Abstract
The remarkable anatomical homeostasis exhibited by complex living organisms suggests that they are inherently reprogrammable information-processing systems offering numerous interfaces to their physiological and anatomical problem-solving capacities. We briefly review data suggesting that the multi-scale competency of living forms affords a new path for biomedicine: managing the innate collective intelligence of tissues and organs. The concept of tissue-level allostatic goal-directedness is already bearing fruit in clinical practice. We sketch a roadmap towards “somatic psychiatry”, using advances in bioelectricity and behavioral neuroscience to design methods that induce self-repair of structure and function. Relaxing the assumption that cellular control mechanisms are static, and exploiting powerful concepts from cybernetics, behavioral science, and developmental biology, may spark definitive solutions to current biomedical challenges.
Keywords: Regenerative medicine, top-down control, bioelectricity, cognition
1. Regenerative Medicine in the 21st Century: towards an anatomical compiler
By 2060, an estimated 48 million people each year (47% of all deaths globally) will suffer greatly at the end of their lives because of both ill health and treatment, which may prolong or exacerbate suffering. The spiral of increasingly expensive interventions to patch up the aging body at the end of life imposes a tremendous burden on health care systems and society at large. Hope for remediation lies in the insight that a very wide range of medical needs boil down to one key capability: control of the anatomical structures that cells build. If we understood how collectives of cells decide what to build and when to stop, we could repair birth defects; regenerate parts lost to traumatic injury, degenerative disease, and aging; and reprogram cancer to normal tissue.
The remarkable complexity of the human body is not directly specified in the genome but arises from the activity of a collection of embryonic blastomeres (Figure 1A). The computational and behavioral subroutines of these cell collectives make anatomical decisions about growth and form, and it is these decisions that we must target and modify when attempting to repair or regenerate missing or damaged organs. Bodies are constructed via a multiscale competency architecture (see Glossary) (Figure 1B, Text Box 1) - each layer processes information to solve problems in physiological, transcriptional, anatomical, and behavioral spaces. Learning to take advantage of their competencies, and manage cellular perceptions [1], memories, and setpoints (not just current biochemical states), provides a powerful roadmap for biomedicine to achieve persistent cures. Thus, the endgame for comprehensive regenerative medicine is the ability to communicate desired tissue- and organ-level outcomes to cell groups.
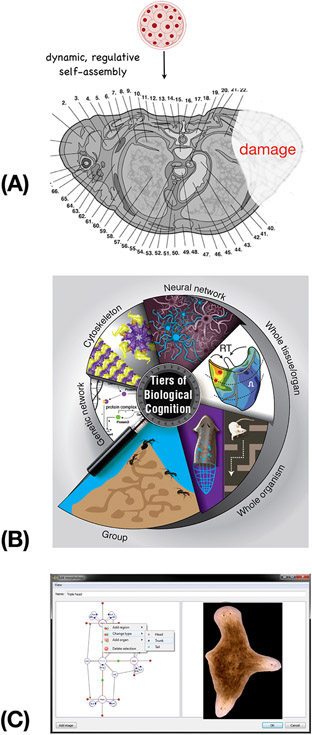
Figure 1: Anatomical compiler and multiscale competency:
(A) The remarkable complexity of the human body (shown here in cross-section through an adult torso) is not specified directly in the genome (which codes for subcellular hardware: proteins) but arises from the activity of a collection of embryonic blastomeres. It is the physiological software implemented by this cellular collective that makes anatomical decisions about growth and form, and it is these decisions that we must target and modify when attempting to repair or regenerate missing or damaged organs. (B) Bodies are constructed via a multiscale competency architecture, where each layer processes information to solve problems in physiological, transcriptional, anatomical, and behavioral spaces. Taking advantage of their competencies is a powerful roadmap for biomedicine. (C) The ultimate goal of this emerging field is to construct an “anatomical compiler” – a system which enables the user to specify any anatomical shape, and converts that to a set of stimuli that must be given to cells to get them to grow it (such as the 3-headed flatworm shown here). Crucially, the anatomical compiler will not be a 3D printer or a device to micromanage gene expression or stem cell fate: it will be in effect a communications device, converting the anatomical goals of the user into a re-specification of the target morphology information in cellular collectives. Images in A,B courtesy of Jeremy Guay of Peregrine Creative. Image in A used with permission from [173]. Image in B used with permission from [14]. Images in C courtesy of Daniel Lobo and Junji Morokuma.
Text Box 1: Multi-scale competency architecture: nested collective intelligences and implications for regenerative medicine.
One remarkable thing about bodies is that problem-solving capacity exists at all scales of organization, acting in different problem spaces. Molecular intelligence manifests in individual molecules that perform chemotaxis [148], and gene-regulatory networks and pathways that form memories driven by previous patterns of stimuli [7, 24, 25, 149, 150]. Cytoskeletal structures can plausibly encode memory [151], and the extracellular matrix is often used by cells as a stigmergic scratchpad to coordinate activity – just as the environment is used by more familiar collective intelligences such as ant colonies [152]. Thus, the ECM can be an attractive target medium in which to manipulate cell perceptions and subsequent behavior in biomedical contexts. Cellular intelligence exhibits degrees of competency, especially in the ability to navigate complex environments (e.g., in vivo tissues or engineered mazes) by integrating and prioritizing cues (from the micro-environment or self-generated [11, 12, 153, 154]) and making decisions based on a history of perceptions [1]. Tissues change gene expression to enable function despite powerful toxins with no direct evolutionary adaptation [22] – an example of problem-solving and generalization based on responses to prior stressors that share some common features. At the level of whole organs, developmental biology abounds with examples of morphogenetic problem-solving by developing and regenerating systems which achieve complex anatomical endpoints despite unexpected changes in chromosome complement, cell size and number, or large-scale injury [6]. All these capabilities, in physiological, transcriptional, and anatomical problem spaces, implement William James’ definition of intelligence as the ability to reach specific goals by diverse means. All competency levels can be exploited for biomedical purposes. At the level of tissues and organs, simple physiological trigger stimuli can induce complex, self-limiting organogenesis for regeneration [69]. At the level of single cell intelligence, time-dependent stimulation protocols can exploit associative conditioning of drugs to break pharmacological habituation of (and thus extend the regenerative efficacy of) therapeutics [138, 150]. At both levels, resetting homeostatic setpoints enables an organism to maintain healthy state without constant intervention [54, 63]. In addition to communicating goals and signals to cells, there is the exciting opportunity of learning from cellular networks - using tools of AI to read out their internal memory and belief states (akin to neural decoding), and building in silico models of the powerful native generalization and problem-solving capacities in cell physiological-transcriptional networks [22] to identify genetic and transcriptional targets relevant to desired outcomes (i.e., using cells’ ability to solve novel problems to help bioengineers solve the inverse problem limiting the use of CRISPR and similar technologies [2]).
This goal can be encapsulated in the following design challenge: to create an anatomical compiler – computer software that accepts an anatomical specification of any desired structure (biological organ or appendage, or even a novel synthetic morphology) and outputs the list of signals needed to coax cells to build exactly that. Such complete control over growth and form is the holy grail of biomedicine and synthetic bioengineering. Crucially, the anatomical compiler is not a 3D printer or some other way of micromanaging the structure and function of cell growth; it will be, in an important sense, a communications device for interfacing to the collective intelligence of cell groups – a translator between human biomedical goals and the setpoints of morphogenetic homeostasis, allostasis, and homeorhesis (Figure 1C). This represents a fundamentally new way to address current limitations in biomedicine.
The high expense and slow progress of drug discovery for complex disease states are due to ubiquitous combinatorial effects, the difficulties of extrapolating from in vitro models to patients, side effects, and drug tolerance/resistance. Current approaches focus on manipulating the cellular ‘hardware’: using CRISPR, gene therapy, protein engineering, and pathway rewiring (mRNA and drugs targeting specific proteins) to change individual components of the cellular machinery and communication systems. This bottom-up approach of micromanaging alterations to individual gene products has a key limitation for complex multiscale living systems - the difficulty of the inverse problem [2]: which genes or molecules need to be tweaked for a desired system-level effect? This problem sets the ceiling for genomic editing approaches just beyond the low-hanging fruit of single-gene, linear disorders, because there is no general procedure for knowing what micro-level interaction rules must be altered to achieve a specific outcome in a highly-emergent, context-sensitive complex system such as a living body.
Current molecular-level approaches are comparable to how computers were programmed in the 1940’s and 1950’s – by physical rewiring. In the information technology revolution, computer science made the remarkable leap to controlling function with stimuli provided over specialized interfaces, to exploit built-in, high-level, modular information-processing capabilities. Recent advances in the fields of diverse intelligence and basal cognition indicate that living creatures, at all scales of organization, come equipped with such modular information-processing capabilities [3-5]. Could biomedicine make a comparable leap from tinkering with the molecular hardware to developing tools to exploit this physiological ‘software’ of life? Emerging understanding of several remarkable features of such real-time decision-making ‘software’ - modularity, top-down control, and multiscale competency - is enabling biomedical workers to tap into a core functionality: the collective intelligence of cells and molecular networks [6, 7].
2. Wisdom of the body: anatomical homeostasis, collective intelligence, and repair driven by the need of function
Ancestral cellular capabilities, tissue-level goals
One of the most important aspects of our evolutionary history, with many implications for medicine, is that we are composed of cells that evolved from independent unicellular organisms. Not just the microbes which live within our bodies, but our actual cells have perception, decision-making, memory, anticipation, and many other capacities [1, 8-12]. Individual cells are superb at handling cell-level goals such as maintaining appropriate metabolic and physiological states despite all sorts of perturbations in their microenvironment. Crucially, they did not lose these capabilities when joining into a multicellular organism. Instead, evolution scaled up their minimal aspirations from the small setpoint landscapes of single cells (proliferation, metabolic states, etc.) to much bigger tissue- and organ-level goals: making specific anatomical structures driven by functional requirements of a multicellular organism [13, 14].
Competency for anatomical homeostasis (often, allostasis) (Figure 2A,B) is what enables accurate development and tissue renewal in the human body, as well as the even more impressive capacities of some organisms to regenerate whole appendages and organs. For example, salamanders regenerate limbs, eyes, jaws, ovaries (Figure 2C). There is even an adult mammalian example: deer regenerate antlers annually, regrowing huge amounts of bone, vasculature, innervation, and skin, with a capacity for forming new morphogenetic memories (Figure 2D) [15]. Cells readily detect deviations from the normal target morphology and rebuild exactly what’s needed, stopping when the correct target morphology is complete. This is also what enables a split early embryo to produce normal and complete monozygotic twins, rather than half-bodies (Figure 2B). Studies tracking how cellular collectives deploy a kind of swarm intelligence in development, regeneration, and cancer suppression indicate that cell groups and organs do not follow a hardwired path during metamorphosis: instead, they move through novel paths until the target anatomy is achieved. For example, when a tadpole’s craniofacial organs are scrambled, normal frogs can still result because these organs will migrate until a correct frog face is constructed (Figure 3A) [16]. This underlying capacity for solving problems in anatomical and physiological state space is the mechanistic underpinning for an extremely powerful component of any intervention: letting the body heal and adapt afterwards.
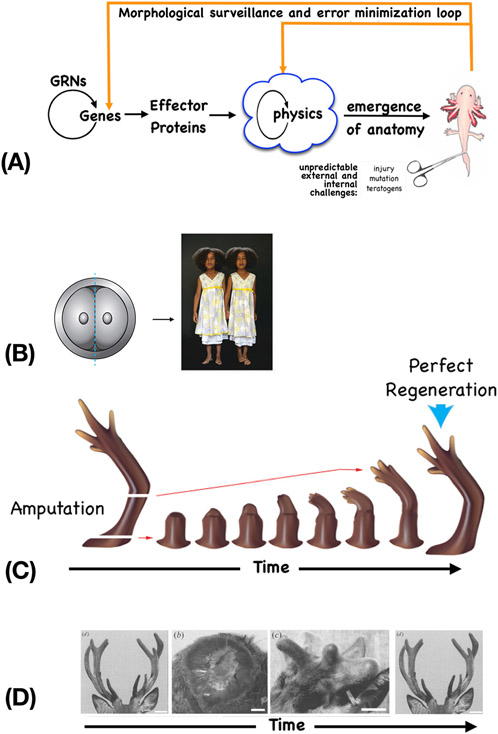
Figure 2: Morphostasis and the control loop of anatomical control.
