Summary
Background:
Timely and accurate intra-operative cryosection evaluations remain the gold standard for guiding surgical treatments for gliomas. However, the tissue freezing process often generates artifacts that make histologic interpretation difficult. In addition, the 2021 WHO Classification of Tumors of the Central Nervous System incorporates molecular profiles in the diagnostic categories, and thus standard visual evaluation of cryosections alone cannot completely inform diagnoses based on the new classification system.
Methods:
To address these challenges, we develop the context-aware Cryosection Histopathology Assessment and Review Machine (CHARM) using samples from 1,524 glioma patients from three different patient populations to systematically analyze cryosection slides.
Findings:
Our CHARM models successfully identified malignant cells (AUROC = 0.98 ± 0.01 in the independent validation cohort), distinguished IDH-mutant tumors from wildtype (AUROC = 0.79 – 0.82), classified three major types of molecularly-defined gliomas (AUROC = 0.88 – 0.93), and identified the most prevalent subtypes of IDH-mutant tumors (AUROC = 0.89 – 0.97). CHARM further predicts clinically important genetic alterations in low-grade glioma, including ATRX, TP53, CIC mutations, CDKN2A/B homozygous deletion, and 1p/19q codeletion via cryosection images.
Conclusions:
Our approaches accommodate the evolving diagnostic criteria informed by molecular studies, provide real-time clinical decision support, and will democratize accurate cryosection diagnoses.
Keywords: 2021 WHO Classification of glioma, cryosection, molecular subtypes, intra-operative diagnoses, machine learning, context-aware neural networks
Graphical Abstract
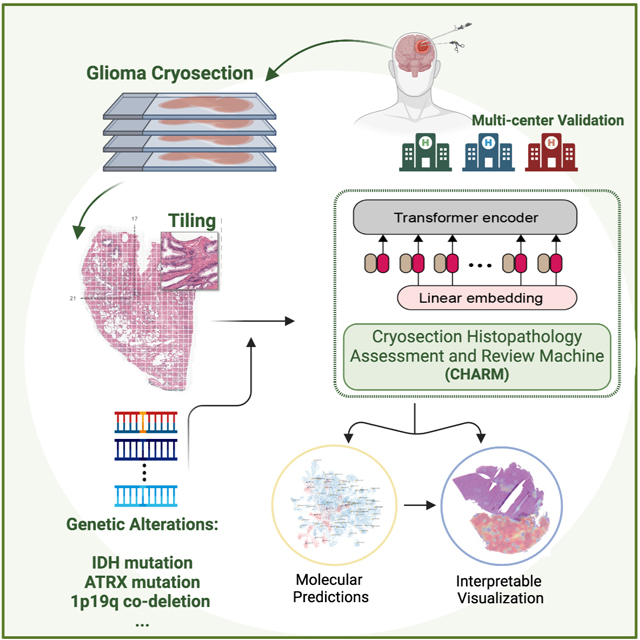
eTOC
Nasrallah et al. established the Cryosection Histopathology Assessment and Review Machine (CHARM), a context-aware machine learning method for glioma diagnosis during surgery. They showed that CHARM identifies pathology imaging patterns indicative of molecular diagnoses of glioma defined by the new WHO Classification.
INTRODUCTION
Advances in understanding the molecular pathogenesis of brain tumors have resulted in the identification of new diagnostic entities and substantial refinement in the diagnosis of infiltrating gliomas, the most common primary tumor of the central nervous system. The integration of molecular and histologic information has not only expanded the recognition of novel tumor types but has also improved prognostication and guided therapeutic decisions. The 2021 WHO Classification of Tumors of the Central Nervous System now recognizes 19 new official tumor types, as well as 3 additional provisional entities, which include molecular features for their classification1. Despite the plethora of entities, adult infiltrating gliomas most often fall into one of three major types: astrocytoma, IDH-mutant; oligodendroglioma, IDH-mutant and 1p/19q-codeleted; and glioblastoma, IDH-wildtype2. These three tumor types have significantly different survival outcomes, therapeutic options, and clinical trial eligibility3.
Establishing the IDH mutational status is essential to making a complete diagnosis given its prognostic significance4. In the absence of an IDH mutation, an infiltrating astrocytoma in an adult is most often a glioblastoma. In the presence of an IDH variant, 1p/19q codeletion status (along with other molecular aberrations) must be established to further classify the tumor as either astrocytoma or oligodendroglioma1. Given the necessity of incorporating molecular information with histologic findings to reach a diagnosis, pathologists are dependent on both for a definitive conclusion as to the nature of a tumor.
Because surgery continues to be the mainstay therapy for infiltrating gliomas, intra-operative examination of frozen tissue, also known as cryosection, is frequently performed to provide a preliminary diagnosis, guide the surgery, and determine treatment strategies6,7. Recent studies suggested that the extent of resection has a differential prognostic impact on patients with different molecular subtypes of glioma defined by the WHO Classification8,9. These findings indicated the need for fast intraoperative glioma diagnosis to guide treatment decisions. Because the WHO guidelines of glioma classification now involve molecular profiles (e.g., IDH mutation and 1p/19q codeletion status), the standard visual evaluation of cryosection tissues alone cannot determine the new diagnostic categories of gliomas, nor guide therapeutic decisions. Due to these limitations, a reliable tool for providing both histologic and molecular analyses at the time of surgery will have significant added value both for hospitals without a neuropathologist and institutions where novel therapies may be initiated at the time of, or shortly after, surgery.
Machine learning has enabled augmented pathology interpretation in recent years10. Advanced methods for analyzing whole-slide pathology images11–15 successfully predicted multi-omics profiles and treatment responses of lung, breast, ovarian, or renal cancers16–20. Gliomas remain one of the most challenging targets in this field because of their significant histologic heterogeneity. While a few studies have attempted to develop machine learning methods for glioma interpretation,21–23 these approaches either attained substantially lower performance compared with other cancer types or required advanced microscopy techniques24. In addition, most of the previous studies have focused their models on permanent slides24–26, which require several days to process and thus are not suitable for timely diagnosis during surgical intervention. Reliable quantitative methods for analyzing cryosection slides for glioma diagnosis and classification are currently lacking28.
In this study, we developed the Cryosection Histopathology Assessment and Review Machine (CHARM), a novel interpretable deep learning framework that incorporates a hierarchical vision transformer (ViT)29, to analyze cryosection samples from multiple patient cohorts. Using digital cryosection whole-slide images, CHARM successfully identified glioma cells, classified the histologic grades, and predicted key molecular alterations that define the 2021 WHO Classification, such as IDH mutation and 1p/19q codeletion status. We validated our results using three large patient cohorts. We showed that CHARM achieved better performance in these diagnostic and prediction tasks than state-of-the-art convolutional neural networks (CNNs). We further interpreted how CHARM arrives at its predictions and correlated these interpretations with the current neuropathology knowledge. Our results demonstrated the potential of interpretable computer vision approaches in analyzing cryosection tissue for real-time, intra-operative decision support for brain tumor diagnosis.
RESULTS
Overview
The machine learning workflow of CHARM is summarized in Figure 1. To minimize the manual efforts required for annotating the digital pathology slides, we employed a weakly-supervised learning approach to analyzing the whole-slide images16,19,29 and assigned the slide-level labels, such as histological grade or genomic biomarkers, to all tiles sampled from this slide. The patient demographics and distributions of genomic profiles of our study cohorts are summarized in Table 1.
Figure 1. Overview of the Cryosection Histopathology Assessment and Review Machine (CHARM).
