Abstract
ARID1A is a subunit of SWI/SNF chromatin remodeling complexes and is mutated in many types of human cancers, especially those derived from endometrial epithelium, including ovarian and uterine clear cell carcinoma (CCC) and endometrioid carcinoma (EMCA). Loss-of-function mutations in ARID1A alter epigenetic regulation of transcription, cell cycle checkpoint control, and DNA damage repair. We report here that mammalian cells with ARID1A deficiency harbor accumulated DNA base lesions and increased abasic (AP) sites, products of glycosylase in the first step of base excision repair (BER). ARID1A mutations also delayed recruitment kinetics of BER long-patch repair effectors. Although ARID1A-deficient tumors were not sensitive to monotherapy with DNA-methylating temozolomide (TMZ), the combination of TMZ with PARP inhibitors (PARPi) potently elicited double strand DNA breaks, replication stress, and replication fork instability in ARID1A-deficient cells. The TMZ and PARPi combination also significantly delayed in vivo growth of ovarian tumor xenografts carrying ARID1A mutations and induced apoptosis and replication stress in xenograft tumors. Together, these findings identified a synthetic lethal strategy to enhance the response of ARID1A-mutated cancers to PARP inhibition, which warrants further experimental exploration and clinical trial validation.
Keywords: PARP inhibitors, Type I ovarian cancer, Temozolomide (TMZ), DNA damage and repair, replication stress
Introduction
Epithelial ovarian cancer (EOC) is among the most aggressive female neoplastic diseases. Additionally, the treatment of advanced ovarian cancer after tumor debulking surgery is complicated because EOC is a constellation of different neoplasms rather than a monolithic disease (1,2). Based on the pathological presentation, molecular features, and tissue of origin, EOC has been broadly classified into Type I and Type II tumors (3,4). Type I EOCs include low-grade serous, clear cell, endometrioid, and mucinous carcinomas, whereas Type II EOCs primarily consist of high-grade serous carcinoma (HGSC). Several distinct molecular genetic features distinguish Type I from Type II EOCs. For example, chromosomal instability and TP53 and genes involved in homologous recombination DNA repair such as BRCA1/2 are frequently mutated in Type II ovarian cancers but rarely in Type I ovarian cancers. On the other hand, somatic mutations involving ARID1A, PTEN/PIK3CA, CTNNB1, and KRAS/BRAF frequently occur in Type I ovarian cancers while somatic mutations involving these genes are uncommon in Type II EOCs, although advanced Type II tumors may develop gene amplifications that magnify signaling of the KRAS-MAPK and PI3K pathways.
Despite having distinct features and disease presentations, for decades, chemotherapy combining carboplatin and paclitaxel has been the first-line therapy for both Type I and Type II EOCs; however, clinical benefits from this regimen are mostly restricted to Type II EOCs, specifically, ovarian HGSC. Many Type I carcinomas, highlighted by the clear cell carcinoma (OCCC) subtype, are intrinsically refractory to conventional chemotherapy, and specialized treatments targeting their tumorigenesis pathways are urgently needed. To provide a basis for precision treatment for Type I EOCs, we have performed cancer genome sequencing to characterize the molecular landscape of Type I EOCs. This research effort led to the discovery of frequent ARID1A-inactivating mutations in OCCCs (5,6). Mechanistically, ARID1A encodes BAF250, a subunit of the SWI/SNF chromatin remodeling complex, which binds to specific chromatin regions, mobilizes nucleosomes through ATP hydrolysis, and modulates global and local chromatin configuration. Studies from independent research groups have shown that ARID1A functions as a tumor suppressor through regulating downstream target genes including p21 and CKDN1A, which regulate cell cycle checkpoints (7,8). Alterations in the transcriptional program through regulatory processes affecting the chromatin landscape provide one of the mechanisms whereby the loss of ARID1A promotes tumorigenesis. In addition, dysregulation of DNA repair mechanisms may also contribute to tumorigenesis driven by the loss of ARID1A. Indeed, yeast studies have documented functional involvement of the SWI/SNF complex in DNA repair (9–11). In mammalian cells, we and others have demonstrated that both non-homologous end joining (NHEJ) and homologous recombination (HR) can be compromised in ARID1A-deficient cells (12–14). Moreover, co-occurrence between ARID1A-loss and mismatch repair (MMR)-deficiency in endometrioid cancers with intact p53 expression (15,16), suggests a promiscuous or broad involvement of SWI/SNF complex in DNA repair.
Based on impaired DNA repair function in ARID1A-deficient tumors, synthetic lethal therapeutic approaches have been successfully developed, which include ionizing radiation in combination with PARP inhibitor or ATR inhibitor (12,17). However, as compared to its therapeutic potency for BRCA-mutated cancers, PARP inhibitor as a single-agent has not yielded adequate efficacy for ARID1A-mutated cancers (12,18). We postulated that exogenously introduced DNA lesions and strand breaks may enhance cellular reliance on PARP-dependent DNA repair function (12). Thus, exogenously-induced DNA lesions would sensitize cancer cells to PARP inhibition while sparing normal cells with intact DNA repair capacity. To test this concept, we screened a panel of genotoxic chemotherapeutic drugs in combination with PARPi and evaluated the ability of the combination treatment to enhance cytotoxicity. We report that Temozolomide (TMZ), an orally active chemotherapeutic drug which methylates (or alkylates) DNA, yields DNA base lesions that cannot be effectively repaired in ARID1A-deficient cancer cells. This “priming” step significantly enhanced responses of ARID1A-deficient cells to PARP inhibitors. These results warrant evaluation in the clinical setting for patients whose tumors harbor ARID1A mutation.
Materials and Methods
Cell lines
The human normal endometrial cell line, hEM3, was generated by immortalization of normal human endometrial epithelium by transduction of lentivirus with a SV40-TAg, then subjected to CRISPR knockout of the ARID1A gene and clonal expansion, as we previously reported (19,20). The hEM3 cells were maintained in RPMI with 15% FBS, 1% Pen/Strep, and 1% NEAA. The hEM3 cells were transferred in RPMI with 10% FBS and 1% Pen/Strep for all experiments and analyses reported in this paper. MCF10a control and ARID1A knockout cells (HD PAR-058 and HD 101–022, Horizon Discovery) were culture in DMEM/F12 with 5% Horse Serum, EGF 20 ng/ml, insulin 10 μg/ml, hydrocortisone 0.5 mg/ml, cholera toxin 100 ng/ml, and 1% Pen/Strep. HCT116 control, the ARID1A knockout counterpart (HD PAR-073 and HD 104–049, Horizon Discovery), ES2, RMG1, HEC1A, OV429, and TOV21G (BCRC Cat# 60407,
RRID:CVCL_3613) were cultured in RPMI with 10% FBS and 1% Pen/Strep. All cell lines were examined for the presence of mycoplasma before usage and re-tested every two months. All cell lines were STR authenticated by the Genetic Resource Core Facility at the Johns Hopkins University. STR similarity search was performed using the CLASTR 1.4.4 web search tool provided by Cellosaurus. Of note, OV429 shows a 98.2% similarity to OV433. The mutation status of ARID1A, KRAS, PIK3CA, TP53, and DNA mismatch repair genes in cancer cell lines used in this study are shown in Table S1.
Antibodies and reagents
The following primary antibodies were used in this study: Phospho-Histone H2A.X (Ser139) (Cell Signaling Technology Cat# 9718, RRID:AB_2118009), RPA32/RPA2 (Cell Signaling Technology Cat# 35869, RRID:AB_2799086), pS345 CHK1 (Cell Signaling Technology Cat# 2348, RRID:AB_331212), pT68 CHK2 (Cell Signaling Technology Cat# 2197, RRID:AB_2080501), CHK2 (Cell Signaling Technology Cat# 6334, RRID:AB_11178526), H2B (Cell Signaling Technology Cat# 12364, RRID:AB_2714167), ARID1A (Cell Signaling Technology Cat# 12354, RRID:AB_2637010), PCNA (Cell Signaling Technology Cat# 13110, RRID:AB_2636979), Cleaved Caspase-3 (Asp175) (Cell Signaling Technology Cat# 9664, RRID:AB_2070042), APE1 (Abcam Cat# ab48832, RRID:AB_867690), pS33 RPA32/RPA2 (ab211877), CHK1 (Abcam Cat# ab32531, RRID:AB_726821), β-Actin (Santa Cruz Biotechnology Cat# sc-47778 HRP, RRID:AB_2714189), anti-CldU (Abcam Cat# ab6326, RRID:AB_305426) and anti-IdU (BD Biosciences Cat# 347580, RRID:AB_10015219). Secondary antibodies against rabbit (Jackson ImmunoResearch Labs Cat# 111–035-144, RRID:AB_2307391) or mouse (Jackson ImmunoResearch Labs Cat# 115–035-166, RRID:AB_2338511) conjugated with HRP were used in this study.
