Abstract
Aim:
Quantify identifiable sources of variability, including key pharmacogenetic variants in oxypurinol pharmacokinetics and their pharmacodynamic effect on serum urate.
Methods:
Hmong participants (n=34) received 100 mg allopurinol twice daily for 7 days followed by 150 mg allopurinol twice daily for 7 days. A sequential population pharmacokinetic pharmacodynamics (PKPD) analysis with non-linear mixed-effects modeling was performed. Allopurinol maintenance dose to achieve target SU was simulated based on the final PKPD model.
Results:
A one-compartment model with first order absorption and elimination best described the oxypurinol concentration-time data. Inhibitory of SU by oxypurinol was described with a direct inhibitory Emax model using steady-state oxypurinol concentrations. Fat-free body mass, estimated creatinine clearance and SLC22A12 rs505802 genotype (0.32 per T allele, 95%CI 0.13, 0.55) were found to predict differences in oxypurinol clearance. Oxypurinol concentration required to inhibit 50% of xanthine dehydrogenase activity was affected by PDZK1 rs12129861 genotype (−0.27 per A allele, 95%CI −0.38, −0.13). Most individuals with both PDZK1 rs12129861 AA and SLC22A12 rs505802 CC genotypes achieve target SU (with at least 75% success rate) with allopurinol below the maximum dose, regardless of renal function and body mass. In contrast, individuals with both PDZK1 rs12129861 GG and SLC22A12 rs505802 TT genotypes would require more than the maximum dose, thus selecting alternative medications.
Conclusion:
The proposed allopurinol dosing guide uses individuals’ fat-free mass, renal function, and SLC22A12 rs505802 and PDZK1 rs12129861 genotypes to achieve target SU.
Keywords: allopurinol, gout, population pharmacokinetics, NONMEM, pharmacometrics
INTRODUCTION
Allopurinol is the first-line urate-lowering therapy (ULT) to prevent gout by lowering serum urate (SU) to a target of 6 mg/dL in all patients who can tolerate the medication.1 Despite the availability of other agents with similar and newer mechanism of actions, allopurinol remains the most widely used agent to manage chronic hyperuricemia and gout worldwide.2 The treat-to-target SU level approach instead of a fixed dose ULT strategy has been recommended by American College of Rheumatology and other organizations.3–5 This approach is supported by an open-label, randomized controlled trial demonstrating that dose escalation resulted in 69% of patients with gout achieved target SU6 comparing to only 20–50% of patients achieve target SU with fixed allopurinol dose.7 Patients who typically fail to achieve SU targets include those who have high SU (>9 mg/dL), moderate-to-severe chronic kidney disease (stage ≥3), or urolithiasis. Patients with aforementioned conditions have a greater risk for gout flares and tophi formation.8,9 Additionally, hyperuricemia (defined as SU ≥6.8 mg/dL) is strongly associated with other chronic conditions, including hypertension,10,11 type 2 diabetes mellitus,12 metabolic syndrome,13 cardiovascular diseases14 and dyslipidemia with elevated low-density lipoprotein cholesterol and hypertriglyceridemia.15
To optimize allopurinol use, several strategies have been proposed. One approach projects an allopurinol maintenance dose based on creatinine clearance (CrCL).16 However, this approach was developed with the specific goal to avoid the allopurinol-induced severe cutaneous adverse reaction (SCAR) and not to achieve target SU. This approach may be sensible because impaired renal function correlated with the development and poor prognosis of allopurinol induced SCAR.17–19 Given this CrCL-based dose approach, it is understandable that only 19% of patients achieved target SU.20 Starting allopurinol dose based on estimated glomerular filtrate rate (eGFR) has been proposed.21 Similarly, the goal was to prevent allopurinol-induced SCAR, with the authors asserting that the starting dose, not the maintenance dose, correlated with the incidence of allopurinol-induced SCAR. Stamp et al21 reported that dose titration is often required to achieve target SU in patients who tolerate allopurinol. An approach that encourages safe targeting of optimal allopurinol dosage to achieve target SU remains elusive. This situation creates a gap in tools that specifically address the goal of dose optimization with the intended purpose of mitigating acute and chronic complications associated with hyperuricemia and gout.
Genome-wide association studies (GWAS) provide insights on how single nucleotide polymorphisms (SNPs) in key transporter genes can impact treatment outcomes. The ABCG2 (BCRP) rs2231142C>A is associated with SU-lowering response to allopurinol22–25 and has been suggested as a guide to improve drug dosage and/or selection by identifying patients in need of alternate therapeutic approaches.26 The SLC22A12 (URAT1) rs505802C>T is not only associated with the risk of hyperuricemia,27 but also importantly associated with the exposure of serum oxypurinol, the active metabolite of allopurinol.28 These two transporters, BRCP and URAT1, may prove to be important when identifying genomic based sources of variability in response to allopurinol.
Several population pharmacokinetics (PK)29–31 and pharmacokinetic-pharmacodynamic (PKPD)32–35 models have been developed. Despite these models identifying that body mass, renal function, and concomitant medications, including diuretics and uricosurics, are important factors, none of the studies illustrated a strong association between SNPs and either PK or PD parameters for oxypurinol. Majority of the aforementioned studies investigated the impact of rs2231142C>A (Q141K) which is sensible due to such missense variants of ABCG2 could decrease oxypurinol renal excretion and thereby lead to higher serum oxypurinol and greater SU-lowering effect22 and multiple observational studies have also suggested a strong association between this variant and SU-lowering response to allopurinol.22–25 Furthermore, most of the studied populations are of European descent. It is, however, plausible that other genetic variants may play important roles in modulating PKPD of allopurinol in populations with different ethnic background.
The aims of this project were to (1) develop a population PKPD model to characterize the relationship between serum oxypurinol and SU, (2) quantify the effects of relevant clinical characteristics and SNPs identified from GWAS on the PKPD effects for oxypurinol, and (3) predict the allopurinol maintenance dose to achieve target SU of <6 mg/dL.
METHODS
Patients and study design
Data from a prospective, open-labeled, genetically-guided, pilot study, Genetics Of HyperUricemia and Gout Therapy in Hmong (GOUT-H) (clinicaltrial.gov, NCT02371421) were analyzed. This study was approved by the Human Research Protection Program at the University of Minnesota Institutional Review Board (IRB #1408M53223). Detailed study design was described in previous publication.28 Briefly, 34 Hmong participants with gout and/or hyperuricemia were screened at the screening visit and enrolled in the study based on the eligibility. After 7 days of allopurinol or febuxostat washout (baseline visit), all the participants took allopurinol 100 mg twice daily for 7 days followed by 150 mg twice daily for 7 days. At the follow-up visit (2 weeks after the baseline visit), participants took the final dose of allopurinol.
Blood samples were collected at the screening, baseline, and follow-up visits to measure SU and serum creatinine after overnight fasting for 10 hours. Additionally, blood and urine samples were collected at 0, 2, 4, and 6-hours post-allopurinol dose at the follow-up visit to measure oxypurinol concentrations.
Oxypurinol and urate assay
Urate concentrations were measured using a Roche COBAS 6000 chemistry analyzer (Roche Diagnostics, Indianapolis, IN) using the enzymatic method with a limit detection of 0.2 mg/dL. The inter-assay coefficient of variation was 1.3% at 5.50 mg/dL and 2.0% at 9.67 mg/dL. Oxypurinol concentrations were measured as described in previous publication.28 None of the SU, serum oxypurinol, and urine oxypurinol concentrations were below the limit of quantification.
Pharmacogenetic testing
Genomic DNA was purified and extracted from saliva samples collected using ORAgene DISCOVER kits (OGR-500, DNA Genotek Inc., Ottawa, ON, Canada) with QIAamp DNA Kit (Qiagen Inc., Germantown, MD, USA) per the manufacturer’s protocol. Nine SNPs were genotyped using the iPLEX Gold method (iPLEX Application, Agena, San Diego, CA, USA). Functionality of each gene and supporting evidence for inclusion were described previously22,27,28,36–38 and are summarized in Supplementary Table S1.
Population PK model
The population analysis for oxypurinol PK was conducted using the nonlinear mixed effects modeling program, NONMEM version 7.4 (ICON Development Solutions, LC, Ellicott City, MD) with the first order conditional estimation method with interaction. Exploratory analyses and diagnostic plots were performed with R and Perl-speaks-NONMEM (PsN) version 5.2.6.39 One and two compartmental PK models with linear elimination and first-order absorption models with and without a time lag were explored. Model derived values of the combined absorption and formation rate constant (Kfm), apparent clearance (CL/fm) and apparent volume (V/fm) for oxypurinol were estimated, where fm represents the fraction of allopurinol dose available as oxypurinol systemically.
The between subject variability (BSV) was assumed to follow a log-normal distribution, described as follow:
where is the model parameter for the individual; is the population mean of the model parameter ; and is a random variable that represents the deviation from the mean of the parameter for the individual; the collection of are assumed to have a mean of zero and variance . The variance of BSV was calculated as a percentage of coefficient of variation (%CV) using the following equation:
The residual unexplained error including additive, proportional, and combined errors were tested.
