Abstract
Background:
Prior predictive models using logistic regression for stillbirth do not leverage the advanced and nuanced techniques involved in sophisticated machine learning methods, such as modeling non-linear relationships between outcomes.
Objective:
To create and refine machine learning models for predicting stillbirth using data available prior to viability (22–24 weeks) and throughout pregnancy using demographic, medical, and prenatal visit data, including ultrasound and fetal genetics.
Study Design:
This is a secondary analysis of the Stillbirth Collaborative Research Network, which included data from pregnancies resulting in stillborn and liveborn infants delivered at 59 hospitals in 5 diverse regions across the US from 2006–2009. The primary aim was creation of a model for predicting stillbirth using data available prior to viability. Secondary aims included refining models with variables available throughout pregnancy and determining variable importance.
Results:
Among 3000 live births and 982 stillbirths, 101 variables of interest were identified. Of the models incorporating data available prior to viability, the Random Forests model had 85.1% accuracy (area under curve [AUC]) and high sensitivity (88.6%), specificity (85.3%), Positive Predictive Value (85.3%), and Negative Predictive Value (84.8%). A Random Forests model using data collected throughout pregnancy resulted in accuracy of 85.0%; this model had 92.2% sensitivity, 77.9% specificity, 84.7% Positive Predictive Value, and 88.3% Negative Predictive Value. Important variables in the pre-viability model included previous stillbirth, minority race, gestational age at earliest prenatal visit and ultrasound, and 2nd trimester serum screening.
Conclusions:
Applying advanced machine learning techniques to a comprehensive database of stillbirths and live births with unique and clinically relevant variables resulted in an algorithm that could accurately identify 85% of pregnancies that would result in stillbirth, before they reached viability. Once validated in representative databases reflective of the United States birthing population and then prospectively, these models may provide effective risk stratification and clinical decision-making support to better identify and monitor those at risk for stillbirth.
Keywords: pre-viability; boosted trees; clinical decision-making; ultrasound; 2nd trimester prenatal screen (Down syndrome risk, unconjugated estriol, maternal serum alpha-fetoprotein); structural racism; prenatal care; factor analysis; random forests
Tweetable statement:
A machine learning algorithm can predict stillbirth risk with data collected prior to viability with 85.1% AUROC.
Introduction
Stillbirth is among the most devastating obstetric outcomes. Evidence-based interventions for preventing stillbirth are not well-developed; thus, identifying those at risk for stillbirth is crucial.1 Previously, logistic regression and other methods relying on linear relationships between variables to achieve classification have been utilized to attempt to predict—and thereby prevent—stillbirth.2–15 While the predictive accuracy for stillbirth from regression has reached up to 88% (based on third trimester cerebroplacental ratio and uterine artery dopplers, in this particular model), these methods do not usually leverage the advanced and nuanced techniques involved in sophisticated machine learning, which includes (but are not limited to) modeling non-linear relationships between covariates in larger datasets that can handle large amounts of variables without overfitting.16,17 More recently, others have attempted to utilize machine learning to predict stillbirth; however, these models are limited by both accuracy and variable availability, relying on measures collected even after the stillbirth has occurred (i.e., fetal weight or gestational age).17–19 The absence of a model combining advanced machine learning techniques with a detailed, clinically accurate database has hampered the creation of an ideal predictive calculator for stillbirth. Similar sophisticated models in obstetrics have improved prediction of vaginal birth after cesarean (VBAC) over traditional logistic regression, for example, proving machine learning to be an invaluable tool.20
The Stillbirth Collaborative Research Network (SCRN) was developed to create a database of stillbirths and representative livebirths to ascertain the causes, incidence, and risk factors for stillbirths.21 Participants in this case-control study completed a comprehensive clinical interview, consented to pathological work-up, and had clinical data abstracted by trained research personnel. The granularity and relative clinical accuracy of this large, comprehensive database of stillbirths and livebirths renders the SCRN dataset well-suited for attempting to develop a machine learning model for predicting stillbirth.
We sought to utilize the SCRN database develop a machine learning model that predicts stillbirth risk with high accuracy and sensitivity. Our primary objective was the production of a pilot model that could predict stillbirth using data available prior to viability (22–24 weeks). Our secondary objectives included further refining that model using variables obtained throughout pregnancy as well as determining variable importance within these models.
Materials and Methods
Sample derivation
Data were derived from SCRN, a multi-site case-control study of stillbirths and representative livebirths conducted from 2006–2009.21 The study included a heterogenous sample of patients who delivered a live or stillborn fetus at >18 weeks gestational age (GA) from five catchment areas across the United States (RI, MA, GA, TX, UT). Hospital locations were chosen such that patients approached for study participation included >90% of stillbirths and livebirths in the geographic region. Design, methods, recruitment experience, and participant inclusion/exclusion criteria have been described in-depth.21 Livebirths <32 weeks gestation (in order to mitigate the large difference in GA between stillbirths and livebirths, on average) and participants of Black race (given the increased stillbirth rates at baseline in Black patients) were oversampled. Data collection included initial maternal interview, chart abstraction, pathology examination and biospecimens, and follow-up interview. Each study site’s Institutional Review Board approved study procedures as well as the Data Coordinating and Analysis Center; participants gave written, informed consent at time of enrollment and for pathologic examinations and follow-up interview.
For this pilot model, participants were included if they (1) delivered a singleton stillbirth or livebirth during the study period and (2) had sufficient available data (defined as ≤80% missing) from which our models could be derived using standard methods for imputing missing data.
Variable selection
The SCRN dataset includes over 6000 measures collected during the initial maternal interview, post-mortem pathologic examination, medical record abstraction, cause of death analysis, and follow-up interview. We selected a series of risk factor variables based on literature review within the following categories: maternal demographics, biologic father of the baby (BFOB) demographics, family history, maternal health history (including mental health history), prenatal laboratory data, 2nd trimester screen (“Quad screen”), prenatal care history, ultrasound, social determinants of health (defined as demographic and economic information extracted from maternal interview, including those intended as markers of underlying structural inequality), and substance use.2–14,22–28 We also sought to reduce the confounding effect of earlier GA at birth in stillbirths in several measures. For example, the raw values of GA at last prenatal visit are skewed higher in livebirths not by virtue of an intrinsic process but rather because, in the parent database, livebirths have a later GA on average than stillbirths; however, the fact that livebirths were born later than stillbirths in the parent database is not valuable to a predictive model. Therefore, instead of using GA at delivery, we calculated ratios of several measures to GA at birth to establish the relative difference in care between stillbirths and livebirths as a potential predictor: GA of last prenatal visit, total number of prenatal visits, and GA of last ultrasound. Several measures were included as surrogates for underlying racism and healthcare inequality, including maternal and BFOB self-reported race (Black, Asian, Pacific Islander, American Indian, Alaskan Native, multiracial, or other race) and ethnicity, income assistance, years of education, and mother or BFOB born in the United States; these measures are intended only as social placeholders for those at higher risk of experiencing implicit bias in their medical care and do not represent an organic difference in participants.