(A) A homeostatic loop schematic depicting the conventional emergence of a set anatomy, and recursive surveillance and adjustment circuits that maintain that precise anatomy over time, despite environmental perturbation. Complex structure and function is the feed-forward result of cells mechanically following local rules, and feedback loops detect error relative to a specified setpoint (a pattern memory of large-scale form, encoded in bioelectric properties – see Figure 5), and induce cells to act to reduce error relative to that setpoint. (B) Anatomical homeostasis implements regulative development, such as for example when an early embryo is cut in half and gives rise to two complete monozygotic twins, not half-bodies. This error minimization strategy also enables regeneration in adults, such as limb regeneration in salamanders (C) and antler regeneration in deer (D). Image in A used with permission from [173]. Photo in B by Oudeschool via Wikimedia commons. Image in C courtesy of Jeremy Guay of Peregrine Creative, used with permission from [174]. Images in D used with permission from [15].
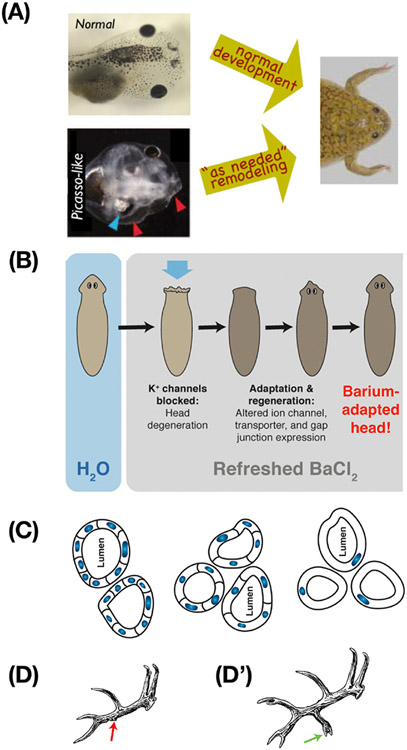
Figure 3: Problem-solving by cells and tissues.
(A) Tadpoles must rearrange their face to become frogs. But, even experimentally-scrambled tadpole faces become normal frog faces as the organs move in novel paths appropriate to their aberrant locations. This illustrates the problem-solving capacity of cellular collectives: they do not merely execute the same hardwired motions each time, but move as needed to reach the frog face target morphology, despite novel circumstances. (B) This ability to navigate a problem space despite novel interventions and scenarios extends to physiological and transcriptional spaces. Planaria soaked in the potassium channel blocker barium experience rapid head degeneration, but soon regenerate new heads that are barium-adapted. This occurs by identifying which genes to turn on and off (only a handful) to enable cells to function despite barium. Since planaria have not evolved under environmental pressure to resist barium, this navigation of gene expression space is a solution to a novel problem. (C) Kidney tubules in newts (seen here in cross-section) develop to a specified diameter regardless of the size of the component cells. When the cells are experimentally forced to be much larger, fewer of them gather to make a tubule of the same size. When they are gigantic, a single cell will bend around itself to generate the lumen. In this case, the solution to a novel problem occurs via different molecular mechanisms depending on cell size (cell:cell communication vs. cytoskeletal bending). (D,D’) Trophic memory is evidenced in seasonal regeneration of deer antlers. Damage in one year (D) alters the stable, stereotypical memory of the antler structure and causes new tines to appear in the same location when the entire rack is re-grown in the subsequent year (D’). Images in A courtesy of Douglas Blackiston and Erin Switzer, used with permission from [71]. Panel B used with permission from [22]. Image in C courtesy of Jeremy Guay of Peregrine Creative, used with permission from [14] and adapted from [164] with permission. Images in D,D’ used with permission from [2].
Beyond homeostasis: cellular problem-solving in novel conditions.
Crucially, cellular collectives can do more than reach the same target morphology despite interventions – they can also improvise solutions to novel problems [6, 17] (Figure 3). Epithelial cells isolated from bodies reboot their multicellularity and create new proto-organisms with unmodified frog or human genomes but entirely different body structures, behaviors, and capabilities [18-21]. This plasticity of structural and functional fate and the associated decision-making competencies of cellular collectives are an unconventional and potentially transformative target for management of complex system-level outcomes.
This autonomy is not only about determining body/organ shape by solving problems in anatomical morphospace. Remarkable examples of creative solutions to new stresses abound in physiological and transcriptional spaces, which are beginning to be understood in model systems. For example, when flatworms are treated with barium (a non-specific blocker of potassium channels), their heads rapidly degenerate; however, if left in barium, they soon re-grow new heads that are barium-insensitive (Figure 3B) [22]. This occurs via induction of a handful of genes that implement a new way to keep an operational head despite abrogated potassium channel function. Since planaria never encounter barium in nature, this is not activation of an evolutionary solution, making it even more remarkable that the tissues identify just the right genes to solve this physiological stressor. Similarly, newt kidney tubules develop to a specified diameter even when the size of the tubule cells is dramatically altered experimentally. When pushed to an extreme, even the mechanism by which cells form the tubule changes to maintain the target morphology (Figure 3C). The full extent of such competencies in physiological and transcriptional spaces is unknown, but they are likely implemented by the known ingredients of all intelligent behavior: diverse forms of learning, which have now been identified in non-neural tissues [7, 23, 24] and even in ubiquitous sub-cellular components of the body such as gene-regulatory networks and signaling pathways [7, 24-27]. These capabilities are now beginning to be exploited in clinical settings (see Clinician’s Corner).
Clinician’s Corner.
Cell-based transplantation using the patient’s own lymph nodes as bioreactors to grow functioning ectopic organs offers a potentially transformative alternative to orthotopic liver transplantation. Preclinical large animal models of ectopic liver have demonstrated exquisite ‘hepatostat’ control, with healthy allogeneic hepatocytes expanding to precisely replace failing native liver, and then adapting to increased or decreased liver mass and function.
Endoscopic Ultrasound (EUS)-guided delivery of hepatocytes into lymph nodes provides a much less invasive option for patients with end-stage liver disease, with a phase 2a clinical trial beginning in 2023 (NCT04496479i).
Targeting electrical decision-making circuits in tissue has shown promise in repairing complex defects of face, brain, heart, and gut in animal models. In addition, spinal cord and limbs have been induced to regenerate in animal models by simple triggers that target the pattern memories of the body. In combination with computational platforms that infer specific interventions, pharmacological or optogenetic (light-based) stimuli can trigger complex repair and remodeling.
In animal models, cancer and precancer margins can be detected by voltage-sensitive fluorescent dyes, and prevented or normalized, even in the presence of powerful oncogenes, by enforcing appropriate bioelectric states (by targeting ion channels).
Ion channel drugs are a powerful set of existing FDA-approved electroceuticals which have proven effective in addressing anatomical defects and cancer in animal models; work is now moving into preclinical studies in human cells and tissues.
Pattern homeostasis and allostasis: a unifying framework for tissue development, maintenance, and regeneration.
Development, regeneration, and cancer suppression can be viewed as facets of the same ubiquitous, dynamic process: pattern homeostasis. This is the drive to maintain a specific set of functional anatomical features, according to an encoded setpoint. When amputated, a salamander’s leg will regenerate, with the cells continuing to proliferate and migrate until a correct leg is completed, at which point they stop. Such continuous, active error minimization carried out by cell collectives has been discussed extensively in the search for organizing principles that enable tissues to resist cancer and aging over decades [28], and in examples where deprivation of information cues leads to disorganization of already formed structures (for example, severing the lingual nerve leads to a disorganization of the papillae of the tongue [29]). Text Box 2 develops this idea in more detail.
Text Box 2: Development and Regeneration as context-sensitive problem-solving.
Embryogenesis is not just an inevitable, emergent working out of mechanical cell-level rules, but a series of regenerative events which “repair” each embryonic stage toward the more correct next stage. This process is guided by rapidly-changing bioelectric prepatterns, which encode target morphology with respect to which error is estimated and minimized [14]. This changing goal state is an example of allostasis [155-160], in which 2nd-order mechanisms progressively alter the setpoint itself, enabling directed change during maturation and metamorphosis. Similarly, planarian flatworms shrink and grow allometrically, continuously remodeling their bodies to scale in perfect proportion given available cell number [161]; vertebrate embryos do the same, expanding limbs and organs as body size increases [162].
Living bodies are incredibly competent at decision-making that optimizes functionality despite huge variability in both external environment and internal components. Planaria are perfect regenerators, as well as immortal and cancer resistant, despite a very chaotic genome and mixoploidy [163]. Salamanders produce correctly-sized body structures even when their cells are made enormous by polyploidy [164]. Whole appendages self-correct: salamander tails grafted to the flank slowly remodel into limbs – a structure more appropriate to the large-scale anatomical context [165]. Thus, large-scale anatomical and physiological specifications drive underlying molecular-biological events.
Central to these phenomena is the context-sensitive, problem-solving capacity of life beyond the genetically-determined protein hardware. Non-genetic pattern memory was discovered in ciliate cortical inheritance [166]; the role of information outside the genome (and its many chromatin modifications), mediated by cytoskeletal structures and other substrates [167], extends to trophic memory in the anatomy of deer antlers [2] (Figure 3D,D’) and planarian regeneration [168]. In mice in which semaphorin (neural guidance cue) proteins are mutated, misrouted dLGN axons use alternate routes to find their way to the visual cortex [154], just like the scrambled craniofacial organs in tadpole metamorphosis [169]. A classic example is that of Slijper’s goat: [170], in which a mammal lacking forelimbs acquired many of the anatomical adjustments needed for bipedal locomotion, on a developmental (not evolutionary) time scale. These dynamic large-scale behaviors are implemented by a physiological layer that performs computations that serve ‘the needs of the organism’ [171, 172]. We return to this physiological layer in section 5, but first provide a clinical example of cell- and tissue-level capacity to respond to changing need of function in the mammalian liver.
This view that cellular collectives actively navigate anatomical, physiological, and transcriptional spaces to achieve a target makes a number of predictions that have been confirmed in vivo. One is the prediction that even major molecular disruptions, such as treatments which perturb the mechanisms that set left-right body asymmetry, are self-corrected over time [30]. This error-correcting capacity is both a potential failure point that leads to complex disease when abrogated by stress, and a very attractive target for modulation.
3. The ‘hepatostat’, a translational example of cellular intelligence and its clinical application for liver disease
The growing global burden of chronic liver disease (in the U.S., it is the 4th leading cause of death among those 45 to 55) and the paucity of therapeutic options [31], make it an important target for innovation in regenerative medicine. The liver has an extraordinary capacity to regenerate, a crucial feature for vertebrate organisms from fish to mammals [32]. In addition to hundreds of functions required for survival, including detoxification, digestion, filtration, metabolism, and protein synthesis; the liver functions as a guardian of the immune system [32]. The largest organ in the human body, the liver’s size is tightly regulated to maintain bodily functions like normal glucose and ammonia levels. The liver is the only solid organ to use its regenerative mechanism to ensure that the liver-to-body weight ratio remains constant. This homeostatic property (Figure 4), the “hepatostat” [33], and its regenerative potential, are unique to the liver and its properties suggest it is an example of cellular intelligence with high clinical relevance.
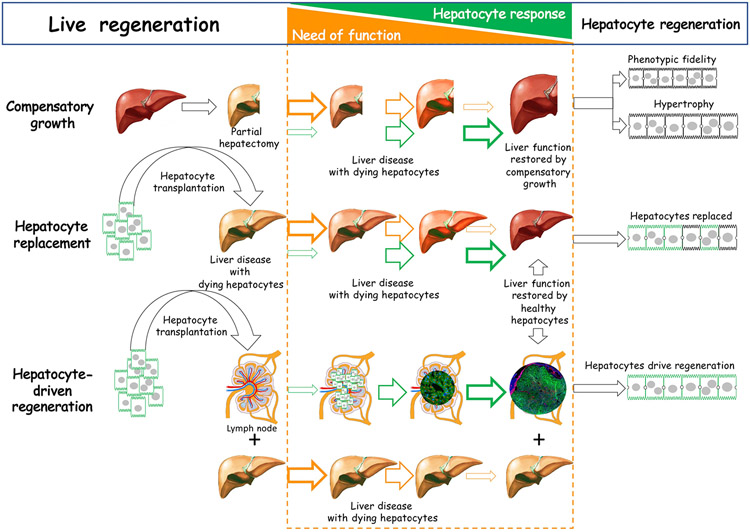
Figure 4: A hepatocyte-centric view for liver regeneration and the need of function.