Population PKPD model
After the final PK model was established, the PKPD model was analyzed with a sequential approach using individual pharmacokinetic parameters with the standard error (IPPSE) method.40,41 Steady-state oxypurinol concentration was linked to the PD model using a direct effect Emax model was tested, using the following equation:
where oxypurinolss is the serum oxypurinol concentration at steady-state; BLurate is the baseline SU; Imax is the maximum inhibitory effect of oxypurinol on xanthine dehydrogenase to inhibit urate production; IC50 is the oxypurinol concentration required to inhibit 50% of the activity of xanthine dehydrogenase; γ is the Hill coefficient for the sigmoid Emax model. The PKPD structural model is depicted in Figure 1.
Figure 1.
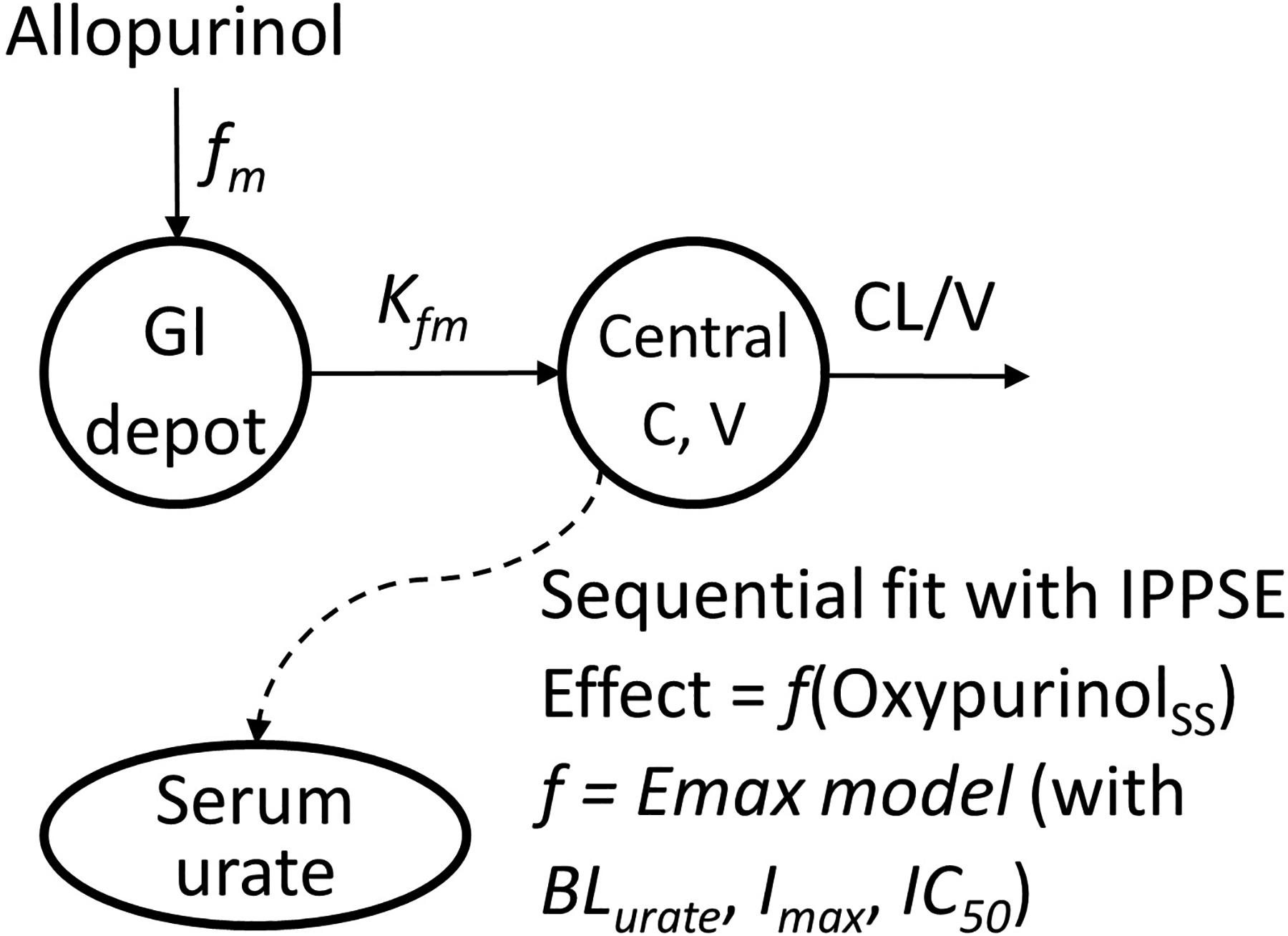
The structural model of the pharmacokinetics and pharmacodynamics effect of allopurinol
BLurate, baseline serum urate; C, serum oxypurinol concentration; CL, apparent oxypurinol clearance; fm, fraction of the allopurinol dose systemically converts to oxypurinol; Kfm, combined absorption and formation rate constant; Imax, maximum inhibitory effect of oxypurinol on xanthine dehydrogenase to inhibit urate production; IC50, oxypurinol concentration required to inhibit 50% of the activity of xanthine dehydrogenase; IPPSE, individual pharmacokinetic parameters with standard error; V, apparent oxypurinol volume of distribution
Covariate model development
Demographics, clinical factors, concomitant medications, and genetic variants were evaluated for their influence on the parameters of PK and PD models. The selection of covariates for testing was based on previous significant findings29,30,33,35,42 and biological plausibility.
Demographic covariates included gender, total body weight (TBW), adjusted body weight (AJBW), and fat-free mass (FFM).43 Renal function was tested as standardized CrCL, estimated from the Cockcroft–Gault equation then normalized to a standard CrCL of a 70 kg human (calculated as observed CrCL*70/ideal body weight) to decorrelate the weight effect on CrCL. Concomitant medications were tested based on participants’ self-reported information. These included drugs that lower SU: losartan,44 HMG-CoA reductase inhibitors (particularly, atorvastatin),45,46 and calcium channel blockers47; and drugs that increase SU: angiotensin converting enzyme inhibitors, angiotensin receptor blockers (but not including losartan), beta-blockers, diuretics, and non-steroidal anti-inflammatory drugs (NSAIDs).47 In addition to testing the effect of each medication type, two categories were also tested: drugs that lower SU and drugs that increase SU.
Nine SNPs related to SU levels or risks of gout development (Supplementary Table S1) were tested. An additive genetic model was assumed for the effect of SNPs on the PKPD parameters.
A stepwise covariant modeling (SCM) approach using the PsN toolkit with the forward and backward thresholds at p < 0.05 and p < 0.01, respectively was used for selecting covariates that contributed to the CL/fm and V/fm for the PK model, and BLurate, Imax, and IC50 for the PD model. The significance of inclusion and elimination of each covariate was tested based on likelihood ratio test that follows the χ2 distribution.
Model selection and qualification
Model selection was dependent on several criteria, including the χ2 (likelihood ratio) test, goodness of fit (GOF) plots. Visual predictive check (VPC) plots (1000 simulations) stratified for significant covariates was used for model qualification. Sampling importance resampling (SIR) procedure48 with five iterations with 1,000, 1,000, 1,000, 2,000, and 2,000 samples (M) and 200, 400, 500, 1,000, and 1,000 resamples (m) were performed to assess precision of the final parameter estimates using PsN. Model development, diagnostics, and graphing were using functions within PsN, Pirana49 and R software (version 4.1.0)50.
Simulations to predict allopurinol maintenance dose
Simulation was performed to examine the impact of important covariates on the serum oxypurinol and urate concentration. Different dosing strategies under a combination of significant covariates in the final PKPD model to achieve target SU of < 6mg/dL were performed for 1000 simulations for a total of 91,584 virtual patients. The distribution of PKPD model parameters were based on the final PKPD model. The model identified maintenance dose was the lowest dose that could achieve the target SU <6mg/dL in at least 75% of the cases. Simulation considerations were based on previous publication33 with a few exceptions. First, the maintenance dose of allopurinol was considered from 50 to 800 mg/day because a maximum of 800 mg/day was approved by the US FDA. Second, creatinine clearance was simulated between 15 to 120 mL/min in 1 mL/min increment then stratified into 15–30 mL/min, 30–60 mL/min, and ≥ 60 mL/min categories. Third, FFM between 50 to 100 kg with 10 kg increment was considered. The impact of SLC22A12 rs505802 CC, CT, TT genotypes on oxypurinol CL/fm and the impact of PDZK1 rs12129861 GG, GA, and AA genotypes on IC50 were considered (see Result section for the rationale for the selection of these covariates). Simulations were performed using R software (version 4.1.0)50.
Nomenclature of Targets and Ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/2051.
RESULTS
Participant characteristics
The 34 participants’ demographics characteristics, clinical features, and self-reported concomitant medications collected at the baseline visit are described in Table 1. Notably, there were only 3 women, and only 1 participant with normal weight, based on the World Health Organization’s Asian criteria-based body-mass index (BMI).52 The dataset included 136 serum oxypurinol, 87 urine oxypurinol, and 102 serum urate concentrations. No genotype information was missing for the 9 SNPs tested. Genotype information and distributions are presented in Supplemental Table 1.
Table 1.