Temozolomide (Cat# S1237), Camptothecin (Cat# S1288), Doxorubicin (Cat# S1208), Topotecan (Cat# S9321), Niraparib (Cat# S7625), Veliparib (Cat# S1004), Ifosfamide (Cat# S1302), WDR5–0103 (Cat# S2184), and HA15 (Cat# S8299) were purchased from Selleckchem. Carboplatin (Cat# HY-17393), Paclitaxel (Cat# HY-B0015), 5-Fluorouracil (Cat# HY-90006), and Olaparib (Cat# HY-10162) were purchased from Med Chem Express. Methyl methanesulfonate (Cat# 129925) and H2O2 (Cat# 1.08597) were purchase from Sigma. Methyl methanesulfonate and H2O2 were directly diluted to working concentration. Carboplatin was dissolved in ddH2O. Other drugs were dissolved and diluted in DMSO to working concentrations.
Cell viability
Cells (1000–1500 cells/well) were plated in 96-well plates and incubated for 24 hours before drug treatment. Each treatment condition was conducted in 4 replicates. Drugs were diluted to indicated concentrations and combinations and were added to plated cells and incubated for 72 hours. The concentration of Olaparib for experiments shown in Fig. 1 was fixed at 5 μM. For experiments shown in Fig. 3A, Olaparib concentrations of 2.5, 5, and 10 μM were used for the combinations with various doses of TMZ (12.5, 25, 50, 100, and 200 μM). After the drug treatments, cell viabilities were measured by incubation of cells with a 10-fold dilution of PrestoBlue (Thermo Fisher) for 2–4 hours after indicated treatments. Viability was determined by fluorescence detection using a POLARstar OPTIMA (BMG LABTECH) plate reader.
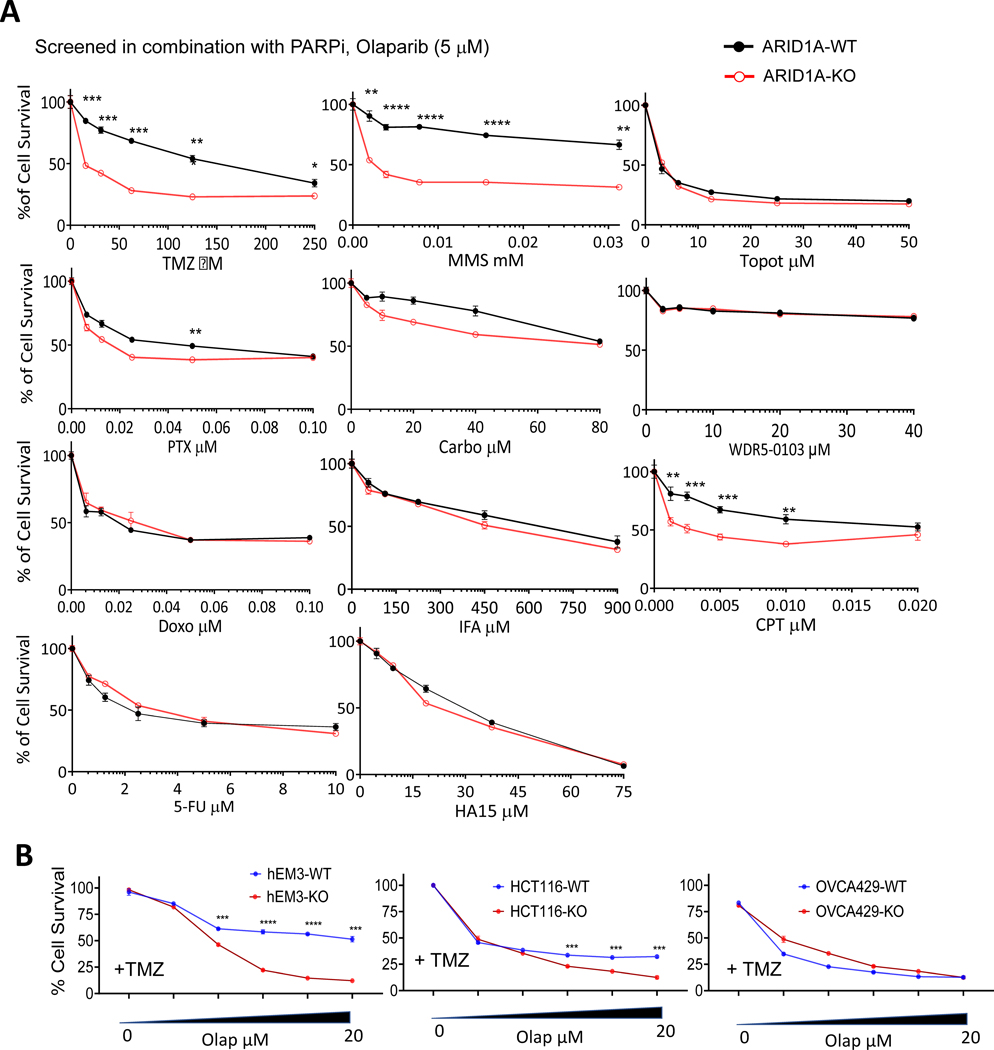
Figure 1. Alkylating agent sensitizes ARID1A-KO cells to Olaparib.
(A) Viabilities of hEM3 ARID1A-WT and ARID1A-KO cells. Cells were treated with the indicated chemotherapeutic drugs agents in the presence of 5 μM Olaparib (Olap). **P < 0.01, ***P < 0.001, ****P < 0.001, Student’s t-test.
(B) Viability of hEM3 and OVCA429 ARID1A-WT or ARID1A-KO cells assessed in the presence of 100 μM temozolomide (TMZ) in combination with serially increased doses of Olaparib. ***P < 0.001, ****P < 0.001, Student’s t-test.
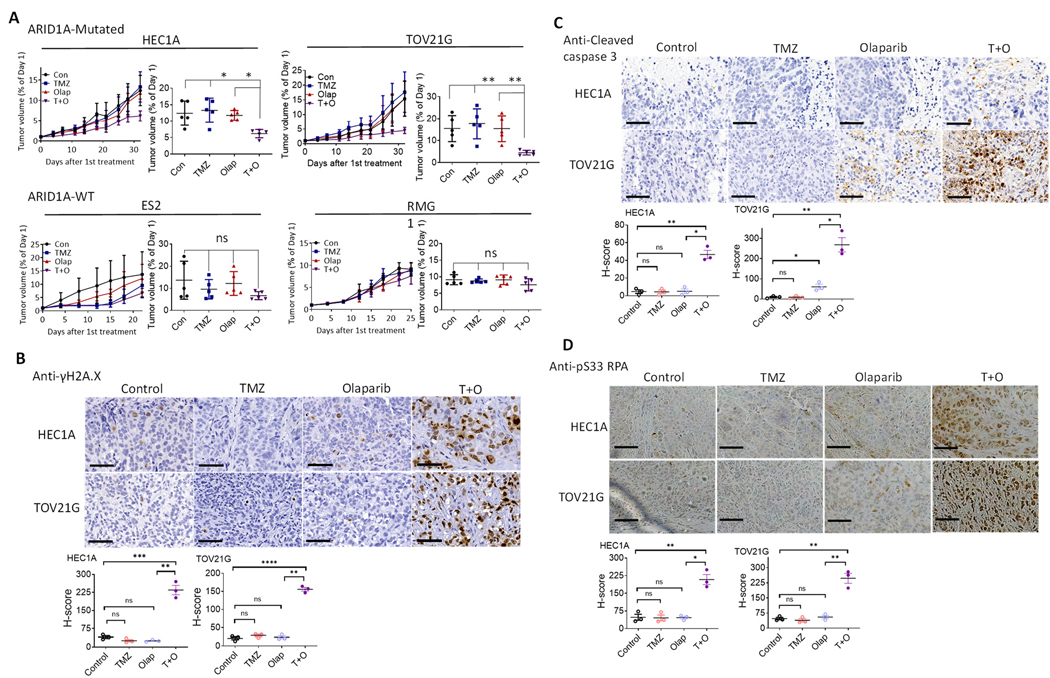
Figure 3. TMZ and Olap combination is effective in treating ovarian xenograft tumors with ARID1A mutations.
(A) In vivo tumor xenografts from ES2, RMG1, HEC1A, and TOV21G cells. Tumor-bearing mice were treated with vehicle (control), TMZ, Olap, or TMZ+Olap (T+O). Tumor growth was measured as tumor volume over a period of 30 days (left) and end point tumor volume compared to day 1 (right). Data are normalized to tumor volume collected at day 1 and presented as mean ± SEM (n = 5). Mann–Whitney test (two-tailed) was used to calculate significance of differences between two comparison groups; *P < 0.01; **P < 0.001.
(B-D) In the two ARID1A-mutant xenograft tumors, HEC1A and TOV21G, the effect of in vivo treatment on replication stress and apoptotic tendency was evaluated using three different markers: γH2A.X (B), cleaved caspase 3 (C), and pS33 RPA (D). (Top) IHC imaging results. Cells expressing the selected markers were immunodetected with DAB. (Bottom) H-score was used to quantify IHC signals of the three markers and is presented as mean ± SEM (n = 3); *P < 05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t-test. Scale bar in each photomicrograph represents 60 μm.
