Abstract
Exercise changes the tumor microenvironment by remodeling blood vessels and increasing infiltration by cytotoxic immune cells. The mechanisms driving these changes remain unclear. Herein, we demonstrate that exercise normalizes tumor vasculature and upregulates endothelial expression of VCAM1 in YUMMER 1.7 and B16F10 murine models of melanoma but differentially regulates tumor growth, hypoxia, and the immune response. We found that exercise suppressed tumor growth and increased CD8+ T-cell infiltration in YUMMER but not in B16F10 tumors. Single-cell RNA sequencing and flow cytometry revealed exercise modulated the number and phenotype of tumor-infiltrating CD8+ T cells and myeloid cells. Specifically, exercise caused a phenotypic shift in the tumor-associated macrophage population and increased the expression of major histocompatibility complex class II transcripts. We further demonstrated that ERK5 S496A knock-in mice, which are phosphorylation deficient at the S496 residue, ‘mimicked’ the exercise effect when unexercised, yet when exercised, these mice displayed a reversal in the effect of exercise on tumor growth and macrophage polarization compared to wild-type mice. Taken together, our results reveal tumor-specific differences in the immune response to exercise and show that ERK5 signaling via the S496 residue plays a crucial role in exercise-induced tumor microenvironment changes.
Keywords: aerobic exercise, melanoma, vascular remodeling, tumor immune infiltrate, single cell RNA sequencing
INTRODUCTION
High levels of physical activity reduce the risk of multiple types of cancer [1, 2], and aerobic exercise improves psychosocial and physiological outcomes including quality of life, fatigue, and physical functioning for patients undergoing cancer treatment [3, 4]. Pre-clinical studies have demonstrated that aerobic exercise modulates the tumor microenvironment by remodeling tumor vasculature, altering hypoxia levels, and increasing cytotoxic immune–cell infiltration [5–8]. However, the mechanisms underlying these changes remain largely unknown. Responses to exercise vary in different tumor models, but heterogeneity in exercise intervention design makes it difficult to determine whether this response variation is due to tumor type, exercise intervention, or both [9, 10]. These context dependent effects are likely related to the complexities of cell-to-cell interactions in the solid tumor microenvironment and the ability of exercise to promote functional changes across many cell types.
The tumor microenvironment is a complex system of multiple cell types, including vascular, immune, stromal, and tumor cells, which interact with one another to dictate tumor growth. Tumor vasculature is dysfunctional with hyper-permeable regions and heterogeneous perfusion patterns. Tumor vessels deliver oxygen, nutrients, and immune cells to solid tumors. However, tumor vessels are unresponsive to cytokines and largely anergic with low levels of expression of the adhesion molecules necessary for leukocyte extravasation [11]. Additionally, poor blood perfusion and overactive tumor cell oxygen consumption creates a hypoxic and hostile environment for cytotoxic immune cells and limits their function. As such, there is a strong link between tumor vascular function and immune suppression in the tumor microenvironment.
Extracellular-regulated protein kinase 5 (ERK5) is a key functional mediator of both endothelial cell (EC) and macrophage inflammation. While there is increasing effort to understand the role of ERK5 within tumor cells, ERK5 is largely unstudied in the cells of the tumor microenvironment. ERK5 is a MAP kinase protein activated in response to various stimuli including shear stress, cytokines, and growth factors [12]. ERK5 has multiple phosphorylation sites, each with different functional implications. Most well described is the “activating” site, an N-terminal TEY motif. TEY phosphorylation results in activation of ERK5 kinase and co-transcriptional activity. In response to cellular stressors, ERK5 is phosphorylated at residue serine 496 (S496) in ECs and macrophages. Phosphorylation at S496 causes inflammatory signaling, including increased EC expression of adhesion molecules like Vascular Cell Adhesion Molecule 1 (VCAM1), Intercellular Adhesion Molecule 1 (ICAM1), and E-selectin (SELE), and macrophage polarization [13, 14].
In this study, we report differences in the response to exercise in two phenotypically and genotypically distinct murine models of melanoma, B16F10 and YUMMER 1.7 (YUMMER). We found that exercise induced similar vascular remodeling yet disparate immune responses in the two models. We identify YUMMER as an “exercise-responsive” tumor in which exercise alone was able to reduce tumor growth. In characterizing the tumor microenvironment of B16F10 and YUMMER with single-cell RNA-sequencing (scRNA-Seq), we uncovered a shift in the phenotype of intratumoral T cells and tumor-associated macrophages (TAMs) in response to exercise. Further, we demonstrated that exercise inhibited ERK5 S496 phosphorylation in TAMs and that prevention of ERK5 S496 phosphorylation mimicked exercise-mediated tumor growth inhibition as well as phenotypic changes in macrophages in the tumor and bone marrow. Overall, a stark contrast was observed between the exercise-induced changes in wild-type mice and mice in which ERK5 could not be phosphorylated at S496.
METHODS
Cell culture
YUMMER 1.7 cells were provided by Dr. Jennifer Wargo (MD Anderson Cancer Center, Department of Genomic Medicine) in 2020 and B16F10 cells by Dr. Michael Davies (MD Anderson Cancer Center, Department of Translational Molecular Pathology) in 2018. Cell lines were cultured in DMEM (Gibco, 11965092) or DMEM/F12 (Gibco, 11320033) media containing 10% fetal bovine serum (Sigma-Aldrich, F2442), 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco, 15140122). YUMMER 1.7 media additionally contained non-essential amino acids. Cells were passaged with Trypsin-EDTA (Corning, 25–051-CI). All cell lines were cultured in an incubator with a stable environment at 37°C, relative humidity at 95%, and CO2 concentration at 5%. Cell lines were routinely confirmed as mycoplasma negative by qRT-PCR, and B16F10 cell line authentication was performed by Short Tandem Repeat profiling. YUMMER cells were not authenticated by our laboratory. Cells were used between passage 2 and 8.
L929 fibroblasts were purchased from ATCC (ATCC CCL-1). L929 cells were grown for three passages from cryogenic storage before seeding for collection of media. Cells were grown in 50 ml of high glucose DMEM containing 10% (vol/vol) FBS, 1 mM L-glutamine (Sigma Aldrich, 25030–081). Cells were kept in an incubator at 37°C with 95% relative humidity and 5% CO2 concentration. The medium was collected after 7 days of culture and replaced with 50 ml of fresh DMEM media for a subsequent 7 days. The two medium collections were then combined and sterile filtered before aliquoting into 50-ml falcon tubes and stored at −20°C.
Animal experiments and exercise regimen
The Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center approved all animal studies. Mice were housed in individually ventilated cages under pathogen-free conditions. Animals had free access to food and water and were kept on a 12-hr light/12-hr dark cycle. C57Bl/6 wild-type mice were purchased from MD Anderson Experimental Radiation Oncology or Jackson Laboratories. The ERK5 S496 knock-in (KI) mice were previously generated on the C57Bl/6 background [13] and were bred for use in this study.
600,000 YUMMER cells or 100,000 B16F10 cells in 100μL phosphate buffered saline (PBS) were injected subcutaneously into the flanks of 8–12 week old male mice. Mixed gender cohorts were used in ERK5 S496 KI experiments. When tumors reached approximately 50mm3 (5–6 days post injection in the YUMMER model and 7 days in the B16F10 model), mice began exercise. Exercise consisted of treadmill running for 45 minutes per day at 12 meters/minute for 12–14 consecutive days. Tumors were measured throughout experiments using calipers. At the end of the experiment, mice were euthanized approximately 12 hours after the final exercise session. PD1-specific antibody for in vivo use was purchased from BioXCell (BE0146, Clone RMP1-14), diluted in PBS, and administered to mice via intravenous injection. Three experiments were performed using the anti-PD1. In the first experiment using YUMMER-bearing mice, 250 μg per mouse were given on days 5, 7, 9, and 11 after palpable tumor was detected. In the next experiment, 100 μg per mouse were given on days 3, 5, and 7 after palpable tumor was detected. In the experiment using B16F10-bearing mice, 500 μg per mouse was given on day 5 after palpable tumor detection followed by 250 μg per mouse given on days 8, 10, and 12.
Tumor vessel hyper-permeability and perfusion assays
Ten minutes prior to mouse euthanasia, tumor-bearing mice were injected via tail vein with 100μl tomato-lectin (2mg/ml, DL-1177-1, VectorLab, Burlingame, CA) to measure vessel perfusion or high molecular weight fluorescein isothiocyanate (FITC)-Dextran (2,000kD mol weight, FD-2000S 10mg/ml; Sigma-Aldrich) to measure tumor vessel hyper-permeability. Tumors were excised and processed for immunofluorescent staining.
Immunofluorescence staining of tumors
YUMMER tumors were frozen directly in OCT. B16F10 tumors were fixed in 10% neutral buffered formalin for 15 minutes at room temperature (RT) and dehydrated at 4°C in 15% and then 30% sucrose for 4 hours and overnight, respectively. B16F10 tumors were then frozen in optimal cutting temperature compound (OCT; 4583, Tissue-Tek). Immunofluorescent staining was performed on 7μm thick frozen tissue slides. Tissue sections were fixed with cold acetone for 8 minutes. Slides were blocked with 4% fish gel blocking solution (CWFS 900.0033, Aurion) for one hour at RT. Primary antibodies were diluted in blocking solution and incubated on slides at 4°C overnight. If required, secondary conjugated antibodies were diluted in blocking solution and incubated on slides for one hour at RT. Slides were mounted with Fluoro-Gel II with DAPI solution (Electron Microscopy Science, 17985–50). Various combinations of the antibodies were used depending on the analysis being performed, see below.
The following primary antibodies were used on frozen tumor sections: rat anti-mouse CD31 (BD Bioscience, 550274, 1:50), rabbit anti-alpha-smooth muscle actin (αSMA, Abcam, ab5694, 1:100), rabbit anti-neural/glial antigen 2 chondroitin sulfate proteoglycan (NG2, AB5320 Millipore, 1:100), anti-mouse VCAM1-APC (Biolegend, 105718, 1:100), anti-mouse F4/80-PE (Biolegend, 123110, 1:100), anti-mouse F4/80-Alexa488 (Biolegend, 123119, 1:100), and anti-rabbit phospho-ERK5 S496 (BioOrbyt, orb5183, 1:50). The following secondary antibodies were used on frozen tumor sections: goat anti-rat Alexa Fluor 488 (Thermo Fisher, A-11006, 1:1000), goat anti-rat Alexa Fluor 594 (Thermo Fisher, A-11007, 1:2000), donkey anti-rabbit Alexa Fluor 647 (Thermo Fisher, A-31573, 1:1000), and goat anti-rabbit Texas Red (Thermo Fisher, T-2767, 1:2000).
Tumor vessel image analyses
Five to ten images were randomly captured at 10x or 20x magnification across a tumor slide stained as described above in varying antibody combinations. Quantifications were performed on each image and all image quantifications corresponding to a sample were averaged to report one value per tumor on graphs. Images were captured using a Leica fluorescence microscope (DMi8; LEICA Microsystems Inc., Morrisville, NC) with LAS X software. Vessel analyses were performed using NIH open-source image software ImageJ (http://rsbweb.nih.gov/ij/).
Perfusion and hyper-permeability
CD31 was co-stained on slides with tumor sections from mice injected with tomato-lectin or FITC-dextran prior to euthanasia. Perfusion was quantified as number of CD31+ vessels co-localizing with tomato-lectin divided by the total number of vessels in the image. Hyper-permeability was quantified as number of CD31+ vessels positive for FITC-dextran signal divided by the total number of vessels in the image.
Mural cell coverage
The mural cell markers αSMA and NG2 were co-stained with CD31 in slides. Mural cell coverage was quantified as the ratio of mural cell–marker area:CD31+ area.
VCAM1 expression in tumor vasculature
The quantification of VCAM1 expression in CD31+ vessel area was performed using a constant signal threshold for the CD31+ channel and a constant signal threshold for the VCAM+ channel. CD31+ area was measured and a mask of the CD31+ area was created and projected onto the corresponding VCAM1 channel image. VCAM1+ area was then visualized and the area of VCAM1+ signal within the CD31+ mask was measured. Last, this VCAM1+CD31+ area was normalized to the total CD31+ area and multiplied by 100 to measure the percent of VCAM1+ area within tumor vessels.
Tumor vessel structure
Tumor vessel structural analysis was performed as described previously [15]. Vessel parameters included the total number of CD31+ vessels, the number of visible open lumens, the average vessel length of all vessels (reported in μm), the number of vessels with a length >100μm (long vessels), and the number of 100μm2 regions per image occupied by CD31+ vessels (microvessel density). Specifically, the microvessel density was calculated by dividing the image area into equal squares (100μm × 100μm) creating a grid. The number of squares in the grid occupied by vasculature was counted and total number of squares are reported as microvessel density.
ERK5 S496 quantification in F4/80+ TAMs in YUMMER tumors by immunofluorescent staining
The same method used to quantify VCAM1+CD31+ area in YUMMER tumors was used to quantify ERK5 S496+F4/80+ area. Briefly, the F4/80+ area was first measured using a set threshold. A mask of the F4/80+ area was created and projected onto the corresponding ERK5 S496 channel image. ERK5 S496+ area was then visualized using a set threshold and the area of ERK5 S496+ signal within the F4/80+ mask was measured. Last, the ERK5 S496+ F4/80+ area was normalized to the total F4/80+ area and multiplied by 100 to measure the percent of ERK5 S496+ area within F4/80+ TAMs. This value was plotted and compared among treatment groups.