After partial hepatectomy from surgical removal or accidental loss of hepatic tissue, hepatocytes respond to the need of function by proliferation, guided by phenotypic fidelity and hypertrophy to restore liver mass. After hepatocyte transplantation directed to the liver with diseased hepatocytes, healthy hepatocytes replace ailing hepatocytes, driven by the need to restore liver function. In hepatocyte transplantation into lymph node, heathy hepatocytes regenerate new liver tissue by proliferating and recruiting other cells to precisely restore lost liver mass and function, a process of complete liver regeneration driven by the need of function. This figure and images were created by Eric Lagasse using Adobe Photoshop and Powerpoint.
Hepatocytes, the specialized epithelial cells of the liver, provide most of the hepatic functions of the body, accounting for over 80% of liver mass. Like planaria, hepatocytes at the cellular level can adapt to new stresses. For example, they are exquisitely sensitive to apoptosis mediated by the Fas receptor, which has been implicated in acute liver injury leading to liver failure and death. However, hepatocytes have also been shown to mount profound cell death resistance during chronic liver injury such as in hereditary tyrosinemia [34], and in response to insults including the monoclonal anti-mouse Fas-activating antibody Jo2, the drug acetaminophen, and induced chronic cholestasis. These represent a distinct, cell-level adaptive response to catastrophic liver injury.
Liver regeneration is complex, thus it is not surprising that multiple regenerative mechanisms have evolved to ensure the stability of liver function [35]. Partial hepatectomy with surgical removal of liver mass has been studied extensively since the 19th century. Unlike regeneration in salamanders or newts, which can regenerate whole appendages and organs, the remnant liver undergoes rapid compensatory hyperplasia to recover the original liver mass, but does not restore the multi-lobular liver anatomy. Furthermore, it displays phenotypic fidelity: hepatocytes generate hepatocytes, endothelial cells generate endothelial cells, etc. Interestingly, compensatory growth is accomplished by a combination of both proliferation and increases in hepatocyte size [36]. After a 70% hepatectomy, 60% of hepatocytes divide, but after 30% partial hepatectomy, none divide: the increase in hepatocyte size alone can compensate for cell loss, and is a faster and more efficient way to increase liver function [36].
Hepatocyte transplantation has long been proposed as a potential alternative to orthotopic liver transplantation (OLT), with engrafted hepatocytes in the native liver restoring missing hepatic functions, an approach that has provided only partial correction to over 100 patients with liver disease [37]. To be successful, transplanted hepatocytes have to replace diseased hepatocytes to compensate for the loss of function, which varies by disorder. Early results in a urokinase plasminogen activator (uPA) transgenic mouse (in which intracellular activation of uPA causes hepatocyte damage and death) showed promise of hepatocyte replacement, but engraftment was reduced by homologous recombination and deletion of the toxic transgene [38], the driver for effective hepatocyte replacement. Better results were obtained in mouse and pig models of Hereditary Tyrosinemia I (HT 1), a human genetic disorder of tyrosine metabolism characterized by progressive liver damage from infancy due to mutations or deletions of the fumarylacetoacetate hydrolase (Fah) gene, which encodes the last enzyme of the tyrosine catabolic pathway [39]. In these models, transplantation of healthy hepatocytes replaced more than 90% of the diseased hepatocytes in the liver within 6 weeks (mice) to three months (pigs), rescuing the animals from lethal liver disease. Thus donor-derived hepatocytes can compete successfully with diseased hepatocytes in the hepatic microenvironment
Cell-based transplantation has been proposed as a therapeutic alternative to OLT or as a bridge for patients who are waiting for OLT [37]. However, transplanted human hepatocytes are generally injected via the splenic artery or the portal vein and for a vast majority of patients, liver cirrhosis/fibrosis and/or portal hypertension make these routes hazardous or impossible, leaving very few options. The notion of an auxiliary liver has been proposed as an alternative strategy. Generally, a healthy liver graft is placed either heterotopically or orthotopically, while leaving all or part of the native diseased liver intact. This approach not only has the potential to bridge patients for transplant, it can also avoid OLT for some by embracing the potential for spontaneous regeneration of the native liver. Although problems were noted in early trials, more recent favorable outcomes are encouraging in cases of acute liver failure [40], metabolic disorder [41-44], and even cirrhotic liver [45, 46].
Tapping the computational capacity of hepatocytes for functional supplementation by ectopic liver
The concept of making an auxiliary liver, “de novo” was discovered after transplanting hepatocytes not to the liver but to different ectopic locations in tyrosinemic mice with induced liver failure: hepatocytes engrafted in lymph nodes regenerated functional auxiliary livers. Analysis showed that hepatocytes were the only donor liver cells engrafted in lymph nodes, subsequently dividing and recruiting other cells from the host. This indicates that hepatocytes act like orchestra conductors, directing a cellular response that includes host cells to generate a functional auxiliary liver. Growth of the auxiliary liver also appeared driven by functional requirement: once it reached the mass sufficient for hepatic function, hepatocyte proliferation ceased [47, 48]. However, if partial hepatectomy is applied to the native liver, the hepatocytes in the lymph node begin to proliferate again until the auxiliary liver achieves normal liver mass and function. Remarkably, without substantial liver disease, hepatocytes transplanted into lymph nodes do not generate liver tissue [48], supporting the need-of-function concept. Direct injection of hepatocytes into a single lymph node in tyrosinemic mice generates around 70% of the normal native liver mass, enough for this ectopic liver to rescue the phenotype. The ectopic liver does not reach 100% of normal native liver mass because the native liver is still present with some hepatic function [47, 48].
Recent transcriptomic analyses comparing auxiliary and native liver revealed selective compensatory expression of hepatic function-controlling genes in auxiliary livers, implying a regulated functional integration between the two livers [49]. As expected, expression of Fah and four other genes coding for enzymes in the tyrosine catabolism pathway were dramatically repressed in native tyrosinemic livers, while auxiliary livers expressed these at levels similar to liver in wild-type controls. Similar results were obtained for genes regulating the coagulation system, urea cycle, and albumin synthesis. These observations reiterate a complex modulation of hepatocyte functions to maintain homeostasis of hepatic function under varying conditions.
Preclinical proof of concept for hepatocyte transplantation into lymph nodes as a cure for liver failure was provided in large animal studies. In the tyrosinemic Fah−/− pig, autologous Fah−/− hepatocytes were isolated, transduced ex-vivo with a lentiviral vector to express human Fah cDNA, and transplanted into mesenteric lymph nodes [50]. Hepatocyte engraftment in the lymph nodes was observed at 6 hours and for over 8 months after transplantation and sufficient liver mass to ameliorate acute liver failure was detected as early as 97 days post-transplantation. At necropsy, auxiliary livers were present in lymph nodes with hepatic lobules and vascularization identified but repopulation of the native liver with Fah-transduced hepatocytes was also detected. Tracking injected transgenic cells revealed that initial expansion of hepatocytes happened in lymph nodes, generating a source of healthy hepatocytes, which subsequently repopulated the diseased liver [50]. Further supporting need-of-function, once the native liver was restored by healthy hepatocytes, the auxiliary liver reduced in size and eventually disappeared, as described previously in tyrosinemic mice [47]. Similar results were obtained in surgically-induced subacute liver failure in an outbred pig model [51], indicating that the “need of function” regulation of ectopic hepatocytes is not specific to tyrosinemia.
Using the lymph node as a site for transplantation, the blueprint of hepatocyte-driven “need of function” and the generation of auxiliary liver has been applied to other organs. Thymus, pancreatic islets, kidney, and several other tissues have shown promise. A key aspect of resolving internally or externally initiated stress and damage is tapping into cellular capability for allostasis. Physicians understand homeostasis very well for scalar properties such as pH or temperature, but the work summarized above and similar evidence suggest that the body’s use of allostasis goes beyond that, providing a promising target for therapeutic manipulation. Rapidly accumulating evidence indicates that all levels of organization of the body include cybernetic agents involved in real-time control and proto-cognitive information processing such as perceptual control loops and minimization of surprise [52, 53]. These systems afford a novel path to biomedical management: targeting the setpoint [54] and cellular perception mechanisms [1, 8, 9, 55, 56] for the desired outcome and letting the homeostatic system do what it does best - reach its goals in a flexible, context-dependent manner.
4. Accessing the wisdom of the body to regulate regeneration: neurobiology and behavior science point the way
Collective intelligence in cellular swarms coordinates cell behaviors that implement complex healthy organs despite cell turnover, injury, and novel stressors. What systems enable this and how might we understand and harness such collective intelligence for biomedicine? In the multiscale competency architecture of life, each level (from subcellular to organ systems to social groups) is solving its own specific problems, with cells and cell collectives intelligently navigating transcriptional and anatomical spaces just as animals navigate 3-dimensional space with diverse degrees of intelligence [57]. How can we capitalize on this insight for biomedical control? The maturing fields of behavior science and neuroscience already offer avenues for understanding, predicting, and manipulating extremely complex systems via inputs and stimuli designed to impact internal decision-making. These fields have a long history of multi-scale approaches, ranging from synaptic pathway machinery to psychiatry and many levels between. These can serve as a template for biomedicine as it begins to expand beyond the current focus on increasingly lower-level targets (genes, proteins, pathways).
Neuroscience addresses problem-solving and intelligence by focusing on the mechanisms and algorithms that process information toward adaptive ends. In contrast, behavioral science works at the other end of the scale. Millennia before we understood anything about the details of the brain, human beings were able to control animals by training, illustrating how estimating a system’s intelligence, coupled with a study of the stimuli and states that serve as rewards and punishments, can enable robust control long before the mechanistic details are clear. Not only does behavioral science offer a path toward system-level outcomes that do not require complete knowledge of underlying details, but it shows the efficiency of this approach: even if it were possible to control an animal’s behavior bottom-up by managing all of the neuronal signals individually, it is much more efficient (and provides more flexible outcomes) to do it top-down by training – exploiting the native perception-memory competency of the system.
Similarly, taking advantage of the body’s own capacities for rearranging its internal state to match desired outcomes is what will enable us to solve the complexity problem plaguing medicine, in which everything is somewhat connected to everything else [58-60], making it hard to derive simple, reliable, linear interventions. Fortunately, cells and tissues offer a built-in system to rearrange their own complex molecular states in the service of a system-level goal: learning. Learning accomplishes the difficult management of myriad molecular details to implement desired organ- or whole body-scale responses. In the short term, current approaches in biomedicine have no hope of this kind of control, which is initiated by high-level experiences at the sensory or receptor layer. This offers a powerful interface for biomedical intervention; it has been argued that many future approaches to complex diseases will take the form of training and manipulating the inputs to pathways, rather than rewiring them [61-63].
5. Developmental bioelectricity: a tractable interface to native high-level anatomical control systems
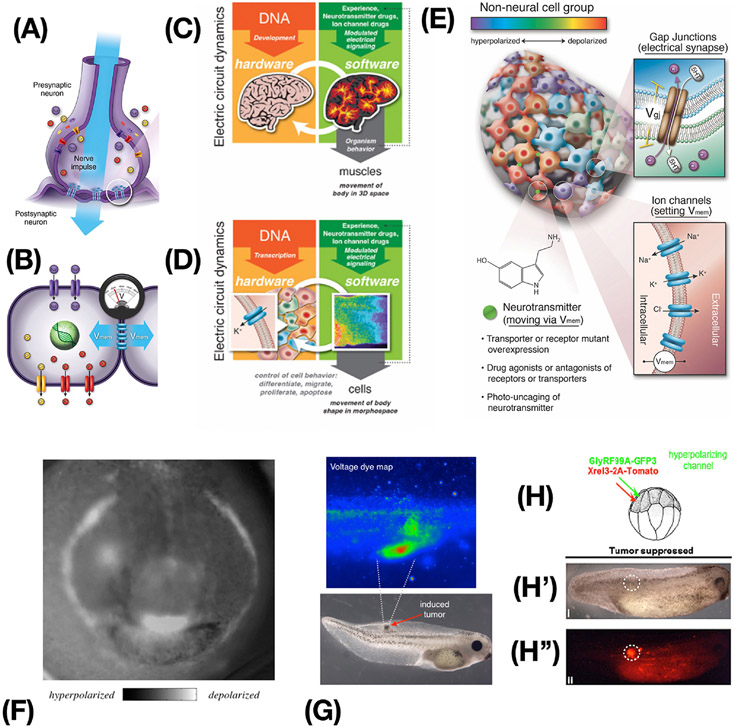
Until recently, learning was considered a unique capacity of the brain and nervous system; why would we expect the same tricks to work elsewhere in the body? Where did the remarkable information processing capacity of brains come from? All of the molecular machinery of the brain – ion channels, electrical synapses, neurotransmitters, etc., as well as many of the algorithms guiding the function of the electrical network - are ancient [64, 65] and used throughout the body (Figures 5A-D). Evolution discovered the computational power of electrical networks around the time of microbial biofilms [66, 67], and has been exploiting it ever since. Ion channels on the cell surface set cellular resting potentials (Vmem), and these can be communicated to neighboring cells via electrical synapses known as gap junctions, creating regional bioelectrical networks (Figure 5B) [68]. Almost every tissue in the body, not just neurons, forms such bioelectrical networks, and it is now known that the outputs of these networks control cell behaviors such as proliferation, differentiation, migration, apoptosis, etc. [69]. Just as neural networks process information to enable bodies to achieve behavioral goals in 3D space by controlling muscle states (Figure 5C), these non-neural bioelectrical networks process information to achieve morphological goals in anatomical morphospace by controlling cell proliferation, migration, and differentiation (Figure 5D) [57]. The similarity between the mechanisms that control electrophysiological states in neurons and in somatic cells allows techniques from neuroscience to be applied to non-neural bioelectric networks (Figure 5E). It is likely that the properties of brains that enable control via highly efficient high-level interactions are not unique, but are rather the sped-up and modified versions of universal cellular capabilities that evolution pivots into diverse problem spaces as needed [14, 70].
