Abstract
Club cells are a type of bronchiolar epithelial cell that serve a protective role in the lung and regenerate damaged lung epithelium. Single-cell RNA sequencing of young adult human prostate and urethra identified cell populations in the prostatic urethra and collecting ducts similar in morphology and transcriptomic profile to lung club cells. We further identified club cell-like epithelial cells by single-cell RNA sequencing of prostate peripheral zone tissues. Here, we aimed to identify and spatially localize club cells in situ in the prostate, including in the peripheral zone. We performed chromogenic RNA in situ hybridization for five club cell markers (CP, LTF, MMP7, PIGR, SCGB1A1) in a series of: 1) non-diseased organ donor prostate; and 2) radical prostatectomy specimens from individuals with prostate cancer. We report that expression of club cell genes in the peripheral zone is associated with inflammation and limited to luminal epithelial cells classified as intermediate cells in proliferative inflammatory atrophy (PIA). Club-like cells were enriched in radical prostatectomy specimens compared to non-diseased prostates and associated with high-grade prostate cancer. We previously reported that luminal epithelial cells in PIA can rarely harbor oncogenic TMPRSS2:ERG (ERG+) gene fusions, and we now demonstrate that club cells are present in association with ERG+ PIA that is transitioning to early adenocarcinoma. Finally, prostate epithelial organoids derived from prostatectomy specimens demonstrate that club-like epithelial cells can be established in organoids and are sensitive to anti-androgen-directed treatment in vitro in terms of decreased androgen signaling gene expression signatures compared to basal or hillock cells. Overall, our study identifies a population of club-like cells in PIA and proposes that these cells play an analogous role to that of club cells in bronchiolar epithelium. Our results further suggest that inflammation drives lineage plasticity in the human prostate and that club cells in PIA may be prone to oncogenic transformation.
Keywords: Prostate cancer, atrophy, club cell, inflammation, intermediate cell, TMPRSS2:ERG, organoid, lineage plasticity
Introduction
Club cells (formerly called Clara cells) are bronchiolar exocrine cells present in narrow bronchioles in the lower respiratory tract. These cells consist of simple, cuboidal epithelium and function as the major secretory cell in the small airway epithelium [1]. Club cells are considered to serve a protective role to the lung, in part due to their secretion of anti-microbial and immunomodulatory proteins such as the anti-inflammatory component of pulmonary surfactant, Secretoglobin Family 1A Member 1 (SCGB1A1) [2]. Notably, club cells are regarded as the airway “second stem cell” due to their regenerative potential for damaged lung epithelium [3,4]. In addition to host defense, single-cell transcriptome analysis of human small airway epithelium identified additional roles for club cells in the lung including xenobiotic metabolism, antiprotease production, and physical barrier function [5].
Recently, single-cell RNA sequencing (scRNA-seq) of the young adult human prostate and urethra identified SCGB1A1-expressing prostate epithelial cells located in the urethra and proximal ductal trunks of the prostate that are similar in morphology and transcriptomic profile to lung club cells [6]. This study likewise identified hillock epithelial cells that are abundant in the prostatic urethra and collecting ducts, as well as the central zone surrounding the ejaculatory ducts [6]. Furthermore, in a study conducted in benign prostatic hyperplasia (BPH) tissues from patients undergoing treatment for prostate enlargement, luminal epithelial cells in 5-alpha reductase inhibitor (5ARI)-induced prostate atrophy were shown to exhibit increased expression of urethral club cell-associated genes including lactoferrin (LTF), Polymeric Immunoglobulin Receptor (PIGR), Olfactomedin 4 (OLFM4) and Secretoglobin Family 3A Member 1 (SCGB3A1) in addition to SCGB1A1 [7]. The authors conclude that luminal epithelial cells in hormonal therapy-induced prostatic atrophy undergo an adaptation from a prostate secretory luminal cell to a club cell-like state [7]. Finally, it has been suggested that the anatomical location and phenotype of prostate club cells indicate that they play an analogous role to pulmonary club cells, e.g. in acting in a protective, anti-microbial function [8]. Aside from their expansion in 5ARI-induced prostate atrophy, club cells are reported to be rare in the prostate apart from the prostatic urethra and collecting ducts [6].
Another recent scRNA-seq analysis conducted on prostate biopsies and radical prostatectomy specimens identified an epithelial cell subset that clustered separately from normal basal and luminal epithelial cells, and that were characterized by expression of PIGR, Matrix Metallopeptidase 7 (MMP7), Ceruloplasmin (CP), and LTF [9]. These cells also expressed SCGB1A1 and their gene expression profile was found to greatly overlap with previously published club cell gene signature scores [9]. These club cell-like epithelial cells were presumably located anatomically distant to the previously reported location of club cells in association with urothelium in normal, non-diseased prostate [6], and in the transition zone in BPH [7]. Rather, these cells were located in the peripheral zone of the prostate and in association with prostatic adenocarcinoma [9]. In the present study, we performed chromogenic RNA in situ hybridization (RISH) for club-cell markers (CP, LTF, MMP7, PIGR, SCGB1A1) in a series of non-diseased organ donor prostate specimens and radical prostatectomy specimens from patients with prostate cancer to identify and spatially localize club cells in the prostate, inclusive of the peripheral zone. We further characterize club-like prostate epithelial cells using prostate epithelial organoids derived from prostate cancer specimens in relation to sensitivity to anti-androgen directed treatment in vitro.
Materials and methods
Patient population and clinical samples
All specimens were acquired using protocols approved by the Johns Hopkins University and University of California, San Francisco Institutional Review Boards. Formalin-fixed paraffin-embedded (FFPE) whole tissue slides (e.g., standard tissue slides) from tissue blocks were obtained from organ donor prostates with no indication of cancer (n=10) and from radical prostatectomy specimens (n=40). One block containing the highest grade index tumor and adjacent benign was chosen from each prostatectomy case. Each slide contained tissue from the peripheral zone, with varying transition zone tissue present. The clinical characteristics of the patient samples are given in Table 1. The patient population for single cell RNA-sequencing analysis has been previously described [9].
Table 1.
Clinical Characteristics of Whole Tissue Section Cohort
| Organ Donor | Radical Prostatectomy | |
|---|---|---|
| No. Patients | 10 | 40 |
| Median age (range, years) | 21 (19–25) | 59 (46–70) |
| Race (No. Patients) | ||
| Black | 3 | 27 |
| White | 7 | 13 |
| Gleason Grade Group (No. Patients) | ||
| 1 | 12 | |
| 2 | 8 | |
| 3 | 0 | |
| 4 | 2 | |
| 5 | 18 | |
| Pathologic Stage (No. Patients) | ||
| pT2 | 23 | |
| pT3 | 17 | |
| N0 | 33 | |
| N1 | 7 |
RNA in situ hybridization (RISH)
RISH was performed using human CP (Cat No. 470231), LTF (Cat. No. 425101), MMP7 (Cat No. 488401), PIGR (Cat No. 472681), and SCGB1A1 (Cat No. 469971) probes (ACD, Newark, CA, USA). RNA quality in all tissues was assessed using peptidylprolyl isomerase B (PPIB) mRNA probe (positive control, ACD, Cat. No. 313901). The ACD RNAscope 2.5 sample preparation for FFPE Tissue kit (ACD, Cat. No. 322380) was used, with the following adjustments: slides were exposed to Pretreat 2 for 8 min, then dipped twice in 1% Tween in deionized water before applying Pretreat 3 for 30 min at 40 °C. The RNAscope 2.0 HD Detection Kit (Brown) (ACD, Cat. No. 322300) was used for signal detection. Positive controls for the RISH assays were performed with the SK-OV-3 cell line for CP [10], the IGROV-1 cell line for MMP7 [11], human intestine for PIGR, and human lung tissues for SCGB1A1. Normal prostate epithelium was used as a negative control (supplementary material, Figure S1). Positive and negative controls for the LTF RISH assay were described previously [12].