Pimonidazole HCl (hypoxyprobe-1) staining and quantification
Mice were injected with 100μL of 18mg/mL Hypoxyprobe-1 (pimonidazole hydrochloride, Hypoxyprobe Inc) intravenously and euthanized after 1hr. For immunofluorescent staining, tumors were rapidly harvested and frozen in OCT. Tumor sections were stained with FITC-MAb1 (Hypoxyprobe Inc, 1:100). Whole tumor tile scan images were captured using Leica LAS X software. The hypoxia area of tumor sections was calculated by normalizing hypoxyprobe-1+ area to total tumor tissue area.
VCAM1 immunofluorescent staining on aorta
Immunofluorescent staining was performed to quantify VCAM1 expression in ECs of the aorta as described previously [16]. Aortas were isolated from the YUMMER tumor–bearing mice that were used to obtain tumor tissue for flow cytometry analysis. The freshly isolated thoracic aorta was fixed with 10% neutral buffered formalin, embedded in OCT, and sectioned at 5μm thickness. Slides were incubated with blocking solution (4% fish gel in PBS) for one hour at RT. Sections were co-incubated with diluted primary antibodies rabbit anti-mouse VCAM1 (Abcam, ab134047, 1:100) and rat anti-mouse CD31 (BD Biosciences, 553370, 1:100) overnight at 4°C. Sections were washed and labeled with fluorescent secondary antibodies for 1.5 hours at RT and mounted with Fluoro-Gel II with DAPI solution. The secondary antibody used to detect VCAM1 was anti-rabbit Alex Fluor 488 (Thermo Fisher, A-11008, 1:200) and the secondary antibody used for CD31 was goat anti-rat Alexa Fluor 594 (Thermo Fisher, A-11007, 1:2000). A negative control was performed in the absence of primary antibodies to determine non-specificity of the secondary antibodies. Images were obtained using the Leica Dmi8 fluorescence microscope with LAS X software and a 10X objective lens. Microscope and camera settings were kept constant during the process of positive signal quantification using NIH open-source image software FIJI (https://hpc.nih.gov/apps/Fiji). All images were converted into 8-bit gray-scale images and the threshold of each channel was set to 50.0. Colocalized regions marked with a white overlay mask on the original green and red channels were considered VCAM1 endothelium colocalization. Colocalized pixels were quantified by the ratio of VCAM1+ to CD31+ area.
Isolation and tissue culture of bone marrow-derived macrophages (BMDM)
BMDMs were isolated and cultured as follows: Bone marrow cells were flushed from the femurs and tibias of C57BL/6 mice or ERK5S496A knock-in mice on the C57BL/6 background, which were tumor bearing and had or had not exercised, using PBS (Sigma-Aldrich P4474). Bone marrow cells were differentiated to macrophages by incubating the cells at a density of 1×106 cells/mL in IMDM (Life Technologies, 12440053) containing 10% FBS 10% (v/v) of the spent medium of L929 cell cultures (generated as described in Cell Culture) as the source of macrophage colony stimulating factor, HEPES (Thermo Fisher, 15630130), and 1% penicillin-streptomycin for 5–8 days at 37°C and 5% CO2 in air.
Western Blot
BMDMs were washed three times in cold PBS and lysed in modified RIPA buffer including 50 mM Tris- HCl, pH 7.4 (Thermo Fisher Scientific, J67501.AK), 150 mM NaCl (Thermo Fisher Scientific, AM9760G), 1 mM ethylenediaminetetraacetic acid (Thermo Fisher Scientific, 17892), 1% Nonidet P-40 (Thermo Fisher Scientific, 85124), 0.1% sodium dodecyl sulfate (Thermo Fisher Scientific,28364), 0.25% sodium deoxycholate (Thermo Fisher Scientific, 89904) with a protease inhibitor cocktail (Sigma Aldrich, p8340), 1 mM phenylmethylsulphonyl fluoride (Thermo Fisher Scientific, 36978), and 10 mM N-ethylmaleimide (Sigma Aldrich, E3876). Cell lysates were then centrifuged (21,000 × g for 15 min) to collect supernatants, proteins were resolved using SDS-PAGE and electro transferred onto nitrocellulose membranes (EMD Biorad, 1620115). The membranes were then immunoblotted with the following antibodies: anti-phospho-ERK5 S496 (BioOrbyt, orb5183), anti-phospho-ERK5 TEY (Cell Signaling Technology, 3371), anti-ERK5 (Cell Signaling Technology, 3372), anti-Trx1 (Proteintech, 14999-1-AP), anti-HO1 (Abcam, ab13243), anti-Gas6 (Sigma Aldrich, T5168), anti-SIRPa (Cell Signaling Technology, 13379), anti-cleaved IL1β (MBL International Corp, PDO39) and anti-Tubulin (Cell Signaling Technology, 3873), then with an HRP-labeled secondary antibody (anti-rabbit, Cell Signaling Technology, 7074, or anti-mouse, Cell Signaling Technology, 7076) and visualized using an enhanced chemiluminescence detection reagent (PerkinElmer, NEL105001EA). Images were captured by ChemiDoc MP imaging system. Tubulin was used as the loading control and was always probed together with the specific protein of interest. In most cases, tubulin immunoblots were not shown to save space. To obtain a fold-increase value, the intensity of each band was measured by Image J, and the mean intensity (integrated optical density) at the baseline (control sample) was calculated from 3 biological samples (n=3), then normalized to tubulin expression and designated as 1. We calculated a fold increase from this control mean intensity with each of the other band intensities.
BMDM efferocytosis assay
Bone marrow mononuclear cells were flushed from wild type, non-tumor bearing mice as described above but were not differentiated into BMDMs. These cells were suspension cultured in IMDM containing 10% FBS (v/v; Hyclone), 1% HEPES, and 1% penicillin- streptomycin. To induce apoptosis in these cells, we treated 1×106 cells/mL in suspension with camptothecin (100nM, Millipore Sigma, PHL89593) for 6hr and labeled them with IncuCyte pHrodo red cell-labeling reagents (250ng/mL, Sartorius, 4649) for 1hr.
For the efferocytosis assay, 1×104 BMDMs, generated as described above from YUMMER tumor-bearing exercised or non-exercised wild type C57BL/6 mice or ERK5S496A knock-in mice on the C57BL/6 background, were plated on a 96-well flat-bottom plate, and 4 × 104 apoptotic cells per well were added to the surface of the BMDMs. After incubating the cells for 1hr, efferocytotic activity was monitored using IncuCyte ZOOM live cell imaging (ESSEN Bioscience, Ann Arbor, MI). Engulfment of pHrodo-labeled cells induces pHrodo fluorescence by the acidic environment of the phagosome, and the fluorescence intensity of the image was determined using IncuCyte ZOOM.
BMDM Cellular ROS assay
Intracellular ROS was measured by using the membrane-permeable oxidation-sensitive fluorescent dye, CM-H2DCF-DA (5-(and-6)-chloromethyl-2’,7’-dichlorodihydrofluorescein diacetate) from Invitrogen (#C6827, Invitrogen, Eugene, OR). BMDM cells generated from C57BL/6 mice or ERK5S496A knock-in mice on the C57BL/6 background, bearing YUMMER tumors, which had or had not exercised, as described above, were seeded overnight in 96-well plates, washed with PBS, and incubated with CM-H2DCF-DA (10μM, 30 min). Fluorescence intensity was measured by a FLUOstar Omega microplate reader (BMG LABTECH spectrofluorometer using excitation at 495 nm and emission at 530 nm.
BMDM Mitochondrial (Mito) ROS assay
BMDM cells generated from C57BL/6 mice or ERK5S496A knock-in mice on the C57BL/6 background, bearing YUMMER tumors, which had or had not exercised, as described above, were washed with PBS and incubated with MitoSOX Red (5μM) (#M36008, Invitrogen, Eugene, OR) at 37°C for 10 minutes. Fluorescence intensity was measured with excitation at 510 nm and emission at 580 nm using a plate reader (Biotek, Winooski, VT) as described in the manufacturer’s instructions (Invitrogen). The mean intensity of MitoSOX fluorescence of BMDMs from exercised mice was normalized against BMDMs from sedentary mice of the same genotype (wild type or ERK5S496A) and the fold change in intensity was calculated.
Flow cytometry
B16F10 or YUMMER tumors from sedentary or exercised mice were harvested and immediately digested with DNase I (200 μg/mL; Roche, 04716728001) and collagenase H (1mg/ml; Sigma-Aldrich, C8051) in RPMI 1460 for 15 minutes at 37°C, and red blood cells (RBCs) were lysed using ammonium chloride lysis buffer (A1049201). Single-cell suspensions were prepared using a 100μm nylon mesh. Cells were blocked for 15 minutes at RT using purified rat anti-mouse CD16/CD32 (BD Biosciences, 553141). Cells were stained with fluorochrome-conjugated surface antibodies for 20 minutes at RT, followed by Ghost Dye UV 450 (Tonbo, 13–0868) for 20 minutes, then fixed, permeabilized, and incubated with intracellular antibodies for 30 minutes on ice. Data was acquired using an LSR Fortessa X-20 flow cytometer (BD Biosciences) and analyzed with FlowJo software version 10. Fluorochrome-conjugated antibodies used for flow cytometry analysis were from Biolegend—CD45 Pacific Blue (103126), F4/80 BV785 (123141), CD3PerCP Cy5.5 (100218), CD206 PE (141705), CD86 PE-Cy7 (105014), CD11b APC (101212), Gr1 APC-Cy7 (108424), MHC II BV786 (107645), Ly6C PE (128008). Ly6G APC-Fire750 (127651), CD11c PE-Cy7 (117318), NK1.1 BV510 (108738), CD69 BV605 (104530), PD1 PE (135205), CD8 BV786 (100750), CD4 APC-Fire 750 (100568), FoxP3 Alexa488 (126406), CTLA4 APC (106310), Tim3/CD366 APC (134007), Lag3/CD223 PE-Cy7 (125225)—and from BD Pharmingen—FoxP3 PE (560414).
Statistical analysis
Statistical analysis was performed using Graphpad Prism 9.0.0 software. Values in graphs are reported as mean ± standard error of the mean or mean ± standard deviation. Normality tests were performed on datasets prior to analysis. Nonparametric data was analyzed using an unpaired Mann Whitney t-test and parametric data was analyzed using unpaired Student’s t-test. Tumor growth curves were analyzed using two-way ANOVAs with post-hoc multiple comparison tests.
scRNA-seq and analysis
Three tumors from sedentary mice and three tumors from exercised mice were dissociated into a single-cell suspension. RBCs were lysed using ammonium chloride lysis buffer. Single-cell libraries were captured and barcoded using the 10X Genomics Single Cell Chromium 3’ V3 chemistry reagents kit (10x Genomics, 1000121) according to the manufacturer’s protocol. The single-cell transcriptome libraries were sequenced using paired-end sequencing in Illumina NovaSeq6000 S2 flowcell at 100 Cycles. Sequencing read lengths of L1, R1, and R2 were 8, 28, and 91, respectively. We demultiplexed the single-cell transcriptome files into FASTQ and aligned them to the mouse genome (mm10) using 10X Genomics Cell Ranger v.6.1.2. We processed the resulting cells from each sample using the Seurat package v.4.0.5 [17] in R software (v.4.2.0) [18] and filtered it according to the following criteria: percent mitochondrial unique molecular identifier (UMI) < 10%, number of UMI > 200, and number of total RNA features > 500. After filtering we merged control and exercise samples for downstream analysis. We normalized samples with SCTransform (ncells = 6000, variable.features.n = 5000) and applied principal component analysis (PCA), extracting the top 50 principal components for downstream Uniform Manifold Approximation and Projection (UMAP) dimensional reduction and shared nearest neighbor (SNN) clustering with Louvain algorithm (resolution = 0.4). We removed empty clusters based on high expression of long noncoding RNAs (lncRNAs) (Malat1, Neat1, Gm26917). We removed doublet clusters based on expression of multiple cell markers known to be mutually exclusive. Clusters with <100 cells were removed and populations were identified according to cell type specific markers. Cell types of interest were subset and re-clustered for further analysis. In the CD3+ T-cell population, we excluded two doublet clusters expressing Cd68 or tumor cell marker Bmp2 and a low-quality cluster with high percentage of mitochondrial genes after which cells were normalized with SCTransform (variable.features.n = 400) and the top 30 PCA components were passed on for UMAP and SNN clustering (resolution = 0.2). In the CD68+ monocyte/macrophage/dendritic cell (DC) population, two doublet clusters were removed which expressed Cd3g or Bmp2, after which cells were normalized with SCTransform (variable.features.n = 800) and the top 40 PCA components were passed on for UMAP and SNN clustering (resolution = 0.3). Differential gene expression was performed using Seurat’s Wilcoxon Rank Sum test and results were plotted using the Enhanced Volcano Plot package [19]. Results from differential expression analysis were used as input for gene set enrichment analysis (GSEA) using package fgsea [20] and the mouse hallmarks of cancer gene set pathways [21] obtained using the msigdbr R package [22]. We performed pseudotime trajectory inference using Slingshot [23] (v.2.4.0) R package. We reclustered the monocyte/macrophage populations separately excluding cycling clusters using Seurat FindClusters function (v.4.1.1) and SCTransform normalization. We converted the Seurat objects to SingleCellExperiment objects and inferred the trajectory graph using the function ‘slingshot’ with default parameters. Trajectories were rooted in the Ly6Clo Mono cluster.