Comet assay
Cells were incubated with indicated treatments and then harvested by resuspension. Cells were embedded in 0.5% low melting point agarose (Promega), and comet assays were performed following the manufacturer’s instructions (Comet Assay Kit, Trevigen). Cells were stained with SYBR™ Gold Nucleic Acid Gel Stain (10,000-fold dilution) (ThermoFisher, S11494), and images were taken from at least 10 independent fields. Quantification of percentage tail DNA was performed by Image J software (ImageJ, RRID:SCR_003070). Comet score and student t-test were used for statistical comparisons.
Combination index
Cells were incubated with Olaparib and TMZ at indicated doses for 72 hours. Cell viabilities were measured, and the Combination Index was calculated using CompuSyn software.
DNA fiber analysis
After treatments, cells were labeled with CldU (8 μg/ml) for 20 minutes followed by ldU (90 μg/ml) for 20 min. Labeled cells were resuspended and mixed 1:4 with unlabeled cells and adjusted to a final concentration of 2.5 ×105 cells/ml. Cell suspensions (2.5 μl) were mixed with 7.5 μl lysis buffer (200 mM Tris pH 7.4, 50 mM EDTA, 0.5% SDS) directly on slides by gently stirring and incubated for 8 minutes at room temperature. Slides were tilted to 30°−45° to allow the lysed cells and chromatin to run down slowly along the slides. Slides were air-dried for at least 2 hours at room temperature and fixed in methanol/acetic acid 1:1 at 4°C overnight. Slides were rehydrated in PBS, incubated with 2.5 M HCl for 1 hour to denature DNA and washed in PBS three times. Slides were flooded with blocking buffer (2% BSA, 0.1% tween 20 in PBS) and incubated with primary antibodies diluted in blocking buffer for 2.5 hours at room temperature. Slides were washed in PBST 0.2% 3 times and incubated with secondary antibodies for 1 hour at room temperature. Slides were washed 3 times in PBST 0.2%, mounted with Gold Antifade Mountant (Thermo Fisher) and analyzed on a Nikon Eclipse 50i microscope.
Chromatin fractionation
Cells were treated or transfected as indicated and pre-extracted as described previously (21), then washed once with pre-extraction buffer. Chromatin fractions were fixed using the Fixation/Permeabilization Solution Kit (BD) for flow cytometry or boiled in Laemmli Sample Buffer (Biorad) for immunoblot.
Flow cytometry
The intact cells were fixed, permeabilized, blocked, and washed according to instructions for the Fixation/Permeabilization Solution Kit (BD) for cell cycle profiling coupled with DNA damage markers. The permeabilized cells were incubated with primary antibody at 4°C overnight. The cells were washed once in wash buffer and incubated with secondary antibodies (1:200 dilution) conjugated with Dylight 488 in Propidium iodide (PI, 5 μg/ml) at RT for 2 hours. The stained cells were washed once in wash buffer prior to analysis.
Chromatin fractions for flow cytometric analysis were prepared as above: first incubated with primary antibody at 4°C overnight, washed, then incubated with secondary antibodies conjugated with Dylight 488 and Dylight 650 in DAPI (1 μg/ml) at RT for 2 hours. Fractions were washed once prior to analysis on an LSR2 or LSR-Fortessa (BD) flow cytometer. Student’s t-test was used for statistical comparisons.
Immunofluorescence staining
Cells were treated as indicated and fixed in 4% paraformaldehyde for 10 minutes at RT, rinsed three times with TBS, blocked and permeabilized in blocking buffer (10% normal goat serum, 0.3% Triton X-100, 100 mM Tris (pH 7.5), 150 mM NaCl) for 1 hour at RT. Primary antibodies in blocking buffer (1:200) against indicated targets were incubated with cells for 2–3 hours at room temperature in a humidified chamber. Cells were washed 4–5 times in TBS and incubated with secondary antibodies (1:200) conjugated with Dylight 488 (Jackson ImmunoResearch) or Dylight 594 (Jackson ImmunoResearch) in blocking buffer for 2 hours at RT. Cells were washed 4–5 times in TBS and mounted using Gold Antifade Mountant (Thermo Fisher).
Dot blot
Genomic DNA was harvested from cells after indicated treatments using a QIAamp DNA Micro Kit (QIAGEN) and dot blotted on positively charged nylon membrane, Amersham™ Hybond™-N+ (GE). Membranes were dried for 2 hours at RT and cross-linked with a UVC 500 Crosslinker (Amersham Biosciences) at 0.25 J/cm2. The membrane was blocked in 5% fat-free milk/TBST (TBS 0.1% tween 20) for 30 minutes and incubated with primary antibodies (1:000) against 3-meC (Active Motif, Cat# 61179, RRID:AB_2793540) and O6-meG (Squarix, Cat# SQM003.1) at 4°C overnight. Membranes were washed three times in TBST and incubated with secondary antibodies (1:10,000) at RT for 2 hours. Membranes were washed three times in TBST, and images were acquired on a ChemiDoc Imaging System (Biorad). Quantifications were performed using Image J.
Micro-irradiation
Cells were plated in Nunc™ Glass Bottom Dishes (Thermo) and transfected with indicated proteins tagged with GFP (OriGene). Cells were incubated in sensitizer (Hoechst 33342; Thermo) for 10 minutes; sensitizer was replaced with phenol red-free RPMI for reduction of background. Micro-irradiation and image collection were performed on a Confocal microscope A1 (Nikon) equipped with a 405 nm diode laser set to 30% (spot irradiation, 1 iteration, zoom 1, and dwell time of 30 μs) at 37°C, 5% CO2 as described for analysis of BER factors (22,23). Quantifications were performed using Image J.
Immunohistochemistry
Deparaffinization and rehydration were performed by incubation in xylene and serial alcohol dilutions. Antigen retrieval was performed by immersion in citrate buffer (Sigma) at 90°C followed by incubation in H2O2 in methanol for 15 minutes at RT to inhibit endogenous peroxidase. Slides were blocked in Antibody diluent (Dako) for 30 minutes at RT. Slides were incubated with primary antibodies against indicated targets overnight at 4°C. Slides were washed three times in TBST for 5 minutes and then incubated with HRP-conjugated secondary antibodies (Dako) for 30 minutes at RT. Slides were washed three times in TBST, chromogen substrate was added, slides were counterstained with hematoxylin (Sigma) and mounted in Cytoseal (Thermo Scientific) mounting medium for analysis.
AP site quantification
Following incubation with indicated treatments, genomic DNA was isolated from cells. An AP site quantification kit (Cat#: STA-324, Cell Biolabs) was used for analyzing the number of AP sites following the manufacturer’s instruction.
Tumor xenografts in mice
HEC1A or TOV21G cells (2 or 4 × 106) suspended in Matrigel were injected subcutaneously into both flanks of 6–8-week-old female Nu/Nu mice. Since ovarian cancer is a female disease, only female mice were used in this study. When tumor sizes reached ~200 mm3, mice were randomly divided into four groups and treated with vehicle (10% DMSO in PBS), TMZ (20 mg/kg), Olaparib (10 mg/kg), or TMZ + Olaparib combination by intraperitoneal (i.p.) injection every other day for three weeks. Measurement of tumor size was initiated on the first day of treatment and was performed twice per week. The experiment was terminated when the total tumor load reached ~2000 mm3. Mice were euthanized, and tumors were fixed in formalin or frozen at −80°C. Immuno-stained slides were evaluated by an experienced gynecological pathologist (IMS) blinded to each in vivo treatment condition. The use of animal has been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
Statistical analyses
Mann–Whitney test (two-tailed) was used for the calculation of significance in experiments when n≥5. Student’s t-test was used when n≤3. Statistical calculations were performed using Prism. Non-linear mixed effects model adjusting for correlation among repeated measures was used for analyzing data in Figure 5. All experiments were performed in at least three biological repeats.