Immunohistochemistry (IHC)
IHC was performed on the Ventana Discovery Ultra IHC/ISH system (Roche Diagnostics, Basel, Switzerland) with the following antibodies: (1) AR - Cat. No. 5153/clone D6F11; 1:400, Cell Signaling Technology, Danvers, MA, USA and (2) CK903 (high molecular weight cytokeratin – HMWK) – Cat. No. ENZ-C34903/clone 34βE12; 1:50, Enzo Life Sciences, Farmingdale, NY, USA. IHC was performed following the manufacturer’s protocols. IHC for ERG (Cat. No. 790-4576/clone EPR3864; prediluted; Roche Diagnostics) and PIN4 (p63 - Cat. No. SKU:163/clone 4A4; 1:50, Biocare Medical, Pacheco, CA, USA + HMWK + alpha-methylacyl-CoA racemase – AMACR - Cat. No. Z2001/clone 13H4; 1:50; Zeta Corporation, Arcadia, CA, USA) were as described previously [13].
Image analysis
Slides were scanned with a 40x objective using a Ventana DP200 (Roche Diagnostics) digital whole slide scanner. Digital image analysis was carried out using HALO 3.1 software (Indica Labs, Albuquerque, NM, USA). Slides were annotated for cancer versus adjacent benign regions and each region (cancer versus benign) was analyzed separately. The Indica Labs Area Quantification v2.1.11 algorithm was adapted to analyze the positive staining in cancer and benign regions on each whole tissue section. Thresholds for the stain were set up with the real-time tune window. The same algorithm was used to calculate the percentage of brown staining-positive tissue per total tissue area across all whole tissue sections. Data were analyzed and graphed using GraphPad Prism Software (version 9; GraphPad Software, Inc., San Diego, CA, USA) and values were considered significant at p < 0.05. Any areas of prostatic urethra present on whole tissue sections were excluded from image analysis.
Single-cell RNA sequencing analysis of prostate epithelial organoids
Organoid domes were grown from single-cell dissociated tumors from two radical prostatectomy specimens using previously established methods [9]. To begin, the organoids were passaged, kept in the control media for one day, and then cultured in one of three conditions. In the control condition, 10 ng/ml dihydrotestosterone (DHT) was present in the media. In the two experimental conditions, either DHT was absent from the media (NODHT) or 10 μM enzalutamide (Enza), an androgen receptor inhibitor, was added to the media. The media was refreshed twice a week, every 3–4 days. Organoids were cultured for a period of two weeks and then dissociated into single cells, captured, and sequenced using the Seq-Well platform. Remaining organoids were grown for an additional three weeks, with one additional passage in between, and then sequenced on the Seq-Well platform [14].
After sequencing, organoids were aligned to the hg19 reference genome using the Dropseq workflow on Terra (https://app.terra.bio/). Downstream analysis of the gene expression matrices was conducted in Seurat [15]. Genes that were expressed by less than 3 cells were removed. Cells were filtered for greater than 500 unique molecular identifiers (UMI), greater than 300 genes, and less than 20% mitochondrial gene content. DecontX [16] was used for ambient RNA decontamination. The filtered count matrix for all cells was normalized scaled based on the top 2,000 most varied genes. The normalized matrix was scaled with a maximum value of 20 without zero-centering. Graphical clustering was performed, and a uniform manifold approximation and projection (UMAP) was generated for visualization. Differentially expressed genes and gene set signatures from previously analyzed non-treated, prostate tumor-derived organoids were used for cell type annotation. Signature scores of previously established androgen response gene sets (HALLMARK_ANDROGEN_RESPONSE, NELSON_RESPONSE_TO_ANDROGEN_UP) were computed using addmodulescore in Seurat [17]. Two-sided Wilcoxon Rank-Sum tests were used for each pair-wise comparison. Gene set enrichment analysis (GSEA) [18] was also performed in GSEA (Ver 4.0.2; https://www.gsea-msigdb.org/gsea/index.jsp) between each treatment condition (NODHT or Enza) and control (DHT) condition.
Results
Club-cell gene expression profile in association with inflammation in atrophic regions of prostate peripheral zone
Based on the results of our single cell analyses, we began by performing RISH for peripheral zone club cell markers (CP, LTF, MMP7, PIGR, SCGB1A1) as defined previously [9] in a series of whole tissue sections from radical prostatectomy specimens (n=9), to identify and spatially localize club cells in the peripheral zone of the prostate. We observed expression of CP and PIGR uniformly along prostatic urothelium, with co-expression of LTF, MMP7, and SCGB1A1 in a subset of urothelial cells and periurethral prostatic glands (Figure 1A). In the peripheral zone, co-localization of the club cell genes was limited to luminal epithelial cells in focal prostatic atrophy (Figure 1B, supplementary material, Figure S2), in regions closely associated with overt inflammatory infiltrates (proliferative inflammatory atrophy or PIA, Figure 1C). PIGR was uniformly expressed in all focal atrophy lesions/PIA, often together with LTF. Furthermore, CP, MMP7, and SCGB1A1 were observed in a subset of luminal epithelial cells in LTF and PIGR-positive focal atrophy (Figure 1C). All five club cell markers were also observed in focal atrophy lesions found in regions where they were admixed with prostate cancer (Figure 1D). Robust expression of all club cell markers except SCGB1A1 was likewise observed in postatrophic hyperplasia (PAH, Figure 2), which is a morphologically distinct subset of focal atrophy that is often closely associated with chronic inflammation in the prostate [19,20]. As with PIA, SCGB1A1 was only expressed in a subset of the cells in PAH that expressed the other club cell markers.
Figure 1.