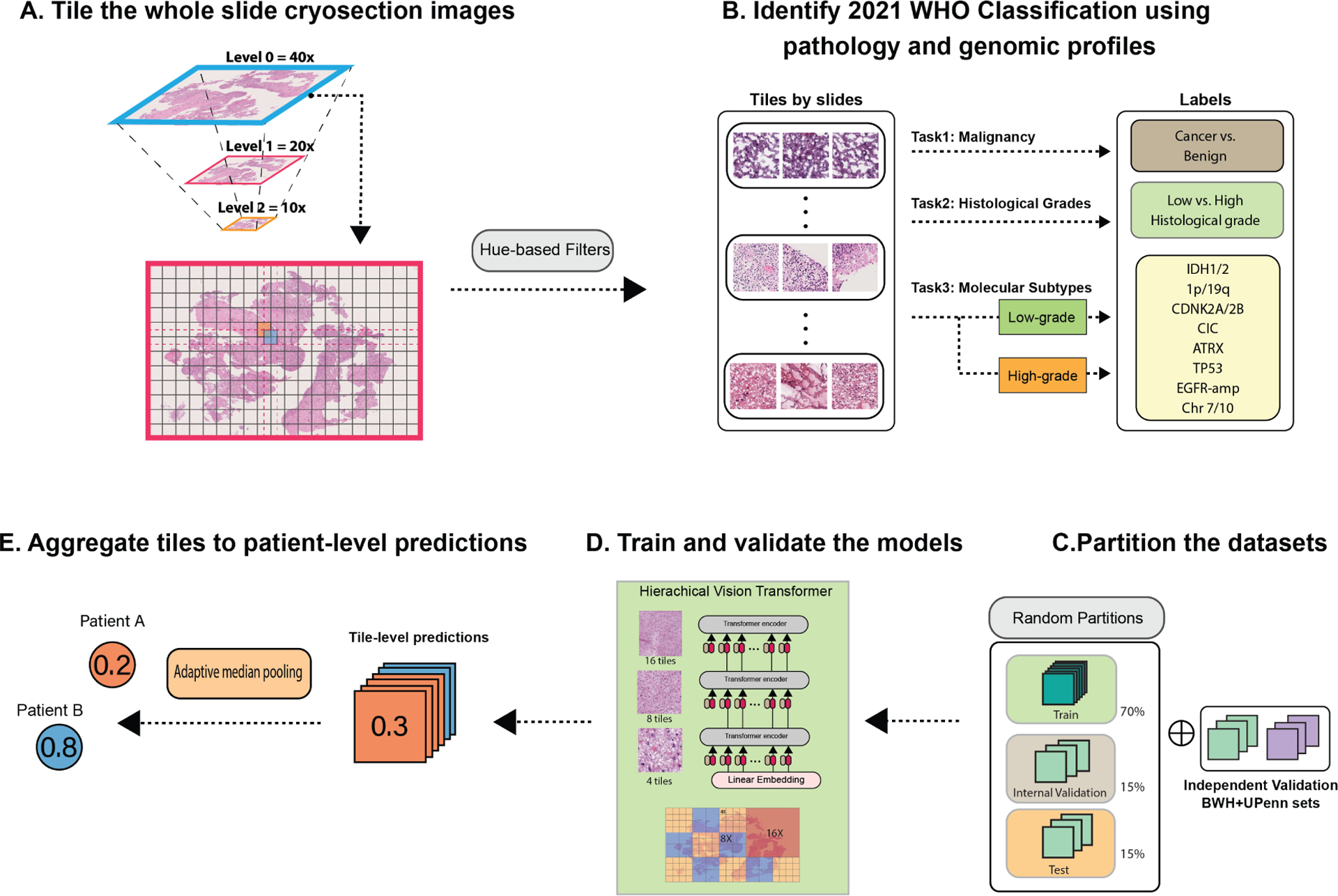
(A) We tiled each gigapixel whole slide cryosection image into patches with 1000 by 1000 pixels and 50 percent overlap between the adjacent patches. (B) We labeled each tile by its histologic grade or molecular profile. For genomic prediction tasks, we trained separate models for patients with different histologic grades (low vs. high) to eliminate the impact of the associations between histologic grades and genomic mutation status. (C) To evaluate the robustness of our models, we first randomly partitioned the dataset for each task in the ratio of 70–15-15 and repeated the random partition process to obtain the standard deviation of our performance metrics. (D) We trained hierarchical vision transformer-based CHARM to predict the molecular and histologic labels and compared the results. (E) After obtaining the tile-level prediction for each patch, we aggregated all tiles from the same slide by adaptive median pooling and generated a slide-level prediction. We further evaluated the models’ performance in the independent test sets and assessed their generalizability in independent patient cohorts.
Table 1.
Patient characteristics of our study cohorts.
| Number of patients | TCGA (n=1,121) | Brigham and Women’s Hospital (n=213) | University of Pennsylvania (n=190) |
|---|---|---|---|
| Age | 49.0 ± 15.7 | 48.8 ± 23.4 | 53.6 ± 15.7 |
| Sex | |||
| Not available | 295 (26.7%) | 23 (10.8%) | 0 (0%) |
| Male | 460 (41.6%) | 108 (50.7%) | 112 (58.9%) |
| Female | 351 (31.7%) | 82 (38.5%) | 78 (41.1%) |
| Histologic grades | |||
| Benign* | 5 (0.4%) | 68 (31.9%) | 0 (0%) |
| Grade 2 | 250 (22.3%) | 65 (30.5%) | 34 (17.9%) |
| Grade 3 | 265 (23.6%) | 14 (6.6%) | 22 (11.6%) |
| Grade 4 | 601 (53.6%) | 66 (30.9%) | 134 (70.5%) |
| Molecular Types of Gliomas | |||
| IDH-mutation, 1p/19q codeletion | 168 (15.0%) | ||
| IDH-mutation, 1p/19q intact | 272 (24.3%) | ||
| IDH-wildtype | 657 (58.6%) | ||
| MGMT | |||
| MGMT (+) | 625 (56.5%) | 45 (21.1%) | 59 (31.1%) |
| MGMT (−) | 295 (26.7%) | 18 (8.5%) | 51 (26.8%) |
| Not Specified | 186 (16.8%) | 150 (70.4%) | 80 (42.1%) |
| de novo cancers with histologic grade 2–3 | |||
| IDH-wildtype | 98 (19.1%) | 17 (32.7%) | 0 (0%) |
| IDH-mutant | 415 (80.9%) | 35 (67.3%) | 41 (100%) |
| de novo cancers with histologic grade 4 | |||
| IDH-wildtype | 559 (95.7%) | 46 (95.8%) | 78 (90.7%) |
| IDH-mutant | 25 (4.3%) | 2 (4.2%) | 8 (9.3%) |
Adjacent benign tissues of brain cancer patients
CHARM Identified Cryosection Diagnoses and Classified the Histologic Grades for Glioma
Confirming whether a sample contains malignant glial cells is the most important task of cryosection diagnoses conducted for suspected gliomas during neurosurgery. However, cryosection slides produced during surgery are difficult to evaluate due to artifacts of the cryosections and the heterogeneity of brain tumors. To address this challenge, we first investigated the performance of the CHARM framework in identifying cancer cells from cryosections. We collected 213 cryosection slides that were obtained intra-operatively from BWH to develop our CHARM model for this task. Among these slides, 145 contain glioma cells (labeled as cancer) while the other 68 do not have any cancer cells (labeled as benign). CHARM achieved an AUROC of 0.94 ± 0.02 in the held-out test set from BWH, while the conventional CNN method achieved an AUROC of 0.80 ± 0.10 (Figure 2A). We further evaluated our models’ generalizability using 1,121 patients in the TCGA cohort. CHARM demonstrated better generalizability (AUROC of 0.98 ± 0.01) compared with CNN (AUROC of 0.88 ± 0.06) in this independent evaluation (Figure S1). We visualized the prediction results of each region using heatmaps and showed that our models correctly identified regions of neoplastic cells and adjacent benign regions (Figures 2D and 2E).
Figure 2. CHARM successfully identified cryosections with malignant tissues and classified the histologic grade of glioma.