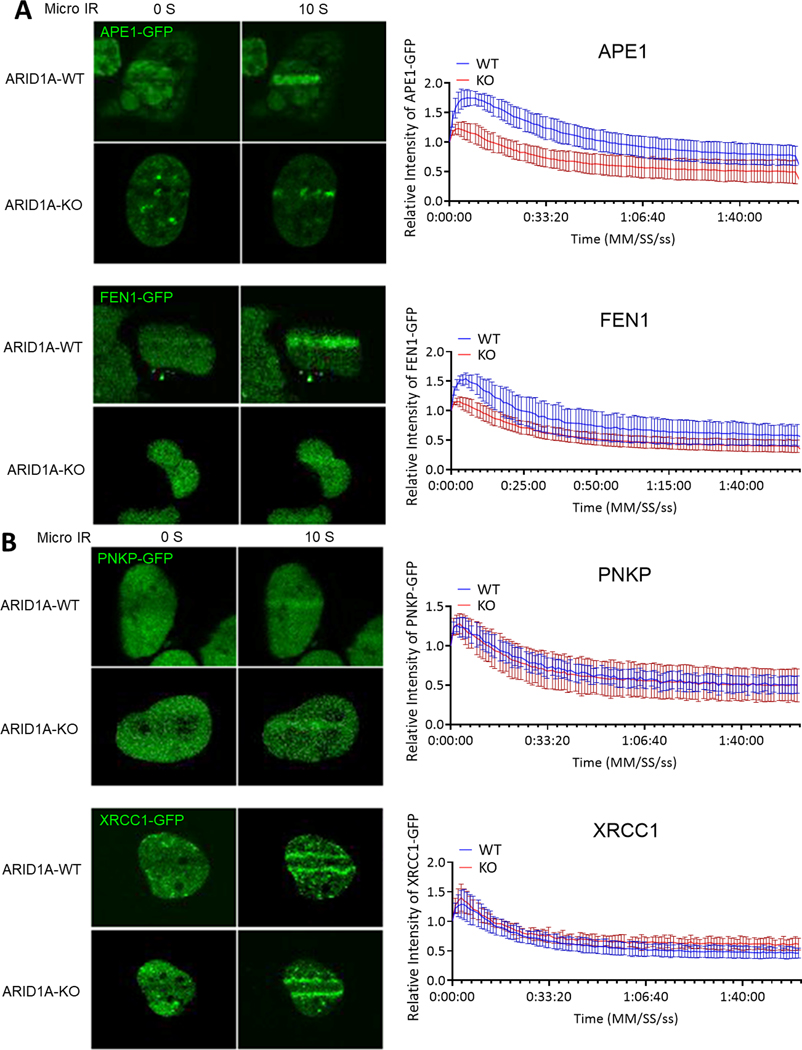
Figure 5. Short patch pathway is not influenced by ARID1A-KO under micro-irradiation (micro-IR).
(A) (Left) Representative live-cell images of hEM3 ARID1A-KO and control cells expressing APE1-GFP and FEN1-GFP at 0 and 10 seconds after micro-IR. (Right) Relative fluorescence intensity at irradiated area measured every second for 2 minutes and plotted as mean ± SEM after normalization to the background fluorescence intensity using Image J. (B) (Left) Representative live-cell images of hEM3 ARID1A-KO and control cells expressing PNKP-GFP and XRCC1-GFP at 0 and 10 seconds after micro-IR. (Right) Relative fluorescence intensity at irradiated area measured every second for 2 minutes and plotted as mean ± SEM after normalization to the background fluorescence intensity using Image J.
Data availability
Data were generated by the authors and included in the article.
Results
ARID1A-deficient mammalian cells are highly sensitive to TMZ/Olaparib combination
PARP inhibitors show significant anti-tumor effects in many Type II ovarian high-grade serous carcinomas, especially those with HR defects including BRCA1 and BRCA2 mutations. However, Type I ovarian cancers are mostly BRCA-wildtype, do not display homologous recombination deficiency signatures nor have high DNA copy number alterations, and their response to PARP inhibitor as a monotherapy or in combination therapy remains to be evaluated (24). As compared to high-grade serous carcinomas, frequent mutations in ARID1A of the SWI/SNF chromatin remodeler complex, were identified in ovarian clear cell carcinoma, a Type I ovarian cancer (5,6). Previous studies in mammalian systems reported promiscuous involvement of ARID1A in DNA damage repairs (12–14). In cancer cells with mutations of key tumor suppressor genes guarding the genome such as TP53 and ARID1A, multiple DNA repair pathways and cellular functions were concurrently impacted, readjusted, and rebalanced to maintain cell survival and support tumor growth (25). This alteration in DNA repair processes is specific for cancer cells, thereby providing opportunities for developing highly specific synthetic lethal treatments.
To identify clinically available genotoxic drugs that may enhance the response of ARID1A-mutated Type I ovarian cancers to PARP inhibitors and thus can be re-purposed for cancer therapy, we tested drug-induced cytotoxic effects in a pair of isogenic cell lines, hEM3, with or without CRISPR-mediated ARID1A knockout (19,20). hEM3 originated from human endometrial epithelium, the cellular origin of ovarian clear cell carcinoma and endometrioid carcinoma, and its application in drug screening and functional studies have been reported previously (19,20). The genotoxic drugs tested here include Topoisomerase II inhibitor: Doxorubicin (Doxo); Topoisomerase I inhibitor: Camptothecin (CPT); DNA cross-linkers: Carboplatin (Carbo) and Ifosfamide (IFA); DNA replication inhibitor: 5-Fluorouracil (5-FU); and DNA alkylating drugs: Temozolomide (TMZ) and Methyl methanesulfonate (MMS). In addition, an experimental drug, WDR5–0103, which inhibits MLL histone methyltransferase activity, as well as HA15, which induces endoplasmic reticulum stress, and Paclitaxel (PTX), which stabilizes microtubules, were also evaluated.
With the exception of CPT, when these drugs were applied singly, none produced a differential cytotoxicity between ARID1A-KO and -WT cells (Fig. S1A). However, when co-applied with PARP inhibitor, alkylating drugs including TMZ and MMS induced significant cell killing of ARID1A-KO cells but not WT cells at most testing doses, (Fig. 1A). TMZ significantly sensitized ARID1A-KO hEM3 to Olaparib (Fig. 1B). Similar observations were made in clonogenic assays performed on another pair of isogenic ARID1A-WT and –KO lines created from MCF10a (Fig. S1B). Clonogenic assays performed on isogenic hEM3 cell lines also confirmed the cytotoxicity data (Fig. S1C). The sensitivity of hEM3 and MCF10a to Olaparib single agent was also assessed. The response to Olaparib did not differ between ARID1A-KO versus -WT cells, except for higher doses of Olaparib (Fig. S2).
When compared to phenotypes observed in the immortalized normal epithelial cells, hEM3 and MCF10a, an endometrioid cancer cell line, OVCA429, with a mutation in MSH6 and HCT116, a colorectal cell line with well-recognized deficiency in DNA mismatch repairs (MMR), were highly sensitive to TMZ/Olap regimen, and ARID1A deficiency displayed a minimal sensitization effect (Fig. 1B).
Since both hEM3 and MCF10a are not MMR-deficient, our results suggest that the alkylating drug, TMZ, significantly enhanced Olaparib sensitivity in MMR-proficient epithelial cells with ARID1A deletion or inactivation, although a future study with a large repertoire of MMR-deficient and -proficient cells is needed to test this hypothesis. We also applied another two PARPi, Veliparib and Niraparib, to verify TMZ/Olap-induced cytotoxicity in ARID1A-KO cells. When co-applied with TMZ, these PARP inhibitors also provoked synergistic cytotoxic responses in ARID1A-KO cells (Fig. S3A).
TMZ has been widely used for treating brain tumors, however, its clinical benefit for solid tumors such as ovarian or lung cancers remains to be established. Recent data from a TMZ/Olaparib combination treatment trial on patients with small cell lung cancer restores the promise of using TMZ to enhance PARPi response in some solid tumors (26).
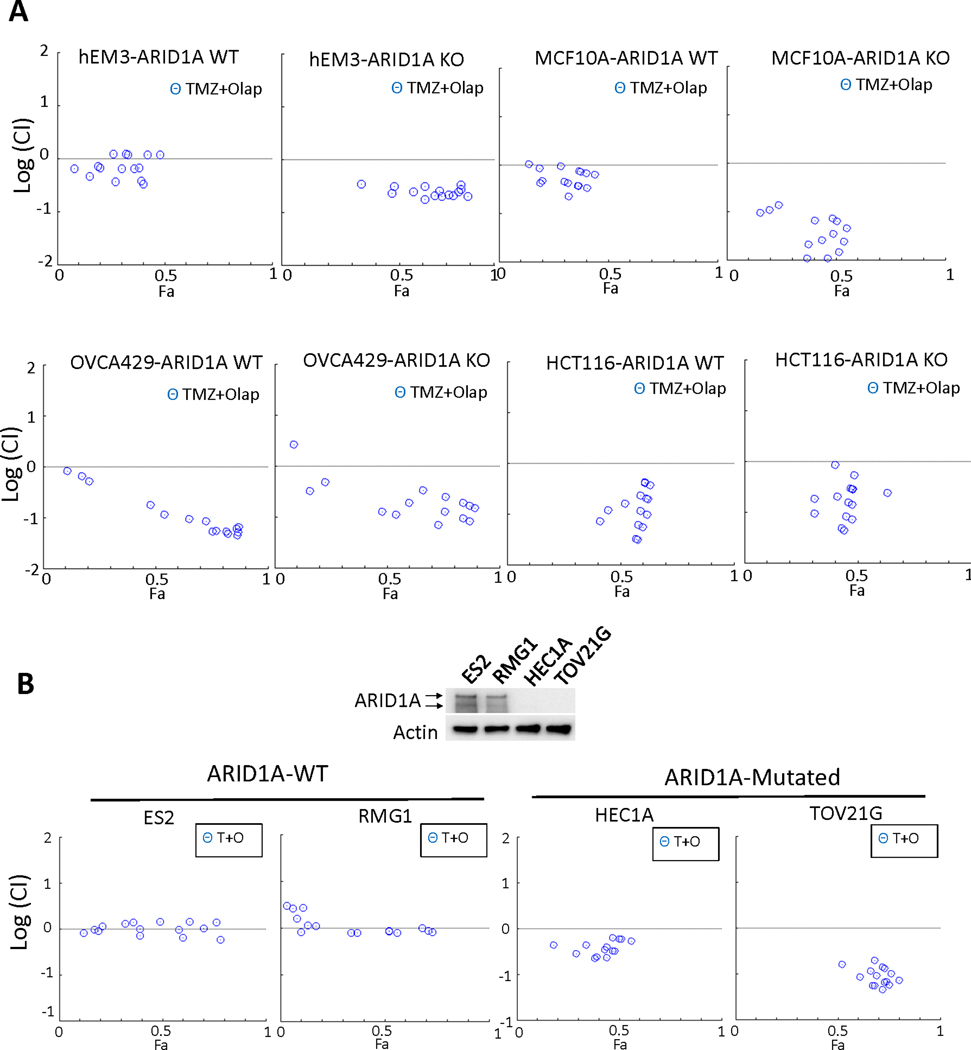
Next, we evaluated TMZ and Olaparib drug-drug interaction in four pairs of isogenic ARID1A-KO and ARID1A-WT cancer cell lines. Among which HCT116 and OVCA429 are characterized by MMR deficiencies, whereas hEM3 and MCF-10a have no other known DNA repair defects. The logarithmic combination index in ARID1A-KO EM3 and MCF-10a cells was significantly lower than 0, indicating a synergistic cytotoxic effect of TMZ/Olap drug combination, whereas the index in their ARID1A-WT counterparts was close to 0, indicating an additive effect (Top, Fig. 2A). Similar synergistic effects in ARID1A-KO hEM3 cells were observed when another alkylating drug, Methyl methanesulfonate (MMS), was co-applied with Olaparib (Fig. S3B). In contrast, MMR-deficient HCT116 and OVCA429 lines showed a synergistic response to the TMZ/Olap combination treatment, irrespective of ARID1A status (Bottom, Fig. 2A).