Data availability
Data generated during this study are included in this article and its Supplementary Data files or are available from the corresponding author upon request. The scRNA-seq data generated in this study are publicly available in NCBI’s Gene Expression Omnibus (GEO) at accession number GSE208205.
RESULTS
Aerobic exercise remodels melanoma tumor vasculature
To evaluate tumor vessel structure and function we compared mice bearing B16F10 or YUMMER melanoma tumors treated with treadmill exercise to control, non-exercised (sedentary) animals bearing the same tumors. We observed structural remodeling after exercise in both models, including a significant increase in elongated vessels in YUMMER tumors (1.4-fold increase, p=0.029, Fig. 1A), and a 19% increase in the average vessel length in B16F10 (p=0.049; Fig. 1B). There was also a significant increase in microvessel density in YUMMER tumors (p=0.038, Fig. 1A) from exercised mice relative to sedentary that was not observed in B16F10 tumors (Fig. 1B). The total vessel number and number of open lumens did not change in response to exercise in either model (Supplementary Fig. S1A–B).
Figure 1. Aerobic exercise remodels melanoma tumor vasculature.
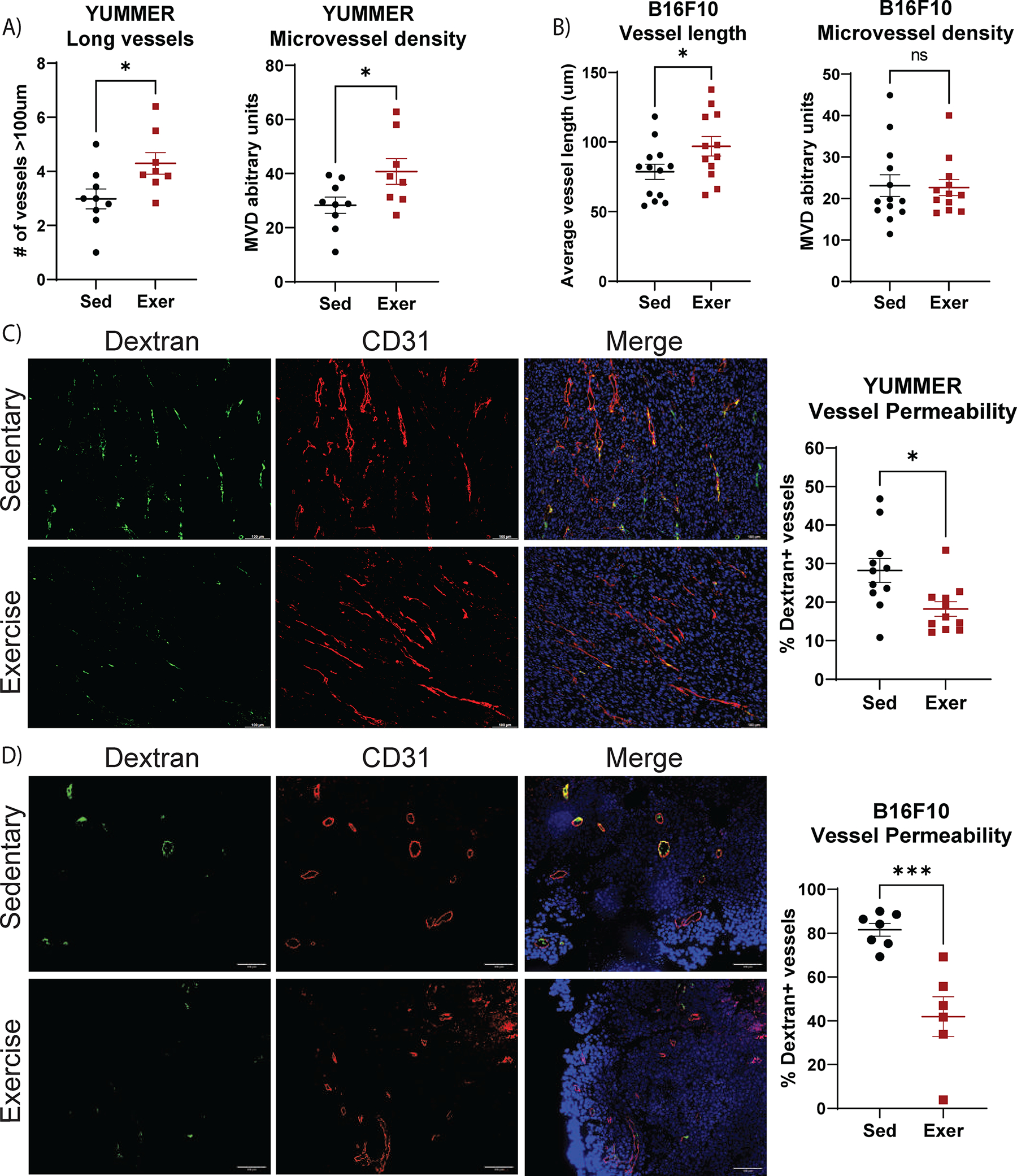
YUMMER and B16F10 tumor–bearing mice performed two weeks of treadmill exercise (Exer) or were non-exercised sedentary controls (Sed). A) YUMMER and B) B16F10 tumor vessel structure was evaluated using CD31 immunofluorescent staining. C,D) FITC-dextran leak (green) and CD31 (red) immunofluorescence with DAPI staining (blue) in C) YUMMER and D) B16F10 tumors; scale bar: 100μm. FITC-dextran leakage quantified as percent of dextran+ vessels in E) YUMMER and F) B16F10 tumors. Graphs are displayed with lines showing the mean +/− standard error of the mean (SEM); each point represents one tumor value obtained by the average of ≥ 5 10x microscopic fields. T-test results are represented on graphs according to the following: ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. The experiment comparing sedentary and exercise-treated mice was repeated four times in the YUMMER model and three times in B16F10 model, with n ≥ 8 mice in each treatment group for each independent experiment.
To determine whether exercise alters vessel function in these tumor models, we measured perfusion and permeability using fluorescently labeled lectin and high molecular weight dextran, respectively. No change was detected in perfusion by exercise in either model (Supplementary Fig. S1C–E). However, dextran leakage was decreased by nearly 40% in YUMMER (p=0.012, Fig. 1C) and 50% in B16F10 tumors (p=0.001, Fig. 1D) from exercised mice relative to sedentary mice. This decrease in permeability appeared to be endothelial cell driven as there were no significant changes in mural cell coverage of vasculature after exercise, as indicated by NG2 and αSMA staining (Supplementary Fig. S1F–K).
Aerobic exercise increases VCAM1 expression in melanoma tumor vasculature
In addition to structural and functional abnormalities, tumor vessels express low levels of adhesion molecules, including VCAM1 and ICAM1, which contributes to poor infiltration by antitumor immune cells such as CD8+ T cells [24–26]. To investigate the relationship between exercise and adhesion molecule expression, immunofluorescent staining for VCAM1 and ICAM1 was performed in tumors. ICAM1 expression was low on tumor vessels in all treatment cohorts, therefore, we focused on VCAM1 expression. We found a significant increase in the expression of VCAM1 on tumor endothelium of exercised mice relative to sedentary mice in YUMMER tumors (p=0.047, Fig. 2A–B), and a similar trend in B16F10 tumors (p=0.089, Fig. 2C–D). No change in VCAM1 expression was detected in the abdominal aorta (Supplementary Fig. S2A), indicating that the effect of exercise on VCAM1 may be different between tumor and healthy vasculature.
Figure 2. Exercise increases VCAM1 expression in melanoma tumor vasculature.
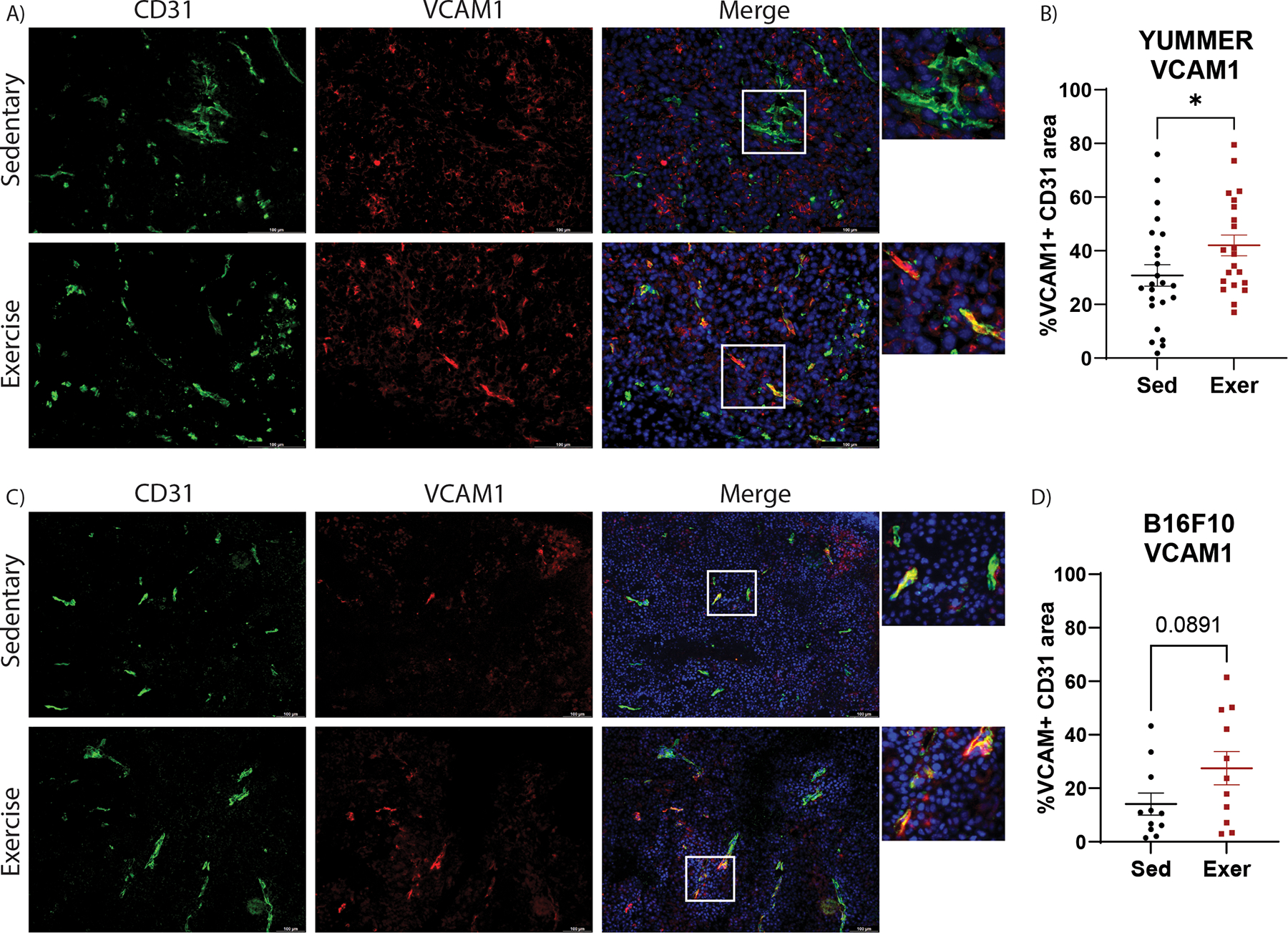
Immunofluorescent images of CD31 (green), VCAM1 (red) and DAPI (blue) in A) YUMMER and C) B16F10 tumors scale bar: 100μm. Images are representative of ≥ 5 images per tumor, ≥ 8 tumors per treatment group. Percent of VCAM1+ CD31+ vessel area was quantified in B) YUMMER and D) B16F10 images. Graphs are displayed with lines showing mean +/− standard error of the mean (SEM); each point represents one tumor value obtained by the average of ≥ 5 10x or 20x microscopic fields. T-test results are represented on graphs according to the following: ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001)
Aerobic exercise differentially affects CD8+ T cells and tumor growth in melanoma tumors
Tumor vasculature delivers immune cells to solid tumors. Additionally, vascular adhesion molecule expression is important for immune cells, like cytotoxic T cells, to extravasate into the tumor tissue[ 27]. Thus, the observation that exercise increases VCAM1 expression in melanoma vasculature prompted us to characterize the effect of aerobic exercise on T-cell infiltration and function in the melanoma models.