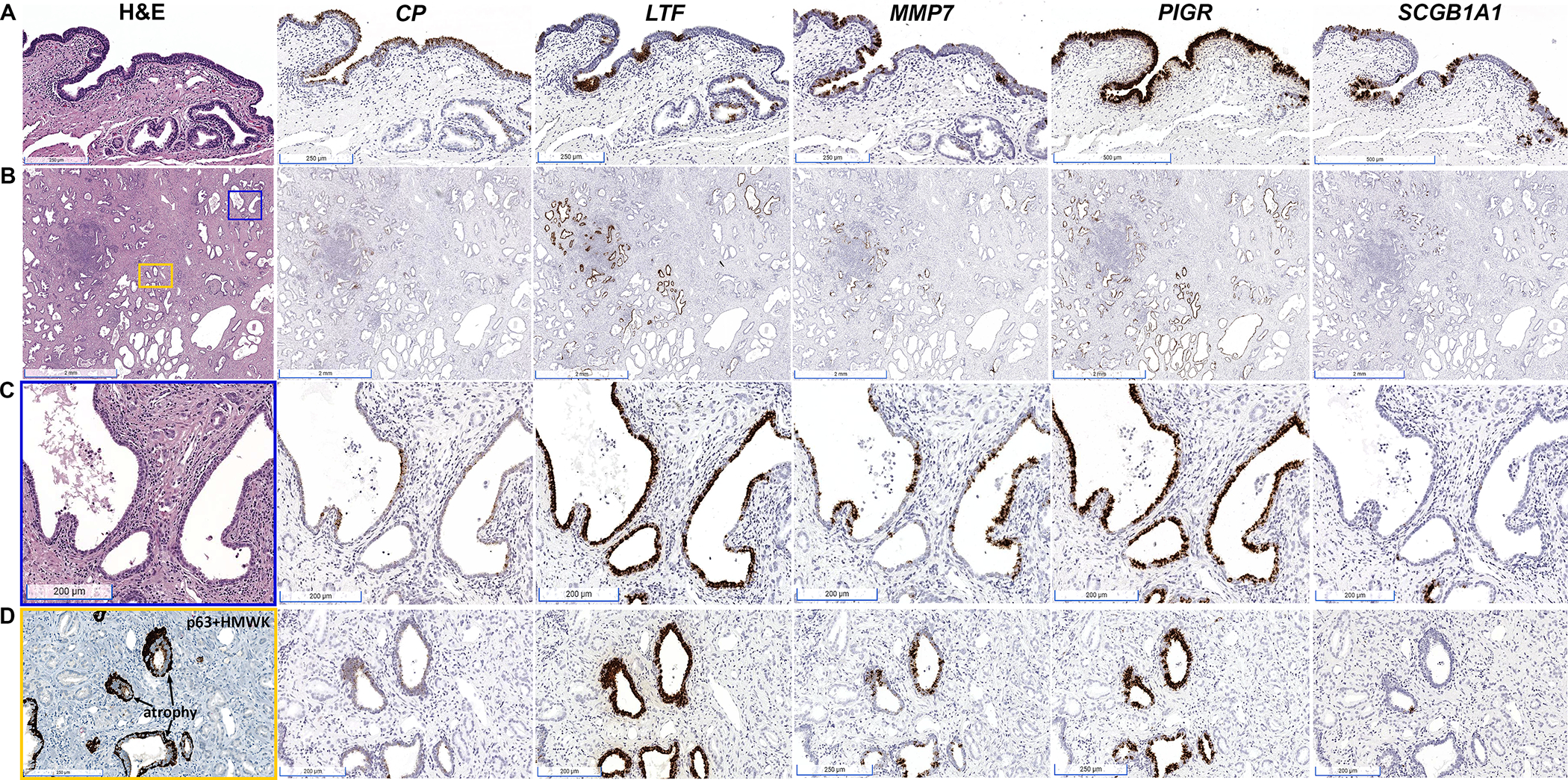
Spatial distribution of mRNA expression of CP, LTF, MMP7, PIGR and SCGB1A1 in human prostate tissues assessed by RISH. (A) Prostatic urothelium uniformly exhibits co-expression of CP and PIGR, with focal expression of LTF, MMP7, and SCGB1A1. (B) Whole tissue section from radical prostatectomy demonstrates co-expression of CP, LTF, MMP7, PIGR, and SCGB1A1 in atrophic glands abundant in prostate peripheral zone (scale bars, 2 mm). (C) Magnified view of blue boxed area in (B) demonstrating inflamed atrophic glands with co-expression of LTF, CP, MMP7, PIGR, and SCGB1A1 in atrophic epithelium (scale bars, 200 μm). (D) Magnified view of yellow boxed area in (B) demonstrating an area of infiltrating prostatic adenocarcinoma admixed with prostatic atrophy. p63 + high molecular weight cytokeratin (HMWK) stain for basal cells denotes the areas of atrophy. The glands that lack the basal cell stain are adenocarcinoma. Co-expression of CP, LTF, MMP7, PIGR, and SCGB1A1 is limited to the luminal cells within the atrophic epithelium and is absent from cancer cells.
Figure 2.
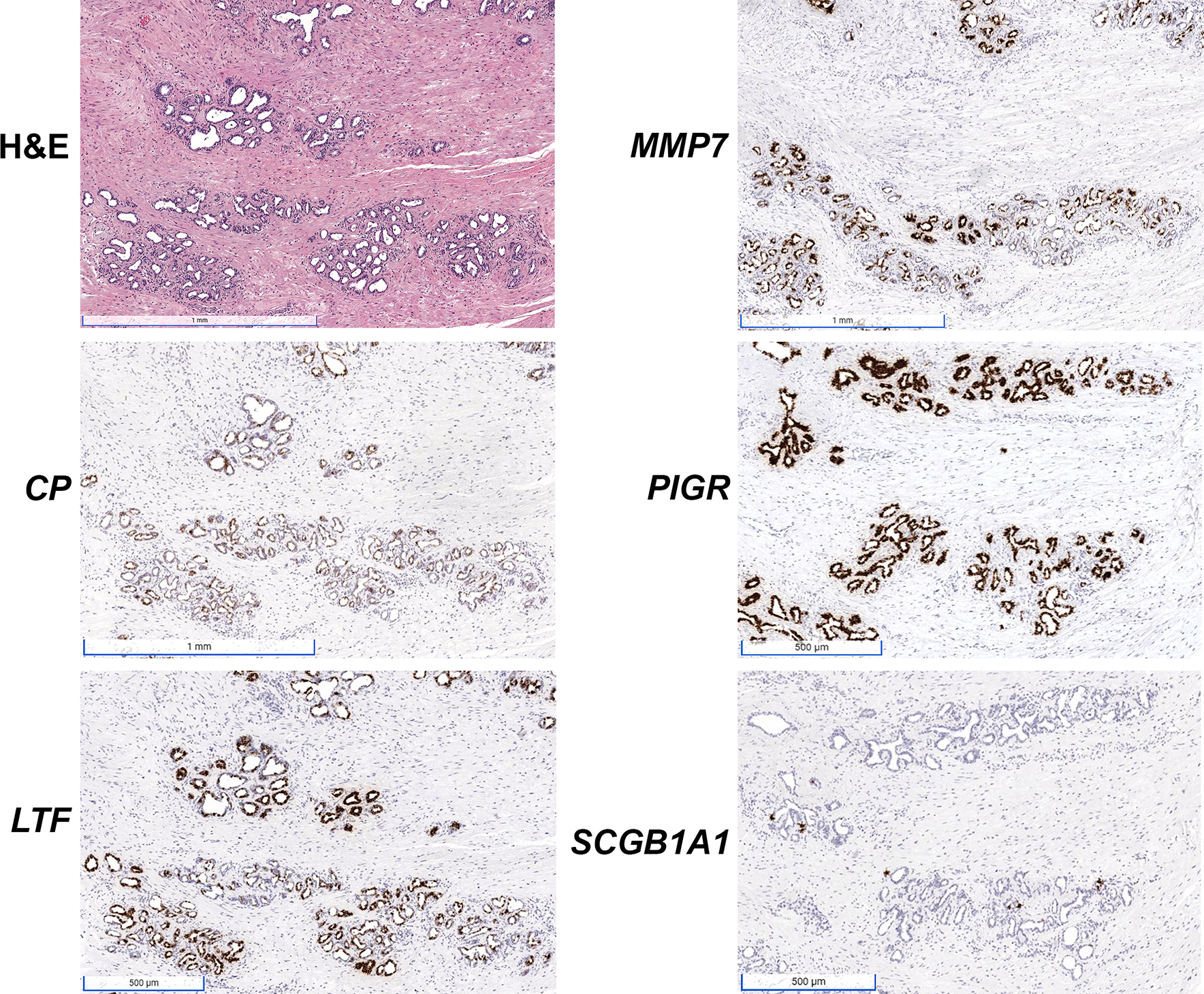
Robust expression of club cell gene markers in postatrophic hyperplasia (PAH) as assessed with RISH for CP, LTF, MMP7, PIGR, and SCGB1A1. SCGB1A1 is only expressed in a subset of the cells positive for the other markers. Scale bars for H&E, MMP7 and CP images: 1 mm. Scale bars for PIGR, LTF and SCGB1A1 images, 500 μm.
Co-expression of the club cell markers was limited to regions of focal atrophy/PIA, however we did observe very focal PIGR positivity in a subset of adenocarcinoma (supplementary material, Figure S3A). Interestingly, these focal areas of PIGR-positive adenocarcinoma were often associated with chronic inflammation (Supp. Figure 3B). SCGB1A1 was also focally present in normal, non-inflamed prostate glands with a staining pattern possibly consistent with infiltrating immune cells (supplementary material, Figure 3C). The staining pattern in benign epithelium that we observed for SCGB1A1 is consistent with RISH analysis in a prior study in cribriform prostate cancer [21].
Figure 3.