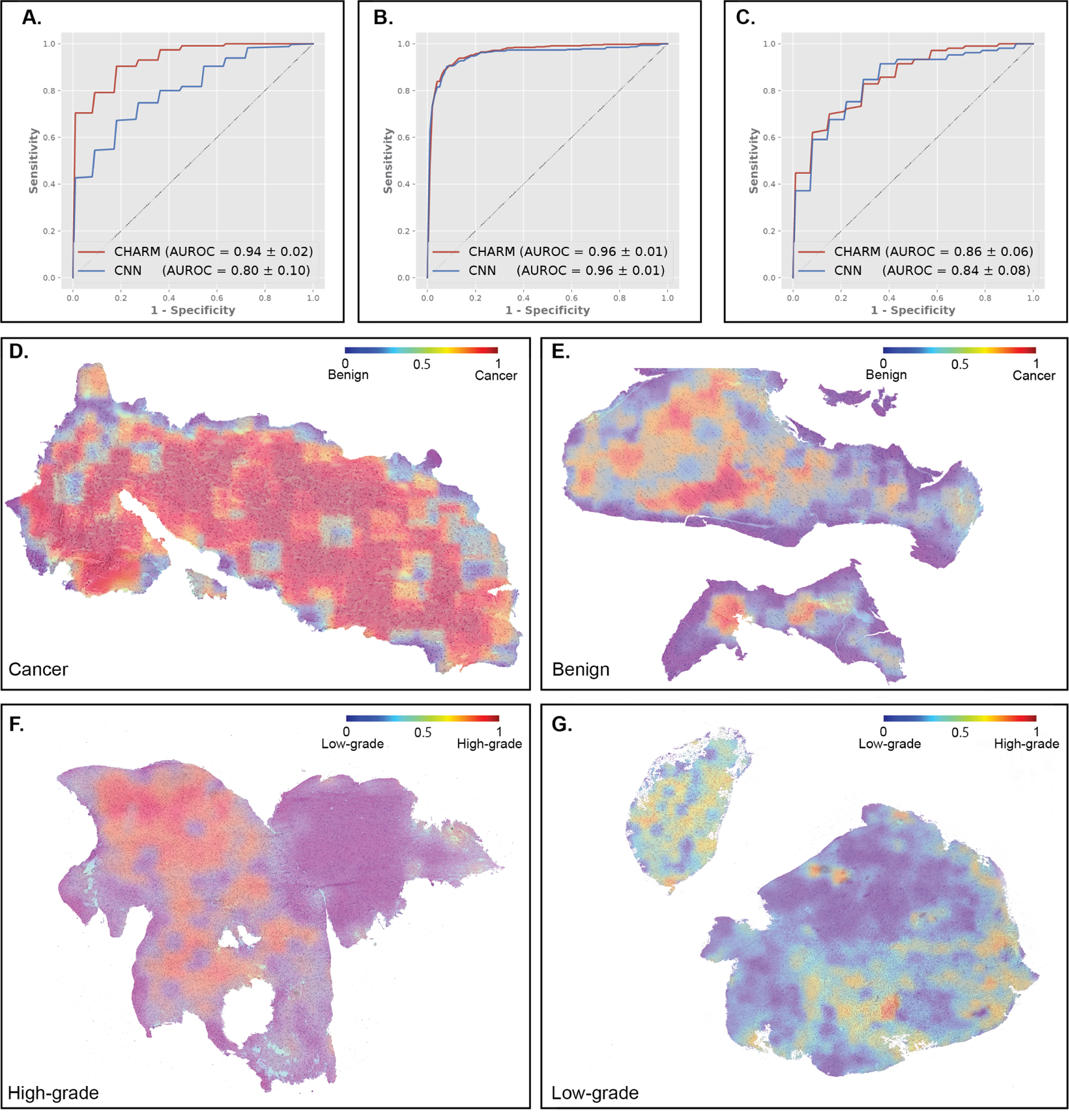
(A) CHARM outperformed convolutional neural network (CNN)-based models in identifying cryosections with malignant tissues, with an AUROC of 0.94 in the hold-out test set. CNN only attained an AUROC of 0.80 in the same task. (B) CHARM successfully classified gliomas with low and high histologic grades in the hold-out test set with an AUROC of 0.96. (C) CHARM demonstrated good generalizability to the independent test cohorts from BWH and UPenn for histologic grade prediction. AUROCs of CHARM and CNN models are 0.84 ± 0.08 and 0.86 ± 0.06, respectively. (D) Visualization of the model’s predictions on glioma tissues. Red indicates predicted malignant regions and blue indicates benign regions. Our prediction models output high probabilities of glioma for regions occupied by malignant cells. (E) Regions of adjacent benign cryosection tissue received low predictive probabilities for neoplasm. (F) Visualization of the model’s predictions on high and low histologic grades. Red indicates predicted high-grade regions and blue indicates predicted low-grade regions. Model predictions of a glioma sample with high histologic grade. Regions with dense glioma and necrosis are predictive of high histologic grade. (G) Model predictions of a glioma sample with low histologic grade.
Next, we examined the CHARM’s capability of classifying the histologic grades of glioma, a pathology evaluation task that assists in determining patients’ prognoses. Each patient was labeled as low histology grade (histologic grade 2 or 3) or high histology grade (histologic grade 4) by neuropathologists. Both CHARM and CNN models were trained using the TCGA data and achieved high performance in the hold-out test set with a mean AUROC of 0.96 ± 0.01 (Figure 2B). When evaluated on independent cohorts from BWH and UPenn, both models demonstrated high prediction performance (CHARM: 0.84 ± 0.08 and CNN: 0.86 ± 0.06; Figure 2C). Figure 2F shows the prediction results of a patient with low-grade glioma, and Figure 2G visualizes a high-grade glioma sample overlaid with our model predictions. These visualization results suggested that CHARM independently discovered that dense areas of tumor cells with necrosis are associated with high histologic grade.
CHARM Predicted IDH Mutational Status
We next employed the CHARM framework to predict IDH mutational status, a key component in the new WHO classification guideline for adult gliomas. To identify additional morphologic patterns indicative of IDH mutational status beyond histologic grades, we built separate models for the two grade groups. In the low histology grade group, results show that CHARM achieved similar performance compared with CNN on TCGA (AUROCs: 0.80 ± 0.074 versus 0.79 ± 0.083; Figure 3A). However, when evaluated with the independent datasets, CHARM outperformed the CNN baseline (0.82 ± 0.11 vs. 0.77 ± 0.08; Figure 3B). Glioma samples in the high histologic grade group were once all considered glioblastoma. The 2021 WHO classification, however, requires an infiltrating glioma to be IDH wildtype to be considered glioblastoma, even in the presence of classic “glioblastoma” histologic features, such as pseudopalisading necrosis and microvascular proliferation. In the high histologic grade group, we further showed that CHARM performed significantly better than CNN in identifying morphological signals indicative of IDH mutational status (AUROCs: 0.79 ± 0.04 versus 0.60 ± 0.05, Figure S2).
Figure 3. CHARM predicted IDH mutational status using cryosection images.
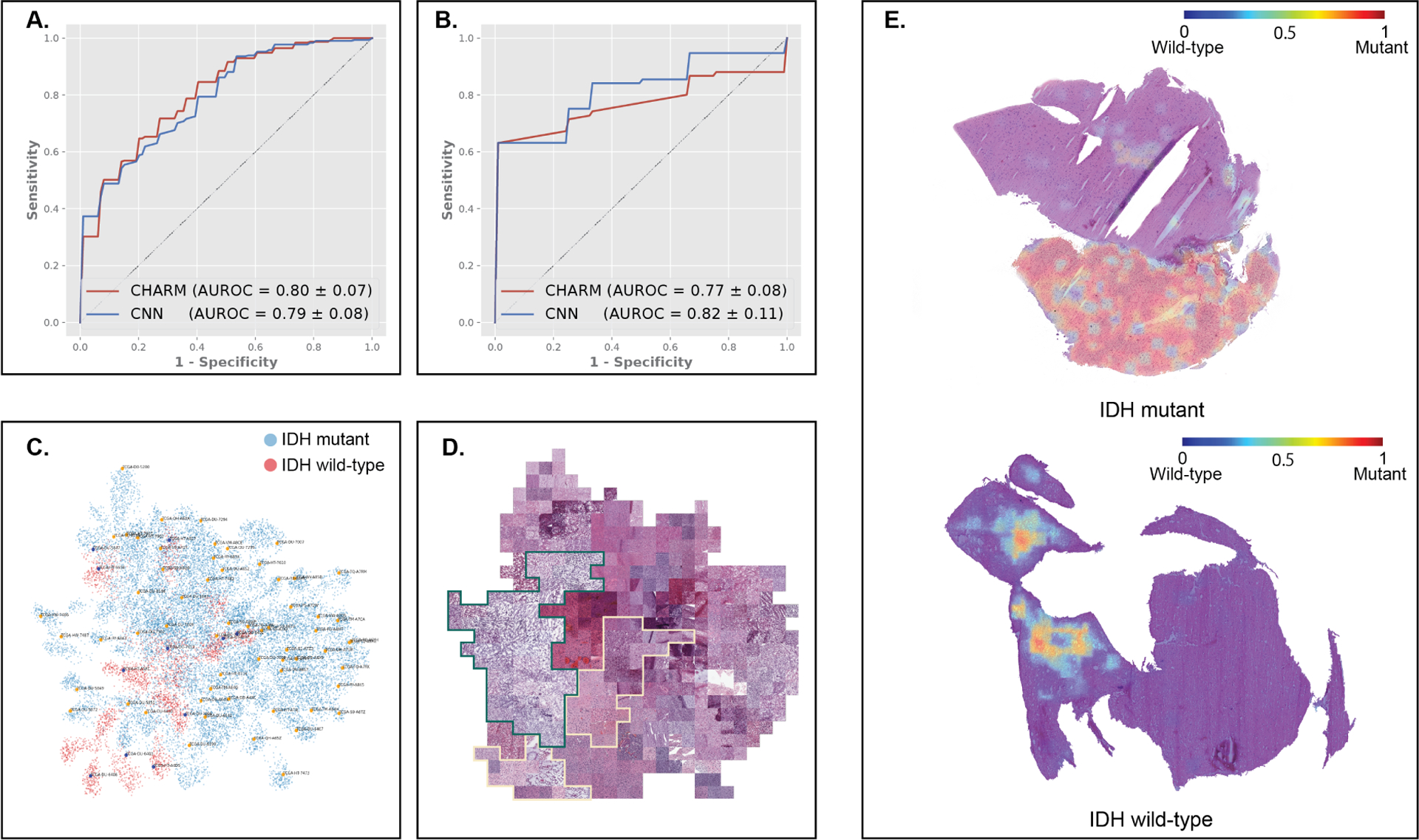
To address confounding by histologic grade, we stratified patients in our datasets into high-and low-histologic-grade groups and developed IDH mutation prediction models for each group. This figure highlights results in the low-grade histology group. (A) CHARM predicted IDH mutation status, with AUROCs of 0.80 and 0.79 on the hold-out test set. (B) Our approaches generalize well to the independent validation sets from BWH and UPenn (AUROCs of CHARM and CNN are 0.82 and 0.77, respectively). (C) UMAP visualizes our model’s learned feature space of histopathology manifestations. IDH-mutant samples from the hold-out test set are shown as blue dots, and those with IDH-wildtype are shown in red. (D) Tile-stitched visualization mapped representative tiles corresponding to the dots in the UMAP plot. The green border circumscribes tiles from IDH-mutant tumors, a majority of which show highly edematous specimens with lower cellularity and fine vessels, and the yellow border circumscribes tiles from IDH-wildtype patients, which lack the coarse edema seen in the IDH-mutant cases, and show greater cellularity and atypia, despite both sets of gliomas having been classified as low-grade within the TCGA set. (E) Heatmaps of model-generated predictions of IDH mutational status. Red indicates regions with high predicted probabilities of being IDH-mutant (top). The adjacent non-neoplastic brain tissue in the sample from an IDH-mutant tumor was correctly ignored by the model. Regions occupied by solid tumors and edematous changes received high attention from the model.