In YUMMER tumors, exercise had no effect on the abundance of CD3+ T cells, CD4+ T cells, or FoxP3+ regulatory T (Treg) cells (Fig. 3A–C). However, there was a significant increase in CD8+ T cells in YUMMER tumors from exercised mice relative to sedentary mice (p=0.039; Fig. 3D, Supplementary Fig. S3A). In addition, exercise increased the number of CD8+ T cells expressing the activation marker CD69 (p=0.046, Fig. 3E, Supplementary Fig. S3B), as well as the number of CD8+ T cells expressing PD1 (p=0.033) and their median fluorescence intensity (MFI; p=0.0527) of PD1 (Fig. 3F, 3G, Supplementary Fig. S3C). PD1 can be expressed across the continuum of T-cell activation to exhaustion. Thus, we also evaluated Lag3 and Tim3, and found no significant difference in expression of these markers between sedentary and exercise groups. Only ~20–25% of PD1+CD8+ cells expressed the terminal exhaustion indicator Tim3, and ~85% of PD1+CD8+ cells expressed Lag3 in either group (Supplementary Fig. S3D–G).
Figure 3. Aerobic exercise suppresses tumor growth and increases CD8+ T-cell infiltration in YUMMER but not B16F10 tumors.
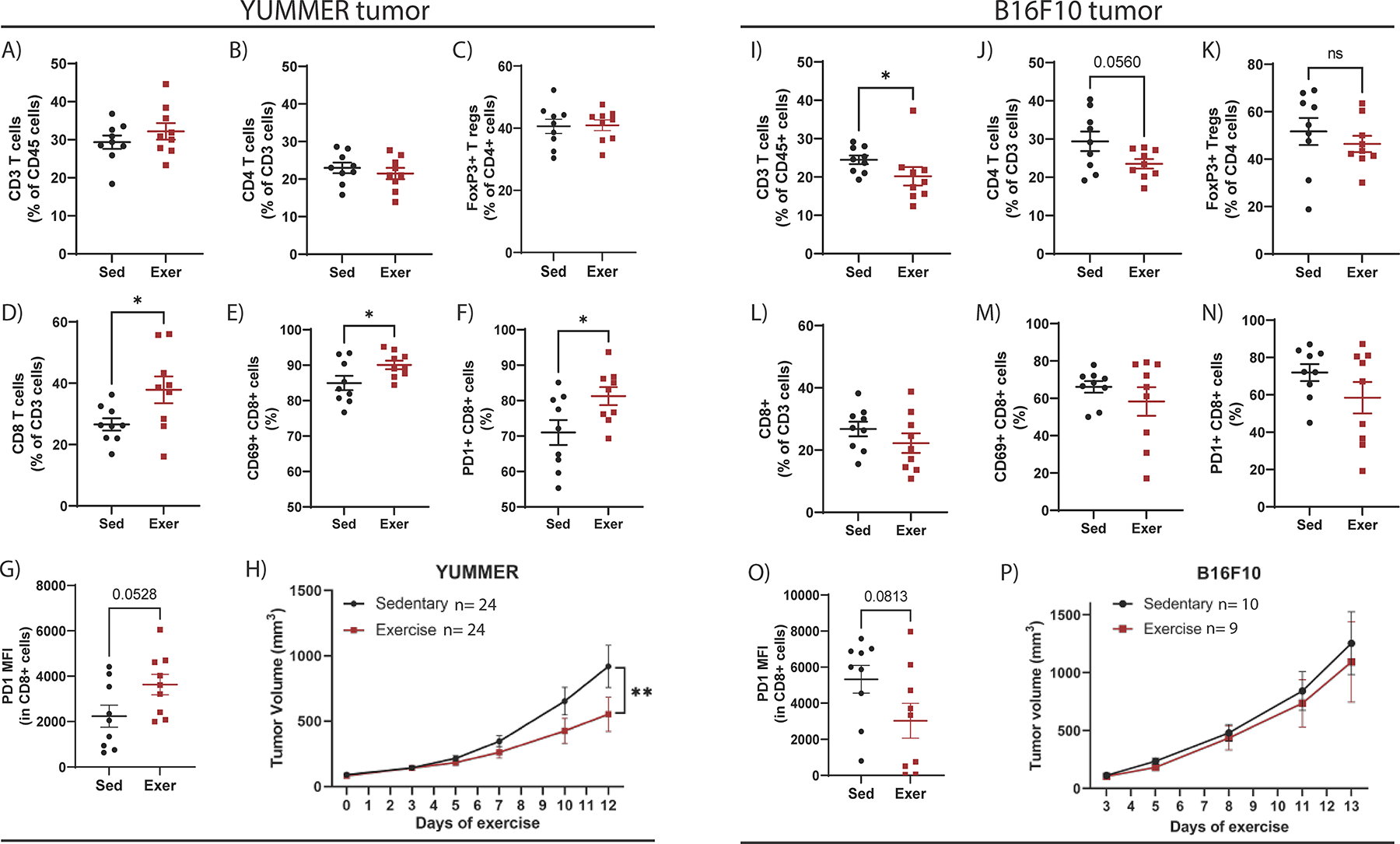
Flow cytometry quantifications of YUMMER tumors to identify the percent of A) CD3+ T cells relative to CD45+ cells, B) CD4+ T cells relative to CD3+ cells, C) FoxP3+ Treg cells relative to CD4+ cells, D) CD8+ T cells relative to CD3+ cells, E) CD69+CD8+ T cells, and F) PD1+CD8+ cells. Flow cytometry quantification on G) YUMMER of PD1 median fluorescence intensity (MFI) in CD8+ T cells. H) YUMMER tumor volumes were monitored with calipers and graphed over time. Tumor growth curve was analyzed using two-way ANOVA (YUMMER two-way ANOVA; Time v Exercise: p=0.0107; Time: p<0.0001; Exercise: p=0.1068) with post-hoc multiple comparisons displayed on graph. Flow cytometry quantifications of B16F10 tumors to identify the percent of I) CD3+ T cells relative to CD45+ cells, J) CD4+ T cells relative to CD3+ cells, K) FoxP3+ Treg cells relative to CD4+ cells, L) CD8+ T cells relative to CD3+ cells, M) CD69+CD8+ T cells, and N) PD1+CD8+ cells. Flow cytometry quantification O) B16F10 tumors of PD1 median fluorescence intensity (MFI) in CD8+ T cells. P) B16F10 tumor volumes were monitored with calipers and graphed over time. Tumor growth curve was analyzed using two-way ANOVA (B16F10 two-way ANOVA; Time v Exercise: p=0.9795; Time: p=0.0002; Exercise: p=0.6706) with post-hoc multiple comparisons displayed on graph. Graphs are displayed with lines showing mean +/− SEM. Each point on a graph represents one tumor analyzed. ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Mouse experiment comparing sedentary and exercise treatment was repeated four times in the YUMMER model and three times in B16F10 model.
In B16F10 tumors, exercise reduced CD3+ T-cell abundance (p=0.023) and CD4+ T cells (p=0.056) but did not change FoxP3+ Treg cells (Fig. 3I–K). In contrast to YUMMER tumors, exercise did not alter CD8+ T-cell abundance or expression of CD69 or PD1 (Fig. 3L–N, Supplementary Fig. S3H–J) and decreased PD1 MFI on CD8+ T cells (p=0.0813, Fig. 3O). Exercise suppressed tumor growth of YUMMER tumors (p=0.004; Fig. 3H) but did not change growth of B16F10 tumors (Fig. 3P).
Given the increased number of CD8+ T cells and increased percentage of those cells that expressed PD1 in YUMMER tumors from exercised mice, we considered whether exercise might enhance the efficacy of anti-PD1 immune checkpoint blockade against YUMMER. Exercise was initiated after tumors were established, and anti-PD1 treatment was begun on day 5 of exercise (Supplementary Fig. S4A). The addition of exercise to anti-PD1 did not significantly increase the antitumor effect relative to anti-PD1 alone (Supplementary Fig. S4B–C). However, there was a trend toward more CD8+ T cells (p=0.097), decreased CD4+ T cells (p=0.004) and FoxP3+ Treg cells (p=0.11), and a significantly increased ratio of CD8+/CD4+ (p=0.003) and CD8+/Treg (p=0.019) cells in YUMMER tumors from mice treated with the combination of exercise and anti-PD1 compared to those treated with only anti-PD1 (Supplementary Fig. S4D–I). Because anti-PD1 alone prevented the growth of several tumors, we considered whether a lower dose of anti-PD1 combined with exercise may reveal a beneficial effect of exercise. However, exercise did not increase the antitumor efficacy of low dose anti-PD1 therapy against YUMMER tumors (Supplementary Fig. S4J–L). Exercise also did not improve anti-PD1 efficacy against B16F10 tumors (Supplementary Fig. S4M–O).
Aerobic exercise reduces hypoxia in YUMMER but not B16F10 tumors
Although vascular remodeling and adhesion molecule upregulation were observed in both models, exercise only increased T-cell abundance and activation in YUMMER tumors. Thus, we considered whether the difference in exercise-induced tumor immune cell changes may be related to hypoxia independent of vascular changes. To assess hypoxia in YUMMER and B16F10 melanoma models, we performed hypoxyprobe-1 (Pimonidazole HCl) staining. Our results revealed an exercise-induced reduction of hypoxic area in YUMMER (Fig. 4A–B) but not B16F10 tumors (Fig. 4C–D).
Figure 4. Exercise reduces hypoxia in YUMMER tumors.
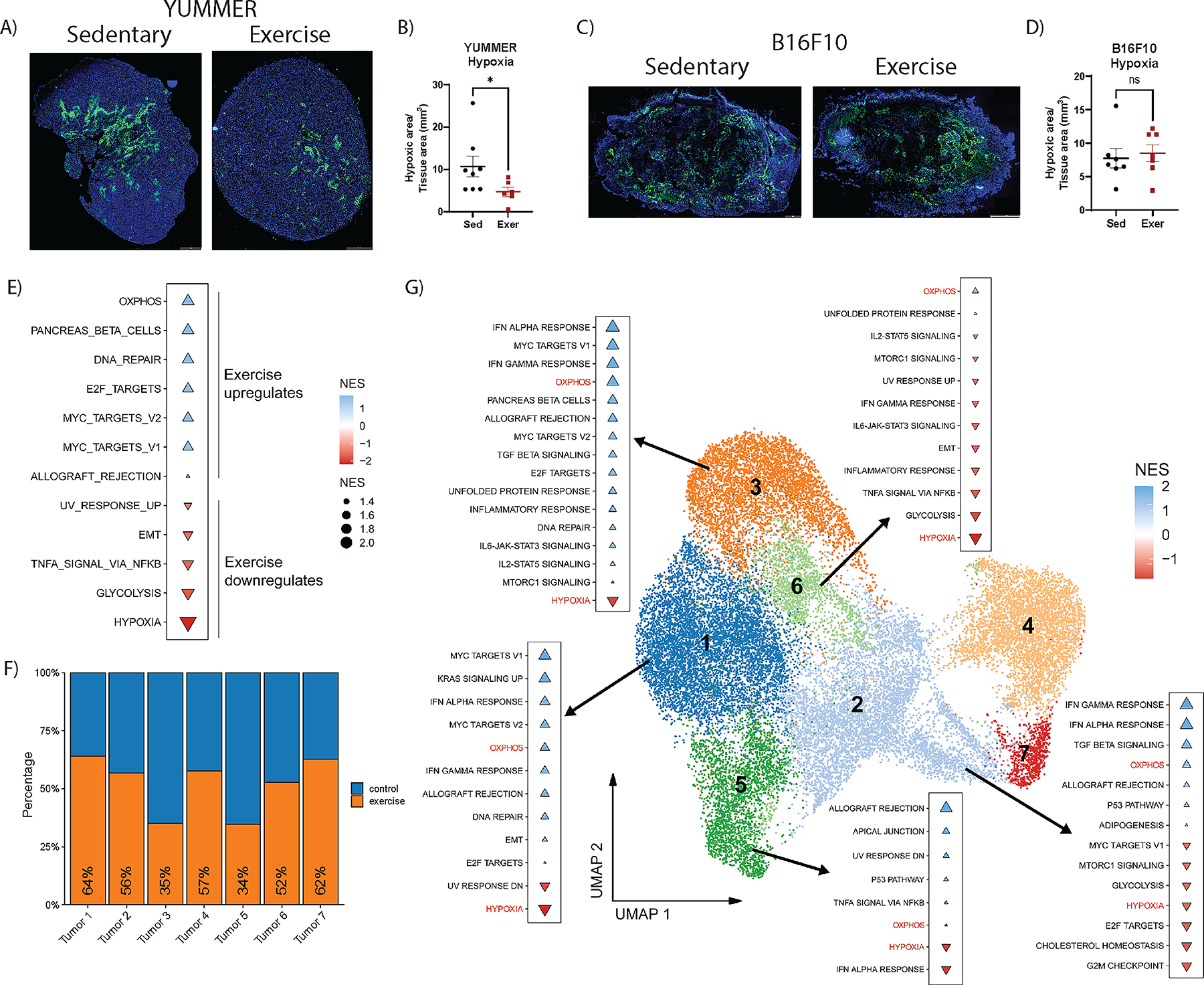
Quantification of hypoxic regions measured by percent of hypoxyprobe-1+ area: total tumor tissue area in tile scan images of A, B) YUMMER and C, D) B16F10 tumors; scale bar: 1mm. Each point on the graphs represents one tumor. Lines on the graph indicate the mean +/− standard error of the mean (SEM). T-test results are indicated by * p<0.05, or ns = non-significant. E) GSEA analysis with significant altered pathways using the hallmarks of cancer gene sets comparing non-cycling tumor cell clusters from scRNA-seq dataset of YUMMER tumors from exercise and sedentary mice (n=3 per group), in which blue triangles represent exercise upregulated pathways and red triangles represent exercise downregulated pathways. Triangle size is proportional to normalized enrichment score (NES). F) Proportion of tumor cells from each cluster from control (blue) samples and exercised (orange) samples. G) UMAP plot of all tumor cells labeled with cluster number. GSEA using the hallmarks of cancer of exercise compared to sedentary samples for clusters Tumor 1, 2, 3, 5, and 6.