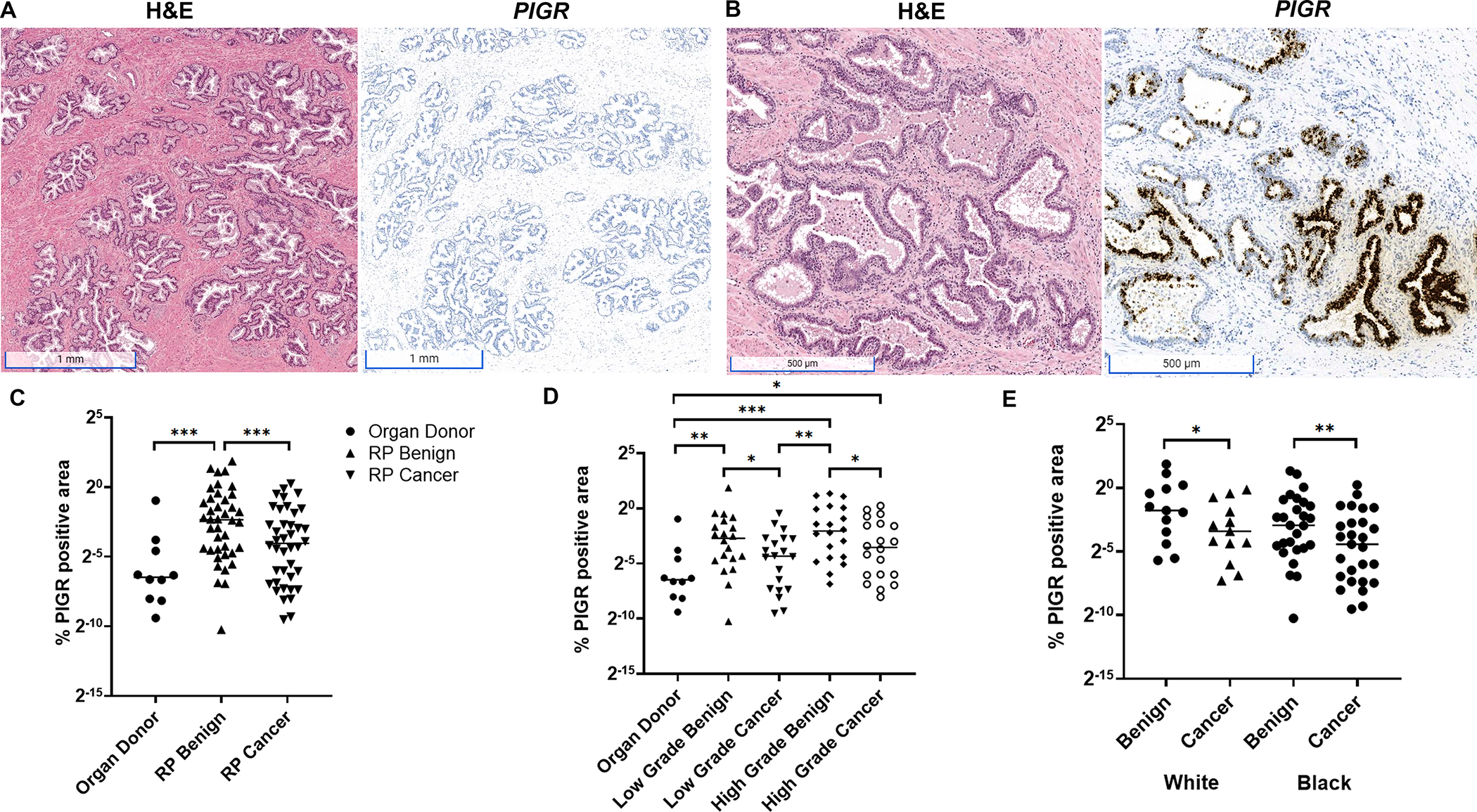
Quantification of PIGR expression in non-diseased organ donor prostate versus radical prostatectomy (RP) tissues. (A) Expression of PIGR was rarely observed in organ donor normal prostate tissues (scale bars, 1 mm) with the exception of (B) rare instances of atrophic epithelium within PIA involved with acute (polymorphonuclear) and chronic (mononuclear) inflammation in organ donor prostate (scale bars, 500 μm). (C) Quantification of PIGR expression across whole tissue sections (annotated for benign versus cancer, urothelium excluded) of radical prostatectomy (RP, n=40) and organ donor prostate (n=10) demonstrates increased expression in benign tissues from radical prostatectomy versus organ donor prostate (Mann–Whitney test) and cancer tissue (Wilcoxon matched-pairs signed rank test). (D) Increased PIGR expression in benign versus cancer tissues was present in both low grade (Grade Group 1,2) and high grade (Grade Group 4,5) prostatectomy tissues (Wilcoxon matched-pairs signed rank test). PIGR expression was likewise higher in both low and high grade benign and high grade cancer versus organ donor prostate (Mann–Whitney test). (E) Higher level expression of PIGR in benign tissues versus cancer in radical prostatectomy was maintained in tissues from both White and Black individuals (Wilcoxon matched-pairs signed rank test). * p < 0.05, ** p < 0.01, *** p < 0.001.
Club-like cells are enriched in radical prostatectomy specimens and associated with high grade prostate cancer
We next questioned whether club epithelial cells could be found in the peripheral zone of non-diseased organ donor prostate or if they were specifically associated with prostate cancer in radical prostatectomy specimens. We performed RISH for PIGR in an expanded series of whole tissue sections from radical prostatectomy (n=40) as well as organ donor specimens (n=10, Table 1). The median age of the individuals for organ donor and radical prostatectomy specimens was 21 and 59, respectively. Whole tissue sections were annotated for cancer versus benign tissues (e.g., cancer-adjacent benign) with exclusion of urothelium. Expression of PIGR was rarely observed in organ donor prostate tissues with the exception of rare instances of inflammation and PIA in organ donor prostate (Figure 3A,B). Quantification of PIGR expression across the whole tissue sections demonstrated increased expression in the cancer-adjacent benign tissues from radical prostatectomy versus organ donor prostate (p= 0.0007) and cancer tissue (p= 0.0005, Figure 3C). Increased PIGR expression in benign versus cancer areas was present in both low grade (Gleason grade group 1,2, p= 0.01) and high grade (Gleason grade group 4,5, p= 0.02) prostatectomy tissues (Figure 3D). PIGR expression was likewise higher in benign regions from cases with both low (p=0.007) and high grade cancer (p=0.0008) and in high grade cancer (p= 0.03) versus organ donor prostate (Figure 3D). Finally, higher expression of PIGR in benign tissue areas versus cancer in radical prostatectomy was maintained in tissues from both self-identified European American (White, p=0.03) and Black/African American individuals (Black, p=0.009, Figure 3E).
Club cell markers are expressed in luminal epithelial cells of PIA defined as intermediate cells in the prostate
Luminal epithelial cells termed “intermediate cells” in PIA display an intermediate phenotype between basal and luminal epithelial cells and have been proposed as tumor initiating cells in prostate cancer [22–24]. Intermediate cells co-express high levels of the typical basal cell markers cytokeratin 5 (KRT5), GSTP1, and BCL2 along with luminal cell keratin KRT18. These cells also express high levels of hepatocyte growth factor receptor MET [22] and low levels of CD38 [23]. We next explored whether the cells that co-express club cell markers would be defined as luminal intermediate cells using previously defined markers. We stained the series of radical prostatectomy tissues that we assessed for club cell markers with CK903 (high molecular weight cytokeratin (HMWK), including KRT5) using IHC. CK903 stains basal cells in the prostate as well as luminal intermediate cells in PIA. We observed almost complete overlap between PIGR-positive luminal epithelial cells and CK903-positive luminal intermediate cells (Figure 4A). Likewise, areas of PIA with co-staining for PIGR and CK903 had lower expression of androgen receptor (AR, Figure 4A), as has been previously described for intermediate cells [22,25].
Figure 4.