Variables with >80% missing data (n = 51) and measures that were only discoverable at delivery and would not be helpful in a predictive model (i.e., GA and fetal weight at delivery) were removed from consideration. Missing data were imputed for the remaining pre-viability variables using Multivariate Imputation by Chained Equations (“mice” package); this method can handle high levels of missingness (up to 80%, which we used as our threshold) by looking to data from other columns and predicting the best estimate for the missing value.29 All variables were confirmed to be “missing at random,” as is required for this imputation method. Iteration number was set to 80 per protocol of one iteration per percent of missing data. Our final literature-based variable set included 101 measures.
After completing variable selection based on literature review, which allows for selection of a wide array of variables, we selected a series of variables using Exploratory Factor Analysis (EFA), as is standard for machine learning methods. This unsupervised machine learning method helps identify which variables best explain the variance within a dataset by creating factors, or groups of variables, that correlate with underlying structures that define the data; using both methods of variable selection (clinically significant variables and variables that explain the most variance in the sample) allows for testing of several different sets of variables.30 Using the “psych” package in R, we first determined the ideal number of factors using the elbow method; using this result, we then computed an EFA model using varimax rotation and a polychoric, heterogenous correlation matrix for binary data (“polycor” package). We then determined factor loadings and selected variables within the factors that simultaneously maintained the largest sample size to avoid overfitting and optimized variance explained.
Machine learning models
We selected several learners based on a priori criteria: (1) utilizes supervised learning; (2) produces variable importance; (3) available from the “caret” package. The following learners were selected: decision trees (tree-based structure defined by a set of if/then statements), boosted trees (a set of decision trees that are reliant on the previous tree, combined in an ensemble for a final prediction), random forests (a set of decision trees that are run independently and combined for a final prediction), and single layer neural networks (simulates connected layered processes, combined for a final prediction). Logistic regression was also completed for each set of variables (pre-viability, all-pregnancy, using EFA, etc.) to allow for comparison with linear statistical methods.
Prior to entry into all models (logistic regression and advanced machine learning), all measures were centered, scaled, and dummy coded. We employed synthetic minority sampling technique (SMOTE) to weight our observations (R package “DMwR”). Data was then partitioned into training and testing sets at a 3:1 ratio with 100 k-fold cross-validation.31 Tuning matrices unique to each model were utilized to optimize probability threshold cutoffs rather than specifying thresholds ad hoc. Confusion matrices were derived from the tested models, reporting accuracy (area under receiver operating characteristic [AUROC]), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), and error rate (Type I and II). Variable importance was also reported for the best model for each set of variables, scaled to 0–100 (0 = least important, 100 = most important).
In pursuit of our primary aim, we included those measures collected prior to viability (22–24 weeks, herein referred to as “pre-viability”). For this primary aim, computational time and memory usage were also calculated to understand future system requirements. For our secondary aims (model refinement), we ran several additional models including measures: (1) available throughout pregnancy (“all-pregnancy”), (2) derived from EFA, and (3) without 2nd trimester serum screening results, which were more commonly utilized at the time of SCRN data collection compared to the present. The best model was chosen as that which optimized AUROC and sensitivity.
Statistical analysis
All analyses were conducted using RStudio (version 2022.07.1, R version 4.1.3). In addition to those statistical measures collected for each model listed above, we also completed sample descriptive analyses comparing stillbirths to livebirths for each measure. We utilized Mann-Whitney tests for continuous variables and Chi-Square tests for categorical variables. P-values <0.05 were considered significant.
Results
Sample description
The SCRN data consisted of 3000 livebirths and 982 stillbirths. Though all were eligible for inclusion in the primary model, 924 births (586 livebirths, 338 stillbirths) did not have sufficient prenatal visit data from later pregnancy and were thus excluded from secondary models that included data from the entirety of pregnancy. Of the 101 measures with <80% missing data assessed, 83 were collected prior to viability (22–24 weeks) and were included in the pre-viability model, and 44 were selected for inclusion in the EFA-derived model. Sample and variable selection are depicted in Supplemental Figure 1.
Measure comparisons between stillbirths and livebirths are depicted in Supplemental Table 1. Notable measures with significantly lower mean ± standard deviation/incidence in stillbirths included positive anemia screen during early pregnancy (defined as hematocrit <33%, per World Health Organization guidelines for anemia during any trimester of pregnancy), preterm labor, anemia during labor/delivery, and ratios of latest prenatal visit and total prenatal visits to birth GA.32 Notable measures with significantly higher mean ± standard deviation/incidence in stillbirths included maternal minority race; maternal and family history of fetal death and/or recurrent pregnancy loss; significant life events 12 months prior to delivery (adapted from the Pregnancy Risk Assessment Monitoring System, which has been validated for use in assessing the contributions of self-reported stressful life events in the year prior to childbirth towards adverse pregnancy outcomes of the index pregnancy); leukocytosis during labor and delivery (defined as white blood cell count > 11×109/L); 2nd trimester maternal serum alpha-fetoprotein (MSAFP) Multiples of the Median (MoM); prenatal fluid loss, preterm labor, reduced fetal movement; ultrasound with fetal anomaly, abnormal fluid, or abnormal growth; and ratio of GA of latest ultrasound to birth GA.33,34
Primary Aim
Our primary aim was achieved via creation of a series of models that distinguished stillbirth from livebirth using data available prior to viability (Figure 1). Results from logistic regression with these variables yielded an AUROC of 0.761. The best pre-viability model utilized a Boosted Trees learner and had AUROC of 0.851, sensitivity of 0.852, specificity of 0.849, PPV of 0.883, and NPV of 0.811. Type I error rate was 14.8%; type II error rate was 15.1%. Model time to completion was variable according to processor speed and required 3.94 kilobytes (kB) of memory.
Figure 1: Model confusion matrices.
