor nucleic acid molecule containing homology arms. However, inserting long contiguous DNA is technically challenging. A grand challenge in genome editing is the highly efficient site-specific insertion, or replacement, of larger DNA cassettes into mammalian genomes. Solving this challenge is important for several fields spanning discovery biology and biotechnology: (i.) generation of transgenic animal models including conditional knockout mice and mice with inducible transcriptional activators [3]–[7]; (ii.) high efficiency insertion of transgenes in cell lines at site-specific locations [8]; (iii.) development of diverse antibody libraries using mammalian cell display [9]; and (iv.) insertion of chimeric antigen receptors in T cells for immunotherapy [10], [11]. More specifically, Easi-CRISPR [7], [12] provides a partial solution to the challenge of efficient DNA cassette insertion in mammalian genome engineering using a long, linear single-stranded DNA (lssDNA) donor as opposed to double stranded DNA [13]–[17] or overlapping short ssDNA molecules.
Current ssDNA synthesis strategies can be complex, expensive, time consuming, prone to high error rates, or rely on specialized equipment, and many of the current methods do not scale well over 2 kilobases in length. Commercial vendors such as IDT offer chemical synthesis of long ssDNA. The traditional method of chemical synthesis involves using phosphoramidite chemistry on solid surfaces [18]. However, chemical synthesis is oftentimes limited to smaller ssDNA fragments. Minev et al demonstrate another method of rapid long linear ssDNA (lssDNA) production called Methanol-Responsive Polymer PCR (MeRPy-PCR) that utilizes polymerase chain reaction with a modified forward primer [19]. The reaction is rapid but requires a two-day primer modification with linear acrylamide under nitrogen. Others have also demonstrated the utility of PCR with modified primers in combination with a variety of selective digestions to synthesize ssDNA [20], [21]. However, reactions that rely heavily on PCR can be prone to moderate error rates, are limited to linear products, and are less conducive to automation as compared to enzymatic techniques. Veneziano et al showed that gene-length lssDNA donors could be produced by enzymatic synthesis techniques such as asymmetric PCR (aPCR) [22]. aPCR involves two amplification primers with one in excess to produce the desired ssDNA, but it can require extensive experimentation to optimize yield of the ssDNA and the technique can frequently result in the creation of unwanted byproducts and nonseparated DNA strands [1], [22]. Alternatively, rolling circle amplification can generate long repeating ssDNA using an isothermal polymerase reaction to continuously anneal deoxynucleotide triphosphates to a DNA primer [23], [24]. While this technique is robust, the reaction time can be long, the amplification efficiency is low, and long plasmid-length DNA encoded includes unwanted bacterial control elements like bacterial origins of replications [1]. There are a wide range of additional techniques for ssDNA synthesis such as transcription and reverse transcription, separation of ssDNA from dsDNA, and a primer exchange reaction [1].
We aimed to develop a robust method to generate long, single-stranded DNA from bacterial-prepped plasmid DNA [7]. The method presented here allows for synthesis of a user-defined long, circular ssDNA (cssDNA) and is cost-effective, simple, and uses readily available materials to generate ssDNA in a short time. By having the user-defined insert of interest (IOI) contained in a plasmid initially and using these straightforward steps, this process can be scaled up for larger microgram ssDNA quantities for downstream applications. Linear ssDNA was demonstrated by endonuclease cleavage, albeit at efficiencies not competitive with existing published protocols involving polymerase replication.
Materials and Methods
Reagents
All DNA modification enzymes, buffers, and reagents were sourced from New England Biolabs (NEB) unless otherwise noted. All primers and oligos were ordered from Integrated DNA Technologies (IDT).
Plasmid Construction
All primer sequences along with any oligos used in this work are listed in Supplementary Table 1, and all plasmids are listed in Supplementary Table 2. Plasmids C16Ex2, C16Ex3, and C16Ex4 were custom synthesized by Azenta life sciences. pIS001 was constructed with Gibson Assembly from plasmid C16Ex2 containing the ROSA-TRE-C16V5-G-pA insert and the pYTK084 backbone [25]. Primers p001 and p002 were used to amplify insert and primers p003 and p004 were used to amplify the backbone, and fragments assembled by Gibson Assembly using NEBuilder® HiFi DNA Assembly Master Mix (NEB; catalog # E2621L). Plasmids pIS002 and pIS003 were also constructed using Gibson Assembly similarly to pIS001, except the inserts were isolated from plasmids C16Ex3 and C16Ex4, respectively, using a restriction digest with BsaI-HFv2 (NEB; catalog # R3733L) in 1X rCutSmart Buffer and subsequent gel extraction. Oxford Nanopore long-read sequencing (Plasmidsaurus) was used to confirm whole and intramolecularly ligated plasmid sequences.
Intramolecular Golden Gate Assembly and Cleanup
All Golden Gate Assembly reactions started with 1 μg of the constructed plasmid (pIS001, pIS002, or pIS003) and BsaI-HFv2 (20 or 60 U from 20 U/μL) (NEB; catalog # R3733L), 400 U of T4 DNA ligase (1 μL of 400 U/μL) (NEB; catalog # M0202L), and 1X T4 DNA ligase buffer (NEB; catalog # B0202S). During optimization, reactions were performed either in 20, 100, or 200 μL reaction volumes. Reaction mixture was placed in a Mastercycler X50s thermocycler for 30 cycles of 37°C for 1 minute followed by 16°C for 1 minute, followed by a 60°C hold for 5 minutes. Following the Golden Gate Intramolecular Ligation Assembly step, the reaction was cleaned and concentrated with a Monarch PCR & DNA Cleanup Kit (NEB catalog # T1030L) and eluted in either 10 or 12 μL of nuclease free H2O. This elution product was then incubated with BsaI-HFv2 (20 or 60 U from 20 U/μL), 1X rCutSmart buffer (10X), nuclease free H20, and an exonuclease (1 μL of either Lambda or Exonuclease III) in a 20 μL cleanup reaction. During optimization, we tested different amounts of Exonuclease III (NEB; catalog # M0206L) (2, 5, 10 U) made by dilution of the stock 100 U /μL using 1X rCutSmart Buffer, or Lambda (NEB; catalog # M0262L) (0.5,5 U), made by 1:10 dilution of the 5 U/μL stock. This reaction was incubated in a thermocycler at 37°C for 30 minutes followed by 80°C for 20 minutes. The Golden Gate intramolecular ligation product for pIS001 was plasmid sequenced using Oxford Nanopore as a custom service from Plasmidsaurus.