To validate the reduced hypoxia identified with hypoxyprobe and understand the mechanisms by which exercise suppressed tumor growth in the YUMMER model, we sought to further characterize the tumor microenvironment and its cell populations. We performed scRNA-seq on six YUMMER whole tumor dissociations, three from sedentary (control) mice (n = 21,329 cells), and three from exercised (exercise) mice (n = 24,308 cells) (Supplementary Fig. S5A). We identified tumor and immune cells using population specific markers (Bmp2 and Ptprc, respectively) and excluded any cluster with dual marker expression or lack of cell specific markers. Joint analysis of single cells from tumors from control and exercised mice was performed in which no significant batch effects were observed (Supplementary Fig. S5B). Our analysis revealed 4 main population groups detected across control and exercise, including 7 tumor clusters (Tumor 1 – Tumor 7), Myeloid Derived Suppressor Cells (MDSCs), Myeloid cells (TAM1, TAM2, and Mono DCs), and lymphocytes (NK, CD4T, CD8 and Cycling CD8T; Supplementary Fig. S5C–D). When comparing proportions of clusters in exercise relative to control, we detected exercise-induced modulation in the abundance of every cluster. These results indicated that exercise influenced multiple cell types in the YUMMER tumor microenvironment (Supplementary Fig. S5E).
To assess the influence of exercise on tumor cell gene expression, we subset the tumor cells clusters and performed GSEA [20] excluding the two Mki67+ cycling clusters (Tumor 4 and Tumor 6; Supplementary Fig. S5D). Gene expression in each cell was compared in exercise relative to control samples using the hallmarks of cancer gene sets [21]. Consistent with the hypoxyprobe-1 staining in YUMMER tumors (Fig. 4A–B), GSEA revealed an exercise-induced downregulation of the hypoxia pathway (Fig. 4E). Further, there was a strong effect of exercise on the expression of genes in metabolic pathways, including a significant upregulation of oxidative phosphorylation (OXPHOS) and downregulation of glycolysis pathways (Fig. 4E).
Differences across tumor populations in exercise and control groups were investigated by comparing the proportions of cells in the distinct tumor clusters. Our data showed a reduction in the abundance of cells from exercise samples in clusters Tumor 3 (35%) and Tumor 5 (34%) while the remaining clusters were enriched in the control group (Fig. 4F). These data suggest that distinct tumor cell populations respond differently to exercise. We further explored the differences between exercise and control groups in the non-cycling clusters with GSEA and the hallmarks of cancer gene set pathway. While the top upregulated and downregulated pathways varied between clusters, we detected consistent upregulation of OXPHOS and downregulation of hypoxia pathways across all clusters, in agreement with a greater oxygen availability in the tumor microenvironment after exercise (Fig. 4G).
scRNA-seq reveals shifts in T-cell populations after exercise in YUMMER tumor
To build upon our flow cytometry observations, we explored the CD3+ T-cell group from the scRNA-seq data of YUMMER tumors after sedentary and exercise treatment. We identified six clusters of CD3+ T cells divided into four CD8+ clusters, one CD4+ cluster, and one CD8−CD4− cluster (Fig. 5A). Each cluster was assigned a name according to the predicted cell function inferred from gene expression patterns (Fig. 5A), namely: cytotoxic CD8+ cells (Cytotoxic), cycling CD8+ cells (Cycling CD8), CD4+ Treg cells, central memory T cells (Cent Mem), a second cluster of cytotoxic CD8+ cells (IFNg+ Cytotoxic), and interferon stimulated CD8+ cells (IFN stimulated; Fig. 5B).
Figure 5. scRNAseq characterization of T-cell populations in YUMMER tumors after sedentary or exercise treatment.
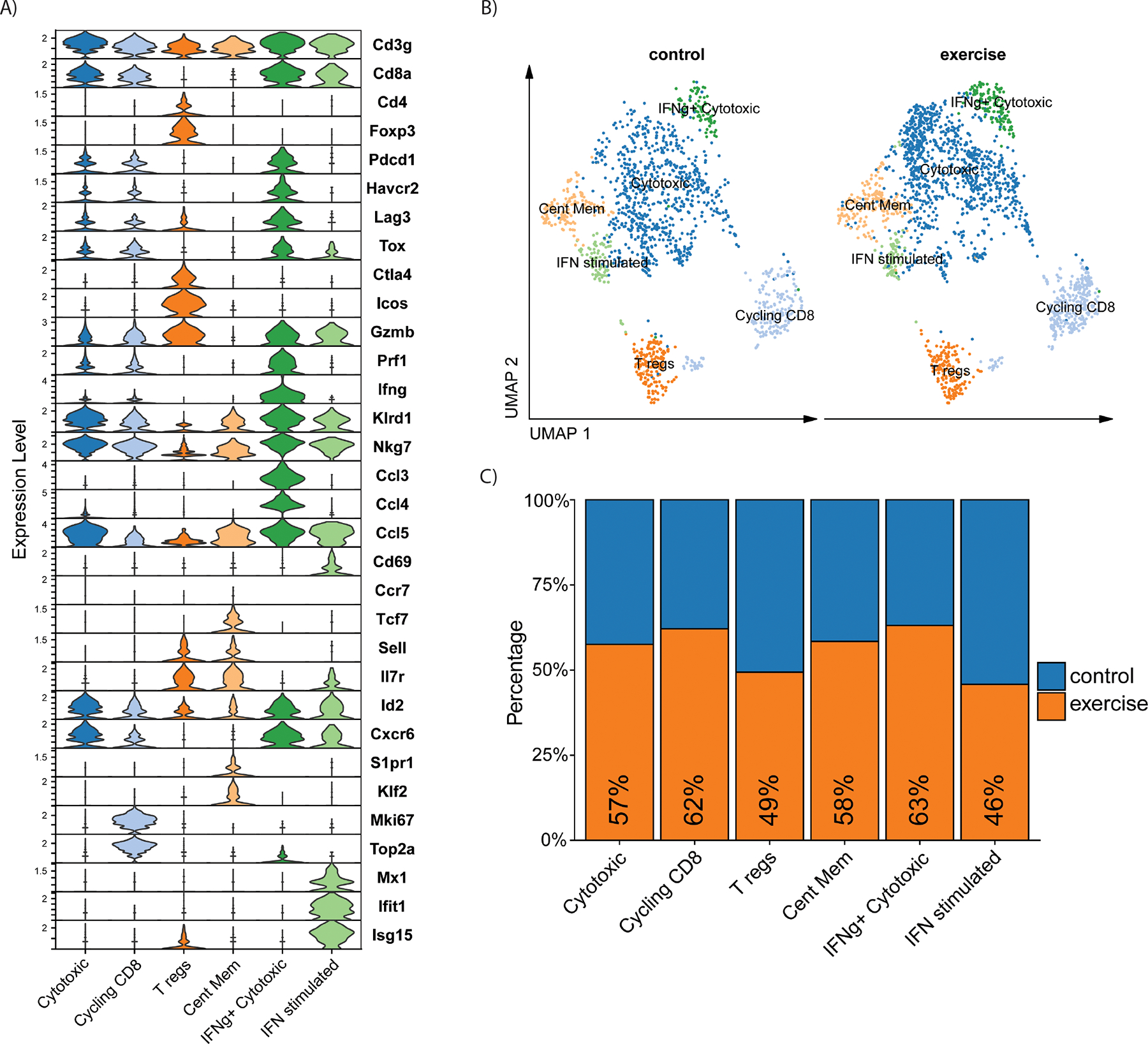
Analysis of the scRNA-seq dataset of YUMMER tumors from exercise and sedentary mice (n=3 per group) from figure 4. A) Stacked violin plot of T-cell gene expression across scRNA-seq T-cell clusters. B) UMAP plot of T cells in YUMMER tumors from control (left) and exercise (right) samples. C) Proportions of the T-cell clusters from control (blue) samples versus exercised (orange) samples
The CD8+ Cytotoxic cluster was the most abundant. The cluster expressed cytotoxic genes (Gzmb, Klrd1, Nkg7), immune checkpoint genes (Pdcd1, Havcr2, Lag3 and Tox), and high levels of Cxcr6, a gene important in intratumoral CD8+ T cells maintaining an effector-like phenotype [28] (Fig. 5A; Supplementary Fig. S6A). The Cent Mem cluster was CD8−CD4− and expressed memory markers, Tcf7, Sell, Il7r, and recirculation markers, S1pr1 and Klf2. Although the Cent Mem cluster did not express Ccr7, the gene signature suggests the cluster may contain naïve T cells. The Cycling CD8+ cluster expressed the same gene pattern as the Cytotoxic cluster in addition to expressing proliferation genes Mki67 and Top2a. CD4 expression was detected only in the Treg cells cluster, which expressed high levels of Icos, Ctla4, and Foxp3. The second CD8+ cytotoxic cluster, named IFNg+ Cytotoxic, displayed the highest levels of cytotoxic genes including Gzmb, Prf1, cytokines Ifng, Ccl3, and Ccl4, and the highest expression of immune checkpoint genes Pdcd1, Havcr2, Lag3 and Tox. Lastly, a small cluster of CD8+ T cells expressed numerous IFN responsive genes including Mx1, Ifit1, and Isg15, indicating IFN stimulation and signaling. The IFN stimulated cluster also expressed effector genes Gzmb, Klrd1, Nkg7, and Cd69 (Fig. 5A; Supplementary Fig. S6A). We observed increased proportions of the clusters Cytotoxic (57%), Cycling CD8 (62%), and IFNg+ Cytotoxic (63%) in tumors from exercised mice (Fig. 5C). The exercise group also had enrichment of the Cent Mem (58%) cluster. Proportions of IFN stimulated (46%) and Treg cell (49%) clusters were similar between sedentary and exercise (Fig. 5C).
Exercise alters myeloid cell phenotypes in YUMMER tumors
Cytotoxic CD8+ T-cell activity is reduced in the tumor microenvironment by immune suppressive cell types like myeloid cells. Thus, we evaluated myeloid cell populations including myeloid-derived suppressor cells (MDSCs), dendritic cells (DCs), and TAMs. Flow cytometry analysis revealed that exercise decreased the frequency of monocytic MDSCs (M-MDSCs, p=0.015) while not changing granulocytic MDSCs (G-MDSCs; Supplementary Fig. S6B–D). Moreover, exercise increased the abundance of major histocompatibility class II (MHCII) antigen-presenting DCs (p=0.01; Supplementary Fig. S6E–F) and F4/80+ TAMs (p=0.002; Fig. 6A). To investigate whether exercise altered TAM polarization, we quantified TAMs expressing the M2-like marker CD206 and the M1-like marker CD86 in sedentary and exercised mice by flow cytometry. There was a significant decrease in the frequency of CD206+CD86− “M2-like” TAMs (p=0.012; Fig. 6B), but no difference in the mean fluorescent intensity (MFI) of CD206 per cell (Fig. 6C) in exercise compared to sedentary tumors. There was no difference in the frequency of CD86+CD206− “M1-like” TAMs, although there was an increase in the MFI of CD86 (p=0.0691; Fig. 6D–F) in exercise compared to sedentary tumors. This is suggestive of an exercise-induced shift in the TAM phenotype away from M2-like, although we did not functionally assess the inflammatory cytokine secreting capacity of TAMs. As commonly described in TAMs, the M1-like and M2-like populations are likely on a polarization continuum with some characteristics of both polarization phenotypes.
Figure 6. Exercise reduces M2-like TAMs and alters the transcriptome of myeloid cells.