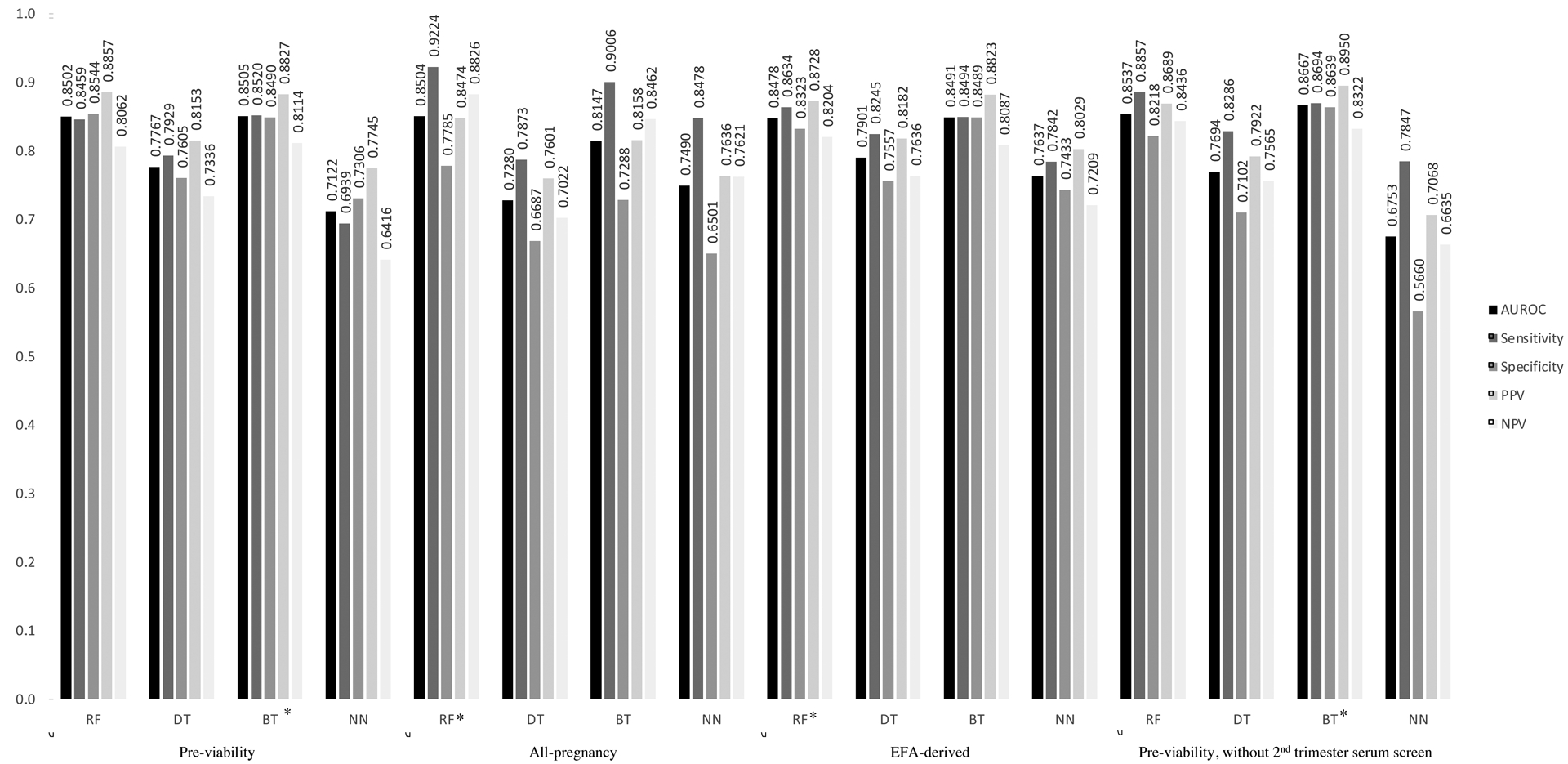
Legend: Area under receiver operating characteristic (AUROC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for each model. Models listed are: Random Forests (RF), Decision Trees (DT), Boosted Trees (BT), and Neural Networks (NN). Models listed including pre-viability variables, all-pregnancy variables, Exploratory Factor Analysis (EFA)-derived variables, and pre-viability variables without 2nd trimester serum screening. Asterisk (*) indicates best model for the given set of variables.
Secondary Aim: alternate models
Compared to AUROC of 0.863 from logistic regression, the best all-pregnancy model utilized a Random Forests learner and had AUROC of 0.850, sensitivity of 0.922, specificity of 0.779, PPV of 0.847, and NPV of 0.883. Compared to AUROC of 0.747 from logistic regression, the best EFA-derived model utilized a Random Forests learner and had AUROC of 0.867, sensitivity of 0.869, specificity of 0.864, PPV of 0.895, and NPV of 0.832. Compared to AUROC of 0.741 from logistic regression, the best pre-viability model without serum screening for genetic aneuploidy utilized a Boosted Trees learner and had AUROC of 0.867, sensitivity of 0.869, specificity of 0.864, PPV of 0.895, and NPV of 0.832.
Secondary Aim: variable importance
Variable importance for pre-viability and all-pregnancy models is shown in Table 1. The following 10 variables were most contributory to AUROC in the pre-viability model (from most to least important): maternal minority race, GA of earliest prenatal visit, paternal minority race, GA of earliest ultrasound, history of intrauterine fetal demise, 2nd trimester human chorionic gonadotropin (hCG), income assistance received, positive anemia screen, BFOB years of education, and BFOB Hispanic ethnicity. Though the most important variables for the pre-viability model are highlighted above, a multitude of other variables, including ultrasound evidence of abnormal fluid (oligo- or polyhydramnios) were important for other models and are listed in Tables 1 and 2.