Figure 2. Synergy between DNA alkylating drug and PARP inhibitor in ARID1A-deficient and ARID1A-proficient cell lines.
(A) Cytotoxicity test reported by logarithmic combination index (CI) plots of DNA alkylating agent Temozolomide (TMZ) in combination with PARP inhibitor, Olaparib, in ARID1A-WT and ARID1A-KO hEM3 cells from endometrial tissue, MCF-10a cells derived from mammary gland epithelium, OVCA429 cells from an ovarian cancer case, and HCT116 cells from a colorectal cancer case. Drugs were applied over a range of concentrations, and the Fa (Fraction Affected) represents the fraction of affected cells by treatment. The horizontal dashed line at Log (CI) = 0 separates synergistic [Log (CI)<0] from antagonistic [Log (CI)>0] and additive [Log (CI)=0] drug-drug interactions. (B) (Top) Immunoblot of ARID1A protein expression in tumor cell lines ES2, RMG1, HEC1A, and TOV21G. ES2 and RMG1 express ARID1A and lack mutations in the ARID1A gene. HEC1A and TOV21G do not display detectable ARID1A expression, and both cells have deleterious mutations in ARID1A. (Bottom) Log (CI) plots showing response to TMZ/Olaparib (T+O) in ARID1A-WT and in ARID1A-mutated cells.
We also evaluated the TMZ/Olaparib combination interaction in four Type I ovarian cancer lines, two of them (HEC1A and TOV21G) harbor ARID1A mutation and the other two (ES2 and RMG1) do not have ARID1A mutation (Fig. 2B). ARID1A protein expression can be detected in the two ARID1A-WT cancer cell lines, ES2 and RMG1, whereas HEC1A and TOV21G, which have ARID1A mutation lose ARID1A protein expression (Top, Fig. 2B). In the drug combination assessment, we observed that TMZ/Olaparib regimen produces a synergistic effect in ARID1A-mutated Type I ovarian cancer cell lines and an additive effect in ARID1A-wildtype ovarian cancer cell lines (analyzed by COMPUSYN software, Fig. 2B). Experiments were also conducted on Type I ovarian cancer cell lines using the same drug combination condition as depicted in Fig. 1A (fixed Olaparib concentration of 5 μM and a serial dose of TMZ). The results demonstrated that Type I cancer cell lines with ARID1A mutations are generally more sensitive to this drug combination treatment than the cell lines without ARID1A mutation (Fig. S3C).
The TMZ and Olaparib combination decreased growth of ARID1A-mutated tumors
To evaluate the efficacy of TMZ/Olap treatment in vivo, we established tumor xenografts from the four Type I ovarian cancer cell lines (HEC1A, TOV21G, ES2, and RMG1) and tested TMZ/Olap either singly or in combination. Significant tumor growth inhibition of HEC1A and TOV21G xenografts, both of which harbor a deleterious ARID1A-mutation, was observed in TMZ/Olap combination-treated mice. Tumor growth inhibition was absent in single agent-treated mice or in mice with ARID1A-wildtype ES2 and RMG1 xenografts (Fig. 3A).
We next assessed expression of the DDR/replication stress markers, γH2A.X and pS33 RPA, and the apoptosis marker, cleaved caspase 3, on tissue sections from these xenograft tumors (Fig. 3B–D). The TMZ/Olaparib combination led to an increased number of γH2A.X-positive, cleaved caspase 3-positive, and pS33 RPA-positive cells in ARID1A-mutated xenograft tumors compared to single agent or vehicle control-treated tumors (Fig. 3B–D). In contrast, we did not detect a significant number of cells positive for either of these markers in ARID1A-wildtype tumor xenografts (Fig. S4A–C).
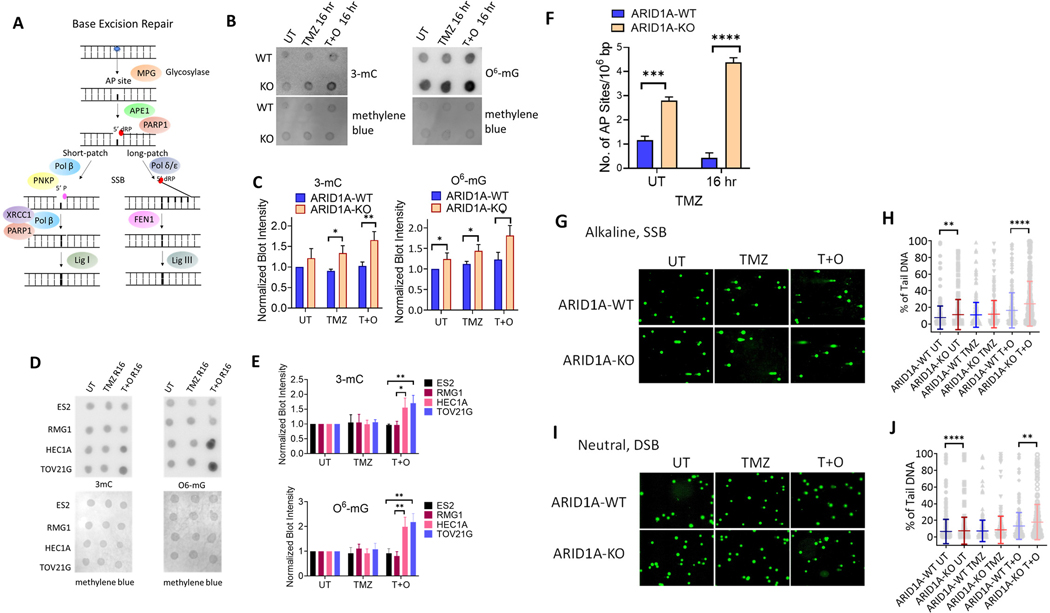
ARID1A deletion compromised base excision repair upon exposure to DNA alkylating agents
Alkylating drugs, such as TMZ, methylate DNA bases at N (80%) and O (8.3%) atoms (27). In response, N-methylations are repaired by DNA base excision repair (BER) or nucleotide excision repair (NER). During normal replication, O6-methylation often occurs on guanines to form O6MeG:T mismatch pairs, which are recognized and repaired by the DNA MMR system. In BER, methylated bases are removed by a DNA glycosylase, resulting in abasic (AP) sites that are recognized by an apurinic/apyrimidinic endonuclease, APE1 (Fig. 4A). In MMR, a gapped duplex is created by incision of the newly replicated strand and a futile repair loop, which creates double-strand break (DSB) intermediates that activate DSB repair pathways or induce apoptosis. Accordingly, methylating agents induce both the DSB repair pathway and G2/M cell cycle arrest.
Figure 4. Defective Base Excision Repair in ARID1A-Deficient Endometrial Epithelial Cells.