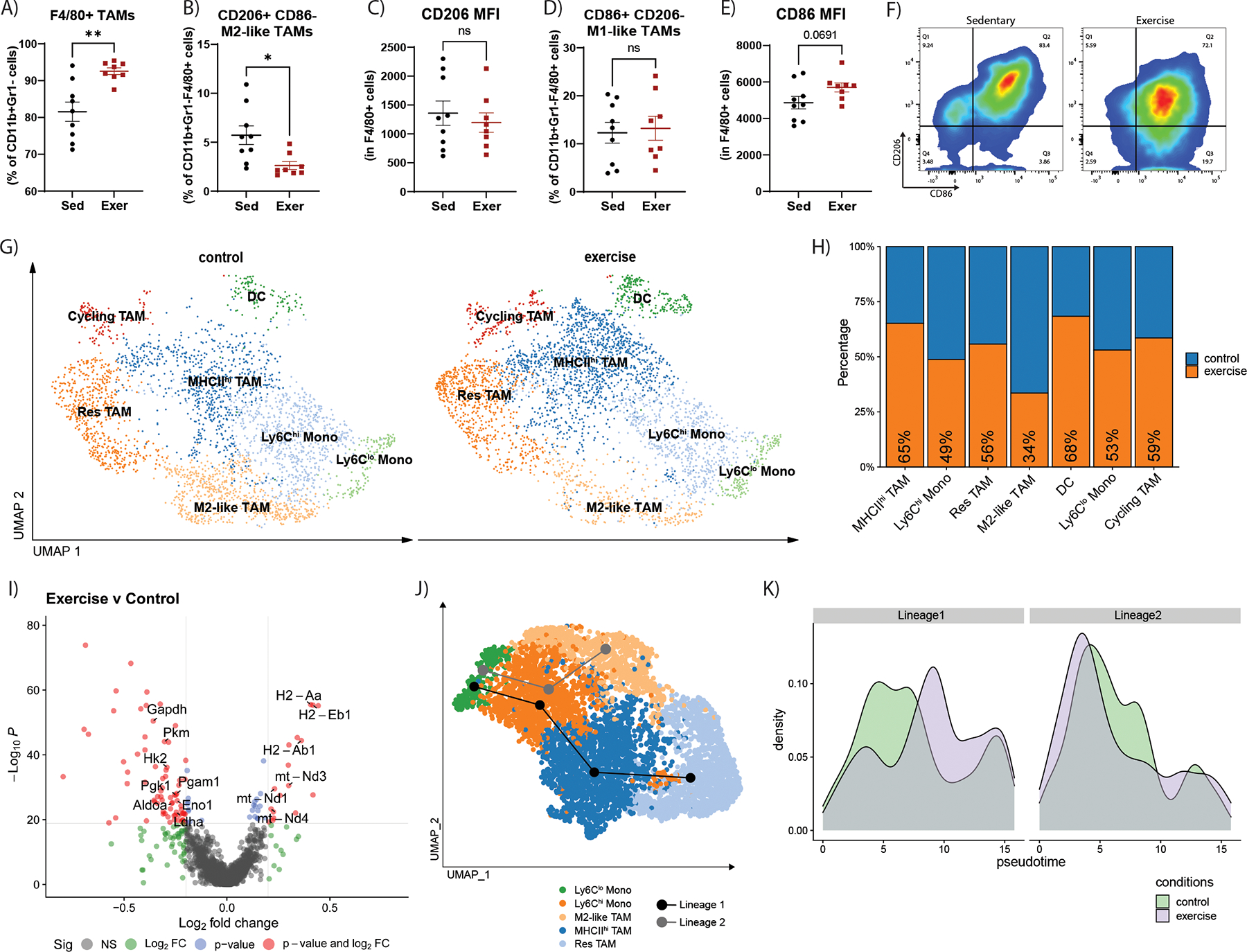
Flow cytometry quantifications of YUMMER tumors of the percent of A) F4/80+ TAMs relative to CD11b+Gr1− cells, B) CD206+CD86− TAMs relative to CD11b+Gr1−F4/80+ cells, C) CD206 MFI in CD11b+Gr1−F4/80+ cells, D) CD86+CD206− TAMs relative to CD11b+Gr1−F4/80+ cells, and E) CD86 MFI in CD11b+Gr1−F4/80+ cells. F) Representative flow cytometry plots of CD206 marker versus CD86 marker in CD11b+Gr1−F4/80+ cells. Graphs are displayed with lines showing mean +/− SEM. Each point on the graphs represents one tumor analyzed. T-test results are represented on graphs according to the following: ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. G-J) Analysis of the scRNA-seq dataset of YUMMER tumors from exercise and sedentary mice (n=3 per group) from figure 4 G) UMAP plot of myeloid cell populations in YUMMER tumors from control (left) and exercise (right) samples. H) Proportions of each cluster from control (blue) samples versus exercised (orange) samples. I) Volcano plot showing differentially expressed genes in myeloid cells. Comparison: exercise versus control. J) Results of pseudotime trajectory analysis performed on non-cycling monocyte and TAM clusters revealed two predicted lineages which are represented with lines on the UMAP plot. Lineage 1: Ly6Clo Monos -> Ly6Chi Monos -> MHCIIhi TAM -> Res TAM (black line) and Lineage 2: Ly6Clo Monos -> Ly6Chi Monos -> M2-like TAM (gray line). K) Control (green) and exercise (purple) sample population density across Lineage 1 or Lineage 2 pseudotime.
To further explore the phenotypes and transcriptional changes occurring in TAMs after exercise, we subset myeloid cell populations from the scRNA-seq data set. Myeloid cells expressing Cd68, including monocyte, macrophage, and DC populations (TAM 1, TAM 2, Mono DCs; Supplementary Fig. S5C), clustered into seven distinct populations classified according to gene markers: MHCIIhi TAM, Lyc6hi Monocytes (Lyc6hi Mono), Resident TAM (Res TAM), M2-like TAM, DC, Ly6Clo Monocytes (Lyc6lo Mono), and Cycling TAM (Fig. 6G, Supplementary Fig. S7A–B). Lyc6lo Mono and Lyc6hi Mono expressed the monocyte markers Ly6c2 and Sell, which encodes CD62L. These populations also expressed the highest levels of the monocyte marker Ccr2. MHCIIhi TAM had high expression of MHCII genes and low expression of Adgre1, which encodes for the macrophage marker F4/80. Res TAMs expressed the highest levels of Adgre1, suggestive of a resident TAM population. M2-like TAMs expressed low levels of MHCII genes and genes associated with immune suppression including Hilpda [29], Atf3 [30], and Ero1l [31], in addition to Vegfa. The DC population expressed MHCII genes in addition to DC marker Cd209a. Lastly, the cycling TAMs expressed macrophage marker Adgre1 and proliferation marker Mki67 (Supplementary Fig. S7A–B).
Comparison of cell proportions revealed that exercise increased clusters expressing high levels of MHCII genes: MHCIIhi TAM (65%) and DC (68%; Fig. 6H). Further, exercise decreased the number of M2-like TAMs (34%; Fig. 6H), in agreement with the flow cytometry experiments (Fig. 6B). Remaining cluster proportions were also altered including an increase in cycling TAMs (59%; Fig. 6H).
To investigate the transcriptional changes caused by exercise in the myeloid populations we performed differential expression analysis comparing exercise to control samples (Fig. 6I). We observed that exercise not only expanded antigen presenting–cell populations but also significantly increased the expression of MHCII genes including H2-Aa, H2-Eb1, and H2-Ab1, particularly in the M2-like TAM cluster (Supplementary Fig. S7C), supporting the idea of an exercise-induced shift in these cells toward antigen presentation. Ingenuity Pathway Analysis of differentially expressed genes in the combined myeloid populations indicated that the top pathways altered by exercise were all downregulated; 3 of the most downregulated had inflammatory signaling functions (Supplementary Fig. S6G), suggesting that overall, exercise increases the antigen presentation capacity of TAMs/myeloid cells without increasing their inflammatory cytokine signaling.
Based on these results we applied the pseudotime lineage branch analysis tool, Slingshot [23], to model lineage trajectories within non-cycling monocytes/macrophages in our dataset. The analysis predicted two lineages across the populations (Lineage 1: Ly6Clo Monos -> Ly6Chi Monos -> MHCIIhi TAM -> Res TAM; Lineage 2: Ly6Clo Monos -> Ly6Chi Monos -> M2-like TAM; Fig. 6J). Population density across predicted pseudotime lineages revealed that cells from exercise samples were more abundant toward the end of lineage 1 compared to control samples (Fig. 6K), suggesting exercise may promote enrichment of MHCIIhi TAM via the lineage 1 trajectory. However, determining if this enrichment is due to cellular differentiation/plasticity within the tumor microenvironment must be validated experimentally in the future.
Of note, numerous metabolic genes were differentially expressed in the myeloid populations. We found exercise increased the expression of OXPHOS-related genes (mt-Nd1, mt-Nd3, mt-Nd4) and decreased the expression of numerous glycolysis-related genes (Gapdh, Pkm, Pgam1, Hk2, Pgk1, Eno1, Aldoa, and Ldha; Fig. 6I). A distinct metabolic gene expression pattern was observed in the M2-like TAM population, which displayed the highest level of glycolysis-related gene expression and the lowest level of OXPHOS-related gene expression, indicating a potential dependency on glycolysis unique to the cluster (Supplementary Fig. S7D–E).
Exercise, in the presence of global ERK5 S496A KI, increases tumor growth and shifts TAM phenotype
Previous studies have focused strongly on T-cell responses to exercise with few investigating the myeloid compartment. Based on our findings that exercise modulated TAM number and phenotype in the exercise responsive YUMMER model, we sought to investigate mechanisms by which exercise alters the tumor microenvironment in YUMMER tumors. Although never studied in the context of a tumor, ERK5 S496 phosphorylation impacts peripheral and bone marrow–derived myeloid cell function by impairing efferocytosis and increasing inflammation, and it impacts transcription of numerous macrophage function–related genes [13, 14], suggesting a potential role for ERK5 signaling via the S496 residue in exercise-induced changes in TAMs. Thus, we assessed ERK5 S496 phosphorylation in TAMs from sedentary and exercise tumor samples.
Consistent with a role for ERK5 S496 signaling in exercise-induced changes in the TAM phenotypic shifts, there was significantly less ERK5 S496 phosphorylation in F4/80+ TAMs in YUMMER tumors from exercised compared to sedentary mice (Fig. 7A–B). To further investigate the role of ERK5 S496 signaling in tumor progression and exercise, we utilized ERK5 S496A KI mice in which serine 496 has been mutated to alanine globally, thus preventing ERK5 phosphorylation at the S496 residue. YUMMER tumor growth was suppressed in sedentary ERK5 S496A KI mice relative to wild-type (WT), similar to the tumor suppressive effect of exercise (Fig. 7C). Also similar to the effects of exercise, tumors from sedentary ERK5 S496A KI mice had significantly fewer CD206+iNOS− M2-like TAMs (p=0.003), and more iNOS+CD206− M1-like macrophages than tumors from sedentary WT mice (p=0.057; Fig. 7D–H). No significant differences were observed between groups in overall number of TAMs, or expression of the M2 marker CD206 or the M1 marker iNOS (Fig. 7D–H).
Figure 7. ERK5 S496A KI alters tumor growth and shifts TAM phenotype differently than with exercise.