Table 1:
Variable importance for pre-viability & all-pregnancy models
| Category | Measure | Pre-viability | All-pregnancy | Pre-viability without 2nd trimester screening |
|---|---|---|---|---|
| Demographics (maternal) | Age (years) | 3.93 | 26.0 | 7.62 |
| Education (years) | 10.9 | 18.7 | 26.4 | |
| Ethnicity (Hispanic) | 1.54 | 14.9 | 3.91 | |
| Race (minority) | 100.0 | 40.7 | 100.0 | |
| Born in United States | 2.99 | 3.83 | 2.07 | |
| Interview conducted in English | 0.00 | 1.96 | 0.23 | |
| Demographics (biologic father of baby) | Ethnicity (Hispanic) | 13.8 | 13.8 | 12.6 |
| Race (minority) | 56.5 | 11.1 | 80.2 | |
| Education (years) | 14.4 | 18.7 | 10.1 | |
| Born in United States | 0.41 | 6.41 | 0.57 | |
| Family history | Chromosomal anomaly | 0.31 | 0.82 | 0.59 |
| Congenital disorder | 0.25 | 1.48 | 0.00 | |
| Developmental delay | 0.77 | 0.75 | 1.21 | |
| Congenital heart disease | 2.39 | 1.50 | 4.18 | |
| Neonatal death | 2.29 | 4.77 | 5.00 | |
| Blood disorder | 0.20 | 1.77 | 0.98 | |
| Neuromuscular disorder | 0.00 | 10.4 | 0.00 | |
| Intrauterine fetal demise | 5.52 | 10.8 | 5.11 | |
| Recurrent pregnancy loss | 1.37 | 1.73 | 2.78 | |
| Diabetes | 0.32 | 2.98 | 0.00 | |
| Maternal health history | Any chronic condition | 0.00 | 13.3 | 4.19 |
| Asthma | 0.55 | 1.87 | 0.24 | |
| Blood disorder | 0.00 | 14.2 | 0.00 | |
| Cardiovascular disease | 0.82 | 12.6 | 1.46 | |
| Gastrointestinal disorder | 0.29 | 6.87 | 0.00 | |
| Hypertension (pre-pregnancy) | 0.80 | 1.83 | 1.58 | |
| Diabetes (pre-pregnancy) | 1.30 | 1.30 | 2.13 | |
| Obesity (body mass index ≥30) | 1.46 | 4.98 | 5.41 | |
| Rheumatologic disease | 0.00 | 11.5 | 0.00 | |
| Kidney disease | 0.59 | 15.5 | 0.54 | |
| Thyroid disease | 0.00 | 0.58 | 0.00 | |
| Mental health condition | 0.00 | 7.59 | 0.00 | |
| Depression | 0.44 | 8.64 | 0.28 | |
| Sexually transmitted infection | 0.46 | 1.62 | 0.22 | |
| Surgery | 0.36 | 8.73 | 2.81 | |
| Urinary tract infection | 0.25 | 3.36 | 7.21 | |
| Obstetric history | Intrauterine fetal demise | 28.8 | 10.9 | 54.3 |
| Fertility treatment | 1.37 | 2.25 | 0.00 | |
| Spontaneous abortion | 3.60 | 3.50 | 3.05 | |
| Total pregnancies (number) | 8.45 | 11.1 | 16.9 | |
| Social history | Received income assistance | 17.5 | 10.5 | 17.0 |
| Significant life events 12 months prior to delivery | 13.5 | 9.83 | 14.9 | |
| Marital status (married) | 3.19 | 16.9 | 1.92 | |
| Moved during pregnancy | 9.64 | 4.84 | 8.08 | |
| Biologic father of baby involved in pregnancy care | 0.00 | 5.23 | 0.00 | |
| Partner desired pregnancy | 2.24 | 10.5 | 2.05 | |
| Patient desired pregnancy | 1.97 | 19.0 | 1.23 | |
| Unstable living situation | 2.44 | 1.13 | 3.02 | |
| Substance use | Smoking during pregnancy | 0.80 | 2.30 | 0.97 |
| Alcohol during pregnancy | 5.03 | 17.2 | 2.75 | |
| Other drug use during pregnancy | 0.00 | 0.43 | 0.00 | |
| Prenatal laboratory data | Anemia screen (positive) | 16.6 | 8.54 | 36.6 |
| Rubella screen (positive) | 0.21 | 0.61 | 0.00 | |
| Chlamydia (positive) | 0.22 | 0.63 | 1.36 | |
| Diabetic screen (positive) | Not in model | 0.98 | Not in model | |
| Anti-D antibodies | 0.00 | 0.90 | 2.20 | |
| Urine culture (positive) | Not in model | 2.52 | Not in model | |
| Bleeding disorder (labor/delivery) | Not in model | 11.0 | Not in model | |
| Anemia (labor/delivery) | Not in model | 20.9 | Not in model | |
| Leukocytosis (labor/delivery) | Not in model | 7.48 | Not in model | |
| Thrombocytopenia (labor/delivery) | Not in model | 1.34 | Not in model | |
| Proteinuria | Not in model | 8.77 | Not in model | |
| 2nd trimester (quad) screen | Down syndrome risk | 3.62 | 23.6 | Not in model |
| hCG (MoM) | 23.1 | 44.3 | Not in model | |
| MSAFP (MoM) | 12.6 | 27.2 | Not in model | |
| UE3 (MoM) | 1.66 | 28.6 | Not in model | |
| Prenatal care | Fluid loss | Not in model | 1.76 | Not in model |
| Abdominal pain (prior to onset of labor) | Not in model | 23.4 | Not in model | |
| Vaginal bleeding | Not in model | 5.15 | Not in model | |
| Hospitalization (prior to delivery) | Not in model | 25.5 | Not in model | |
| Preterm labor (prior to delivery) | Not in model | 3.42 | Not in model | |
| Reduced fetal movement | Not in model | 13.4 | Not in model | |
| Elevated blood pressure | Not in model | 8.05 | Not in model | |
| Influenza during pregnancy | Not in model | 9.30 | Not in model | |
| Earliest prenatal visit (GA) | 67.7 | 57.0 | 64.8 | |
| Ratio of latest prenatal visit to bGA | Not in model | 92.9 | Not in model | |
| Ratio of total prenatal visits to bGA | Not in model | 26.0 | Not in model | |
| GA at which pregnancy discovered | 11.7 | 27.2 | 24.5 | |
| Ultrasound | Fetal anomaly | 6.88 | 1.53 | 12.0 |
| Sex (female) | 5.64 | 29.9 | 9.85 | |
| 10-week ultrasound (completed) | 2.80 | 9.70 | 0.75 | |
| Anatomy ultrasound (completed) | 0.00 | 17.6 | 9.95 | |
| Abnormal fluid (oligohydramnios or polyhydramnios) | Not in model | 16.1 | Not in model | |
| Abnormal growth (large [>90th percentile] or small [<10th percentile] for gestational age) | Not in model | 1.13 | Not in model | |
| Placental anomalies | Not in model | 1.23 | Not in model | |
| Earliest ultrasound (GA) | 46.0 | 60.7 | 66.4 | |
| Ratio of latest ultrasound to bGA | Not in model | 100.0 | Not in model |
Values are given as variable importance (range 0–100). Values >10 (higher importance to model accuracy) indicated in bold.