Figure 5: Bioelectricity as an ancient somatic control mechanism.
(A) The remarkable problem-solving capacities of brains arise as a function of physiological dynamics of computations mediated by voltage states propagating through networks of neurons. (B) Somatic cells also set resting potentials via ion channels and most cells can communicate voltage to their neighbors. (C,D) There is a deep isomorphism between the actions of the nervous system to control goal-directed movement by controlling muscles (C), and the action of non-neural bioelectric networks to achieve anatomical setpoints by controlling downstream cell behavior and navigating anatomical morphospace (D). (E) Techniques borrowed from neuroscience can be used to control network topology (target gap junctional electrical synapses) and node state (resting potential of each cell) via optogenetic, genetic, and pharmacological techniques. (F) The “electric face” in a frog embryo [71], is required for normal development and encodes the target morphologies to which cells will build (regulating gene expression, cell migration, differentiation, etc.). (G) Pathological bioelectric patterns, such as the depolarizations induced by oncogenes, can be targeted with optogenetic or mRNA-based ion channel misexpression strategies (H) to control the voltage state and force cells to participate in the electrical network’s project of normal tissue maintenance instead of tumorigenesis. In this example of a tadpole injected with a human oncogene, there is no tumor (H’) despite the very strong presence of a red fluorescence protein-labeled oncoprotein (H”). Images in A-E courtesy of Jeremy Guay of Peregrine Creative. Images in A, B used with permission from [69]. Images in C,D used with permission from [175]. Image in E used with permission from [176]. Image in F used with permission from [71]. Images in G,H,H’,H” used with permission from [87].
Endogenous bioelectrical prepatterns guiding subsequent gene expression and anatomy, such as “the electric face” in a frog embryo (Figure 5F) [71], are required for normal development and encode the target morphologies to which cells will build. Long-range bioelectrical decision-making has been implicated in the formation of appendages, induction of specific organs, size control, and alignment of major body axes [72-74]. Because of this, manipulating the bioelectric interface has been effective in animal models for induction of wound healing [75-77], appendage regeneration [78], and repair of birth defects. Existing computational methods can predict effective treatments with human-approved ion channel drugs (electroceuticals [79, 80]) that can trigger repair of complex malformations of the brain, heart, and gut in animal models [81-83]. Remarkably, this repair works not only for malformations induced by chemical teratogens, but also those induced by genetic disruptions such as mutations of the key regulator gene Notch [81, 83]. These results demonstrate that even some genetic hardware defects with pleiotropic and complex effects can be fixed “in software”, by relatively simple trigger stimuli. A similar overriding of genetic default states is seen in work on cancer [84-87], where ion channel modulation can drive cells towards normal phenotypes, normalizing or preventing tumors in vitro and in vivo, despite the presence of powerful carcinogenic mutations such as KRAS variants (Figure 5G,H).
Why do bioelectric states control complex structure and function, and why do the workhorse tools of neuroscience (such as optogenetics, active inference framework, psychoactive compounds, etc.) work outside the brain in similar ways? Recent work suggests that bioelectricity is a kind of cognitive glue that binds individual cells to common organ-level purpose throughout the body [13, 88]; bioelectric networks not only store setpoints for anatomical homeostasis but also implement the distributed communication and computations needed for tissues to recognize large-scale errors and follow a path to normalization. It has been shown that pattern memories (storing the homeostatic setpoint toward which morphogenesis will try to build) are stored as stable bioelectric patterns that can be visualized, interpreted, and re-written [72]. Thus, it is possible, for example, to edit the stored target morphology of a flatworm to read “2 heads” instead of the default “1 head”, and produce a line of permanently-regenerating 2-headed flatworms without altering their genome.
Crucially, as in the brain, the electrophysiological states of somatic cells offer privileged access to the computational medium that makes large-scale decisions. Progress in this emerging field reveals that outcomes are not purely feed-forward emergent but actually a cybernetic process with an explicitly encoded, tractable goal state, providing a way to resolve the complexity, non-linearity, and degeneracy problems that limit bottom-up interventions [2]). Thus, bioelectricity provides a physiological control layer between the microlevel molecular hardware specified by the genome and large-scale anatomical outcomes, offering an ideal target for high-level biomedical intervention.
6. What is possible? Transformative regenerative medicine by exploiting tissue intelligence.
The proposed research program offers to unify the sharp divides in the community, between molecular and synthetic biologists who view their medium as a machine, and the organicist, holistic view of systems biology, ecology, and human-centered medicine. Living bodies are “machines”, not in the naively mechanistic sense, which robs them of their obvious intelligence and agency, but in the sense that a “machine" is something that can be rationally understood, repaired, improved, and created if one finds the appropriate level of agency for interacting with it [14], the effective interface(s), and the set of stimuli that is internally meaningful to it.
This view of bodies as versatile, multiscale problem-solving agents amenable to the tools of behavioral and cognitive sciences – neither dumb and mechanical, nor inscruitably mysterious - offers to redefine our view of what is possible in radical regenerative medicine. First, by providing the regenerative medicine worker with a new set of affordances as targets for biomedical manipulations, including modular multiscale control, allostatic loops, internal informational states of (unconventional) active agents, and highly competent problem-solving mechanisms. Existing examples of structural defects that can be repaired by transient signals are just the tip of the iceberg for a coming regenerative medicine roadmap in which interventions will leverage all the innate subroutines and capabilities of living matter. From physiological diseases such as diabetes (viewed as a kind of learning disorder [89]) to birth defects [81], cancer [85], and regenerative capacity [90], we propose that a very diverse set of conditions can be addressed by triggering, redirecting, modifying, and someday enhancing the body’s self-monitoring and self-repair capabilities.
Planaria – a creature with a true centralized brain, bilateral symmetry, and learning capacity - offer a glimpse of what is possible. Not only are they highly regenerative and maintain memories across head regeneration, but they also reveal a critical gap in the current paradigm. Their chaotic, mixoploid bodies should, under current theories of cancer and aging as due to accumulation of genetic errors, be rife with tumors and senescence. Instead, they are highly resistant to cancer and apparently immortal [91, 92]. We have proposed that this phenomenon [93] is due to an evolutionary focus on physiological algorithms that are under pressure to produce a highly adaptive, functional body despite extreme noise at the molecular level. In contrast to the current theory that undifferentiated (i.e., regeneration-competent) cells are a cancer risk for long-lived animals, we suggest that anatomical plasticity control mechanisms and algorithms are what enable long-term healthspan and recovery from injury and stress. Planarians’ ability to adjust to novel internal and external [22] challenges is extreme, but all living beings have this to some degree – the ability to work towards a coherent solution despite unpredictable circumstances. Understanding and harnessing this capability is the frontier of regenerative medicine.
Tapping into cellular intelligence offers unique advantages for biomedicine
Glimpses of this future medicine have been seen in preclinical models, using physiological signals to reprogram tumors (Figure 5H) [94-96] and induce appendage regeneration. Figure 6 shows examples of the ability to initiate complex cell-behavioral subroutines by manipulating electrophysiological state in precise ways, including inducing ectopic eyes in Xenopus tadpoles (Figure 6A), and regeneration of tadpole tails and frog limbs (Figures 6B,C). Especially striking in the former is that ectopic eyes are functional even if induced in the tail, where they connect to the spinal cord rather than the brain [97, 98]. In addition, when the number of cells treated to induce an eye is small, a functional eye still results, and the resulting lens includes both treated cells and unmanipulated neighboring cells recruited to help complete the job. This capacity to achieve size control by hacking other cells is a native competency of tissue that does not have to be engineered or micromanaged, but can be exploited biomedically.
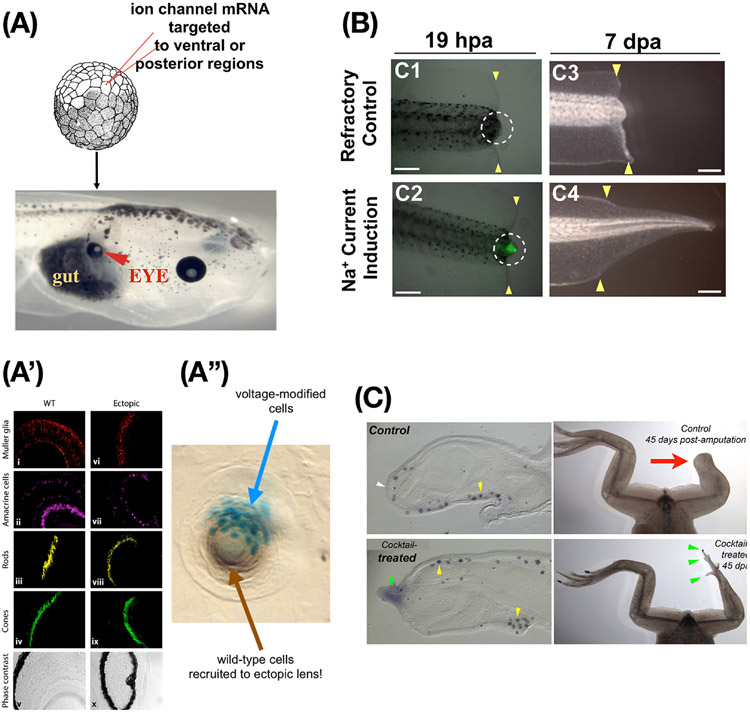
Figure 6: Bioelectric repair strategies.
(A) Misexpressing potassium channels in specific cells to induce the bioelectric prepattern corresponding to a native eye (Fig. 5F) can produce an ectopic eye (red arrowhead) in locations where the “master eye gene” Pax6 cannot do so (posterior to the head); thus, novel differentiation and morphogenesis competencies of cells are revealed by using higher-level (bioelectric) prompts instead of biochemical transcription factor machinery. (A’) Immunohistochemistry confirms all the correct tissue contents of an eye are present despite the very simple nature of the trigger. We did not specify what genes to turn on or how to make an eye; rather, we specified a bioelectric signal that said “make an eye here” and relied on the competency of the cells to do the rest. (A”) When too few cells (labeled in cyan) are injected, the resulting lens includes unmanipulated neighboring cells recruited to help complete the job. (B) Regeneration of the tadpole tail can be induced in non-regenerative conditions by a brief, 1-hour soak in monensin – a sodium ionophore which drives a depolarized wound state (green signal), inducing a tail regrowth program that lasts almost 2 weeks. (C) The same monensin signal induces MSX-1-positive blastema cells and eventually leg regeneration in froglets at a normally non-regenerative stage, showing not only that bioelectric states can induce complex appendage repair in vertebrates, but also that the initial signal does not have to contain much information about the organ to be restored – the host body contains that information, which can be triggered via this interface. Images in A,A’ used with permission from [177]. Image in A”” used with permission from [14]. Images in B used with permission from [90]. Images in C used with permission from [78].