To investigate the morphological features employed by CHARM in IDH mutation prediction, we visualized the feature space extracted by our IDH classification models (Figure 3C). We found that most adult patients with IDH-wildtype infiltrating astrocytoma (i.e., the new “glioblastoma” category based on the 2021 WHO Classification) formed a distinct cluster in the two-dimensional projected feature space. Based on the data distribution in the UMAP plot, we further investigate the morphological differences between IDH-wildtype and IDH-mutant samples (Figure 3D). We found that our models focused on tumor regions with high cellularity when predicting IDH mutation status, while ignoring adjacent non-neoplastic tissues (Figure 3E). In IDH-wildtype patients, the regions receiving high attention levels from the model displayed signs of edematous changes.
CHARM Identified Three Major Types of Gliomas in the 2021 WHO Classification
In the 2021 WHO Classification, most adult infiltrating gliomas belong to one of the three major categories: IDH-mutant with 1p/19q co-deletion (molecular oligodendroglioma), IDH-mutant with intact 1p/19q (molecular astrocytoma) and IDH-wildtype (molecular glioblastoma). These molecular subtypes of gliomas have different clinical prognoses and management strategies30. To address this clinical need, we employed CHARM to predict these new classifications.
Our results show that both CHARM and the CNN baseline models performed well in classifying patients into the major molecular subtypes of glioma. The AUROCs of CHARM for identifying these molecular categories are 0.88 ± 0.03, 0.90 ± 0.02, and 0.93 ± 0.02. The AUCs of CNN for the same diagnostic classifications are 0.86 ± 0.02, 0.87 ± 0.03, and 0.92 ± 0.02 (Figure 4A). Figures 4B–D visualize our model’s attention for classifying these tumor types. Overall, CHARM relies on atypical nuclei and edematous regions for differentiating the molecular groups. In addition, the extent of cellularity helps to distinguish between tumor types, as indicated by the placement of the reddest contours around regions of high cellular density.
Figure 4. CHARM classified major types of gliomas defined by the 2021 WHO Classification of Tumors of the Central Nervous System.
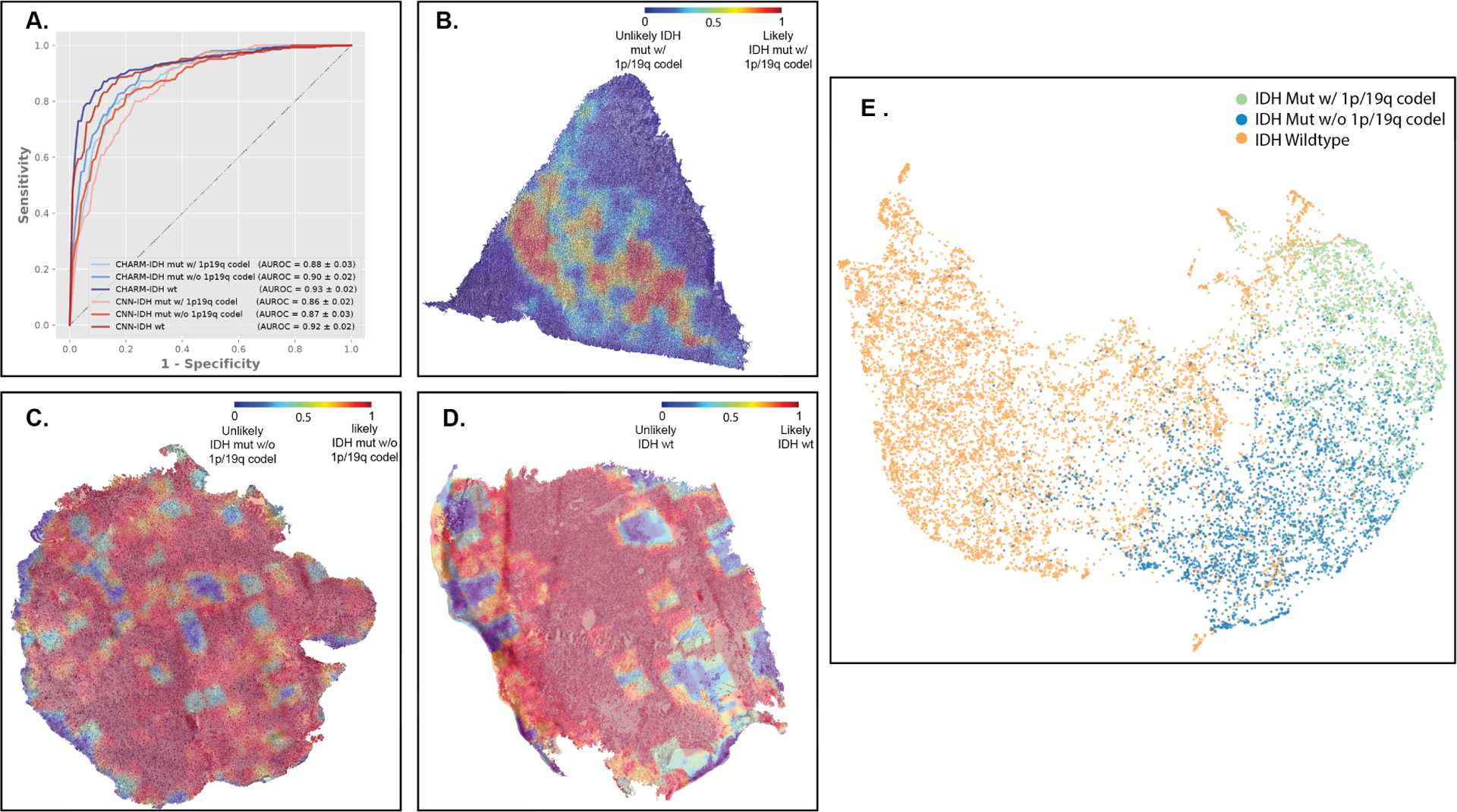
(A) Both CHARM and CNN-based models successfully classified glioma subtypes based on the 2021 WHO Classification. Here we focused on the three most prevalent types of adult gliomas: molecular oligodendroglioma with IDH mutation and 1p/19q codeletion (CHARM AUROC = 0.88 ± 0.03; CNN = 0.86 ± 0.02), molecular astrocytoma with IDH mutation but without 1p/19q codeletion (CHARM: 0.90 ± 0.02; CNN: 0.87 ± 0.03), and IDH-wildtype glioblastoma (CHARM: 0.93 ± 0.02; CNN: 0.92 ± 0.02). (B) Visualization of the regions of interest identified by our models for distinguishing the three molecular types. The color value “1” in red indicates the regions most relevant to the corresponding glioma types. The red areas on the heatmap for oligodendrogliomas with IDH mutation and 1p/19q codeletion highlight edema and foci of the most clearly seen tumor nuclei. (C) The astrocytoma tissue with an IDH mutation exhibits homogeneous and atypical but relatively lower cellularity in contrast to glioblastoma. CHARM’s consistent use of these morphological features leads to a predominance of redness in the tissue. (D) Regions receiving high attention from CHARM for IDH wild-type prediction (highlighted in red) align with hypercellular and atypical morphologies presented in the glioblastoma tissue. The regions in blue, where CHARM pays less attention, favor the histological artifacts. (E) Summarized feature spaces extracted by our machine learning models. IDH-wildtype gliomas (orange dots) cluster in a space distinct from IDH-mutant samples. While IDH mutant gliomas with and without 1p/19q co-deletion occupy neighboring regions in this feature space, aggregations among samples from the same groups are observed.
To further characterize the morphological feature space CHARM utilized for differentiating these molecular types, we extracted imaging features from each sample and applied UMAP to project them to a two-dimensional visual space (Figure 4E). Our results indicate that patients with molecular glioblastoma (IDH-wildtype) form a cluster distinct from the IDH-mutated gliomas. Among the IDH-mutated gliomas, we can further identify the two clusters related to the presence and absence of 1p/19q co-deletion. Moreover, quantitative measurements of the proximity of imaging features32 also suggest that patients with IDH-wildtype gliomas are distinct from IDH-mutant tumor patients with or without 1p/19q codeletion (cosine similarity scores = 0.897 and 0.906, respectively). In comparison, the morphological difference between samples with and without 1p/19q codeletion in the IDH mutation group is less prominent (cosine distance = 0.958). These observations are consistent with the insights from the 2021 WHO Classification that IDH mutation and 1p/19q codeletion plays a pivotal role in differentiating tumors and is crucial for determining appropriate treatment plans.