Non-contributory to model (importance <1.0): Family history of cystic fibrosis, neural tube defect; maternal health history of cancer, seizures; history of emotional, physical, or sexual abuse; positive gonorrhea, syphilis, or antibody screen
Abbreviations: hCG = human chorionic gonadotropin; MoM = multiples of median; MSAFP = maternal serum alpha-feto protein; UE3 = unconjugated estriol; GA = gestational age; bGA = gestational age at birth/delivery
Table 2:
Variable importance for exploratory factor analysis-derived model
| Category | Measure | EFA-derived |
|---|---|---|
| Demographics (maternal) | Age (years) | 47.5 |
| Ethnicity (Hispanic) | 16.5 | |
| Race (minority) | 61.7 | |
| Born in United States | 7.48 | |
| Interview conducted in English | 4.34 | |
| Demographics (biologic father of baby) | Ethnicity (Hispanic) | 23.2 |
| Race (minority) | 28.0 | |
| Education (years) | 43.1 | |
| Born in United States | 8.17 | |
| Maternal health history | Any chronic condition | 7.18 |
| Blood disorder | 4.13 | |
| Cardiovascular disease | 3.84 | |
| Gastrointestinal disorder | 4.55 | |
| Rheumatologic disease | 4.94 | |
| Kidney disease | 3.65 | |
| Mental health condition | 5.09 | |
| Depression | 6.26 | |
| Sexually transmitted infection | 5.75 | |
| Surgery | 11.1 | |
| Social history | Received income assistance | 11.7 |
| Significant life events 12 months prior to pregnancy | 21.2 | |
| Marital status (married) | 8.81 | |
| Biologic father of baby involved in pregnancy care | 5.45 | |
| Partner desired pregnancy | 6.22 | |
| Patient desired pregnancy | 13.3 | |
| Substance use | Alcohol during pregnancy | 10.1 |
| Other drug use during pregnancy | 2.23 | |
| Prenatal laboratory data | Anemia screen (positive) | 13.0 |
| Chlamydia (positive) | 1.52 | |
| Bleeding disorder (labor/delivery) | 11.2 | |
| Anemia (labor/delivery) | 11.8 | |
| Leukocytosis (labor/delivery) | 11.0 | |
| Proteinuria | 16.1 | |
| Prenatal care | Abdominal pain (prior to onset of labor) | 7.34 |
| Elevated blood pressure | 7.43 | |
| Influenza during pregnancy | 7.22 | |
| Earliest prenatal visit (GA) | 97.3 | |
| Ratio of total prenatal visits to bGA | 51.3 | |
| GA at which pregnancy discovered | 55.8 | |
| Ultrasound | 10-week ultrasound (completed) | 6.71 |
| Anatomy ultrasound (completed) | 11.1 | |
| Earliest ultrasound (GA) | 100.0 |
Values are given as variable importance (range 0–100). Values >10 (higher importance to model accuracy) indicated in bold.
Abbreviations: GA = gestational age; bGA = gestational age at birth/delivery
Comment
Principal Findings
With 85.1% AUROC, we were able to utilize machine learning to identify pre-viable participants at risk for stillbirth in a nationwide dataset. Using measures collected throughout pregnancy results in AUROC of 85.0%. Our analyses identified variables obtained prior to viability associated with increased risk of developing stillbirth, which included maternal minority race, prior stillbirth, GA of earliest prenatal visit and ultrasound, and 2nd trimester serum screening. Overall, our pilot models had high AUROC and sensitivity; in the context of preventing stillbirth, the bias towards sensitivity is preferred as this model is unlikely to miss those at risk for such a devastating event. These findings provide an important first step in developing a widespread risk calculator that will allow providers to accurately triage those at increased risk of stillbirth.
Results in the Context of What is Known
Predicting risk of stillbirth has been a longtime aim in clinical obstetric research, resulting in many publications utilizing simple modeling methods such as logistic regression.3,5–14,23,25,26,28,35,36 A more limited selection of literature has focused on utilizing sophisticated machine learning methods, which offer the ability to address non-linear relationships between measures in large datasets. Koivu et al. were able to achieve accuracy of 73–74% utilizing information from maternal medical and obstetric history, while Malacova et al. achieved maximum accuracy of 84% using measures collected from current pregnancy and maternal medical and obstetric history.17,18 Neither study included ultrasound or fetal genetic information in their models, even though data obtained via prenatal ultrasound and fetal genetic screening or testing can be pivotal in increasing risk of subsequent stillbirth.37 Khatibi et al. utilized sophisticated feature selection (via K-means cluster analysis) to identify variables of interest and enter these into machine learning classifiers.19 While these authors were able to achieve high model accuracy (up to 90.6%), they did so using measures such as GA at delivery, which is not clinically relevant in predicting cases other than intrapartum stillbirths.
As mentioned previously, our study included ultrasound and fetal genetic information, ubiquitous diagnostic tools that will likely continue to advance, offering us the opportunity to expand our model as technology becomes more widely utilized. Furthermore, unlike other machine learning models, we sought to identify patients at risk for stillbirth using data available prior to viability, rather than at or around delivery. Though delivery-based prediction is valuable in expediting delivery, a pre-viability model offers the opportunity to intervene earlier (at or before the usual initiation of antenatal testing, 28 weeks) with less invasive methods such as fetal surveillance for patients at highest risk, hopefully preventing subsequent stillbirth as early as the extreme prematurity period. However, before this or other machine learning algorithms can be employed in clinical settings for in vivo risk prediction, these algorithms must first be validated prospectively. Then, if high rates positive prediction persist in prospective studies, clinical guidelines can be developed that incorporate these models to predict risk of stillbirth at the level of individual patients.
Clinical Implications
Our model offers a highly accurate tool for predicting stillbirth pre-viability, which, when validated and refined, could be easily implemented as a risk calculator for clinicians during prenatal visits. While concerns exist regarding computational requirements for running such models, machine learning tools have been successfully implemented in the clinical setting in a number of fields, including radiology and critical care.38,39 Implementation of such models have, furthermore, been associated with improvements in clinical workflow despite possible increased work burden or system requirements.40 Our pre-viability model requires 3.94kB of memory, which is reasonable and feasible to use in most microcontrollers and larger devices for patient-level computations.41 Though automated data extraction from the electronic health record to provide real-time risk prediction poses its own challenges, several risk calculators—such as the Sequential Organ Failure Assessment to provide real-time identification for organ failure and a risk calculator for automatic detection of people at risk for psychosis—have been successfully developed and implemented.42,43 Once this calculator has been prospectively validated in a novel database, we will aim to incorporate the streamlined, embedded risk predictor into commonly used electronic health records.