(A) The mechanism of Base Excision Repair (BER). Alkylating agents such as Temozolomide (TMZ) can induce methylation of DNA bases, which is repaired through BER. When such lesions are induced, the DNA glycosylase MPG will remove the entire base from the DNA strand resulting in an abasic (AP) site. Subsequently, AP endonuclease (APE1) cleaves the AP site where a 5’ deoxyribose phosphate (dRP) end is exposed. From here, this pathway may diverge into two DNA repair routes: short patch repair or long patch repair. In the short patch repair pathway, the dirty ends are cleaned up by Pol β or PNKP. PARP1 first binds to damaged DNA and initiates recruitment of XRCC1 and DNA polymerase beta (Pol β). They collectively form a complex and replace a complementary nucleotide using the sister strand as a template. The newly synthesized DNA is ligated by DNA ligase I (Lig I). In the long patch repair pathway, a longer strand (2–10 bases) is synthesized by DNA polymerase delta/epsilon (Pol δ/ε) to replace the redundant 5’ dRP end, which is then removed by the flap endonuclease (FEN1). The newly synthesized DNA strand is ligated by DNA ligase III (Lig III).
(B) Dot blot showing levels of alkylated DNA in hEM3 ARID1A-WT and ARID1A-KO cells exposed to the indicated treatment regimens for 4 hrs, then recovery for 16 hrs. (Left) Methylcytosine (3-mC), (Right) O6-Methylguanine (O6-mG). Untreated (UT), Temozolomide (TMZ), combination of Temozolomide and Olaparib (T+O). Methylene blue staining is used as a loading control.
(C) Normalized blot intensity calculated from result shown in B. Student’s t-test was used to calculate significance and to normalize the result. Data are presented as mean ± SEM. n=3; *P < 0.05; **P < 0.01.
(D) Dot blot showing levels of alkylated DNA assessed in Type I ovarian cancer cell lines with (HEC1A, TOV21G) or without (ES2, RMG1) ARID1A mutation exposed to the indicated condition. Methylene blue staining is used as a loading control. *P < 0.05; **P < 0.01 (Multiple unpaired t-test).
(E) Normalized blot intensity calculated from results shown in D. **P < 0.01 (Multiple unpaired t-test).
(F) Number of apurinic (AP) sites per 106 base pair (bp) in hEM3 ARID1A-WT and ARID1A-KO cells at 0 hr and 16 hr after TMZ drug treatment. AP sites result from DNA glycosylase, an upstream effector in the BER pathway. The value was measured using an AP site quantification kit (Cell Biolabs). Student t-test was used to calculate significance. Data are presented as mean ± SEM. n=3; ***P < 0.001, ****P < 0.0001 (Multiple unpaired t-test).
(G) Alkaline comet assay was performed on hEM3 cells (ARID1A-WT and -KO) 16 hours after the treatment with TMZ, TMZ+O, or vehicle control (UT).
(H) Quantification of data from (G). Data are presented as mean ± SEM, **P < 0.01, ****P < 0.0001 (Mann-Whitney test). (I) Neutral comet assay was performed on hEM3 cells (ARID1A-WT and ARID1A-KO) 16 hours after the treatment with TMZ, TMZ+O, or vehicle control.
(J) Quantification of data from (I). Data are presented as mean ± SEM; **P < 0.01, ****P < 0.0001 (Mann-Whitney test).
Because DNA base methylation adducts introduced by TMZ can be repaired by the BER mechanism, we first evaluated BER capacity in ARID1A-KO cells versus wildtype controls. The levels of methylated DNA bases including Methylcytosine (3-mC) and O6-Methylguanine (O6-mG) were assessed by dot blot assay using antibodies against each specific methylated base (28) (Fig. 4B). As expected, we detected higher levels of alkylated DNA bases in ARID1A-KO cells than in ARID1A-WT cells. Furthermore, the differences were more pronounced in TMZ/Olap-treated cells (Fig. 4C). We also verify the findings in Type I ovarian cancer cell lines, HEC1A and TOV21G, with deleterious ARID1A mutation and in ES2 and TOV21G ovarian cancer cell lines without ARID1A mutation (Fig. 4D–E). These results are in agreement with the above findings in which cancer cells with ARID1A mutation after alkylating drug and PARPi combined treatment accumulated more abundant DNA base lesions than cancer cells without ARID1A mutation.
We also measured the number of apurinic/apyrimidinic sites (AP) sites generated by BER or spontaneous depurination (29) and found a higher number of endogenous AP sites in ARID1A-KO cells than in ARID1A-WT cells. Moreover, the number of AP sites in ARID1A-KO cells was increased by drug treatment involving either TMZ or PARPi. Since AP sites are generated by glycosylase, the data suggest both KO and WT cells likely have active glycosylase activity. However, since drug-induced AP sites returned to basal levels in ARID1A-WT cells within 16 hours, but did not return in ARID1A-KO cells, KO cells likely have a delayed BER activity, downstream of glycosylase (Fig. 4F).
DNA comet assays in alkaline or neutral conditions were also performed to assess ssDNA and dsDNA breaks, respectively (Fig. 4G–J). A marginal increase in ssDNA breaks, comet tail lengths in alkaline condition, can be observed in the hEM3 ARID1A-KO as compared to the ARID1A-WT cells. When cells were exposed to TMZ/PARPi, both ssDNA and dsDNA breaks were increased in ARID1A-KO as compared to the ARID1A-WT cells (Fig. 4H & 4J). The increased endogenous ssDNA breaks in ARID1A-KO cells are congruent with the greater number of AP sites generated by glycosylase in these KO cells (data shown in Fig. 4F).
To determine whether there is a hindrance of BER in ARID1A-KO, we assessed recruitment kinetics of BER effectors after micro-irradiation-induced base lesions and single-strand DNA breaks in ARID1A-KO or –WT cells (23,30). Plasmids containing GFP-tagged XRCC1 or PNKP (short-patch BER repair) or APE1 or FEN1 (long-patch BER repair) were transfected into ARID1A-KO or ARID1A-WT cells. The relative intensity curve of APE1 and FEN1 peaked at a significantly higher rate in the ARID1A-WT cells than the ARID1A-KO cells (P= 6.5251e-09 and P= 1.3029e-09, respectively, Fig. 5A). The peak relative intensity was 1.8043 ± 0.1444 from the APE1transfected WT cells as compared to 1.2537 ± 0.1021 from the APE1transfected KO cells (p= 8.6058e-07). For FEN-transfected cells, the peak relative intensity was 1.5663 ± 0.1218 from the ARID1A-WT cells, compared to 1.1716 ± 0.0709 in the KO cells (P= 6.2096e-06) (Fig. 5A). In contrast, recruitment kinetics of short-patch BER effectors, XRCC1 and PNKP, were not affected by the ARID1A status (Fig. 5B). There was no difference in the peak intensity or rate to reach peak between the ARIDA-WT and ARID1A-KO cells. Collectively, the data suggest that ARID1A influences long-patch BER effectors but perhaps does not affect short-patch BER effectors (Fig. 4A).
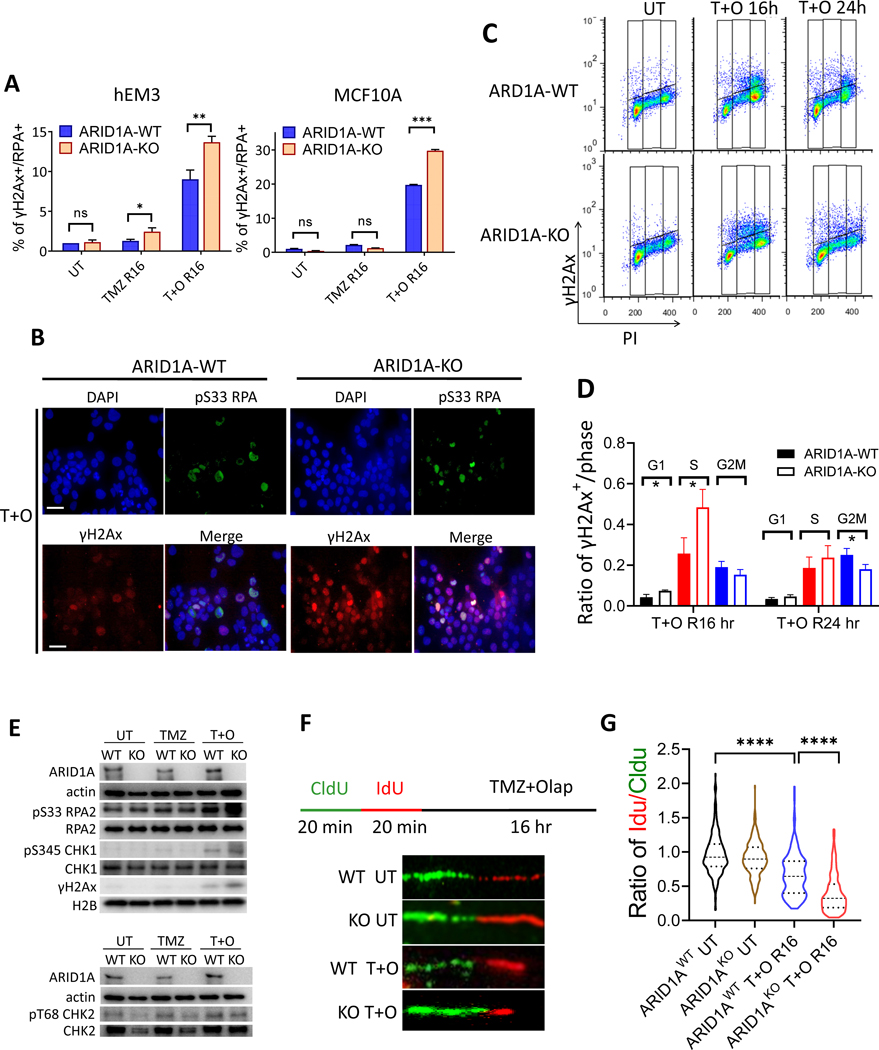
TMZ/Olaparib combination elicits replication stress and hampers replication fork stability in ARID1A-KO cells
Delayed or hindered recruitment of DNA repair effectors to sites of single stranded DNA damage can result in accumulation of unrepaired ssDNA lesions to dsDNA breaks during DNA replication and can cause replication stress followed by cell death if the lesions are not swiftly resolved. To determine the levels of replication stress, we used the following approaches. First, we directly measured RPA loading onto chromatin coupled with γH2A.X levels (31) and observed a higher percentage of chromatin-bound RPA and γH2A.X in TMZ treated ARID1A-KO hEM3 and MCF-10a cells compared to their ARID1A-WT counterparts (Fig. 6A). Adding Olaparib to TMZ further increased replication stress in ARID1A-KO cells (Fig. 6A). Immunofluorescence analysis of the replication stress markers γH2A.X and pS33 RPA2 showed significantly upregulated nuclear expression of both markers in TMZ/Olaparib-treated ARID1A-KO hEM3 cells (Fig. 6B). In contrast, single agent- or vehicle control-treatment did not elicit significant upregulation of either marker in these cell lines.