Template Prep Step for Production of cssDNA
All template preparation reactions occurred in 25 μL and were comprised of the 20 μL cleanup step, 1.5 μL of nuclease free water, 10 U of either Nb.BbvCI (NEB; catalog # R0631L) or Nt.BbvCI (NEB; catalog # R0632L (1 μL of 10 units/μL), 10 U of Exonuclease III (1 μL of a 1:10 dilution in 1X rCutSmart Buffer of 100 U/μL stock), 20 U of Exonuclease I (1μL of 20 U/μL) (NEB; catalog # M0293L), and 0.5 μL of 10X rCutSmart Buffer. This reaction was placed in the thermocycler at 37°C for 60 minutes followed by 80°C for 20 minutes. The results were column purified with the Monarch PCR & DNA Cleanup Kit and eluted in either 10 or 12 μL of nuclease free water.
sgRNA Production and Cas9-sgRNA Cleavage Experiments
To make the sgRNA, the EnGen sgRNA Synthesis Kit, S. pyogenes (NEB; catalog # E3322V) was used with a DNA primer as a template (Supplementary Table 1). The linearization of the cssDNA was performed in three steps: a pre-cycle, a cutting step, and inactivation of Cas9. In the pre-cycle step, 3 μL of 300 nM sgRNA diluted in NFH2O, 1 μL of 1 μM Cas9 Nuclease (diluted from 20 μM stock in 1X NEBuffer r3.1) (NEB; catalog # M0386T), and 1X NEBuffer 3.1 (NEB; catalog # B6003S) were combined in a 20 μL reaction volume, and the solution was placed in the thermocycler at 25°C for 10 minutes. In the digestion step, 1 μg of M13mp18 ssDNA (NEB; catalog # N4040S) was added to the pre-cycle results for a total reaction volume of 30 μL. This reaction was placed in the thermocycler for 60 minutes at 37°C. For the inactivation step, we added 0.8 U of Proteinase K (1 μL of 0.8 U/ μL) and incubated at room temperature for 10 minutes, or alternatively tested a heat inactivation of 65°C for 15 minutes.
Gel Imaging and Reaction Conditions
To visualize the results of the procedure, the dsDNA was run on 1% w/v agarose gel in 0.5X Tris-acetate-EDTA (TAE) buffer (made from a 50X comprising 24.2% w/v tris base, 5.71% v/v glacial acetic acid, and 10% v/v 0.5M EDTA, pH8.0) at a range of 100 – 120 V. To run the gels, we used a 1 kb Plus DNA Ladder for Safe Stains (NEB; catalog # N0559S), Gel Loading Dye, Purple (6X) (NEB; catalog # B7024S), and SYBR™ Safe DNA Gel Stain (ThermoFisher Scientific; catalog # S33102). Prior to running on gels, ssDNA for the linearization experiments was denatured by adding 60% v/v formamide solution (formamide with 0.5% v/v EDTA buffered to pH 8.0 with NaOH, filtered through a 0.22μm filter and autoclaved) to the reaction followed by heating at 70°C for 5 minutes and then on ice for 5 minutes [26]. ssDNA was run on a 1.5% w/v agarose gel in 0.5X TAE.
Quantification of DNA with Gel Densitometry and Fluorescent Assay
To quantify the DNA yield at various steps in the process, ImageJ [27], [28] was used to analyze gels and obtain intensity values from pictures of the bands on the gels. The intensity of the bands is proportional to the mass of the DNA present, and we used this to calculate the percent of a specific DNA product compared to the total DNA mass in the reaction.
The cssDNA yield for pIS001 and pIS002 was calculated using a Quibit ssDNA Assay kit from Invitrogen (ThermoFisher Scientific; catalog # Q10212). cssDNA generated from the template preparation step was column purified using a Monarch PCR & DNA Cleanup Kit and eluted into 11 μL. The cleaned ssDNA samples were prepared according to the ssDNA Assay kit instruction by dilution in 189 μL of Qubit working solution (1:200 dilution of Qubit ssDNA Reagent in Qubit ssDNA buffer) for a total reaction volume of 200 μL. The relative fluorescence units (RFU) of each sample were obtained using a Qubit 3.0 Fluorometer. By comparing the RFU values to a standard curve generated with ssDNA of known concentrations, we quantified the yield nanograms of ssDNA generated. We calculated the maximum theoretical yield as the percentage of IOI in the whole plasmid divided by two to account for the ssDNA product, and calculated the percent yield using these theoretical values.
Results
Our initial overall goal was to develop a robust, efficient method that can prepare long circular and linear ssDNA containing a user defined insert of interest (IOI) from plasmid dsDNA. Our constraints for any new method were that it be easily scalable for longer IOI segments, require no specialized equipment or reagents, be robust towards different insert sequences, have high yield of ssDNA products, remove the plasmid backbone section from the final ssDNA product, and be a single-day procedure.