CHARM Identified 2021 WHO Subtypes within IDH-Mutant Gliomas
In the 2021 WHO Classification, IDH-mutant gliomas are further classified into multiple tumor subtypes by their histologic and molecular profiles. To address this clinical need, we further employed CHARM to predict three major subtypes with relatively high prevalence: (1) oligodendroglioma with IDH mutation and 1p/19q codeletion (category 1), (2) lower grade astrocytoma with IDH mutation but without CDKN2A/2B homozygous deletion (HD) (category 2), and (3) IDH-mutant grade 4 astrocytoma (category 3). TableS1 presents the number of patients in each category. Our results show that both CHARM and CNN are capable of classifying patients into these three categories, especially for identifying patients from category 1 (AUROC of CHARM = 0.97 ± 0.02; AUROC of CNN = 0.95 ± 0.03) and category 2 (CHARM: 0.94 ± 0.03; CNN: 0.96 ± 0.03). CHARM has slightly better performance for detecting patients in category 3 (0.89 ± 0.07 versus CNN: 0.84 ± 0.07), but the difference is not statistically significant (Wilcoxon signed-rank test P=0.273; Figure S3A). While these models directed attention to edematous tissue for the less aggressive tumor (oligodendroglioma, category 1), hypercellularity with prominent vasculature and incipient necrosis with microvascular proliferation is highlighted in samples of categories 2 and 3 (Figures S3B–D).
In addition to these established glioma categories, previous studies indicate the clinical importance of CDKN2A/2B and the worse prognosis for IDH-mutant glioma with homozygous deletion of these genes2. Therefore, we investigated a new group of patients with IDH-mutant low-grade astrocytoma with CDKN2A/2B HD (category 4). Only 11 patients have this subtype in our study cohort. In the image feature space projected by UMAP (Figure S5E), patients in category 3 are mapped into two clusters (blue); ten out of eleven patients in category 4 occupy locations close to one of these two blue clusters. Additionally, the proximity of imaging features computed by cosine similarity scores32 among patients in categories 3 and category 4 suggests that these two groups of patients possess similar morphological profiles (cosine distance = 0.997 between categories 4 and 3; cosine distance = 0.928 between categories 4 and 2). These quantitative findings are consistent with clinical evidence showing that these two subgroups of tumors are more aggressive and have worse clinical outcomes than other subtypes32. Similar analytical approaches can investigate the heterogeneity of neoplasm samples within an established tumor group.
CHARM Predicted Diagnostic Genetic Biomarkers of Glioma
We further evaluated CHARM for predicting mutations in multiple genetic biomarkers (ATRX, TP53, CDKN2A/B HD, CIC mutations, 1p/19q codeletion, chromosome 7/10 alteration, and EGFR amplification) related to glioma diagnostics and treatment options. We conducted separate tests for the two histology grade groups (low-grade versus high-grade), respectively, to identify the morphological patterns indicative of these molecular profiles, in addition to histologic grade. CHARM successfully predicted ATRX mutation (AUROC = 0.79 ± 0.052), TP53 mutation (AUROC = 0.87 ± 0.041), CDKN2A/B HD (AUROC = 0.80 ± 0.063), CIC mutation (AUROC = 0.79 ± 0.03), and 1p/19q codeletion (AUROC =0.82 ± 0.047) in the low histologic grade group using cryosection images (Figure 5A). Additionally, CHARM reached an AUROC of 0.71 ± 0.031 for predicting EGFR amplification (Figure S4A), which is frequently present in glioblastoma (Table S2). Figures 5B–E and S4B–C visualize the attention weights that CHARM employed to predict each of these molecular biomarkers. For example, CHARM associated classic histologic features, such as round nuclei and intermediate cellularity, with oligodendrogliomas, employed cortical infiltration for predicting ATRX mutation, and highlighted areas of prominent vascularity when identifying EGFR amplification.
Figure 5. CHARM predicts key genomic markers of low-grade gliomas employed by the 2021 WHO Classification of Tumors of the Central Nervous System.
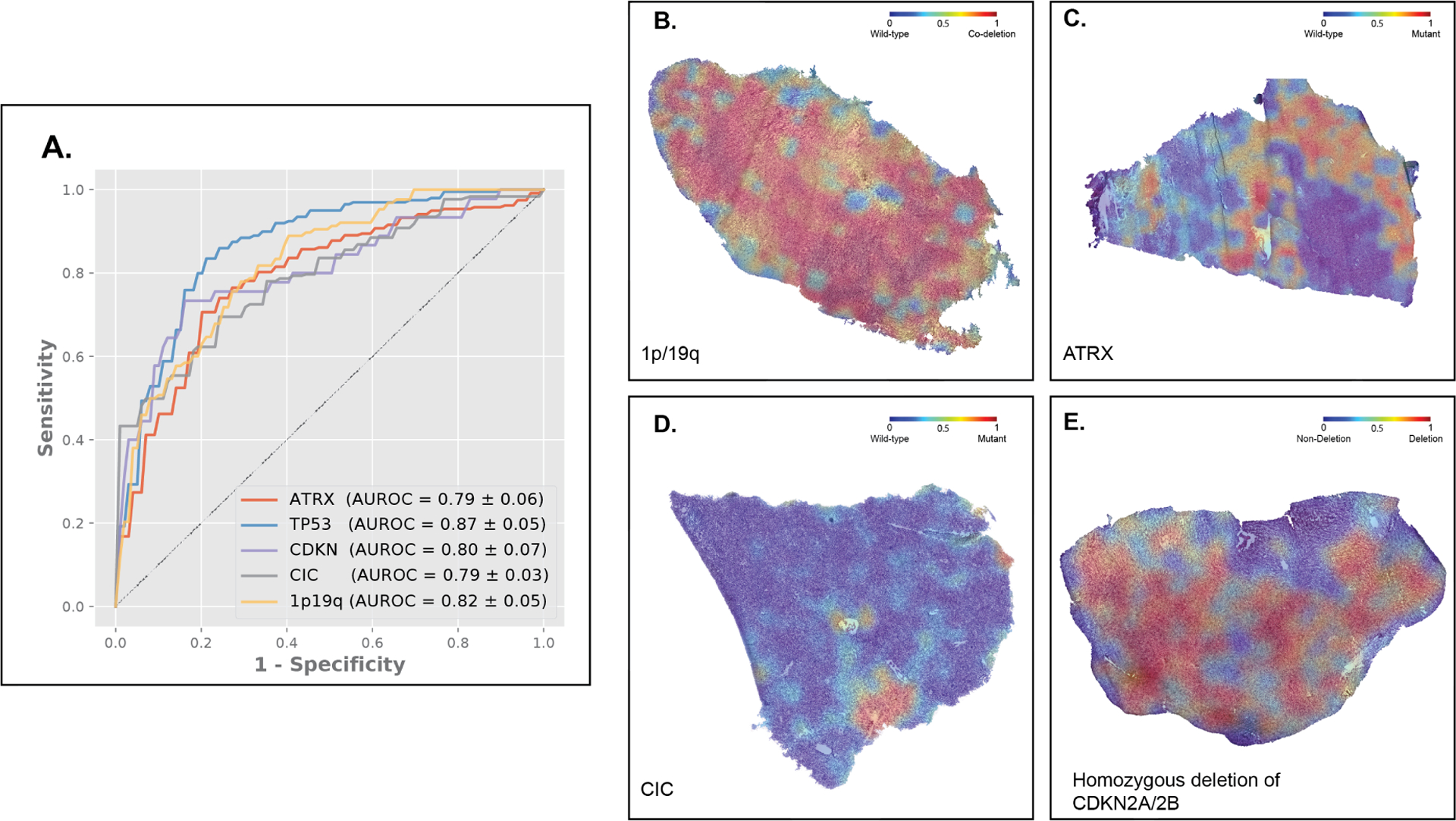
(A) CHARM successfully predicted clinically important genomic markers, including ATRX, TP53, CIC mutations, CDKN 2A/2B homozygous deletion, and 1p19q codeletion. (B) The model visualization heatmap shows that classic histopathology features, including round nuclei, intermediate cellularity, and edema receive high attention in predicting 1p/19q codeletion. (C) The ATRX heatmap shows greater attention was paid to areas of cortical infiltration by the tumor, in parts of the tissue least affected by freezing artifact. (D) The high-attention regions of a CIC mutated tissue show a homogeneous infiltrating glioma with edema and minimal atypia. (E) In glioma tissues with CDKN2A/2B homozygous deletion, regions receiving high attention show greater atypia and cellularity, relative to the ATRX mutant tissue.