Recent attention has been drawn to the effect of involving predictive modeling on clinical counseling and patient experience. Several recent presentations showed that patients had varying feelings surrounding the use of a calculator to predict successful VBAC.44,45 Moreover, clinicians cited use of such a calculator more often to discourage rather than encourage a trial of labor.46 Given that stillbirth is a devastating and anxiety-provoking event, it would be important to consider the effect of receiving any result from a predictive stillbirth model on patient care. Stillbirth counseling is often multidisciplinary, and such counseling would have to be adjusted in the setting of a new predictive calculator.47 Further studies using our algorithm will assess the necessary support structures for patients and clinicians in order for them to utilize a stillbirth calculator.
The results of our variable importance analyses (secondary aim) indicate that certain demographic and pregnancy history measures are more important in predicting stillbirth than others. One of the most important measures was history of past stillbirth, which is to be expected: those with prior stillbirth have 2–10 times higher risk of recurrent stillbirth in a subsequent pregnancy compared to those without this history.47 Self-reported maternal minority race was important in the accuracy of our model. Black women have a two-fold higher risk for stillbirth even when controlling for other contributing factors, largely attributable to structural factors that perpetuate healthcare inequities according to race, especially structural racism.48–51 GA at first/earliest prenatal visit was significant within our model; when examining the variance between stillbirths and livebirths in this measure, stillbirths had an earlier GA at first prenatal visit than livebirths. Past literature has suggested a relationship between underutilization of prenatal care and risk for stillbirth; though our stillbirth cohort presented earlier to care than our livebirth cohort, this does not necessarily indicate more adherence to recommended prenatal care schedules, which may impact stillbirth risk.52–55 It is also possible that these patients presented earlier to care due to early pregnancy complications that predisposed them to ultimately having a stillbirth such as bleeding, pregnancy of unknown location, or hyperemesis gravidarum. None of these datapoints were included in our analyses as they were not comprehensively collected in the parent database. However, the impact of such conditions on prenatal care utilization and subsequent stillbirth risk will be investigated in future iterations of this work.56
Research Implications
The future directions for this work are expansive given the opportunities such an accurate model provides. The model first must be validated in a database with representative stillbirth rates (as the SCRN database oversampled for stillbirths) and then prospectively using an external dataset; this process will ensure that its predictive abilities are as strong in other populations. Once validated, the model may be implemented in the setting of a randomized control trial, which will yield further information for researchers not just regarding the utility of the model, but also patient and clinician perspectives on this predictive score and possibly the most effective interventions for preventing stillbirth (e.g., content/quality of prenatal care, antenatal testing, serial ultrasounds, or timing of induction of labor).
Strengths and Limitations
Our model has several strengths of note. The SCRN dataset is unusually granular, allowing us to include high-quality metrics such as ultrasound data that are not incorporated into other predictive models.17,19,37 Our methodology is also balanced between sophisticated (utilizing a systematic approach to identify the best learner from a series of models) and accessible (easily implemented using limited memory). Our model is also interpretable: by producing results that include variable importance, we avoid the “black box” of machine learning wherein results are derived without an understanding of contributing factors, which can be valuable in a clinical setting.57
Our model also has several limitations. First, the SCRN study enrollment intentionally oversampled stillbirths, and controls were also biased towards higher-risk population (including a large proportion of preterm livebirths with obstetric complications). Use of this model in low-risk cohorts may result in lower PPV, although AUROC, sensitivity, and specificity should be maintained. Therefore, validation of the model in an external, generalizable cohort remains our primary next step; this validation will also allow us to assess externally whether any of our models were overfit. Second, the SCRN data included complete 2nd trimester (quad) screen, which was a routine part of prenatal care in the United States when the SCRN database was created but is less commonly utilized now. Therefore, we ran additional models without this screening panel, which yielded similar model metrics. Current American College of Obstetricians and Gynecologists guidelines suggest that non-invasive prenatal testing for aneuploidy should be offered to all patients regardless of age or genetic risk.58 While our primary model is applicable for patients and communities where quad screening is still performed, inclusion of the current schema (cell-free DNA ± nuchal translucency ± MSAFP) is a high-priority for future iterations of this work.59 We expect this to increase the predictive accuracy of our model, especially if metrics such as fetal fraction or even results of whole genome sequencing are included.60 We also recognize that several novel biomarkers, including soluble fms-like tyrosine kinase 1 (sFlt-1) to placental growth factor (PlGF) ratio, have been identified for predicting adverse pregnancy outcomes; these may have utility for inclusion in future iterations of this model as well.61
It is possible that some variables included in our model—such as leukocytosis—may occur more often among stillbirth pregnancies due to presence of an infection that may have contributed to fetal demise or as an inflammatory response to a retained stillbirth. However, this possibility is less likely give the rates of leukocytosis among those with stillbirth and livebirth were clinically similar in the parent database (32.7% livebirths versus 38.7% of stillbirths). Finally, though our dataset is robust, including stillbirths of many etiologies and gestational ages, it does not account for the clinical decision making that providers may have already been doing to prevent stillbirth in high-risk patients, which may explain why clinical factors with clear associations with stillbirth were not found to be significant in our model. For example, pregnancies with known fetal growth restriction may have undergone close fetal antenatal surveillance, thereby reducing their risk of stillbirth.
Conclusions
Using a comprehensive dataset of stillbirths and livebirths, sophisticated machine learning techniques created a pilot model with 85.1% AUROC for predicting stillbirth using data available prior to viability. Once validated in generalizable datasets and prospectively, such a model holds great promise for serving as a clinical decision-making tool, allowing providers to assess an individual patient’s risk of stillbirth and design an individual surveillance plan in the hope of preventing this devastating pregnancy outcome.
Supplementary Material
AJOG at a glance:
A. Why was this study conducted?
Prior models for predicting stillbirth do not leverage the nuanced techniques involved in sophisticated machine learning methods and incorporate data points after viability.
Using data derived from the Stillbirth Collaborative Research Network, we sought to develop a machine learning model for predicting stillbirth using data available prior to viability.
B. What are the key findings?
Using data derived prior to viability, our model predicted stillbirth had high predictive ability to identify a patient that would go on to have a stillbirth versus a livebirth (area under the curve [AUC] 85.1%; sensitivity 85.2%, specificity 84.9%, positive predictive value [PPV] 88.3%, negative predictive value [NPV] 81.1%), likelihood ratio [LR] 5.64.
Machine learning models that incorporated variables collected throughout pregnancy—including after viability—resulted in AUC of 85.0%.
C. What does this study add to what is already known?
Few sophisticated machine learning models for predicting stillbirth risk have been created.