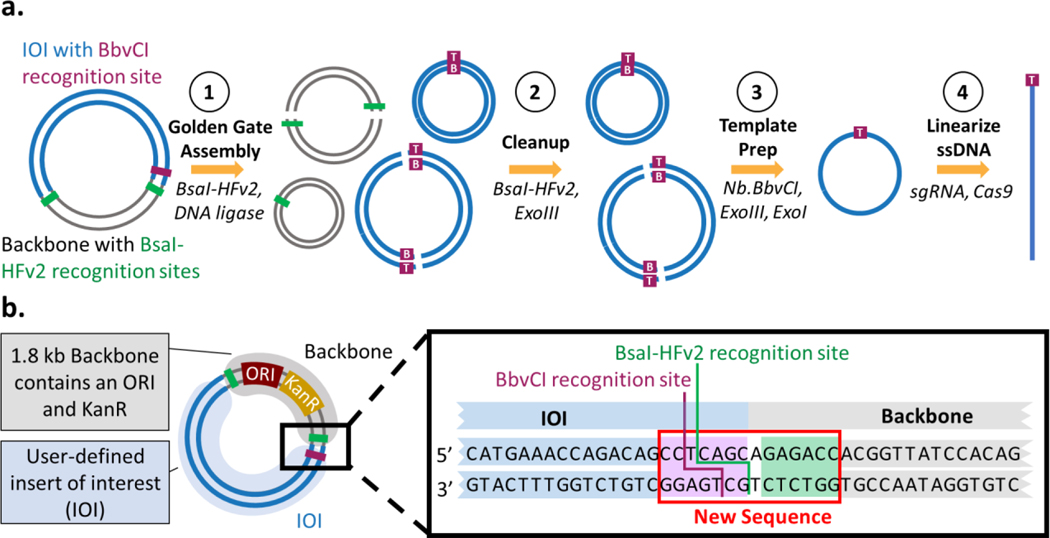
In order to meet the design criteria, we developed a strategy involving a Golden Gate intramolecular ligation of the insert (Figure 1a). In the first step, Golden Gate assembly generates, among other products, a circular dsDNA molecule that contains only the insert through intramolecular ligation. In the second step, a cleanup step involving restriction digestion and exonuclease treatment removes unwanted byproducts such as the linearized DNA and circular plasmid backbone. In the third step, we generate IOI-specific circular ssDNA (cssDNA) using a nickase and exonuclease digestion; this process leaves a scar site (< 7 nt at either the 5′ or 3 ′ end) introduced to accommodate the nicking process. In the final step, cssDNA is linearized at the scar site or any selected region using an sgRNA-Cas9 construct, resulting in user-defined linear ssDNA.
Figure 1: Protocol overview for generating custom cssDNA and lssDNA from plasmid DNA. a. Process for generation of long, linear ssDNA.
Starting from a plasmid containing the insert of interest (IOI; blue), Golden Gate assembly is used to generate an intramolecular ligation product of the IOI. Undesired reaction products like backbone (gray) and concatemer plasmids of the insert are shown (Step 1). The two strands of the plasmid are labelled as T (top) and B (bottom), referencing the preference of the enzyme BbvCI. A cleanup step using BsaI-HFv2 and Exonuclease III removes undesired backbone reaction products (step 2). Then, circular ssDNA can be created from dsDNA using a nickase followed by exonuclease degradation (step 3). Finally, the cssDNA is linearized by Cas9 cleavage at the BbvCI recognition site, leaving a maximum of a 7 nt 5’ or 3’ scar (step 4). b. Plasmid design. The IOI (blue) is ligated into a backbone (grey) containing a high copy number ORI, a KanR resistance cassette, a BbvCI recognition site, and flanking BsaI-HFv2 recognition sites. The inset shows the sequence design of the adjacent BbvCI and BsaI-HFv2 recognition sites.
A plasmid compatible with this process would include (i.) a minimal backbone enabling selection and replication in E. coli; (ii.) a BsaI restriction site adjacent to the insert for intramolecular ligation in the Golden Gate assembly; and (iii.) a flanking BbvCI restriction site for nickase digestion (Figure 1b). The insert cannot contain any BsaI recognition sequence, and any BbvCI restriction sites must occur in the same 5’-->3’ orientation to ensure selective cutting of a single strand of the dsDNA during the template prep step. Key to the success of the Golden Gate assembly step is the use and directionality of the Type IIS enzyme BsaI, as the ability of this enzyme to cleave outside of its recognition site allows for isolation of the IOI. Plasmid pIS001 (5160 bp), a test plasmid compatible with the strategy, was prepared by Gibson assembly [29] of the pYTK084 backbone [25] containing a high copy number ORI and a kanamycin resistance cassette with the 3379bp insert ROSA-TRE-C16V5-G-pA. This insert contains 5’ and 3’ homology arms of mouse ROSA26 locus, TRE Tight, CEACAM16 coding sequence V5 tag, EGFP, and BGH polyA.
To optimize the process of ssDNA synthesis, we individually tested each of the four steps for functionality, identified byproducts, and tested variable conditions on distinct steps. Optimization for steps one (Golden Gate intramolecular ligation) and two (reaction clean-up) were performed on pIS001. For step three (formation of cssDNA) we used an existing nicking/digesting protocol that is part of the nicking mutagenesis technique previously developed by our lab [30] which was performed without optimization. Finally, step four (lssDNA generation) was performed on the bacteriophage DNA M13mp18 as both dsDNA and cssDNA are commercially available.