Lastly, because TMB is a known predictor of patients’ responses to immune checkpoint blockade in many cancer types33, we developed prediction models for estimating TMB using whole-slide images. Our model that integrates cryosection imaging and demographic profiles achieved a significantly better prediction than a demographic-based baseline model (Spearman’s correlation coefficient: histopathologyempowered = 0.58 ± 0.042, baseline model = 0.35 ± 0.041, Wilcoxon signed-rank test P-value = 0.04; Figure S5).
DISCUSSION
This is the first study that systematically examines the relationship between cryosection histology and the molecular profiles of glioma. We developed the CHARM framework to provide accurate diagnostic and subtype prediction, and we validated our machine learning models in multiple patient cohorts, showing the reliability of our approaches. In addition, we visualized our models’ attention scores for identifying IDH mutational status from cryosection histopathology, which illuminated the histologic patterns potentially indicative of these important molecular aberrations. Given the fast inference time (less than one second per image tile) of our automated approach, this methodology can be practically applied to intra-operative diagnoses of glioma subtypes, which can enhance intra-operative surgical decisions, expedite enrolling eligible patients into clinical trials, and provide up-to-date diagnostic classification in resource-limited regions.
The updated molecular-based WHO classification improves our understanding and prognostic assessments of gliomas. With new molecular profiling techniques and emerging treatments, the WHO regularly updates its guidelines for categorizing cancers, including a major overhaul of CNS cancer classification in 202134. However, patients living in areas without access to genetic sequencing could not fully benefit from the new classification criteria35–39. In addition, obtaining molecular diagnostic results requires days to weeks, which cannot provide real-time decision support during the surgical removal of glioma tissues. CHARM addresses these issues by enabling fast and low-cost evaluations based on cryosection slides, attains significantly better prediction performance compared with previous studies40,41, and differentiated the molecular subtypes of glioma defined by the 2021 WHO Classification. CHARM further associated histologic features with molecular profiles. For example, round nuclei, edema, and intermediate cellularity were identified in oligodendrogliomas defined by IDH mutation and 1p/19q codeletion. Hence, CHARM demonstrated the potential for data-driven approaches to identify morphological characteristics related to novel molecular biomarkers. Our machine learning-based methods allow researchers to reevaluate morphological patterns and tumor microenvironments related to the new diagnostic guidelines.
In addition, our quantitative analyses provide new insights into the morphology associated with key genetic aberrations and illuminate morphological similarities between cancers with similar clinical behavior. For example, CHARM revealed the morphological similarity between IDH-mutant low-grade astrocytomas with CDKN2A/2B HD, and IDH-mutant grade 4 astrocytomas. This pathology finding is consistent with clinical evidence showing these two subgroups of tumors are more aggressive than other subtypes.
The hierarchical ViT architecture of CHARM preserves the global contextual connections between image features remotely scattered in a slide43. In contrast, CNN-based models used convolution kernels that emphasize the local interaction between regions nearby. In practice, pathologists evaluate a slide using both local and global contextual findings, which is aligned with how the hierarchical vision transformer processes images43 This observation may explain why CHARM achieved better performances in most tasks compared with CNNs. When training on smaller datasets (e.g., n=25 for detecting IDH-mutant samples within the high histological grade group), ViT’s capability of learning useful embeddings is more limited43. Overall, our results suggested that CHARM with ViT performs better in most glioma classification tasks, while CNN-based models may be useful for scenarios with limited training samples.
Another critical clinical challenge CHARM overcomes is the variable quality of cryosection images. Previous pathology studies12 usually focused on permanent slides because they are less likely to contain poor-quality imaging regions, such as tissues of unequal thickness or fragmented tissues. Nevertheless, processing permanent section slides takes days, and thus it is not applicable for intra-operative pathology evaluation that guides surgical operations. Other studies have employed T2-weighted MR images to predict glioma subtypes45,46. However, MRI-based approaches require more time to acquire and process the images, do not achieve better performance compared with our methods, and cannot provide morphological understanding and visualization at the cell level. Another recent study employed stimulated Raman histology images to predict the molecular subtypes of glioma24. Our methods achieved similar prediction performance and do not require special microscopy techniques.
In summary, our CHARM platform successfully identified glioma cells, classified histologic grade, and predicted clinically important molecular profiles using hematoxylin and eosin (H&E)-stained cryosection images, which enables fast intra-operative diagnoses. Our visualization framework further empowers pathologists to identify the morphological patterns associated with molecular profiles and clinical outcomes. Taken together, CHARM demonstrated the possibility of extracting untapped biomedical signals from cryosection slides and facilitated the development of real-time precision oncology.
Limitations of Study
As treatment modalities evolve, novel molecular and treatment response-based disease features and new disease classifications are expected to arise. With new revisions of glioma classifications in the future, we will need to adjust or retrain our data-driven models to accommodate the new classification systems. Because our informatics approaches are fully automated, fast and objective re-analyses can be tailored to the evolving diagnostic guidelines within hours. In addition, our study cohorts were based in North America and may not represent all populations worldwide. Furthermore, our current approach does not reduce the time needed for preparing digital images of cryosections. Recent developments in rapid molecular diagnosis may complement the standard cryosection-based tissue evaluation48,49. Future studies can extend our machine learning framework to images generated by faster digitizing methods and incorporate molecular profiling results from rapid diagnostics.
STAR METHODS
Resource Availability
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kun-Hsing Yu, Kun-Hsing_Yu@hms.harvard.edu.
Materials Availability:
This study did not generate new unique reagents.
Data and Code Availability:
This paper analyzes TCGA data, which is available at https://portal.gdc.cancer.gov/
All original code has been deposited at https://github.com/hms-dbmi/charm and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Human Subjects
We obtained 2,334 cryosection whole slide images from 1,524 patients from three independent study sites. The Cancer Genome Atlas (TCGA) contains 1,121 patients from 46 hospitals nationwide, and the slides were scanned using Aperio scanners at 40x magnification, with molecular profiles obtained from whole-exome sequencing. In addition, we built independent institutional cohorts by collecting data from 213 patients from Brigham and Women’s Hospital (BWH) and 190 patients from the University of Pennsylvania (UPenn). Cryosection slides from BWH and UPenn were scanned using a Hamamatsu S210 scanner at 40x magnification, and patients’ molecular profiles were obtained from targeted sequencing. No patient overlaps were found between these cohorts. Written informed consent was obtained from the patients at the time of enrollment.
In the BWH and the UPenn cohorts, patients’ clinical information was based on clinician-reported data. The clinical information of TCGA cohort was obtained from the National Cancer Institute Genomic Data Commons. 71.4% of the patients in the TCGA cohort have race information. Among them, 91.9% were White, 6.6% were Black or African American, and 1.4% were Asian. We do not have racial information for other patients who participated in this study. Socioeconomic status was not available for all patients.
Method Details
Overview of Machine Learning Approaches in CHARM
We employed a hierarchical vision transformer43 to build the backbone of CHARM. For each task, we selected patients (the development cohort) from one of the three cohorts to develop models. We randomly split patients in the development cohort into one of the training, validation, or hold-out test sets using a 70%−15%−15% ratio. During the training process, class-balanced batch sampling and class-balanced loss function were implemented to alleviate instance imbalance of the datasets. In the prediction phase, each tile received a separate prediction from the model. The median value of all available tiles of a patient was employed as the prediction for this patient. This aggregation approach requires less parameterization, making it more robust and generalizable for deep learning models43.
After the models were finalized, we first evaluated the models’ internal validity using the hold-out test set of the development cohort (internal test set). To further demonstrate the generalizability of our models, we evaluated our models in the independent test cohort that did not participate in the model development process. We repeated the random splitting of the development cohort, training, and external testing five times and calculated the corresponding mean and standard deviation for all performance metrics for each task. We compared CHARM with EfficientNet-B5, a stateof-the-art CNN with high performance in analyzing digital pathology43. Figure 1 summarizes our machine learning approach, and Data S1 is our method checklist.
Cancer Identification using Cryosection Images
We first trained our CHARM model for identifying glioma using cryosection slides from BWH because TCGA and UPenn did not have sufficient benign samples for training (TCGA has only five benign WSIs while UPenn did not collect pathology slides of adjacent benign tissue). We reported the areas under the receiver operating characteristics curves (AUROCs) of CHARM and the baseline EfficientNet-B551 CNN models. Furthermore, we evaluated the generalizability of the trained models using patients in TCGA and visualized the performance of our models. Models derived from the BWH dataset were directly applied to all images from TCGA, and AUROCs with standard deviations were computed.
Histologic Grade Prediction using Cryosection Images
Given its sample diversity, we employed the TCGA cohort to develop prediction models for the histologic grading of glioma tissues using cryosection images. We classified the samples into two groups: histologic grade 4 versus lower-histologic grade (grades 2 and 3) groups due to their treatment implications. We first assessed the models on the hold-out test set in TCGA. Furthermore, samples from BWH and UPenn were combined as an independent test cohort because both datasets were digitized by the same scanner and scanning parameters. To develop a domain adaptable model, we partitioned the independent cohort into 70%−15%−15% fine-tuning, validation, and independent test sets.