GOUT-H participants characteristics
| Characteristicsa | N = 34b |
|---|---|
| Age (years)b | 43 ± 13 (24–68) |
| Gender, malec | 31 (91%) |
| Height (cm)b | 160 ± 7 (146–179) |
| Weight (kg)b | 84 ± 17 (54–134) |
| BMI (kg/m2)b,d | 32.5 ± 5.5 (21.6–47.0) |
| Normalc | 1 (2.9%) |
| Overweightc,d | 4 (12%) |
| Obesityc, | 29 (85%) |
| Estimated CrCL (mL/min)b,e, | 87 ± 31 (25–165) |
| 15 ≤ Estimated CrCL < 30 | 1 (3%) |
| 30 ≤ Estimated CrCL < 60 | 8 (24%) |
| Estimated CrCL ≥ 60 | 25 (74%) |
| Baseline serum urate (mg/dL)b | 9.61 ± 1.67 (5.8–13.0) |
| Post-treatment serum urate (mg/dL)b | 5.4 ± 1.1 (3.1 – 7) |
| Steady-state serum oxypurinol0hr (mg/L)f | 10.6 [7.8, 16.3] (4.3–30.4) |
| Steady-state serum oxypurinol6hr (mg/L)f | 12.6 [9.4, 18.1] (6.4–28.4) |
| Self-reported medications related to SU/gout c,g | |
| Drugs that lower serum urate | 10 (29%) |
| Losartan | 1 (2.9%) |
| HMG-CoA inhibitors | 5 (15%) |
| Calcium channel blockers | 5 (15%) |
| Drugs that increase serum urate | 22 (65%) |
| Angiotensin converting enzyme inhibitors | 5 (15%) |
| Angiotensin receptor blockers (not losartan) | 1 (2.9%) |
| Beta-blockers | 6 (18%) |
| Diuretics | 4 (12%) |
| Non-steroidal anti-inflammatory drugs | 16 (47%) |
BMI, body mass index; CrCL, creatinine clearance; SU, serum urate
Characteristics were assessed at the baseline study visit after 10 days washout period.
Mean ± standard deviation (range)
n (%)
Overweight was defined as BMI 23.0–27.5 kg/m2; obesity was defined as BMI > 27.5 kg/m2 based on World Health Organization Asian criteria-based BMI52.
Estimated CrCL was calculated using Cockcroft-Gault Equation with adjusted body weight.
Median [interquartile range] (range)
Only medications that may impact serum urate are listed.
Final PK model
A one compartment PK model with first order absorption/conversion and elimination with proportional residual error model provided the best fit to the observed serum oxypurinol-time data. Using a two-compartment PK model or other residual error models provided similar fits, so the simpler model was retained. Covariance between BSV for CL/fm and V/fm was tested but this resulted in similar BSV estimates; therefore, the covariance was not included.
Model development steps for the oxypurinol PK model are summarized in Supplemental Table 2. The final model included FFM on CL/fm and V/fm allometric scaled using the theoretical value (0.75 for CL/fm and 1 for V/fm), renal function using estimated CrCL, and SLC22A12 rs505802C>T. Using TBW as a covariate on CL/fm improved the model fit but failed to improve the fit when used as covariate on V/fm. On the other hand, using either AJBW or FFM as a covariate on both CL/fm and V/fm improved the fit. The selection of FFM as a covariate instead of AJBW was based on previous findings that FFM was also found to be a significant covariate.30,33 HMG-CoA reductase inhibitors and drugs that decrease SU reduced CL/fm by about 48% and 30%, respectively, but the effect was not statistically significant in the SCM step (Supplemental Table 2). In addition to SLC22A12 rs505802C>T as a covariate on CL/fm, CARMIL1 rs742132A>G and PDZK1 rs12129861G>A were found to be significant during the forward selection step but were excluded during backward elimination step. The combined absorption and formation rate constant (Kfm) and its BSV were fixed to initial estimates (which is similar to the value, 0.92 h−1 reported in the literature53) due to insufficient data to support the parameter estimates and high shrinkage.54
The BSV in CL/fm decreased from 42.8% to 28.3%, and V/fm decreased from 40.7% to 32.4% after including significant covariates. The results of the base and final (including covariate) PK models are summarized in Table 2, and the final estimates for CL/fm and V/fm for oxypurinol are given by:
Table 2.
Parameter estimates for the base (without covariates) and final population pharmacokinetic/pharmacodynamic models using sequential fit
| Parameter | Base model | Final model | SIR, median (95%CI) |
|---|---|---|---|
| Fixed parameters | |||
| CL/fm (L/h) | 1 | 1.05 | 1.05 (0.92, 1.22) |
| V/fm (L) | 47.7 | 59.3 | 58.8 (50.8, 70.7) |
| Kfm (/h) | 1.1 (fixed) | 1.1 (fixed) | 1.1 (fixed) |
| BLurate (mg/dL) | 9.3 | 9.0 | 9.0 (8.5, 9.5) |
| Imax (mg/dL) | 6.1 | 7.6 | 7.7 (5.3, 11.3) |
| IC50 (mg/L) | 8.0 | 17.6 | 18.2 (7.5, 32.8) |
| Effects of covariates on CL/f m | |||
| Standardized creatinine clearance (power) | - | 0.45 | 0.45 (0.18, 0.73) |
| SLC22A12 rs505802 T allelea | - | 0.32 | 0.32 (0.13, 0.55) |
| Effects of covariates on SU | |||
| Standardized creatinine clearance (power) on baseline SU | - | −0.18 | −0.18 (−0.33, −0.036) |
| PDZK1 rs12129861 A allele on IC50a | - | −0.27 | −0.27 (−0.38, −0.13) |
| Random effect parameters, CV% (RSE%) [shrinkage] | |||
| BSV CL/fm | 42.8 [0%] | 28.3 [0%] | 28.3 (22.6, 34.5) |
| BSV V/fm | 40.7 [25%] | 32.4 [30%] | 31.6 (19.5, 43.0) |
| BSV Kfm | 27.9 (fixed) | 27.9 (fixed) | 27.9 (fixed) |
| BSV BLurate | 11.1 [11%] | 13.7 [6] | 13.8 (10.1, 17.7) |
| BSV Imax | 32.4 (fixed) | 32.4 (fixed) | 32.4 (fixed) |
| BSV IC50 | 71.8 (fixed) | 71.8 (fixed) | 71.8 (fixed) |
| Residual error | |||
| Serum oxypurinol, proportional (CV%) [shrinkage] | 5.2 [23.2%] | 5.2 [21.3%] | 5.4 (4.5, 6.0) |
| Serum urate, additive (mg/dL) [shrinkage] | 0.90 [16%] | 0.69 [18%] | 0.69 (0.49, 1.03) |
BLurate, baseline serum urate; fm, fraction of the allopurinol systemically available as oxypurinol; CL/fm, apparent clearance of oxypurinol; V/fm, apparent volume of distribution of oxypurinol; Kfm, combined absorption and formation rate constant; Imax, maximum inhibitory effect of oxypurinol on xanthine dehydrogenase to inhibit urate production; IC50, oxypurinol concentration at half maximum inhibitory effect; CrCL, creatinine clearance calculated using ideal body weight; FFM, fat free mass; Cssoxy, steady-state plasma oxypurinol concentration; SU, serum urate
An additive genetic model was assumed for the effect of SNPs on the PKPD parameters. The fractional effect of genotype was calculated as 1 + estimated effect per allele.
To estimate the renal and non-renal CL/fm of oxypurinol, a PK model with both serum and urine oxypurinol data was fitted. The renal CL/fm was 0.77 L/h (77%) and non-renal CL/fm was 0.23 L/h (23%) (Supplemental Table 3). Similar to the PK model with serum oxypurinol, the estimated CrCL and SLC22A12 rs505802C>T were found to be significant with renal CL/fm. NSAIDs, CARMIL1 rs742132A>G, and SLC2A9 rs1014290C>T were found to be significant in the forward selection step but not in the backward elimination step on renal CL/fm. No covariates were found to be significant with non-renal CL/fm.
Final PD model
A direct effect Emax model with BLurate, Imax, and IC50 with additive residual error model provided an adequate model to the SU data. The Hill coefficient could not be demonstrated to be different from 1.0 and was subsequently fixed to unity. The BSV for Imax, and IC50 was fixed to estimates from the base model due to insufficient data to estimate the precision of these parameters and high shrinkage in the final model.54
Model development steps for the PD model are summarized in Supplemental Table 4. The final model included estimated CrCL on BLurate and PDZK1 rs12129861G>A on IC50. The results of the base and final (including covariate) PD models are summarized in Table 2, and the final estimates for SU response are given by:
Model evaluation
The median parameter estimates with its 95%CI using SIR were comparable to the parameter estimate for the final PKPD models suggesting the PKPD model is stable (Table 2). The covariance step for base and final models presented in Table 2 and Supplementary Table 3 was successful but the results were not shown because SIR provided a better estimate for the precision of parameters. The GOF plots for the final PKPD models also showed no visual or statistical bias for the model prediction (Figure 2).
Figure 2.