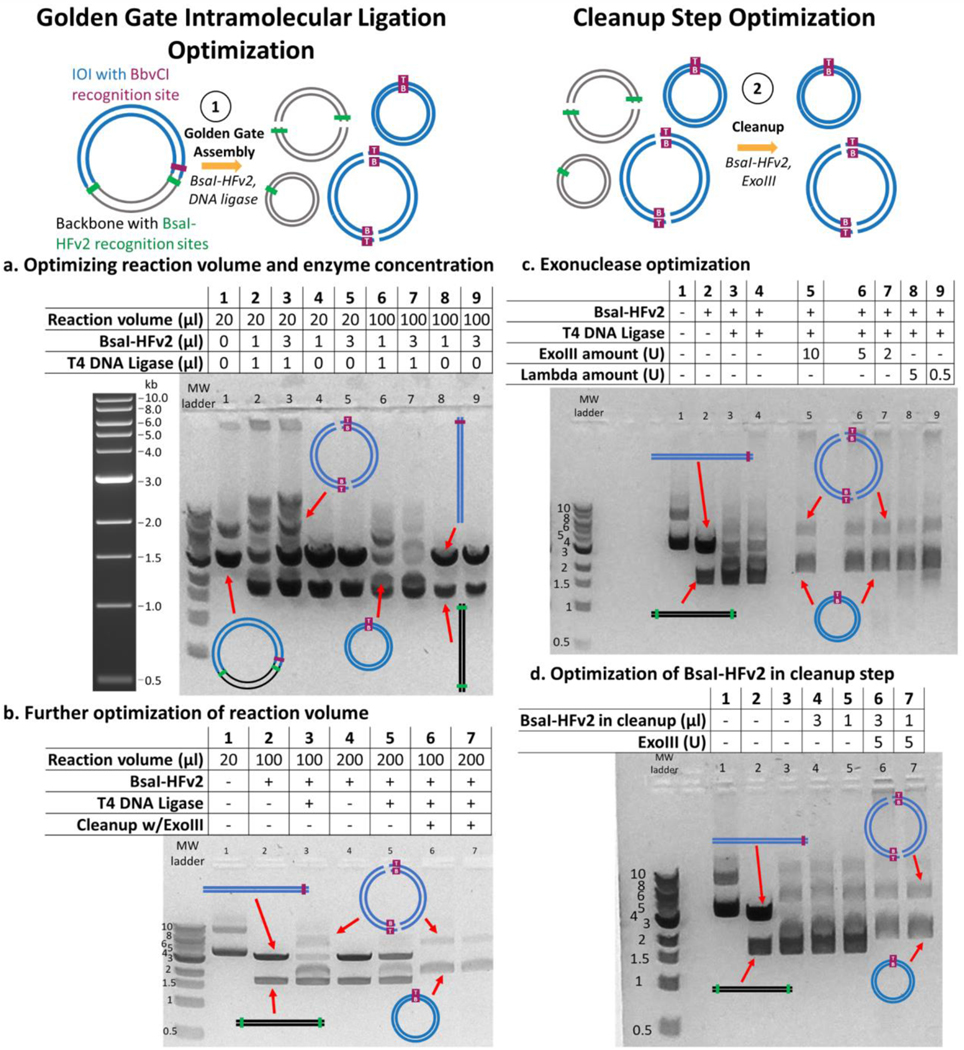
We first confirmed the Golden Gate intramolecular ligation of the ROSA-TRE-C16V5-G-pA insert in a 20 μL reaction volume performed in 1X T4 DNA ligase buffer containing 1 μg pIS001, BsaI-HFv2 (1 μL of a 20 U/μL stock), and T4 DNA ligase. The reaction was subjected to 30 cycles of 37°C incubation for one minute followed by 16°C for one minute, and then a single 60°C incubation for 5 minutes. Digestion with only BsaI-HFv2 resulted in two distinct bands at the approximate sizes of backbone (1.8 kb) and insert (3.4 kb) (Fig 2a). When T4 DNA ligase was added to the reaction mixture, it generated additional bands (Fig 2a) presumably corresponding to monomers and concatemers of different insert and backbone ligation products. We hypothesized that the band at circa 2 kb corresponded to the desired intramolecular ligation product of the insert and sought to optimize its formation by testing different reaction volumes (20 mL, 100 mL) and BsaI-HFv2 amounts (1, 3 μL of a 20 U/μL stock). We observed a lower abundance of the linearized insert at the higher BsaI-HFv2 amount of 3 μL (Figure 2a). With respect to increased reaction volume, we hypothesized that decreasing the overall concentration of linearized IOI fragments would result in fewer concatemers and more of the putative desired intramolecular ligated insert. Our experiments supported this hypothesis: we found that 100 μL reaction volume resulted in higher concentration of circularized insert monomer as judged by gel densitometry (Figure 2a). Increasing the reaction volume to 200 μL did not appreciably increase the concentration of monomer but it did qualitatively decrease the formation of higher order concatemers (Figure 2b). Thus, a reaction volume of 200 μL and a BsaI-HFv2 amount of 60 U (3 μL of 20 U/μL) was used for all further experiments. One disadvantage of the larger reaction volume is that the downstream steps work in a smaller reaction volume, and so an additional DNA concentration column clean-up is necessary between steps one and two.
Figure 2: Optimization of the intramolecular ligation by Golden Gate Assembly and of the cleanup step.
All lanes showing reaction intermediates contain a starting amount of 1 μg of pIS001 dsDNA. a. Optimization of reaction volumes of 20 μl and 100 μl and two different BsaI-HFv2 concentrations (1 μl or 3 μl of a 20 U/μl stock). Higher BsaI-HFv2 and the larger volume resulted in less linearized IOI, and lower concatemers than other conditions. Sample 1 kb ladder from NEB shown for reference. b. A larger reaction volume of 200 μl resulted in similar formation of monomeric linear plasmid and formation of concatemers when 3 μl BsaI-HFv2 was included. Lanes 6 and 7 show removal of linear DNA using ExoIII. c. ExoIII and lambda exonucleases were tested for functionality in the cleanup step at different dilutions using Golden Gate assembly conditions optimized in panel b. Both exonucleases were able to remove linearized DNA while not appreciably degrading circularized IOI at all concentrations evaluated. d. Optimization of BsaI-HFv2 in cleanup step (1 μl or 3 μl of a 20 U/μl stock).
Following column clean-up, exonuclease digestion removed the linear backbone, resulting only in intramolecular ligation products that are monomers and concatemers of insert sequences as validated by whole plasmid sequencing using Oxford nanopore.
Optimization was performed on the cleanup step by evaluating different concentrations of exonucleases (Lambda, ExoIII) able to degrade linear dsDNA with a variety of end geometries. Both exonucleases worked sufficiently in cleaning up any remaining linear DNA and any backbone concatemers, leaving only the circular dsDNA plasmid of the insert (Figure 2c). In the optimized procedure we used 5 U of ExoIII (1 μL of a 1:20 dilution in 1X CutSmart Buffer of 100 U /μL in a 20 μL reaction volume), although we note that 2 U (1:50 dilution) ExoIII does not show qualitative differences in circularized insert yield and presence of linear contaminants. Two different concentrations of BsaI-HFv2 were tested in the cleanup step (1, 3 μL of a 20 U/μL stock), with the lower concentration sufficient to remove any circularized backbone as judged by gel densitometry (Figure 2d). Finally, we used analytical gel densitometry to estimate the mass percentage of monomers to the total DNA mass observed on the gels in the cleanup step. The average percent mass of monomers (over total mass including concatemers) was calculated over four independent gel runs using pIS001. Based on gel densitometry, 75 ± 7% (n = 4, error represents 1 s.d.) of the DNA visible by gel electrophoresis after the cleanup step is the desired dsDNA monomer IOI product.