IDH Mutation Status Prediction using Cryosection Images
We employed similar machine learning approaches to identify the associations between histopathology image patterns and IDH mutation status. Considering the known association between histologic grades and IDH mutation status, we stratified patients into “high” and “low” histologic grade groups and trained separate models for these two groups. This stratified approach eliminated the confounding from histologic grades in predicting IDH status. We trained our models with TCGA cohort and first reported the model performance in the hold-out test set. We next evaluated the model generalizability on BWH and UPenn using the same procedure as that of the histological classification task.
Molecular Classification of Glioma Using Cryosection Images
To enable glioma diagnoses based on the updated WHO Classification, we evaluated the performance of our approaches in classifying the three major molecular types of gliomas defined in the 2021 WHO Classification of Tumors of the Central Nervous System: (1) gliomas with IDH mutation and 1p/19q codeletion (oligodendroglioma), (2) gliomas with IDH mutation but without 1p/19q codeletion (astrocytoma), and (3) IDH-wildtype (glioblastoma). We developed models in the TCGA cohort given their complete molecular profiles. Due to the significant role played by CDKN2A/2B homozygous deletion (HD) in astrocytoma, we further employed CHARM to classify low-grade gliomas into three finer groups: (1) oligodendroglioma with IDH mutation and 1p/19q codeletion, (2) astrocytoma with IDH mutation but without CDKN2A/2B HD, (3) IDH-mutant astrocytoma with grade 4 histology and/or CDKN2A/2B homozygous deletion. After developing CHARM for these three categories, we investigated the imaging feature space of these diagnostic groups and that of astrocytoma histology, IDH mutation, and CDKN2A/2B HD (category 4).
Finally, we employed CHARM to identify cryosection samples with key genomic biomarkers involved with the 2021 WHO Classification of Tumors of the Central Nervous System. These include ATRX, TP53, CIC mutations, CDKN2A/B homozygous deletion, 1p/19q codeletion, EGFR amplification, and chromosome 7/10 alteration.
Interpretation and Visualization of the Machine Learning Models
To better interpret the morphological patterns employed by our data-driven models, we (1) visualized the model-learned latent feature space by projecting it into a two-dimensional embedding space and (2) devised heatmaps based on model output to characterize model predictions at the slide level.
In the molecular classification task, we used CHARM to extract 1024-dimensional latent feature vectors to represent tile images and characterize the similarities and differences between the three major types of gliomas. We applied Uniform Manifold Approximation and Projection (UMAP)52 to project these latent vectors to a two-dimensional space for visualization, given its capability of balancing local and global distances.
We further visualized the model predictions in heatmaps with a standard colormap, where regions with high predicted probabilities of the positive class (e.g., high-grade glioma, and mutations in IDH, ATRX, and CIC genes) were in red and those with low predicted probabilities were in blue. We applied a Gaussian kernel to average and smooth the color weights. Neuropathologists with more than 40 years of combined practice experience (M.P.N. and J.A.G.) independently evaluated the heatmaps to connect pathology descriptions of the tumor microenvironment with computer-generated model visualization.
Quantification and Statistical Analyses
We calculated AUROCs and the Spearman correlation coefficients using scikit-learn 0.23.2. We employed the Wilcoxon rank-sum test to identify the most informative features that contributed to distinguishing IDH mutated glioma with or without 1p/19q codeletion from IDH-wildtype gliomas.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| The Cancer Genome Atlas | National Cancer Institute Genomic Data Commons | https://portal.gdc.cancer.gov/ |
| Software and algorithms | ||
| CHARM | Nasrallah et al. | DOI: 10.5281/zenodo.7972059 |
| PyTorch | Meta AI | https://pytorch.org/ |
| Tensorflow | https://www.tensorflow.org/ | |
| scikit-learn | David Cournapeau | https://github.com/scikit-learn/scikit-learn |
| timm | Ross Wightman | https://github.com/huggingface/pytorch-image-models |
Highlights.
We established the Cryosection Histopathology Assessment and Review Machine (CHARM).
CHARM predicted IDH mutation and 2021 WHO Classification of glioma using cryosections.
CHARM provided quantitative evidence supporting the reclassification of astrocytoma.
CHARM facilitated real-time cryosection diagnoses and can be extended to other cancers.
Context and significance.
Real-time pathology diagnosis of tissue samples is crucial for the optimal treatment of brain cancers. Nonetheless, samples prepared for real-time evaluation (i.e., cryosection samples) are of variable quality and difficult to evaluate even for specialized doctors. In addition, the new WHO guidelines for brain cancer diagnosis include genetic information in addition to the known pathology patterns described in the medical literature. To address this challenge, Nasrallah et al. developed the Cryosection Histopathology Assessment and Review Machine (CHARM), a context-aware artificial intelligence (AI) system for brain cancer diagnosis. CHARM successfully identified cancer cells, predicted genetic mutation status, and classified WHO-defined cancer subtypes using cryosection samples. This advanced AI method could facilitate real-time cancer diagnoses during surgery.
Acknowledgments:
We thank Alexander Bruce for his assistance with slide scanning at the Digital Imaging Facility, Department of Pathology, Brigham and Women’s Hospital, and Kristina C. Grieco, Raquel Arias-Camison, Nina Thakur, Dominique Ballinger, and Alexa Craig for their assistance in collecting and curating the pathology slides. We thank Shannon Gallagher and Faith McDonald for their administrative support. We thank the Microsoft Azure for Research Award, AWS Cloud Credits for Research, Google Cloud Platform research credit program, the NVIDIA GPU Grant Program, and the Extreme Science and Engineering Discovery Environment (XSEDE) at the Pittsburgh Supercomputing Center (allocation TG-BCS180016) for their computational support. This work was conducted with support from the Digital Imaging Facility, Department of Pathology, Brigham and Women’s Hospital, Boston, MA. This work is supported in part by the National Institute of General Medical Sciences grant R35GM142879, the Department of Defense Peer Reviewed Cancer Research Program Career Development Award, the Google Research Scholar Award, the Blavatnik Center for Computational Biomedicine Award (to KHY), the Partners’ Innovation Discovery Grant, and the Schlager Family Award for Early Stage Digital Health Innovations (to JAG and KHY).
Funding:
This work is supported in part by the National Institute of General Medical Sciences grant R35GM142879, the Google Research Scholar Award, the Blavatnik Center for Computational Biomedicine Award, the Partners’ Innovation Discovery Grant, and the Schlager Family Award for Early Stage Digital Health Innovations.