Our pilot model uses novel variables, including ultrasound and fetal genetics, and provides a framework for prospective validation that may, should results remain similar to those produced by the current model presented here, help to accurately predict stillbirths using data available prior to viability.
Once externally validated, our model may be used for clinical decision-making support.
Funding:
Eunice Kennedy Shriver National Institute of Child Health and Human Development: U10-HD045953 Brown University, Rhode Island; U10-HD045925 Emory University, Georgia; U10-HD045952 University of Texas Medical Branch at Galveston, Texas; U10-HDO45955 University of Texas Health Sciences Center at San Antonio, Texas; U10-HD045944 University of Utah Health Sciences Center, Utah; and U01-HD045954 RTI International, RTP. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure statement: The authors report no conflicts of interest.
Presentation: 43rd Annual Pregnancy Meeting of the Society for Maternal-Fetal Medicine. San Francisco, CA. February 6 – 11, 2023
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tess E.K. CERSONSKY, Warren Alpert Medical Medical School of Brown University/Women & Infants Hospital of Rhode Island, Providence, RI.
Nina K. AYALA, Warren Alpert Medical Medical School of Brown University/Women & Infants Hospital of Rhode Island, Providence, RI.
Halit PINAR, Warren Alpert Medical Medical School of Brown University/Women & Infants Hospital of Rhode Island, Providence, RI.
Donald J. DUDLEY, University of Virginia, Charlottesville, VA.
George R. SAADE, University of Texas Medical Branch, Galveston, TX.
Robert M. SILVER, University of Utah Health, Salt Lake City, UT.
Adam K. LEWKOWITZ, Warren Alpert Medical Medical School of Brown University/Women & Infants Hospital of Rhode Island, Providence, RI.
References
- 1.Ota E et al. Antenatal interventions for preventing stillbirth, fetal loss and perinatal death: an overview of Cochrane systematic reviews. Cochrane Database of Systematic Reviews 2020, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishak M & Khalil A Prediction and prevention of stillbirth: Dream or reality. Curr Opin Obstet Gynecol 33, 405–411 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Smith GCS et al. Maternal and biochemical predictors of antepartum stillbirth among nulliparous women in relation to gestational age of fetal death. BJOG 114, 705–714 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Allen RE et al. Predictive accuracy of second-trimester uterine artery Doppler indices for stillbirth: a systematic review and meta-analysis. Ultrasound in Obstetrics & Gynecology 47, 22–27 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Townsend R et al. Can risk prediction models help us individualise stillbirth prevention? A systematic review and critical appraisal of published risk models. BJOG 128, 214–224 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Townsend R et al. Prediction of stillbirth: an umbrella review of evaluation of prognostic variables. BJOG 128, 238–250 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Conde-Agudelo A, Bird S, Kennedy SH, Villar J & Papageorghiou AT First- and second-trimester tests to predict stillbirth in unselected pregnant women: a systematic review and meta-analysis. BJOG 122, 41–55 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kayode GA et al. Predicting stillbirth in a low resource setting. BMC Pregnancy Childbirth 16, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trudell AS, Tuuli MG, Colditz GA, Macones GA & Odibo AO A stillbirth calculator: Development and internal validation of a clinical prediction model to quantify stillbirth risk. PLoS One 12, e0173461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasupathy D & Smith GCS The analysis of factors predicting antepartum stillbirth. Minerva Ginecol 57, 397–410 (2005). [PubMed] [Google Scholar]
- 11.Reddy UM Prediction and prevention of recurrent stillbirth. in Stillbirth 65–74 (CRC Press, 2010). doi: 10.3109/9781841847191-7. [DOI] [PubMed] [Google Scholar]
- 12.Smith GCS Predicting antepartum stillbirth. Curr Opin Obstet Gynecol 18, 625–630 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Smith GCS Predicting antepartum stillbirth. Clin Obstet Gynecol 53, 597–606 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Akolekar R, Tokunaka M, Ortega N, Syngelaki A & Nicolaides KH Prediction of stillbirth from maternal factors, fetal biometry and uterine artery Doppler at 19–24 weeks. Ultrasound Obstet Gynecol 48, 624–630 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Mboya IB, Mahande MJ, Mohammed M, Obure J & Mwambi HG Prediction of perinatal death using machine learning models: a birth registry-based cohort study in northern Tanzania. Paediatrics 10, e040132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidey-Gibbons JAM & Sidey-Gibbons CJ Machine learning in medicine: a practical introduction. BMC Med Res Methodol 19, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koivu A & Sairanen M Predicting risk of stillbirth and preterm pregnancies with machine learning. Health Inf Sci Syst 8, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malacova E et al. Stillbirth risk prediction using machine learning for a large cohort of births from Western Australia, 1980–2015. Scientific Reports 2020 10:1 10, 1–8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatibi T, Hanifi E, Sepehri MM & Allahqoli L Proposing a machine-learning based method to predict stillbirth before and during delivery and ranking the features: nationwide retrospective cross-sectional study. BMC Pregnancy Childbirth 21, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipschuetz M et al. Prediction of vaginal birth after cesarean deliveries using machine learning. Am J Obstet Gynecol 222, 613.e1–613.e12 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Parker CB et al. Stillbirth Collaborative Research Network: design, methods and recruitment experience. Paediatr Perinat Epidemiol 25, 425–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephansson O, Dickman PW, Johansson A & Cnattingius S Maternal Hemoglobin Concentration During Pregnancy and Risk of Stillbirth. JAMA 284, 2611–2617 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Berhie KA & Gebresilassie HG Logistic regression analysis on the determinants of stillbirth in Ethiopia. Maternal Health, Neonatology and Perinatology 2016 2:1 2, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalafat E, Ozturk E, Sivanathan J, Thilaganathan B & Khalil A Longitudinal change in cerebroplacental ratio in small-for-gestational-age fetuses and risk of stillbirth. Ultrasound in Obstetrics & Gynecology 54, 492–499 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Familiari A et al. Mid-pregnancy fetal growth, uteroplacental Doppler indices and maternal demographic characteristics: role in prediction of stillbirth. Acta Obstet Gynecol Scand 95, 1313–1318 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Allotey J et al. External validation of prognostic models to predict stillbirth using International Prediction of Pregnancy Complications (IPPIC) Network database: individual participant data meta-analysis. Ultrasound in Obstetrics & Gynecology 59, 209–219 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Khalil A et al. Value of third-trimester cerebroplacental ratio and uterine artery Doppler indices as predictors of stillbirth and perinatal loss. Ultrasound in Obstetrics & Gynecology 47, 74–80 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Muin DA, Windsperger K, Attia N & Kiss H Predicting singleton antepartum stillbirth by the demographic Fetal Medicine Foundation Risk Calculator—A retrospective casecontrol study. PLoS One 17, e0260964 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Buuren S & Groothuis-Oudshoorn K mice: Multivariate Imputation by Chained Equations in R. J Stat Softw 45, 1–67 (2011). [Google Scholar]
- 30.Cudeck R Exploratory Factor Analysis. in Handbook of Applied Multivariate Statistics and Mathematical Modeling (eds. Tinsley HEA & Brown SD) 265–296 (Academic Press, 2000). doi: 10.1016/B978-012691360-6/50011-2. [DOI] [Google Scholar]
- 31.Xiong Z et al. Evaluating explorative prediction power of machine learning algorithms for materials discovery using k-fold forward cross-validation. Comput Mater Sci 171, 109203 (2020). [Google Scholar]
- 32.WHO. Iron deficiency anaemia: assessment, prevention and control. (2001).