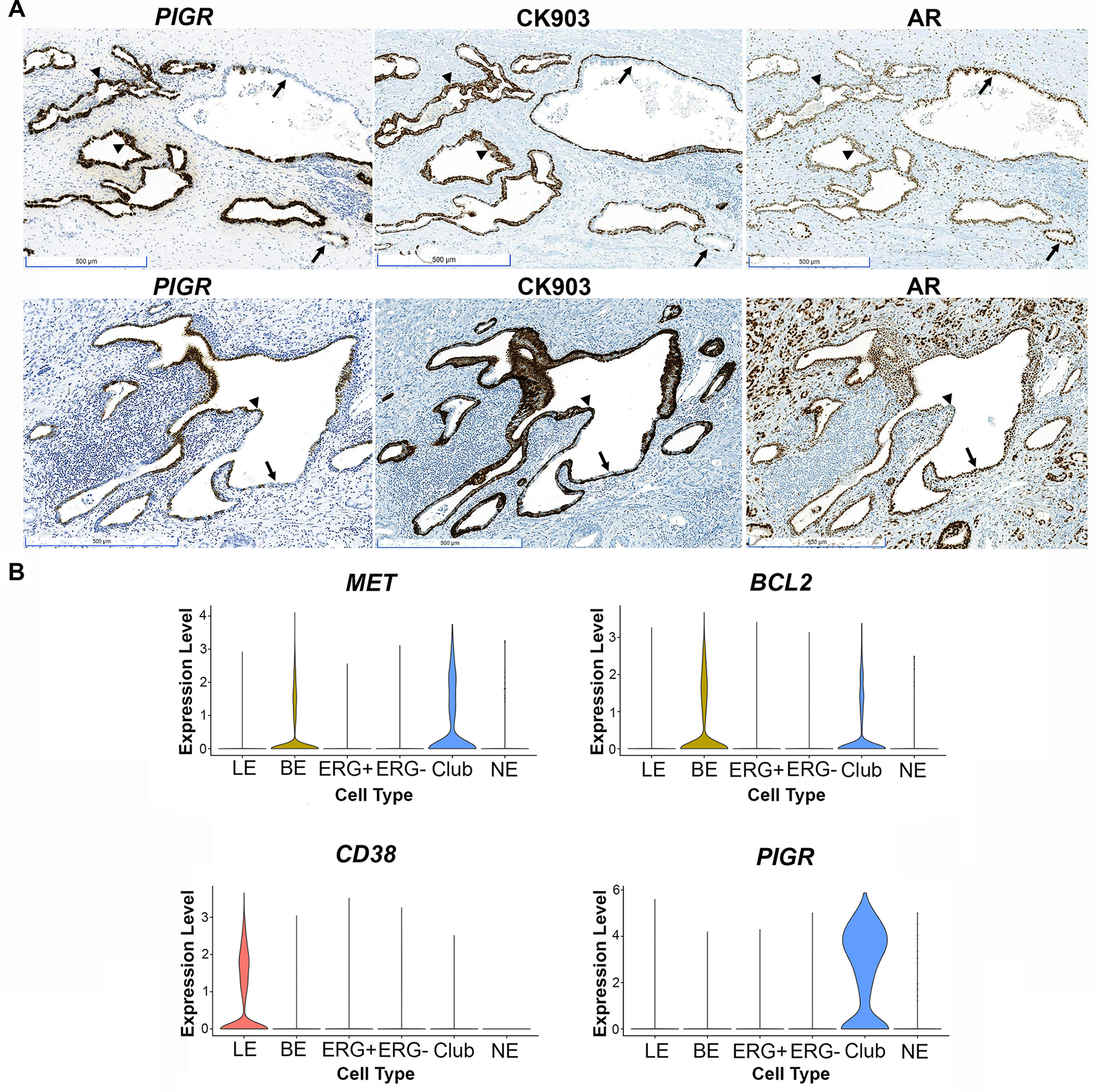
Club-like cells in prostate peripheral zone are luminal intermediate cells in PIA. (A) RISH for PIGR and IHC using CK903 and AR antibodies in PIA lesions (scale bars, 500 μm). Note decreased AR expression in areas positive for PIGR and luminal CK903 (arrowheads) and increased AR expression in areas negative for PIGR and with only basal cell staining for CK903 (arrows). (B) Single cell RNA sequencing conducted in peripheral zone tissues separated by cell type: luminal epithelial (LE), basal epithelial (BE), ERG positive (ERG+) and negative (ERG-) cancer, club cells (club), and neuroendocrine cells (NE). Note expression of the intermediate cell markers MET, BCL2, and lack of expression of CD38 in club cells.
Analysis of our previously published single cell RNA sequencing data [9] confirmed that cells that expressed a club cell gene signature were positive for MET and BCL2 and negative for CD38 (Figure 4B), further supporting that club cells in the peripheral zone of the prostate would be classified as luminal intermediate cells found in PIA.
Club-like cells are present in association with putative prostate cancer precursor lesions
We previously reported the identification of TMPRSS2:ERG fusion positive (ERG+) PIA or low grade prostatic intraepithelial neoplasia (LGPIN) in a series of radical prostatectomy cases that harbored active bacterial infections [13]. In a subset of these cases, ERG+ PIA/LGPIN was found to be in association with early invasive adenocarcinoma (microadenocarcinoma), suggesting that TMPRSS2:ERG fusion events can be initiated within PIA or LGPIN lesions that then progress to early invasive cancer. Since intermediate cells in PIA (which we now demonstrate are club cells) have been proposed as tumor initiating cells in prostate cancer [22–24], we tested whether club cells are present in association with ERG+ PIA lesions. Co-expression of luminal p63/HMWK, PIGR, and LTF was observed in PIA regions adjacent to ERG+ PIA (Figure 5 and supplementary material, Figure S4). Of interest, PIGR expression was maintained in ERG+ PIA, but LTF was absent. PIGR expression was also found to be variably maintained in the adjacent microadenocarcinoma (Figure 5 and supplementary material, Figure S4).
Figure 5.