The potential to access and trigger complex outcomes without having to micromanage intractable complexity is one of the unique and powerful strengths of harnessing collective cellular intelligence for biomedicine. Specifically, by recognizing the encoding with which target states are stored and interpreted, whole organs such as eyes and limbs can be induced by simple triggers of the downstream machinery (scaling and orientation to rest of body, correct internal structures, and self-limiting growth). Another novel and promising example is appendage (tail and limb) regeneration, in which very transient exposures [90, 99] are sufficient to change downstream decision-making. One of the key advantages of this approach is the minimization of unintended consequences (e.g., the side effects of current molecular-level approaches). By offloading the control complexity onto the biological system itself and exploiting native, highly coordinated programs instead of trying to micromanage all necessary details, the many unforeseen consequences of local tweaks can likely be avoided. The essential next step toward clinical therapeutics is to understand the full hierarchy of information-processing capacities in vivo – to learn what cell collectives measure, how they distribute attention across physical modalities, and what other competencies exist that can be harnessed.
Interoperability: a potent consequence of the conservation of cellular problem-solving capacities.
The impact of understanding this phenomenon goes well beyond repair of standard organ form and function. The ability of cells and tissues to solve novel problems underlies the interoperability of life – it’s what makes possible a wide range of chimeric, hybrid, bioengineered, synthetic, and cyborg living technology [17]. Algae inserted into a tadpole brain enable it to function via a photosynthetic mode [100, 101], smart implants [102, 103] readily integrate into tissues, closed-loop control systems enable novel living beings to operate in virtual worlds [104, 105], and Xenopus tadpoles perform well in visual training when eyes are cued to develop on their tails [97, 98]. None of these require evolutionary adaptation in the traditional, genetically-mediated and selected-for sense; the biological communication interface and problem-solving capacity is sufficiently generic that it works immediately, across kingdoms (neurons and bacteria, brains and algae, etc.) and across evolved natural forms and designed inorganic constructs [106, 107]. Future bioengineering and biomedicine will undoubtedly exploit this powerful universal design principle, in the context of ethics considerations which include both imperatives of improving health and broader issues of the impacts of synthetic bioengineering [17, 108, 109].
7. The mind of the body: towards biomedicine as somatic psychiatry
A number of frameworks for understanding the complexity of living beings [14] have suggested a range of computational capacity which includes not only mechanical systems, but also those with homeostatic, allostatic, learning, and more advanced capabilities. This forms a continuum of persuadability – a spectrum on which several classes of conceptual and empirical tools can be deployed to induce desired changes (Figure 7A,B). We cannot simply guess where the components of living bodies fit on this continuum: we must do experiments and be prepared for surprises about behavioral sophistication, with respect to discovery of novel cell capabilities and of ways in which cells resist efforts to manipulate them. One striking example is signaling pathways and gene regulatory networks. While these deterministic, simple systems look like paradigm cases of mechanical clockwork that must be re-wired to modify, they have been shown to exhibit six kinds of learning [25, 26]. Further, these networks can be efficiently controlled in the same way as brainy animals – using training protocols such as Pavlovian conditioning [24-26]. What other affordances do living tissues offer for effective prediction and control, beyond the homeostatic goal resetting strategies described above?
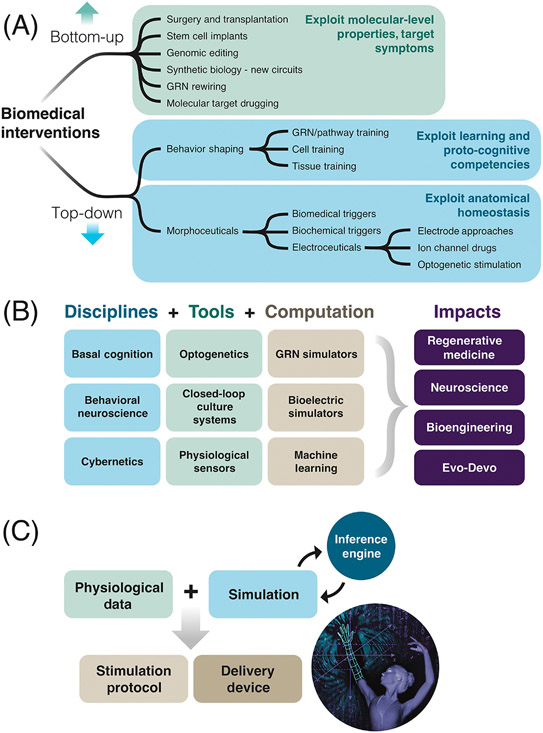
Figure 7: The landscape of emerging biomedical interventions.
(A) Biomedical interventions can be classified into two main approaches. Bottom-up: conventional approaches such as genomic editing, molecular medicine, and stem cell biology seek to control outcomes by focusing on the micro-level hardware. Aside from successes in targeting low-agency invaders (antibiotics, surgery, and chemotherapy), permanent repair is very hard to accomplish: symptoms tend to recur when the intervention is stopped, and emergent system complexity makes it very hard to know which genes or proteins to target for a desired large-scale outcome. Top-down: these novel strategies leverage the host body’s native competencies, treating it as a computational, goal-directed navigational system and targeting its memories, assessment of current state, and effector subroutines. These include shaping cell and tissue behavior via experiences or via signals provided by morphoceuticals (interventions that target the decision-points of anatomical homeostasis) and a subclass – electroceuticals. (B) Tools, concepts, and computational frameworks from several different fields can be used to develop new ways to reprogram biological behaviors at different levels to advance regenerative medicine, neuroscience, synthetic bioengineering, and basic evolutionary developmental biology. (C) A basic workflow in top-down control for biomedicine consists of a simulation platform being fed physiological data, which can predict novel interventions (time-dependent stimuli) to shift system goals to states that effect long-term repair. These must be coupled with protocols for applying such interventions, and a delivery technology (e.g., a wearable bioreactor or smart implant). Images from Jeremy Guay of Peregrine Creative, used with permission.
“Somatic psychiatry” describes how once we recognize body tissues as collective agents with navigational competencies for diverse problem spaces, we can consider techniques from behavioral and cognitive science as interventions. Recent advances at the intersection of high-level patient beliefs and psychopharmacological pathways [110-113] are beginning to show the importance of top-down controls over body biochemistry. This multiscale nature of biological controls is not limited to placebo effects. Even voluntarily getting out of bed in the morning is a process in which executive function (a high-level cognitive state) ends up controlling the membrane voltage potentials of muscle cells (a molecular-level biophysical property). Thus, mind-body medicine (Text Box 3) [114-116] is not a rare phenomenon – rather, it is an everyday occurrence essential to adaptive behavior, and offers deep opportunity for transformative approaches to health and disease. Behavioral neuroscience provides a framework for understanding multi-scale phenomena in which high level mental states and physical states interact: it has approaches suitable for dealing with levels of organization from synaptic proteins to social psychiatry, and precedents for cross-level frameworks [117, 118].
Text Box 3: From mind-body medicine to AI as interpreter of our body-mind.
“Words and drugs have the same mechanism of action.”
– Fabrizio Benedetti
The focus of today’s approaches to biomedicine and bioengineering are largely bottom-up: via implementation of specific functions by control of the lowest-level components (proteins, DNA sequences, etc.). But biology uses an integrated, multiscale competency architecture where higher levels of organization make decisions about the kinds of system-level outcomes we would like to control: large-scale shape and complex physiological states. The ultimate example of this is in the nervous system, where cognitive states (goals, beliefs, hopes, intentions, etc.) must connect to the functionality of the body. Recent and classic work on biofeedback, mind-body medicine (e.g., gene expression changes in the brain following meditative practices or exposure to music), psychoneuroimmunology, and placebo/nocebo effects have clearly shown that physiological and genetic states can be controlled by high-level nodes. It is crucial to note that mind-body control is not some unusual corner case relegated to exceptional circumstances like hypnotic states. Every time one gets out of bed in the morning to begin a day of tasks, what allows it to happen is a multiscale transduction mechanism that converts executive-level metacognitive intent into depolarization of muscle cell resting potential. Thus, our embodied minds already have the capacity to control complex molecular events and harness them toward adaptive actions without each level knowing the details of the levels below and above it. The work of pioneers such as Fabrizio Benedetti [110-113], who showed that the same mechanisms are activated by drug exposure and by expectation of drug, demonstrate a critical aspect of our evolved architecture that can be exploited therapeutically. This ability of cognitive states to implement complex downstream changes is not a unique feature of brains – rather, intelligence and distributed control are baked into all somatic cells and tissues, and are potential therapeutic targets. The native bioelectric interface linking complex goals (e.g., “grow an organ of the appropriate size and shape) to the molecular implementation machinery opens a transformative possibility for artificial intelligence to serve as a powerful GPS that can guide control of living tissue to navigate transcriptional, physiological, and anatomical landscapes. New advances such as large language models offer the possibility of literally being translators between our minds (and their goals of inducing health) and the primitive intelligence of the body, helping to derive stimuli, training protocols, and experiences as therapeutics that shape the behavior of physiological and anatomical subsystems to increase healthspan.
Specifically, we suggest that tissues, organs, and molecular pathways be increasingly examined for diverse types of learning and proto-cognitive capabilities. It is likely that the literature on basal cognition and single-cell training [119, 120] can be developed into protocols to train cells for desired gene expression levels, physiological states, or anatomical outcomes. There have already been efforts to understand diabetes as a learning disorder [89] and to model cardiac memory as bona fide memory [121]. Moreover, it is possible that the existing interface between cognitive levels amenable to top-down control could be exploited to control physiological and biochemical body states [122-124]. To some extent this has already begun, via efforts in biofeedback [125], placebo/nocebo research [126, 127], and hypnodermatology [128]. In all of these, high-level cognitive information is transduced into changes at the cellular and molecular level.
Since the time of Pavlov, it has been known that physiological circuits can be trained by experience [129], or by persistent physiological states that propagate or extend over distance in the organism. One example is that the contralateral healthy leg rapidly shows the same bioelectric state, in the same location, as an injured leg [130], revealing the propagation of quite specific status information throughout the body. Regenerative medicine approaches could parallel this ability of physical damage and stressors to set up persistent physiological states, using temporary physiological stimuli to control cell behavior and induce regenerative repair. As described above, work in animal and cell models has already demonstrated that the bioelectric control system that orchestrates the behavior of cell collectives in embryogenesis and regeneration is a tractable interface for reversing neoplasm [84, 85]. Because of the similarities between the somatic bioelectric system and the one operating in the brain, it is possible that targeting the bioelectric system will offer even more powerful methods to control bodily processes than re-writing setpoints. This would enable techniques from behavioral science (targeting beliefs, memories, and anticipatory self- and world-models) to control complex, system-level outcomes that are resistant to existing approaches. The conceptual tools used to understand cognition, such as active inference [131, 132], are already being used to understand morphogenesis and the scaling of cellular capacities in vivo [52, 133].
The future is likely to involve a tight combination of mainstream molecular pharmacology, computationally-derived new ways to use existing drugs [25, 26], and a new “somatic psychiatry” [62, 134]. Existing computational tools can already predict treatment paradigms for solving problems such as pharmaceutical resistance in patients by treating it as a form of habituation, instead of developing new drugs or targeting genes [25, 26]. It is now clear that anatomy, like physiological states, is not static – it needs to be actively maintained [28, 135-137]. Mature tissue structures disorganize after denervation, pointing to constant informational upkeep. Identifying these mechanisms and learning to work with them (in addition to micromanaging the details) may ultimately help resolve not only repair of large-scale injury and carcinogenic transformation, but aging itself. We suggest that cybernetics, control theory, and behavioral neuroscience can provide biomedicine with an entirely new toolkit for enlarging the healthspan, by targeting not the molecular and physiological landscape of the body, but the perceptions, decision-making, memories, problem-solving competencies, setpoints, and preferences of the tissue-level agent that navigates those landscapes. The sciences of behavior and adaptive embodiment include many useful conceptual frameworks including active inference (and perceptual control theory in general), dynamic learning and representation, first-person perspective of systems that make decisions and prioritize resources, goal-driven behavior in homeostatic and allostatic circuits, and integrated information. These are defined in substrate-independent ways that readily suggest porting to non-neural somatic contexts in biomedicine, using communication strategies that do not reduce living things to hardwired mechanism but rather take advantage of their cooperation as active agents engaged in the common task of improving health. The tools to implement these strategies in regenerative medicine are analogous to those used in behavioral science to communicate with, and thus predictably impact, a wide range of agents, from single cells to whole human beings. Specifically, pulsed drug stimulation, optogenetics, and acoustics can be exploited to reset goal states (anatomical setpoints), train cells to associate desired complex responses with simple trigger stimuli, re-write current state information (the informational priors of tissues exposed to stressors), control organ-level stress responses, alter the energy landscape in transcriptional and anatomical decision-making, and re-draw the borders of functional modules (via control of gap-junctional borders in tissue compartments) [138].