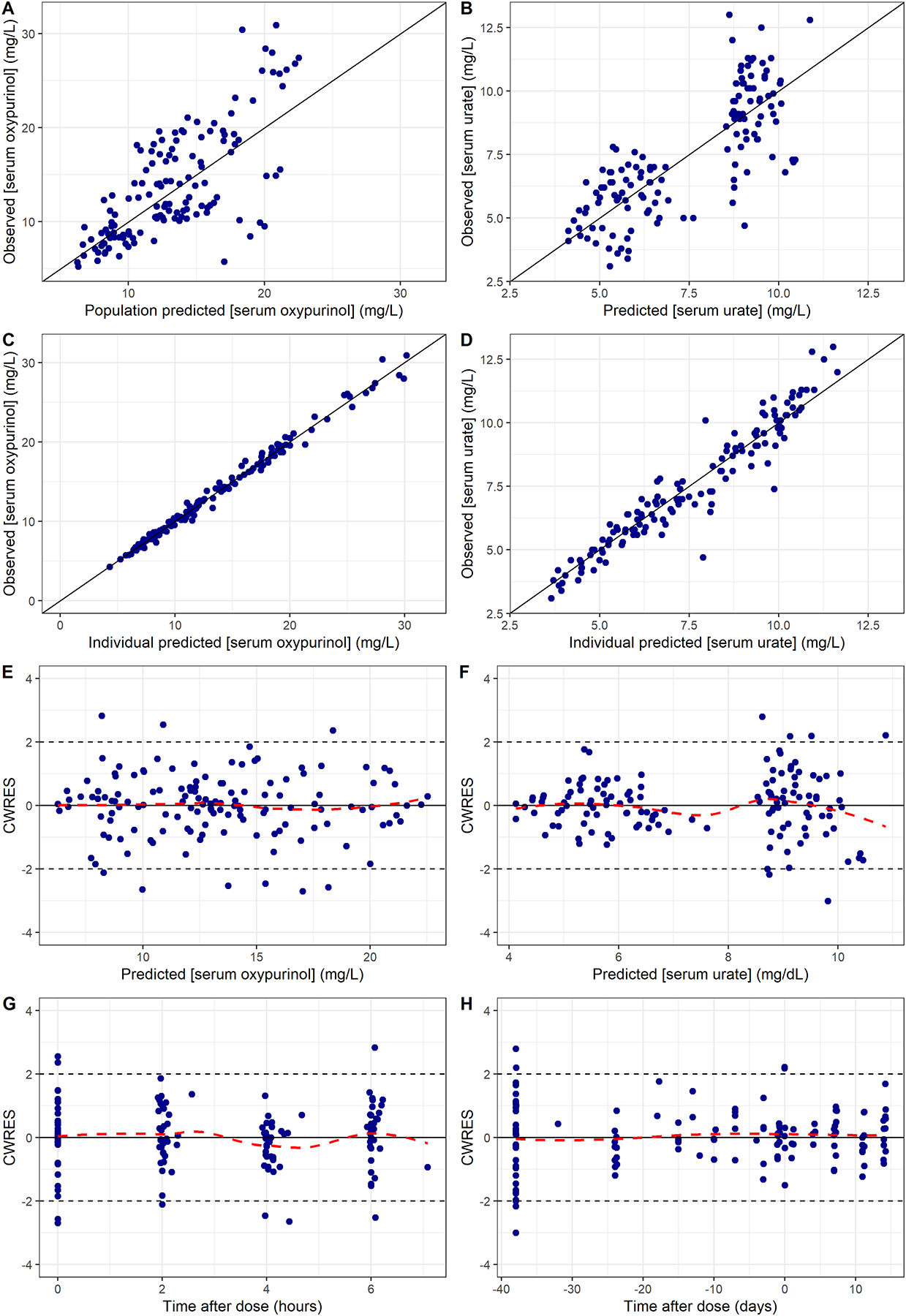
Goodness-of-fit plots of the final PKPD models for oxypurinol.
A and B, observed versus population-predicted concentration for serum oxypurinol and serum urate.
C and D, observed versus individual-predicted concentration for serum oxypurinol and serum urate.
E and F, conditional weighted residuals versus population-predicted concentration for serum oxypurinol and serum urate.
G and H, conditional weighted residuals versus time after dose for serum oxypurinol and serum urate.
CWRES = conditional weighted residuals.
The VPC plot was stratified by SLC22A12 rs505802C>T for the PK model and stratified by PDZK1 rs12129861G>A for the PD model, presented in Figure 3A. The VPC for the serum oxypurinol showed the median, 5th, and 95th percentiles of the model predicted serum oxypurinol concentrations followed the observed data in SLC22A12 rs505802 CC and CT genotypes. Due to the small sample size in TT genotype group, the 95%CI of the predicted oxypurinol concentrations overlapped and the small sample size limited the utility of VPC. The VPC plots for the PD model showed some inadequacy in capturing SU at the screening visit (time between −40 to 0 days) in PDZK1 rs12129861 GG genotype group. Nonetheless, the predicted SU followed the observed SU data well at the baseline and the follow-up visits in all three genotype groups (Figure 3B).
Figure 3.
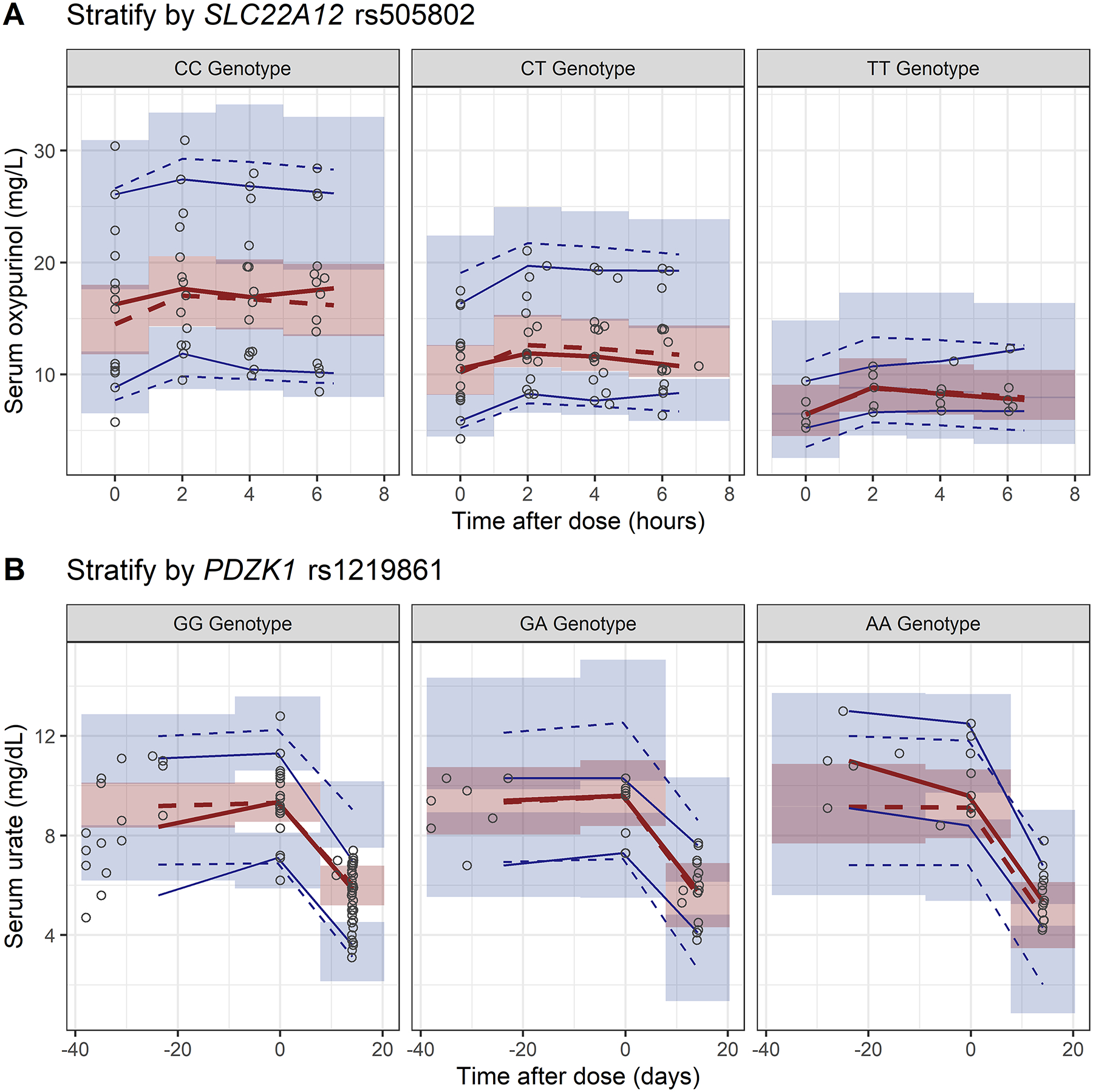
Visual predictive checks of the final PKPD models for oxypurinol.
A, serum oxypurinol concentration stratified by SLC22A12 rs505802 genotypes.
B, serum urate concentration stratified by PDZK1 rs12129861 genotypes.
The open circles represent the observed data. The blue (5th and 95th) and red (50th) solid and dashed lines represent the percentiles of the observed and simulated data, respectively. The shaded areas are the 95% confidence intervals of the simulated concentrations for the corresponding percentile values.