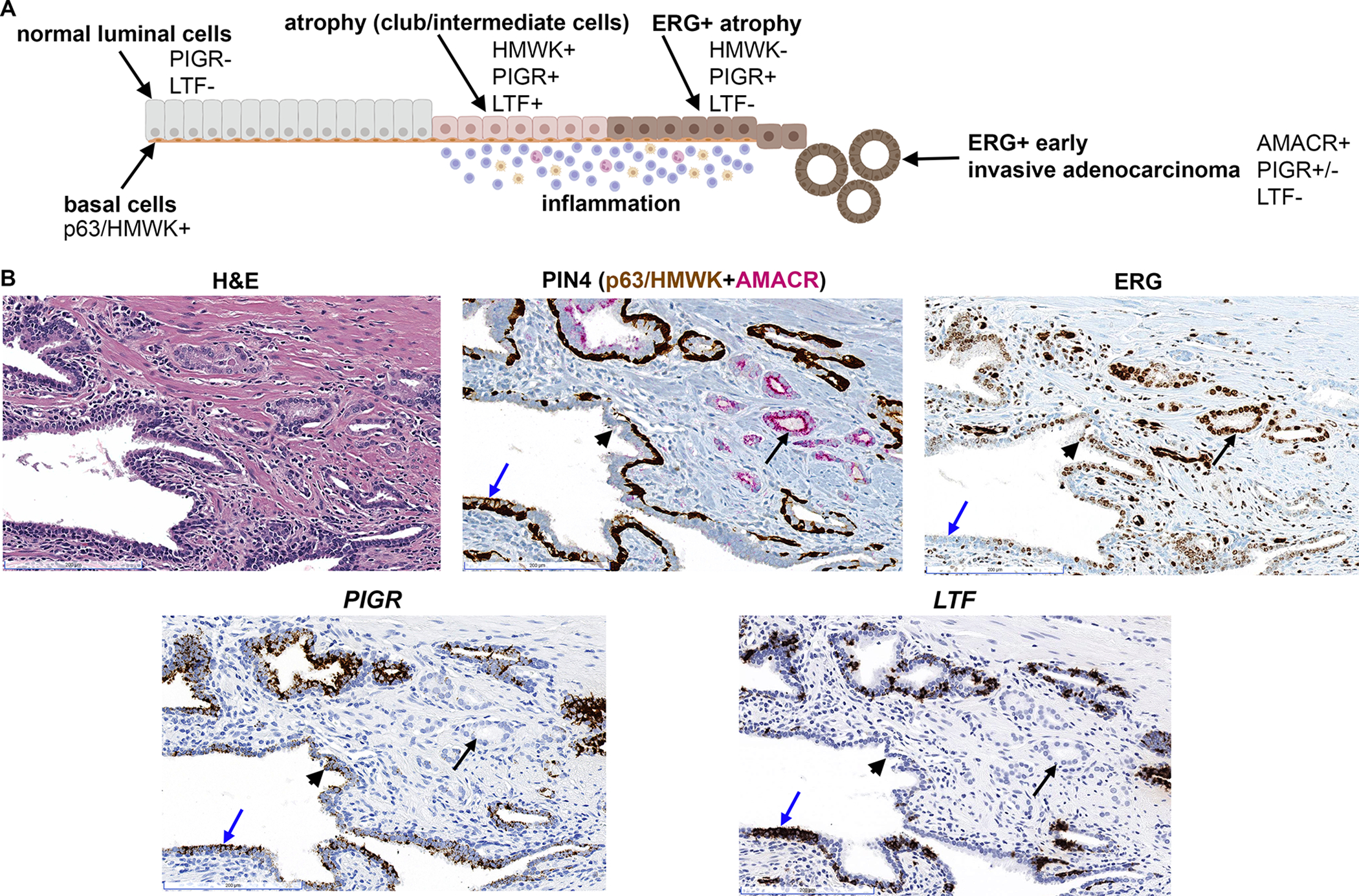
Club-like cells in association with ERG+ PIA and early invasive cancer. (A) Schematic model for the observed expression of club cell genes (PIGR, LTF) in normal prostate epithelium, PIA (atrophy), ERG+ atrophy, and ERG+ early invasive adenocarcinoma (microadenocarcinoma). Made using BioRender. (B) Club-like cells (PIGR+, LTF+) are observed in intermediate cells in PIA lesion (blue arrow) that is transitioning with ERG+ PIA (black arrowhead). Co-expression of PIGR and absence of LTF is observed in ERG+ PIA (black arrowhead). LTF is absent and PIGR is variably expressed (see also supplementary material, Figure S4) in associated ERG+ microadenocarcinoma (black arrow). PIGR and LTF assessed by RISH. ERG and PIN4 assessed by IHC, scale bars, 200 μm.
Club-like epithelial cells are established in organoids and are responsive to anti-androgen directed treatment in vitro
Based on our identification of club cells in prostate cancer tissues, we previously identified club-like cells in prostate epithelial organoids derived from prostate cancer specimens [9]. To further characterize the behavior of these club-like epithelial cells in vitro, we derived epithelial organoids from two localized prostate cancer patients (one White, one Black individual). Following a two-week treatment period with enzalutamide (Enza), androgen deprivation (NODHT), or synthetic androgen (DHT), we isolated a total of a total of 12,502 cells from these organoids. Using signature gene sets from previously analyzed prostate epithelial organoids and gene markers from normal prostate epithelial cell types, four cell populations including basal epithelial cells, hillock cells, club cells, and dividing cells were characterized among all treatment conditions. Overall, compared to cells from freshly isolated prostate epithelial tissues [9], the cells from these organoid populations exhibited more homogeneous gene expression profiles, resulting in a UMAP without clear separation of hillock from basal or club cells (Figure 6A). Similarly, we observed no distinctive separation within the three treatment conditions (Figure 6B), nor by patients (Figure 6C). Furthermore, we computed androgen response signature scores using two previously established signature gene sets HALLMARK_ANDROGEN_RESPONSE and NELSON_RESPONSE_TO_ANDROGEN_UP for comparison between the treatment conditions and cell types. Compared to basal epithelial and hillock cells, club cells exhibited the largest decrease in the two treatment conditions (NODHT and Enza) from control (DHT) condition (two-sided Wilcoxon rank-sum test, *** p < 0.001) (Figure 6D). This finding was also confirmed by gene set enrichment analysis (GSEA), in which the HALLMARK_ANDROGEN_RESPONSE was found to be the most significantly downregulated gene set in the HALLMARK gene set collection (msigdb.com) in the NODHT/Enza condition for club cells compared to the control (DHT) condition.
Figure 6.
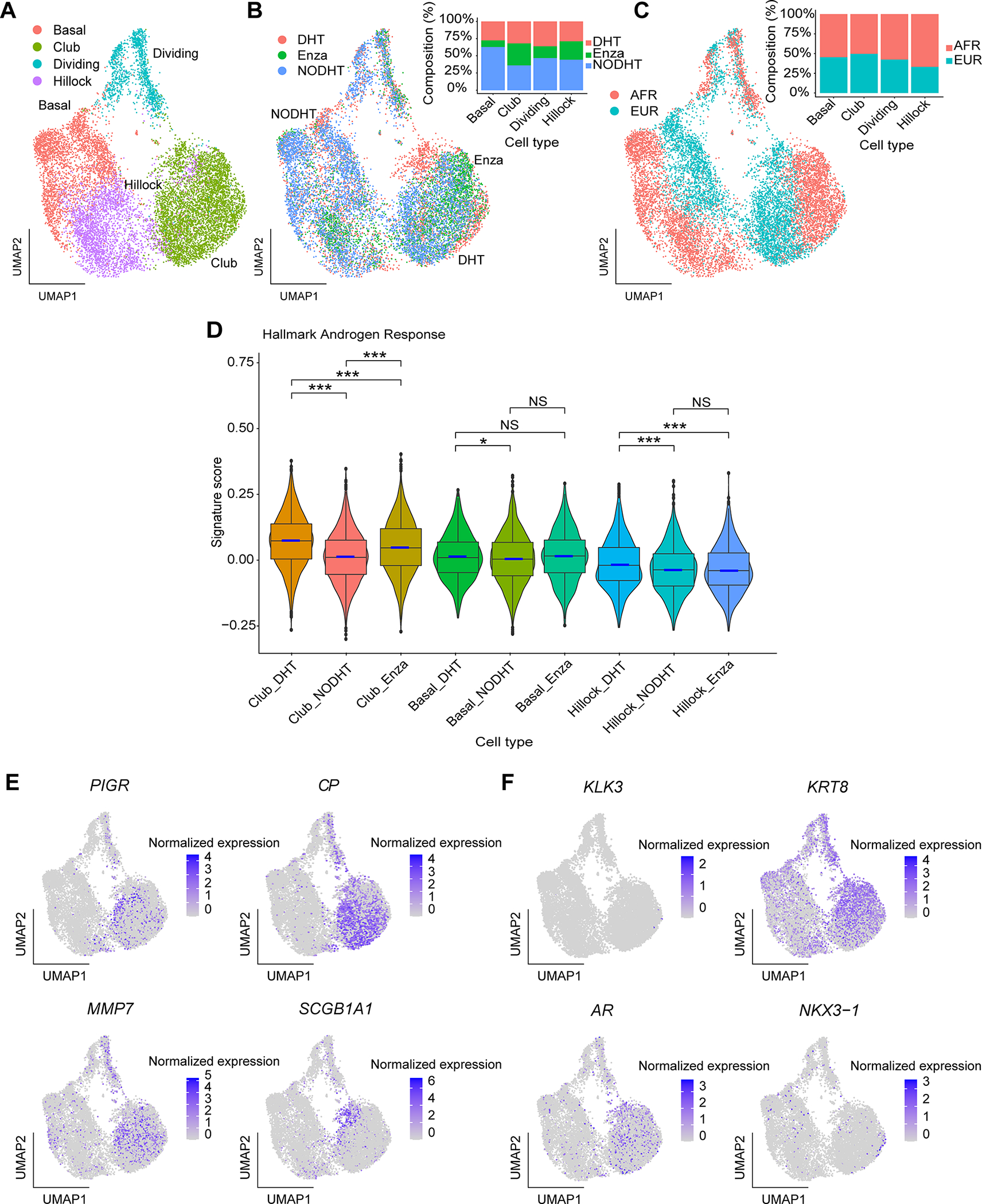
Club cells are present in epithelial cell organoids and are responsive to anti-androgen directed treatment in vitro. (A) Uniform Manifold Approximation and Projection (UMAP) of all cells captured from the organoid samples. Cells are colored based on the cell type annotations. (B) UMAP of all organoid cells colored by the three treatment conditions (DHT, NODHT, and Enza). Cell type compositions from the three conditions are shown in the bar chart. (C) UMAP of all organoid cells colored by patient ancestry groups (Black and White). Cell type compositions are shown in the bar chart. (D) Box plots of Hallmark Androgen Response signature scores across three cell types and three treatment conditions. The box plots present the 25th percentile, the median, the 75th percentile, and outlying or extreme values. The whiskers of the box plots extend to a maximum of 1.5 times the size of the interquartile range. Violin bodies showing the data distribution are super-imposed on the box plots. Statistical significance levels from pair-wise two-sided Wilcoxon rank sum tests are shown (NS: not significant; *: p < 0.05; ***: p < 0.001). (E) Feature plots of four club cell markers in the organoid cells. (F) Feature plots of four luminal cell markers in the organoid cells. Normalized expression levels are shown in the color bars for the feature plots.