8. Concluding Remarks
Interestingly, the most successful current biomedical interventions, such as drugs and surgery, target “invaders” in the body: microbes (antibiotics), parasites (anti-parasitics), and cellular defectors comprising cancer (chemotherapy). These interventions effect actual cures – stable return to health after the intervention ceases. Meanwhile, treatments that definitively and reliably induce permanent repair to host organism structure and function are few. Instead, most existing drug interventions address symptoms, which return when drug administration ends. How can we learn to push physiological circuits into stable, healthy states, and overcome limitations of pharmacological resistance and side effects?
The current medical approach has two main poles: trying to address symptoms by drugging molecular targets, and waiting for the body to “heal” after various procedures. We suggest that instead, we need to develop interventions that target the healing systems, specifying their target states by reading and writing physiological memories in vivo, and augmenting their capabilities with the somatic equivalent of nootropics (cognitive enhancers) (Figure 7A).
Bacteria [139] and parasites [140-142] have been hacking the higher levels of biology for eons; so can we, by realizing that cells and tissues are an agential material [143] that affords many powerful methods of prediction and control left on the table by current mechanistic approaches (Figure 7B). We know the body uses proto-cognitive architectures and the benefits thereof; but we have only scratched the surface of understanding the capabilities, goal states, and affordances each medically-relevant subsystem offers (see Outstanding Questions). As with any novel being in behavioral science [144], we need to know how much and what kind of behavioral plasticity the body has – its competencies and motivational currency– but, akin to animal training, we don’t need to know all of the mechanistic details that implement the behavior in order to control it.
Outstanding questions.
In the context of leveraging collective intelligence and decision-making for biomedicine, how do we understand functional concepts such as instigator, determinant, and the hardware/software distinction as applied to genetics, physiology, and anatomy? Theoretical advances required for translation include a deep revision of ideas on causation and the encoding/interpretation of biological signals.
What is needed to move towards clinical practice? The BioElectric tissue simulation engine (BETSE), which models ion channel and gap junction activity to predict bioelectric dynamics, has predicted successful interventions to correct developmental defects and normalize tumors in preclinical models. How can the same perspective be incorporated into work on biochemical and biomechanical signaling? Advanced computational tools are necessary to predict and infer interventions in bioelectric networks, trainable pathways, active inference systems, and other networks to identify candidate electroceuticals.
What other affordances do living tissues offer for effective prediction and control beyond the homeostatic goal resetting strategies described above?
What technologies will offer the best methods for delivery of high-level master regulator stimuli in vitro and in vivo? What technologies can be used to overcome the depth limitations of light in physiological sensors?
How much do the policies of collective intelligence generalize across organ systems, organisms, and pathologies? Although there are diverse examples, to answer this question we will need to construct datasets that integrate living-state bioelectrical and other physiological information across stages, individuals, and health-disease states.
What are the crucial factors that determine whether a 'need-of-function'/lymph node-as-bioreactor approach can be applied to augment/replace/repair organ systems beyond the liver?
The story of the body is, fundamentally, the story of mind emerging from physics. Each of us was once a quiescent oocyte – a blob of chemistry that became a complex human metacognitive consciousness. The same mechanisms that enabled cells to solve physiological problems at the dawn of life were pivoted to solve morphogenetic problems during development, and then to solve behavioral problems via the nervous system [57]. Thus insights about problem-solving competencies can follow bidirectionally between neurocognitive science and molecular physiology of somatic health and disease [63]. Developing means of communication with pathways, cells, organ subsystems, and the human microbiome [13, 145], will be central to progress in biomedicine (Figure 7C). Thus, the medicine of the future will look more like psychiatry than like chemistry (albeit delivered through novel bioengineering interfaces [17, 146, 147]). By taking seriously the continuity of evolutionary and developmental biology, and borrowing broadly from the information and control sciences as much as from chemistry and biophysics, truly transformative vistas in health and augmented human capacity come within reach.
Highlights.
Recent advances in diverse intelligence leverage insights from cognitive neuroscience and collective computation to offer a new approach to system-level health: communication and behavior-shaping of the activity of cellular swarms
Regenerative medicine can exploit the native problem-solving of molecular and cellular networks to induce complex organ repair and cancer reprogramming
Bioelectric networks formed by somatic cells offer a highly tractable interface to exploit the collective intelligence of cells and tissues. Pre-clinical studies, many with FDA-approved drugs, demonstrate applications for birth defects, appendage regeneration, and cancer suppression
Implanted hepatocytes build an ectopic liver exactly tuned to replace lost function, offering the prospect of clinical application for end-stage organ failure, and a vision for further therapies based on inherent cell capabilities
Acknowledgements:
We thank Susan Lewis and Julia Poirier for assistance with the manuscript. We apologize to all authors whose relevant work could not be cited here due to journal rules limiting the number, type, and age of citations. M.L. gratefully acknowledges support from the Templeton World Charity Foundation (TWCF0606), the John Templeton Foundation (62212), and the Guy Foundation (103733-00001). E.L. is supported by the NIH (DK114282 and NS113810) and the State of Pennsylvania.
Glossary
- allostasis:
the process by which the body responds to stressors to regain appropriate or adaptive states
- anatomical (or pattern) homeostasis:
the morphogenetic capacity for a collection of cells to undergo novel activities (e.g. proliferation, migration, differentiation, apoptosis) to regain a specific anatomical state
- basal cognition:
the evolutionarily ancient and ubiquitous capacity of individual cells and cell groups to have certain types of memory, problem-solving, and other behavioral skills
- collective intelligence:
the memories, preferences, goals, and computational skills that are an emergent feature of a group of active agents (e.g., cells) and do not belong to any agent individually
- developmental bioelectricity:
the use of bioelectric networking, via voltage-sensitive ion channel and gap junction properties, by all tissues of the body to solve problems of anatomical control
- electroceuticals:
therapeutic agents or interventions (ion channel targeting drugs, bioelectronic devices) that work by altering electrical state of cells and tissues to trigger endogenous healing or corrective functions
- inverse problem:
the difficulty of inferring which low-level properties (e.g., DNA) must be changed to give a specific system-level outcome. The inverse problem occurs because molecular states (e.g., genes) do not directly code for form and function; the relationship between local rules and final outcome is highly complex and emergent
- machine:
a physical system that is understandable to some extent from the 3rd-person perspective. It has features that allow other observers (parasites, conspecifics, its own cellular parts, and bioengineers/medical workers) to predict and manipulate its behavior using appropriate interfaces and the right model of its level of agency and cognitive capacity. All agents, on the wide and diverse spectrum from simple mechanical devices to the human organism, have machine-like aspects which can be used to optimally interface with it in specific settings, using tools that range from hardware rewiring (e.g., genomic editing) to training and complex communication protocol. Those aspects, at multiple levels of organization, include well-defined behavioral and cognitive features that, while tractable to medical manipulation, also provide the system’s remarkable, agential quality
- morphospace:
a virtual, multidimensional space of possible anatomical configurations for an organ or organism
- multiscale competency architecture:
the principle that each layer of organ systems, organs, tissues, cells, and molecular networks in an organism is a problem-solving system with goals and memories which cooperate and compete with each other
- need of function:
the driver necessary for cells to restore function, e.g., during liver regeneration when hepatocytes transplanted ectopically will drive cells to rebuild a functional liver
- OLT:
orthotopic liver transplantation involves the replacement of the diseased native liver with a normal healthy liver taken from a deceased or living donor
- somatic psychiatry:
emerging field in which tools of communication are used to control tissue. Much as a therapist effects major life changes using a verbal interface, somatic psychiatry exploits ways to target memory, decision-making, preferences, encoded goal states, cellular perceptions and informational states in the body to improve health
- top-down control:
strategy that targets large-scale features of the body: information flows, memories, encoded goal states, and other representations of large-scale features. Top-down control targets cellular collectives’ representations of states which do not exist at the molecular level, e.g. “number of fingers”
Footnotes
Competing Interests: M.L. is a co-founder of Morphoceuticals, a company working in the regenerative medicine space, and is supported by Astonishing Labs, a company that includes the targeting of computational capacities of pathways among its goals. E.L. is a co-founder of LyGenesis, a clinical-stage cell therapy company that transforms a patient’s lymph nodes into bioreactors capable of growing functioning ectopic organs.
Resources
Registered at clinicaltrials.gov
References
- 1.Bugaj LJ et al. (2017) Interrogating cellular perception and decision making with optogenetic tools. J Cell Biol 216 (1), 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobo D et al. (2014) A linear-encoding model explains the variability of the target morphology in regeneration. Journal of the Royal Society, Interface / the Royal Society 11 (92), 20130918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baluska F et al. (2022) Cellular sentience as the primary source of biological order and evolution. Biosystems 218, 104694. [DOI] [PubMed] [Google Scholar]
- 4.Baluska F et al. (2022) Cellular and evolutionary perspectives on organismal cognition: from unicellular to multicellular organisms. Biological Journal of the Linnean Society. [Google Scholar]
- 5.Reber AS and Baluska F (2021) Cognition in some surprising places. Biochem Biophys Res Commun 564, 150–157. [DOI] [PubMed] [Google Scholar]
- 6.Levin M (2023) Collective Intelligence of Morphogenesis as a Teleonomic Process. In Evolution “on Purpose” : Teleonomy in Living Systems (Corning PA, Kauffman SA, Noble D, Shapiro JA, Vane-Wright RI, Pross A ed), pp. 175–198, MIT Press. [Google Scholar]
- 7.Csermely P et al. (2020) Learning of Signaling Networks: Molecular Mechanisms. Trends Biochem Sci 45 (4), 284–294. [DOI] [PubMed] [Google Scholar]
- 8.Antebi YE et al. (2017) Combinatorial Signal Perception in the BMP Pathway. Cell 170 (6), 1184–1196 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell A and Lim W (2016) Cellular perception and misperception: Internal models for decision-making shaped by evolutionary experience. Bioessays 38 (9), 845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MZ et al. (2017) Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Mol Cell 67 (5), 757–769 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tweedy L et al. (2020) Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown. Science 369 (6507). [DOI] [PubMed] [Google Scholar]
- 12.Tweedy L and Insall RH (2020) Self-Generated Gradients Yield Exceptionally Robust Steering Cues. Front Cell Dev Biol 8, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin M (2019) The Computational Boundary of a “Self”: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition. Frontiers in Psychology 10 (2688), 2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin M (2022) Technological Approach to Mind Everywhere: An Experimentally-Grounded Framework for Understanding Diverse Bodies and Minds. Frontiers in Systems Neuroscience 16, 768201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price J and Allen S (2004) Exploring the mechanisms regulating regeneration of deer antlers. Philos Trans R Soc Lond B Biol Sci 359 (1445), 809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Racovita A et al. (2022) Engineered gene circuits capable of reinforcement learning allow bacteria to master gameplaying. bioRxiv, 2022.04.22.489191. [Google Scholar]
- 17.Clawson WP and Levin M (2022) Endless forms most beautiful 2.0: teleonomy and the bioengineering of chimaeric and synthetic organisms. Biological Journal of the Linnean Society. [Google Scholar]
- 18.Blackiston D et al. (2021) A cellular platform for the development of synthetic living machines. Science Robotics 6 (52), eabf1571. [DOI] [PubMed] [Google Scholar]