Allopurinol maintenance dose prediction
Table 3 presents the predicted allopurinol daily maintenance dose to achieve serum urate of <6 mg/dL with 75% of success rate. In general, individuals with lower FFM or higher CrCL require lower allopurinol dose. Individuals with SLC22A12 rs505802 T allele or PDZK1 rs12129861 G allele require a higher allopurinol dose. Individuals with chronic kidney disease (CrCL <60 mL/min) who carry both SLC22A12 rs505802 TT and PDZK1 rs12129861 GG genotypes require a higher than maximum dose, and hence would be candidates for alternative medications.
Table 3.
Predicted allopurinol daily maintenance dose to achieve serum urate of <6 mg/dL with 75% of success rate, considering genetic variants of SLC22A12 rs505802 and PDZK1 rs12129861
| Fat Free Mass (FFM) | |||||||
|---|---|---|---|---|---|---|---|
| CrCL (mL/min) | 50 kg | 60 kg | 70 kg | 80 kg | 90 kg | 100 kg | |
| PDZK1 rs12129861 GG | SLC22A12 rs505802 CC | ||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 500 | 650 | 700 | 700 | 800 | Alternative | |
| ≥60 | 400 | 450 | 450 | 550 | 550 | 600 | |
| SLC22A12 rs505802 CT | |||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 700 | 750 | Alternative | Alternative | Alternative | Alternative | |
| ≥60 | 500 | 550 | 600 | 650 | 750 | 800 | |
| SLC22A12 rs505802 TT | |||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥60 | 600 | 650 | 750 | Alternative | Alternative | Alternative | |
| PDZK1 rs12129861 GA | SLC22A12 rs505802 CC | ||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 400 | 450 | 500 | 600 | 600 | 650 | |
| ≥60 | 250 | 300 | 350 | 400 | 450 | 450 | |
| SLC22A12 rs505802 CT | |||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 550 | 600 | 650 | 750 | 800 | Alternative | |
| ≥60 | 350 | 400 | 450 | 500 | 550 | 550 | |
| SLC22A12 rs505802 TT | |||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 650 | 750 | Alternative | Alternative | Alternative | Alternative | |
| ≥60 | 450 | 500 | 550 | 650 | 700 | 750 | |
| PDZK1 rs12129861 AA | SLC22A12 rs505802 CC | ||||||
| ≥15 and <30 | 550 | 600 | 700 | 800 | 800 | Alternative | |
| ≥30 and <60 | 250 | 300 | 350 | 400 | 400 | 450 | |
| ≥60 | 200 | 200 | 250 | 250 | 300 | 300 | |
| SLC22A12 rs505802 CT | |||||||
| ≥15 and <30 | 750 | 800 | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 350 | 400 | 400 | 450 | 500 | 550 | |
| ≥60 | 250 | 250 | 300 | 350 | 350 | 350 | |
| SLC22A12 rs505802 TT | |||||||
| ≥15 and <30 | Alternative | Alternative | Alternative | Alternative | Alternative | Alternative | |
| ≥30 and <60 | 400 | 500 | 500 | 600 | 650 | 700 | |
| ≥60 | 300 | 350 | 350 | 400 | 450 | 450 | |
“Alternative” indicates an alternative medicine is preferred over allopurinol, given that the target serum urate was not achieved despite the maximum dose of allopurinol (800 mg/day). CrCL, creatinine clearance.
DISCUSSION
Allopurinol is the first-line ULT; however, many patients fail to achieve target SU on allopurinol. We developed a population PKPD model and identified the importance of clinical variables on the PKPD parameters in Hmong participants with gout and/or hyperuricemia. Body mass (FFM), renal function (estimated CrCL), and SLC22A12 rs505802C>T are key determinants to the PK of oxypurinol. Baseline SU, estimated CrCL, and PDZK1 rs12129861G>A are important covariates to the PD of oxypurinol. When determining the minimum allopurinol maintenance dose to achieve target SU, all of the aforementioned clinical factors need to be considered.
The final estimated population oxypurinol clearance [CL/fm of 1.05 L/h (95%CI 0.88–1.31)] was similar to a previous study (1.32 L/h)33 where the study participants had similarly estimated CrCL (70 mL/min versus 87 mL/min in GOUT-H). The final estimated population oxypurinol volume of distribution [V/fm of 59.3 L (95%CI 51.3–71.9)] was higher than the aforementioned study (41.6 L), possibly due to the older mean age of participants in their study (60-year-old versus 43-year-old in GOUT-H) and their approach to adjust for body mass (TBW versus FFM in GOUT-H). Given that the plasma protein binding for oxypurinol is negligible, the distribution of oxypurinol is similar to water content.55 Since elderly typically have 10–15% less total body water compared to younger individuals,56 the higher observed volume of oxypurinol (V/fm) in our population is expected.
The final estimated population parameters for the PD model (BLurate: 9 mg/dL, Imax: 7.6 mg/dL, IC50: 17.6 mg/L) were similar to participants with gout and/or hyperuricemia (BLurate: 8.5 mg/dL or 0.511 mmol/L, Imax: 6.87 mg/dL or 0.409 mmol/L, IC50: 14.1 mg/L or 83.9 μmol/L)33 but different from the healthy participants (BLurate: 4.6 mg/dL, Imax: 1 mg/dL, IC50: 2.59 mg/L).35 Higher Imax value observed in patients with hyperuricemia suggests the maximum SU lowering effect of allopurinol depends on the baseline SU level. The considerably higher IC50 in patients with gout and/or hyperuricemia indicates that a higher dose of allopurinol to achieve the same effect compared to non-hyperuricemic adults. This is likely due to the competitive inhibition of SU on xanthine dehydrogenase.
Similar to previous findings30,33, we found that FFM predicts oxypurinol clearance and volume of distribution better than TBW. Since the majority of our study participants were either overweight or obese, FFM approximates the lean body weight better43 and better reflects the true volume of distribution of oxypurinol. Renal function also plays a critical role in both PK and PD of oxypurinol, which has been demonstrated in previous population PKPD analyses and clinical studies.57–59 Contrary to a clinical observation that a lower allopurinol dose is needed to achieve target SU in patients with renal impairment (CrCl≤60 ml/min) compared with patients with CrCl >60 ml/min,57 we predicted that a higher allopurinol dose is required in patients with renal impairment. Although estimated CrCL is positively associated with both CL/fm and BLurate in the PKPD model, the overall contribution of renal function is larger in BLurate. This observation was consistent with previous published PKPD model33 where a higher allopurinol dose was required in patients with renal impairment compared to those without renal impairment if patients were taking diuretics. This relationship, which would appear to be counterintuitive, is likely under-appreciated by clinicians and clinical pharmacologists.
Drugs that may impact SU were not important factors in the final PKPD model. This contrasts with other studies that clearly demonstrated that people taking diuretics have a 25–30% lower oxypurinol clearance compared to those not taking diuretics.29,30,33 We did not observe this relationship in our study, likely due to our modest count of participants (n=4) who were taking various diuretics (hydrochlorothiazide, triamterene/hydrochlorothiazide, furosemide, and bumetanide). The association of loop, thiazide, and thiazide-like (but not potassium-sparing) diuretics with increased SU and higher incidence of gout, are well documented from both clinical observations60–63 and in vitro studies64–66. The proposed mechanisms for this observation includes either inhibition of urate efflux transporters, such as MRP4 (ABCC4)65 and NPT1 (SLC17A1)66, or increased urate reabsorption secondary to extracellular fluid volume depletion from diuresis.64 On the other hand, the evidence of how diuretics impact the PK of oxypurinol is less clear with some previous studies implicating loop diuretics, particularly furosemide, to be associated with increased plasma oxypurinol concentrations.57,67 Despite not being statistically significant, we found that patients taking HMG-CoA reductase inhibitors were associated with 52% decrease in oxypurinol CL/fm. The majority of the participants were taking atorvastatin (4/5, 80%), which suggests the potential impact of atorvastatin on the clearance of oxypurinol.45,46
SLC22A12 rs505802C>T was found to be a key determinant of oxypurinol clearance CL/fm. This association is plausible because oxypurinol undergoes extensive reabsorption through URAT1 encoded by SLC22A12,68 such that URAT1 dysfunction would impact the disposition of oxypurinol. Although the association between ABCG2 rs2231142C>A and SU-lowering response to allopurinol has been established in GWAS and replicated in other observational studies,22–25 no studies have shown a clear association between this SNP (rs2231142) and the PK parameters of allopurinol or oxypurinol. However, we cannot rule out the importance of ABCG2 rs2231142C>A, particularly in patients with extrarenal underexcretion hyperuricemia. Since a larger portion of the GOUT-H Hmong participants were overproduction hyperuricemia, instead of extrarenal underexcretion hyperuricemia,28 the impact of ABCG2 rs2231142C>A may be diminished in our study population.
An interesting finding was the impact of PDZK1 rs12129861G>A on IC50 in the inhibitory Emax model of oxypurinol. Of note, it is possible that PDZK1 rs12129861G>A may impact Imax given similar magnitude reduction in objective function value (OFV). However, the genetic effect on the folding protein is more likely to affect the drug binding affinity and thus impact the potency (IC50) rather than affecting the maximal effect (Imax). PDZK1 is a key component of urate-transporting molecular complex for URAT1 and OAT4.69,70 The PDZK1 rs12129861 A allele was also associated with a lower SU level27 and a decrease risk of gout.71,72 We found individuals with AA genotype have almost half of the IC50 as GG genotype (8.1 versus 17.6 mg/L) suggesting a higher affinity of oxypurinol with individuals with AA genotype. However, since this SNP is in the upstream region of PDZK1, a causal SNP has yet to be determined; a mechanistic study needs to be performed to elucidate the impact of PDZK1 on oxypurinol SU-lowering effect.