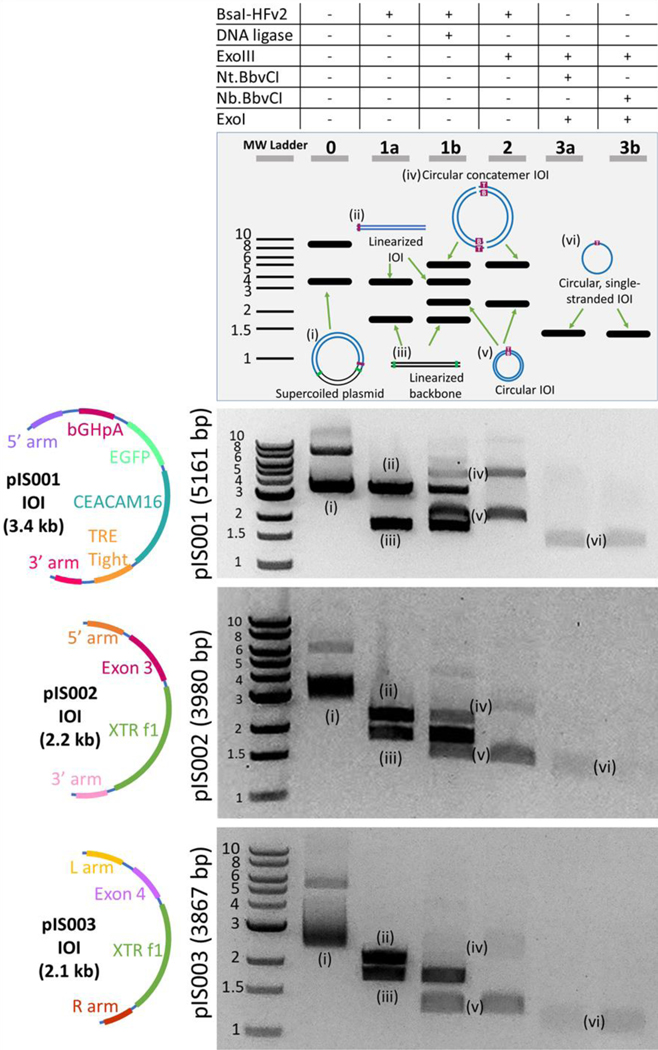
Given our optimization of steps one and two, and previous optimization of cssDNA production (step 3) [30], we applied the complete protocol to pIS001 to produce cssDNA for the ROSA-TRE-C16V5-G-pA insert (Figure 3). We digested reaction products from step two with either Nt.BbvCI or Nb.BbvCI in order to demonstrate that cssDNA could be produced in either the sense or antisense direction (Figure 3). This overall procedure from a 1 μg plasmid DNA input took a minimum of 4.5 h: 1.5 h for the Golden Gate intramolecular ligation, 0.25 h for column clean-up, 1.25 h for the cleanup step, and 1.5 h for the template preparation step.
Figure 3. cssDNA productions for three separate inserts.
The optimized protocol was tested on the three different plasmids that were constructed (pIS001, pIS002, and pIS003), with inserts shown in the first column. The results of each step of the process for 1 μg of pIS001 (5160 bp), pIS002 (3980 bp), and pIS003 (3867 bp) are shown. Lanes correspond to the steps in the protocol listed in Figure 1. Lane 0: undigested plasmid DNA containing supercoiled and linearized plasmids. Lane 1a: plasmid is digested with BsaI-HFv2. Lane 1b: Golden Gate Assembly including BsaI-HFv2 and DNA ligase. Lane 2: Cleanup step following Golden Gate assembly. Lanes 3a-b: cssDNA is visible when either the Nt.BbvCI (3a) or Nb.BbvCI (3b) nickase is used for template prep. ssDNA bands do not appear as brightly as dsDNA in part because the SYBR-Safe dye used has a lower quantum yield when bound to ssDNA than dsDNA at the wavelength used for imaging.
To demonstrate that this method robustly produces cssDNA for distinct insert sequences, we prepared two additional plasmids: pIS002 (3980 bp) contains a 5’ arm, CEACAM16 Exon3, XTR cassette [31], and 3’ arm, and pIS003 (3867 bp) contains a 5’ arm, CEACAM16Exon4, XTRf1, and 3’ arm. pIS002 also contained an additional BbvCI site, and the nicking step was performed with the additional BbvCI site in the same direction as the existing site so that this did not impact the results. For both plasmids, our method could successfully produce insert cssDNA in either the sense or antisense configurations (Figure 3). The cssDNA production step appeared to remove higher order concatemers, which we attribute to incomplete ligation of the concatemers by the T4 DNA ligase. For example, a two piece concatemer has four nicks to seal, whereas a monomer only has two, and any nicks on the strand complementary to the nickase would result in full degradation of dsDNA. Finally, we performed three technical replicates of our protocol for both pIS001 and pIS002 and evaluated the cssDNA yield using a Qubit ssDNA Assay Kit. For an input of 1 μg of plasmid dsDNA, we observed 109 and 120 ng of cssDNA for pIS001 and pIS002, respectively, resulting in a theoretical yield of 33±2% (pIS001) to 43±2% (pIS002) (Table 1; n=3, error represents 1 s.d.). Note that the quantification assay required an additional concentration step that may reduce yield by up to 30%, so the true yield is likely marginally higher than what is reported.