- 19.Kriegman S et al. (2020) A scalable pipeline for designing reconfigurable organisms. Proc Natl Acad Sci U S A 117 (4), 1853–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriegman S et al. (2021) Kinematic self-replication in reconfigurable organisms. Proc Natl Acad Sci U S A 118 (49). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumuskaya G et al. (2022) Motile Living Biobots Self-Construct from Adult Human Somatic Progenitor Seed Cells. bioRxiv, 2022.08.04.502707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emmons-Bell M et al. (2019) Regenerative Adaptation to Electrochemical Perturbation in Planaria: A Molecular Analysis of Physiological Plasticity. iScience 22, 147–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baluška F and Levin M (2016) On Having No Head: Cognition throughout Biological Systems. Front Psychol 7, 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watson RA et al. , Associative memory in gene regulation networks, Artificial Life Conference XII, Odense, Denmark, 2010, pp. 194–201. [Google Scholar]
- 25.Biswas S et al. (2021) Gene Regulatory Networks Exhibit Several Kinds of Memory: Quantification of Memory in Biological and Random Transcriptional Networks. iScience 24 (3), 102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas S et al. (2023) Learning in Transcriptional Network Models: Computational Discovery of Pathway-Level Memory and Effective Interventions. International Journal of Molecular Sciences 24 (1), 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freddolino PL et al. (2018) Stochastic tuning of gene expression enables cellular adaptation in the absence of pre-existing regulatory circuitry. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin H (2006) What keeps cells in tissues behaving normally in the face of myriad mutations? BioEssays 28 (5), 515–24. [DOI] [PubMed] [Google Scholar]
- 29.Nagato T et al. (1995) Effect of denervation on morphogenesis of the rat fungiform papilla. Acta Anat (Basel) 153 (4), 301–9. [DOI] [PubMed] [Google Scholar]
- 30.McDowell G et al. (2016) From cytoskeletal dynamics to organ asymmetry: a nonlinear, regulative pathway underlies left-right patterning. Philos Trans R Soc Lond B Biol Sci 371 (1710), 20150409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao J et al. (2020) Epidemiological Realities of Alcoholic Liver Disease: Global Burden, Research Trends, and Therapeutic Promise. Gene Expr 20 (2), 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delgado-Coello B (2021) Liver regeneration observed across the different classes of vertebrates from an evolutionary perspective. Heliyon 7 (3), e06449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalopoulos GK (2017) Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology 65 (4), 1384–1392. [DOI] [PubMed] [Google Scholar]
- 34.Vogel A et al. (2004) Chronic liver disease in murine hereditary tyrosinemia type 1 induces resistance to cell death. Hepatology 39 (2), 433–43. [DOI] [PubMed] [Google Scholar]
- 35.Michalopoulos GK and Bhushan B (2021) Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 18 (1), 40–55. [DOI] [PubMed] [Google Scholar]
- 36.Miyaoka Y and Miyajima A (2013) To divide or not to divide: revisiting liver regeneration. Cell Div 8 (1), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iansante V et al. (2018) Human hepatocyte transplantation for liver disease: current status and future perspectives. Pediatr Res 83 (1-2), 232–240. [DOI] [PubMed] [Google Scholar]
- 38.Rhim JA et al. (1994) Replacement of diseased mouse liver by hepatic cell transplantation. Science 263 (5150), 1149–52. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura K et al. (2007) Animal models of tyrosinemia. J Nutr 137 (6 Suppl 1), 1556S–1560S; discussion 1573S-1575S. [DOI] [PubMed] [Google Scholar]
- 40.Faraj W et al. (2010) Auxiliary liver transplantation for acute liver failure in children. Ann Surg 251 (2), 351–6. [DOI] [PubMed] [Google Scholar]
- 41.McKiernan P (2013) Liver transplantation and cell therapies for inborn errors of metabolism. J Inherit Metab Dis 36 (4), 675–80. [DOI] [PubMed] [Google Scholar]
- 42.Shanmugam NP et al. (2011) Auxiliary liver transplantation: a form of gene therapy in selective metabolic disorders. J Clin Exp Hepatol 1 (2), 118–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rela M et al. (1999) Auxiliary partial orthotopic liver transplantation for Crigler-Najjar syndrome type I. Ann Surg 229 (4), 565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burdelski M and Rogiers X (1999) Liver transplantation in metabolic disorders. Acta Gastroenterol Belg 62 (3), 300–5. [PubMed] [Google Scholar]
- 45.Dokmak S et al. (2013) Auxiliary liver transplantation with a small deceased liver graft for cirrhotic liver complicated by hepatocellular carcinoma. Transpl Int 26 (11), e102–4. [DOI] [PubMed] [Google Scholar]
- 46.Ren W et al. (2012) Integrating repopulation and regeneration of the auxiliarily transplanted small liver graft: the solution for organ shortage and immunosuppression. Med Hypotheses 79 (2), 241–5. [DOI] [PubMed] [Google Scholar]
- 47.Hoppo T et al. (2011) Rescue of lethal hepatic failure by hepatized lymph nodes in mice. Gastroenterology 140 (2), 656–666 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komori J et al. (2012) The mouse lymph node as an ectopic transplantation site for multiple tissues. Nat Biotechnol 30 (10), 976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han B et al. (2022) Fat-associated lymphoid clusters as expandable niches for ectopic liver development. Hepatology 76 (2), 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicolas CT et al. (2020) Ex Vivo Cell Therapy by Ectopic Hepatocyte Transplantation Treats the Porcine Tyrosinemia Model of Acute Liver Failure. Mol Ther Methods Clin Dev 18, 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fontes P et al. (2020) Development of Ectopic Livers by Hepatocyte Transplantation Into Swine Lymph Nodes. Liver Transpl 26 (12), 1629–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuchling F et al. (2020) Morphogenesis as Bayesian inference: A variational approach to pattern formation and control in complex biological systems. Phys Life Rev 33, 88–108. [DOI] [PubMed] [Google Scholar]
- 53.Calvo P and Friston K (2017) Predicting green: really radical (plant) predictive processing. J R Soc Interface 14 (131). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pezzulo G and Levin M (2016) Top-down models in biology: explanation and control of complex living systems above the molecular level. J R Soc Interface 13 (124). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kramer BA et al. (2022) Multimodal perception links cellular state to decision-making in single cells. Science 377 (6606), 642–648. [DOI] [PubMed] [Google Scholar]
- 56.Toettcher JE et al. (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155 (6), 1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fields C. and Levin M (2022) Competency in Navigating Arbitrary Spaces as an Invariant for Analyzing Cognition in Diverse Embodiments. Entropy (Basel) 24 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyle EA et al. (2017) An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169 (7), 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolodkin A. et al. (2012) Understanding complexity in neurodegenerative diseases: in silico reconstruction of emergence. Frontiers in physiology 3, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waliszewski P et al. (1998) On the holistic approach in cellular and cancer biology: nonlinearity, complexity, and quasi-determinism of the dynamic cellular network. J Surg Oncol 68 (2), 70–8. [DOI] [PubMed] [Google Scholar]
- 61.Mathews J and Levin M (2018) The body electric 2.0: recent advances in developmental bioelectricity for regenerative and synthetic bioengineering. Curr Opin Biotechnol 52, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pio-Lopez L. et al. (2022) Active inference, morphogenesis, and computational psychiatry. Front Comput Neurosci 16, 988977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pezzulo G and Levin M (2015) Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs. Integr Biol (Camb) 7 (12), 1487–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koshland DE (1983) The Bacterium as a Model Neuron. Trends in Neurosciences 6 (4), 133–137. [Google Scholar]
- 65.Morimoto BH and Koshland DEJ (1991) Short-term and long-term memory in single cells. Faseb J 5 (7), 2061–7. [DOI] [PubMed] [Google Scholar]
- 66.Yang CY et al. (2020) Encoding Membrane-Potential-Based Memory within a Microbial Community. Cell Syst 10 (5), 417–423 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prindle A et al. (2015) Ion channels enable electrical communication in bacterial communities. Nature 527 (7576), 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathews J and Levin M (2017) Gap junctional signaling in pattern regulation: Physiological network connectivity instructs growth and form. Dev Neurobiol 77 (5), 643–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levin M and Martyniuk CJ (2018) The bioelectric code: An ancient computational medium for dynamic control of growth and form. Biosystems 164, 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fields C. et al. (2020) Morphological Coordination: A Common Ancestral Function Unifying Neural and Non-Neural Signaling. Physiology 35 (1), 16–30. [DOI] [PubMed] [Google Scholar]
- 71.Vandenberg LN et al. (2011) V-ATPase-dependent ectodermal voltage and pH regionalization are required for craniofacial morphogenesis. Dev Dyn 240 (8), 1889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levin M (2021) Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer. Cell 184 (4), 1971–1989. [DOI] [PubMed] [Google Scholar]
- 73.Harris MP (2021) Bioelectric signaling as a unique regulator of development and regeneration. Development 148 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bates E. (2015) Ion Channels in Development and Cancer. Annu Rev Cell Dev Biol 31, 231–47. [DOI] [PubMed] [Google Scholar]
- 75.Zhao S. et al. (2020) Biomedical applications of electrical stimulation. Cell Mol Life Sci 77 (14), 2681–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reid B and Zhao M (2014) The Electrical Response to Injury: Molecular Mechanisms and Wound Healing. Adv Wound Care (New Rochelle) 3 (2), 184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao M. et al. (2012) Electrical signaling in control of ocular cell behaviors. Progress in retinal and eye research 31 (1), 65–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tseng A and Levin M (2013) Cracking the bioelectric code: Probing endogenous ionic controls of pattern formation. Communicative & Integrative Biology 6 (1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathews J. et al. (2022) Ion Channel Drugs Suppress Cancer Phenotype in NG108-15 and U87 Cells: Toward Novel Electroceuticals for Glioblastoma. Cancers (Basel) 14 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Churchill CDM et al. (2018) EDEn – Electroceutical Design Environment: An Ion Channel Database with Small Molecule Modulators and Tissue Expression Information. iScience 11, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pai VP and Levin M (2022) HCN2 channel-induced rescue of brain, eye, heart and gut teratogenesis caused by nicotine, ethanol and aberrant notch signalling. Wound Repair Regen. [DOI] [PubMed] [Google Scholar]
- 82.Pai VP et al. (2018) HCN2 Rescues brain defects by enforcing endogenous voltage pre-patterns. Nature Communications 9 (1), 998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pai VP et al. (2015) Endogenous Gradients of Resting Potential Instructively Pattern Embryonic Neural Tissue via Notch Signaling and Regulation of Proliferation. The Journal of Neuroscience 35 (10), 4366–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chernet BT and Levin M (2013) Transmembrane voltage potential is an essential cellular parameter for the detection and control of tumor development in a Xenopus model. Disease models & mechanisms 6 (3), 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chernet BT et al. (2016) Use of genetically encoded, light-gated ion translocators to control tumorigenesis. Oncotarget 7 (15), 19575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chernet BT and Levin M (2014) Transmembrane voltage potential of somatic cells controls oncogene-mediated tumorigenesis at long-range. Oncotarget 5 (10), 3287–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chernet BT and Levin M (2013) Endogenous Voltage Potentials and the Microenvironment: Bioelectric Signals that Reveal, Induce and Normalize Cancer. Journal of clinical & experimental oncology Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levin M (2021) Bioelectrical approaches to cancer as a problem of the scaling of the cellular self. Prog Biophys Mol Biol 165, 102–113. [DOI] [PubMed] [Google Scholar]
- 89.Goel P and Mehta A (2013) Learning theories reveal loss of pancreatic electrical connectivity in diabetes as an adaptive response. PLoS One 8 (8), e70366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tseng AS et al. (2010) Induction of vertebrate regeneration by a transient sodium current. J Neurosci 30 (39), 13192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oviedo NJ and Beane WS (2009) Regeneration: The origin of cancer or a possible cure? Semin Cell Dev Biol 20 (5), 557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahu S. et al. (2017) Secrets from immortal worms: What can we learn about biological ageing from the planarian model system? Semin Cell Dev Biol 70, 108–121. [DOI] [PubMed] [Google Scholar]
- 93.Shreesha L and Levin M (2023) Cellular Competency during Development Alters Evolutionary Dynamics in an Artificial Embryogeny Model. Entropy 25 (1), 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tlsty TD and Hein PW (2001) Know thy neighbor: stromal cells can contribute oncogenic signals. Curr Opin Genet Dev 11 (1), 54–9. [DOI] [PubMed] [Google Scholar]