Finally, we examined club cell and luminal cell markers in these organoid samples and found that previously documented club cell marker expression (PIGR, CP, MMP7 and SCGB1A1) [6,9] was mostly limited to the club cell population (Figure 6E). Compared to our previous scRNA-seq study in localized prostate cancer tumor tissues [9] as well as our observations in prostatectomy tissues using RISH in the present study, LTF expression was not detected in club cells in organoids in vitro. We also investigated luminal cell marker expression such as KLK3, KRT8, NKX3-1 and AR within these organoid samples (Figure 6F) and found a higher percentage of KRT8+ club cells and AR+ club cells compared to basal and hillock cells.
Discussion
scRNA-seq of the young adult human prostate identified epithelial cells located in the urethra and proximal ductal trunks of the prostate that are similar in morphology and transcriptomic profile to club cells [6]. Likewise, a study of BPH found luminal epithelial cells in 5ARI-induced prostate atrophy that exhibited increased expression of urethral club cell-associated genes [7]. Aside from the prostatic urethra and collecting ducts and their expansion in 5ARI-induced prostate atrophy, club cells are reported to be rare in the prostate [6]. Our recent scRNA-seq analysis conducted in cells isolated from the peripheral zone of the prostate challenges this dogma, and identified an epithelial cell subset that expressed characteristic club cell markers that we called club cells [9]. In the present study, we have now spatially localized these peripheral zone club cells in prostate tissues and identified them in the luminal compartment of focal atrophy/PIA. Furthermore, these club cells expressed characteristic “intermediate cell” markers including MET, BCL2, and lower expression of CD38. Importantly, prostate intermediate cells with similar gene expression profiles have been reported to exhibit regenerative potential and progenitor-like characteristics both in our prior study [9] and in previous reports, e.g. CD38lo luminal cells [23], and luminal 2 club cells [26]. This finding would be consistent with our observation here that club cells are present in association with ERG+ PIA lesions that are progressing to early invasive adenocarcinoma. Intriguingly, we found that whereas club cells that are adjacent to/transitioning with ERG+ PIA express LTF, both ERG+ PIA and ERG+ microadenocarcinoma are LTF-negative (Figure 5 supplementary material, Figure S4). We previously reported that LTF is silenced by CpG island hypermethylation in prostate adenocarcinoma [12]. Our finding that LTF is absent in ERG+ PIA suggest that this gene silencing may occur very early in the carcinogenic process. Collectively, our study adds to a growing body of evidence that PIA (and, hence, peripheral zone club cells) may serve as a risk factor lesion for prostate cancer development.
We report that prostate club cells are not normally present in the peripheral zone, since they were largely absent in young organ donor prostate tissues in the absence of inflammation. We therefore conclude that the club cell phenotype is induced in the adult prostate in association with cellular injury and/or inflammation, and possibly with aging, in the development of PIA. In addition to similar gene expression profiles to lung club cells, prostate club cells are also histologically similar to lung club cells. Whereas the luminal epithelial cells in normal prostate acini are tall columnar cells, the club-luminal epithelial cells in PIA are shortened and cuboidal [22], akin to club cells in the lung [1]. This intriguing similarity, both in terms of gene expression and phenotype, between lung club cells and prostate club cells, suggest that prostate club cells may play a similar functional role to club cells in the lung, e.g., a protective, anti-microbial role. We speculate that the presence of club cells in the prostatic urethra and budding prostatic ducts may signify a protective and anti-microbial function for these cells, especially related to regulation of the urinary microbiome and potential urinary pathogens [27]. The induction of this same cell type in the peripheral zone in PIA may likewise indicate that PIA has formed in response to a pathogen insult. Indeed, the classical club cell genes studied here such as the anti-microbial innate immune protein LTF [12,28], the extracellular pathogen neutralizer and non-specific microbial scavenger PIGR [29,30], and the anti-bacterial peptide activator MMP7 [31] are all involved in pathogen defense.
We report that club cells in PIA, as assessed by quantification of PIGR-expressing cells in whole tissue sections, are more abundant in cancer-adjacent benign prostate tissues than in cancer regions. This is not unexpected, as both inflammation and PIA are more prevalent within benign regions of the peripheral zone than admixed with prostate cancer [32]. However, whereas low grade cancer was quantitatively similar to organ donor prostate, high grade cancer had significantly higher quantification of PIGR-positive areas. Of interest, of the five club cell markers that we assessed in this study (CP, LTF, MMP7, PIGR, SCGB1A1), PIGR was the only marker that was focally expressed in a subset of adenocarcinoma (supplementary material, Figure S3A,B). Future studies will assess whether this focal expression of PIGR is specifically associated with higher grade prostate cancer.
In a prior BPH study [7], gene expression profiling showed that 5ARI-induced atrophy of prostate luminal cells correlated with reduced androgen receptor signaling and increased expression of club cell genes. Prostate luminal cells within hormonal atrophy acini adapted to decreased DHT conditions by increasing NF-κB signaling and BCL2 expression, and exhibited increased survival. The club cells that we identified in the peripheral zone also expressed increased BCL2 (Figure 4B). It has been postulated that focal atrophy/PIA is morphologically distinct from diffuse/hormonal atrophy that is induced after androgen withdrawal or androgen receptor blockade [33]. The results of the present study imply a potential overlap between atrophy induced by hormonal treatments and focal atrophy/PIA. Additional study is needed to clarify the similarities and differences between these two types of atrophy. Whether peripheral zone club cells associated with PIA exhibit increased survival with anti-androgen treatment remains to be determined.
Finally, it should be noted that both our RISH staining of club cell markers in prostatectomy tissues (Figure 1) and single cell profiling of prostate epithelial cell organoids (Figure 6) indicate a heterogeneity to club cell gene expression profiles, such that not all cells that we classify as club cells express all club cell markers. While our characterization of these cells in the luminal compartment of PIA as prostate club cells may be an oversimplification, we assess that multiple cell states likely exist in these lesions, and suggests the transdifferentiation potential of prostate luminal cells. Future studies will clarify these cellular states and their functional role in prostate carcinogenesis.
Supplementary Material
Figure S1. Positive and negative controls for CP, MMP7, PIGR, and SCGB1A1 RISH assays.
Figure S2. Expression of CP, LTF, MMP7, PIGR and SCGB1A1 in PIA assessed by RISH. Scale bars, 200 μm.