Table 1.
cssDNA yield calculations for the optimized cssDNA protocol. Error bars report 1 s.d., n=3 technical replicates.
| Plasmid | Yield (ng/1 μg input DNA) | Maximum Theoretical Yield (ng/ 1 μg input DNA) | Yield (% theoretical) |
|---|---|---|---|
| pIS001 | 109±7 | 327 | 33±2 |
| pIS002 | 120±6 | 276 | 43±2 |
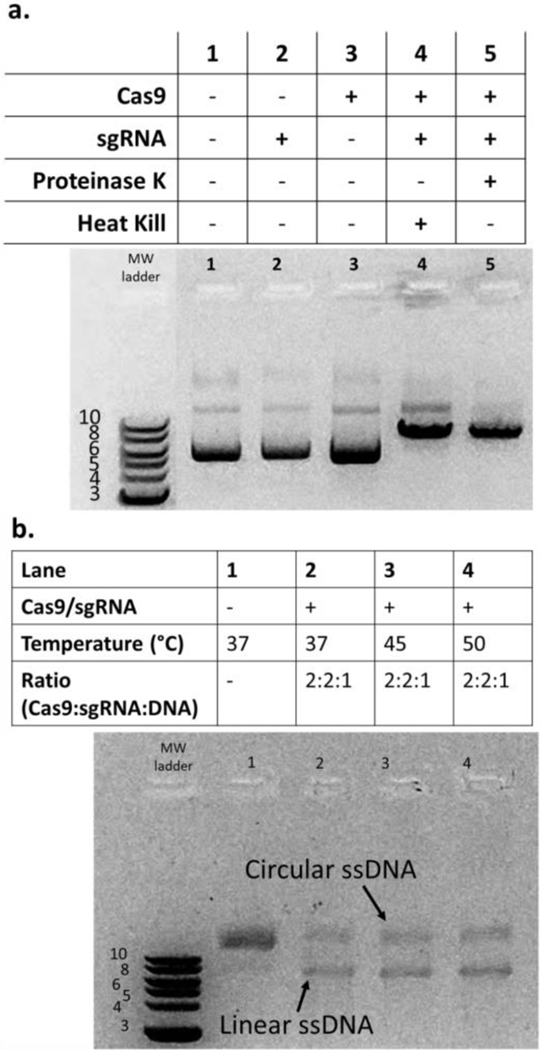
For producing linear ssDNA from cssDNA, we sought a commercially available endonuclease capable of site-specific cleavage of ssDNA. While CRISPR-Cas9 functions predominantly on dsDNA, Sternberg and Ma reported that diverse Cas9 proteins could cleave ssDNA, albeit at a reduced yield [32], [33]. In particular, they reported that the common and commercially available SpyCas9 cleaves ssDNA at approximately 50% yield on short ssDNA constructs. To evaluate SpyCas9, we used M13mp18 (7249 nts) circular ssDNA and dsDNA, as both are commercially available. We produced a sgRNA specific for M13mp18 and first addressed whether the sgRNA-SpyCas9 complex could cleave M13mp18 dsDNA (Figure 4a). We found cleavage only when both sgRNA and SpyCas9 were present in the reaction mixture, confirming sgRNA functionality. Next, we tested sgRNA-SpyCas9 functionality on circular ssDNA (Figure 4b). We observed appreciable cleavage at all incubation temperatures tested (Figure 4b) but no improvement at any temperature over the approximately 50% yield similar to the observations by Sternberg (2014) and Ma (2015). Through gel densitometry, we estimated a 52 ± 8% linearization efficiency (n = 3, error represents 1 s.d.). We also tested various Cas9:sgRNA:DNA molar ratios (5:5:1; 10:10:1; data not shown), but the optimization results did not yield any further improvements in lssDNA production.
Figure 4. sgRNA- SpyCas9 complex cleaves cssDNA to produce lssDNA.
The linearization of the DNA using sgRNA-SpyCas9 complexes was tested using 1 μg of M13mp19 dsDNA or ssDNA. a. Testing functionality of sgRNA. SpyCas9 and designed sgRNA could cleave dsDNA only when both were included in the reaction (lanes 4–5). Proteinase K or 60oC incubation was used to dissociate the RNP from DNA. b. sgRNA- SpyCas9 complex cleaves cssDNA with moderate efficiency. ssDNA cleavage using the sgRNA-Cas9 complex was successful but had poor yield. In order to properly visualize and differentiate linear ssDNA from circular ssDNA, the DNA was run on a 1.5% w/v gel with the addition of 60% v/v formamide followed by heating at 70°C for 5 minutes and then on ice for 5 minutes (to help denature the ssDNA and result in clearer bands). Variable temperatures did not increase yield.
Discussion & Conclusion
Our method presented here can be used to prepare cssDNA at a theoretical yield of up to 43±2% using commercially available reagents and in less than 5 h. The robustness of the method was demonstrated using three differently sized inserts 2.1–3.4 kb in length. Based on these results, we hypothesize, but did not demonstrate, that we could accommodate larger nucleotide inserts as well. Follow-up work could test the upper size limits of this protocol. One potential limitation of our cssDNA protocol for downstream applications like genome editing is the introduction of a 7 nt scar site on the insert. In theory, this scar site can be removed if the insert already contains an endogenous BbvCI recognition site in an opportune location. It remains to be demonstrated whether this 7 nt scar impacts insertion efficiencies using long 5’ and 3’ homology arms.
The template prep step in this procedure which uses BbvCI nicking and exonuclease digestion to generate ssDNA from dsDNA is identical to the first two steps of a procedure used by Wrenbeck et al [30] to generate user-defined mutations. Wrenbeck et al generate cssDNA as part of a larger mutagenesis protocol called nicking mutagenesis. As the template prep step is already optimized, we modified this step only to accommodate the volume change caused by previous steps in the ssDNA generation procedure. The major advance in this paper is optimization of an intramolecular ligation step to remove unwanted plasmid DNA from the final cssDNA insert.