Limitations
A number of limitations should be noted. First, the small sample size (n=34) limits the ability to identify important covariates that could further explain the BSV in PKPD parameters of oxypurinol. In addition, the SCM with modest type I error control for the forward (p <0.05) and backward steps (p <0.01) with multiple testing may result in false positive findings given the limited number of participants.73 For example, the exponent of CrCL on baseline SU was −0.18 (95%CI −0.33, −0.036) suggesting CrCL was not a major determinant of SU despite a significant reduction in OFV when including CrCL in the model (Supplementary Table 4). However, the inclusion of renal function on SU was mainly driven by the physiology given more than 2/3 of serum urate is eliminated by kidney. In addition, the significant association between SLC22A12 rs505802 genotype and oxypurinol clearance, and PDZK1 rs12129861 genotype with IC50 in this population but not in other populations highlight the importance of including diverse populations in clinical studies. In other words, these observations may be unique to the Hmong population studied. Secondly, PK sampling scheme only covered half of the dosing interval that may negatively impact the accuracy of oxypurinol PK parameters estimate. This was a design feature suggested by the Hmong Genomics Board based on respecting the practical limitations of our participants. Given oxypurinol likely exhibits one compartmental PK behavior that is in concordance with previous studies29,30,33,35 and the maximum oxypurinol concentration observed in our study was at 2 hours, these provide confidence in our estimates. Thirdly, although we identified SLC22A12 rs505802 and PDZK1 rs12129861 are key determinants for PKPD response of oxypurinol, these SNPs are in the non-coding region, thus the causal SNPs for the differences observed in CL/fm and IC50 among individuals with different genotypes require further investigation. Forth, the imprecision of the PD parameters, such as IC50 and CrCL exponent on baseline SU, was larger than the PK parameters. In addition, the BSV for IC50 was fixed at a large value based on initial model fitting due to limited data available. These factors should be considered when interpreting simulation results and be aware that the actual variability will be higher than the prediction as the simulations were based on typical values.
Due to the intentional inclusion of southeast Asians of Hmong ancestry, we caution overinterpretation of the findings of this study to other populations. This caution is based both from the perspective of limited information concerning what is known about the relative role of renal function and uric acid disposition in this population relative to others as well as the observed differences in the prevalence of allele frequencies found in Hmong relative to other southeast Asian populations or populations of non-southeast Asian ancestry. As this was a pilot study in this unique population, the proposed allopurinol maintenance dose to achieved target SU requires validation in a prospective clinical study in a larger Hmong population.
CONCLUSION
In summary, we developed a population PKPD model for oxypurinol in Hmong participants with gout and/or hyperuricemia who take allopurinol. Body mass and renal function are key determinants for oxypurinol clearance and baseline SU, which aligns with previous findings. We also identified SNPs that can impact the oxypurinol clearance and its SU-lowering effect, which could have clinical importance. Considering all the important covariates, we propose a maintenance dose scheme of allopurinol to achieve target SU in the Hmong population that could help to better manage gout in this population, which exhibits a high prevalence of gout.74,75 The validity of this dosing scheme will require further study. However, we believe this study represents an important step in demonstrating the value of clinical trials including unique, under-represented populations who are at high risk for clinical consequences from hyperuricemia and gout and could benefit from effective ULT.
Supplementary Material
What is already known
Allopurinol exhibits large variability in pharmacokinetics and pharmacodynamics.
Patient characteristics and concomitant medications have been identified as sources of the variability but have not accounted for all of it.
The impact of genetic variants has been explored but no significant association has been established with population pharmacokinetics and pharmacodynamics analysis.
What does this study add
Genetic variants in SLC22A12 were associated with oxypurinol clearance and variants in PDZK1 were associated with urate-lowering effect of oxypurinol.
The allopurinol maintenance dose to achieve target serum urate level depends on patients’ body mass, renal function, and genetic variants in SLC22A12 and PDZK1.
What is the clinical significance
An individualized dosing approach is proposed to optimize allopurinol for Hmong adults with gout and/or hyperuricemia
Acknowledgment:
The authors wish to thank the support from all the Hmong participants, the Hmong GOUT-H Research Board, and Minnesota Community Care.
Funding:
This work was supported by the University of Minnesota, Office of Community Engagement of the Clinical and Translational Science Institute for its funding via 2014 Collaborative Pilot Grants No. 22556 and National Center for Advancing Translational Sciences of the National Institutes of Health Award Number UL1TR000114. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- AJBW
adjusted body weight
- BLurate
baseline serum urate
- BSV
between subject variability
- CL/fm
apparent clearance
- CrCL
creatinine clearance
- CV
coefficient of variation
- FFM
fat-free mass
- fm
the fraction of allopurinol dose available as oxypurinol systemically
- γ
the Hill coefficient for the sigmoid Emax model
- GOF
goodness of fit
- GOUT-H
Genetics Of HyperUricemia and Gout Therapy in Hmong
- GWAS
Genome-wide association studies
- IC50
the oxypurinol concentration required to inhibit 50% of the activity of xanthine dehydrogenase
- Imax
the maximum inhibitory effect of oxypurinol on xanthine dehydrogenase to inhibit urate production
- IPPSE
individual pharmacokinetic parameters with the standard error
- Kfm
rate constant
- NSAIDs
non-steroidal anti-inflammatory drugs
- Oxypurinolss
the serum oxypurinol concentration at steady-state
- PK
pharmacokinetics
- PKPD
pharmacokinetic-pharmacodynamic
- SCAR
severe cutaneous adverse reaction
- SCM
stepwise covariant modeling
- SIR
sampling importance resampling
- SNPs
single nucleotide polymorphisms
- SU
serum urate
- TBW
total body weight
- ULT
urate-lowering therapy
- V/fm
apparent volume
- VPC
visual predictive check
Footnotes
Conflict of interest disclosure: All listed authors declare no conflicts of interest.
Ethics approval: This study was approved by the Human Research Protection Program at the University of Minnesota IRB (# 1408M53223).
Patient consent: Consent was obtained for all the study participants prior to any study related activities.
ClinicalTrials.gov Identifier: NCT02371421
Data availability:
Raw data is not available to the public due to lack of patient consent for data sharing. NONMEM control files for pharmacokinetics and pharmacodynamics modeling and R script for allopurinol dose simulation are available in the supplemental text 1–4.