Figure S3. Expression of PIGR in prostate adenocarcinoma and SCGB1A1 in association with normal, non-inflamed prostate epithelium. (A) Focal expression of PIGR in prostate adenocarcinoma (scale bars, 500 μm). (B) PIGR expression in adenocarcinoma was often associated with inflammation (scale bars, 250 μm). (C) SCGB1A1 positive cells (arrows) in association with normal, non-inflamed prostate epithelium (scale bars, 500 μm).
Figure S4. Club-like cells in association with ERG+ PIA and early invasive cancer. PIGR and LTF assessed by RISH. ERG and PIN4 assessed by IHC, 200X magnification. Co-expression of PIGR and absence of LTF is observed in ERG+ PIA (black arrowheads). LTF is absent and PIGR is variably expressed (see also Figure 5) in associated ERG+ microadenocarcinoma (black arrow). Scale bars, 200 μm.
Acknowledgements
This work was supported by Prostate Cancer Foundation Challenge Award 19CHAS03 (F.W.H. and K.S.S.) and NCI grant 5U19CA214253 (F.W.H. and K.S.S.).
Footnotes
No conflicts of interest were declared.
Data availability statement
The R code and dataset for the single-cell RNA sequencing analysis can be found in Github repository https://github.com/angelussong/Clubcell_Organoid.
References
- 1.Dean CH, Snelgrove RJ. New rules for club development: New insights into human small airway epithelial club cell ontogeny and function. Am J Respir Crit Care Med 2018; 198: 1355–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu M, Yang W, Wang X, et al. Lung secretoglobin Scgb1a1 influences alveolar macrophage-mediated inflammation and immunity. Frontiers in Immunology 2020; 11: 584310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid AT, Sutanto EN, Chander-Veerati P, et al. Chapter 3 - Ground zero—the airway epithelium. In: Rhinovirus Infections. Bartlett N, Wark P, Knight D, (ed)^(eds). Academic Press, 2019; 61–98. [Google Scholar]
- 4.Kathiriya JJ, Brumwell AN, Jackson JR, et al. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell 2020; 26: 346–358.e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo W-L, Shenoy SA, Li S, et al. Ontogeny and biology of human small airway epithelial club cells. American Journal of Respiratory and Critical Care Medicine 2018; 198: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry GH, Malewska A, Joseph DB, et al. A cellular anatomy of the normal adult human prostate and prostatic urethra. Cell reports 2018; 25: 3530–3542.e3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joseph DB, Henry GH, Malewska A, et al. 5-alpha reductase inhibitors induce a prostate luminal to club cell transition in human benign prostatic hyperplasia. J Pathol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baures M, Dariane C, Tika E, et al. Prostate luminal progenitor cells: from mouse to human, from health to disease. Nature Reviews Urology 2022; 19: 201–218. [DOI] [PubMed] [Google Scholar]
- 9.Song H, Weinstein HNW, Allegakoen P, et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat Commun 2022; 13: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CM, Lo H-W, Shao R-P, et al. Selective activation of ceruloplasmin promoter in ovarian tumors. Potential Use for Gene Therapy 2004; 64: 1788–1793. [DOI] [PubMed] [Google Scholar]
- 11.Scotton CJ, Wilson JL, Scott K, et al. Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Cancer Research 2002; 62: 5930–5938. [PubMed] [Google Scholar]
- 12.Porter CM, Haffner MC, Kulac I, et al. Lactoferrin CpG island hypermethylation and decoupling of mRNA and protein expression in the early stages of prostate carcinogenesis. The American journal of pathology 2019; 189: 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha E, Coulter JB, Guzman W, et al. Oncogenic gene fusions in nonneoplastic precursors as evidence that bacterial infection can initiate prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 2021; 118: e2018976118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes TK, Wadsworth MH 2nd, Gierahn TM, et al. Second-strand synthesis-based massively parallel scRNA-Seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity 2020; 53: 878–894.e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019; 177: 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S, Corbett SE, Koga Y, et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biology 2020; 21: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016; 352: 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Marzo AM, Platz EA, Epstein JI, et al. A working group classification of focal prostate atrophy lesions. The American journal of surgical pathology 2006; 30: 1281–1291. [DOI] [PubMed] [Google Scholar]
- 20.Shah R, Mucci NR, Amin A, et al. Postatrophic hyperplasia of the prostate gland: neoplastic precursor or innocent bystander? The American journal of pathology 2001; 158: 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong HY, Sheng Q, Hesterberg AB, et al. Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease. Nat Commun 2022; 13: 6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Leenders GJ, Gage WR, Hicks JL, et al. Intermediate cells in human prostate epithelium are enriched in proliferative inflammatory atrophy. Am J Pathol 2003; 162: 1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Grogan TR, Hieronymus H, et al. Low CD38 identifies progenitor-like inflammation-associated luminal cells that can initiate human prostate cancer and predict poor outcome. Cell Rep 2016; 17: 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007; 7: 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Marzo AM, Marchi VL, Epstein JI, et al. Proliferative inflammatory atrophy of the prostate: implications for prostatic carcinogenesis. Am J Pathol 1999; 155: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karthaus WR, Hofree M, Choi D, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science 2020; 368: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrestha E, White JR, Yu SH, et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. The Journal of urology 2018; 199: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sfanos KS, Wilson BA, De Marzo AM, et al. Acute inflammatory proteins constitute the organic matrix of prostatic corpora amylacea and calculi in men with prostate cancer. Proceedings of the National Academy of Sciences 2009; 106: 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrier C, Sprenger N, Corthésy B. Glycans on secretory component participate in innate protection against mucosal pathogens. Journal of Biological Chemistry 2006; 281: 14280–14287. [DOI] [PubMed] [Google Scholar]
- 30.Phalipon A, Cardona A, Kraehenbuhl JP, et al. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity 2002; 17: 107–115. [DOI] [PubMed] [Google Scholar]
- 31.Burke B The role of matrix metalloproteinase 7 in innate immunity. Immunobiology 2004; 209: 51–56. [DOI] [PubMed] [Google Scholar]
- 32.Sfanos KS, Yegnasubramanian S, Nelson WG, et al. The inflammatory microenvironment and microbiome in prostate cancer development. Nature Reviews Urology 2018; 15: 11–24. [DOI] [PubMed] [Google Scholar]
- 33.De Marzo AM. The pathology of human prostatic atrophy and inflammation. In: P rostate Cancer: Biology, Genetics, and the New Therapeutics. Chung LWK, Isaacs WB, Simons JW, (ed)^(eds). Humana Press: Totowa, NJ, 2007; 33–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Positive and negative controls for CP, MMP7, PIGR, and SCGB1A1 RISH assays.
Figure S2. Expression of CP, LTF, MMP7, PIGR and SCGB1A1 in PIA assessed by RISH. Scale bars, 200 μm.
Figure S3. Expression of PIGR in prostate adenocarcinoma and SCGB1A1 in association with normal, non-inflamed prostate epithelium. (A) Focal expression of PIGR in prostate adenocarcinoma (scale bars, 500 μm). (B) PIGR expression in adenocarcinoma was often associated with inflammation (scale bars, 250 μm). (C) SCGB1A1 positive cells (arrows) in association with normal, non-inflamed prostate epithelium (scale bars, 500 μm).
Figure S4. Club-like cells in association with ERG+ PIA and early invasive cancer. PIGR and LTF assessed by RISH. ERG and PIN4 assessed by IHC, 200X magnification. Co-expression of PIGR and absence of LTF is observed in ERG+ PIA (black arrowheads). LTF is absent and PIGR is variably expressed (see also Figure 5) in associated ERG+ microadenocarcinoma (black arrow). Scale bars, 200 μm.
Data Availability Statement
The R code and dataset for the single-cell RNA sequencing analysis can be found in Github repository https://github.com/angelussong/Clubcell_Organoid.