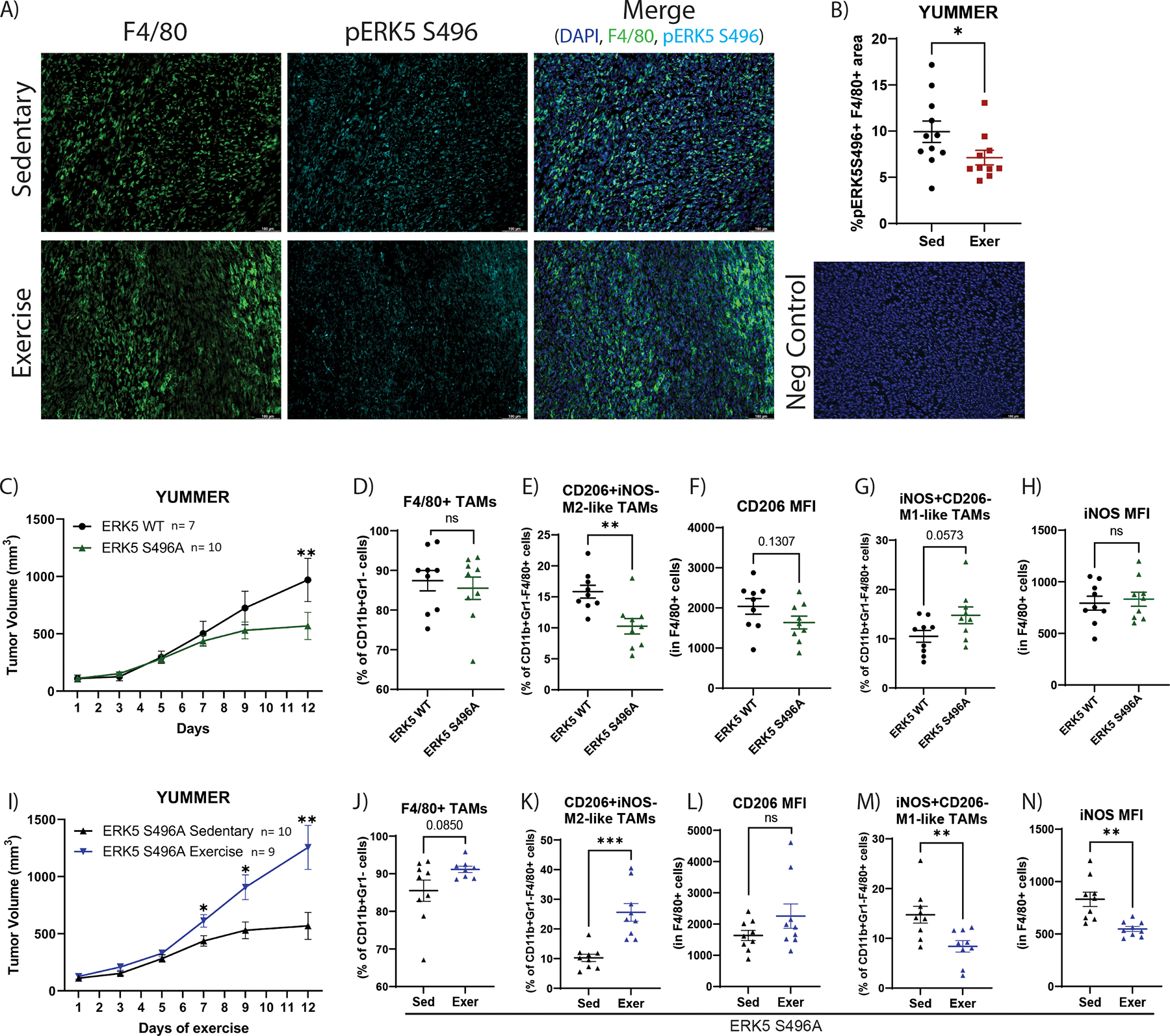
YUMMER tumor–bearing mice performed approximately two weeks of treadmill exercise (Exer) or were non-exercised sedentary controls (Sed). A) Immunofluorescent images of F4/80 (marker for TAMs; green), phospho-ERK5 S496 (cyan), and DAPI (blue) on YUMMER tumors, representative of >5 microscopic fields per tumor and 7 –10 tumors per group; scale bar: 100μm. B) Quantification of percent of phospho-ERK5 S496+F4/80+ area in YUMMER tumors from sedentary or exercised mice. Graphs are displayed with mean +/− standard error of the mean (SEM); each point represents one tumor value obtained by the average of ≥ 5 10x microscopic fields. C) YUMMER tumor volume in ERK5 WT (C57Bl/6) mice and ERK5 S496A KI mice was monitored with calipers. Tumor growth curves were analyzed using two-way ANOVA (Time v ERK5S496A: p=0.0122; Time: p<0.0001; Exercise: p=0.0.2119) with post-hoc multiple comparisons displayed on graphs. ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Flow cytometry quantification of the percent of D) F4/80+ TAMs relative to CD11b+Gr1− cells, E) CD206+iNOS− TAMs relative to CD11b+Gr1−F4/80+ cells, F) CD206 MFI in CD11b+Gr1−F4/80+ cells, G) iNOS+CD206− TAMs relative to CD11b+Gr1−F4/80+ cells, H) iNOS MFI in CD11b+Gr1−F4/80+ cells. I) YUMMER tumor volume in sedentary or exercised ERK5 S496A KI mice was monitored with calipers. Tumor growth curves were analyzed using two-way ANOVA (Time v Exercise: p<0.0001; Time: p<0.0001; Exercise: p=0.0061) with post-hoc multiple comparisons displayed on graphs. ns= p > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Flow cytometry quantification of the percent of J) F4/80+ TAMs relative to CD11b+Gr1− cells, K) CD206+iNOS− TAMs relative to CD11b+Gr1−F4/80+ cells, L) CD206 MFI in CD11b+Gr1−F4/80+ cells, M) iNOS+CD206− TAMs relative to CD11b+Gr1−F4/80+ cells, N) iNOS MFI in CD11b+Gr1−F4/80+ cells. Mouse experiments utilizing ERK5 S496A KI mice were independently repeated two times. Each point on a graph represents one tumor analyzed.
When treated with exercise, tumors from ERK5 S496A KI mice had a reversed phenotype compared to WT mice. In ERK5 S496A KI mice, exercise increased growth of tumors (Fig. 7I). In addition, in contrast to the effects in WT mice, exercise increased TAM abundance in ERK5 S496A KI mice (p=0.085; Fig. 7J), as well as increased M2-like TAMs (p<0.001; Fig. 7K). No change in expression of M2 marker CD206 was observed (Fig. 7L). We detected decreased levels of M1-like TAMs (p=0.007; Fig. 7M) and expression of the M1 marker iNOS (p=0.001; Fig. 7N) in ERK5 S496A KI exercised mice relative to sedentary.
Finally, we took advantage of the global nature of the ERK5 S496A mutation and investigated changes in vascular inflammation because ERK5 S496 phosphorylation increases inflammation via adhesion molecule expression in endothelial cells [32, 33]. We found that ERK5 S496A KI mice had significantly higher VCAM1 expression on tumor vasculature (p= 0.034), again similar to the effect of exercise alone, while the expression of VCAM1 in ERK5 S496A exercised mice was lower than the ERK5 S496A KI alone (p =0.079, Supplementary Fig. S8A–B).
The stark contrast between the exercise-induced changes in WT mice and ERK5 S496A KI mice suggests a critical role for ERK5 in mediating exercise-induced changes in the tumor microenvironment. The complex interactions between the multiple factors changed by exercise and the effects of inhibiting ERK5 S496 signaling are not clear. We used evaluation of BMDMs isolated from tumor-bearing sedentary and exercised mice to understand whether ERK5 and exercise-regulated signaling may impact the immune response, specifically macrophages, systemically. Exercise decreased ERK5 S496 phosphorylation and increased ERK5 TEY phosphorylation in WT BMDMs. ERK5 TEY phosphorylation was similarly increased in ERK5 S496A KI BMDMs, but it was suppressed by exercise in these mice (Supplementary Fig. S8C–E). BMDMs from both exercised WT or sedentary ERK5 S496A KI mice displayed increased expression of antioxidants, Trx1 and HO1, and Gas6 (“Eat-me” signal) relative to sedentary WT, while the combination of exercise and ERK5 S496A KI reversed this increase (Supplementary Fig. S8F–I). SIRPα (“Don’t eat-me” signal) expression was decreased in exercised WT and sedentary ERK5 S496A KI mice compared to sedentary WT, however, this change was not significantly reversed in exercised ERK5 S496A KI mice (Supplementary Fig. S8J).
Finally, exercise of WT mice or ERK5 S496A KI decreased cellular ROS and efferocytosis in BMDMs relative to sedentary WT, while the combination of exercise and ERK5 S496A KI increased cellular ROS and efferocytosis (Supplementary Fig. S8K–M). These data consistently suggest that exercise and loss of ERK5 S496 phosphorylation both promote antioxidant responses and phagocytic function of macrophages, while exercise combined with loss of ERK5 S496 phosphorylation causes an opposite effect of either alone.
DISCUSSION
Aerobic exercise influences multiple systems of the body concurrently by improving cardiovascular health, altering systemic cytokines, and mobilizing immune cells [34] in addition to changing tumor biology by numerous mechanisms [35]. The current study characterizes the effect of aerobic exercise on the microenvironment of two murine melanoma models. We demonstrate that exercise improves tumor vasculature and upregulates VCAM1 in both models, but differentially regulates tumor hypoxia, tumor growth, and the immune response in YUMMER and B16F10 models. Moreover, we uncover the role of ERK5 S496 as an exercise-responsive residue with functions in modulating adhesion molecule expression and TAM polarization.
One way tumor vessels participate in immune evasion is by downregulating immune adhesion molecules critical for immune entry to the tumor tissue [24]. Strategies to increase VCAM1 expression in tumor vasculature have proven efficacious, causing increased CD8+ T-cell presence in tumor tissue and decreased tumor volume [36]. Our data suggest that aerobic exercise is a novel method to increase adhesion molecules while simultaneously increasing immune cell mobilization into the tumor. However, increased VCAM1 expression cannot explain the differences in the immune response to exercise between YUMMER and B16F10 tumors, as exercise increased VCAM1 in both models.
Despite similar tumor vessel remodeling, exercise reduced hypoxia in YUMMER but not B16F10 tumors. Thus, the reduced hypoxia in YUMMER tumors may be due to tumor cell intrinsic changes in oxygen consumption. Tumor cells without proper oxygen availability depend on anaerobic metabolic pathways, including glycolysis and lactate production. This creates a hypoxic, acidic environment detrimental to the antitumoral function of immune cells [37]. Consistent with this, we found lower levels of hypoxia and an increased number of active CD8+ T cells after exercise in YUMMER tumors. In contrast, CD8+ T-cell abundance and activation remained unchanged in B16F10 tumors. Of note, we did not experimentally test a causative relationship between hypoxia and the tumor immune response. While our data is consistent with the concept of an inverse relationship between tumor hypoxia and an antitumor immune response by cytotoxic T cells, no causation between hypoxia and immune response is demonstrated by our data.
In agreement with the increased frequency of CD8+ T cells in YUMMER but not B16F10 tumors of exercised mice, exercise reduced the growth of YUMMER but not B16F10 tumors. Of note, we previously found that exercise increases B16F10 growth [6]. These conflicting results may stem from B16F10 cell stocks acquired from different sources, C57Bl/6 mice bred in different facilities with varying microbiomes and immune systems, and the use of male mice in the current study compared to mixed sex cohorts in our previous publication. Our most recent data are consistent with the observations of several independent groups that have reported that exercise performed after tumor implantation does not alter B16F10 growth [7, 38].
In the YUMMER tumor model, exercise increased PD1 expression on intratumoral CD8+ T cells, suggesting that exercise may enhance the efficacy of immune checkpoint blockade in melanoma. However, we did not observe a significant improvement in anti-PD1 efficacy when combined with exercise to treat YUMMER, although an increase in CD8+/Treg cell ratio was detected. Nevertheless, exercise may still be a useful adjuvant to immune checkpoint blockade for the treatment of melanoma. The timing of exercise in relation to immune checkpoint blockade administration, tumor size at treatment initiation, and appropriate dosing of anti-PD1 and exercise are all likely critical for the combination of the two to succeed and warrant further study.
Preclinical studies investigating the effect of exercise in tumor immune compartments have largely focused on lymphocytes without sufficient evaluation of the myeloid cells. We found that exercise increases the frequency of DCs while reducing that of M-MDSCs and M2-like TAMs, as assessed by flow cytometry. Additionally, scRNA-seq revealed an increase in TAMs expressing MHCII indicating that exercise may increase antigen presentation in the tumor microenvironment, which could contribute to T-cell activation. Moreover, our data demonstrate that exercise decreases glycolysis-related genes and increases OXPHOS-related genes in myeloid-cell populations. Macrophage phenotypes are linked to their metabolic states. M1 macrophages rely on glycolysis and M2 macrophages on OXPHOS. However, TAM phenotypes are more complex, with studies revealing potential reliance on glycolysis, fatty acid oxidation, or OXPHOS [39]. We report that in YUMMER tumors, metabolic gene expression was heterogenous in TAMs with M2-like TAMs having high glycolytic gene expression and low OXPHOS gene expression relative to the other myeloid populations, suggesting a dependency on glycolysis. A caveat of our findings regarding exercise regulation of metabolism and lineage trajectories in TAMs is that we did not validate the scRNAseq-based bioinformatics conclusions at the protein or functional level. Additional studies are required to investigate the role of exercise in TAM metabolism and polarization.
There is no disputing that aerobic exercise has robust effects on tumor biology. However, our knowledge of the mechanisms responsible for these effects is limited. Here we illustrate the importance of the MAPK protein, ERK5, in the tumor response to aerobic exercise. Specifically, we uncover a role for ERK5 S496 phosphorylation in tumor vessel adhesion molecule expression and TAM phenotype. The tumor microenvironment changes caused by exercise were largely mimicked by ERK5 S496A KI mice. However, the effects of exercise in ERK5 S496A KI mice were largely reversed relative to WT mice, with exercise resulting in a notable increase in M2-like macrophages in tumors from ERK5 S496A KI mice. Based on these findings, we evaluated the role of ERK5 S496A and exercise in BMDMs revealing that depletion of ERK5 S496 phosphorylation and exercise promotes efferocytosis and reduces oxidative stress in BMDMs illustrating a striking systemic shift in macrophage function.
ERK5 is a complex protein [40] and the reversal of the exercise phenotype, particularly in macrophage polarization, may be explained by compensatory signaling via other ERK5 regulatory modifications (such as TEY phosphorylation or sumoylation). ERK5 phosphorylation site–specific signaling should be interrogated further and would benefit from the use of cell specific instead of global ERK5 S496A KI. More studies are needed to fully characterize the effects of the ERK5 S496 residue on the tumor microenvironment and in the context of exercise.
Limitations of this study include our lack of treadmill acclimation. Treadmill exposure can cause stress in mice and alter responses to exercise. Additionally, although sedentary mice were handled for the same amount of time as exercising mice, they were never placed on the treadmill. These shortcomings in study design should be addressed in future studies. Further, another limitation is that some conclusions (i.e., metabolic changes in TAMs and tumor cells by exercise) are supported only by scRNAseq data and not validated at the protein level. Finally, our analysis of T cells within tumors is limited by the fact that we did not perform any functional assessments and therefore cannot make conclusions about whether exercise changed tumor infiltrating T-cell function.
This study reveals context dependencies of exercise in two murine models of the same tumor type, melanoma. This, in turn, demonstrates the need for more in-depth characterization of exercise across different cancers and sub-types within a cancer type. Our findings illustrate a multitude of changes that occur in a solid tumor in response to aerobic exercise and suggest a critical role for ERK5 in mediating exercise-induced changes in the tumor microenvironment.
Supplementary Material
Synopsis.
The mechanisms underlying exercise-induced changes in the tumor microenvironment are incompletely understood. The authors demonstrate that aerobic exercise alters intratumoral T- and myeloid-cell function and that ERK5 S496 phosphorylation is key in exercise-dependent changes in these immune cells.