- 95.Maffini MV et al. (2005) Stromal regulation of neoplastic development: age-dependent normalization of neoplastic mammary cells by mammary stroma. The American journal of pathology 167 (5), 1405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Telerman A et al. (2010) Tumor reversion holds promise. Oncotarget 1 (4), 233–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blackiston DJ and Levin M (2013) Ectopic eyes outside the head in Xenopus tadpoles provide sensory data for light-mediated learning. The Journal of experimental biology 216 (Pt 6), 1031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blackiston DJ et al. (2017) Serotonergic stimulation induces nerve growth and promotes visual learning via posterior eye grafts in a vertebrate model of induced sensory plasticity. npj Regenerative Medicine 2 (1), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murugan NJ et al. (2022) Acute multidrug delivery via a wearable bioreactor facilitates long-term limb regeneration and functional recovery in adult Xenopus laevis. Sci Adv 8 (4), eabj2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ozugur S. et al. (2022) Transcardial injection and vascular distribution of microalgae in Xenopus laevis as means to supply the brain with photosynthetic oxygen. STAR Protoc 3 (2), 101250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ozugur S. et al. (2021) Green oxygen power plants in the brain rescue neuronal activity. iScience 24 (10), 103158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Magisetty R and Park SM (2022) New Era of Electroceuticals: Clinically Driven Smart Implantable Electronic Devices Moving towards Precision Therapy. Micromachines (Basel) 13 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan TH et al. (2022) Odd dynamics of living chiral crystals. Nature 607 (7918), 287–293. [DOI] [PubMed] [Google Scholar]
- 104.Chao ZC et al. (2008) Shaping embodied neural networks for adaptive goal-directed behavior. PLoS computational biology 4 (3), e1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kagan BJ et al. (2022) In vitro neurons learn and exhibit sentience when embodied in a simulated game-world. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehrali M et al. (2018) Blending Electronics with the Human Body: A Pathway toward a Cybernetic Future. Adv Sci (Weinh) 5 (10), 1700931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Staufer O et al. (2016) Functional fusion of living systems with synthetic electrode interfaces. Beilstein J Nanotechnol 7, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levin M et al. (2020) Applications and ethics of computer-designed organisms. Nat Rev Mol Cell Biol 21 (11), 655–656. [DOI] [PubMed] [Google Scholar]
- 109.Heyd D (2012) Is there anything unique in the ethics of synthetic biology? Perspect Biol Med 55 (4), 581–9. [DOI] [PubMed] [Google Scholar]
- 110.Evers AWM et al. (2018) Implications of Placebo and Nocebo Effects for Clinical Practice: Expert Consensus. Psychother Psychosom 87 (4), 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Piedimonte A and Benedetti F (2016) Words and Drugs: Same Mechanisms of Action? Journal of Contemporary Psychotherapy 46 (3), 159–166. [Google Scholar]
- 112.Lui F et al. (2010) Neural bases of conditioned placebo analgesia. Pain 151 (3), 816–824. [DOI] [PubMed] [Google Scholar]
- 113.Benedetti F et al. (2007) When words are painful: unraveling the mechanisms of the nocebo effect. Neuroscience 147 (2), 260–71. [DOI] [PubMed] [Google Scholar]
- 114.Saatcioglu F (2013) Regulation of gene expression by yoga, meditation and related practices: a review of recent studies. Asian J Psychiatr 6 (1), 74–7. [DOI] [PubMed] [Google Scholar]
- 115.Agnati LF et al. (2012) Aspects on the integrative actions of the brain from neural networks to "brain-body medicine". Journal of receptor and signal transduction research 32 (4), 163–80. [DOI] [PubMed] [Google Scholar]
- 116.Taylor AG et al. (2010) Top-down and bottom-up mechanisms in mind-body medicine: development of an integrative framework for psychophysiological research. Explore 6 (1), 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lu HY et al. (2021) Multi-scale neural decoding and analysis. J Neural Eng 18 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Betzel RF and Bassett DS (2017) Multi-scale brain networks. Neuroimage 160, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gershman SJ et al. (2021) Reconsidering the evidence for learning in single cells. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nilsonne G et al. (2011) Learning in a simple biological system: a pilot study of classical conditioning of human macrophages in vitro. Behav Brain Funct 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zoghi M (2004) Cardiac memory: do the heart and the brain remember the same? J Interv Card Electrophysiol 11 (3), 177–82. [DOI] [PubMed] [Google Scholar]
- 122.Rogers MP et al. (1983) Conditioned immunosuppression? Am J Psychiatry 140 (8), 1110–1. [DOI] [PubMed] [Google Scholar]
- 123.Rogers MP et al. (1979) The influence of the psyche and the brain on immunity and disease susceptibility: a critical review. Psychosom Med 41 (2), 147–64. [DOI] [PubMed] [Google Scholar]
- 124.Rogers MP et al. (1976) Behaviorally conditioned immunosuppression: replication of a recent study. Psychosom Med 38 (6), 447–51. [DOI] [PubMed] [Google Scholar]
- 125.Miller NE (1978) Biofeedback and visceral learning. Annu Rev Psychol 29, 373–404. [DOI] [PubMed] [Google Scholar]
- 126.Ongaro G and Kaptchuk TJ (2019) Symptom perception, placebo effects, and the Bayesian brain. Pain 160 (1), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beauregard M and O'Leary D (2008) Believing can make it so: the neuroscience of the placebo effect. Adv Mind Body Med 23 (1), 14–8. [PubMed] [Google Scholar]
- 128.Mason AA (1952) A case of congenital ichthyosiform erythrodermia of Brocq treated by hypnosis. Br Med J 2 (4781), 422–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mathews J et al. (2022) Cellular Signaling Pathways as Plastic, Proto-cognitive Systems: Implications for Biomedicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Busse SM et al. (2018) Cross-limb communication during Xenopus hindlimb regenerative response: non-local bioelectric injury signals. Development 145 (19). [DOI] [PubMed] [Google Scholar]
- 131.Friston K (2010) The free-energy principle: a unified brain theory? Nat Rev Neurosci 11 (2), 127–38. [DOI] [PubMed] [Google Scholar]
- 132.Badcock PB et al. (2019) The hierarchically mechanistic mind: A free-energy formulation of the human psyche. Phys Life Rev 31, 104–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ramstead MJD et al. (2019) Variational ecology and the physics of sentient systems. Phys Life Rev 31, 188–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Adams RA et al. (2016) Computational Psychiatry: towards a mathematically informed understanding of mental illness. J Neurol Neurosurg Psychiatry 87 (1), 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rubin H. (2007) Ordered heterogeneity and its decline in cancer and aging. Advances in cancer research 98, 117–47. [DOI] [PubMed] [Google Scholar]
- 136.Rubin H (1992) Mechanisms for enduring biological change. Am J Physiol 262 (1 Pt 1), L111–3. [DOI] [PubMed] [Google Scholar]
- 137.Rubin H (1990) On the nature of enduring modifications induced in cells and organisms. Am J Physiol 258 (2 Pt 1), L19–24. [DOI] [PubMed] [Google Scholar]
- 138.Mathews J et al. (2023) Cellular signaling pathways as plastic, proto-cognitive systems: Implications for biomedicine. Patterns (N Y) 4 (5), 100737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Williams K et al. (2020) Regulation of axial and head patterning during planarian regeneration by a commensal bacterium. Mech Dev 163, 103614. [DOI] [PubMed] [Google Scholar]
- 140.Eberhard W et al. (2014) Zombie bugs? The fungus Purpureocillium cf. lilacinum may manipulate the behavior of its host bug Edessa rufomarginata. Mycologia 106 (6), 1065–72. [DOI] [PubMed] [Google Scholar]
- 141.Elya C. et al. (2018) Robust manipulation of the behavior of Drosophila melanogaster by a fungal pathogen in the laboratory. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loreto RG and Hughes DP (2019) The metabolic alteration and apparent preservation of the zombie ant brain. J Insect Physiol 118, 103918. [DOI] [PubMed] [Google Scholar]
- 143.Davies J and Levin M (2022) Synthetic morphology via active and agential matter. Nature Bioengineering. [Google Scholar]
- 144.Abramson CI and Levin M (2021) Behaviorist approaches to investigating memory and learning: A primer for synthetic biology and bioengineering. Commun Integr Biol 14 (1), 230–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Strassmann JE and Queller DC (2010) The social organism: congresses, parties, and committees. Evolution 64 (3), 605–16. [DOI] [PubMed] [Google Scholar]
- 146.Rotenberg MY and Tian BZ (2018) Talking to Cells: Semiconductor Nanomaterials at the Cellular Interface. Advanced Biosystems 2 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Belwafi K et al. (2021) Embedded Brain Computer Interface: State-of-the-Art in Research. Sensors (Basel) 21 (13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krist KT et al. (2021) A simple theory for molecular chemotaxis driven by specific binding interactions. J Chem Phys 155 (16), 164902. [DOI] [PubMed] [Google Scholar]
- 149.McGregor S. et al. (2012) Evolution of associative learning in chemical networks. PLoS Comput Biol 8 (11), e1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Biswas S. et al. (2022) Learning in Transcriptional Network Models: Computational Discovery of Pathway-Level Memory and Effective Interventions. Int J Mol Sci 24 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Craddock TJ et al. (2012) Cytoskeletal signaling: is memory encoded in microtubule lattices by CaMKII phosphorylation? PLoS computational biology 8 (3), e1002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Metzcar J et al. (2022) A novel model of multicellular communication through extracellular matrix microstructure. bioRxiv, 2022.11.21.514608. [Google Scholar]
- 153.Sarris M and Sixt M (2015) Navigating in tissue mazes: chemoattractant interpretation in complex environments. Curr Opin Cell Biol 36, 93–102. [DOI] [PubMed] [Google Scholar]
- 154.Little GE et al. (2009) Specificity and plasticity of thalamocortical connections in Sema6A mutant mice. PLoS Biol 7 (4), e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.McEwen BS (1998) Stress, adaptation, and disease. Allostasis and allostatic load. Ann N Y Acad Sci 840, 33–44. [DOI] [PubMed] [Google Scholar]
- 156.Zimmer C et al. (2022) Information theory in vertebrate stress physiology. Trends Endocrinol Metab 33 (1), 8–17. [DOI] [PubMed] [Google Scholar]
- 157.Tschantz A et al. (2022) Simulating homeostatic, allostatic and goal-directed forms of interoceptive control using active inference. Biol Psychol 169, 108266. [DOI] [PubMed] [Google Scholar]
- 158.Deans C (2021) Biological Prescience: The Role of Anticipation in Organismal Processes. Front Physiol 12, 672457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Colditz IG (2020) A consideration of physiological regulation from the perspective of Bayesian enactivism. Physiol Behav 214, 112758. [DOI] [PubMed] [Google Scholar]
- 160.Schulkin J and Sterling P (2019) Allostasis: A Brain-Centered, Predictive Mode of Physiological Regulation. Trends Neurosci 42 (10), 740–752. [DOI] [PubMed] [Google Scholar]
- 161.Oviedo NJ et al. (2003) Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn 226 (2), 326–33. [DOI] [PubMed] [Google Scholar]
- 162.Cooke J (1981) Scale of body pattern adjusts to available cell number in amphibian embryos. Nature 290 (5809), 775–8. [DOI] [PubMed] [Google Scholar]
- 163.Levin M et al. (2018) Planarian regeneration as a model of anatomical homeostasis: Recent progress in biophysical and computational approaches. Semin Cell Dev Biol 87, 125–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fankhauser G (1945) Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. Journal of Experimental Zoology 100 (3), 445–455. [DOI] [PubMed] [Google Scholar]
- 165.Farinella-Ferruzza N (1956) The transformation of a tail into a limb after xenoplastic transformation. Experientia 15, 304–305. [Google Scholar]
- 166.Pilling OA et al. (2017) Insights into transgenerational epigenetics from studies of ciliates. Eur J Protistol 61 (Pt B), 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fields C and Levin M (2017) Multiscale memory and bioelectric error correction in the cytoplasm–cytoskeleton-membrane system. Wiley Interdisciplinary Reviews: Systems Biology and Medicine 10 (2), e1410-n/a. [DOI] [PubMed] [Google Scholar]
- 168.Oviedo NJ et al. (2010) Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev Biol 339 (1), 188–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vandenberg LN et al. (2012) Normalized shape and location of perturbed craniofacial structures in the Xenopus tadpole reveal an innate ability to achieve correct morphology. Developmental Dynamics 241 (5), 863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Slijper EJ (1942) Biologic anatomical investigations on the bipedal gait and upright posture in mammals - With special reference to a little goat born without forelegs II. Proceedings of the Koninklijke Nederlandse Akademie Van Wetenschappen 45 (1/5), 407–415. [Google Scholar]
- 171.Kozo-Polyansky BM (1924) Symbiogenesis: A New Principle of Evolution, Harvard University Press. [Google Scholar]
- 172.Noble D (2022) Modern physiology vindicates Darwin's dream. Exp Physiol 107 (9), 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Levin M. (2023) Darwin's agential materials: evolutionary implications of multiscale competency in developmental biology. Cell Mol Life Sci 80 (6), 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Levin M et al. (2017) Endogenous Bioelectric Signaling Networks: Exploiting Voltage Gradients for Control of Growth and Form. Annu Rev Biomed Eng 19, 353–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Levin M (2023) Bioelectric networks: the cognitive glue enabling evolutionary scaling from physiology to mind. Anim Cogn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pio-Lopez L and Levin M (2023) Morphoceuticals: Perspectives for discovery of drugs targeting anatomical control mechanisms in regenerative medicine, cancer and aging. Drug Discov Today 28 (6), 103585. [DOI] [PubMed] [Google Scholar]
- 177.Pai VP et al. (2012) Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development 139 (2), 313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]