References
- 1.FitzGerald JD, Dalbeth N, Mikuls T, et al. 2020 American College of Rheumatology Guideline for the Management of Gout. Arthritis & Rheumatology. 2020/05/10 2020;n/a(n/a)doi: 10.1002/art.41247 [DOI] [PubMed] [Google Scholar]
- 2.Dalbeth N, Gosling AL, Gaffo A, Abhishek A. Gout. Lancet. May 15 2021;397(10287):1843–1855. doi: 10.1016/S0140-6736(21)00569-9 [DOI] [PubMed] [Google Scholar]
- 3.Khanna D, Fitzgerald JD, Khanna PP, et al. 2012 American College of Rheumatology guidelines for management of gout. Part 1: systematic nonpharmacologic and pharmacologic therapeutic approaches to hyperuricemia. Arthritis care & research. Oct 2012;64(10):1431–1446. doi: 10.1002/acr.21772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richette P, Doherty M, Pascual E, et al. 2016 updated EULAR evidence-based recommendations for the management of gout. Annals of the rheumatic diseases. Jan 2017;76(1):29–42. doi: 10.1136/annrheumdis-2016-209707 [DOI] [PubMed] [Google Scholar]
- 5.Jordan KM, Cameron JS, Snaith M, et al. British Society for Rheumatology and British Health Professionals in Rheumatology guideline for the management of gout. Rheumatology. Aug 2007;46(8):1372–4. doi: 10.1093/rheumatology/kem056a [DOI] [PubMed] [Google Scholar]
- 6.Stamp LK, Chapman PT, Barclay ML, et al. A randomised controlled trial of the efficacy and safety of allopurinol dose escalation to achieve target serum urate in people with gout. Annals of the rheumatic diseases. Sep 2017;76(9):1522–1528. doi: 10.1136/annrheumdis-2016-210872 [DOI] [PubMed] [Google Scholar]
- 7.Khanna P, Hagerty D, Mischler R, Morlock R. FRI0397 Adherence to EULAR recommendations for the treatment of GOUT. Annals of the rheumatic diseases. 2014;71(Suppl 3):448–449. doi: 10.1136/annrheumdis-2012-eular.2854 [DOI] [Google Scholar]
- 8.Shoji A, Yamanaka H, Kamatani N. A retrospective study of the relationship between serum urate level and recurrent attacks of gouty arthritis: evidence for reduction of recurrent gouty arthritis with antihyperuricemic therapy. Arthritis and rheumatism. Jun 15 2004;51(3):321–5. doi: 10.1002/art.20405 [DOI] [PubMed] [Google Scholar]
- 9.Dalbeth N, House ME, Horne A, Taylor WJ. Reduced creatinine clearance is associated with early development of subcutaneous tophi in people with gout. BMC musculoskeletal disorders. Dec 21 2013;14:363. doi: 10.1186/1471-2474-14-363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feig DI. Hyperuricemia and hypertension. Advances in chronic kidney disease. Nov 2012;19(6):377–385. doi: 10.1053/j.ackd.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 11.Kuwabara M, Hisatome I, Niwa K, et al. Uric Acid Is a Strong Risk Marker for Developing Hypertension From Prehypertension: A 5-Year Japanese Cohort Study. Hypertension. Jan 2018;71(1):78–86. doi: 10.1161/HYPERTENSIONAHA.117.10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sluijs I, Beulens JW, van der AD, Spijkerman AM, Schulze MB, van der Schouw YT. Plasma uric acid is associated with increased risk of type 2 diabetes independent of diet and metabolic risk factors. The Journal of nutrition. Jan 2013;143(1):80–85. doi: 10.3945/jn.112.167221 [DOI] [PubMed] [Google Scholar]
- 13.Chen LY, Zhu WH, Chen ZW, et al. Relationship between hyperuricemia and metabolic syndrome. Journal of Zhejiang University Science B. Aug 2007;8(8):593–8. doi: 10.1631/jzus.2007.B0593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehmet Kanbay MS, Baris Afsar, et al. The role of uric acid in the pathogenesis of human cardiovascular desease. Heart. 2013;99:759–756. [DOI] [PubMed] [Google Scholar]
- 15.Jensen T, Niwa K, Hisatome I, et al. Increased Serum Uric Acid over five years is a Risk Factor for Developing Fatty Liver. Sci Rep. Aug 6 2018;8(1):11735. doi: 10.1038/s41598-018-30267-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hande KR, Noone RM, Stone WJ. Severe Allopurinol Toxicity: Description and Guidelines for Prevention in Patients with Renal Insufficiency. The American journal of medicine. 1984;76:47–56. [DOI] [PubMed] [Google Scholar]
- 17.Chung WH, Chang WC, Stocker SL, et al. Insights into the poor prognosis of allopurinol-induced severe cutaneous adverse reactions: the impact of renal insufficiency, high plasma levels of oxypurinol and granulysin. Annals of the rheumatic diseases. Dec 2015;74(12):2157–2164. doi: 10.1136/annrheumdis-2014-205577 [DOI] [PubMed] [Google Scholar]
- 18.Yang CY, Chen CH, Deng ST, et al. Allopurinol Use and Risk of Fatal Hypersensitivity Reactions: A Nationwide Population-Based Study in Taiwan. JAMA Intern Med. Sep 2015;175(9):1550–7. doi: 10.1001/jamainternmed.2015.3536 [DOI] [PubMed] [Google Scholar]
- 19.Hung SL, Chung WH, Liou LB, et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. P Natl Acad Sci USA. Mar 15 2005;102(11):4134–4139. doi: 10.1073/pnas.0409500102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalbeth N, Kumar S, Stamp L, Gow P. Dose adjustment of allopurinol according to creatinine clearance does not provide adequate control of hyperuricemia in patients with gout. J Rheumatol. Aug 2006;33(8):1646–50. [PubMed] [Google Scholar]
- 21.Stamp LK, Taylor WJ, Jones PB, et al. Starting dose is a risk factor for allopurinol hypersensitivity syndrome: a proposed safe starting dose of allopurinol. Arthritis and rheumatism. Aug 2012;64(8):2529–36. doi: 10.1002/art.34488 [DOI] [PubMed] [Google Scholar]
- 22.Wen CC, Yee SW, Liang X, et al. Genome-wide association study identifies ABCG2 (BCRP) as an allopurinol transporter and a determinant of drug response. Clin Pharmacol Ther. May 2015;97(5):518–525. doi: 10.1002/cpt.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RL, Wallace MC, Phipps-Green AJ, et al. ABCG2 loss-of-function polymorphism predicts poor response to allopurinol in patients with gout. Pharmacogenomics J. Mar 2017;17(2):201–203. doi: 10.1038/tpj.2015.101 [DOI] [PubMed] [Google Scholar]
- 24.Wallace MC, Roberts RL, Nanavati P, et al. Association between ABCG2 rs2231142 and poor response to allopurinol: replication and meta-analysis. Rheumatology. Apr 1 2018;57(4):656–660. doi: 10.1093/rheumatology/kex467 [DOI] [PubMed] [Google Scholar]
- 25.Brackman DJ, Yee SW, Enogieru OJ, et al. Genome-Wide Association and Functional Studies Reveal Novel Pharmacological Mechanisms for Allopurinol. Clin Pharmacol Ther. Sep 2019;106(3):623–631. doi: 10.1002/cpt.1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright DFB, Dalbeth N, Phipps-Green AJ, et al. The impact of diuretic use and ABCG2 genotype on the predictive performance of a published allopurinol dosing tool. British journal of clinical pharmacology. May 2018;84(5):937–943. doi: 10.1111/bcp.13516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolz M, Johnson T, Sanna S, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. Jun 2009;5(6):e1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roman YM, Culhane-Pera K, Lo M, et al. The Impact of rs505802 for SLC22A12 on Oxipurinol and Uric Acid Disposition in Hmong Patients on Allopurinol from the Genetics of Hyperuricemia Therapy in Hmong (GOUT-H) Study. Clinical Pharmacology & Therapeutics. 2017;101(S1;PI-110):S5–S99. doi: 10.1002/cpt.570 [DOI] [Google Scholar]
- 29.Stocker SL, McLachlan AJ, Savic RM, et al. The pharmacokinetics of oxypurinol in people with gout. British journal of clinical pharmacology. Sep 2012;74(3):477–89. doi: 10.1111/j.1365-2125.2012.04207.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright DF, Stamp LK, Merriman TR, Barclay ML, Duffull SB, Holford NH. The population pharmacokinetics of allopurinol and oxypurinol in patients with gout. Eur J Clin Pharmacol. Jul 2013;69(7):1411–21. doi: 10.1007/s00228-013-1478-8 [DOI] [PubMed] [Google Scholar]
- 31.Pilon MO, Leclair G, Oussaid E, et al. An association study of ABCG2 rs2231142 on the concentrations of allopurinol and its metabolites. Clin Transl Sci. Jun 10 2022;doi: 10.1111/cts.13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham GG, Kannangara DR, Stocker SL, et al. Understanding the dose-response relationship of allopurinol: predicting the optimal dosage. British journal of clinical pharmacology. Dec 2013;76(6):932–8. doi: 10.1111/bcp.12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright DF, Duffull SB, Merriman TR, Dalbeth N, Barclay ML, Stamp LK. Predicting allopurinol response in patients with gout. British journal of clinical pharmacology. Feb 2016;81(2):277–89. doi: 10.1111/bcp.12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannangara DRW, Graham GG, Wright DFB, et al. Individualising the dose of allopurinol in patients with gout. British journal of clinical pharmacology. Sep 2017;83(9):2015–2026. doi: 10.1111/bcp.13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vora B, Brackman DJ, Zou L, et al. Oxypurinol pharmacokinetics and pharmacodynamics in healthy volunteers: Influence of BCRP Q141K polymorphism and patient characteristics. Clin Transl Sci. Jul 2021;14(4):1431–1443. doi: 10.1111/cts.12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kottgen A, Albrecht E, Teumer A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. Feb 2013;45(2):145–154. doi: 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada Y, Sim X, Go MJ, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. Aug 2012;44(8):904–9. doi: 10.1038/ng.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitart V, Rudan I, Hayward C, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. Apr 2008;40(4):437–42. doi: 10.1038/ng.106 [DOI] [PubMed] [Google Scholar]