- 33.Hall KS, Dalton VK, Zochowski M, Johnson TRB & Harris LH Stressful life events around the time of unplanned pregnancy and women’s health: Exploratory findings from a national sample. Matern Child Health J 21, 1336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mank V, Azhar W & Brown K Leukocytosis. in StatPearls (StatPearls Publishing, 2023). [PubMed] [Google Scholar]
- 35.Whitney E, Phatak A & Pereira G Predicting stillbirth using LASSO with structured penalties. Proceedings of the 34th International Workshop on Statistical Modelling, Volume II 405–409 (2020). [Google Scholar]
- 36.Goy J, Dodds L, Rosenberg MW & King WD Health-risk behaviours: examining social disparities in the occurrence of stillbirth. Paediatr Perinat Epidemiol 22, 314–320 (2008). [DOI] [PubMed] [Google Scholar]
- 37.American College of Obstetricians and Gynecologists. Obsetric Care Consensus 10: Management of Stillbirth. Obstetrics & Gynecology 135, e110–e132 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Wei X & Eickhoff C Embedding Electronic Health Records for Clinical Information Retrieval. (2018) doi: 10.48550/arxiv.1811.05402. [DOI] [Google Scholar]
- 39.Lee TC, Shah NU, Haack A & Baxter SL Clinical Implementation of Predictive Models Embedded within Electronic Health Record Systems: A Systematic Review. Informatics 2020, Vol. 7, Page 25 7, 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sendak M, Gao M, Nichols M, Lin A & Balu S Machine Learning in Health Care: A Critical Appraisal of Challenges and Opportunities. eGEMs 7, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha SS, Sandha SS & Srivastava M Machine Learning for Microcontroller-Class Hardware: A Review. IEEE Sens J 22, 21362–21390 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver D et al. Real-world implementation of precision psychiatry: Transdiagnostic risk calculator for the automatic detection of individuals at-risk of psychosis. Schizophr Res 227, 52–60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aakre C et al. Prospective validation of a near real-time EHR-integrated automated SOFA score calculator. Int J Med Inform 103, 1–6 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Hamm RF, Wang E, Szymczak JE & Levine LD Implementation of a calculator to predict cesarean during labor induction: the patient perspective. Am J Obstet Gynecol 228, S66–S67 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubashkin N “You don’t really know until you try”: VBAC prediction from the patient perspective. Am J Obstet Gynecol 226, S531 (2022). [Google Scholar]
- 46.Thornton PD, Liese K, Adlam K, Erbe K & McFarlin BL Calculators Estimating the Likelihood of Vaginal Birth After Cesarean: Uses and Perceptions. J Midwifery Womens Health 65, 621–626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy UM Management of pregnancy after stillbirth. Clin Obstet Gynecol 53, 700–709 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Leisher SH Exploring racial disparity in stillbirth rates through structural racism and methylation of stress-related genes: From systemic to epigenetic. Columbia Academic Commons (2023). doi: 10.7916/3BM5-0867. [DOI] [Google Scholar]
- 49.Williams AD, Wallace M, Nobles C & Mendola P Racial residential segregation and racial disparities in stillbirth in the United States. Health Place 51, 208–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willinger M, Ko CW & Reddy UM Racial disparities in stillbirth risk across gestation in the United States. Am J Obstet Gynecol 201, 469.e1–469.e8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arechvo A et al. Maternal Race and Stillbirth: Cohort Study and Systematic Review with Meta-Analysis. J Clin Med 11, 3452 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Partridge S, Balayla J, Holcroft CA & Abenhaim HA Inadequate prenatal care utilization and risks of infant mortality and poor birth outcome: A retrospective analysis of 28,729,765 U.S. deliveries over 8 years. Am J Perinatol 29, 787–793 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Faiz AS, Demissie K, Rich DQ, Kruse L & Rhoads GG Trends and risk factors of stillbirth in New Jersey 1997-2005. 10.3109/14767058.2011.596593 25, 699–705 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Reime B et al. Does underutilization of prenatal care explain the excess risk for stillbirth among women with migration background in Germany? Acta Obstet Gynecol Scand 88, 1276–1283 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Cersonsky TEK et al. Adherence to recommended prenatal visits and stillbirth risk: a Stillbirth Collaborative Research Network secondary analysis. Am J Obstet Gynecol 228, S238–S239 (2023). [Google Scholar]
- 56.Fiaschi L, Nelson-Piercy C, Gibson J, Szatkowski L & Tata LJ Adverse Maternal and Birth Outcomes in Women Admitted to Hospital for Hyperemesis Gravidarum: a Population-Based Cohort Study. Paediatr Perinat Epidemiol 32, 40–51 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence 2019 1:5 1, 206–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.American College of Obstetricians and Gynecologists. Practice Bulletin Number 226: Screening for Fetal Chromosomal Abnormalities. Obstetrics & Gynecology 136, e48–e69 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Schmidt A & Shanks A Quadruple Screening in the Age of Cell-Free DNA: What are We Losing? OBM Genetics 2021, Vol. 5, 138 5, 1–15 (2021). [Google Scholar]
- 60.Konuralp Atakul B et al. Could high levels of cell-free DNA in maternal blood be associated with maternal health and perinatal outcomes? 10.1080/01443615.2019.1671324 40, 797–802 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Zeisler H et al. Predictive Value of the sFlt-1:PlGF Ratio in Women with Suspected Preeclampsia. N Engl J Med 374, 13–22 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