We were unable to demonstrate a high efficiency protocol for preparation of lssDNA using an endonuclease capable of cleaving ssDNA; competing, published methods using polymerase reaction are currently superior to the enzymatic method developed here. Using commercially available SpyCas9 with a sgRNA specific for bacteriophage ssDNA, we were able to achieve a cleavage efficiency of approximately 50%, and neither higher temperature nor higher molar ratios of SpyCas9:sgRNA compared with ssDNA resulted in appreciable increases in cleavage efficiency. It is known that the sgRNA-SpyCas9 complex can bind to ssDNA, but the efficiency of the cleavage step is dramatically reduced relative to dsDNA [32]. We calculated with gel densitometry that 52 ± 8% of the bacteriophage M13mp18 cssDNA was cleaved to linear ssDNA. Sternberg et al., showed that other Cas9 proteins, like CdiCas9, could cleave ssDNA at higher yields. CdiCas9 and other high-yield ssDNA cutting Cas9 were not explored in depth here due to the criterion of the reagents being readily available; this should be a prime focus in future work. An alternative future direction is to test rare type II restriction enzymes that have been reported to cleave ssDNA [34].
The ssDNA produced in this study can be used for a variety of biotechnology applications. One major application of ssDNA is for inserting long contiguous DNA into animal models using Easi-CRISPR. Easi-CRISPR uses long ssDNA donors injected with crRNA + tracrRNA + Cas9 ribonucleoprotein (ctRNP) complexes into mouse zygotes to efficiently perform genome engineering. SsDNA can also be implemented in other variations of templated homology directed repair (HDR) for genome editing because ssDNA increases the likelihood of HDR with reduced cellular toxicity [11], [22], [35]–[37]. Another potential application includes DNA origami which is limited by the availability of diverse single-stranded DNA scaffolds that are required for the method. Self-assembling DNA nanostructures can be designed and constructed using DNA origami-folding, which involves using single-stranded DNA to direct the folding of a long ssDNA scaffold [38]. To this end, previous reports show a DNA origami potential yield of 195 mg origami [39] from a high cell density fermentation at the 1.9 L pilot scale. We can calculate a yield of 570 mg from a 1.9 L high density fermentation: an OD600 of 200 times one trillion bacteria per OD600 L time 1.9 Liters times 500 plasmids per bacteria times 2.3e6 g plasmid dsDNA per mol plasmid divided by Avogadro’s number. From this plasmid dsDNA yield, we can calculate that the amount of cssDNA we would have a theoretical maximum of 186 mg (using pIS001 as an example template). Our actual yield, following the reported values in the optimized process (Table 1), would be 80 mg. Thus, our cssDNA yield is competitive with respect to yield for smaller lssDNA produced for DNA origami.
In conclusion, we have demonstrated that user-defined circular ssDNA can be easily and cost-effectively generated using widely available reagents and basic lab techniques. We were unable to demonstrate conversion of cssDNA to linear ssDNA at a higher efficiency, but there is room for further optimization for this step and it is possible that with further experimentation, a high efficiency linear ssDNA can be synthesized using this method. Regardless, the method proposed here for cssDNA synthesis could be easily scaled up as needed to produce larger quantities of ssDNA at a reasonable cost and promises to make ssDNA creation a more affordable and accessible process for researchers.
Supplementary Material
Acknowledgements
We thank Channabasavaiah B. Gurumurthy and Rolen M. Quadros at the University of Nebraska, Omaha for support in design and acquisition of the plasmid inserts and for regular helpful discussions. Author contributions: Designed Golden Gate constructs: P.J.S., I.K.S., Z.T.B.; Designed experiments: I.K.S., P.J.S., M.S.N., Z.T.B, T.A.W.; Performed experiments: I.K.S.,M.S.N., Z.T.B.; Wrote the manuscript: I.K.S., T.A.W.; Edited the manuscript: all co-authors.
Funding Statement
This work was supported by NIH NIGMS under Award Number R21GM129559-01 to T.A.W.
Footnotes
Conflict of Interest Statement
The authors declare no conflicts of interest.
Data Availability
All raw gel images used in this study are available in the supporting information. All Golden Gate plasmids used in this study are available from AddGene (accession # pending publication).
References
- [1].Hao M, Qiao J, and Qi H, “Current and Emerging Methods for the Synthesis of Single-Stranded DNA,” Genes, vol. 11, no. 2, p. 116, Jan. 2020, doi: 10.3390/genes11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Doudna JA and Charpentier E, “The new frontier of genome engineering with CRISPR-Cas9,” Science, vol. 346, no. 6213, p. 1258096, Nov. 2014, doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- [3].Lloyd KCK, “A knockout mouse resource for the biomedical research community,” Ann. N. Y. Acad. Sci, vol. 1245, no. 1, pp. 24–26, 2011, doi: 10.1111/j.1749-6632.2011.06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].“A Mouse for All Reasons,” Cell, vol. 128, no. 1, pp. 9–13, Jan. 2007, doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- [5].Gurumurthy CB and Lloyd KCK, “Generating mouse models for biomedical research: technological advances,” Dis. Model. Mech, vol. 12, no. 1, p. dmm029462, Jan. 2019, doi: 10.1242/dmm.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Miura H, Gurumurthy CB, Sato T, Sato M, and Ohtsuka M, “CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA,” Sci. Rep, vol. 5, no. 1, Art. no. 1, Aug. 2015, doi: 10.1038/srep12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Quadros RM et al. , “Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins,” Genome Biol., vol. 18, no. 1, p. 92, May 2017, doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jacobi AM et al. , “Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes,” Methods San Diego Calif, vol. 121–122, pp. 16–28, May 2017, doi: 10.1016/j.ymeth.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pogson M, Parola C, Kelton WJ, Heuberger P, and Reddy ST, “Immunogenomic engineering of a plug-and-(dis)play hybridoma platform,” Nat. Commun, vol. 7, no. 1, Art. no. 1, Aug. 2016, doi: 10.1038/ncomms12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Srivastava S and Riddell SR, “Engineering CAR-T cells: Design concepts,” Trends Immunol, vol. 36, no. 8, pp. 494–502, Aug. 2015, doi: 10.1016/j.it.