- 39.Lindbom L, Pihlgren P, Jonsson EN. PsN-Toolkit--a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed. Sep 2005;79(3):241–57. doi: 10.1016/j.cmpb.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 40.Lacroix BD, Friberg LE, Karlsson MO. Evaluation of IPPSE, an alternative method for sequential population PKPD analysis. J Pharmacokinet Pharmacodyn. Apr 2012;39(2):177–93. doi: 10.1007/s10928-012-9240-x [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Beal SL, Sheinerz LB. Simultaneous vs. sequential analysis for population PK/PD data II: robustness of methods. J Pharmacokinet Pharmacodyn. Dec 2003;30(6):405–16. doi: 10.1023/b:jopa.0000012999.36063.4e [DOI] [PubMed] [Google Scholar]
- 42.Stocker SL, Graham GG, McLachlan AJ, Williams KM, Day RO. Pharmacokinetic and pharmacodynamic interaction between allopurinol and probenecid in patients with gout. J Rheumatol. May 2011;38(5):904–910. doi: 10.3899/jrheum.101160 [DOI] [PubMed] [Google Scholar]
- 43.Janmahasatian S, Duffull SB, Ash S, Ward LC, Byrne NM, Green B. Quantification of lean bodyweight. Clinical pharmacokinetics. 2005;44(10):1051–65. doi: 10.2165/00003088-200544100-00004 [DOI] [PubMed] [Google Scholar]
- 44.Enomoto A, Kimura H, Chairoungdua A, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. May 23 2002;417(6887):447–52. doi: 10.1038/nature742 [DOI] [PubMed] [Google Scholar]
- 45.Milionis HJ, Kakafika AI, Tsouli SG, et al. Effects of statin treatment on uric acid homeostasis in patients with primary hyperlipidemia. American heart journal. Oct 2004;148(4):635–40. doi: 10.1016/j.ahj.2004.04.005 [DOI] [PubMed] [Google Scholar]
- 46.Kose E, An T, Kikkawa A, Matsumoto Y, Hayashi H. Effects on serum uric acid by difference of the renal protective effects with atorvastatin and rosuvastatin in chronic kidney disease patients. Biol Pharm Bull. 2014;37(2):226–31. doi: 10.1248/bpb.b13-00418 [DOI] [PubMed] [Google Scholar]
- 47.Choi HK, Soriano LC, Zhang Y, Rodriguez LA. Antihypertensive drugs and risk of incident gout among patients with hypertension: population based case-control study. BMJ. 2012;344:d8190. doi: 10.1136/bmj.d8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dosne AG, Bergstrand M, Harling K, Karlsson MO. Improving the estimation of parameter uncertainty distributions in nonlinear mixed effects models using sampling importance resampling. J Pharmacokinet Pharmacodyn. Dec 2016;43(6):583–596. doi: 10.1007/s10928-016-9487-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keizer RJ, Karlsson MO, Hooker A. Modeling and Simulation Workbench for NONMEM: Tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst Pharmacol. Jun 26 2013;2:e50. doi: 10.1038/psp.2013.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2021. [Google Scholar]
- 51.Alexander SP, Christopoulos A, Davenport AP, et al. THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: G protein-coupled receptors. Br J Pharmacol. Oct 2021;178 Suppl 1:S27–S156. doi: 10.1111/bph.15538 [DOI] [PubMed] [Google Scholar]
- 52.Consultation WHOE. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. Jan 10 2004;363(9403):157–63. doi: 10.1016/S0140-6736(03)15268-3 [DOI] [PubMed] [Google Scholar]
- 53.Wright DF, Doogue MP, Barclay ML, et al. A population pharmacokinetic model to predict oxypurinol exposure in patients on haemodialysis. Eur J Clin Pharmacol. Jan 2017;73(1):71–78. doi: 10.1007/s00228-016-2133-y [DOI] [PubMed] [Google Scholar]
- 54.Savic RM, Karlsson MO. Importance of shrinkage in empirical bayes estimates for diagnostics: problems and solutions. AAPS J. Sep 2009;11(3):558–69. doi: 10.1208/s12248-009-9133-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnheim K, Krivanek P, Oberbauer R. Pharmacokinetics and pharmacodynamics of allopurinol in elderly and young subjects. British journal of clinical pharmacology. Oct 1999;48(4):501–9. doi: 10.1046/j.1365-2125.1999.00041.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andres TM, McGrane T, McEvoy MD, Allen BFS. Geriatric Pharmacology: An Update. Anesthesiol Clin. Sep 2019;37(3):475–492. doi: 10.1016/j.anclin.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 57.Stamp LK, Barclay ML, O’Donnell JL, et al. Relationship between serum urate and plasma oxypurinol in the management of gout: determination of minimum plasma oxypurinol concentration to achieve a target serum urate level. Clin Pharmacol Ther. Sep 2011;90(3):392–8. doi: 10.1038/clpt.2011.113 [DOI] [PubMed] [Google Scholar]
- 58.Stamp LK, Chapman PT, Barclay M, et al. Relationships Between Allopurinol Dose, Oxypurinol Concentration and Urate-Lowering Response-In Search of a Minimum Effective Oxypurinol Concentration. Clin Transl Sci. Jan 2020;13(1):110–115. doi: 10.1111/cts.12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamp LK, Chapman PT, Barclay ML, et al. How much allopurinol does it take to get to target urate? Comparison of actual dose with creatinine clearance-based dose. Arthritis research & therapy. Nov 16 2018;20(1):255. doi: 10.1186/s13075-018-1755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McAdams DeMarco MA, Maynard JW, Baer AN, et al. Diuretic use, increased serum urate levels, and risk of incident gout in a population-based study of adults with hypertension: the Atherosclerosis Risk in Communities cohort study. Arthritis and rheumatism. Jan 2012;64(1):121–9. doi: 10.1002/art.33315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savage PJ, Pressel SL, Curb JD, et al. Influence of long-term, low-dose, diuretic-based, antihypertensive therapy on glucose, lipid, uric acid, and potassium levels in older men and women with isolated systolic hypertension: The Systolic Hypertension in the Elderly Program. SHEP Cooperative Research Group. Arch Intern Med. Apr 13 1998;158(7):741–51. doi: 10.1001/archinte.158.7.741 [DOI] [PubMed] [Google Scholar]
- 62.Choi HK, Atkinson K, Karlson EW, Curhan G. Obesity, weight change, hypertension, diuretic use, and risk of gout in men: the health professionals follow-up study. Arch Intern Med. Apr 11 2005;165(7):742–8. doi: 10.1001/archinte.165.7.742 [DOI] [PubMed] [Google Scholar]
- 63.Bruderer S, Bodmer M, Jick SS, Meier CR. Use of diuretics and risk of incident gout: a population-based case-control study. Arthritis Rheumatol. Jan 2014;66(1):185–96. doi: 10.1002/art.38203 [DOI] [PubMed] [Google Scholar]
- 64.Weinman EJ, Eknoyan G, Suki WN. The influence of the extracellular fluid volume on the tubular reabsorption of uric acid. J Clin Invest. Feb 1975;55(2):283–91. doi: 10.1172/JCI107931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.El-Sheikh AA, van den Heuvel JJ, Koenderink JB, Russel FG. Effect of hypouricaemic and hyperuricaemic drugs on the renal urate efflux transporter, multidrug resistance protein 4. Br J Pharmacol. Dec 2008;155(7):1066–75. doi: 10.1038/bjp.2008.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jutabha P, Anzai N, Kitamura K, et al. Human sodium phosphate transporter 4 (hNPT4/SLC17A3) as a common renal secretory pathway for drugs and urate. The Journal of biological chemistry. Nov 5 2010;285(45):35123–32. doi: 10.1074/jbc.M110.121301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stamp LK, Barclay ML, O’Donnell JL, et al. Furosemide increases plasma oxypurinol without lowering serum urate--a complex drug interaction: implications for clinical practice. Rheumatology. Sep 2012;51(9):1670–6. doi: 10.1093/rheumatology/kes091 [DOI] [PubMed] [Google Scholar]
- 68.Iwanaga T, Kobayashi D, Hirayama M, Maeda T, Tamai I. Involvement of Uric Acid Transporter in Increased Renal Clearance of the Xanthine Oxidase Inhibitor Oxypurinol Induced by a Uricosuric Agent, Benzbromarone. Drug metabolism and disposition: the biological fate of chemicals. 2005;33(12):1791–1795. [DOI] [PubMed] [Google Scholar]
- 69.Anzai N, Miyazaki H, Noshiro R, et al. The multivalent PDZ domain-containing protein PDZK1 regulates transport activity of renal urate-anion exchanger URAT1 via its C terminus. The Journal of biological chemistry. Oct 29 2004;279(44):45942–50. doi: 10.1074/jbc.M406724200 [DOI] [PubMed] [Google Scholar]
- 70.Miyazaki H, Anzai N, Ekaratanawong S, et al. Modulation of renal apical organic anion transporter 4 function by two PDZ domain-containing proteins. Journal of the American Society of Nephrology : JASN. Dec 2005;16(12):3498–506. doi: 10.1681/ASN.2005030306 [DOI] [PubMed] [Google Scholar]
- 71.Higashino T, Matsuo H, Sakiyama M, et al. Common variant of PDZ domain containing 1 (PDZK1) gene is associated with gout susceptibility: A replication study and meta-analysis in Japanese population. Drug Metab Pharmacokinet. Dec 2016;31(6):464–466. doi: 10.1016/j.dmpk.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 72.Li M, Li Q, Li CG, et al. Genetic polymorphisms in the PDZK1 gene and susceptibility to gout in male Han Chinese: a case-control study. International journal of clinical and experimental medicine. 2015;8(8):13911–8. [PMC free article] [PubMed] [Google Scholar]
- 73.Ribbing J, Jonsson EN. Power, selection bias and predictive performance of the Population Pharmacokinetic Covariate Model. J Pharmacokinet Pharmacodyn. Apr 2004;31(2):109–34. doi: 10.1023/b:jopa.0000034404.86036.72 [DOI] [PubMed] [Google Scholar]
- 74.Portis AJ, Laliberte M, Tatman P, et al. High prevalence of gouty arthritis among the Hmong population in Minnesota. Arthritis care & research. Oct 2010;62(10):1386–1391. doi: 10.1002/acr.20232 [DOI] [PubMed] [Google Scholar]
- 75.Roman YM, Lor K, Xiong T, Culhane-Pera K, Straka RJ. Gout prevalence in the Hmong: a prime example of health disparity and the role of community-based genetic research. Per Med. May 2021;18(3):311–327. doi: 10.2217/pme-2020-0107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data is not available to the public due to lack of patient consent for data sharing. NONMEM control files for pharmacokinetics and pharmacodynamics modeling and R script for allopurinol dose simulation are available in the supplemental text 1–4.