Figure 6. Elevated replication stress in ARID1A-KO cells after combination treatment with TMZ+Olap.
(A) Level of replication stress in response to indicated drug treatment is represented by percentage of cells positive for both γH2A.X signaling (γH2A.X+) and RPA chromatin binding (RPA+) in ARID1A-WT and ARID1A-KO cell lines derived from hEM3, MCF-10a, HCT116, and OVCA429. n=3; *P < 0.05. **P < 0.01; ***P < 0.005 (Multiple unpaired t-test).
(B) Immuno-fluorescence images of hEM3 ARID1A-WT and ARID1A-KO cells treated with TMZ+Olap for 4 hrs then recovery for 16 hrs. Anti-pS33 RPA antibody is tagged with a red fluorophore and anti-γH2A.X with a green fluorophore. Scale bar in each photomicrograph is 10 μm.
(C) Flow cytometric analysis of cell cycle distribution and γH2A.X expression in hEM3 ARID1A-WT and ARID1A-KO cells at 0, 16, and 24 hours after TMZ+Olap treatment.
(D) Quantification of γH2A.X+ cells normalized to each cell cycle stage. Data are presented as mean ± SEM, n = 3; *P < 0.05 (Multiple unpaired t-test).
(E) Expression levels of replication markers assessed by immunoblots. CHK1 and RPA are phosphorylation substrates of ATR, whereas CHK2 is the phosphorylation substrate of ATM.
(F) Top: Schema showing cells incubated with thymidine analogue CIdU for 20 min followed by IdU for 20 min, and then treated with TMZ+Olap for 16 h. CIdU (green) and IdU (red) incorporation into DNA tracks was visualized by immunofluorescence. Bottom: DNA fiber analysis showing DNA tracks with incorporation of CIdU (green) and IdU (red) analogues.
(G) Length of each labeled track (CIdU and IdU) in DNA combing assay was measured using Image J and quantified by violin plot after normalization of IdU to CIdU. Mann Whitney test was used calculate significance. Approximately 300 tracks were counted for each group; ****P < 0.0001.
We further evaluated DNA damage patterns, assessed by γH2A.X positivity, throughout the cell cycle. Delayed progression of γH2A.X+ in S phase was seen at 16 h after TMZ/Olaparib treatment ARID1A-KO cells (Fig. 6C & 6D), but by 24 h the percentage of γH2A.X+ cells in S phase was similar between ARID1A-KO and -WT cells (Fig. 6C & 6D). Cell cycle distribution was assessed simultaneously in this experiment. In the first 4–8 h, the percentages of cell populations in G1, S, and G2/M phases were similar between ARID1A-KO and ARID1A-WT cells (Fig. S5A). By 16–24 h, G2/M arrest became apparent in the ARID1A-WT cells. At the same time, G2/M arrest also occurred in ARID1A-KO cells, but to a lesser extent than in the ARID1A–WT cells (Fig. S5A).
Significantly increased numbers of γH2A.X+ cells in S phase were also observed in MCF-10a ARID1A-KO cells 16 h after TMZ+Olap treatment (Fig. S5B). However, S phase repair in MCF-10a ARID1A-KO cells was slower than in hEM3, as a significant fraction of γH2A.X+ cells were still in S phase 24 h after TMZ+Olap treatment.
Replication stress was further assessed on the basis of expression of pS345 CHK1, pS33 RPA2, and γH2A.X, all of which are ATR kinase substrates. Western blots demonstrated increased phosphorylation levels in ATR-related proteins including pS345 CHK1 and pS33 RPA2 alongside elevated γH2A.X levels in ARID1A-KO cells upon exposure to TMZ/Olaparib (Fig. 6E, Western blot quantification in Fig. S6). However, we did not observe a similar effect on phosphorylation of the ATM-related protein, pT68 CHK2. Taken together, the results indicate that the TMZ/Olaparib combination aggravated replication stress in cells lacking ARID1A.
Since replication stress portends replication fork abnormalities, we evaluated replication fork stability by DNA fiber analysis in cells subjected to different treatments (Fig. 6F). Shortening of the second track (red) relative to the first track (green) in the treated cells reflects degradation of the stalled forks. In the absence of drug treatment, the ratio of IdU/CIdU tracks was close to 1.0 (Fig. 6G). TMZ/Olaparib treatment was associated with a shorter second track in both ARID1A-KO and -WT cells but the track shortening was most pronounced in ARID1A-KO cells (Fig. 6G).
Discussion
It has been well recognized that one of the most celebrated advances in treating advanced ovarian cancer in recent years is the introduction of PARP inhibitors as the first-line maintenance therapy (32). PARP inhibitors were initially used for treating patients with ovarian high-grade serous carcinomas having germline mutations in BRCA1 and BRCA2 or other genes of the homologous recombination DNA repair pathway for mending double-strand DNA breaks (32). Subsequent studies showed that patients without such mutations could also benefit from PARP inhibitors, suggesting their broader applications beyond the initial intended use focusing on cancers with a BRCAness genotype (33). While the development of future generation PARP inhibitors is in sight, exploring other therapeutic agents to enhance PARP inhibitor response or to sensitize PARP refractory cancers and to identify cancer biomarkers to predict treatment response for PARP inhibitors will open new avenues for the next era of ovarian cancer treatment.
The results of this study are significant in the following accounts. First, we identified a new synthetic lethal strategy to enhance responses of ARID1A-mutated or deficient cancer cells to PARP inhibitors. We demonstrated that cancer cells with ARID1A-deficiency became highly vulnerable to PARP inhibitors when alkylating drugs were co-administrated. Second, because ovarian clear cell carcinomas are intrinsically refractory to conventional platinum-based chemotherapy (34–36), there is an unmet need to develop novel and effective treatments for women with this subtype of ovarian cancers. The genetic-specific targeting strategy reported here warrants clinical evaluation in this population of patients. Third, our findings suggest that well-established cancer drugs can be repurposed to treat cancer types that have not been evaluated previously. In support of this view, TMZ, which was discovered more than two decades ago as a treatment for glioblastoma multiforme, was recently found to have substantial clinical activity for relapsed small cell lung cancer when combined with the PARP inhibitor, Olaparib (26).
ARID1A-deficient mammalian cells exhibit modestly increased DNA strand breaks while maintaining a comparable proliferation rate as ARID1A-expressing cells, suggesting some level of dysregulated DNA repair function. In cancer cells, mutations in ARID1A gene may cause dysregulated or “rebalanced” DNA repair mechanisms, rendering them susceptible to genotoxic insults and PARP inhibitors. In terms of mechanism, we speculate that DNA base lesions elicited by an alkylating drug could not be swiftly and efficiently resolved in endometrial epithelial cells or Type I ovarian cancer cells with ARID1A mutation. This could be attributed to dysregulated expression of one or more BER pathway members or compromised recruitment of BER factors to DNA lesions. As a result, ARID1A-deficient cells largely rely on the PARP1-dependent DNA repairs including NER and alternative NHEJ to safeguard their survival. Therefore, PARP inhibition aggravates DNA repair defects associated with ARID1A deficiency, causing replication fork stalling, fork collapse, and cancer cell death. In the DNA fiber assays, we indeed observed that end resection of the replication fork, resulting from prolonged stall, was significantly enhanced in TMZ plus Olaparib treated ARID1A-deficient cells.