Inclusion and diversity:
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of interests:
K.-H. Y. is an inventor of U.S. Patent 10,832,406, which was assigned to Harvard University. K-H.Y. was a consultant of Curatio.DL. All other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, et al. (2021). The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 23, 1231–1251. 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu VM, O’Connor KP, Shah AH, Eichberg DG, Luther EM, Komotar RJ, and Ivan ME (2020). The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol 148, 221–229. 10.1007/s11060-020-03528-2. [DOI] [PubMed] [Google Scholar]
- 3.Pekmezci M, Rice T, Molinaro AM, Walsh KM, Decker PA, Hansen H, Sicotte H, Kollmeyer TM, McCoy LS, Sarkar G, et al. (2017). Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol 133, 1001–1016. 10.1007/s00401-017-1690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, Gilbert MR, and Yang C (2020). IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. British Journal of Cancer 122, 1580–1589. 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han W, Qin L, Bay C, Chen X, Yu K-H, Miskin N, Li A, Xu X, and Young G (2020). Deep transfer learning and radiomics feature prediction of survival of patients with high-grade gliomas. American Journal of Neuroradiology 41, 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanai N, and Berger MS (2008). GLIOMA EXTENT OF RESECTION AND ITS IMPACT ON PATIENT OUTCOME. Neurosurgery 62, 753–766. 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- 7.Lawler SE, Speranza MC, Cho CF, and Chiocca EA (2017). Oncolytic Viruses in Cancer Treatment: A Review. JAMA Oncol 3, 841–849. 10.1001/jamaoncol.2016.2064. [DOI] [PubMed] [Google Scholar]
- 8.Hervey-Jumper SL, Zhang Y, Phillips JJ, Morshed RA, Young JS, McCoy L, Lafontaine M, Luks T, Ammanuel S, Kakaizada S, et al. (2023). Interactive Effects of Molecular, Therapeutic, and Patient Factors on Outcome of Diffuse Low-Grade Glioma. J Clin Oncol 41, 2029–2042. 10.1200/JCO.21.02929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molinaro AM, Hervey-Jumper S, Morshed RA, Young J, Han SJ, Chunduru P, Zhang Y, Phillips JJ, Shai A, Lafontaine M, et al. (2020). Association of Maximal Extent of Resection of Contrast-Enhanced and Non-Contrast-Enhanced Tumor With Survival Within Molecular Subgroups of Patients With Newly Diagnosed Glioblastoma. JAMA Oncol 6, 495–503. 10.1001/jamaoncol.2019.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K-H, Beam AL, and Kohane IS (2018). Artificial intelligence in healthcare. Nature biomedical engineering 2, 719–731. [DOI] [PubMed] [Google Scholar]
- 11.Yu KH, Berry GJ, Rubin DL, Ré C, Altman RB, and Snyder M (2017). Association of Omics Features with Histopathology Patterns in Lung Adenocarcinoma. Cell Syst 5, 620–627.e623. 10.1016/j.cels.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, and Fuchs TJ (2019). Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nature Medicine 25, 1301–1309. 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ektefaie Y, Yuan W, Dillon DA, Lin NU, Golden JA, Kohane IS, and Yu KH (2021). Integrative multiomics-histopathology analysis for breast cancer classification. NPJ Breast Cancer 7, 147. 10.1038/s41523-021-00357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, and Mahmood F (2021). Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng 5, 555–570. 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai P-C, Lee T-H, Kuo K-C, Su F-Y, Lee T-LM, Marostica E, Ugai T, Zhao M, Lau MC, and Väyrynen JP (2023). Histopathology images predict multi-omics aberrations and prognoses in colorectal cancer patients. Nature Communications 14, 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K-H, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, and Snyder M (2016). Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nature Communications 7, 12474. 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naik N, Madani A, Esteva A, Keskar NS, Press MF, Ruderman D, Agus DB, and Socher R (2020). Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nature Communications 11, 5727. 10.1038/s41467-020-19334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu K-H, Hu V, Wang F, Matulonis UA, Mutter GL, Golden JA, and Kohane IS (2020). Deciphering serous ovarian carcinoma histopathology and platinum response by convolutional neural networks. BMC Medicine 18, 236. 10.1186/s12916-020-01684-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marostica E, Barber R, Denize T, Kohane IS, Signoretti S, Golden JA, and Yu KH (2021). Development of a Histopathology Informatics Pipeline for Classification and Prediction of Clinical Outcomes in Subtypes of Renal Cell Carcinoma. Clin Cancer Res 27, 2868–2878. 10.1158/10780432.CCR-20-4119. [DOI] [PubMed] [Google Scholar]
- 20.Yu K-H, Wang F, Berry GJ, Re C, Altman RB, Snyder M, and Kohane IS (2020). Classifying non-small cell lung cancer types and transcriptomic subtypes using convolutional neural networks. Journal of the American Medical Informatics Association 27, 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ertosun MG, and Rubin DL (2015). Automated Grading of Gliomas using Deep Learning in Digital Pathology Images: A modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc 2015, 1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 22.Jin L, Shi F, Chun Q, Chen H, Ma Y, Wu S, Hameed NUF, Mei C, Lu J, Zhang J, et al. (2021). Artificial intelligence neuropathologist for glioma classification using deep learning on hematoxylin and eosin stained slide images and molecular markers. Neuro Oncol 23, 44–52. 10.1093/neuonc/noaa163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei L, Jones KA, Shboul ZA, Chen JY, and Iftekharuddin KM (2021). Deep Neural Network Analysis of Pathology Images With Integrated Molecular Data for Enhanced Glioma Classification and Grading. Front Oncol 11, 668694. 10.3389/fonc.2021.668694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollon T, Jiang C, Chowdury A, Nasir-Moin M, Kondepudi A, Aabedi A, Adapa A, Al-Holou W, Heth J, Sagher O, et al. (2023). Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat Med 29, 828–832. 10.1038/s41591023-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noorbakhsh J, Farahmand S, Foroughi Pour A, Namburi S, Caruana D, Rimm D, Soltanieh-Ha M, Zarringhalam K, and Chuang JH (2020). Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. Nat Commun 11, 6367. 10.1038/s41467-020-20030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalsa SSS, Hollon TC, Adapa A, Urias E, Srinivasan S, Jairath N, Szczepanski J, Ouillette P, Camelo-Piragua S, and Orringer DA (2020). Automated histologic diagnosis of CNS tumors with machine learning. CNS Oncol 9, CNS56. 10.2217/cns-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong AH, Sharmanska V, Limbӓck-Stanic C, and Grech-Sollars M (2020). Optimization of deep learning methods for visualization of tumor heterogeneity and brain tumor grading through digital pathology. Neurooncol Adv 2, vdaa110. 10.1093/noajnl/vdaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mat Zin AA, and Zulkarnain S (2019). Diagnostic Accuracy of Cytology Smear and Frozen Section in Glioma. Asian Pac J Cancer Prev 20, 321–325. 10.31557/APJCP.2019.20.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. (2021). An Image is Worth 16×16 Words: Transformers for Image Recognition at Scale. arXiv [cs.CV] [Google Scholar]
- 30.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, et al. (2019). Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25, 1054–1056. 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, Bendszus M, Balana C, Chinot O, Dirven L, et al. (2021). EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 18, 170–186. 10.1038/s41571-020-00447-z.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singhal A (2001). Modern Information Retrieval: A Brief Overview. IEEE Data Eng. Bull 24, 35–43. [Google Scholar]
- 33.Appay R, Dehais C, Maurage CA, Alentorn A, Carpentier C, Colin C, Ducray F, Escande F, Idbaih A, Kamoun A, et al. (2019). CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol 21, 1519–1528. 10.1093/neuonc/noz124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, and Kurzrock R (2017). Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 16, 2598–2608. 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorgulescu JB, Torre M, Harary M, Smith TR, Aizer AA, Reardon DA, Barnholtz-Sloan JS, and Perry A (2019). The Misclassification of Diffuse Gliomas: Rates and Outcomes. Clin Cancer Res 25, 2656–2663. 10.1158/1078-0432.CCR-18-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, et al. (2015). Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372, 2481–2498. 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eskilsson E, Røsland GV, Solecki G, Wang Q, Harter PN, Graziani G, Verhaak RGW, Winkler F, Bjerkvig R, and Miletic H (2018). EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol 20, 743–752. 10.1093/neuonc/nox191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez FJ, Vizcaino MA, and Lin MT (2016). Recent Advances on the Molecular Pathology of Glial Neoplasms in Children and Adults. J Mol Diagn 18, 620–634. 10.1016/j.jmoldx.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, Ringel M, and Schork N (2019). Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med 2, 69. 10.1038/s41746-019-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, and Wick W (2013). ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol 126, 443–451. 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 41.Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A, Yates LR, Jimenez-Linan M, Moore L, and Gerstung M (2020). Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer 1, 800–810. 10.1038/s43018-020-0085-8. [DOI] [PubMed] [Google Scholar]
- 42.Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, Krause J, Niehues JM, Sommer KAJ, Bankhead P, et al. (2020). Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 1, 789–799. 10.1038/s43018-020-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, and Guo B (2021). Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv [cs.CV] [Google Scholar]
- 44.Ghaffari Laleh N, Muti HS, Loeffler CML, Echle A, Saldanha OL, Mahmood F, Lu MY, Trautwein C, Langer R, Dislich B, et al. (2022). Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Medical Image Analysis 79, 102474. 10.1016/j.media.2022.102474. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Yang S, Zhang J, Wang M, Zhang J, Huang J, Yang W, and Han X (2021). TransPath: Transformer-Based Self-supervised Learning for Histopathological Image Classification. In (Springer International Publishing; ), pp. 186–195. 10.1007/978-3-030-87237-3_18. [DOI] [Google Scholar]
- 46.Choi YS, Bae S, Chang JH, Kang SG, Kim SH, Kim J, Rim TH, Choi SH, Jain R, and Lee SK (2021). Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol 23, 304–313. 10.1093/neuonc/noaa177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nalawade S, Murugesan GK, Vejdani-Jahromi M, Fisicaro RA, Bangalore Yogananda CG, Wagner B, Mickey B, Maher E, Pinho MC, Fei B, et al. (2019). Classification of brain tumor isocitrate dehydrogenase status using MRI and deep learning. J Med Imaging (Bellingham) 6, 046003. 10.1117/1.JMI.6.4.046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larjavaara S, Mäntylä R, Salminen T, Haapasalo H, Raitanen J, Jääskeläinen J, and Auvinen A (2007). Incidence of gliomas by anatomic location. Neuro Oncol 9, 319–325. 10.1215/15228517-2007-016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH, et al. (2014). Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc Natl Acad Sci U S A 111, 11121–11126. 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shankar GM, Francis JM, Rinne ML, Ramkissoon SH, Huang FW, Venteicher AS, Akama-Garren EH, Kang YJ, Lelic N, Kim JC, et al. (2015). Rapid Intraoperative Molecular Characterization of Glioma. JAMA Oncol 1, 662–667. 10.1001/jamaoncol.2015.0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan M, and Le QV (2020). EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv [cs.LG] [Google Scholar]
- 52.McInnes L, Healy J, Saul N, and Großberger L (2018). UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software 3, 861. 10.21105/joss.00861. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes TCGA data, which is available at https://portal.gdc.cancer.gov/
All original code has been deposited at https://github.com/hms-dbmi/charm and is publicly available as of the date of publication.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