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roth TL et al. , “Reprogramming human T cell function and specificity with non-viral genome targeting,” Nature, vol. 559, no. 7714, pp. 405–409, Jul. 2018, doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Miura H, Quadros RM, Gurumurthy CB, and Ohtsuka M, “Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors,” Nat. Protoc, vol. 13, no. 1, Art. no. 1, Jan. 2018, doi: 10.1038/nprot.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Inui M et al. , “Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system,” Sci. Rep, vol. 4, no. 1, Art. no. 1, Jun. 2014, doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ma Y et al. , “Generating rats with conditional alleles using CRISPR/Cas9,” Cell Res, vol. 24, no. 1, Art. no. 1, Jan. 2014, doi: 10.1038/cr.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Horii T and Hatada I, “Challenges to increasing targeting efficiency in genome engineering,” J. Reprod. Dev, vol. 62, no. 1, pp. 7–9, 2016, doi: 10.1262/jrd.2015-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen B et al. , “Generation of gene-modified mice via Cas9/RNA-mediated gene targeting,” Cell Res, vol. 23, no. 5, Art. no. 5, May 2013, doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Quadros RM, Harms DW, Ohtsuka M, and Gurumurthy CB, “Insertion of sequences at the original provirus integration site of mouse ROSA26 locus using the CRISPR/Cas9 system,” FEBS Open Bio, vol. 5, pp. 191–197, Mar. 2015, doi: 10.1016/j.fob.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kosuri S and Church GM, “Large-scale de novo DNA synthesis: technologies and applications,” Nat. Methods, vol. 11, no. 5, Art. no. 5, May 2014, doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Minev D et al. , “Rapid in vitro production of single-stranded DNA,” Nucleic Acids Res, vol. 47, no. 22, pp. 11956–11962, Dec. 2019, doi: 10.1093/nar/gkz998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Inoue YU et al. , “An Optimized Preparation Method for Long ssDNA Donors to Facilitate Quick Knock-In Mouse Generation,” Cells, vol. 10, no. 5, Art. no. 5, May 2021, doi: 10.3390/cells10051076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Noteborn WEM, Abendstein L, and Sharp TH, “One-Pot Synthesis of Defined-Length ssDNA for Multiscaffold DNA Origami,” Bioconjug. Chem, vol. 32, no. 1, pp. 94–98, Jan. 2021, doi: 10.1021/acs.bioconjchem.0c00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Veneziano R, Shepherd TR, Ratanalert S, Bellou L, Tao C, and Bathe M, “In vitro synthesis of gene-length single-stranded DNA,” Sci. Rep, vol. 8, no. 1, Art. no. 1, Apr. 2018, doi: 10.1038/s41598-018-24677-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johne R, Müller H, Rector A, van Ranst M, and Stevens H, “Rolling-circle amplification of viral DNA genomes using phi29 polymerase,” Trends Microbiol, vol. 17, no. 5, pp. 205–211, May 2009, doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- [24].Monsur Ali M et al. , “Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine,” Chem. Soc. Rev, vol. 43, no. 10, pp. 3324–3341, 2014, doi: 10.1039/C3CS60439J. [DOI] [PubMed] [Google Scholar]
- [25].Lee ME, DeLoache WC, Cervantes B, and Dueber JE, “A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly,” ACS Synth. Biol, vol. 4, no. 9, pp. 975–986, Sep. 2015, doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- [26].Masek T, Vopalensky V, Suchomelova P, and Pospisek M, “Denaturing RNA electrophoresis in TAE agarose gels,” Anal. Biochem, vol. 336, no. 1, pp. 46–50, Jan. 2005, doi: 10.1016/j.ab.2004.09.010. [DOI] [PubMed] [Google Scholar]
- [27].Schindelin J et al. , “Fiji: an open-source platform for biological-image analysis,” Nat. Methods, vol. 9, no. 7, Art. no. 7, Jul. 2012, doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schneider CA, Rasband WS, and Eliceiri KW, “NIH Image to ImageJ: 25 years of image analysis,” Nat. Methods, vol. 9, no. 7, Art. no. 7, Jul. 2012, doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, and Smith HO, “Enzymatic assembly of DNA molecules up to several hundred kilobases,” Nat. Methods, vol. 6, no. 5, Art. no. 5, May 2009, doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- [30].Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KEJ, and Whitehead TA, “Plasmid-based one-pot saturation mutagenesis,” Nat. Methods, vol. 13, no. 11, Art. no. 11, Nov. 2016, doi: 10.1038/nmeth.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Robles-Oteiza C et al. , “Recombinase-based conditional and reversible gene regulation via XTR alleles,” Nat. Commun, vol. 6, no. 1, Art. no. 1, Nov. 2015, doi: 10.1038/ncomms9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sternberg SH, Redding S, Jinek M, Greene EC, and Doudna JA, “DNA interrogation by the CRISPR RNA-guided endonuclease Cas9,” Nature, vol. 507, no. 7490, pp. 62–67, Mar. 2014, doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ma E, Harrington LB, O’Connell MR, Zhou K, and Doudna JA, “Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes,” Mol. Cell, vol. 60, no. 3, pp. 398–407, Nov. 2015, doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nishigaki K, Kaneko Y, Wakuda H, Husimi Y, and Tanaka T, “Type II restriction endonucleases cleave single-stranded DNAs in general.,” Nucleic Acids Res, vol. 13, no. 16, pp. 5747–5760, Aug. 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen F et al. , “High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases,” Nat. Methods, vol. 8, no. 9, Art. no. 9, Sep. 2011, doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cong L et al. , “Multiplex Genome Engineering Using CRISPR/Cas Systems,” Science, vol. 339, no. 6121, pp. 819–823, Feb. 2013, doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Davis L and Maizels N, “Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair,” Proc. Natl. Acad. Sci, vol. 111, no. 10, pp. E924–E932, Mar. 2014, doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Han D, Pal S, Nangreave J, Deng Z, Liu Y, and Yan H, “DNA Origami with Complex Curvatures in Three-Dimensional Space,” Science, vol. 332, no. 6027, pp. 342–346, Apr. 2011, doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- [39].Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, and Dietz H, “Biotechnological mass production of DNA origami,” Nature, vol. 552, no. 7683, Art. no. 7683, Dec. 2017, doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw gel images used in this study are available in the supporting information. All Golden Gate plasmids used in this study are available from AddGene (accession # pending publication).