It is worth mentioning that genome-wide sequencing performed on Type I and Type II ovarian cancers clearly demonstrated that uniquely dysregulated DNA repair mechanisms stratify subtypes of ovarian cancers (37). Mutation signatures or genomic scars as a consequence of defective DNA repair/guarding mechanisms and exposure to endogenous or exogenous factors were categorized, and the patterns were used to indicate mutational processes contributing to the development of specific tumor types (38). Studies using this approach have demonstrated that approximately 50% of ovarian HGSCs display homologous recombination deficiencies, attributed to inactivation of the BRCA pathway (37). On the other hand, OCCCs predominately display mutation signatures corresponding to dysregulated APOBEC and aging. Mutation signatures/genomic scars associated with smoking and transcription-coupled DNA repair, likely related to BER and NER, were also identified in OCCC genomes. Collectively, dysregulated DNA repair/DNA guarding mechanisms in OCCCs predicted by their unique mutation signatures may shed light on OCCC etiology and pathogenesis. Mutation signatures can also be exploited for correlation with treatment response and disease prognosis, thus their roles and clinical significance in OCCCs warrant future active investigation.
The synthetic lethal approach reported here is expected to be specific for tumor cells, because normal cells are mutation-free and have intact DNA repair programs. The results are also congruent with our previous report showing that priming ARID1A mutated ovarian cancer cells by low-dose ionizing radiation, which induces simple DNA lesions and simple DNA breaks, sensitized these cancer cells to PARP inhibitors (12). The increased DNA base lesions and DNA breaks induced by TMZ or IR in cancer cells increase their reliance on PARP-dependent DNA repairs. Consequently, we see a marked synergistic effect with combined treatment of TMZ or IR and PARP inhibition in ARID1A-deficient cancers.
Collectively, the preclinical data reported herein provide a promising synthetic lethal strategy for treating cancer patients whose tumors harbor inactivating mutations in ARID1A while sparing normal cells and tissues which should not harbor such mutations. Clinically, the FDA-approved alkylating agent, TMZ, has long been used for cancer treatment. Similarly, at least four PARP inhibitors have been approved to date for cancer first-line or maintenance therapy. The combination of TMZ and PARPi which shows synergistic effects in ARID1A-deficient cancers reported here merits further clinical evaluation.
Supplementary Material
Significance.
The combination of temozolomide and PARP inhibitors exploits the specific DNA damage repair status of ARID1A-inactivated ovarian cancers to suppress tumor growth.
Acknowledgments
This study was supported by NIH grants P50CA228991 (IM Shih, TL Wang, P Huang, P Oberdoerffer, S Gaillard), R01CA215483 (IM Shih), R01CA260628 (IM Shih, TL Wang), and the Collaborative Research Development Grant (IM Shih, TL Wang) from the Ovarian Cancer Research Alliance.
The authors would like to thank Dr. Shyam K. Sharan, National Institutes of Health, and Dr. Yun Chen, Johns Hopkins University, for their protocol and technical assistance.
Footnotes
The authors declare that they have no competing interest.
References
- 1.Kurman RJ, Shih Ie M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am J Pathol 2016;186:733–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol 2009;4:287–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol 2010;34:433–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int 2014;2014:934261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010;330:228–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu RC, Wang TL, Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 2014;15:655–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res 2011;71:6718–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohm KA, Hodges AJ, Czaja W, Selvam K, Smerdon MJ, Mao P, et al. Distinct roles for RSC and SWI/SNF chromatin remodelers in genomic excision repair. Genome Res 2021;31:1047–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czaja W, Mao P, Smerdon MJ. Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae. DNA Repair (Amst) 2014;16:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers AL, Downs JA. The RSC and INO80 chromatin-remodeling complexes in DNA double-strand break repair. Prog Mol Biol Transl Sci 2012;110:229–61 [DOI] [PubMed] [Google Scholar]
- 12.Park Y, Chui MH, Suryo Rahmanto Y, Yu ZC, Shamanna RA, Bellani MA, et al. Loss of ARID1A in Tumor Cells Renders Selective Vulnerability to Combined Ionizing Radiation and PARP Inhibitor Therapy. Clin Cancer Res 2019;25:5584–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res 2014;74:2465–75 [DOI] [PubMed] [Google Scholar]
- 14.Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov 2015;5:752–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allo G, Bernardini MQ, Wu RC, Shih Ie M, Kalloger S, Pollett A, et al. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol 2014;27:255–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiegand KC, Lee AF, Al-Agha OM, Chow C, Kalloger SE, Scott DW, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol 2011;224:328–33 [DOI] [PubMed] [Google Scholar]
- 17.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun 2016;7:13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G, Tu W, Yang L, Peng G, Yang L. ARID1A deficiency and immune checkpoint blockade therapy: From mechanisms to clinical application. Cancer Lett 2020;473:148–55 [DOI] [PubMed] [Google Scholar]
- 19.Park Y, Jung JG, Yu ZC, Asaka R, Shen W, Wang Y, et al. A novel human endometrial epithelial cell line for modeling gynecological diseases and for drug screening. Lab Invest 2021;101:1505–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suryo Rahmanto Y, Jung JG, Wu RC, Kobayashi Y, Heaphy CM, Meeker AK, et al. Inactivating ARID1A Tumor Suppressor Enhances TERT Transcription and Maintains Telomere Length in Cancer Cells. J Biol Chem 2016;291:9690–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013;155:1088–103 [DOI] [PubMed] [Google Scholar]
- 22.Campalans A, Kortulewski T, Amouroux R, Menoni H, Vermeulen W, Radicella JP. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res 2013;41:3115–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gassman NR, Wilson SH. Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair (Amst) 2015;31:52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konstantinopoulos PA, Lheureux S, Moore KN. PARP Inhibitors for Ovarian Cancer: Current Indications, Future Combinations, and Novel Assets in Development to Target DNA Damage Repair. Am Soc Clin Oncol Educ Book 2020;40:1–16 [DOI] [PubMed] [Google Scholar]
- 25.Williams AB, Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb Perspect Med 2016;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farago AF, Yeap BY, Stanzione M, Hung YP, Heist RS, Marcoux JP, et al. Combination Olaparib and Temozolomide in Relapsed Small-Cell Lung Cancer. Cancer Discov 2019;9:1372–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syro LV, Rotondo F, Camargo M, Ortiz LD, Serna CA, Kovacs K. Temozolomide and Pituitary Tumors: Current Understanding, Unresolved Issues, and Future Directions. Front Endocrinol (Lausanne) 2018;9:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahrer J, Frisch J, Nagel G, Kraus A, Dorsam B, Thomas AD, et al. DNA repair by MGMT, but not AAG, causes a threshold in alkylation-induced colorectal carcinogenesis. Carcinogenesis 2015;36:1235–44 [DOI] [PubMed] [Google Scholar]
- 29.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem 1999;68:255–85 [DOI] [PubMed] [Google Scholar]
- 30.Tampere M, Mortusewicz O. DNA Damage Induction by Laser Microirradiation. Bio-protocol 2016;6:e2039 [Google Scholar]
- 31.Teloni F, Michelena J, Lezaja A, Kilic S, Ambrosi C, Menon S, et al. Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability. Mol Cell 2019;73:670–83 e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat Rev Clin Oncol 2018;15:564–76 [DOI] [PubMed] [Google Scholar]
- 33.Harter P, Mouret-Reynier MA, Pignata S, Cropet C, Gonzalez-Martin A, Bogner G, et al. Efficacy of maintenance olaparib plus bevacizumab according to clinical risk in patients with newly diagnosed, advanced ovarian cancer in the phase III PAOLA-1/ENGOT-ov25 trial. Gynecol Oncol 2022;164:254–64 [DOI] [PubMed] [Google Scholar]
- 34.Takano M, Kikuchi Y, Yaegashi N, Kuzuya K, Ueki M, Tsuda H, et al. Clear cell carcinoma of the ovary: a retrospective multicentre experience of 254 patients with complete surgical staging. Br J Cancer 2006;94:1369–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho CM, Chien TY, Shih BY, Huang SH. Evaluation of complete surgical staging with pelvic and para-aortic lymphadenectomy and paclitaxel plus carboplatin chemotherapy for improvement of survival in stage I ovarian clear cell carcinoma. Gynecol Oncol 2003;88:394–9 [DOI] [PubMed] [Google Scholar]
- 36.Ho CM, Huang YJ, Chen TC, Huang SH, Liu FS, Chang Chien CC, et al. Pure-type clear cell carcinoma of the ovary as a distinct histological type and improved survival in patients treated with paclitaxel-platinum-based chemotherapy in pure-type advanced disease. Gynecol Oncol 2004;94:197–203 [DOI] [PubMed] [Google Scholar]
- 37.Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, Ha G, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet 2017;49:856–65 [DOI] [PubMed] [Google Scholar]
- 38.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were generated by the authors and included in the article.