Acknowledgments
We would like to acknowledge the MD Anderson CPRIT Single Core (CPRIT RP180684) for processing and sequencing the scRNA-seq samples in this paper. Thank you to Nicholas Navin and all the members of the core including Tuan Tran, Anna Casasent, Jerome Lin, and Jianzhou Li for their input in experimental design and data interpretation. We would like to thank Aislyn Schalck for help guiding and interpreting single cell immune cell data. Supplementary Fig. S4A was created with BioRender.com.
Funding:
This work was supported in part by funding from the Cancer Prevention Research Institute of Texas (CPRIT RP190256), which supported HS, RB, MI, JA, and KS. KS and JL were supported in part by funding from a Career Enhancement Program award of The University of Texas MD Anderson Cancer Center SPORE in Melanoma provided by the National Cancer Institute (5P50CA221703-04). HS was supported by CPRIT RP170067 during the completion of this work. HS and DCM were supported by a fellowship from the Schissler Foundation. SP was funded by the Pauline Altman-Goldstein Foundation Discovery Fellowship. JA was supported in part by grants from the National Institutes of Health (AI-156921 and CA016672). The Advanced Cytometry & Sorting Core Facility and Flow Cytometry and Cellular Imaging Facility at MD Anderson, utilized for flow cytometry experiments, are supported by NCI P30CA016672 and CA016672, respectively. MS is funded by the CPRIT Training Program (RP210028).
Footnotes
Author disclosures:
The authors have report nothing to disclose.
References
- 1.Moore SC, Lee IM, Weiderpass E, Campbell PT, Sampson JN, Kitahara CM, … Patel A. v. (2016). Association of Leisure-Time Physical Activity With Risk of 26 Types of Cancer in 1.44 Million Adults. JAMA Internal Medicine, 176(6), 816–825. 10.1001/JAMAINTERNMED.2016.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mctiernan A, Friedenreich CM, Katzmarzyk PT, Powell KE, Macko R, Buchner D, … Piercy KL (2019). Physical Activity in Cancer Prevention and Survival: A Systematic Review. Medicine and science in sports and exercise, 51(6), 1252–1261. 10.1249/MSS.0000000000001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerritsen JKW, & Vincent AJPE (2016). Exercise improves quality of life in patients with cancer: a systematic review and meta-analysis of randomised controlled trials. British Journal of Sports Medicine, 50(13), 796–803. 10.1136/BJSPORTS-2015-094787 [DOI] [PubMed] [Google Scholar]
- 4.Segal R, Zwaal C, Green E, Tomasone JR, Loblaw A, Petrella T, & Group E for P. with C. G. D. (2017). Exercise for people with cancer: a systematic review. Current Oncology, 24(4), e290. 10.3747/CO.24.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betof AS, Lascola CD, Weitzel D, Landon C, Scarbrough PM, Devi GR, … Dewhirst MW (2015). Modulation of Murine Breast Tumor Vascularity, Hypoxia, and Chemotherapeutic Response by Exercise. JNCI: Journal of the National Cancer Institute, 107(5). 10.1093/jnci/djv040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schadler KL, Thomas NJ, Galie PA, Bhang DH, Roby KC, Addai P, … Ryeom S (2016). Tumor vessel normalization after aerobic exercise enhances chemotherapeutic efficacy. Oncotarget, 7(40), 65429–65440. 10.18632/oncotarget.11748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen L, Idorn M, Olofsson GH, Lauenborg B, Nookaew I, Hansen RH, … Hojman P (2016). Voluntary Running Suppresses Tumor Growth through Epinephrine- and IL-6-Dependent NK Cell Mobilization and Redistribution. Cell Metabolism, 23(3), 554–562. 10.1016/j.cmet.2016.01.011 [DOI] [PubMed] [Google Scholar]
- 8.Rundqvist H, Veliça P, Barbieri L, Gameiro PA, Bargiela D, Gojkovic M, … Johnson RS (2020). Cytotoxic t-cells mediate exercise-induced reductions in tumor growth. eLife, 9, 1–25. 10.7554/ELIFE.59996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buss LA, Williams T, Hock B, Ang AD, Robinson BA, Currie MJ, & Dachs GU (2021). Effects of exercise and anti-PD-1 on the tumour microenvironment. Immunology letters, 239, 60–71. 10.1016/J.IMLET.2021.08.005 [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Sanderson SM, Zessin A, Ashcraft KA, Jones LW, Dewhirst MW, … Hsu DS (2018). Exercise inhibits tumor growth and central carbon metabolism in patient-derived xenograft models of colorectal cancer. Cancer & Metabolism 2018 6:1, 6(1), 1–11. 10.1186/S40170-018-0190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huijbers EJM, Khan KA, Kerbel RS, & Griffioen AW (2022). Tumors resurrect an embryonic vascular program to escape immunity. Science Immunology, 7(67), 6388. 10.1126/SCIIMMUNOL.ABM6388/ASSET/DBFA8AF6-1395-4453-99BC-D9036AFA5994/ASSETS/IMAGES/LARGE/SCIIMMUNOL.ABM6388-F3.JPG [DOI] [PubMed] [Google Scholar]
- 12.Nithianandarajah-Jones GN, Wilm B, Goldring CEP, Müller J, & Cross MJ (2014). The role of ERK5 in endothelial cell function. Retrieved from https://research.aston.ac.uk/portal/files/19202509/ERK5_in_endothelial_cell_function.pdf [DOI] [PubMed]
- 13.Kotla S, Zhang A, Imanishi M, Ko KA, Lin SH, Gi YJ, … Abe J ichi. (2021). Nucleus-mitochondria positive feedback loop formed by ERK5 S496 phosphorylation-mediated poly (ADP-ribose) polymerase activation provokes persistent pro-inflammatory senescent phenotype and accelerates coronary atherosclerosis after chemo-radiation. Redox Biology, 47. 10.1016/J.REDOX.2021.102132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh M. v., Kotla S, Le NT, Ko KA, Heo KS, Wang Y, … Abe JI (2019). Senescent Phenotype Induced by p90RSK-NRF2 Signaling Sensitizes Monocytes and Macrophages to Oxidative Stress in HIV-Positive Individuals. Circulation, 139(9), 1199–1216. 10.1161/CIRCULATIONAHA.118.036232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmonti E, Savage H, Zhang A, Bedoya CAF, Morrell MG, Harden A, … Schadler K (2020). Modulating sphingosine-1-phosphate receptors to improve chemotherapy efficacy against Ewing sarcoma. International Journal of Cancer, ijc.32862. 10.1002/ijc.32862 [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Hong J, Umetani M, Lavoy EC, Kim JH, & Park Y (2020). Vascular Protection by Exercise in Obesity: Inflammasome-associated Mechanisms. Medicine and science in sports and exercise, 52(12), 2538–2545. 10.1249/MSS.0000000000002419 [DOI] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, … Satija R (2021). Integrated analysis of multimodal single-cell data. Cell, 184(13), 3573–3587.e29. 10.1016/J.CELL.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. (2022). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/. [Google Scholar]
- 19.Blighe K, Rana S, & Myles L (2021). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.12.0. https://github.com/kevinblighe/EnhancedVolcano. [Google Scholar]
- 20.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, & Sergushichev A (2021). Fast gene set enrichment analysis. bioRxiv, 060012. 10.1101/060012 [DOI] [Google Scholar]
- 21.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, & Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems, 1(6), 417–425. 10.1016/J.CELS.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolgalev I (2022). msigdbr: MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format. R package version 7.5.1. https://igordot.github.io/msigdbr. [Google Scholar]
- 23.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, … Dudoit S (2018). Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics, 19(1), 1–16. 10.1186/S12864-018-4772-0/FIGURES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijbers EJM, Khan KA, Kerbel RS, & Griffioen AW (2022). Tumors resurrect an embryonic vascular program to escape immunity. Science Immunology, 7(67), 6388. 10.1126/SCIIMMUNOL.ABM6388/ASSET/DBFA8AF6-1395-4453-99BC-D9036AFA5994/ASSETS/IMAGES/LARGE/SCIIMMUNOL.ABM6388-F3.JPG [DOI] [PubMed] [Google Scholar]
- 25.Wu NZ, Klitzman B, Dodge R, & Dewhirst MW (1992). Diminished Leukocyte-Endothelium Interaction in Tumor Microvessels1 | Cancer Research | American Association for Cancer Research. Cancer Research, 52(15), 4265–4268. Retrieved from https://aacrjournals.org/cancerres/article/52/15/4265/497819/Diminished-Leukocyte-Endothelium-Interaction-in [PubMed] [Google Scholar]
- 26.Griffioen AW, Damen CA, Martinotti S, Blijham GH, & Groenewegen G (1996). Endothelial intercellular adhesion molecule-1 expression is suppressed in human malignancies: the role of angiogenic factors. Cancer Research, 56(5), 1111–1117. Retrieved from https://pubmed.ncbi.nlm.nih.gov/8640769/ [PubMed] [Google Scholar]
- 27.Harjunpää H, Asens ML, Guenther C, & Fagerholm SC (2019). Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Frontiers in Immunology, 10(MAY), 1078. 10.3389/FIMMU.2019.01078/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.di Pilato M, Kfuri-Rubens R, Pruessmann JN, Ozga AJ, Messemaker M, Cadilha BL, … Mempel TR (2021). CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell, 184(17), 4512–4530.e22. 10.1016/J.CELL.2021.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Zhou X, Zeng H, Wu D, & Liu L (2021). HILPDA Is a Prognostic Biomarker and Correlates With Macrophage Infiltration in Pan-Cancer. Frontiers in Oncology, 11. 10.3389/FONC.2021.597860/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sha H., Zhang D., Zhang Y., Wen Y., & Wang Y. (2017). ATF3 promotes migration aand M1/M2 polarization of macrophages by activating tenascin-C via Wnt/β-catenin pathway. Molecular medicine reports, 16(3), 3641–3647. 10.3892/MMR.2017.6992 [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Wang C, Li S, Qu Y, Xue P, Ma Z, … Wang J (2021). ERO1L Is a Novel and Potential Biomarker in Lung Adenocarcinoma and Shapes the Immune-Suppressive Tumor Microenvironment. Frontiers in Immunology, 12. 10.3389/FIMMU.2021.677169/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vu HT, Kotla S, Ko KA, Fujii Y, Tao Y, Medina J, … Abe J-I (2018). Ionizing Radiation Induces Endothelial Inflammation and Apoptosis via p90RSK-Mediated ERK5 S496 Phosphorylation. Frontiers in Cardiovascular Medicine, 5. 10.3389/fcvm.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le N-T, Heo K-S, Takei Y, Lee H, Woo C-H, Chang E, … Abe J -i. (2013). A Crucial Role for p90RSK-Mediated Reduction of ERK5 Transcriptional Activity in Endothelial Dysfunction and Atherosclerosis. Circulation, 127(4), 486–499. 10.1161/CIRCULATIONAHA.112.116988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiuza-Luces C, Santos-Lozano A, Joyner M, Carrera-Bastos P, Picazo O, Zugaza JL, … Lucia A (2018). Exercise benefits in cardiovascular disease: beyond attenuation of traditional risk factors. Nature Reviews Cardiology 2018 15:12, 15(12), 731–743. 10.1038/s41569-018-0065-1 [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Casado A, Martín-Ruiz A, Pérez LM, Provencio M, Fiuza-Luces C, & Lucia A (2017). Exercise and the Hallmarks of Cancer. Trends in Cancer, 3(6), 423–441. 10.1016/j.trecan.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 36.Nakajima K, Ino Y, Yamazaki-Itoh R, Naito C, Shimasaki M, Takahashi M, … Hiraoka N (2020). IAP inhibitor, Embelin increases VCAM-1 levels on the endothelium, producing lymphocytic infiltration and antitumor immunity. OncoImmunology, 9(1). 10.1080/2162402X.2020.1838812/SUPPL_FILE/KONI_A_1838812_SM7258.ZIP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalaora S, Nagler A, Wargo JA, & Samuels Y (2022). Mechanisms of immune activation and regulation: lessons from melanoma. Nature Reviews Cancer 2022 22:4, 22(4), 195–207. 10.1038/s41568-022-00442-9 [DOI] [PubMed] [Google Scholar]
- 38.Buss LA, Ang AD, Hock B, Robinson BA, Currie MJ, & Dachs GU (2020). Effect of post-implant exercise on tumour growth rate, perfusion and hypoxia in mice. PLoS ONE, 15(3). 10.1371/JOURNAL.PONE.0229290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Wang J, Yadav DK, Bai X, & Liang T (2021). Glucose Metabolism: The Metabolic Signature of Tumor Associated Macrophage. Frontiers in Immunology, 12, 2525. 10.3389/FIMMU.2021.702580/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin ECK, Amantea CM, Nomanbhoy TK, Weissig H, Ishiyama J, Hu Y, … Rosenblum JS (2016). ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proceedings of the National Academy of Sciences of the United States of America, 113(42), 11865–11870. 10.1073/PNAS.1609019113/SUPPL_FILE/PNAS.1609019113.SFIG03.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during this study are included in this article and its Supplementary Data files or are available from the corresponding author upon request. The scRNA-seq data generated in this study are publicly available in NCBI’s Gene Expression Omnibus (GEO) at accession number GSE208205.


